Abstract
Pasteurization is a common method for dairy products, typically heating at 72 °C for 15 s or 63 °C for 30 min. The 17 samples of commercial pasteurized milk were divided into three groups according to the shelf life: group A (1–5 days), group B (6–10 days) and group C (11–15 days), and the diversity composition of microbial communities in the samples was analyzed. Among all groups, Proteobacteria, Firmicutes, and Bacteroidetes were the dominant bacterial phyla. The lactic acid bacteria (LAB) were mostly Streptococcus, Weissella and Lactobacillus, and there were high proportions of Streptococcus thermophilus in group A, Weissella paramesenteroides in group B, and Lactobacillus plantarum in group C. Furthermore, a strain of Enterococcus faecium SFM2 was isolated from the A2 sample, which showed better temperature tolerance compared to the E. faecium SFM1 of oral origin. After treatment at 50 °C for 2 h, the survival rates of E. faecium SFM1 and SFM2 were 28.20 ± 0.04% and 82.58 ± 0.01%, respectively. This study investigated the diversity of microorganisms in pasteurized milk, providing effective information for analyzing the potential microbiota of commercial pasteurized milk. Meanwhile, it provided new ideas for expanding the resource pool of thermoresistant LAB.
Keywords: Microbial diversity, Pasteurized milk, Thermoresistant, Lactic acid bacteria
Subject terms: Bacteria, Microbial communities
Introduction
As one of the most widely consumed beverages, milk contains various essential nutrients like proteins and vitamins. Meanwhile, the high nutrient content provides an ideal environment for the growth of many microorganisms, including lactic acid bacteria (LAB). The analysis of the biodiversity in the milk environment and the isolation and identification of microorganisms from milk are of great scientific significance, and there are many studies have analyzed the microbial populations in different raw milk. For instance, Yuan et al.1 analyzed the variation in raw milk microbiota from three farms by 16S rRNA amplicon sequencing, providing new information on the ecology of raw milk microbiota at the farm level and contributing to the understanding of the variation in raw milk microbiota in China. Luoyizha et al.2 sequenced the bacterial populations in donkey milk from different regions and revealed possible harmful pathogens and probiotics, which also provided a new way for potential future use of microbial resources.
In order to reduce the number of live microorganisms in raw milk, liquid dairy products are usually sterilized by ultra-high temperature sterilization (UHT) treatments and pasteurization. A typical UHT treatment is 135–150 °C for 1–10 s, which inactivates almost all microorganisms, and the treated product could be stored at room temperature for 6–9 months3. The low temperature long time (LTLT, 63 °C, 30 min) treatment and high temperature short time (HTST, 72 °C, 15 s) treatment are two common methods of pasteurization4. Pasteurization could effectively eliminate the vast majority of microorganisms in dairy products, but some heat-resistant microorganisms still survive after treatment. Khalil et al.5 reported that after pasteurization, the content of Enterobacteriaceae bacteria in goat milk decreased significantly, but there were still some LAB in the samples, such as Lactobacillus kefiranofaciens and Lactobacillus kefiranofaciens. Rashtchi et al.6 found that the number of Escherichia coli in cheese prepared with pasteurized milk as raw material was significantly lower than that in cheese fermented with raw milk, while the number of LAB decreased significantly in the early fermentation period and remained stable after that. Müller et al.7 conducted 16S rRNA sequencing on pasteurized and UHT milk, which showed that there was still a higher level of LAB in milk after pasteurization, while the content of LAB in milk after UHT was very low. Because there are still some heat-resistant microorganisms in pasteurized samples, the enrichment of thermoresistant LAB can be realized after the treatment of this method7,8.
LAB are generally accepted as a group of Gram-positive, nonmotile, non-spore-forming bacteria that can ferment hexose carbohydrates under microaerophilic to strictly anaerobic conditions to produce mainly lactic acid9. LAB are widely distributed in milk, vegetables, and meat products. Some strains of LAB have potential probiotic effects, including regulating blood pressure, improving lactose intolerance, and lowering serum cholesterol10. LAB are usually prepared into bacterial powder by freeze-drying or spray-drying for applications in food, medicine and other fields. The productivity of freeze-drying technology is one-fifth that of spray-drying11, and its high cost and long drying time limit its application12. In contrast, the cost of spray drying is more than 10 times lower than that of freeze drying, and it has the advantages of rapid, efficient and continuous production13. However, the high temperature during spray drying will reduce the survival rate of LAB14. In addition, the high-temperature environment in which Lactobacillus plantarum and Lactobacillus fermentum et al. participate in various fermentation processes also has a certain impact on the activity of LAB, thus affecting the quality of fermented food. Therefore, it is necessary to obtain thermoresistant LAB. Ramírez-Chavarín et al.15 and Pérez-Chabela et al.16 reported that a variety of natural thermoresistant LAB are applied to the preservation and fermentation of meat products as biological protective agents, which can improve the safety of cooked meat products. Chen et al.17 suggested that the improved thermostable Lactobacillus plantarum can also be used as a silage additive to promote lignocellulose degradation and improve fermentation quality.
The main means to improve the heat resistance of LAB are mutagenesis treatment or adding protective agents. Although the success rate of ultraviolet mutagenesis or other mutagenesis techniques is high, the experiment period is long and the operation is complicated. The cost of various types of protective agents is slightly higher, and their safety is still to be verified. Using pasteurized milk as a sample to isolate thermoresistant LAB, this method does not require a long domestication process or gene editing, thus it can be used in the food industry. In this study, 17 kinds of commercial pasteurized milk were divided into three groups according to the shelf life: A (1–5 d), B (6–10 d), and C (11–15 d). The sequence information of the 16S rRNA V3–V4 region was amplified by molecular biological methods to analyze the relationship between bacterial diversity and shelf life. Furthermore, microbial isolation and culture technology were used to screen potential heat-resistant LAB in pasteurized milk and determine their thermoresistant properties. The objective of this study was to enrich the biodiversity information of pasteurized milks, thus providing a new way and idea for enriching the resource base of heat-tolerant LAB.
