Abstract
Mutation of the von Hippel-Lindau tumor suppressor gene (vhl) causes the von Hippel-Lindau cancer syndrome as well as sporadic renal clear cell carcinoma. To pursue our study of the intracellular localization of VHL protein in relation to its function, we fused VHL to the green fluorescent protein (GFP) to produce the VHL-GFP fusion protein. Like VHL, VHL-GFP binds to elongins B and C and Cullin-2 and regulates target gene product levels, including levels of vascular endothelial growth factor and glucose transporter 1. VHL-GFP localizes predominantly to the cytoplasm, with some detectable nuclear signal. Inhibition of transcription by actinomycin D or 5,6-dichlorobenzimidazole riboside (DRB) causes VHL to be redistributed to the nucleus. A cellular fusion assay was used to demonstrate that inhibition of transcription induces a decrease in the nuclear export rate of VHL. The dependence of transcription for trafficking is lost with a deletion of exon 2, a region with a mutation causing a splice defect in the VHL gene in sporadic renal clear cell carcinoma. Addition of a strong nuclear export signal (NES) derived from the Rev protein results in complete nuclear exclusion and abrogates the redistribution of VHL-GFP-NES into the nucleus upon inhibition of transcription. Leptomycin B, which inhibits NES-mediated nuclear export, reverts the distribution of VHL-GFP-NES to that of VHL-GFP and restores sensitivity to actinomycin D and DRB. Uncoupling of VHL-GFP trafficking to transcription either by an exon 2 deletion or fusion to NES abolishes VHL function. We suggest that VHL function requires not only nuclear or cytoplasmic localization, but also exon 2-mediated transcription-dependent trafficking between these two cellular compartments.
The von Hippel-Lindau gene (vhl) was identified in 1993 as a tumor suppressor gene whose germ line mutations are associated with the inherited von Hippel-Lindau cancer syndrome (22, 28, 30, 34). This disorder is characterized by development of multiple benign and malignant tumors in many organs, including the kidneys, retina, central nervous system, pancreas, and adrenal gland (5, 8, 17, 38, 65). Further genetic analysis has revealed that the VHL gene is clearly inactivated in over 80% of sporadic renal clear cell carcinomas (RCC), the most frequent form of kidney cancer (14, 18, 21, 59).
The VHL gene, which contains three exons, codes for a 213-amino-acid protein of 25 kDa with no similarity to other known proteins, thus giving no clues about its function. Immunoprecipitation experiments have shown that VHL forms a stable heterotetrameric complex with elongins B and C as well as human Cullin-2 (VHL–elongin BC–Hs-Cul-2) (2, 9, 10, 25, 36, 51). Two of these proteins, elongin C and Hs-Cul-2, share structural homology with yeast (Saccharomyces cerevisiae) SKP1 and Cdc53, respectively (3, 26, 39, 67). SKP1 and Cdc53 are part of a complex that is involved in the targeting of pro teins for ubiquitination. Studies of the effect of reintroduction of VHL in VHL-negative RCC have indicated that it may play a role in the posttranscription regulation of the stability of a specific class of mRNAs that include vascular endothelial growth factor (VEGF), glucose transporter 1 (Glut-1), and other mRNAs (19, 24, 27, 36, 58). It has been suggested that the VHL–elongin BC–Hs-Cul-2 complex may target ubiquitination proteins involved in the regulation of the stability of VEGF and other mRNAs (36). There are several naturally occurring mutations in VHL patients within exon 3 that abrogate assembly with elongins B and C and Hs-Cul-2, suggesting a functional role for this complex (36, 51). However, most inactivating point mutations in sporadic RCC lie outside of the elongin BC–Hs-Cul-2 binding domain, suggesting that these sequences code for VHL tumor suppression functions which are still unknown.
Very little is known about the regulatory mechanisms underlying VHL cellular functions. In an attempt to answer these questions, we and others have studied the intracellular localization by indirect immunofluorescence or immunochemistry and have shown that VHL localizes predominantly to the cytoplasm, with some detectable nuclear signal (7, 9, 23, 31, 37, 64). Several proteins differentially localize between the nucleus and the cytoplasm, some of which require continuous nuclear-cytoplasmic shuttling to achieve their cellular functions (48, 52–54, 56, 70). This prompted us to hypothesize that VHL may need to traffic between the nucleus and the cytoplasm to perform its functions. We also reasoned that because VHL has been linked to the posttranscriptional regulation of specific mRNAs, ongoing transcription might be required for the shuttling process to occur.
We decided to further examine these possibilities by fusing VHL to green fluorescent protein (GFP) to form the hybrid protein VHL-GFP. Fusion to GFP enables detection in living cells without requiring fixation, antibodies, and permeabilization, all sources of common artifacts (35, 63). We report here that VHL-GFP reproduces the known biochemical, functional, and cellular localization characteristics of VHL. VHL-GFP can be detected predominantly in the cytoplasm, with some nuclear signal in all cell lines studied, regardless of expression levels. We present evidence that VHL-GFP participates in a dynamic RNA polymerase II transcription-dependent shuttling process between the cytoplasm and the nucleus. Transcription-dependent trafficking is mediated by sequences encoded by exon 2, which is the site of frequent mutation in human RCC. Uncoupling of transcription dependence to VHL-GFP localization and trafficking, either by an exon 2 deletion or fusion to a strong nuclear export signal (NES) such as Rev NES, disrupts VHL function. These results support a model in which VHL-GFP requires transcription-dependent nuclear-cytoplasmic shuttling to perform its functions.
MATERIALS AND METHODS
Cell culture.
The 786-0 (VHL-negative) renal carcinoma cells, Cos-7 African green monkey kidney cells, and HeLa cells were obtained from the American Type Culture Collection (Rockville, Md.). All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS) in a 37°C, humidified, 5% CO2-containing-atmosphere incubator.
Expression vectors.
The human VHL cDNA, which codes for a 213-amino-acid VHL protein, was subcloned into either pcDNA3.1(−) (Invitrogen), the retrovirus pHIT111 (kindly provided by A. Kingsman), or pSX, a modified version of the mammalian expression vector pCDL-SRα (9). A Flag (F) epitope tag (DYKDDDDK) or a hemagglutinin (HA) epitope was added to the N terminus of the VHL cDNA open reading frame to produce the FVHL or HA-VHL construct, respectively. A cDNA coding for an enhanced fluorescence version (Fred25) (63) of the GFP was subcloned at the C terminus of FVHL to produce the FVHL-GFP fusion protein. The VHL version without the Flag epitope tag was also cloned into the three vectors to produce the VHL-GFP fusion protein. Deletion mutants were produced by the PCR and cloned between the Flag tag and GFP to replace the wild-type VHL cDNA. The deletion mutants are referred to as ΔN for N-terminal truncations, ΔC for C-terminal truncations, and Δ for internal truncations. FVHL-GFP-NES and FVHL-GFP-NESM fusions were produced by fusion of FVHL-GFP at its C terminus to the strong NES of HIV Rev, LPPLERLTL (NES) (11), or to a mutated form of NES (NESM), LPPAERATL (underlining indicates mutated amino acids). All constructs were verified by standard DNA sequencing.
Transfections and establishment of stable 786-0 cell lines.
Stable cell lines were established by different strategies. First, a transient three-plasmid system was used to produce high-titer stocks of pHITVHL-GFP, a VHL-GFP-producing retrovirus vector derived from pHIT (60). Briefly, 293T cells were cotransfected with 3.4 μg of pHIT60 (Moloney murine leukemia virus gag-pol expression), 0.2 μg of pHCMV-G (vesicular stomatitis virus G protein), and 14.2 μg of pHITVHL-GFP per 60-mm-diameter petri dish by the calcium phosphate coprecipitation method. The medium (DMEM with 10% FCS) was changed after 10 h. Supernatant was harvested 48 h posttransfection, filtered (0.45-μm pore diameter), and frozen at −80°C. A virus titer of >1 × 107 infectious particles/ml was obtained. For infection, 4 × 105 786-0 cells were plated in a 60-mm-diameter petri dish. After 12 h, cells were washed and incubated with 2 ml of the virus stock, including 16 μl of Polybrene (1 mg/ml) for 2 h. Three milliliters of DMEM (10% fetal calf serum [FCS]) was added. After 24 h, cells were washed, trypsinized, and plated in a 75-cm2 cell culture flask. On the fourth day after infection, cells were incubated under selection with 0.5 mg of G418 per ml. A noninfected control was put under selection at the same time. While in the control cells there was extensive cell death after 2 weeks, the infected cells showed no signs of G418-induced death, indicating a high proportion of transduced cells. Second, 786-0 stable cell lines expressing FVHL-GFP or mutants cloned into pcDNA3 were also produced by a standard calcium phosphate method followed by G418 selection. Cells positive for FVHL-GFP, VHL-GFP, and VHL-GFP mutants were enriched by fluorescence-activated cell sorting. Cells were trypsinized, resuspended in phenol red-free DMEM containing 2% FCS, and sorted in a Becton Dickinson FacsStar at 488 nm. The brightest 3% of the cells were collected and put back into culture. We were able to obtain stable populations of cells which contained at least 99% GFP-positive cells by using this approach. Transient transfections were performed either by the calcium phosphate technique or electroporation as described elsewhere (31). The 786-0 cells expressing a VHL cDNA fused to an HA tag (WT-7) were a kind gift from William G. Kaelin, Jr. (Dana-Faber Cancer Institute, Harvard University).
