Abstract
Distal cholangiocarcinoma is a rare and highly aggressive malignant tumor. The inherent tumor characteristics and growth pattern of cancer cells pose a challenge for diagnosis and treatment. Chemotherapy resistance leads to limited treatment options for patients with advanced cholangiocarcinoma. However, drug resistance studies in cholangiocarcinoma are often limited by the use of preclinical models that do not accurately replicate the essential features of the disease. In this study, we established and characterized a primary multidrug-resistant distal cholangiocarcinoma cell line, CBC3T-6. STR profiling indicated no evidence of cross-contamination. This cell line remains stable during long-term in vitro culture and is characterized by short doubling times and rapid subcutaneous tumor formation in mice. In addition, among the first-line anticancer drugs for cholangiocarcinoma, CBC3T-6 cells showed varying degrees of resistance to gemcitabine, oxaliplatin, cisplatin, and 5-FU. Whole exome sequencing analysis revealed that CBC3T-6 cells contained a variety of potentially pathogenic somatic cell mutations, such as TP53 and KRAS mutations. ABCB1 mutation as a possible therapeutic target for multidrug resistance. In conclusion, CBC3T-6 cells can be used as a useful tool to study the mechanism of cholangiocarcinoma and develop new therapeutic strategies for multidrug resistance.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81392-0.
Keywords: Distal cholangiocarcinoma, Cell line, Multidrug resistance, KRAS mutations, TP53 mutations
Subject terms: Cancer, Cell biology, Cancer
Introduction
Cholangiocarcinoma (CCA) is a malignant tumor originating from the intrahepatic tubules to the major extrahepatic bile ducts. Anatomically, CCA can be divided into intrahepatic cholangiocarcinoma (iCCA), hilar cholangiocarcinoma (hCCA) and distal cholangiocarcinoma (dCCA)1. The prevalence of different types of CCA varies widely, with hCCA accounting for 50–60% of all CCA, followed by dCCA (20–30%), and the rarest being iCCA (10–20%)2,3. Global CCA mortality is on the rise worldwide4. Surgical resection remains the mainstay of treatment for dCCA. However, the 5-year survival rate of patients after complete resection has been reported to be between 18% and 54% 5–7. A major problem in current chemotherapy for human cancers is the development of chemoresistance, especially multidrug resistance, as is the case for CCA8,9. Many chemotherapeutic agents, such as 5-fluorouracil (5-FU), gemcitabine, cisplatin, oxaliplatin, and paclitaxel, have low response rates and short median survival times in the treatment of patients with CCA10–12.
During embryogenesis, CCA originating from different sites are highly heterogeneous and have different tumorigenic processes13. Therefore, CCA cell models from different anatomical sites can provide better research value. The low prevalence of dCCA and the lack of validated preclinical models have resulted in a lack of high-level evidence to substantiate the effectiveness of treatment. Clarifying the mechanism underlying tumor chemotherapeutic resistance is highly important, and the search for low cytotoxicity and efficient combination chemotherapy strategies will provide a potential direction for overcoming the bottleneck of tumor chemotherapeutic resistance. Patient-derived models are fundamental to the study of cancer and have led to significant advances in drug development, basic research and clinical applications14. Cell lines, patient-derived xenografts (PDXs), and organoids are currently the three major patient-derived models studied in vitro, with cell lines being the most common15–17. Cell line models have contributed greatly to the study of key molecular mechanisms involved in the progression of CCA and the development of strategies to overcome drug resistance.
Therefore, for the first time, we report a novel multidrug-resistant primary dCCA cell line, CBC3T-6, which exhibits varying degrees of resistance to first-line chemotherapeutic agents for CCA.
Results
Establishment and authentication of the CBC3T-6 cell line
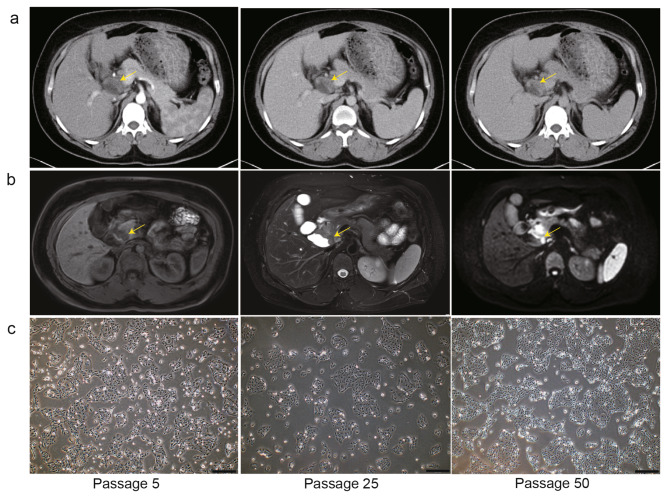
The donor patient was a 44-year-old Chinese woman with a diagnosis of dCCA confirmed by computed tomography (CT) and magnetic resonance imaging (MRI) (Fig. 1a, b). We successfully established a new dCCA cell line, CBC3T-6, from the surgically resected primary site, which has been maintained for more than 50 generations to date. CBC3T-6 cells grew in an adherent monolayer and were fusiform or polygonal in shape. In addition, CBC3T-6 cells grew rapidly and could accumulate pile up when they filled the culture dish, indicating the loss of contact inhibition. From P5 to P50, the morphology of the cells remained almost the same (Fig. 1c). The CBC3T-6 cell line was identified by analyzing short tandem repeat (STR) at 21 loci (Table 1). The STR profile of CBC3T-6 cells was essentially consistent with that of the source tissue and did not match any of the cell lines in the ATCC, DSMZ or CELLOSAURUS databases, indicating that CBC3T-6 is a novel cell line.