Materials and methods
Sample collection and preparation
Pasteurized cow milk was purchased from local supermarkets in Jinan, Shandong, China. The products originate from Beijing, Tianjin, Shanghai, Inner Mongolia Autonomous Region, Jiangsu Province, Zhejiang Province, Guangdong Province, Sichuan Province, Hubei Province, Shandong Province, Anhui Province, Henan Province, and Heilongjiang Province. The samples were placed into sterile tubes in the laboratory for metagenomics DNA extraction as follows. Pasteurized cow milk of 2 mL was mixed thoroughly and centrifuged at 10,000×g for 10 min. The supernatant was discarded, and genomic DNA was extracted from the remaining pellet using the Easy-DNA-Extraction Kit (Invitrogen, California, USA). The extracted DNA was quantified using a Qubit 2.0 spectrometer (Invitrogen, California, USA). PCR amplification of the bacterial 16S rRNA genes V3–V4 region was performed using the forward primer 338F (5ʹ-ACTCCTACGGGAGGCAGCA-3ʹ) and the reverse primer 806R (5ʹ-GGACTACHVGGGTWTCTAAT-3ʹ). PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2 × 250 bp sequencing was performed using the Illlumina NovaSeq platform with NovaSeq 6000 SP Reagent Kit (500 cycles) at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China).
Sequence analysis
Microbiome bioinformatics were performed with QIIME2 2019.418 with slight modification according to the official tutorials (https://docs.qiime2.org/2019.4/tutorials/). Briefly, raw sequence data were demultiplexed using the demux plugin following by primers cutting with cutadapt plugin19. Sequences were then quality filtered, denoised, merged and chimera removed using the DADA2 plugin20. Non-singleton amplicon sequence variants (ASVs) were aligned with mafft21.
Isolation and identification of microorganisms in pasteurized milk
The pasteurized milk samples were inoculated (20%, v/v) into LB and MRS liquid medium (Haibo, Qingdao, China), respectively. After cultivation for 24 h, 1 mL of the sample was mixed with 99 mL of sterile distilled water. Appropriate dilutions of 10–1 to 10–6 were carried out in the sterilized saline water. The LAB and other bacteria were isolated on the MRS and LB agar medium, respectively. After incubation at 37 °C, the predominant, morphologically distinct, and well-isolated colonies were transferred to the corresponding medium for further enrichment.
The strains were cultured in the MRS and LB medium at 37 °C for 24 h, after which the cells of the strains were harvested by centrifugation and the genomic DNA of the strains was extracted using a genome DNA isolation kit (Vazyme, Nanjing, China). Further, the 16S rRNA gene was amplified using the universal primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and primer 1492R (5′-TACGGYTACCTTGTTACGACTT-3′). PCR was performed by applying the 2× SanTaq PCR Mix (Yeasen, Shanghai, China). For the amplification reaction, 1 mixture was prepared in a 200-µL tube that contained 9.5 µL of sterilized ddH2O, 1 µL of primer 27F, 1 µL of 1492R, 1 µL of genomic DNA, and 12.5 µL of 2× SanTaq PCR Mix. The PCR amplification program was performed as follows: denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 10 min). Amplified 16S rRNA genes were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China), and the 16S rRNA gene sequence was identified using the BLAST program developed by the NCBI.
The optimum temperature and temperature tolerance of Enterococcus faecium
After overnight incubation, E. faecium SF1 and E. faecium SF2 were inoculated (2%, v/v) into MRS liquid medium, and statically cultured at 30 °C, 35 °C, 40 °C, 45 °C, and 50 °C, respectively. After cultivation for 24 h, the OD600 values of E. faecium were measured by ultraviolet spectrophotometer. To determine the temperature tolerance of E. faecium, E. faecium SFM1 and E. faecium SFM2 were collected by centrifugation after 12 h of cultivation. The bacterial cells were suspended in an equal volume of PBS buffer solution (NaCl, 8 g/L; KCl, 0.2 g/L; Na2HPO4, 1.44 g/L; KH2PO4, 0.24 g/L, pH 7.4). After incubation at 30 °C, 35 °C, 40 °C, 45 °C, and 50 °C for 2 h, the number of viable cell counts was counted to obtain the survival rates of the strain.
Safety evaluation
Hemolytic activity of E. faecium SFM1 and E. faecium SFM2 was assessed by streaking each isolate on Columbia CNA blood agar plates (Haibo, Qingdao, China). Plates were incubated at 37 °C for 24 h. Isolates surrounded by a transparent halo were classified as β-hemolytic, and those presenting no halo were classified as γ-hemolytic. Staphylococcus aureus ATCC6538 was used as a positive control (β-hemolytic).
The antimicrobial resistance profile of E. faecium SFM1 and E. faecium SFM2 was determined using the disc diffusion method. Resistance to penicillin (10 μg), chloramphenicol (30 μg), erythromycin (15 μg), vancomycin (30 μg), rifampicin (5 μg), ciprofloxacin (5 μg), and tetracycline (30 μg) was tested using discs from Oxoid, UK. The antibiotic tablets were placed on MRS plates containing 4.5 × 107 CFU/mL E. faecium and incubated at 37 °C for 24 h. The mean (M) and standard deviation (SD) of the inhibition halo diameter (x) of E. faecium SFM1 and E. faecium SFM2 were calculated for each antimicrobial, and isolates were classified as follows: x ≤ 15 mm—resistant; 16 mm < x < 20 mm—intermediate; x ≥ 21 mm—susceptible.
Characterization of probiotic potential
The simulated gastric fluid and intestinal fluid were prepared using the method of Son et al.22 with modification, and all digestive fluids were filtered through a 0.22-μm filter for sterilization. After overnight incubation, E. faecium SFM1 and E. faecium SFM2 were inoculated (2%, v/v) into MRS liquid medium and cultured statically at 37 °C, respectively. After cultivation for 12 h, 1 mL suspension of E. faecium SFM1 and E. faecium SFM2 was mixed with 9 mL of artificial gastric fluid (0.2% NaCl, 0.3% pepsin, pH 2.5), and incubated at 37 °C, respectively. After incubation for 1 h and 2 h, the cells were collected by centrifugation at 2000×g for 10 min, washed three times with PBS buffer, and then re-suspended in an equal volume of PBS buffer. 100 μL of the re-suspension was applied to MRS solid medium, the viable bacteria numbers were counted, and the survival rates of the strains in gastric fluid were calculated. Furthermore, 1 mL of the above resuspended cell solution was mixed with 9 mL of artificial intestinal fluid (0.1% trypsin, 0.6% bile salt, pH 8), and incubated at 37 °C for 8 h. The survival rates in artificial intestinal fluid were determined by the same method.