Immunoprecipitation and immunoblotting.
Stable 786-0 cells or transiently transfected Cos-7 cells were plated overnight in DMEM–10% FCS. Transfected cells were incubated either with or without labeling medium, which included DMEM without methionine and cysteine, 10% FCS, and 0.1 mCi of Trans-35S label per ml, for 6 h at 37°C. Cells were lysed in a mixture of 20 mM Tris-HCl (pH 8.0), 137 mM NaCl, 1% Triton X-100, 10% (vol/vol) glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin per ml, 1 μg of aprotinin per ml, 1 mM NaF, and 1 mM sodium orthovanadate. Lysates were immunoprecipitated with the anti-Flag M2 monoclonal antibody (MAb) (1 μg/ml), anti-HA MAb (HA-11) (1 μg/ml), or anti-VHL MAb (PharMingen) (10 μg/ml). Precipitates were washed five times with the same buffer and analyzed on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels. For total cell lysates, cells were washed several times in phosphate-buffered saline (PBS), scraped from the petri dishes, centrifuged, and resuspended in 4% SDS in PBS (31). The samples were boiled for 5 min, and the DNA was sheared by passage of lysates through 19-gauge needles. Protein concentration was determined by the bicinchoninic acid method (Pierce) and was used to normalize protein loading in whole-cell immunoblot assay.
RT-PCR.
VEGF mRNA expression level in 786-0 cells was measured by a quantitative reverse transcription (RT)-PCR approach (44). Briefly, RNA was isolated and reverse transcribed with random hexamers and avian myeloblastosis virus reverse transcriptase. The cDNA was amplified with primers specific for VEGF (VEGF-S, GAGCCTTGCCTTGCTGCTCT; VEGF-A, GCACACAGGATGGCTTGAAGATGTAC). The sense primer was radiolabelled with 32P. After 20 cycles of amplification with AmpliTaq (Perkin-Elmer), the products were separated on a denaturing 6% acrylamide–urea sequencing gel. The gel was dried and exposed on a Phosphoimager screen (Fuji). Signals were quantified on a FujiBas Phosphoimager. As an internal control for the amount of RNA, primers for porphobilinogen deaminase (PBGD) were used. To ensure the linearity of the amplification, three dilutions of the cDNA were amplified with the VEGF-specific primers. The signal intensity of the bands was plotted against the dilution, and a linear regression was determined for each sample. The r2 values ranged from 0.97 to 0.99, indicating linearity of the amplification reaction.
Fluorescence analysis and image processing.
GFP analysis was generally performed 16 to 24 h after transient transfection or plating of stable cell lines. Live-cell image analysis was performed as described previously (6). Briefly, for each image, data were collected in a single imaging session with identical filters, exposure, and illumination settings. Images were manipulated with IPlab Spectrum, NIH-Image, and Adobe Photoshop software on a Macintosh computer. Nuclear and cytoplasmic values and the nuclear/cytoplasmic ratio were measured with IPlab Spectrum as follows. Total cellular signal was measured by multiplying the area of the cell by the integrated pixel intensity within the cell and subtracting the background value (obtained by measuring pixel intensity from a cell-free region of the image). Nuclear signal was obtained by measuring the pixel intensity in the nucleus subtracted by the mean background multiplied by the area of the nucleus. Cytoplasmic signal equals total cellular signal minus nuclear signal. Finally, the nuclear/cytoplasmic ratio was measured by dividing nuclear signal by cytoplasmic signal. All pixel values were measured well below the saturation limits, even though some of the prints may appear saturated by the signals. This is because images obtained with a slow-scan charge-coupled device (CCD) camera have a depth of 12 bits. Therefore, every pixel can assume an intensity value ranging between 0 and 4095. Since current monitor and printing technology only allows the presentation of 8-bit information (256 gray values), the 12-bit data have to be assigned to an 8-bit range. In IPlab Spectrum, this is done by normalization (i.e., defining a minimal and maximal cutoff value (pixels below are black, pixels above are white), followed by a contrast enhancement step which favors the pixels carrying important information. This manipulation does not alter the original 12-bit data, and quantitations are done with the original 12-bit images. This can lead to a situation in which brighter images may appear saturated compared to weaker signals, while they are, in fact, well below saturation levels (e.g., Fig. 4A, donor nucleus −DRB and +DRB at 10 min).
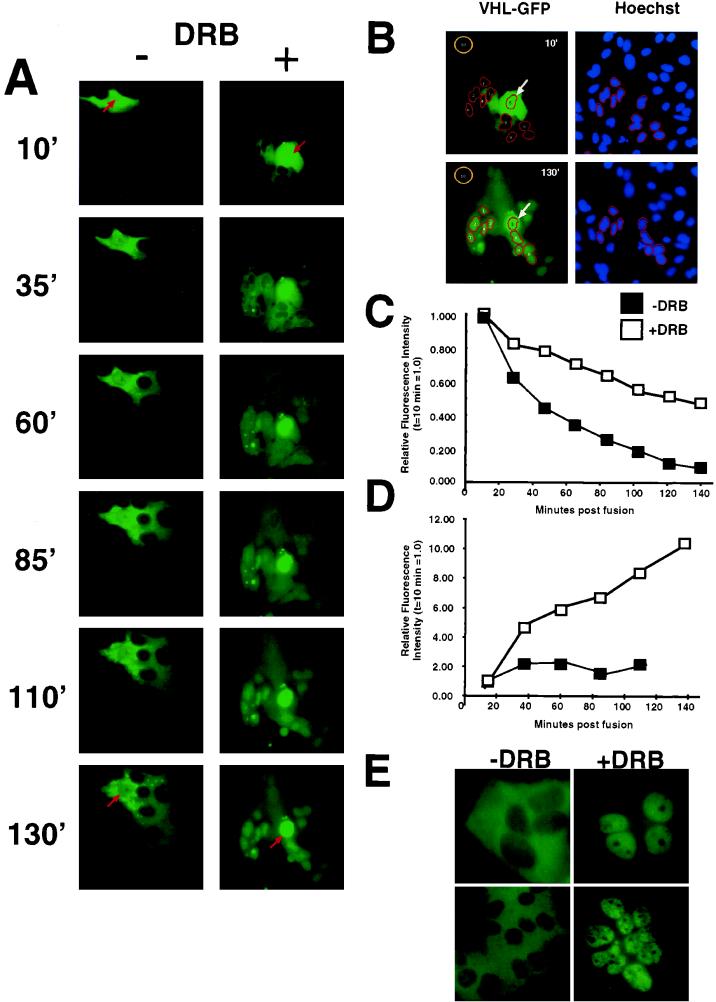
FIG. 4.
Effect of transcription inhibition on nucleocytoplasmic trafficking of VHL-GFP in a cellular fusion assay. HeLa cells were transfected with VHL-GFP and fused with nontransfected HeLa cells as described in Materials and Methods. Cells were transferred to the incubation chamber mounted on the microscope and time-lapse images taken at subsaturating illumination conditions. (A) Two typical polykaryon formations over time are shown. Arrows point to the donor nuclei of the polykaryons. The left panel shows the shuttling behavior of VHL-GFP in the absence of DRB with a high export rate from the donor nucleus (red arrow) and slow accumulation of VHL-GFP in the acceptor nuclei. The right panel shows a fusion in which DRB was added prior to and during fusion, after which VHL-GFP is exported from the donor nucleus at a much lower rate and the acceptor nuclei acquire VHL-GFP at a much higher rate. (B) Example of quantitative measurements of VHL-GFP signal intensity in donor and acceptor nuclei after PEG fusion. The first and last time point were chosen to demonstrate how quantitations were done. Counterstaining of cells with Hoechst 33342 dye (2 μg/ml for 10 min) provided nuclear dimensions. The brightness information from the blue image taken at a 400-nm wavelength was used to define nuclear regions. The donor nucleus (arrow, region 5) and the acceptor nuclei without any detectable signal at 13 min postfusion, which accumulated VHL-GFP after 133 min, were selected for this example. The regions for the selected acceptor nuclei were transferred to the green image taken at 488 nm. Green images were taken with identical exposure times and under nonsaturated conditions. Images were normalized and contrast was enhanced with identical settings with IPLab Spectrum; this allows direct visual comparison of signals between frames. The pixel intensity values per se are not affected by this step. By using IPLab’s “measure segments” command, the intensity values of the selected nuclei were determined. Region 10 was used to determine the background fluorescence in the field. Mean pixel intensities per pixel were determined as total pixel intensity/number of pixels, and background was subtracted. The donor nuclei and the average of the acceptor nuclei were determined and used for panels C and D. All measurements were performed under the nonsaturating conditions of the CCD camera and the screen, although the printing paper may appear saturated. (C) The intensity decrease of VHL-GFP from the donor nucleus was quantified in separate experiments either with or without DRB (each line represents the mean of two experiments). Signal intensity at the first time point (10 min postfusion) was set to 1, and the successive values were set in relation to that. There was clearly a slower decrease in intensity of the nuclear VHL from the DRB-treated donor nucleus. (D) The signal intensity in the acceptor nuclei was measured over time to determine the rate of net import. The +DRB line represents two experiments (mean values of 19 nuclei in total), and the −DRB line represents two experiments (mean value of 16 nuclei). A faster increase in VHL-GFP fluorescence in acceptor nuclei is observed in the presence of DRB. (E) VHL-GFP is able to shuttle in a transcription-dependent manner between the different nuclei and the large cytoplasm of an polykaryon. Two examples are shown. −DRB indicates that the fusion was performed in the absence of DRB, where VHL-GFP can be detected predominantly in the cytoplasm. +DRB indicates that the fusion was performed without DRB for 130 min, and at that time, DRB was added for 2 h. Notice that VHL-GFP is now predominantly in the nuclei. All of the experiments presented in Fig. 4 were performed in the presence of cycloheximide. These results indicate that VHL-GFP is able to traffic between the different nuclei via the cytoplasm of the fusions.
