Abstract
Background:
Breast cancer (BRCA), the hormone related malignant tumor, is well-known for poor prognosis. ZNF208 mainly acts as a transcription factor in various tumors, and the single nucleotide polymorphisms (SNPs) of ZNF208 are related to telomere length. Nevertheless, its role in breast tumorigenesis is largely unknown.
Methods:
We systematically investigated the gene expression, prognostic value, and promoter methylation of ZNF208 in BRCA with Gene Expression Profiling Interactive Analysis (GEPIA) and DNA Methylation Interactive Visualization Database (DNMIVD). Meanwhile, we clarified the association of ZNF208 with tumor-infiltrating immune cells (TICs) from Tumor Immune Estimation Resource (TIMER). Furthermore, we determined the biological process and functional enrichment from Cancer single-cell state atlas (CancerSEA). Finally, we verified our results with prognostic analysis and immunohistochemistry (IHC) assay.
Results:
We discovered that ZNF208 was downregulated in breast cancer, and low expression of ZNF208 predicted worse prognosis of BRCA patients. The promoter methylation level of ZNF208 was obviously increased, and ZNF208 was associated with TlCs in BRCA. In addition, ZNF208 could inhibit the metastasis and invasion biological processes, and regulate the MAPK and RAS signaling pathways in BRCA.
Conclusion:
Our findings illustrate that ZNF208 can function as a tumor suppressor and predict prognosis of breast cancer.
Keywords: BRCA, ZNF208, prognosis, immune infiltration, tumor suppressor
Introduction
Breast cancer (BRCA) is one of the most malignant tumors among women in the world, especially in Europe and the United States. 1 The incidence rate of BRCA is about 2.26 million women per year in the world, while 680 000 women eventually died of BRCA.2,3 The typical characteristics of BRCA are rapid growth, poor prognosis, high metastasize, and high recurrence. 4 Breast is the target organ of many endocrine hormones, among which estrone and estradiol are directly related to the incidence of breast cancer. 5 Thus, BRCA is mainly histologically classified as progesterone receptor (PR+/-), estrogen receptor (ER+/-), and human epidermal growth factor receptor 2 (HER2+/-). 6 Based on this histological stratification, Luminal A, Luminal B, HER2-amplified, basal-like, and normal-like subtypes are the major subtypes of BRCA. Remarkably, the king of breast cancer is triple negative breast cancer (TNBC), namely, ER, PR, and HER2 all negatively expressed in BRCA, indicating the hormone therapy is noneffective. 7 The epigenetic changes of BRCA are complicated, including BRCA 1/2 mutation, p53 mutation, HER2 amplification, and so on. 6 These malignant molecular events constitutively activate the MAPK/ERK, PI3K/AKT, and Wnt/β-catenin signaling pathways in BRCA. 8 However, the efficiency of surgery, radiation therapy, chemotherapy, hormone therapy, and immunotherapy therapies are extremely poor. 9 Therefore, it is pressing to explore the key molecular mechanism and discover effective therapeutic target for BRCA.
Zinc finger proteins (ZNFs), the transcription factor family, regulates the transcription and translation of targets genes, through sequence-specific binding to target molecules DNA, RNA, and other zinc finger proteins. 10 ZNF208 locates in chromosome 19. 11 The single nucleotide polymorphisms (SNPs) of ZNF208 are reported as telomere length associated gene from a genome-wide association study (GWAS), which contributed to many diseases, such as coronary artery disease, 12 chronic obstructive pulmonary disease, 13 HIV, TB, 14 fertile activity, 15 and longevity. 16 Moreover, ZNF208 is widely discovered participating in multiple myeloma, 17 laryngeal lancer, 18 non-small-cell lung cancer, 19 pancreatic neuroendocrine neoplasms, 11 pancreatic adenocarcinoma, 20 glioma, 21 esophageal cancer, 22 neuroblastoma and childhood cancers, 23 chronic lymphocytic leukemia, 24 and NF1-plexiform neurofibroma. 25 However, the exact function of ZNF208 in tumors is poorly illustrated without detailed mechanisms, especially in breast cancer. Consequently, it’s urgent to clearly elucidate the biological function and the mechanism of ZNF208 in BRCA.
In this study, we evaluated the expression of ZNF208 in BRCA through the utilization of online software. In addition, we investigated the prognostic relevance of ZNF208 in BRCA, as well as conducted an analysis to investigate the correlation between ZNF208 expression and its promoter methylation level in BRCA. We also clarified the association of ZNF208 with TlCs with online software. Furthermore, we analyzed the biological process and functional enrichment of ZNF208 involving in BRCA. Finally, we verified the expression and prognostic correlation of ZNF208 in BRCA using TCGA database and immunohistochemistry (IHC) assay. We aimed to establish a theoretical basis that supports the potential conclusion that ZNF208 may be a molecular target for the precise treatment of BRCA.
Materials and Methods
Expression analyses of ZNF208 in pan-cancer
The University of Alabama at Birmingham Cancer data analysis Portal (UALCAN) (https://ualcan.path.uab.edu/) is an interactive web resource for analyzing cancers and can identify biomarkers form The Cancer Genome Atlas (TCGA) data, Genotype-Tissue Expression (GTEx) data, and Cancer Cell Line Encyclopedia (CCLE) data. 26 We performed pan-cancer gene expression analyses with it.
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) provides researchers with customizable functionalities based on TCGA and GTEx data, 27 and was used to compare the expression of ZNF208 in different cancers.
The expression of ZNF208 in BRCA
The Cancer Genome Atlas Program (TCGA) database (https://portal.gdc.cancer.gov/) covers genome, transcriptome, epigenetics, proteome, and other omics data of 33 types of cancer. According to the optional cut-off value of ZNF208, the mRNA expression profiles were classified into high-expression and low-expression groups. 28 We compared the expression of ZNF208 in BRCA with it.
The prognostic value of ZNF208 for BRCA
Kaplan-Meier Plotter database (http://kmplot.com/analysis/) can assess the correlation between genes expression and the survival in more than 30 000 kinds of cancer samples. We drew the recurrence free survival (RFS) of ZNF208 in BRCA. 29
OncoLnc (https://www.oncolnc.org/) links TCGA survival data to various RNA expression levels, 30 and the prognosis curve of ZNF208 in BRCA was drew with it.
Breast Cancer Gene-Expression Miner v5.0 (bc-GenExMiner v5.0) (http://bcgenex.ico.unicancer.fr/) is a statistical mining tool of published annotated breast cancer transcriptomic data (DNA microarrays and RNA-seq). 31 We observed the ZNF208 expression in BRCA with different clinicopathological characteristics with it.
The promoter methylation level of ZNF208 in BRCA
DNMIVD (http://119.3.41.228/dnmivd/index/), a database for DNA methylation profile of human cancers with high throughput microarray data from TCGA and GEO data. 32 MethHC (http://methhc.mbc.nctu.edu.tw/php/index.php), a database of DNA methylation and gene expression in human cancers. 33 The association of ZNF208 Fragments Per Kilobase of transcript per Million mapped reads (FPKM) with ZNF208 promoter methylation level in BRCA was taken from these database.
Methsurv (https://biit.cs.ut.ee/methsurv/), a web tool to perform multivariable survival analyses using DNA methylation data. 34 We analyzed the relationship of ZNF208 promoter methylation with the survival of BRCA patients with it.
Immune cells infiltration and immune checkpoints
The Human Protein Atlas (HPA) (https://www.proteinatlas.org/), a comprehensive website to study the protein localization and levels in common human organs, tissues, and cells. 35 On this basis, we explored the immune infiltration different single-cell types in BRCA.
Tumor Immune Estimation Resource (TIMER) (https://cistrome.shinyapps.io/timer/), a tool to systematically analyzing immune infiltration of 10 897 cancer samples from 32 types of cancer. We investigated the relationship between ZNF208 expression and different leukocyte. 36
Gene alterations analyses of ZNF208
The cBio cancer genomics portal (cBioPortal) (https://www.cbioportal.org/) a multi-dimensional cancer gene set data, including somatic mutations, DNA copy-number alterations (CNAs), methylation, RNA expression, and protein enrichment. 37 On this basis, we showed the gene alterations analyses of ZNF208 gene.