Determination of carbon sources of Enterococcus faecium
To detect the growth of E. faecium SFM1 (isolated from human oral) and E. faecium SFM2 (isolated from pasteurized milk) with different carbon sources, the glucose in MRS medium was replaced with 2% (w/v) fructose, sucrose, lactose, galactose, galacto-oligosaccharides (GOS), and stevioside to prepare MRS-fructose, MRS-sucrose, MRS-lactose, MRS-galactose, MRS-GOS, and MRS-stevioside medium. E. faecium SFM1 and SFM2 were inoculated (2%, v/v) into different kinds of MRS with different carbon sources cultured statically, and the sugar-free MRS was used as the control group. After cultivation for 24 h, the OD600 value was determined by a UV-5500 Ultraviolet–Visible spectrophotometer (Shanghai, China).
Statistical analysis
Each experiment was repeated at least three times. The results were statistically compared and analyzed with SPSS 22.0 software. This included calculating the mean and standard deviation of different parallel experiments. The p value was calculated by using the Student’s t testing method.
Results
Sequence quality control
A total of 2,329,929, including 1,908,493 high-quality 16S rRNA gene sequences were obtained using 16S rRNA sequencing (Table S1). A total of 33,783,415 ASVs (amplicon sequence variants) were finally obtained using a 99% sequence similarity threshold. The Good’s coverage was 99% on average, further indicating that it was sufficient for pasteurized milk microbiota analysis at the current sequencing depth.
Alpha and Beta diversities of milk microbiota based on shelf life
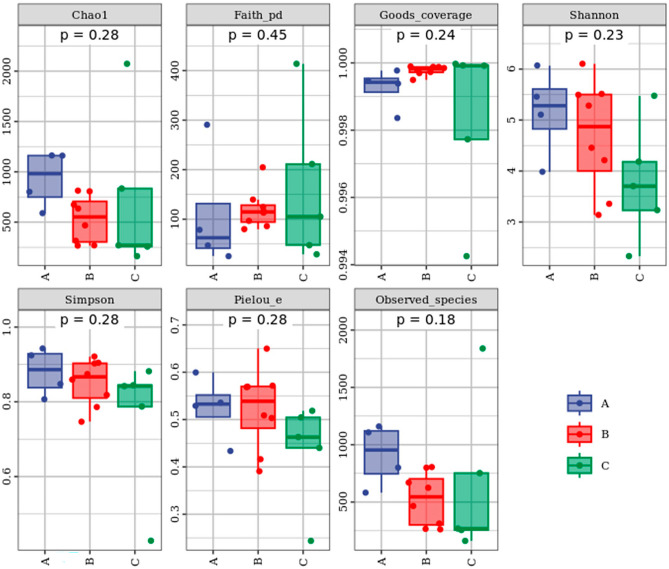
17 samples were divided into three groups according to the shelf-life: group A (1–5 days), group B (6–10 days), and group C (11–15 days). The α-diversity analysis of the three groups with different shelf life was shown in Fig. 1. Overall, bacterial community richness and diversity were generally higher in group A.
Fig. 1.
Alpha analysis metrics of pasteurized milk according to different shelf lives. Richness of microbiota in pasteurized milk was evaluated according to Chao1, Observed species, bacterial evenness and community coverage were evaluated by Pielou’s evenness, Shannon, and Simpson index. The evolutionary diversity and the degree of coverage were evaluated by Faith’s PD and Good’s coverage.
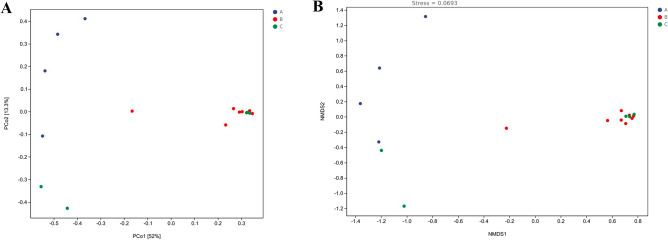
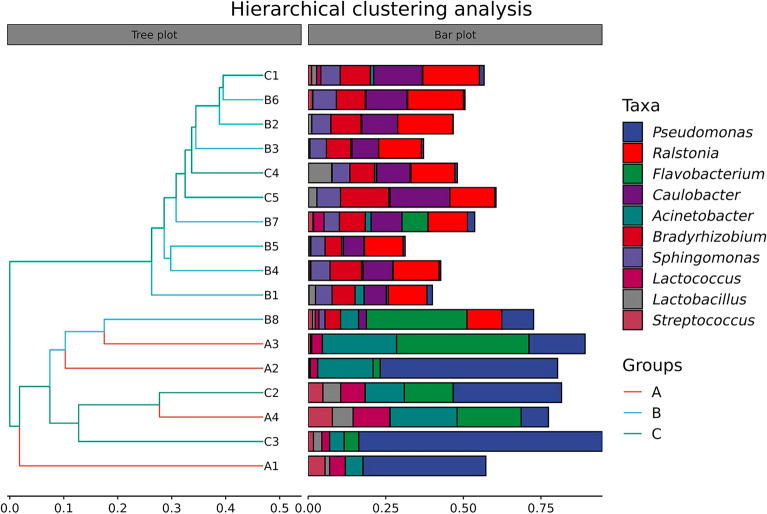
Changes in the microbial composition of pasteurized milk in different shelf life were compared by β-diversity analysis (Fig. 2A,B). The results showed that the microbial composition of group A differed significantly from that of group B and group C. PCoA at the genus level showed significant differences in microbial community composition between the three groups. Hierarchical cluster analysis revealed the similarities and differences among the three groups at the genus level (Fig. 3). It suggested that the community composition of group B and group C was similar, which was consistent with the analysis results of PCoA and NMDS. Hierarchical cluster analysis represented the top 10 genera in the sample in terms of abundance, which revealed that Pseudomonas, Flavobacterium, and Acinetobacter in group A had a high abundance, while Lactococcus microbe was present in some samples. The species composition of group B and group C was similar, which mainly included Ralstonia, Caulobacter, Bradyrhizobium, Sphingomonas and other microorganisms at the genus level. PCoA, NMDS and hierarchical cluster maps were drawn to evaluate beta diversity. The results showed that the species composition of group A was significantly different from that of the other two groups, while the species composition of group B and group C was similar.