Polykaryon assay.
HeLa cells were transfected by a standard calcium phosphate-coprecipitation technique and inspected by 488-nm fluorescence microscopy for VHL-GFP expression. Usually between 10 and 30% of the cells presented strong fluorescence. The cells were trypsinized 24 h after transfection and mixed with nontransfected HeLa cells in a ratio of 1 to 10. The cell mixture was plated at a concentration of 1.5 × 106 cells per 60-mm-diameter dish and left overnight for reattachment. Approximately 90% of the transfected cells reattached under these conditions. The confluent cell layer was visually inspected for even distribution of GFP-producing cells surrounded by nontransfected cells. Approximately 30% of reattached fluorescent cells appeared as doublets, indicating a recent cell division event. Cells were washed twice with prewarmed PBS and fused for 2 min by addition of a prewarmed 50% solution of polyethylene glycol (PEG) 4000 in PBS (Gibco BRL no. 14030-035). PEG was removed thoroughly by four washes with prewarmed PBS, and the cells were incubated with warm DMEM (10% FCS plus antibiotics). Cells were observed under phase-contrast microscopy and scanned for fusion events involving one donor cell with surrounding acceptor cells. Briefly, cells were observed on an inverted microscope with indirect fluorescence equipment (Zeiss Axiovert 135TV). Culture dishes were put on a heated stage within a mounted controlled-environment chamber (Zeiss) providing a temperature of 37°C, high humidity, and 5% CO2. Images of any given field were taken automatically every few minutes by a computer-controlled, high-resolution, slow-scan Quantix CCD camera (Photometrics, Tucson, Az.). Care was taken to integrate individual frames under subsaturating conditions (i.e., with no pixels reaching an intensity value of 4,095). Frames of such a series were taken with identical integration times, usually between 1 and 3 s/frame at a 2 by 2 binning with an electronic gain of 2. Images were stored and analyzed on a Macintosh PowerPC 7600 computer with IPLab Spectrum software (Scanalytics, Vienna, Va.). Frame series were put together to create quicktime movies of the events. Selected time points were chosen for quantitation. In some experiments, cells were stained before fusion with Hoechst 33342 dye (1 μg/ml) for 10 min to allow identification of acceptor nuclei. Cells were pretreated with cycloheximide (10 μg/ml, a condition found to completely inhibit protein synthesis) 2 h before fusion and kept in the presence of cycloheximide after fusion. DRB was added at 25 μg/ml as indicated. An example of quantitation is presented in Fig. 4B.
RESULTS
Function of FVHL-GFP and VHL-GFP fusion proteins.
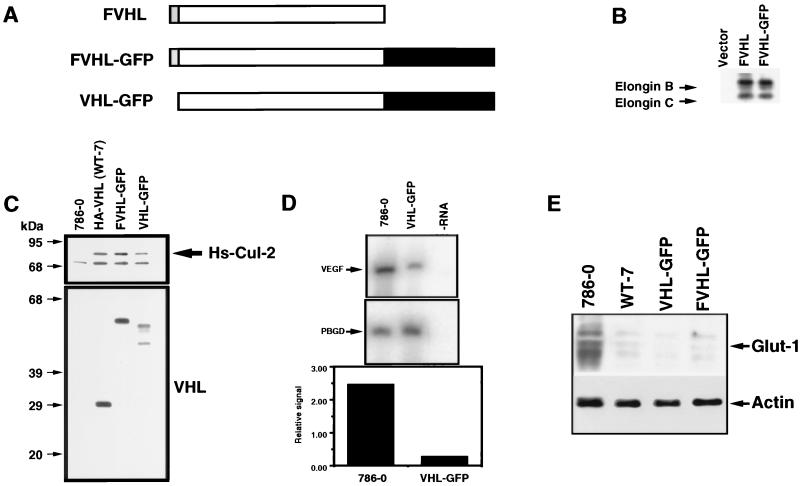
A full-length human VHL coding sequence modified by an amino-terminal Flag (F) epitope tag was fused at the carboxy terminus to a full-length open reading frame encoding a strong autofluorescing mutant of the GFP (62) (Fig. 1A). This construct, termed FVHL-GFP, a fusion protein lacking the amino-terminal epitope (VHL-GFP), and various VHL mutants fused to GFP were cloned into several expression vectors (see Materials and Methods). Expression vectors were transfected into various cell lines, and the proteins produced were analyzed with functional assays. FVHL-GFP was shown to assemble with elongins B and C to levels equivalent to those of FVHL, as demonstrated by immunoprecipitation of transiently transfected Cos-7 cells (Fig. 1B) (2, 9, 10, 25). VHL-negative RCC 786-0 cell lines stably expressing FVHL-GFP or VHL-GFP were also established by using retrovirus vectors expressing these fusions. We chose 786-0 RCC, because one VHL allele is deleted, and the other one codes for a VHL protein truncated at amino acid 104. This provides a suitable system with which to study the effect of reintroduction of wild-type VHL in a VHL-negative background (24, 27, 36, 58). FVHL-GFP-stably-expressing 786-0 cells were also established by standard calcium phosphate transfection. The protein product of FVHL-GFP was of the predicted molecular weight by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 1C). VHL-GFP gave two products consistent with the alternative use of the first two in-frame methionines in the predicted VHL open reading frame, a phenomenon sometimes observed for endogenous VHL (Fig. 1C) (23, 25). Western blotting with an anti-VHL antibody indicated that VHL-GFP and FVHL-GFP fusion proteins in the 786-0 stable cell lines accumulated to levels 1- to 10-fold over those of endogenous VHL detected in different cell lines, such as HEK293, Cos-7, Jurkat, and HeLa (data not shown). FVHL-GFP and VHL-GFP coprecipitated similar levels of Hs-Cul-2 (Fig. 1C) compared to an HA-tagged VHL (36, 51), demonstrating that fusion to GFP did not affect VHL complex formation with the known associated proteins.
FIG. 1.
Functional comparison of wild-type VHL and VHL linked to the GFP. (A) Schematic diagram of VHL fusion to the GFP. The VHL cDNA codes for a 213-amino-acid protein (open box). A Flag-tagged epitope (F) was fused at the N-terminus (shaded box). The GFP was fused at the C terminus (black box [not in scale]), resulting in FVHL-GFP. A version without the Flag tag was also produced (VHL-GFP). (B) FVHL-GFP assembles with elongins B and C. Cos-7 cells were transfected with 5 μg of plasmid DNA, plated for 16 h, incubated for 6 h in the presence of [35S]Met, and immunoprecipitated with the M2 anti-Flag antibody as described in Materials and Methods. Vector, pSX. (C) FVHL-GFP and VHL-GFP assemble with Hs-Cul-2. Stable VHL-negative RCC 786-0 cells (786-0) expressing either HA-tagged VHL (WT-7), Flag-tagged VHL-GFP to force expression from the first methionine of VHL (FVHL-GFP), or a GFP fusion without the Flag tag (VHL-GFP) to produce fusion proteins initiated by two methionines were lysed and immunoprecipitated with a monoclonal anti-VHL antibody. Precipitated proteins were run on SDS-PAGE (12% polyacrylamide) and transferred on a nitrocellulose membrane. The membrane was incubated in the presence of rabbit anti-Hs-Cul-2 (top panel) or rabbit anti-VHL (bottom panel) antibodies. Notice that FVHL-GFP and VHL-GFP assemble with Hs-Cul-2, as does HA-VHL (WT-7) (an arrow points at Hs-Cul-2). FVHL-GFP produces a single protein with a size of approximately 55 kDa, whereas VHL-GFP produces two proteins presumably initiated from two methionines, a phenomenon sometimes observed with endogenous VHL and a VHL cDNA with an epitope tagged at the C terminus (9). (D) VHL-GFP inhibits the production of VEGF mRNA. RT-PCR was performed as described in Materials and Methods with primers specific for VEGF mRNA and PBGD mRNA as a control. Also shown is a quantification of the VEGF/PBGD signal ratios. −RNA, RT-PCR was performed in buffer without RNA. (E) FVHL-GFP and VHL-GFP downregulate levels of Glut-1 protein. Total cell extracts obtained from 786-0, WT-7 (expressing HA-VHL), FVHL-GFP, and VHL-GFP were run on SDS-PAGE (10% polyacrylamide) gel and transferred to a nitrocellulose membrane, and the membrane was incubated in the presence of an anti-Glut-1 antibody. Notice the high levels of Glut-1 in 786-0 cell extract and that FVHL-GFP and VHL-GFP downregulate Glut-1 levels, as does HA-VHL (WT-7). Blots were also incubated with actin as a loading control.