Functions analyses of ZNF208 in BRCA
Cancer single-cell state atlas (CancerSEA) (http://biu.edu.cn/CancerSEA/) is the database decoding 14 distinct functional states (such as metastasis, stemness, invasion, and proliferation) of 25 cancer types at single-cell resolution. Functions of ZNF208 in BRCA were found in this database. 38
HOME for Researchers (https://www.home-for-researchers.com/) is used to analyze the gene expression, prognosis, and immunity between human cancer samples and paired normal tissues. We drew a scatter diagram of TIMER Scors of the different leukocytes, explored relationship between ZNF208 expression and different leukocyte, and checked immune checkpoint expression of the leukocytes.
Enrichment analyses
ZNF208 related differentially expressed genes (DEGs) were screened from cBioPortal database and were shown in supplementary analytical tables. P < .05, Spearman’s correlation > .5 was considered statistically significant.
Sangerbox database (http://sangerbox.com/home.html), a platform provides interactive customizable analysis tools, including various kinds of correlation analyses, pathway enrichment. Sangerbox was used to analyze the functional roles of DEGs. P < .05 and False discovery rate (FDR) < 0.25 were considered statistically significant.
Functional Enrichment analysis tool (FunRich) (http://funrich.org/download), an independent software tool for functional enrichment and interaction network analysis of genes and proteins. 39 We explored the biological processes, biological pathways, and molecular functions of the ZNF208 DEGs in BRCA.
STRING (https://cn.string-db.org/), the protein-protein interaction (PPI) networks functional enrichment analysis database, covering more than 20 million proteins from more than 5000 species organisms. The PPI of ZNF208 in BRCA was taken. 40
IHC staining evaluation
Formalin-fixed human BRCA tissue specimens and the adjacent peritumoral tissues specimens were collected from the Second Affiliated Hospital of Shaanxi University of Chinese Medicine between January 2023 and January 2024. Patients received no preoperative therapy and had signed an informed consent. The inclusion criteria are that more than 2 senior neuropathologists confirmed the histopathological diagnosis is breast cancer according to the classification of World Health Organization (WHO). Using IHC assay, we evaluated the protein levels of ZNF208 proteins with anti-ZNF208 antibody (TP74767, Abmart). The detailed protocol was performed as described previously. 37
Statistical analysis
Statistical details were available in the figure legends. Unpaired/paired Student’s t test (2-tailed) were used to analyze differences between two groups. The Kaplan-Meier method was used to perform the survival curves, and the Log-rank test was used to conduct the statistical analysis. Fisher’s exact test was used for the IHC quantification of ZNF208 expression in BRCA tissues and normal tissues. The data were shown as mean ± standard deviation (SD) unless otherwise noted. All statistical analysis were performed in GraphPad Prism 7. P value < .05 was considered significantly.
Results
Downregulation of ZNF208 in BRCA
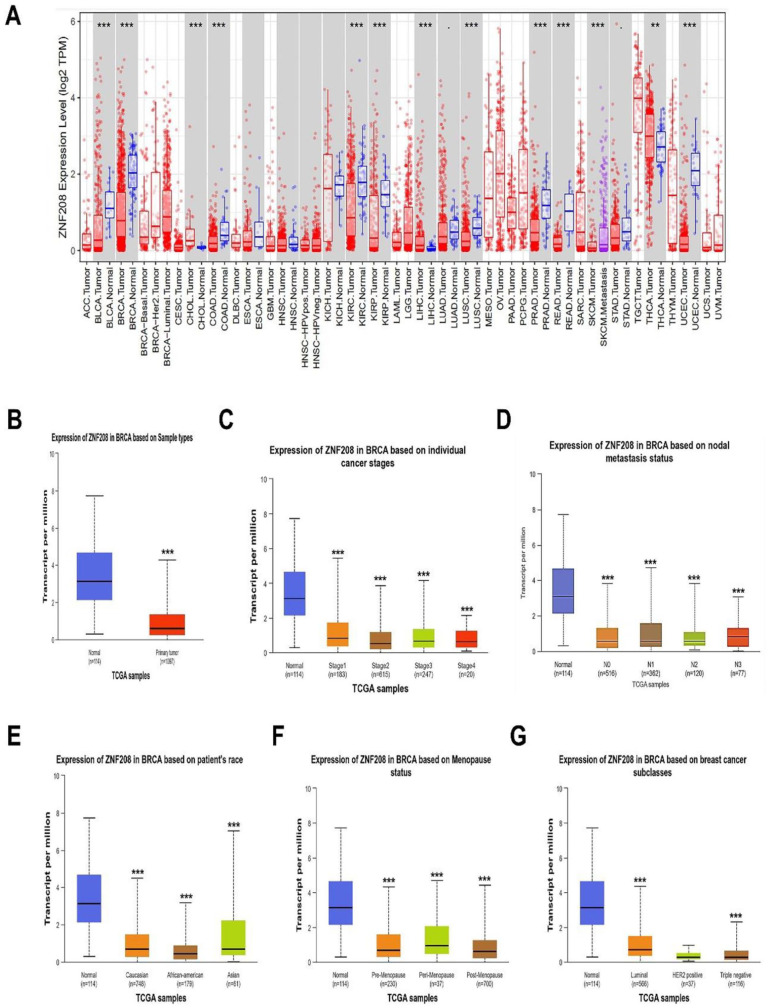
Studies show that ZNF208 is involved in maintenance of telomere length and transcriptional regulations, and the SNP variants of ZNF208 were reported to be associated with the risk of tumors in GWAS. 21 To explore the role of ZNF208 protein in various cancer, we first analyzed the mRNA levels of ZNF208 in pan-cancers from TIMER database, and the result indicated that transcript per million (TPM) of ZNF208 was obviously downregulated in 9 types of cancers compared with the adjacent normal tissues, including bladder urothelial carcinoma (BLCA), BRCA, colon adenocarcinoma (COAD), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), and uterine corpus endometrial carcinoma (UCEC) (Figure 1A). We further analyzed the mRNA levels of ZNF208 in different cancer types from UALCAN and GEPIA databases. The results showed that the mRNA level of ZNF208 were obviously downregulated in pan-cancers, especially in BLCA, BRCA, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, KIRP, PRAD, READ, UCEC, and uterine carcinosarcoma (UCS) (Supplementary Fig. S1A and B). Correspondingly, ZNF208 was found to be significantly decreased in the BRCA tissues than in normal tissues from UALCAN database (Figure 1B). Next, we explored the expression of ZNF208 in BRCA patients with other clinical characteristics from UALCAN database. The expression of ZNF208 was clearly downregulated in stage 1 to stage 4 BRCA tissues than normal tissues (Figure 1C). Compared with BRCA tissues with or without regional lymph node metastasis, the normal tissues expressed more mRNA of ZNF208 (Figure 1D). Moreover, the BRCA patients of Caucasian, African American, and Asian all showed fewer mRNA level of ZNF208 than normal tissues (Figure 1E). In addition, the ZNF208 expression of BRCA tissues during premenopause, perimenopause, and postmenopause were lower than normal tissues (Figure 1F). As well, compared with normal tissues, the luminal and triple negative subtypes of BRCA tissues showed lower ZNF208 mRNA expression than normal tissues (Figure 1G). Moreover, we also found that the ZNF208 mRNA levels were higher in normal tissues than in BRCA patients of different ages or genders (Supplementary Fig. S1C and D). These data confirmed that the ZNF208 mRNA levels were low in BRCA regardless of clinical characteristics.
Figure 1.