Fig. 2.
Beta diversity was visualized with principal components analysis (PCoA) (A) and NMDS analysis (B). Dots with different colors represent the samples of different shelf lives. The horizontal and vertical axes represent the first and second principal coordinates, respectively, and the percentages on the horizontal and vertical axes are the contribution of that principal coordinate to the difference of the sample matrix data. The closer the projection distance between two points on the coordinate axis, the more similar the community composition.
Fig. 3.
Hierarchical clustering analysis. The similarity between samples was shown in the form of a hierarchical tree. The closer the projection distance between two branches, the higher similarity of the community composition between the samples.
Differences in microbial composition of pasteurized milk between different shelf-life groups
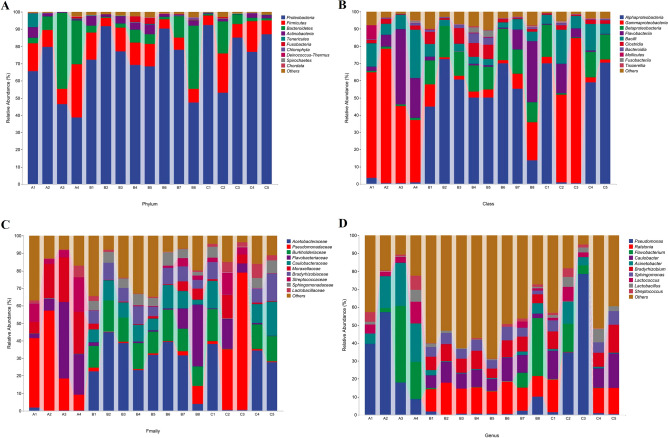
As shown in Fig. 4A, Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla, with relative abundance accounting for 94.54 ± 4.22% of the total bacteria. The most common classes in pasteurized milk were Alphaproteobacteria, Gammaproteobacteria, and Flavobacteria (Fig. 4B). At the family level, Acetobacteraceae, Ralstonia, and Burkholderiaceae were the dominant families (Fig. 4C). The most common genus in pasteurized milk were Pseudomonas, Ralstonia, and Flavobacterium (Fig. 4D). The relative abundance of microbial communities at the class, family, and genus levels varied significantly among samples with different shelf lives. The distribution of Gammaproteobacteria in group A samples was higher than that in group B and group C. The samples of group A contained little or no Acetobacteraceae and Burkholderiaceae, while the samples of group B and group C contained a higher number. The dominant bacteria in group A were Pseudomonas and Flavobacterium, while Ralstonia was the dominant bacteria in group B and group C. Lactococcus had the highest proportion in group A, and Lactobacillus had the highest number in group B and group C.
Fig. 4.
Relative abundance of bacteria in pasteurized milk at the level of phylum (A), class (B), family (C), and genus (D). Genera of the top ten abundant microorganisms were listed and unclassified were marked as others.
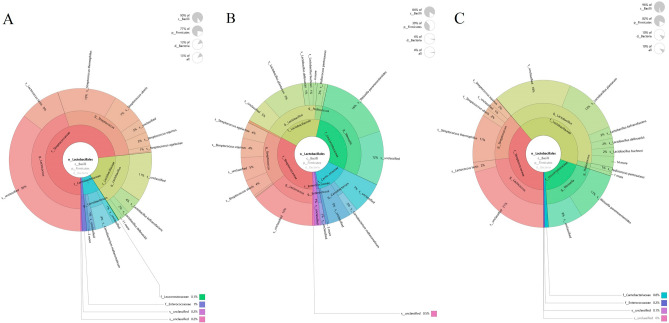
The Krona species composition map showed the species distribution of LAB in the three groups of samples with different shelf lives. The microorganisms with the highest abundance in the three groups all included the genus of Streptococcus, and there was a high proportion of Lactococcus lactis and Streptococcus thermophilus present in group A samples (Fig. 5A). In group B, the microorganisms with a higher proportion were Lactobacillus plantarum and Weissella paramesenteroides (Fig. 5B). The Lactobacillus plantarum and Weissella paramesenteroides in group C samples accounted for a relatively high proportion (Fig. 5C).
Fig. 5.
The Krona species composition map of LAB in group A (A), group B (B), and group C (C) based on different shelf lives.
Isolation and identification of bacteria in pasteurized milk
The 16S rRNA genes of isolated strains were amplified and sequenced for further genetic identification. The obtained 16S rRNA gene fragments were aligned via the BLAST program in NCBI, and the results indicated that Staphylococcus saprophyticus, Streptococcus parauberis, Enterococcus faecium, and Bacillus subtilis were isolated from group A. Staphylococcus, Enterobacter and Enterococcus were mostly isolated from group B. In group C, Enterobacter cloacae, Exiguobacterium acetylicum, and Microbacterium oxydans were isolated (Table 1).
Table 1.
The strains isolated from LB medium and MRS medium.
| Samples | Strains isolated in LB medium | Strains isolated in MRS medium |
|---|---|---|
| A1 | Staphylococcus saprophyticus | Streptococcus parauberis |
| A2 | Enterococcus faecium | – |
| A3 | – | – |
| A4 | Bacillus subtilis | – |
| B1 | Staphylococcus epidermidis | Staphylococcus epidermidis |
| B2 | – | – |
| B3 | Enterobacter ludwigii | Enterobacter cloacae |
| B4 | Enterobacter cloacae | Enterobacter cloacae |
| B5 | Enterococcus gallinarum | Enterococcus casseliflavus |
| B6 | Enterobacter cloacae | – |
| B7 | – | – |
| B8 | Enterobacter hormaechei | Enterobacter hormaechei |
| C1 | Enterobacter cloacae | – |
| C2 | – | – |
| C3 | – | – |
| C4 | – | – |
| C5 | Exiguobacterium acetylicum | Microbacterium oxydans |
“–” indicated that data was not detected.