To test the function of FVHL-GFP and VHL-GFP fusion proteins, we utilized the ability of wild-type VHL to repress levels of VEGF and Glut-1 when reintroduced in the VHL-negative 786-0 RCC (19, 24, 36, 58). The levels of VEGF mRNA were measured by quantitative RT-PCR with PBGD mRNA as a control. The 786-0 cells expressing VHL-GFP showed an approximately 10-fold-lower level of VEGF mRNA compared to nonexpressing 786-0 RCC cells (Fig. 1D), consistent with previously published results (19, 24, 27, 36, 58). HA-VHL, FVHL-GFP, and VHL-GFP were all able to downregulate levels of Glut-1 proteins in stable RCC 786-0 cells (Fig. 1E) (36). We concluded that the VHL-GFP fusion proteins are functional and can be used to study the localization and other properties of VHL.
Intracellular localization of VHL-GFP.
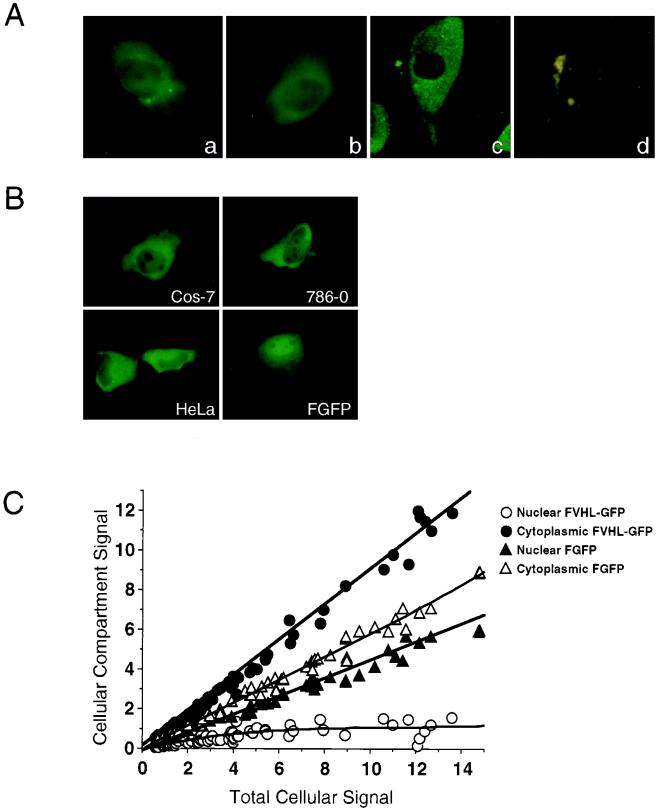
To determine the intracellular localization of VHL-GFP, we first examined the distribution of FVHL-GFP and VHL-GFP in stably expressing 786-0 cells. These cell lines express small amounts of the fusion proteins which are comparable to those of endogenous VHL in VHL-positive cells. Therefore, the observed GFP fluorescence is weak and requires sensitive imaging techniques to be detected. We observed a predominantly cytoplasmic localization of FVHL-GFP (Fig. 2Aa) and VHL-GFP (Fig. 2Ab). The presence of VHL-GFP in the nucleus was demonstrated by confocal microscopy (Fig. 2Ac) and detected in the nucleoplasm, excluding the nucleoli. Because of the very low level of expression of the fusion proteins in the stable cell lines, the autofluorescence of 786-0 cells was readily detectable as cytoplasmic green or yellow dots and should not be confused with diffuse distribution of FVHL-GFP and VHL-GFP (Fig. 2Ad). Flow cytometry and quantitative CCD imaging indicated that a fraction of cells expressed up to sixfold-less VHL-GFP compared to the mean average of a stably expressing 786-0 polyclonal population without any significant difference in the observed cellular distribution.
We also wanted to test the cellular distribution of the GFP fusion proteins in transiently transfected cells expressing higher levels than those of the stable cell lines, making the detection of GFP fluorescence much easier. Essentially identical patterns were observed in transiently transfected Cos-7, 786-0, and HeLa cells (Fig. 2B), as well as several other cell lines (data not shown) expressing from 4-fold-less to 20-fold-higher levels than those of the stable 786-0 cells, as measured with a CCD camera. Cos-7 cells (Fig. 2B, FGFP) or other cell lines (data not shown) were also transiently transfected with Flag epitope-tagged GFP (no VHL sequences). FGFP has a much higher nuclear/cytoplasmic ratio than that of the same cells expressing FVHL-GFP. In Fig. 2C, we compared the nuclear versus cytoplasmic levels of either FGFP or FVHL-GFP in relation to the total amount of cellular signal. Cells expressing VHL-GFP show a strong cytoplasmic signal accumulation compared to cells expressing GFP alone. This is true for the whole range of expression levels obtained with transient transfections. Thus, the addition of VHL to GFP results in prominent (but not complete) exclusion of the chimeric protein from the nucleus. The predominantly cytoplasmic distribution of VHL-GFP presented here reproduces the reported steady-state localization of endogenous VHL in kidney cells and of ectopically expressed VHL in cultured cells (7, 9, 23, 31, 37, 64).
FIG. 2.
Cellular localization of VHL-GFP and FVHL-GFP in live transfected cells. (A) VHL-GFP localizes predominantly to the cytoplasm, with some nuclear signal in stably expressing RCC 786-0 cells. (a) FVHL-GFP picture obtained by CCD camera. (b) VHL-GFP picture obtained by CCD camera. (c) VHL-GFP picture obtained by confocal microscopy. (d) 786-0 not expressing GFP fusion proteins (picture obtained by CCD camera). (B) FVHL-GFP localize predominantly to the cytoplasm with some nuclear signal in different transiently overexpressing cells. Shown are transiently transfected Cos-7, 786-0, and HeLa cells expressing FVHL-GFP and Cos-7 cells expressing FGFP (without VHL). (C) Quantitative comparison of the nuclear and cytoplasmic distributions of FVHL-GFP and FGFP in transiently transfected Cos-7 cells expressing different amounts of the fusion proteins (total cellular signal). CCD images were quantitated as described in Materials and Methods. A value of 1 for the total cellular signal is approximately the same as that for the stable 786-0 cell lines expressing either FVHL-GFP or VHL-GFP. Notice that VHL restricts the nuclear accumulation of the reporter GFP regardless of the cellular expression levels.
Inhibition of transcription redistributes VHL-GFP to the nucleus.
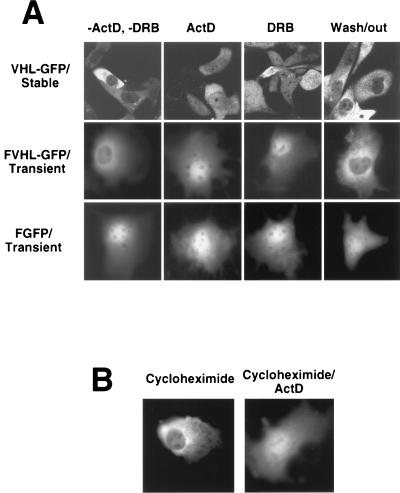
Several proteins involved in posttranscriptional regulation of mRNA shuttle in a transcription-dependent manner between the nucleus and the cytoplasm (48–57). VHL cellular distribution and its link to mRNA stability prompted us to suggest that the function of VHL may also be linked to its ability to engage in a transcription-dependent nuclear-cytoplasmic shuttle. 786-0 cells stably expressing VHL-GFP were examined by confocal microscopy before and after treatment with either actinomycin D (ActD) or 5,6-dichlorobenzimidazole riboside (DRB) (Fig. 3A, first row, VHL-GFP/Stable). Untreated cells (−ActD and −DRB) exhibited a predominantly cytoplasmic signal. After 3 h of ActD treatment (ActD), we detected a clear shift of VHL-GFP most readily appreciated as an increase in nuclear fluorescence. The same effect was observed after treatment with the RNA polymerase II-specific inhibitors DRB (Fig. 3A, first row, DRB) and α-amanitin (not shown). Removal of DRB resulted in a redistribution similar to that of the untreated state indicating that the phenomenon is reversible (Fig. 3A, first row, Wash/out). Identical results were observed with the RCC 786-0 cells stably expressing FVHL-GFP (data not shown). The same effects of these two drugs were observed in Cos-7 cells transiently transfected with FVHL-GFP (Fig. 3A, second row, FVHL-GFP/Transient). CCD camera quantitation demonstrated a threefold increase in nuclear signal without a change in total cellular fluorescence levels after stable or transiently transfected cells achieved their new steady state (data not shown). In contrast, neither ActD nor DRB had any effect on the distribution of FGFP (Fig. 3A, third row, FGFP/Transient) (62). Inhibition of protein synthesis with cycloheximide had no effect on the distribution of FVHL-GFP in transiently transfected Cos-7 cells (Fig. 3B) or stably expressing RCC 786-0 cells (data not shown). Ongoing protein synthesis was not required for the redistribution effect of the transcriptional inhibitors (Fig. 3B). It is likely that the effect of ActD or DRB is the result of inhibition of new RNA synthesis and is not mediated by the failure to synthesize a product from the transcribed RNA.