ZNF208 is downregulated in BRCA. (A) The expression levels of ZNF208 in different cancer types from the TCGA database were analyzed with TIMER database. **P < .01, ***P < .001. (B) Different expression of ZNF208 between BRCA tissues and normal tissues were analyzed from UALCAN database. ***P < .001. (C, D) Different expression of ZNF208 between BRCA tissues and normal tissues based on individual tumor stages and nodal metastasis status were analyzed from UALCAN database. No means no regional lymph node metastases. N1 means metastases in 1 to 3 axillary lymph nodes. N2 means metastases in 4 to 9 axillary lymph nodes. N3 means metastases in 10 or more axillary lymph nodes. ***P < .001. (E, F) Different expression of ZNF208 between BRCA tissues and normal tissues based on patient’s race and menopause status were analyzed from UALCAN database. ***P < .001. (G) Different expression of ZNF208 between BRCA tissues and normal tissues based on breast cancer subclasses were analyzed from UALCAN database. ***P < .001.
Association of decreased expression of ZNF208 with poor prognosis of BRCA patients
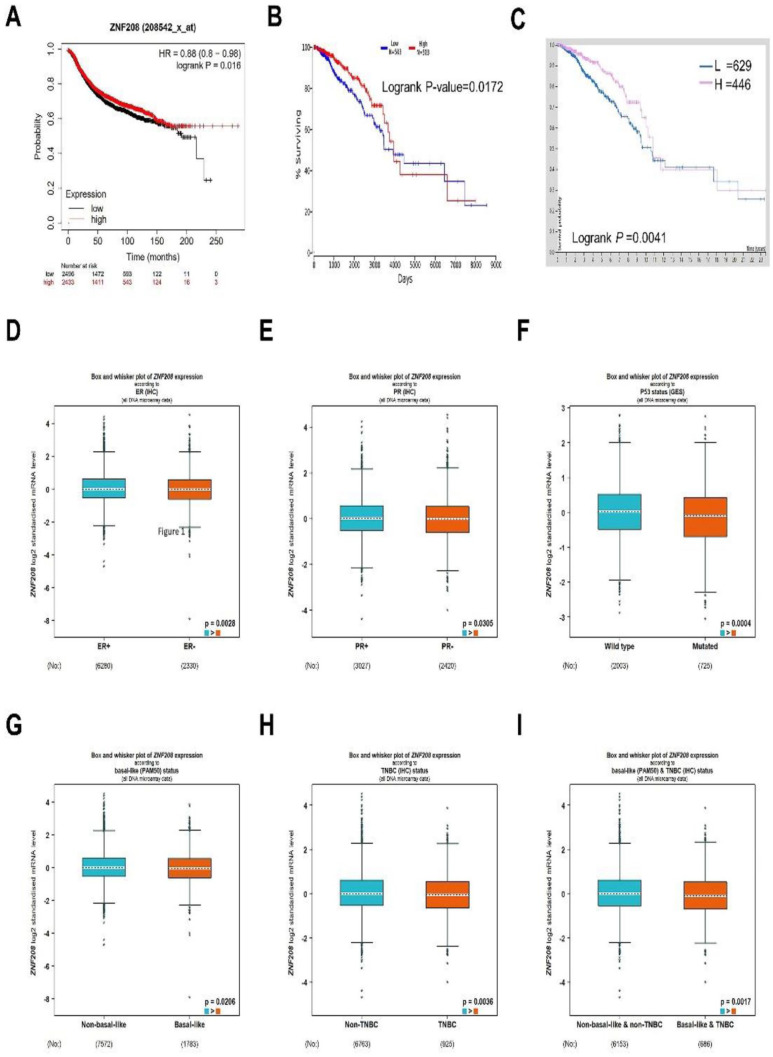
We next pretended to evaluate the correlation between the expression of ZNF208 and the prognosis of BRCA patients. As shown in the Kaplan-Meier Plotter, OncoLnc, and HPA databases, low expression of ZNF208 was closely related to poor prognosis in patients with BRCA (Figure 2A to C). Then, we tried to analyze the expression of ZNF208 in BRCA tissues with clinicopathological characteristics from bc-GenExMiner v5.0 database, and we found that ZNF208 was low expressed in ER/PR-negative BRCA tissues than control ER/PR positive BRCA tissues (Figure 2D and E). As well, the wild type P53 BRCA tissues showed more ZNF208 mRNA expression than mutated P53 BRCA tissues (Figure 2F). At the same time, the non-basal-like BRCA tissues expressed more ZNF208 mRNA than the basal-like BRCA tissues (Figure 2G). We also found that the non-TNBC BRCA tissues showed more ZNF208 mRNA expression than TNBC BRCA tissues (Figure 2H). In addition, the non-basal-like and non-TNBC BRCA tissues expressed more ZNF208 mRNA than basal-like and TNBC BRCA tissues (Figure 2I). While there was no difference in ZNF208 expression between HER2-negative and HER2 positive BRCA tissue (Supplementary Fig. S2A), and the BRCA tissue with wild type BRCA2 showed no difference in ZNF208 expression compared with BRCA tissues with mutated BRCA2 (Supplementary Fig. S2B). Besides, the BRCA tissues with nodal metastasis expressed lower ZNF208 mRNA than without nodal metastasis (Supplementary Fig. S2C). We also found that there was no difference in ZNF208 expression of BRCA patients older than 51 years old compared with those 51 years old or younger (Supplementary Fig. S2D). Conclusively, we conclude that low expression of ZNF208 is associated with clinicopathological characteristics which indicated poor prognosis of BRCA.
Figure 2.
The relationship between ZNF208 expression and prognosis in breast cancer. (A to C) The relationship of ZNF208 expression with the survival of BRCA patients from TCGA database were analyzed from Kaplan-Meier Plotter, OncoLnc, and HPA database. (D, E) Box and whisker plot of ZNF208 expression according to ER expression status and PR expression status of BRCA patients from bc-GenExMiner v5.0 database. (F to H) Box and whisker plot of ZNF208 expression according to P53 status, basal-like status, and TNBC status of BRCA patients from bc-GenExMiner v5.0 database. (I) Box and whisker plot of ZNF208 expression according to basal-like and TNBC status of BRCA patients from bc-GenExMiner v5.0 database.
Upregulation of ZNF208 promoter methylation level in BRCA
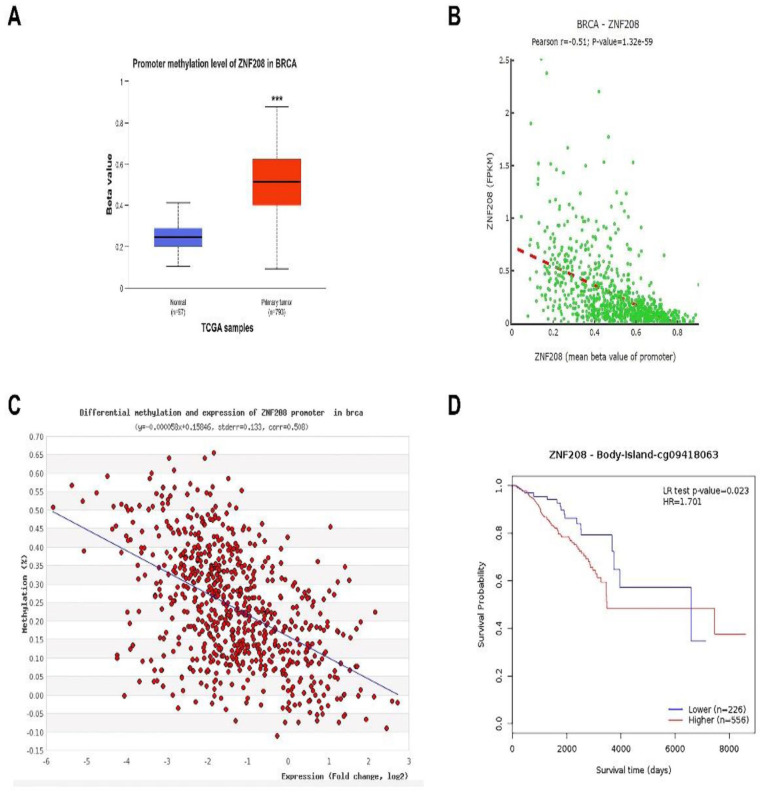
Subsequently, we intended to explain the expression aberration of ZNF208 from UALCAN and DNMIVD databases, and the ZNF208 promoter methylation level was higher in BRCA tissue than in normal tissue (P < .001) (Figure 3A and Supplementary Fig. S3A). Moreover, the ZNF208 FPKM were respectively negatively correlated with ZNF208 promoter methylation level from DNMIVD database using Pearson Correlation (r = −.51, P < .001) and Spearman Correlation (r = −.59, P < .001) (Figure 3B and Supplementary Fig. S3B). The MethHC database also confirmed the negative correlation between ZNF208 FPKM and ZNF208 promoter methylation in BRCA (Figure 3C). Using Methsurv data with cg09418063, the Kaplan-Meier plot clarified that patients with lower level of ZNF208 promoter methylation predicted better survival prognosis of BRCA (Figure 3D). All these data elucidate that the methylation level of ZNF208 is upregulated and indicating poor prognosis of BRCA.