Heat resistance of Enterococcus faecium
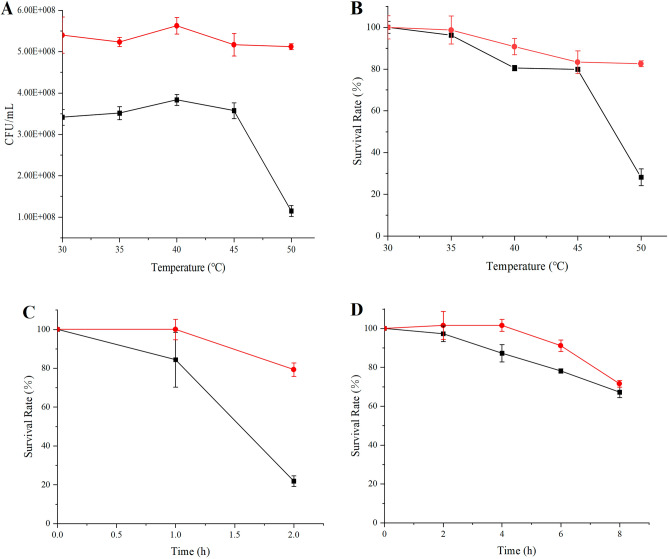
In this study, a strain of E. faecium SFM2 was isolated from the A2 sample. Furthermore, the optimum temperature and temperature tolerance of E. faecium SFM2 were explored and compared with the human oral source E. faecium SFM1 preserved in our laboratory. The optimal temperature of E. faecium SFM1 and E. faecium SFM2 was 35 °C, and the viable cell counts of E. faecium SFM2 were higher than those of E. faecium SFM1 at different temperatures (Fig. 6A). The temperature tolerance of these two strains was determined at 50 °C. After incubation at 50 °C for 2 h, the survival rates of E. faecium SFM1 and E. faecium SFM2 reached 28.20 ± 0.04% and 82.58 ± 0.01%, respectively (Fig. 6B).
Fig. 6.
(A) The viable cell counts of E. faecium cultured at different temperatures for 24 h. (B) The survival rates of E. faecium treated at different temperatures for 2 h. (C) The survival rates of E. faecium in simulated gastric fluid. (D) The survival rates of E. faecium in simulated intestinal fluid. E. faecium SFM1 was represented by black solid lines, and E. faecium SFM2 was represented by red solid lines.
Safety evaluation and assessment of probiotic potential of Enterococcus faecium
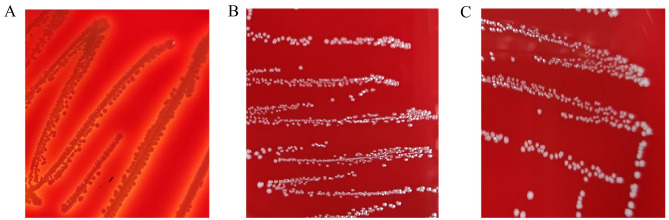
The safety evaluation results showed that both E. faecium SFM1 and E. faecium SFM2 presented no halo in Columbia CNA blood agar plate, and were classified as γ-hemolytic (Fig. 7). Another important safety concern is the presence of mobile antimicrobial resistance genes. The antibiotic sensitivity of E. faecium to 7 commonly used antibiotics was examined. As shown in Table 2, E. faecium SFM1 was sensitive to penicillin, chloramphenicol, vancomycin, rifampicin, ciprofloxacin, and tetracycline, while E. faecium SFM2 was sensitive to chloramphenicol, vancomycin, and rifampicin.
Fig. 7.
Hemolytic activity of S. aureus ATCC6538 (A), E. faecium SFM1 (B), and E. faecium SFM2 (C).
Table 2.
Antibiotic susceptibility profile of E. faecium SFM1 and E. faecium SFM2.
| Strains | Antibiotic | Bacteriostatic zone diameter (mm) | Antibiotic susceptibility |
|---|---|---|---|
| E. faecium SFM1 | Penicillin | 27.67 ± 0.58 | S |
| Chloramphenicol | 28.67 ± 1.53 | S | |
| Erythromycin | 13.67 ± 1.53 | R | |
| Vancomycin | 22.67 ± 1.53 | S | |
| Rifampicin | 31.67 ± 1.15 | S | |
| Ciprofloxacin | 22.67 ± 1.15 | S | |
| Tetracycline | 28.67 ± 0.58 | S | |
| E. faecium SFM2 | Penicillin | 18.00 ± 1.00 | I |
| Chloramphenicol | 26.67 ± 1.53 | S | |
| Erythromycin | 11.67 ± 0.58 | R | |
| Vancomycin | 22.67 ± 1.53 | S | |
| Rifampicin | 30.33 ± 0.58 | S | |
| Ciprofloxacin | 17.33 ± 0.58 | I | |
| Tetracycline | 12.33 ± 0.58 | R |
Values are presented as mean ± standard deviation of three independent experiments (n = 3).
“S” represented sensitive, “I” represented intermediate, and “R” represented resistance.
Furthermore, the tolerance of E. faecium SFM1 and E. faecium SFM2 in simulated gastric fluid and simulated intestinal fluid was shown in Fig. 6C,D. The survival rates of E. faecium SFM1 and E. faecium SFM2 were 21.88 ± 0.03% and 79.31 ± 0.03% (p < 0.001) after gastric fluid treatment for 2 h. After 8 h of intestinal fluid treatment, the survival rates of E. faecium SFM1 and E. faecium SFM2 reached 67.19 ± 0.03% and 71.57 ± 0.02% (p < 0.001), respectively. Therefore, E. faecium SFM2 isolated from pasteurized milk represented better temperature as well as gastrointestinal tolerance.
The carbon sources of Enterococcus faecium
The utilization of different carbon sources by E. faecium SFM1 and SFM2 was shown in Table 3. The carbon source spectra of the two strains were similar, both showing the highest growth in sucrose. After cultivation for 24 h, the OD600 value of E. faecium SFM1 and E. faecium SFM2 reached 1.58 ± 0.00 and 1.77 ± 0.00 (< 0.05), respectively. In addition, compared with E. faecium SFM1, E. faecium SFM2 exhibited higher biomass in glucose, fructose, lactose, and galactose.
Table 3.