Fig. 1.
Clinical information and biological features of CBC3T-6 cells. a, b Computed tomography (CT) and magnetic resonance imaging (MRI) showing distal bile duct tumors. c Bright field morphology of CBC3T-6 cells at 5th, 25th and 50th passages (scale bars, 200 μm).
Table 1.
STR profile of tumor tissue and cell line.
| Marker | CBC3T-6 | Tumor tissue |
|---|---|---|
| D19S433 | 14, 14.2 | 14, 14.2 |
| D5S818 | 11 | 11 |
| D21S11 | 30, 31 | 30, 31 |
| D18S51 | 13.3, 14.3 | 14, 15 |
| D6S1043 | 19 | 19 |
| AMEL | X | X |
| D3S1358 | 15, 16 | 15, 16 |
| D13S317 | 8, 10 | 8, 10 |
| D7S820 | 11, 13 | 11, 13 |
| D16S539 | 9, 11 | 9, 11 |
| CSF1PO | 10 | 10 |
| Penta D | 11, 15 | 11, 15 |
| D2S441 | 11, 15 | 11, 15 |
| vWA | 14 | 14 |
| D8S1179 | 12, 12 | 12, 14 |
| TPOX | 9, 11 | 9, 11 |
| Penta E | 12, 14 | 12, 14 |
| TH01 | 8, 9.3 | 8, 9.3 |
| D12S391 | 20, 22 | 20, 22 |
| D2S1338 | 22, 23 | 22, 23 |
| FGA | 21, 25 | 21, 25 |
Characteristics of CBC3T-6 cells
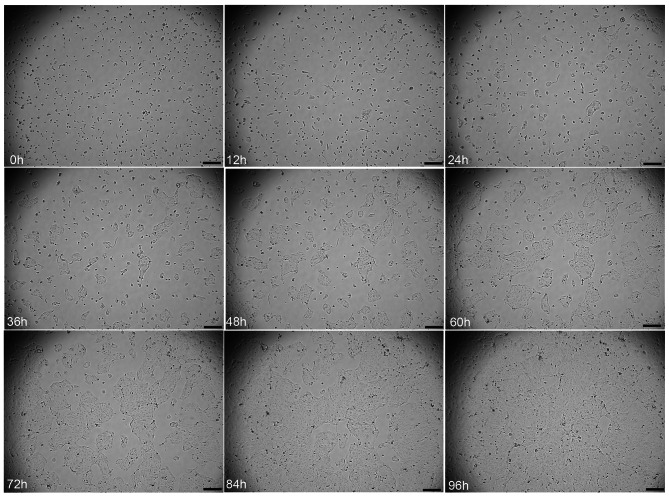
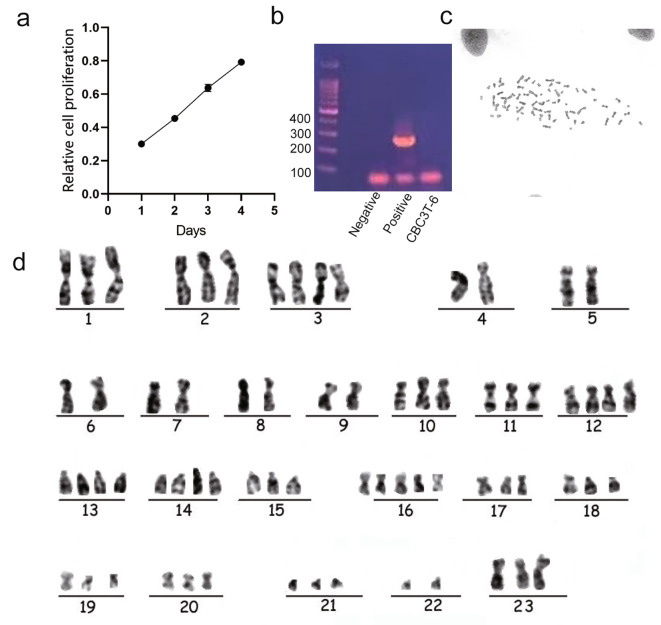
We used live cell imaging to observe the growth status of CBC3T-6 cells in real time. The cells exhibited aggregation and formed clumps of growth after apposition to the wall (Fig. 2). Moreover, by analyzing the growth curve of CBC3T-6 cells, we calculated the doubling time of these cells to be approximately 50 h (Fig. 3a). CBC3T-6 cells were not contaminated with mycoplasma (Fig. 3b). G-banding karyotype analysis of a representative single cell of CBC3T-6 showed that this cell line is a nearly triploid line with complex chromosome number and structural abnormalities and a typical karyotype number of 68 (Fig. 3c, d).
Fig. 2.
Live cell imaging analysis of CBC3T-6 cells (0, 12, 24, 36, 48, 60, 72, 84 and 96 h). Scale bars, 200 μm.
Fig. 3.