FIG. 3.
Effect of transcription and translation inhibition on the cellular distribution of VHL-GFP. (A) Comparison of the nuclear-cytoplasmic distributions of VHL-GFP in stably expressing RCC 786-0 cell lines (top row), FVHL-GFP in transiently expressing Cos-7 cells (middle row), and FGFP (without VHL) in transiently expressing Cos-7 cells (bottom row) before (−ActD and −DRB) and after the addition of either ActD (transcription inhibitor; 5 μg/ml for 3 h) or DRB (RNA polymerase II-specific inhibitor; 25 μg/ml for 3 h). Wash/out indicates that pictures were taken 16 h after washing out of DRB from the cultures. (B) Comparison of the nuclear-cytoplasmic distribution of FVHL-GFP in transiently transfected Cos-7 cells incubated either in the presence of cycloheximide (translation inhibitor; 100 μg/ml for 3 h) or for 1 h in the presence of cycloheximide followed by the addition of ActD for 3 h (Cycloheximide/ActD).
Study of the dynamic trafficking properties of VHL-GFP in a cellular fusion assay.
An established method for measuring the ability of a nuclear protein to engage nuclear-cytoplasmic shuttling is to monitor its accumulation into the nuclei of a newly formed polykaryon (52, 53). However, VHL is a predominantly cytoplasmic protein, making the demonstration of shuttling by this technique more difficult. Therefore, we used our controlled-environment CCD imaging setup to study the quantitative changes in fluorescence signal intensity in both donor and acceptor nuclei in polykaryons over time. This approach allows the study of the dynamic cellular trafficking ability of VHL-GFP, in contrast to the static analysis offered by immunofluorescence techniques. A single transiently transfected HeLa cell expressing VHL-GFP was fused with an excess of untransfected HeLa cells, and the fluorescence intensity in the “donor” nucleus was quantified as a function of time in either the absence or presence of DRB (Fig. 4). The nuclear areas were identified after Hoechst 33342 staining (Fig. 4B). All measurements were performed well under the saturation limits of the CCD camera (see the detailed explanation in Materials and Methods [“Fluorescence analysis and image processing” and “Polykaryon assay” sections]). These experiments were performed in the presence of cycloheximide to prevent new protein synthesis. In the absence of DRB, the fluorescence signal rapidly left the donor nucleus, with 50% of the initial signal lost after less than 40 min (Fig. 4A and C, −DRB). The loss of nuclear VHL-GFP is markedly slower in the presence of DRB, with which more than 140 min was required to see a 50% drop in signal from the donor nucleus (Fig. 4A and C, +DRB). We also measured the effect of DRB on the accumulation of VHL-GFP in acceptor nuclei. In the absence of DRB, most of the signal is distributed throughout the cytoplasm of the large polykaryon, with some nuclear signal present (Fig. 4A, −DRB at 130′ min, and D, −DRB). Although the steady-state distribution is highly cytoplasmic, VHL-GFP is still able to traffic into and out of the nuclei of the polykaryon, as demonstrated by the addition of DRB 130 min after synctium formation, which caused acute nuclear accumulation (Fig. 4E). In contrast, the presence of DRB resulted in progressive accumulation in the recipient nuclei, as described in single cells (Fig. 4A, compare −DRB and +DRB at 130 min, and D, +DRB). These results indicate that DRB (or ActD) acts by slowing down the nuclear export rate, which causes an increased nuclear residency time of VHL-GFP.
Localization of the region of VHL required for ActD-dependent nuclear accumulation.
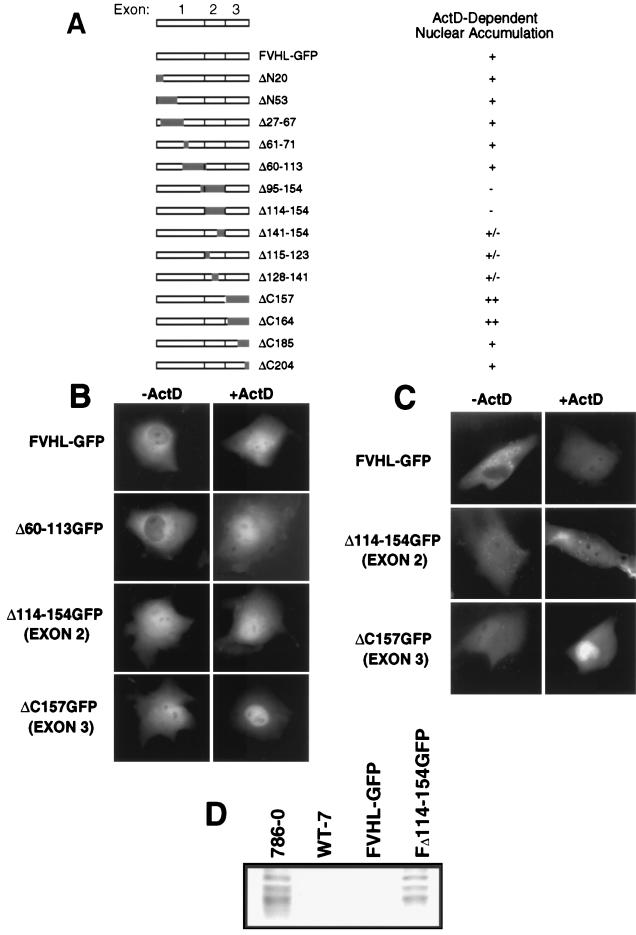
Data presented in Fig. 3 and 4 indicated that VHL localization is sensitive to ongoing RNA polymerase II activity, and the addition of transcription inhibitors caused a partial redistribution of VHL-GFP to the nucleus. The next step was to map a domain in VHL that mediates nuclear accumulation upon treatment with ActD. Furthermore, we wanted to identify which domain of VHL prevents the complete nuclear redistribution of VHL-GFP in cells treated with ActD. For these reasons, we studied the effect of ActD treatment on several VHL mutants (Fig. 5A). Some of these mutants showed different levels of nuclear and cytoplasmic accumulation; therefore, ActD responsiveness was defined as a clear increase in nuclear signal after treatment. A large internal-truncation mutant, Δ60–113GFP, was indistinguishable from VHL-GFP before and after addition of ActD (Fig. 5A and B). An exon 2 deletion mutant (Δ114–154GFP) showed increased nuclear localization without ActD, but did not accumulate more in the nucleus upon arrest of transcription (Fig. 5A and B). Mutants with small deletions of exon 2 (Δ115–123GFP, Δ128–141GFP, and Δ141–154GFP) each showed a partial loss of ActD responsiveness. Interestingly, ΔC157GFP (deletion of exon 3), whose distribution is similar to that of Δ141–154GFP in untreated cells (higher nuclear content), displayed a very strong additional redistribution to the nucleus upon ActD treatment, resulting in almost exclusive nuclear localization (Fig. 5A and B). We concluded that exon 2 sequences are required for ActD-dependent nuclear redistribution, whereas assembly with elongin BC–Hs-Cul-2 (through exon 3 sequences) is not.
FIG. 5.
Effect of arrest of transcription on deletion mutants of VHL. Cos-7 cells were transfected with 5 μg of plasmid DNA encoding either FVHL-GFP or deletion mutants of VHL. (A) Schematic diagram of the VHL cDNA that contains three exons, FVHL-GFP, and the different deletion mutants. On the right is the measured effect of ActD compared to the steady-state distribution of the mutants without ActD. −, no detectable nuclear accumulation; +/−, slight nuclear accumulation; +, strong nuclear accumulation; ++, very strong nuclear accumulation. (B) Cellular localization of FVHL-GFP and the three key deletion mutants of exon 1 (Δ60–113GFP), exon 2 (Δ114–154GFP), and exon 3 (ΔC157GFP) before (−ActD) and after (+ActD) the addition of ActD as assessed by transient transfection of Cos-7 cells. (C) Cellular localization before and after addition of ActD of FVHL-GFP and two key deletion mutants of exon 2 (Δ114–154GFP) and exon 3 (ΔC157GFP) in stably expressing RCC 786-0 cells. Dots observed in stable cell lines are 786-0 autofluorescence. (D) An exon 2 mutant is unable to downregulate levels of Glut-1 protein. Whole-cell extracts (∼30 μg of protein/lane) from RCC 786-0, RCC 786-0 stably expressing HA-tagged VHL (WT-7), FVHL-GFP, and a truncation mutant of exon 2 (Δ114–154GFP) were run on SDS-PAGE (10% polyacrylamide) gel and immunoblotted with an anti-Glut-1 polyclonal antibody.