The utilization of different carbon sources by E. faecium SFM1 and E. faecium SFM2.
| E. faecium SFM1 | E. faecium SFM2 | |
|---|---|---|
| Control | 0.79 ± 0.02i | 0.77 ± 0.02i |
| Glucose | 1.44 ± 0.02d | 1.67 ± 0.02b |
| Fructose | 1.21 ± 0.03f | 1.40 ± 0.00d |
| Sucrose | 1.58 ± 0.00c | 1.77 ± 0.00a |
| Lactose | 1.41 ± 0.06d | 1.69 ± 0.02b |
| Galactose | 1.06 ± 0.00g | 1.34 ± 0.05e |
| GOS | 1.36 ± 0.02e | 1.35 ± 0.01e |
| Stevioside | 0.98 ± 0.01h | 0.96 ± 0.03h |
Values are presented as mean ± standard deviation of three independent experiments (n = 3).
Different lowercase superscript letters indicated statistically differences at P < 0.05.
Discussion
Metagenomic sequencing is a commonly used method to analyze species diversity in samples. Nowadays, microbial diversity analysis is used to explore the microbiota of the milk environment, which provides the latest information on the changes in microbiota, the control of food-borne pathogens, and the discovery of beneficial microorganisms. Yuan et al.1 used 16S rRNA amplicon sequencing to analyze the variation in raw milk microbiota throughout the year from three farms in China, providing new information on the ecology of the raw milk microbiota in pastures and contributing to understanding the changes in the Chinese milk microbiota. Luoyizha et al.2 used high-throughput sequencing technology to determine bacterial communities in donkey milk from two counties in eastern and western China, providing the microbial profiles of pathogens and spoilage bacteria that need to be controlled, and proposing the possible utilization of beneficial microbial resources for the future. Delgado et al.8 assessed the microbial diversity of raw milk treated by pasteurization (63 °C, 30 min), and sequencing results showed that S. thermophilus accounted for more than 98% of the reads in IPM samples (incubation at 42 °C for 24 h after pasteurization). This study provided the possibility to isolate new thermophilic LAB strains from pasteurized milk. In our study, 16S rRNA amplification sequencing was performed on 17 commercial pasteurized milk samples with different shelf lives, and the distribution of microorganisms in pasteurized milk with different shelf lives provided effective information for analyzing the potential microbiota of pasteurized milk and controlling foodborne pathogenic microorganisms.
Pasteurization is usually heated at 62.5 °C for 30 min or at 72–75 °C for 15 s, and the shelf life of the sample may be related to the sterilization time and temperature. Therefore, we thought that the shorter the shelf life, the lower the pasteurization temperature or the shorter the processing time. The heat treatment temperature of group A samples with the shortest shelf life may be lower, and the viable cell counts of sample A4 were the highest, so some thermoresistant LAB (such as E. faecium SFM2) could survive. The shelf life of group C was the longest, and the content of pathogenic bacteria such as Staphylococcus epidermidis was significantly lower than that of group B. The results of microbial isolation and identification showed that the number of viable cell counts in 17 kinds of commercial pasteurized milk was less than 104 CFU/mL. In addition, there were two samples that did not meet the conditions for building a library and did not participate in the diversity analysis, which may be the samples that also used ceramic filtration technology after pasteurization, so that the number of bacteria in the product was lower than the standard of the database (data not shown).
The diversity of microorganisms in raw milk may come from a variety of sources, such as milk teats, milking equipment, feeding environment, feed and manure, and even after a short period of high-temperature treatment, a small amount of microorganisms may remain in the finished product23. S. epidermidis was isolated both in group A and group B. S. epidermidis is a normal human microbiota and is widely distributed on the skin surface and intestinal tract of humans and other animals, but it is also a conditional pathogen, easy to cause cross infection in hospitals, and a pathogen that causes mastitis in dairy cows24, Fernandes25. The microorganisms isolated from group B samples mostly belong to the genera of Enterobacter and Enterococcus, such as Ec. gallinarum and Eb. cloacae, which are widely present in human and animal feces and soil. Besides, they are a common contaminating strain of dairy products and are also pathogens causing mastitis in cattle26,27.
LAB is a natural and important microbial community in the milk environment. Different studies have reported LAB in the milk environment. For example, camel milk is typically dominated by Lactobacillus helveticus, Lactiplantibacillus plantarum and Lacticaseibacillus casei28, cow milk contains a significant LAB population that includes Lactobacillus, Streptococcus and Enterococcus1, goat milk is also typically dominated by LAB, including genera of Lactococcus and Lactobacillus29, buffalo milk contains a large population of LAB, including Lactococci and Lactobacilli30, and LAB are also present in breast milk, including Lactobacillus and Bifidobacterium23. In this study, metagenomic sequencing results also showed that the cow milk contained a high abundance of LAB. In addition, LAB strains were also screened in culture experiments.
This study also provided a new idea for expanding the resource pool of thermoresistant LAB strains. The application range of thermoresistant LAB is very wide, leading researchers to obtain thermoresistant LAB strains through different methods. For example, Dorau et al.31 used a setup where the temperature was gradually increased over time, and isolated two evolved Lactococcus lactis strains (RD01 and RD07) better able to tolerate high growth temperatures. Jeon et al.32 developed a new Lactobacillus acidophilus strain with improved heat resistance while retaining the existing beneficial properties through the adaptive laboratory evolution (ALE) method. Yang et al.33 found that Mg2+ at 10–50 mmol/L could improve the survival rate of Lactobacillus plantarum and Lactobacillus rhamnosus under high-temperature environments. Gong et al.13 and Moreira et al.34 used skim milk powder, sucrose, etc. as protective agents to increase cell viability. In addition, gene editing could also improve the stress resistance of the strain. Desmond et al.35 overexpressed groEL in Lactobacillus paracasei and Lactococcus lactis, and the recombinant strains were cultured at 60 °C and 54 °C, the heat-adapted cultures maintained the highest level of viability (5-log-unit increase, approximately). Through the results of this study, pasteurized milk is a good source for isolating thermoresistant LAB strains, and E. faecium SFM2 was obtained.