Characterization of CBC3T-6 cells. a Cumulative growth curve of CBC3T-6 cells. b Mycoplasma test results. c, d Karyotyping of CBC3T-6 cells.
Tumorigenicity in vivo
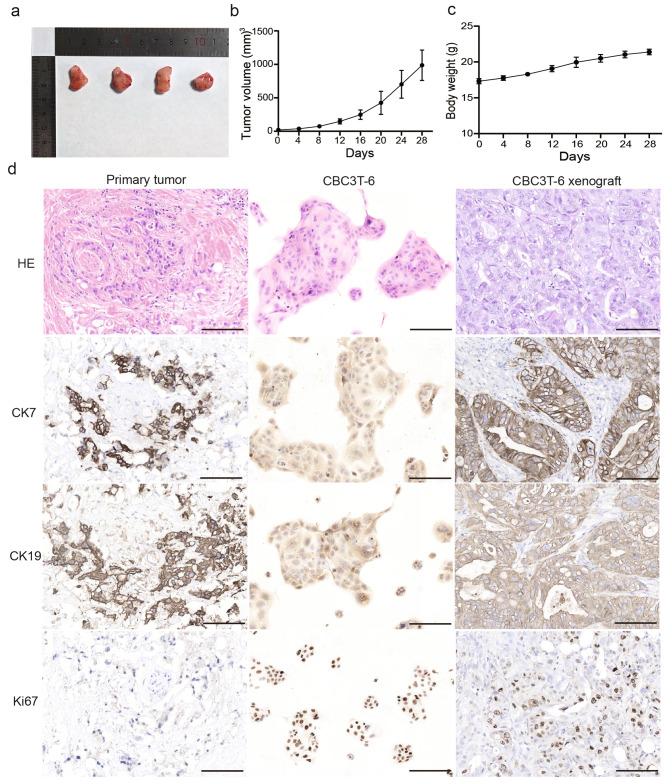
To determine whether CBC3T-6 cells are tumorigenic in vivo, we performed xenograft transplantation. One week after subcutaneous injection of the cells, an increase in the size of the subcutaneous tumor was visible to the naked eye, and the tumor was excised 28 days after inoculation. The results showed a 100% tumorigenicity rate (n = 4) for CBC3T-6 and there was no significant change in the body weight of the mice, indicating that this cell line is suitable for cell line-derived xenograft models (Fig. 4a-c). H&E and IHC showed concordance between primary tumor tissue, xenograft tumor tissue, and CBC3T-6 cells. In addition, the in vitro cultured cell line CBC3T-6 showed positive expression of the proliferation marker Ki67 as well as the biliary markers CK7 and CK19. (Fig. 4d). These results indicate that the CBC3T-6 cell line cultured in vitro faithfully recapitulates and maintains the expression status of the primary tumor and is highly tumorigenic, which provides a valuable model for mechanistic studies of CCA.
Fig. 4.
Tumorigenicity of CBC3T-6 in NOD/SCID mice. a Tumor masses formed subcutaneously in mice. b Tumor volume growth curves formed by CBC3T-6 cells. c The body weight curve of the mice. d H&E staining and immunohistochemical (CK19, CK7, Ki67) results of primary tumors, CBC3T-6 xenografts and CBC3T-6 cells.
Sensitivity to anticancer agents
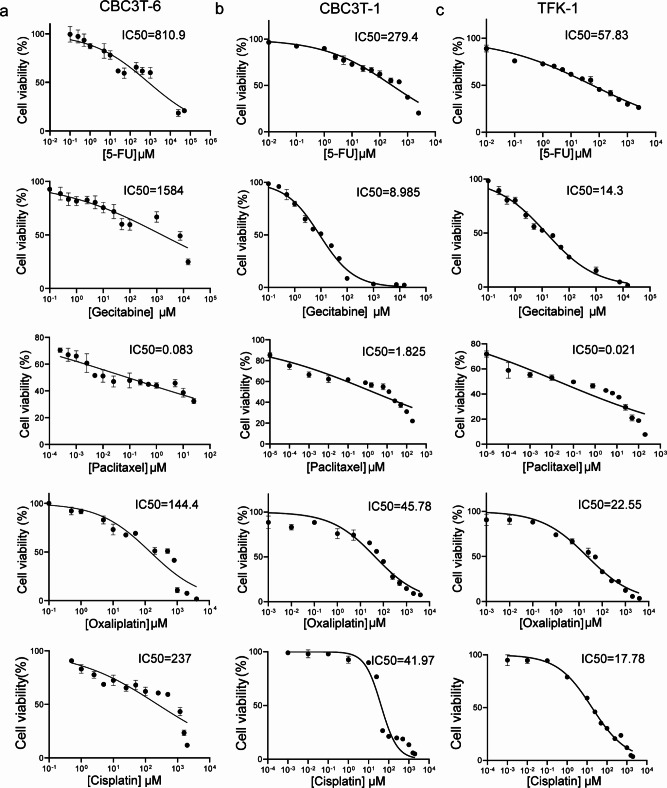
Most patients with CCA have metastatic or locally progressive disease at presentation. Chemotherapy is the standard treatment for patients with metastatic and unresectable disease or recurrent CCA, with gemcitabine in combination with cisplatin recognized as the standard chemotherapy regimen18,19. We assessed the sensitivity of CBC3T-6 cells to first-line anticancer drugs for the treatment of CCA. The IC50 values of gemcitabine, 5-FU, cisplatin, oxaliplatin and paclitaxel were 1584 µM, 810.9 µM, 237 µM, 144.4 µM, and 0.083 µM, respectively (Fig. 5a). The results showed that CBC3T-6 cells were resistant to gemcitabine, oxaliplatin, 5-FU and cisplatin to varying degrees and were more sensitive to paclitaxel. In addition, CBC3T-6 cells were less sensitive to gemcitabine, 5-FU, cisplatin, and oxaliplatin drugs compared to CBC3T-1 cells and TFK-1 cells (Fig. 5b, c). Resistance limits the use of these first-line chemotherapeutic agents in clinical treatment. The construction of this multidrug-resistant cell line contributes to further understanding the resistance mechanism of CCA, thereby improving the efficacy of chemotherapy and prolonging survival.