Figure 3.
The methylation level of ZNF208 in breast cancer. (A) Different promoter methylation level of ZNF208 between BRCA tissues and normal tissues was analyzed from UALCAN database. (B) Linear regression analysis was performed to investigate association of the ZNF208 FPKM with ZNF208 promoter methylation level from DNMIVD database. (C) Linear regression analysis was performed to investigate association of the ZNF208 expression with ZNF208 promoter methylation level from MethHC database using Pearson correlation. (D) The relationship of ZNF208 promoter methylation with the survival of BRCA patients from Methsurv database.
The relationship between ZNF208 single-cell analyses and immune infiltrates’ abundances in BRCA
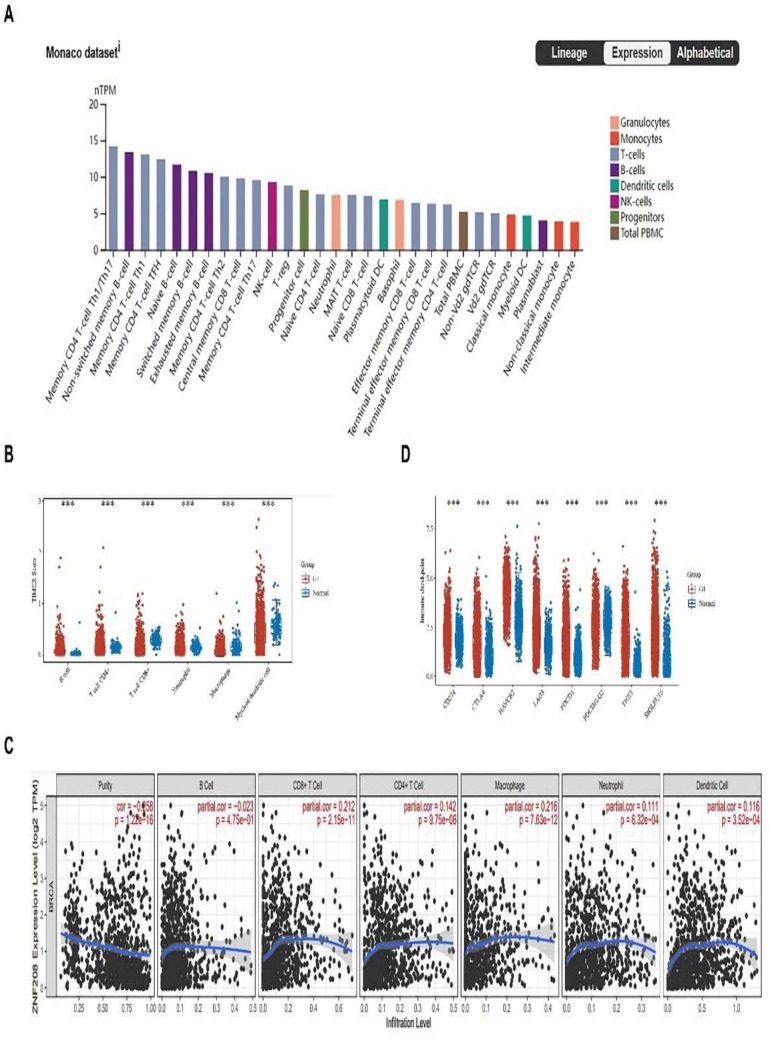
The immune checkpoints on immune cells suppress the immunity and promote tumorigenesis. 41 Based on the HPA database, breast tissue could be divided into 30 clusters by single cell sequencing analyses. In these clusters, ZNF208 was highly expressed in various types of T cells, B cells, and monocytes cells (Figure 4A). The TIMER Scors of immune landscape from HOME for Researchers database indicated that CD4 + T cell, CD8 + T + cell, neutrophil, macrophage, and myeloid dendritic cell were relatively higher in normal tissues than BRCA tissues, while the B cell was relatively lower in normal tissues (Figure 4B). We use Timer 2.0 data to visualize the correlation between ZNF208 expression and immune infiltration levels in breast cancer. The results showed that ZNF208 expressions were positively associated with CD8 + T cell (r = .212, P < .001), CD4 + T cell (r = .142, P < .001), macrophage (r = .216, P < .001), neutrophil (r = .111, P < .001), and dendritic cell (r = .116, P < .001) (Figure 4C). The data from HOME for Researchers database further confirmed that ZNF208 expressions were positively correlated with different immune cells (Supplementary Fig. S4). This suggests that ZNF208 may facilitate the microenvironment, mainly through T lymphocyte and monocytes to inhibit the occurrence of breast cancer. Then, we analyzed the immune checkpoint of ZNF208 in BRCA, and the results showed that Programmed Cell Death 1 Ligand 1 (PDL1/CD274), Cytotoxic T Lymphocyte-Associated Protein 4 (CTLA4), Hepatitis A Virus Cellular Receptor 2 (HAVCR2), Lymphocyte Activating 3 (LAG3), Programmed Cell Death 1 (PD1/PDCD1), T Cell Immunoreceptor With Ig And ITIM Domains (TIGIT), and Sialic Acid Binding Ig-Like Lectin (SIGLEC15) genes were relatively higher in BRCA tissues than normal tissues, while Programmed Cell Death 1 Ligand 2 (PDL2/PDCD1LG2) (P < .001) was relatively higher in normal tissues (Figure 4D). Consequently, immune checkpoint molecules are enriched in BRCA tissues.
Figure 4.
Analyses of immune infiltration, immune checkpoints. (A) The relationship between ZNF208 expression and different single-cell types in breast tissues from HPA database. Different colors columns represent different types of cells. (B) A scatter diagram of TIMER Scors comparing the expression of the ZNF208 associated leukocytes between BRCA and the normal tissues from HOME for Researchers. ***P < .001. (C) Infiltration level of ZNF208 associated B cell, CD8 + T cell, CD4 + T cell, macrophage, neutrophil, and dendritic cell infiltration from Timer database. (D) A scatter diagram of immune checkpoint of the leukocytes CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT, and SIGLEC15 gene expression between the normal tissues and BRCA tissues from HOME for Researchers. ***P < .001.