E. faecium is a type of LAB, widely present in the feces of healthy infants and long-lived elderly people, and it could be used as a starter culture or probiotic culture in animal feed additives36. Aspri et al.37 isolated E. faecium from donkey milk, which showed good antibacterial activity against S. aureus and Listeria monocytogenes. Quintela-Baluja et al.38 performed a proteomic analysis of two bacteriocin-producing E. faecium strains and found that they hold great promise in terms of bioengineering and biotechnology properties. Rajput et al.39 isolated E. faecium GMB16 from goat and sheep milk, which showed significant probiotic potential and immunological properties. In this study, a strain of E. faecium SFM2 was isolated from the A2 sample. The safety evaluation results showed that E. faecium SFM2 presented no halo in Columbia CNA blood agar plate and was classified as γ-hemolytic. Hemolytic ability is a relevant virulence factor that can be present in pathogenic microorganisms. To be safe for use, probiotic strains must be nonhemolytic. Besides, E. faecium SFM2 was sensitive to chloramphenicol, vancomycin, and rifampicin. Furthermore, the physiological and biochemical characteristics of E. faecium SFM2 were explored and compared with the human oral source E. faecium SFM1 preserved in the laboratory, including sugar utilization, optimal temperature, temperature tolerance, and the survival rates in a simulated gastrointestinal environment. The results showed that E. faecium SFM1 and E. faecium SFM2 had better utilization capacity for sucrose, glucose, and lactose. The optimal temperature of E. faecium SFM1 and E. faecium SFM2 was 35 °C, and the OD600 of E. faecium SFM2 was higher than that of E. faecium SFM1. After incubation at different temperatures for 2 h, E. faecium SFM2 showed higher survival rates in the temperature range of 30–50 °C. Besides, E. faecium SFM2 showed stronger gastrointestinal tolerance than E. faecium SFM1. This study provided a new idea for enriching the resource library of high-temperature resistant LAB strains.
Conclusion
According to the shelf life, all samples were divided into three groups: group A (1–5 d), group B (6–10 d), and group C (11–15 d). The analysis of bacterial diversity in pasteurized milk showed that Proteobacteria, Firmicutes and Bacteroidetes accounted for the highest proportion in the three groups. The species composition of group A was significantly different from that of group B and group C, while the latter two groups were similar. Most of the LAB in the three groups were Lactobacillus and Lactococcus, and the top five LAB all contained Lc. raffinosa and Lb. salivarius. Furthermore, a strain of E. faecium SFM2 was isolated from the A2 sample, which showed better temperature tolerance compared to the E. faecium SFM1 of oral origin. After treatment at 50 °C for 2 h, the survival rates of E. faecium SFM1 and SFM2 were 28.20 ± 0.04% and 82.58 ± 0.01%, respectively. Through this study, we provided some information on the microbial diversity in pasteurized milk, and pointed out that pasteurized milk is an important source of thermoresistant LAB. In the future, we can try to isolate more thermoresistant LAB from pasteurized milk and study their probiotic-related properties.
Supplementary Information
Acknowledgements
This work was supported by Natural Science Foundation of Shandong Province ZR2021QC160.
Author contributions
Jiancun Zhao: Data curation, Formal analysis, Writing—original draft; Jian Gong: Data curation, Formal analysis; Wanjie Liang: Formal analysis; Susu Zhang: Funding Acquisition, Supervision, Writing—Review & Editing.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80947-5.
References
- 1.Yuan, H. et al. Microbial properties of raw milk throughout the year and their relationships to quality parameters. Foods11(19), 3077. 10.3390/foods11193077 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luoyizha, W. et al. Compared analysis of microbial diversity in donkey milk from Xinjiang and Shandong of China through High-throughput sequencing. Food Res. Int.137, 109684. 10.1016/j.foodres.2020.109684 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Sunds, A. V., Rauh, V. M., Sørensen, J. & Larsen, L. B. Maillard reaction progress in UHT milk during storage at different temperature levels and cycles. Int. Dairy J.77, 56–64. 10.1016/j.idairyj.2017.08.008 (2018). [Google Scholar]
- 4.Chatterton, D. E. W. et al. Bioactive proteins in bovine colostrum and effects of heating, drying and irradiation. Food Funct.11(3), 2309–2327. 10.1039/c9fo02998b (2020). [DOI] [PubMed] [Google Scholar]
- 5.Khalil, R. A. et al. Artisanal household milk pasteurization is not a determining factor in structuring the microbial communities of labneh Ambaris: A pilot study. Foods11(23), 3874. 10.3390/foods11233874 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashtchi, P., Bazmi, A., Noshirvani, N. & Moosavy, M. H. Comparison of the microbial, physicochemical, and sensorial properties of raw and pasteurized Lighvan cheeses during ripening time. Food Sci. Nutr.9(10), 5527–5535. 10.1002/fsn3.2511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller, T., Lunardi, L. M., Maciel, M. J. & Rempel, C. Microbiological profile of raw refrigerated and processed bovine milk at dairy industries from Vale do Taquari, Rio Grande do Sul, Brazil. Arq. Inst. Biol.89, e00302021. 10.1590/1808-1657000302021 (2022). [Google Scholar]
- 8.Delgado, S. et al. Diversity of thermophilic bacteria in raw, pasteurized and selectively-cultured milk, as assessed by culturing, PCR-DGGE and pyrosequencing. Food Microbiol.36(1), 103–111. 10.1016/j.fm.2013.04.015 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Wu, C., Huang, J. & Zhou, R. Genomics of lactic acid bacteria: Current status and potential applications. Crit. Rev. Microbiol.43(4), 393–404. 10.1080/1040841x.2016.1179623 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Liu, J. et al. Soybean protein isolate treated with transglutaminase (TGase) enhances the heat tolerance of selected lactic acid bacteria strains to spray drying. Food Chem.404(Pt B), 134676. 10.1016/j.foodchem.2022.134676 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Huang, S. et al. Spray drying of probiotics and other food-grade bacteria: A review. Trends Food Sci. Technol.63, 1–17. 10.1016/j.tifs.2017.02.007 (2017). [Google Scholar]