Fig. 5.
Cell viability of CBC3T-6 cells, CBC3T-1 cells, and TFK-1 cells after 48 h of exposure to anticancer drugs. a-c Dose‒response curves of CBC3T-6 cells, CBC3T-1 cells and TFK-1 cells treated with 5-FU, gemcitabine, cisplatin, paclitaxel, and oxaliplatin.
Whole exome sequencing
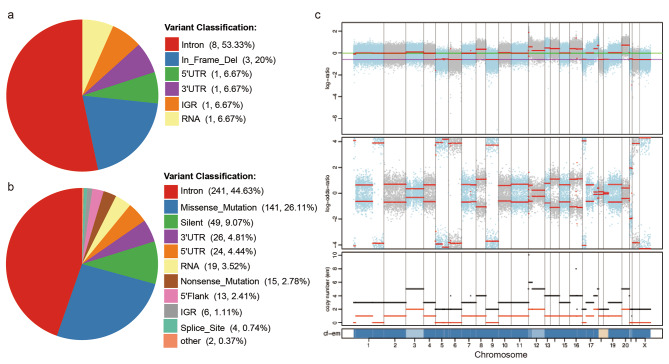
The evolutionary hypothesis of cancer suggests that cancer is the result of an accumulation of mutations in somatic genes. CCA contains a highly heterogeneous landscape of genomic mutations associated with poor patient prognosis20–22. To clarify the genetic alterations in the CBC3T-6 cell line, we performed whole exome sequencing (WES) analysis on cultured CBC3T-6 cells and tumor-matched normal tissue. On average, we identified 17 somatic Insertions/deletions (InDels) and 540 somatic single nucleotide variants (SNVs) through WES (Fig. 6a, b). somatic copy number variation (CNV) profiles were also compared with those of matched normal tissue (Fig. 6c). 47 somatic CNVs were identified, including 16 deletions and 31 duplications. Furthermore, we compared somatic mutations with known driver genes in databases and the literature and screened known driver genes in CBC3T-6 cells (Supplementary Table 1). Among these known driver genes, TP53 and KRAS mutations are most common in dCCA. KRAS is a proto-oncogene, and KRAS mutations can lead to hyperactivation of downstream pathways and promote tumorigenesis, including cell proliferation, migration and metastasis23. Previous work has suggested that KRAS mutations are more prevalent in hCCA, whereas TP53 mutations are more prevalent in dCCA24. ABCB1 is involved in the transport of anticancer drugs such as paclitaxel and vincristine25. Study reports that overexpression of P glycoprotein (P-gp) plays a key role in the progression of multidrug resistance in cancer26,27. 5-FU treatment induced ABCB1 expression in two CCA cell lines, KKU-M055 and KKU-M214, and the 5-FU-resistant cell line M214-5FUR showed up to 16-fold expression of ABC transporter proteins compared to parental cells28. Inhibition of the expression of the transporter protein ABCB1 sensitizes CCA cells to chemotherapeutic agents, thereby increasing the antitumor activity of chemotherapeutic agents29. Mutations in the ABCB1 gene may contribute to drug resistance in CBC3T-6 cells.
Fig. 6.
Whole exome sequencing of CBC3T-6 cells and matched normal tissues. a, b Distribution of somatic Insertions/deletions (InDels) and somatic single nucleotide variants (SNVs) annotation results. c Profile of somatic copy number variations (CNVs) results for the CBC3T-6 cell line.
CIViC is a specialized database for the clinical annotation of somatic variants in tumors that includes all kinds of genetic variants and somatic variants related to treatment, prognosis, diagnosis and susceptibility. First-line anticancer drug screening identified CBC3T-6 cells as a multidrug-resistant cell line. Cancer heterogeneity is now recognized as an important factor contributing to treatment resistance. Substantial genetic variations suggest a complex heterogeneity in CCA. To address its resistance mechanisms, we used the CIViC database to annotate the identified somatic mutation KRAS G12D to predict response to targeted therapies (Supplementary Table 2). Overall, most driver alterations and genetic pathway alterations were statistically similar between hCCA and dCCA, but dCCA tended to have a higher rate of TP53/KRAS-G12D alterations30,31.