ZNF208 regulates many biological processes of BRCA
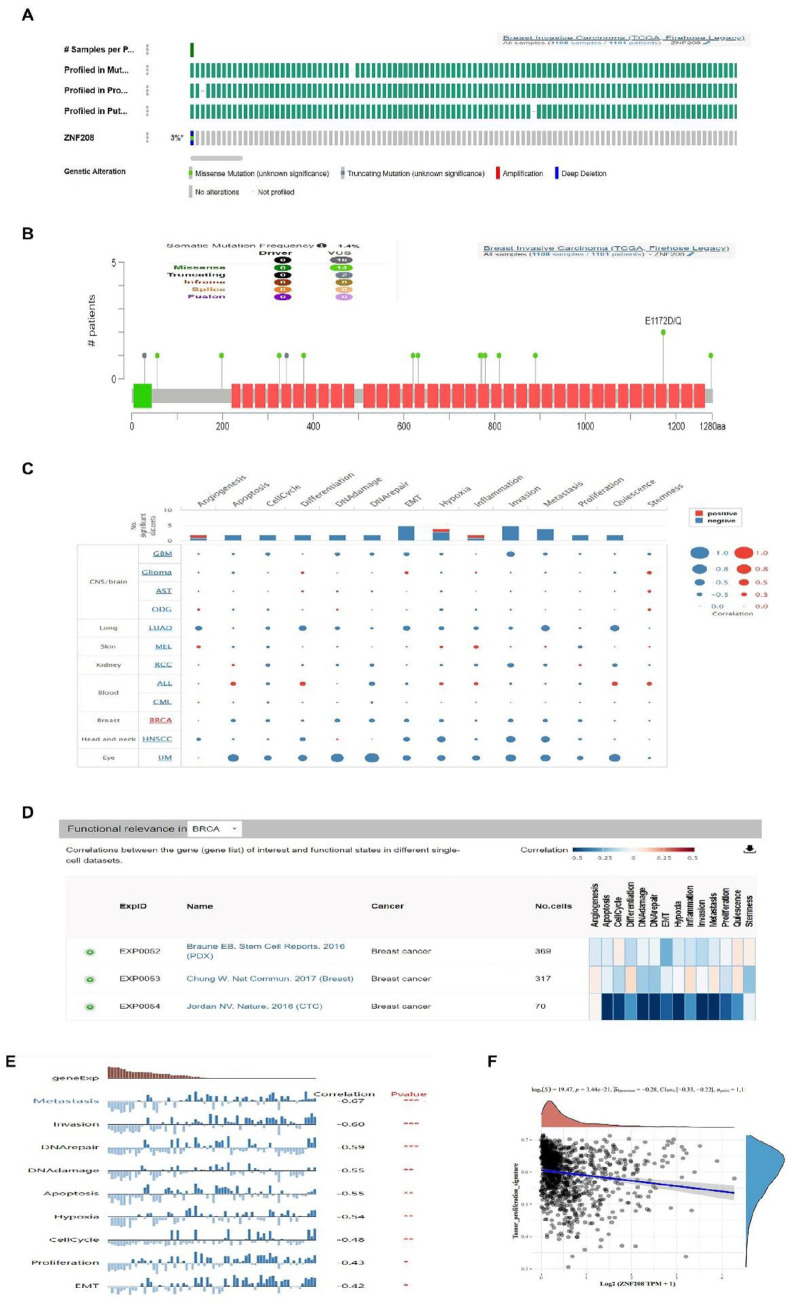
To explicit the gene genetic character, we used the cBioPortal database of 1108 or 1084 patients for cancer genomics database to determine ZNF208 gene alterations and mutations. The proportions of ZNF208 gene alterations from high to low were no alterations, deep deletion, and missense mutation (Figure 5A and Supplementary Fig. S5A). In addition, the missense mutations of ZNF208 with unknown significance were the most frequent somatic mutations in BRCA (Figure 5B and Supplementary Fig. S5B). These meant that ZNF208 gene was partially deleted in breast cancer. We suggested that ZNF208 may be a tumor suppressor in BRCA previously. Thus, we analyze the biofunction of ZNF208 in cancers from CancerSEA database, and the interactive bubble chart shown the different correlations in cancers (Figure 5C). Moreover, the results indicating that the expression of ZNF208 were mainly targeted in metastasis, invasion, hypoxia, DNA damage, apoptosis, cell cycle, and proliferation biological processes in BRCA (Figure 5D and E). As well, the linear regression showed that the ZNF208 gene expression were significantly negatively correlated with the metastasis (r = −.67, P < .001) and invasion (r = −.60, P < .001) biological processes in BRCA (Supplementary Fig. S5C). In addition, the linear regression also showed the ZNF208 gene expression were obviously negatively correlated with the proliferation (r = −.43, P < .05) and epithelial-mesenchymal transition (EMT) (r = −.42, P < .05) biological processes in BRCA (Supplementary Fig. S5D). Correspondingly, the TPM of ZNF208 was respectively negatively correlated with tumor proliferation signature (P = 3.44e-21; P^Spearman = -0.28) in BRCA from HOME for Researchers database (Figure 5F). Thus, we conclude that ZNF208 could inhibit the metastasis, invasion, proliferation and EMT processes of BRCA.
Figure 5.
The biological processes of ZNF208 in BRCA. (A, B) The cBioPortal database was used to analyze ZNF208 gene alterations and mutations in BRCA of 1108 patients. (C) Functional states of ZNF208 and its association with 12 different types of cancers from the CancerSEA database, shown was the interactive bubble chart. (D) The correlations between ZNF208 and functional states in different single-cell data sets of BRCA. (E) 9 functional states that are significantly related to ZNF208 expression of EXP0054 (GSE75367) in BRCA. *P < .05, **P < .01, ***P < .001. (F) Spearman regression analysis was performed to investigate the association of ZNF208 TPM with tumor proliferation signature in BRCA from HOME for Researchers database.
Potential molecular mechanism of ZNF208 in BRCA
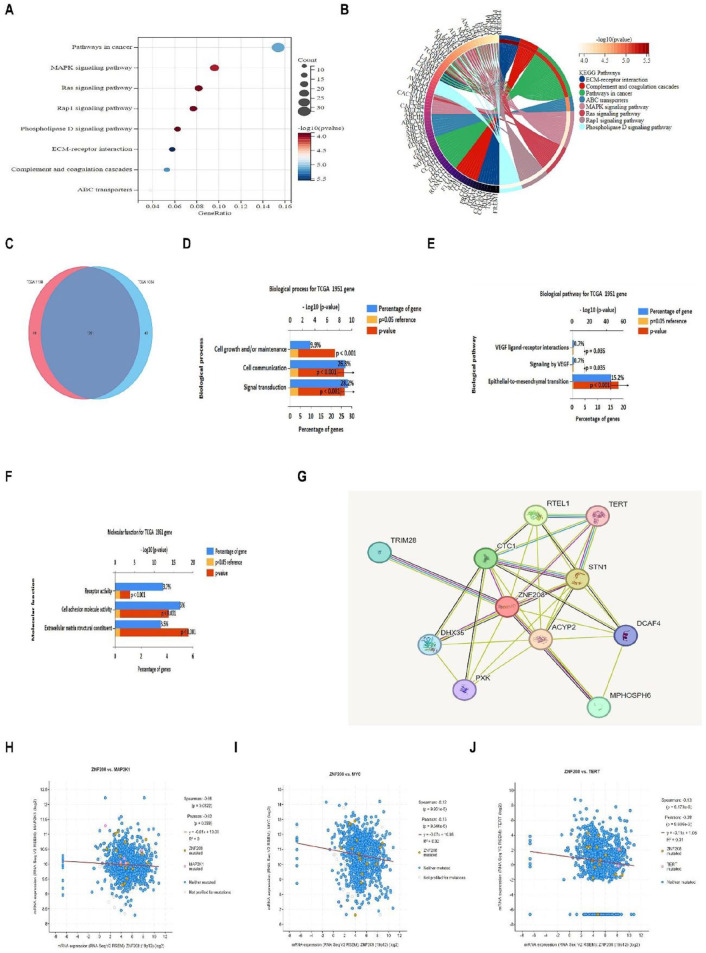
Next, we investigate the possible functional pathways of ZNF208 involved in BRCA from Sangerbox database, and the top 8 significant KEGG pathway analyses were sorted in ascending order of FDR. We found the DEGs of ZNF208 mainly function on cancer pathways, MAPK and RAS signaling pathways in BRCA (Figure 6A and Supplementary Fig. S6A). Then, the circle diagram partially showed the DEGs of ZNF208 which enriched in different pathways (Figure 6B and Supplementary Fig. S6B). Furthermore, we analyzed the number of commonly upregulated or downregulated genes among top 2000 DEGs of ZNF208 from FunRich database, and the Venn diagram showed the 1951 coregulated genes (Figure 6C). In addition, we conducted the gene enrichment from FunRich database, and identified the top 3 biological processes for coregulated genes, such as signal transduction, cell communication, and cell growth and/or maintenance (fold change > 1.3, P < .05) (Figure 6D). We also discovered the 3 biological pathways coregulated genes mostly involved, such as epithelial-to-mesenchymal (EMT) transition, VEGF ligand-receptor interactions, and signaling by VEGF (fold change > 5, P < .05) (Figure 6E). Moreover, gene enrichment showing the top 3 molecular function for coregulated genes in BRCA, such as extracellular matrix structural constituent, cell adhesion molecule activity, and receptor activity (fold change > 1.9, P < .05) (Figure 6F). These results indicated that DEGs of ZNF208 mainly impact the signal transduction of EMT transition and extracellular matrix structural constituent in BRCA. Finally, we explored the PPI network of ZNF208 protein, and the networks showed the telomerase reverse transcriptase (TERT) and regulator of telomere elongation helicase 1 (RTEL1) were tightly connected with ZNF208 based on experimentally determined results and text mining from STRING database (Figure 6G). To confirm our previous data, we analyzed the coexpression genes of 1108 and 1084 BRCA patients from cBioPortal database and found the significant negative correlations between the mRNA level of ZNF208 and the mRNA level of MEK1, MYC and TERT in BRCA (Figure 6H to J and Supplementary Fig. S6C-E). These results further validate that the transcription factor ZNF208 can regulate MAPK signaling pathways, RAS signaling pathways, EMT-associated signaling pathways, and telomere length.