Because the screening of the mutants was performed with Cos-7 cells overexpressing the fusion proteins, we wondered if we could reproduce the data in stable RCC 786-0 cells expressing near-endogenous levels of VHL. As shown in Fig. 5C, similar results were obtained in stable cells expressing low levels of the fusion proteins.
We then monitored the ability of an exon 2 mutant that failed to be redistributed more to the nucleus upon ActD treatment to repress levels of Glut-1 protein in RCC VHL-negative 786-0 cells. These cells express high levels of Glut-1 protein, which can be significantly lowered by the reintroduction of wild-type VHL (36). The introduction of FVHL-GFP into these cells is as effective as HA-VHL in repressing Glut-1 levels (Fig. 5D [see also Fig. 1E]). The naturally occurring, cancer-causing splicing mutant (Δ114–154GFP [exon 2 mutant]), which abrogated transcription-dependent trafficking, failed to repress Glut-1 levels (Fig. 5D). Therefore, ActD-dependent nuclear accumulation of FVHL-GFP is mediated by sequences encoded by exon 2, the loss of which resulted in the inability to regulate Glut-1 protein levels. This provides the first clue as to the function of this domain of VHL, which is frequently mutated in sporadic RCC.
Uncoupling transcription dependency from localization causes a partial loss of VHL-GFP function.
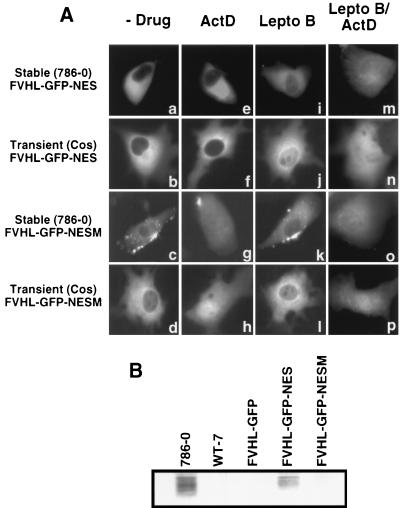
Is transcription-dependent nuclear-cytoplasmic trafficking of VHL required for its function? To answer this question, we constructed a VHL fusion protein whose localization and trafficking properties are insensitive to ongoing transcription. FVHL-GFP was fused to a strong heterologous NES and tested for nuclear accumulation in the presence of ActD. FVHL-GFP was fused at the C terminus of GFP to the Rev NES (LPPLERLTL (11) to produce the FVHL-GFP-NES fusion protein. A mutated NES (LPPAERATL), whose capacity to convey rapid nuclear export should be greatly diminished (69), was also produced (FVHL-GFP-NESM). When the NES-containing protein was expressed either stably at a low level in 786-0 or transiently in Cos-7 cells, the GFP signal was exclusively cytoplasmic, regardless of expression levels (Fig. 6Aa and Ab). The mutated NESM-containing fusion protein had a distribution indistinguishable from that of FVHL-GFP (Fig. 6Ac and Ad [also compare with Fig. 2A and B]). When ActD was added to these cells, we did not detect any shift in the distribution of FVHL-GFP-NES protein to the nucleus (Fig. 6Ae and Af) indicating that NES has the ability to override transcription-dependent trafficking of VHL-GFP. FVHL-GFP-NESM responded to the addition of ActD with a clear shift to the nucleus (Fig. 6Ag and Ah), as observed for FVHL-GFP (Fig. 3). To show that VHL-GFP-NES is still able to shuttle, albeit in a transcription-independent manner, we inhibited the NES pathway by adding leptomycin B. Leptomycin B has been shown to block the interaction of NES with its cognate CRM-1 receptor and thereby prevent the export of NES-containing protein from the nucleus (47, 68). After 2 to 6 h of incubation in 20 nM leptomycin B, there was a marked redistribution of FVHL-GFP-NES to the nucleus resembling the distribution of FVHL-GFP (Fig. 6Ai and Aj). This demonstrated that FVHL-GFP-NES is able to shuttle in and out of the nucleus, even if shuttling is insensitive to ActD. In contrast, either FVHL-GFP-NESM (fig. 6Ak and Al) or FVHL-GFP (data not shown) failed to redistribute upon addition of leptomycin D. FVHL-GFP-NES showed nuclear accumulation in response to ActD only in the presence of leptomycin B (Fig. 6Am and An), while the effect of ActD on VHL-GFP-NESM is unaltered by the presence of leptomycin B (Fig. 6Ao and Ap). These results suggest that VHL export is unrelated to the CRM-1 pathway.
The fact that Rev NES abolished ActD-dependent nuclear accumulation provided us with the means to test whether transcription-dependent trafficking is required for VHL-GFP function. With this in mind, we tested the ability of FVHL-GFP-NES to downregulate levels of Glut-1 protein in 786-0 cells. The addition of Rev NES to VHL resulted in a marked loss of VHL function (Fig. 6B, FVHL-GFP-NES) compared to the level of HA-VHL and FVHL-GFP function (Fig. 6B). FVHL-GFP-NESM, which showed trafficking sensitive to ActD, has activity similar to that of FVHL-GFP (Fig. 6B). These results demonstrate that uncoupling transcription with trafficking by fusion to a strong NES, or by a naturally occurring cancer causing exon 2 deletion, alters VHL function and suggest that the described trafficking pattern is required for VHL tumor suppression function.
FIG. 6.
Cellular distribution and function of FVHL-GFP fused to the strong nuclear export signal of human immunodeficiency virus (HIV) Rev. (A) FVHL-GFP was fused at its C terminus to the NES of HIV Rev (LPPLERLTL) to produce the FVHL-GFP-NES fusion protein. FVHL-GFP was also fused to a mutated form of NES (LPPAERATL) to produce the FVHL-GFP-NESM fusion protein. Stably expressing RCC 786-0 cells (first and third panels from the top) or transiently expressing Cos-7 cells (second and fourth panels from the top) were treated without drugs (−Drug), with ActD for 3 h (ActD), with leptomycin B for 2 to 6 h (Lepto B), or with leptomycin B for 2 to 6 h followed by the addition of ActD for 3 h (Lepto B/ActD). Dots observed in FVHL-GFP-NESM are 786-0 autofluorescence. (B) FVHL-GFP-NES shows a defect in downregulation of Glut-1 protein levels. Whole-cell extracts (∼30 μg of protein/lane) from RCC 786-0 and RCC 786-0 stably expressing HA-tagged VHL (WT-7), FVHL-GFP, FVHL-GFP-NES, and FVHL-GFP-NESM were run on SDS-PAGE (10% polyacrylamide) gel and immunoblotted with an anti-Glut-1 polyclonal antibody.
DISCUSSION
We have fused VHL to GFP to study the intracellular localization, dynamic trafficking properties, and function of VHL. We performed several control experiments to ascertain the validity of using these fusion proteins. First, the GFP fusion proteins, with or without an additional epitope tag (Flag), were equally capable of assembling with the known biochemical partners of VHL. Second, several different measures of VHL function in cells, including the regulation of VEGF and the Glut-1 gene, could be reproduced with the GFP fusion proteins. Third, the steady-state distributions of VHL-GFP were similar in VHL-negative cells and other cell lines expressing different levels of VHL. Importantly, the steady-state distribution of VHL-GFP reproduced the reported localization of endogenous VHL in human kidney cells as well as in overexpressed VHL in cultured cells by immunochemistry or immunofluorescence techniques (7, 9, 23, 31, 37, 64). This indicates that fusion to GFP and expression levels do not influence the cellular distribution and function of the VHL moiety. Taken together, these data strongly suggest that VHL-GFP is mimicking the biochemical, functional, and cellular localization attributes of VHL. By the GFP approach, we were able to demonstrate that VHL-GFP dynamically traffics in a transcription-dependent manner between the nucleus and the cytoplasm. Transcription-dependent shuttling is mediated by sequences encoded by exon 2, the site of frequent mutations in RCC. We suggest that VHL requires not only nuclear-cytoplasmic localization, but also transcription-dependent trafficking between these two compartments in order to function.
Nuclear-cytoplasmic trafficking of VHL.