Discussion
Most patients with CCA have metastatic or locally progressive disease at presentation. Chemotherapy is the standard treatment for patients with metastatic and unresectable disease or recurrent CCA32. Treatment options for patients with CCA are limited due to primary or acquired resistance to existing chemotherapeutic agents and the lack of new drugs available for clinical application. New preclinical research models that summarize the heterogeneity of CCA may help screen CCA patients for effective drugs. Although patient-derived cell line-based models do not fully represent the complex tumor microenvironment, their simplicity, rapidity and cost-effectiveness make them important tools for drug discovery and understanding of disease processes33. According to the Cellosaurus (https://web.expasy.org/cellosaurus/), only a limited number of cell lines are available from public biobanks34. To elucidate the molecular molecules responsible for drug resistance in CCA and to develop new therapeutic approaches, patient-derived drug-resistant cell models are crucial. Here, we report a novel primary multidrug-resistant cell line, CBC3T-6, derived from the tumor tissues of dCCA patients.
In this study, we comprehensively characterized the CBC3T-6 cell line according to cancer-associated phenotypes and genomic alterations. This cell line remains stable during long-term in vitro culture and is characterized by rapid proliferation and a short doubling time. It is suitable for use as an in vitro model in the basic research of CCA. Karyotype analysis revealed a tumor polyploid structure with chromosome number and structural abnormalities. In addition, injection of the newly derived cells into the subcutis of mice led to tumor formation within a short period of time. The maintenance of primary tumor characteristics and the reliability of the subcutaneous graft tumor model were confirmed in the immortalized CBC3T-6 cell line. Therefore, we believe that this new cell line can be used as a tool for in vivo scientific research.
The most prominent advantage of CBC3T-6 is primary multidrug resistance, with varying degrees of resistance to gemcitabine, oxaliplatin, cisplatin, and 5-FU, the first-line anticancer drugs for CCA. The mechanisms of drug resistance in CCA cells can be divided into the following main categories: decreased drug uptake and increased drug export35,36; reduction in the proportion of active drug in the cell37,38; changes in the molecular targets of antitumor drugs28; activation of anti-apoptotic pathways or inactivation of pro-apoptotic pathways39; and interactions with the tumor microenvironment9. A common approach to study the mechanisms of drug resistance in CCA is to analyze differentially expressed genes between wild-type and drug-resistant cell lines. These genes are generated by repeated and increased drug treatment, or by genetic manipulation, such as gene overexpression, knockout, or point mutations, to establish drug-resistant cell lines9. Previous studies established 5-FU and gemcitabine-resistant CCA cell lines KKU-213FR and KKU-213GR, and screened MET, LAMB1, ITGA3, NOTCH2, CDH2, and NDRG1 using comparative proteomics methods. Furthermore, the functions of these proteins are closely related to the epithelial-mesenchymal transition (EMT) signaling pathway40. Conventional chemotherapy has limited efficacy in the treatment of CCA. Therefore, there is a clinical need for multidrug combinations for different types of CCA to improve patient prognosis. Primary drug-resistant cell lines have an advantage over acquired drug-resistant cells due to the retention of their original tumor properties. In addition, WES analysis revealed that CBC3T-6 cells contained multiple somatic mutations, and the most common TP53 and KRAS mutations in CCA were also included. KRAS allele variants are highly prevalent in hCCA and dCCA, with the highest prevalence of variant G12D. In addition, ABCB1 gene mutations are associated with one of the causes of multidrug resistance in CBC3T-6 cells.
In conclusion, we established a novel primary multidrug-resistant human distal cholangiocarcinoma cell line that better recapitulates the characteristics of the tumor. This model could provide a useful preclinical tool for studying the development of cholangiocarcinoma and the mechanism of cross-resistance.
Materials and methods
Patient background
The patient had elevated tumor marker levels (CA199, 276.0 U/mL), imaging techniques revealed limited cystic dilatation of the intrapancreatic segment of the common bile duct with eccentric irregular wall thickening, and a diagnosis of dCCA was made. No preoperative radiotherapy or chemotherapy was administered. The patient underwent pancreaticoduodenectomy, and the postoperative pathologic diagnosis was low-differentiated adenocarcinoma of the bile duct. Written informed consent was obtained from the patient for the use of her clinical data and pathological specimen in accordance with the Declaration of Helsinki.
Primary cell culture
The CBC3T-6 primary cell line was established as described in Bai et al.41. Briefly, the tumor tissue was cut into 1 mm3 pieces and digested using collagenase II for 15 min. The dissociated tissues were filtered through a 100-µm cell strainer. The cell suspension was centrifuged (300 g/5 min) and resuspended in DMEM/F-12 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and 100 mg/mL Primocin™ (InvivoGen, USA). The cells were then cultured in a humidified incubator at 37 °C with 5% CO2. All subsequent experiments were performed after 30 generations.
Short tandem repeat (STR) profiling
The CBC3T-6 cell line and the tumor sample were authenticated by the CCTCC. Briefly, genomic DNA was extracted from cells and tissues using the QIAamp DNA Mini Kit (Qiagen, Netherlands). The alleles of 21 loci in CBC3T-6 cells were analyzed using an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, USA). The STR profiles were then compared to the profiles in the ATCC, DSMZ and CELLOSAURUS databases.
Mycoplasma detection by PCR
Culture supernatants from CBC3T-6 cells were collected and assayed according to the mycoplasma detection kit (Invitrogen, USA). DNA fragments were imaged under UV irradiation.