Figure 6.
Functional enrichment analyses of ZNF208. (A) The cBioPortal database was used to analyze the differentially expressed genes of ZNF208 in BRCA of 1108 patient, and the top 500 genes were entered into the Sangerbox analysis tool to enrich them onto the corresponding KEGG pathways. Shown were the top eight KEGG pathways enrichment analyses of DEGs (P < .001). (B) Shown was the circle diagram of the pathways and the corresponding DEGs (P < .001). (C) Venn diagrams from FunRich database showing the coregulated genes among the top 2000 DEGs of ZNF208 in BRCA. (D to F) Gene enrichment from FunRich database showing the top 3 biological process, top 3 biological pathways, and top 3 molecular functions for coregulated 1951 genes of ZNF208. P < .05. (G) The STRING database was used to draw the protein-protein interaction networks, shown were the proteins interacted with ZNF208. P < .05. (H to J) Spearman linear regression analyses were performed to investigate the association of ZNF208 gene mRNA expression with MAP2K1 (r = −.06, P < .05), MYC (r = −.12, P < .001), TERT (r = −.13, P < .001) expression 1108 BRCA patients from cBioPortal database.
Validation of ZNF208 expression in BRCA
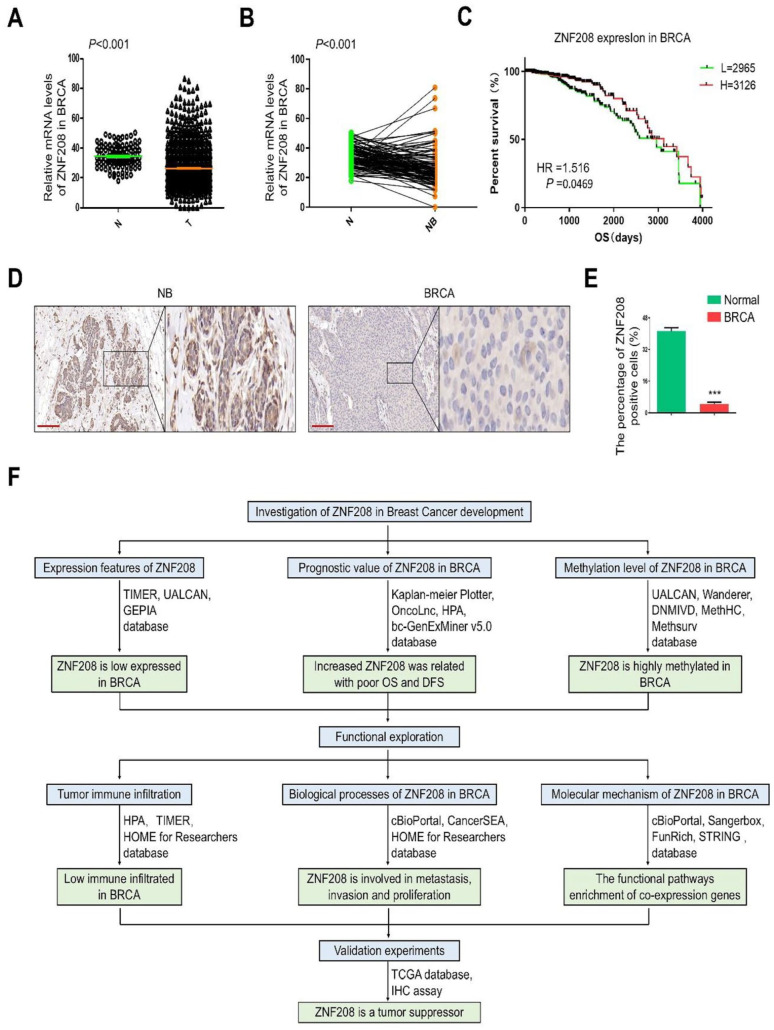
To validate the expression of ZNF208 protein in BRCA, we analyzed the mRNA levels of ZNF208 in BRCA and non-cancerous breast tissues from TCGA database, and the BRCA tissues showed lower ZNF208 mRNA expression (Figure 7A). Moreover, the paired t test also showed that the mRNA level of ZNF208 were significantly downregulated in BRCA than paired adjacent peritumoral tissues (Figure 7B). Then, we evaluated the correlation between the expression of ZNF208 and the prognosis of BRCA patients. The Kaplan-Meier survival analysis suggested that patients with high expression of ZNF208 had a long Overall Survival (OS) in BRCA (HR = 1.516, P = 0.0469) (Figure 7C). As well, we verified the ZNF208 protein expression with IHC assay, the representative IHC images shows that ZNF208 was mainly localized in intracellular of the specimens and the staining was predominant in the adjacent peritumoral tissues than the tumor tissues (Figure 7D). The histogram confirmed that the numbers of ZNF208 positively expressed cells were significantly lower in the cancer specimens than in the noncancerous breast tissues (Figure 7E). These results further validate that ZNF208 is low expressed in BRCA tissues and predicting worse prognosis of BRCA patients. The overall workflow diagram of this study is summarized in Figure 7F. We systematically illustrated the gene expression features, prognostic value, promoter methylation levels, tumor immune infiltration, biological processes, and molecular mechanism of ZNF208 in BRCA with bioinformatics analyses. The validation experiments were conducted with TCGA database and IHC assay. In summary, ZNF208 may become a valuable prognostic marker and potential therapeutic target for breast cancer.
Figure 7.
Validating the expression level and prognostic value of ZNF208 in BRCA. (A) The mRNA levels of ZNF208 in BRCA tissues and in normal tissues were analyzed from TCGA database. ***P < .001. (B) The expression levels of ZNF208 in BRCA tissues and the adjacent peritumoral tissues were analyzed from TCGA database by paired t test. NB means the adjacent peritumoral tissues. ***P < .001. (C) Differences of OS between ZNF208 high-expression and low-expression groups were determined with the Kaplan-Meier curve from TCGA database. (D) Representative IHC images of ZNF208 in tumors and adjacent peritumoral tissues. Scale bars, 50 µm. (E) The quantitative analysis of the protein levels of ZNF208 in IHC images using densitometry (right panels). Data were shown as mean ± SD. ***P < .001 (n = 3). (F) A schematic overview of different steps taken to examine the association between ZNF208 and BRCA.