- 12.Kiepś, J. & Dembczyński, R. Current trends in the production of probiotic formulations. Foods11(15), 2330. 10.3390/foods11152330 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong, P. et al. Sampling method for studying the activity of lactic acid bacteria during spray drying. Dry. Technol.36(10), 1236–1242. 10.1080/07373937.2017.1396226 (2018). [Google Scholar]
- 14.Rokka, S. & Rantamäki, P. Protecting probiotic bacteria by microencapsulation: Challenges for industrial applications. Eur. Food Res. Technol.231(1), 1–12. 10.1007/s00217-010-1246-2 (2010). [Google Scholar]
- 15.Ramírez-Chavarín, N. L., Wacher-Rodarte, C. & Pérez-Chabela, M. L. Characterization and identification of thermotolerant lactic acid bacteria isolated from cooked sausage as bioprotective cultures. J. Muscle Foods21(3), 585–596. 10.1111/j.1745-4573.2009.00206.x (2010). [Google Scholar]
- 16.Pérez-Chabela, M. L., Lara-Labastida, R., Rodriguez-Huezo, E. & Totosaus, A. Effect of spray drying encapsulation of thermotolerant lactic acid bacteria on meat batters properties. Food Bioprocess Technol.6(6), 1505–1515. 10.1007/s11947-012-0865-y (2013). [Google Scholar]
- 17.Chen, C. et al. Effects of Acremonium cellulase and heat-resistant lactic acid bacteria on lignocellulose degradation, fermentation quality, and microbial community structure of hybrid elephant grass silage in humid and hot areas. Front. Microbiol.12(9), 1164. 10.3389/fmicb.2022.1066753 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolyen, E. et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr.6, e272952. 10.7287/peerj.preprints.27295v2 (2018). [Google Scholar]
- 19.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J.17, 10–12. 10.14806/ej.17.1.200 (2011). [Google Scholar]
- 20.Callahan, B. J. et al. Dada2: High-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583. 10.1038/nmeth.3869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh, K., Misawa, K., Kuma, K. & Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res.30, 3059–3066. 10.1093/nar/gkf436 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Son, S. H. et al. Potential probiotic Lactobacillus plantarum Ln4 from kimchi: Evaluation of β-galactosidase and antioxidant activities. LWT Food Sci. Technol.85, 181–186. 10.1016/j.lwt.2017.07.018 (2017). [Google Scholar]
- 23.Quigley, L. et al. The complex microbiota of raw milk. FEMS Microbiol. Rev.37(5), 664–698. 10.1111/1574-6976.12030 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol.7(8), 555–567. 10.1038/nrmicro2182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dos Santos, F. F. et al. Presence of mecA-positive multidrug-resistant Staphylococcus epidermidis in bovine milk samples in Brazil. J. Dairy Sci.99(2), 1374–1382. 10.3168/jds.2015-9931 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Mukuna, W., Mafiz, A., Pokharel, B., Tobenna, A. & Kilonzo-Nthenge, A. Antibiotic resistant Enterobacteriaceae in milk alternatives. Foods10(12), 3070. 10.3390/foods10123070 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam, H. M. et al. Antimicrobial resistance of enterococci isolated from mastitic bovine milk samples in Korea. Zoonoses Public Health57(7–8), e59–e64. 10.1111/j.18632378.2009.01307.x (2010). [DOI] [PubMed] [Google Scholar]
- 28.Khedid, K., Faid, M., Mokhtari, A., Soulaymani, A. & Zinedine, A. Characterization of lactic acid bacteria isolated from the one humped camel milk produced in Morocco. Microbiol. Res.164, 81–91. 10.1016/j.micres.2006.10.008 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Nikolic, M. et al. Characterization of lactic acid bacteria isolated from Bukuljac, a homemade goat’s milk cheese. Int. J. Food Microbiol.122, 162–170. 10.1016/j.ijfoodmicro.2007.11.075 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Han, B. Z. et al. A survey on the microbiological and chemical composition of buffalo milk in China. Food Control18, 742–746. 10.1016/j.foodcont.2006.03.011 (2007). [Google Scholar]
- 31.Dorau, R., Chen, J., Liu, J., Ruhdal Jensen, P. & Solem, C. Adaptive laboratory evolution as a means to generate Lactococcus lactis strains with improved thermotolerance and ability to autolyze. Appl. Environ. Microbiol.87(21), e0103521. 10.1128/aem.01035-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon, S. et al. Complete genome sequence of the newly developed Lactobacillus acidophilus strain with improved thermal adaptability. Front. Microbiol.12, 697351. 10.3389/fmicb.2021.697351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y. et al. Complete genome sequence of Lactobacillus salivarius AR809, a probiotic strain with oropharyngeal tract resistance and adhesion to the oral epithelial cells. Curr. Microbiol.79(9), 280. 10.1007/s00284-022-02963-w (2022). [DOI] [PubMed] [Google Scholar]
- 34.Moreira, M. T. C. et al. Challenges associated with spray drying of lactic acid bacteria: Understanding cell viability loss. Compr. Rev. Food Sci. Food Saf.20(4), 3267–3283. 10.1111/1541-4337.12774 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Desmond, C., Fitzgerald, G. F., Stanton, C. & Ross, R. P. Improved stress tolerance of GroESL-overproducing Lactococcus lactis and probiotic Lactobacillus paracasei NFBC 338. Appl. Environ. Microbiol.70(10), 5929–5936. 10.1128/aem.70.10.5929-5936.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suvorov, A. et al. Evaluation of the efficacy of Enterococcus faecium L3 as a feed probiotic additive in chicken. Probiot. Antimicrob. Proteins15(5), 1169–1179. 10.1007/s12602-022-09970-0 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Aspri, M. et al. Application of bacteriocin-producing Enterococcus faecium isolated from donkey milk, in the bio-control of Listeria monocytogenes in fresh whey cheese. Int. Dairy J.73, 1–9. 10.1016/j.idairyj.2017.04.008 (2017). [Google Scholar]
- 38.Quintela-Baluja, M. et al. Rapid proteomic characterization of bacteriocin-producing Enterococcus faecium strains from foodstuffs. Int. J. Mol. Sci.23(22), 13830. 10.3390/ijms232213830 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajput, K., Dubey, R. C. & Kumar, A. Probiotic potential and immunomodulatory properties in Enterococcus faecium GMB24 and Enterococcus hirae SMB16 isolated from goat and sheep milk. Arch. Microbiol.204(10), 619. 10.1007/s00203-022-03217-w (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.