Cell counting kit (CCK)-8 cell growth analysis
CBC3T-6 cells were seeded into 96-well plates at a density of 4 × 103 cells per well. After incubation for 24, 48, 72, and 96 h, CCK8 reagent (APExBio, USA) was added to the cells, which were then incubated for 2 h, after which the absorbance at 450 nm was measured. Doubling time was calculated using the online software Doubling Time Computing Version 3.1.0.
Live cell imaging
Cells were inoculated in 96-well plates until they attached to the surface. Images were captured by a Cytation 1 imaging system (Biotek, USA) under a 4x objective. Label-free live cell proliferation measurements were achieved by capturing two high-contrast brightfield images at each 2-hour time point. Images were processed with Gen 5.
Chromosome karyotype analysis
Karyotyping of CBC3T-6 cells was performed by G banding. When the cells reached 70% confluence, mitotic arrest was induced with 0.4 µg/ml colchicine for 2 h. Then, the cells were collected and incubated with a hypotonic solution (0.075 M KCl) for 30 min (37 °C) and fixed 3 times with methanol: acetic acid (3:1) at room temperature for 10 min. Slides were then prepared and stained with Giemsa.
Tumorigenicity in NOD/SCID mice
In vivo, the tumorigenicity of CBC3T-6 cells was assessed by the ability of the cells to form tumors by subcutaneous injection in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice. Four NOD/SCID female mice (4 to 6 weeks old) were purchased from GemPharmatech Co. Ltd (Nanjing, China). A total of 2 × 106 cells were injected subcutaneously into the right subscapular region of NOD/SCID mice. The animals were kept in a laminar flow cabinet under specific pathogen-free conditions. Subsequently, the mice were continuously monitored for tumor size and body weight. After 28 days, the mice were euthanized by carbon dioxide inhalation. Tumor tissues were then collected, measured and weighed, fixed in 10% formalin, embedded in paraffin, stained with hematoxylin and eosin (H&E) and subjected to immunohistochemistry (IHC). The animal experiment was performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of China. The protocol was approved by the Ethics Committee of the First Hospital of Lanzhou University. The authors complied with the ARRIVE guidelines.
H&E staining and IHC
Resected tumor, CBC3T-6 cell and subcutaneously transplanted tumor samples from the mice were embedded in paraffin and cut into 4 μm thick sections. The sections were dewaxed in xylene, hydrated in a graded alcohol series, and washed with phosphate-buffered saline for H&E staining. For IHC analysis, the slides were deparaffinized in xylene and dehydrated using a graded alcohol series. The slides were then heated in a pressure cooker with 10 mM sodium citrate (pH 6.5) for 15 min. The nonspecific antigens were blocked with catalase enzyme body for 10 min, and 10% normal goat serum was added for 10 min. The slides were incubated overnight at 4 °C with anti-CK7 (MAB-0828, 1:400, MaiXin, China), anti-CK19 (MAB-0829, 1:400, MaiXin, China) and anti-Ki67 (MAB-0672, 1:1000, MaiXin, China) antibodies. Excess primary antibodies were removed by washing with PBS, and then the slices were incubated with secondary antibodies (KIT-5001, MaiXin, China) for 30 min for DAB color development. Finally, counterstaining was performed with hematoxylin.
Sensitivity to anti‑cancer drugs
Currently, the main first-line anticancer drugs for CCA are gemcitabine, oxaliplatin, cisplatin, 5-FU and paclitaxel. Anticancer drugs were purchased from MedChemExpress (MCE, China). Cells were inoculated at 5 × 103 per well in 96-well plates and cultured for 24 h. Different concentrations of first-line anticancer drugs were added. After 48 h of treatment, cell proliferation was detected using a CCK-8 assay kit. IC50 values were calculated using GraphPad Prism 8 (GraphPad Software, Inc.).
Whole‑exome sequencing (WES) of CBC3T-6 cells
WES was performed at BGI (Wuhan, China). Briefly, genomic DNA was extracted from CBC3T-6 cells and tumor-matched normal tissue. Library construction and whole‑exome capture of genomic DNA were performed using SureSelect Human All Exon V6 (Agilent) and the captured DNA library was sequenced on the DNBSEQ platform. Sequencing reads were aligned to the reference human genome (GRCh 38) using the Burrows‒Wheeler Aligner. Somatic mutations were detected by comparing CBC3T-6 cells and tumor-matched normal tissue, filtering out germline mutations in normal tissue and retaining only those somatic mutations found in tumor cells during the analysis. Analysis of WES data for somatic mutations, including somatic single nucleotide variants (SNVs), somatic INDELs and somatic copy number variations (CNVs).
We compared somatic mutations with known driver genes in databases and the literature and screened out known driver genes in CBC3T-6 cells. The reference data sources used were the Integrative OncoGenomics (IntOGen), Cancer Gene Census (CGC), three highly cited articles42–44 and pan-cancer data45.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
M.B., R.W., H.Z. and W.M. proposed the idea and designed the research; M.B. and R.W. performed the in vitro cell validation experiments; The animal experiments were conducted by M.B., R.Z. and C.H.; M.B. and R.W. wrote the draft of the manuscript; N.J. and H.Y. collected the clinical specimens and data; W.F., L.G., Y.J., H.M., J.C., C.Z., Y.Z. and P.Y. contributed to the manuscript revision; Y.L., Q.J. and N.M. performed the data analysis; H.Z. and W.M. supervised the research and were responsible for the integrity of the data analysis. All authors read and approved the submitted version.