Discussion
Breast cancer, which is hormone-related and mainly occurs around the perimenopause, is known for the strong malignancy and poor prognosis in females. 5 The complex molecular subtypes of BRCA includes Luminal A, Luminal B, HER2-amplified, basal-like, normal-like, and the TNBC subtypes, indicating the therapies and efficacy are limited. 7 The transcription factor ZNF208 mainly regulates the transcription of downstream target genes through sequence-specific binding to target molecules. 10 As reported, the genetic variants of ZNF208 are involved in telomere length regulation and associated with various diseases, such as coronary artery disease, 12 chronic obstructive pulmonary disease, 24 non-small-cell lung cancer, 19 and pancreatic adenocarcinoma. 20 However, the role of ZNF208 in BRCA is totally unclear.
In this study, we explored the function of ZNF208 in breast cancer tumorigenesis. By analyzing the online databases, we found that the mRNA level of ZNF208 was significantly lower in tumors than in normal tissues, especially in BLCA, BRCA, COAD, KIRP, PRAD, READ, and UCEC. Thus, we conclude that ZNF208 expression is downregulated in many cancers. The clinical characteristics such as ages, genders, races have some effect on the gene expression of BRCA. 6 Then, we analyzed the expression of ZNF208 with the clinical characteristics in BRCA. The results showed that ZNF208 expression was significantly downregulated in stage 1 to stage 4 BRCA tissues, in regional lymph node metastasis BRCA tissues, in different races of BRCA tissues, in perimenopause BRCA tissues, in luminal and triple negative subtypes of BRCA tissues, in different ages of BRCA tissues, and in different genders of BRCA tissues than in normal tissues. As well, no correlation was shown among the clinical characteristics with ZNF208 expression. Conclusively, these results indicate that ZNF208 is significantly downregulated in BRCA tissues.
To explore the correlation between ZNF208 expression and the prognosis of BRCA patients, we drew the survival curves from online databases, and low expression of ZNF208 predicting the worse prognosis of BRCA patients. The overall survival of BRCA patients is attributed to various significant clinical and genetic factors, such as age, tumor grade, nodal metastasis status, histological types, ER/PR/HER2 expression status, P53 mutations, and BRCA2 mutations. 9 Then, we analyzed the correlation between ZNF208 expression and the genetic characteristics of BRCA patients. The results showed that ZNF208 were downregulated in ER/PR-negative BRCA tissues, in mutated P53 BRCA tissues, in basal-like BRCA tissues, in basal-like and TNBC BRCA tissues, in TNBC BRCA tissues and in nodal metastasis BRCA tissues than the control tissues. Considering that the ER/PR-negative status, P53 mutation, basal-like subtypes and TNBC subtypes of breast cancers predicting the worse clinical prognosis, we suggest that the downregulated expression of ZNF208 is closely connected with short overall survivals of BRCA patients. In conclusion, we conclude that ZNF208 may be a potential prognostic marker and therapeutic target of BRCA.
It is widely demonstrated that the epigenetic alterations of promoter hypomethylation or hypermethylation inducing the aberrations of gene expression in tumors. 42 Thus, we used the online databases to explore the expression aberration of ZNF208, and we found that ZNF208 promoter methylation level was higher in BRCA tissue than in normal tissue. Hypermethylation can prevent transcription factors from binding to DNA, thereby hindering gene transcription. 43 As well, we explained the relationship between ZNF208 expression and ZNF208 promoter methylation level, and found that ZNF208 FPKM were respectively negatively correlated with ZNF208 promoter methylation level in BRCA tissue. Moreover, we found the high level of ZNF208 promoter methylation predicted worse survival prognosis of BRCA. Conclusively, the ZNF208 promoter methylation level is high in BRCA.
Tumor immune microenvironment is the specific microenvironment around tumor cells. In this microenvironment, various cells and components interact and coordinate with each other, affecting the growth of tumors. 44 Therefore, we wonder to know the proportions of immune cells in breast cancer tissues, and the online database showed that ZNF208 TPM is enriched in T cells, B cells, and monocytes cells. Furthermore, we intended to explain the relationship between the expression of ZNF208 and immune infiltration levels of BRCA, and ZNF208 expression was significantly positively associated with CD8 + T cell, CD4 + T cell, macrophage, neutrophil, and dendritic cell. These results suggest that ZNF208 mainly improves the ratios of T lymphocyte and monocytes cells in breast cancers. It is worth to mention that immune checkpoint is a class of immunosuppressive molecules, and the overexpression or over function of these molecules suppress the immunity and trigger tumors and other diseases. 41 Thus, we compared the immune checkpoint of ZNF208 in BRCA with normal tissues from online database, and found the CD274, CTLA4, HAVCR2, LAG3, PDCD1, TIGIT, and SIGLEC15 were highly expressed in BRCA than normal tissues. We conclude that immune checkpoint molecules are highly expressed in BRCA, and immunotherapy may be a potent therapy for BRCA.
The results from cBioPortal database showed that ZNF208 gene was partially deleted in BRCA. Given that the ZNF208 was suggested to be a tumor suppressor in BRCA, we analyzed the biofunction of ZNF208 in BRCA from the online database. Consequently, we found that there were negative correlations between ZNF208 expression and the metastasis, invasion, proliferation, and EMT processes of breast cancers. In summary, the ZNF208 may inhibit the malignant processes of tumorigenesis in BRCA.
In tumor cells, abnormal cell signaling pathways trigger important biological processes such as cell proliferation, metastasis, and invasion. 45 We intended to illustrate the potential molecular mechanism of ZNF208 in BRCA and the enrichment analyses showed the DEGs of ZNF208 mainly involved in cancer pathways, MAPK and RAS signaling pathways in BRCA. In addition, the gene enrichment was conducted with coregulated 1951 DEGs of ZNF208 in BRCA, showing the most affected biological processes were signal transduction, cell communication, and cell growth and/or maintenance. As well, the most crucial biological pathways were epithelial-to-mesenchymal transition, VEGF ligand-receptor interactions and signaling by VEGF. Moreover, the most obviously changed 3 molecular functions of coregulated genes were extracellular matrix structural constituent, cell adhesion molecule activity and receptor activity. All these suggests that DEGs of ZNF208 are mainly involved in the EMT associated signaling pathways. As TERT and RTEL1 are indispensable for telomere length maintenance, 46 we draw the PPI network and exhibited the clear interaction between ZNF208 and TERT or RTEL1. As known, the mitogen-activated protein kinase kinase 1 (MAP2K1/MEK1) and proto-oncogene c-Myc (MYC) are crucial molecules in MAPK and RAS signaling pathways. 47 Finally, the spearman linear regression analyses showed that ZNF208 were negatively correlated with MEK1, MYC and TERT in BRCA. In conclusion, ZNF208 can regulate the MAPK, RAS signaling pathways, EMT associated signaling pathways, and telomere length.
To further validate our study, we carried out the gene expression and prognosis analysis of ZNF208 in BRCA. We found that ZNF208 mRNA expression was low in BRCA tissues than normal controls, and the low expression of ZNF208 predicting the worse prognosis of BRCA patients. As well, ZNF208 protein level was lower in BRCA tissues than adjacent peritumoral tissues. These results further support our previous conclusion that ZNF208 may act as a tumor suppressor in BRCA.
Nevertheless, our study had not explored the specific mechanism of ZNF208 acting on BRCA, and the mechanism can be further studied by series in vivo and in vitro experiments. Conclusively, we found that high expression of ZNF208 predicts better prognosis and suppresses the tumorigenesis of BRCA. Then, preliminary pathological detection of the ZNF208 protein expression can be performed on BRCA patients to further clarify the degree of tumor malignancy. Furthermore, exploring certain agonists trigger the expression of ZNF208, producing the corresponding protein molecules of ZNF208 and immunotherapy may be new ideas and methods for the treatment of BRCA.
Conclusions
In summary, our results revealed that the expression of ZNF208 is lower in breast cancer relative to matched-normal tissues, and ZNF208 could be a prognostic biomarker in breast cancer. Based on the functional analysis, we speculate that ZNF208 may inhibit the progression of breast cancer by regulating MAPK, RAS signaling pathways.