With the experiments reported in this study, the VHL tumor suppressor gene product should be added to the growing list of proteins that exhibit dynamic trafficking between the nucleus and the cytoplasm (20, 46, 49). The most striking aspect of the intracellular dynamics of VHL-GFP is the nuclear accumulation in response to inhibition of RNA polymerase II-mediated transcription. The incomplete redistribution of VHL to the nucleus upon ActD treatment is reminiscent of the partial redistribution of nuclear proteins, such as Rev, heterogeneous nuclear ribonucleoprotein particle A (hnRNP A), and ICP27 (just to name a few), to the cytoplasm or cytoplasmic molecules to the nucleus [poly(A)+ binding protein] upon treatment with ActD (1, 3, 41, 52, 53, 57, 62). One possibility is that shuttling of VHL-GFP (or another known shuttling protein) may still occur even in the presence of ActD. This argument is supported by data obtained with the cellular fusion assay shown in Fig. 4. Although there is a dramatic decrease in the nuclear export rate of VHL-GFP in the presence of DRB (RNA polymerase II inhibitor), there is still a small fraction of VHL-GFP that is able to export from the donor nucleus. This may occur through a nuclear export mechanism independent of ongoing transcription or might be due to some diffusion of VHL-GFP from the nucleus.
We have identified one deletion mutant (ΔC157GFP) that is able to almost completely be redistributed to the nuclei of all cells. It is conceivable that exon 3 sequences regulate the shuttling ability of VHL-GFP through undefined signals interfering with the complete redistribution of VHL-GFP to the nucleus. One possibility is that a fraction of VHL assembles with cytoplasmic structures and is unavailable to engage in shuttling during the 3-h time frame of the experiments. Exon 3 sequences would mediate assembly with cytoplasmic structures, and deletion of these sequences would render the mutant VHL free to continuously shuttle and completely accumulate in the nucleus upon treatment with ActD. Supporting this conclusion is our finding that VHL mutants in exon 3 have lost the ability to be cross-linked to cytoplasmic structures upon fixation with formaldehyde (data not shown). More work will be required for us to understand why exon 3 sequences are able to hinder the complete nuclear redistribution of VHL in ActD-treated cells.
Regardless of the mechanisms that impede the complete nuclear redistribution of VHL-GFP upon ActD treatment, the increase in nuclear VHL-GFP is most likely the consequence of impairment of its nuclear export, as shown by the quantitative cellular fusion experiments. By fusion of a cell expressing VHL-GFP with nontransfected cells, we introduced a strong negative gradient immediately after fusion initiation. This allows an estimation of initial rates of export from the donor nucleus even for predominantly cytoplasmic proteins, such as VHL-GFP. Using this approach, we demonstrated that the initial rate of loss of VHL-GFP from the donor nucleus was markedly decreased (by greater than threefold) when transcription is inhibited. This is sufficient to explain the steady-state shift of VHL-GFP in the presence of either ActD or DRB of about threefold in the ratio of nuclear to cytoplasmic protein in single cells. The marked increase in accumulation of VHL-GFP in the acceptor nuclei is also noteworthy. The increased nuclear accumulation may be explained as follows. In untreated polykaryons, immediately after fusion initiation, cytoplasmic VHL-GFP is diluted by the surrounding cytoplasm. VHL-GFP is also rapidly exported from the donor nucleus into the cytoplasm. VHL-GFP is imported into and rapidly reexported from the acceptor nuclei to eventually establish a steady-state distribution that is largely cytoplasmic (as with VHL-GFP in single cells). In polykaryons treated with DRB, nuclear VHL-GFP enters the cytoplasm at about one-third the rate of the untreated cells. VHL-GFP molecules that do export from the donor nucleus as well as the molecules already present in the cytoplasm are taken up by recipient nuclei, where the export rate is markedly reduced, resulting in a higher level of accumulation into the multiple nuclei of the polykaryon. The threefold decrease in the nuclear export rate upon arrest of transcription is sufficient to explain the observed changes in the steady-state distribution of VHL-GFP in single cells. The fusion experiments strongly support our conclusion that VHL has the ability to shuttle between the nucleus and the cytoplasm and that shuttling requires ongoing RNA polymerase II transcription.
Transcription-dependent nuclear-cytoplasmic trafficking: a new function for the clinically relevant exon 2 of VHL?
The information required for ActD-dependent nuclear accumulation appears to be contained within the second exon of VHL. While we have not precisely identified the exon 2 sequences required for the putative transcription-dependent nuclear export, the partial inhibition of the effect in several nonoverlapping exon 2 deletions suggests that the effect is due to a relatively extended sequence. A significant fraction of naturally occurring missense mutations in VHL syndrome patients and, more commonly, in sporadic RCC occur in exon 2 (18). Moreover, it has been shown that a splicing defect of exon 2, which results in VHL lacking amino acids 114 to 154, causes sporadic RCC (18). The functions inactivated by these cancer-predisposing mutations are still completely unknown. Elongins B and C and Hs-Cul-2, currently the only known components of the VHL complex, are obviously not required for nuclear accumulation in ActD-treated cells. This is demonstrated by the fact that exon 3 deletions, which result in the loss of elongin B and C and Hs-Cul-2 assembly, do not abrogate ActD-dependent nuclear accumulation. It will be interesting to test some of these missense mutations for their impact on transcription-dependent nuclear export. Nonetheless, this is the first demonstration of any biological activity of exon 2-encoded sequences in VHL.
The VHL export pathway is distinct from the CRM-1–exportin 1 pathway.
Great progress has been made recently in elucidating the components of nuclear export for proteins that contain a short, leucine-rich export signal (11, 15, 16, 55, 66). The leucine-rich NES sequence has recently been shown to bind to the CRM-1 protein in the presence of Ran-GTP (13, 15, 29, 45, 50). The Streptomyces product leptomycin B is a potent inhibitor of CRM-1-mediated nuclear export (47, 68). The addition of the Rev NES to FVHL-GFP, which resulted in nuclear exclusion, abrogated the effect of ActD and caused a partial defect in Glut-1 regulation. Leptomycin B had no discernible effect on the distribution of VHL-GFP, but did restore the sensitivity of VHL-GFP-NES to ActD. Our interpretation is that the Rev NES functions through an export pathway different from and probably independent of the one normally utilized by VHL. This NES–CRM-1 pathway seems to be dominant in the NES-containing VHL-GFP fusion protein, perhaps because the added NES is more potent than the VHL putative export signal. Alternatively, NES may chaperone VHL away from a rate-limiting transcription-dependent step in VHL nuclear export. It is therefore likely that VHL utilizes a different type of export pathway, such as that used for other nuclear-cytoplasmic trafficking proteins (12, 41–43). Illumination of the VHL export pathway will require the identification of nuclear proteins that specifically recognize the relevant exon 2 sequences. In addition, the data obtained with the NES fusion demonstrate that transcription-dependent trafficking between the nucleus and the cytoplasm is required for VHL function.
Transcription-dependent nuclear-cytoplasmic trafficking and VHL function.
The mechanism by which inhibition of transcription reduces the postulated nuclear export ability of VHL is not known. Several RNA-binding proteins, such as hnRNP A, hnRNP K, Rev, ICP27, and some SR proteins require ongoing transcription for nuclear-cytoplasmic cycling. However, these are all nuclear proteins which fail to undergo nuclear import in the absence of ongoing transcription, leading to their cytoplasmic accumulation (4, 40–42, 52, 53, 57). VHL, in contrast, does appear to behave like another predominantly cytoplasmic protein, the poly(A)+ binding protein, which assembles with RNA and requires ongoing transcription for nuclear export (1). That VHL shares trafficking characteristics with the poly(A)+ binding protein supports the hypothesis that it has the ability to escort transcripts out of the nucleus. This raises the possibility that VHL-GFP is exported along with newly transcribed mRNA molecules. While there is no direct experimental evidence for this or for any interaction between VHL or its partners with RNA, the fact that VHL has been shown to influence the half-life of target mRNAs may point to a still undemonstrated ability of VHL to indirectly interact with RNA (19, 24, 32, 33). An interesting hypothesis, suggested by Kaelin and collaborators (36), is that the VHL–elongin BC–Hs-Cul-2 complex may play a role in ubiquitin-mediated degradation of proteins involved in regulating the stability of specific mRNAs. To do so, VHL–elongin BC–Hs-Cul-2 might need to assemble with target mRNAs in the nucleus as part of an RNP. The RNP would then be rapidly exported toward the cytoplasm, providing an explanation for the cytoplasmic localization of VHL-GFP at steady state. If this is the case, transcription-inhibiting drugs would act by reducing the amount of target mRNAs required for VHL–elongin BC–Hs-Cul-2 export. Once in the cytoplasm, VHL–elongin BC–Hs-Cul-2 would function to control the fate of the mRNAs. If this model is correct, the assembly with the RNPs may be specified by exon 2, and the deletion of these sequences would explain the loss of ActD sensitivity.