Funding
This work was supported by funds from the National Natural Science Foundation of China (82060551), Natural Science Foundation of Gansu Province of China (24JRRA323), Gansu Province Health Care Industry Research Program Projects (GSWSKY2020-11), Lanzhou Science and Technology Bureau (2019-4-43), and Gansu Province Young Science and Technology Talents Support Project Program (GXH20210611-).
Data availability
The datasets used and/or analyzed in this study are available from the corresponding authors upon reasonable request.
Declarations
Ethical declarations
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the First Hospital of Lanzhou University, Lanzhou, Gansu, China (Approval number: LDYYLL2024-296).
Informed consent
The participant provided written informed consent for the use of her clinical and pathological specimens.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingzhen Bai and Ruoshui Wang contributed equally to this work.
Contributor Information
Hengwei Zhang, Email: ldyy_zhanghw@lzu.edu.cn.
Wenbo Meng, Email: mengwb@lzu.edu.cn.
References
- 1.Brindley, P. J. et al. Cholangiocarcinoma. Nat. Reviews Disease Primers7, 65. 10.1038/s41572-021-00300-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banales, J. M. et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol.17, 557–588. 10.1038/s41575-020-0310-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeOliveira, M. L. et al. Cholangiocarcinoma - Thirty-one-year experience with 564 patients at a single institution. Ann. Surg.245, 755–762. 10.1097/01.sla.0000251366.62632.d3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadamuro, M. et al. Targeted therapies for extrahepatic cholangiocarcinoma: preclinical and clinical development and prospects for the clinic. Expert Opin. Investig. Drugs. 30, 377–388. 10.1080/13543784.2021.1880564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komaya, K. et al. Recurrence after resection with curative intent for distal cholangiocarcinoma. Br. J. Surg.104, 426–433. 10.1002/bjs.10452 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Courtin-Tanguy, L. et al. Multicentre study of the impact of factors that may affect long-term survival following pancreaticoduodenectomy for distal cholangiocarcinoma. Hpb20, 405–410. 10.1016/j.hpb.2017.10.016 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Zhou, W. W. et al. Prognostic factors and patterns of recurrence after curative resection for patients with distal cholangiocarcinoma. Radiother Oncol.147, 111–117. 10.1016/j.radonc.2020.03.017 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Emran, T. B. et al. Multidrug Resistance in Cancer: understanding Molecular mechanisms, Immunoprevention and therapeutic approaches. Front. Oncol.12, 891652. 10.3389/fonc.2022.891652 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng, Q. F., Zhang, B., Li, C. F. & Zhang, X. W. Overcome Drug Resistance in Cholangiocarcinoma: New Insight Into Mechanisms and Refining the Preclinical Experiment Models. Frontiers in Oncology 12. https://doi.org/ARTN 85073210.3389/fonc.850732 (2022). (2022). [DOI] [PMC free article] [PubMed]
- 10.Khosla, D. et al. Recent advances in Molecular Pathobiology and therapeutic approaches. Cancers (Basel). 16. 10.3390/cancers16040801 (2024). Cholangiocarcinoma. [DOI] [PMC free article] [PubMed]
- 11.Ramirez-Merino, N., Aix, S. P. & Cortes-Funes, H. Chemotherapy for cholangiocarcinoma: an update. World J. Gastrointest. Oncol.5, 171–176. 10.4251/wjgo.v5.i7.171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin, H. J., Zheng, G., Li, Q. & Shen, L. Y. Metabolic reprogramming induced by DCA enhances cisplatin sensitivity through increasing mitochondrial oxidative stress in cholangiocarcinoma. Front Pharmacol 14. https://doi.org/ARTN 112831210.3389/fphar.1128312 (2023). (2023). [DOI] [PMC free article] [PubMed]
- 13.Roskams, T. & Desmet, V. Embryology of extra- and intrahepatic bile ducts, the ductal plate. Anat. Rec. 291, 628–635. 10.1002/ar.20710 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Hou, X. et al. Opportunities and challenges of patient-derived models in cancer research: patient-derived xenografts, patient-derived organoid and patient-derived cells. World J. Surg. Oncol.20, 37. 10.1186/s12957-022-02510-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couzin-Frankel, J. Hope in a mouse. Science346, 28–29. 10.1126/science.346.6205.28 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Tuveson, D. & Clevers, H. Cancer modeling meets human organoid technology. Science364, 952–955. 10.1126/science.aaw6985 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kim, S. Y. et al. Patient-derived cells to Guide targeted therapy for Advanced Lung Adenocarcinoma. Sci. Rep.9, 19909. 10.1038/s41598-019-56356-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y. H., Song, Y. H. & Liu, S. L. The new insight of treatment in Cholangiocarcinoma. J. Cancer. 13, 450–464. 10.7150/jca.68264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macias, R. I. R., Rimassa, L. & Lamarea, A. The promise of precision medicine: how biomarkers are shaping the future of cholangiocarcinoma treatment. Hepatobil Surg. Nutr.12, 457–461. 10.21037/hbsn-23-215 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jusakul, A. et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of Cholangiocarcinoma. Cancer Discov.7, 1116–1135. 10.1158/2159-8290.Cd-17-0368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montal, R. et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J. Hepatol.73, 315–327. 10.1016/j.jhep.2020.03.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng, M. J. et al. Proteogenomic characterization of cholangiocarcinoma. Hepatology77, 411–429. 10.1002/hep.32624 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merz, V. et al. Targeting KRAS: the Elephant in the room of epithelial cancers. Front. Oncol.11, 638360. 10.3389/fonc.2021.638360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simbolo, M. et al. Molecular characterization of extrahepatic cholangiocarcinoma: perihilar and distal tumors display divergent genomic and transcriptomic profiles. Expert Opin. Ther. Tar. 25, 1095–1105. 10.1080/14728222.2021.2013801 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Ambudkar, S. V. et al. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol.39, 361–398. 10.1146/annurev.pharmtox.39.1.361 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Lagas, J. S. et al. Breast Cancer resistance protein and P-glycoprotein limit Sorafenib Brain Accumulation. Mol. Cancer Ther.9, 319–326. 10.1158/1535-7163.Mct-09-0663 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Tepsiri, N. et al. Drug sensitivity and drug resistance profiles of human intrahepatic cholangiocarcinoma cell lines. World J. Gastroentero. 11, 2748–2753. 10.3748/wjg.v11.i18.2748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon, H. et al. L1 cell adhesion molecule and epidermal growth factor receptor activation confer cisplatin resistance in intrahepatic cholangiocarcinoma cells. Cancer Lett.316, 70–76. 10.1016/j.canlet.2011.10.024 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Seubwai, W. et al. Inhibition of NF-kappaB activity enhances sensitivity to anticancer drugs in Cholangiocarcinoma Cells. Oncol. Res.23, 21–28. 10.3727/096504015X14424348426071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston, W. A. et al. Extrahepatic Cholangiocarcinoma: genomic variables Associated with Anatomic Location and Outcome. JCO Precis Oncol.8, e2400206. 10.1200/PO.24.00206 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffat, G. T. et al. KRAS allelic variants in biliary tract cancers. JAMA Netw. Open.7, e249840. 10.1001/jamanetworkopen.2024.9840 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khosla, D. et al. Cholangiocarcinoma: Recent Advances in Molecular Pathobiology and Therapeutic Approaches. Cancers 16. https://doi.org/ARTN 80110.3390/cancers16040801 (2024). [DOI] [PMC free article] [PubMed]
- 33.Langhans, S. A. Three-Dimensional in Vitro Cell Culture models in Drug Discovery and Drug Repositioning. Front. Pharmacol.9, 6. 10.3389/fphar.2018.00006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isidan, A. et al. Development and characterization of human primary Cholangiocarcinoma Cell lines. Am. J. Pathol.192, 1200–1217. 10.1016/j.ajpath.2022.05.007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Becerra, P. et al. No correlation between the expression of FXR and genes involved in Multidrug Resistance phenotype of primary liver tumors. Mol. Pharmaceut. 9, 1693–1704. 10.1021/mp300028a (2012). [DOI] [PubMed] [Google Scholar]
- 36.Pongmaneratanakul, S., Tanasanvimon, S., Pengsuparp, T. & Areepium, N. Prevalence of CTR1 and ERCC1 polymorphisms and response of biliary Tract Cancer to Gemcitabine-Platinum Chemotherapy. Asian Pac. J. Cancer Prev.18, 857–861. 10.22034/APJCP.2017.18.3.857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahnvajanawong, C. et al. Orotate phosphoribosyl transferase mRNA expression and the response of cholangiocarcinoma to 5-fluorouracil. World J. Gastroentero. 18, 3955–3961. 10.3748/wjg.v18.i30.3955 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeekpudsa, P., Kukongviriyapan, V., Senggunprai, L., Sripa, B. & Prawan, A. Suppression of NAD(P)H-quinone oxidoreductase 1 enhanced the susceptibility of cholangiocarcinoma cells to chemotherapeutic agents. Journal of Experimental & Clinical Cancer Research 33. https://doi.org/Artn 1110.1186/1756-9966-33-11 (2014). [DOI] [PMC free article] [PubMed]
- 39.Razumilava, N. et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology55, 465–475. 10.1002/hep.24698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerdkumthong, K. et al. Proteomics and Bioinformatics Identify Drug-Resistant-Related Genes with Prognostic Potential in Cholangiocarcinoma. Biomolecules 14. https://doi.org/ARTN 96910.3390/biom14080969 (2024). [DOI] [PMC free article] [PubMed]
- 41.Bai, M. et al. Establishment and characterization of a novel hilar cholangiocarcinoma cell line, CBC3T-1. Hum. Cell. 37, 364–375. 10.1007/s13577-023-01003-4 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey, M. H. et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 173, 371-+. (2018). https://doi.org/ARTN385.e110.1016/j.cell.02.060 (2018). [DOI] [PMC free article] [PubMed]
- 43.Vogelstein, B. et al. Cancer Genome Landscapes Sci.339, 1546–1558. 10.1126/science.1235122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamborero, D. et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep.3, 2650. 10.1038/srep02650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature502, 333–. 10.1038/nature12634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed in this study are available from the corresponding authors upon reasonable request.