We have shown that all perturbations that abrogated transcription-dependent nuclear-cytoplasmic trafficking of VHL diminish its ability to function. These include exon 2 deletions and, interestingly, the addition of a functional NES which exports the protein in a way that is no longer dependent on ongoing transcription. The fact that an exon 2 mutant can still accumulate in the nucleus and the cytoplasm and that the NES fusion shuttles in a transcription-independent manner establishes that the function of VHL requires not only localization within the nucleus and the cytoplasm, but also transcription-dependent trafficking. To explain these phenomena in detail will require further work. Whatever the details, the abundance of naturally occurring mutations in exon 2, especially in sporadic RCC, may well be explained by the loss of transcription-dependent nuclear export that appears to characterize both the fate and function of the VHL tumor suppressor gene product.
ACKNOWLEDGMENTS
S.L. and M.N. contributed equally to this work.
We sincerely thank Heather Kontaxis, Rebecca Begley, and Marie-Christine Fournier for their excellent technical contribution to this work. We also thank Juan Bonifacino and Mary Dasso for critical reviews and comments.
M.N. is a fellow of the AIDS-Stipendienprogramm des Bundesministeriums fuer Bildung und Forschung, Germany. A.P. is supported by a fellowship from the Deutsche Forschungsgemeinschaft. This work was supported in part by NCI through a contract with ABL.
REFERENCES
- 1.Afonina E, Stauber R, Pavlakis G N. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 2.Aso T, Lane W S, Conaway J W, Conaway R C. Elongin (SIII). A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 3.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 4.Caceres J F, Screaton G R, Krainer A R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, Kishida T, Yao M, Hustad T, Glavac D, et al. Germline mutations in the von Hippel-Lindau disease tumor suppressor gene: correlations with phenotype. Hum Mutat. 1995;5:66–75. doi: 10.1002/humu.1380050109. [DOI] [PubMed] [Google Scholar]
- 6.Cole N B, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corless C L, Kibel A, Iliopoulos O, Kaelin W G. Immunostaining of the von Hippel-Lindau gene product in normal and neoplastic human tissues. Hum Pathol. 1997;28:459–464. doi: 10.1016/s0046-8177(97)90035-6. [DOI] [PubMed] [Google Scholar]
- 8.Crossey P A, Richards F M, Foster K, Green J S, Prowse A, Latif F, Lerman M I, Zbar B, Affara N A, Ferguson-Smith M A, Maher E R. Identification of intragenic mutations in the von Hippel-Lindau disease tumour suppressor gene and correlation with disease phenotype. Hum Mol Genet. 1994;3:1303–1308. doi: 10.1093/hmg/3.8.1303. [DOI] [PubMed] [Google Scholar]
- 9.Duan D R, Humphrey J S, Chen D Y T, Weng Y, Sukegawa J, Lee S, Gnarra J R, Linehan W M, Klausner R D. Characterization of the VHL tumor suppressor gene product: localization, complex formation, and the effect of natural inactivating mutation. Proc Natl Acad Sci USA. 1995;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 11.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNA. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U, Michael W M, Luhrmann R, Dreyfuss G. Signal-mediated nuclear export pathways of protein and RNAs. Trends Cell Biol. 1996;6:290–294. doi: 10.1016/0962-8924(96)20030-3. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 14.Foster K, Prowse A, van den Berg A, Fleming S, Hulsbeek M M F, Crossey P A, Richards F M, Cairns P, Affara N A, Ferguson-Smith M A, Buys C H, Maher E R. Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum Mol Genet. 1994;3:2169–2173. doi: 10.1093/hmg/3.12.2169. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 17.Gnarra J R, Duan D R, Weng Y, Humphrey J S, Chen D Y T, Lee S, Pause A, et al. Molecular cloning of the von Hippel-Lindau tumor suppressor gene and its role in renal carcinoma. Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 18.Gnarra J R, Tory K, Weng Y, Schmidt L, Wei M H, Li H, Latif F, Liu F S, Chen F, Duh F M, et al. Mutations of the VHL tumor suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 19.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassiliou E O, Idfield E H, Klausner R D, Linehan W M. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 21.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D R, Gnarra J R, Linehan W M, Baylin S B. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinds P W, Weinber R A. Tumor suppressor genes. Curr Opin Genet Dev. 1994;4:135–141. doi: 10.1016/0959-437x(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 23.Iliopoulos O, Kibel A, Gray S, Kaelin W G. Tumor suppression by the von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 24.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Goldberg M A. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 26.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 27.Knebelmann B, Ananth S, Cohen H T, Sukhatme V P. Transforming growth factor alpha is a target for the von Hippel-Lindau tumor suppressor. Cancer Res. 1998;58:226–231. [PubMed] [Google Scholar]
- 28.Knudson A G. Antioncogenes and human cancer. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29749. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 30.Latif F, Tory K, Gnarra J R, Yao M, Dub F, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 31.Lee S, Chen D Y T, Humphrey J S, Gnarra J R, Linehan W M, Klausner R D. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy A P, Levy N S, Goldberg M A. Post-transcriptional regulation of vascular endothelial growth factor by hypoxia. J Biol Chem. 1996;271:2746–2753. doi: 10.1074/jbc.271.5.2746. [DOI] [PubMed] [Google Scholar]
- 33.Levy A P, Levy N S, Goldberg M A. Hypoxia-inducible protein binding to vascular endothelial growth factor mRNA and its modulation by the von Hippel-Lindau protein. J Biol Chem. 1996;271:25492–25497. doi: 10.1074/jbc.271.41.25492. [DOI] [PubMed] [Google Scholar]
- 34.Linehan W M, Lerman M I, Zbar B. Identification of the von Hippel-Lindau (VHL) gene. Its role in renal cancer. JAMA. 1995;273:564–570. [PubMed] [Google Scholar]
- 35.Lippincott-Schwartz, J. Uses of GFP in mammalian cells. In M. Chalfic et al. (ed.), Applications of green fluorescent proteins, in press. John Wiley and Sons, New York, N.Y.
- 36.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaelin W G., Jr Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Los M, Jansen G H, Kaelin W G, Lips C J, Blijham G H, Voest E E. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab Investig. 1996;75:231–238. [PubMed] [Google Scholar]
- 38.Lubensky I A, Gnarra J R, Bertheau P, Walther M M, Linehan W M. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am J Pathol. 1996;149:2089–2095. [PMC free article] [PubMed] [Google Scholar]
- 39.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 41.Michael W M, Choi M D, Dreyfuss G. Nuclear export signal in hnRNP A1: a signal-mediated temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 42.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- 44.Neumann M, Harrison J, Saltarelli M, Hadziyannis E, Erfle V, Felder B K, Pavlakis G N. Splicing variability in HIV-1 revealed by quantitative RNA-PCR. AIDS Res Hum Retroviruses. 1994;10:1531–1542. doi: 10.1089/aid.1994.10.1531. [DOI] [PubMed] [Google Scholar]
- 45.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 46.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 47.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 48.Nix D A, Beckerle M C. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 50.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 51.Pause A, Lee S, Worrell R A, Chen D Y T, Burgess W H, Linehan W M, Klausner R D. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- 53.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 54.Pollard V W, Micheal W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 55.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus Rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roux P, Blanchard J M, Fernandez A, Lamb N, Jeanteur P, Piechaczyk M. Nuclear localization of c-Fos, but not v-Fos proteins, is controlled by extracellular signals. Cell. 1990;63:341–351. doi: 10.1016/0092-8674(90)90167-d. [DOI] [PubMed] [Google Scholar]
- 57.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seimeister G, Weidel K, Mohrs K, Barleon B, Martiny-Baron G, Marne D. Reversion of deregulated expression of vascular endothelial growth factor in human renal carcinoma cells by von Hippel-Lindau tumor suppressor gene. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 59.Shuin T, Kondo K, Torigoe S, Kishida T, Kubota Y, Hosaka M, Nagashima Y, Kitamura H, Latif F, Zbar B, Lerman M I, Yao M. Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res. 1994;54:2852–2855. [PubMed] [Google Scholar]
- 60.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 62.Stauber R, Gaitanaris G A, Pavlakis G N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 63.Stauber R H, Horie K, Carney P, Hudson E A, Tarasova N I, Gaitanaris G A, Pavlakis G N. Development and applications of enhanced green fluorescent protein mutants. BioTechniques. 1998;24:462–466. doi: 10.2144/98243rr01. [DOI] [PubMed] [Google Scholar]
- 64.Tsuchiya H, Iseda T, Hino O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- 65.Walther M M, Lubensky I A, Venzon D, Zbar B, Linehan W M. Prevalence of microscopic lesions in grossly normal renal parenchyma from patients with von Hippel-Lindau disease, sporadic renal cell carcinoma and no renal disease: clinical implications. J Urol. 1995;154:2010–2014. [PubMed] [Google Scholar]
- 66.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 67.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 68.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 69.Zhang M J, Dayton A I. Tolerance of diverse amino acid substitutions at conserved positions in the nuclear export signal (NES) of HIV-1 Rev. Biochem Biophys Res Commun. 1998;243:113–116. doi: 10.1006/bbrc.1997.8070. [DOI] [PubMed] [Google Scholar]
- 70.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110:1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]