Supplemental Material
Supplemental material, sj-docx-1-onc-10.1177_11795549241301341 for High Expression of ZNF208 Predicts Better Prognosis and Suppresses the Tumorigenesis of Breast Cancer by Jing Wei, Fangzheng Jiao, Xiaoya Wang, Yifan Qiao, Zihan Yuan, Fang Liu, Yan Fang and Yanfang Pan in Clinical Medicine Insights: Oncology
Supplemental material, sj-docx-2-onc-10.1177_11795549241301341 for High Expression of ZNF208 Predicts Better Prognosis and Suppresses the Tumorigenesis of Breast Cancer by Jing Wei, Fangzheng Jiao, Xiaoya Wang, Yifan Qiao, Zihan Yuan, Fang Liu, Yan Fang and Yanfang Pan in Clinical Medicine Insights: Oncology
Acknowledgments
We appreciate the support of the Second Affiliated Hospital of Shaanxi University of Traditional Chinese Medicine.
Footnotes
Author Contributions: Conceived and designed the analysis: JW and YF. Collected the data: JW, FL, and YP. Contributed data or analysis tools: FJ and XW. Performed the analysis: XW, YQ, and ZY. Wrote the article: JW and YF.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Key Research and Development Projects of Shaanxi Province (No. 2023-YBSF-556), the National Natural Science Foundation of China (No. 82003806).
Availability of Data and Materials: All the data and materials can be acquired from corresponding author by a reasonable request.
Consent for Publication: All participant and authors consent for publication.
Ethics Approval and Consent to Participate: This research was conducted using publicly available data from databases. This study was approved by the Ethics Committee of the Second Affiliated Hospital of Shaanxi University of Chinese Medicine on March 4, 2023 (protocol code SUCMDL20230304001). All methods in the study were performed in accordance with relevant institutional/national/international guidelines. All individuals signed informed consent to participate in the study. The privacy rights of human subjects are always respected.
ORCID iD: Jing Wei  https://orcid.org/0009-0008-5216-8899
https://orcid.org/0009-0008-5216-8899
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel R, Miller K, Fuchs H, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2. Tang Y, Tian W, Zheng S, et al. Dissection of FOXO1-induced LYPLAL1-DT impeding triple-negative breast cancer progression via mediating hnRNPK/β-catenin complex. Research (Wash D C). 2023;6:0289. doi: 10.34133/research.0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Z, Yang L, Wu P, et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. 2022;21:29. doi: 10.1186/s12943-022-01498-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y, Tan Y, Yuan J, et al. circLIFR-007 reduces liver metastasis via promoting hnRNPA1 nuclear export and YAP phosphorylation in breast cancer. Cancer Lett. 2024;592:216907. doi: 10.1016/j.canlet.2024.216907 [DOI] [PubMed] [Google Scholar]
- 5. Harbeck N, Gnant M. Breast cancer. Lancet (London, England). 2017;389:1134-1150. doi: 10.1016/s0140-6736(16)31891-8 [DOI] [PubMed] [Google Scholar]
- 6. Barzaman K, Karami J, Zarei Z, et al. Breast cancer: biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535. doi: 10.1016/j.intimp.2020.106535 [DOI] [PubMed] [Google Scholar]
- 7. Yeo SK, Guan JL. Breast cancer: multiple subtypes within a tumor. Trends Cancer. 2017;3:753-760. doi: 10.1016/j.trecan.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neuzillet C, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. MEK in cancer and cancer therapy. Pharmacol Ther. 2014;141:160-171. doi: 10.1016/j.pharmthera.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 9. Hanker A, Sudhan D, Arteaga C. Overcoming endocrine resistance in breast cancer. Cancer Cell. 2020;37:496-513. doi: 10.1016/j.ccell.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vilas CK, Emery LE, Denchi EL, Miller KM. Caught with one’s zinc fingers in the genome integrity cookie jar. Trends Genet. 2018;34:313-325. doi: 10.1016/j.tig.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gentiluomo M, Capurso G, Morelli L, et al. Genetically determined telomere length is associated with pancreatic neuroendocrine neoplasms onset. Neuroendocrinology. 2022;112:1168-1176. doi: 10.1159/000524659 [DOI] [PubMed] [Google Scholar]
- 12. Song Y, Yan M, Li J, Li J, Jin T, Chen C. Association between TNIP1, MPHOSPH6 and ZNF208 genetic polymorphisms and the coronary artery disease risk in Chinese Han population. Oncotarget. 2017;8:77233-77240. doi: 10.18632/oncotarget.20432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding Y, Zhou X, Wu C, et al. Telomere length, ZNF208 genetic variants and risk of chronic obstructive pulmonary disease in the Hainan Li population. J Gene Med. 2018;20:e3061. doi: 10.1002/jgm.3061 [DOI] [PubMed] [Google Scholar]
- 14. Wang S, Chang E, Byanyima P, et al. Association between common telomere length genetic variants and telomere length in an African population and impacts of HIV and TB. J Hum Genet. 2019;64:1033-1040. doi: 10.1038/s10038-019-0646-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gentiluomo M, Luddi A, Cingolani A, et al. Telomere length and male fertility. Int J Mol Sci. 2021;22:3959. doi: 10.3390/ijms22083959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao X, Liu X, Zhang A, et al. The correlation of copy number variations with longevity in a genome-wide association study of Han Chinese. Aging. 2018;10:1206-1222. doi: 10.18632/aging.101461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roy Choudhury S, Ashby C, Zhan F, van Rhee F. Epigenetic deregulation of telomere-related genes in newly diagnosed multiple myeloma patients. Cancers. 2021;13:6348. doi: 10.3390/cancers13246348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang S, Wen X, Zhao R, Bai Y. Genetic variation in the ZNF208 gene at rs8103163 and rs7248488 is associated with laryngeal cancer in the Northwestern Chinese Han male. Front Genet. 2022;13:813823. doi: 10.3389/fgene.2022.813823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu B, Li H, Liu X, et al. CircZNF208 enhances the sensitivity to X-rays instead of carbon-ions through the miR-7-5p /SNCA signal axis in non-small-cell lung cancer cells. Cell Signal. 2021;84:110012. doi: 10.1016/j.cellsig.2021.110012 [DOI] [PubMed] [Google Scholar]
- 20. Zhang W, Shang S, Yang Y, et al. Identification of DNA methylation-driven genes by integrative analysis of DNA methylation and transcriptome data in pancreatic adenocarcinoma. Exp Ther Med. 2020;19:2963-2972. doi: 10.3892/etm.2020.8554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cui Y, Li G, Yan M, et al. The effects of gene polymorphisms on glioma prognosis. J Gene Med. 2017;19:345-352. doi: 10.1002/jgm.2989 [DOI] [PubMed] [Google Scholar]
- 22. Wang H, Yu J, Guo Y, et al. Genetic variants in the ZNF208 gene are associated with esophageal cancer in a Chinese Han population. Oncotarget. 2016;7:86829-86835. doi: 10.18632/oncotarget.13468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walsh KM, Whitehead TP, de Smith AJ, et al. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis. 2016;37:576-582. doi: 10.1093/carcin/bgw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ojha J, Codd V, Nelson CP, et al. Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev. 2016;25:1043-1049. doi: 10.1158/1055-9965.Epi-15-1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirbe A, Dahiya S, Miller C, et al. Whole exome sequencing reveals the order of genetic changes during malignant transformation and metastasis in a single patient with NF1-plexiform neurofibroma. Clin Cancer Res. 2015;21:4201-4211. doi: 10.1158/1078-0432.Ccr-14-3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649-658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. doi: 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J, Lichtenberg T, Hoadley K, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400-416.e11. doi: 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725-731. doi: 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 30. Zheng H, Zhang G, Zhang L, et al. Comprehensive review of web servers and bioinformatics tools for cancer prognosis analysis. Front Oncol. 2020;10:68. doi: 10.3389/fonc.2020.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jézéquel P, Gouraud W, Azzouz FB, et al. [Interest of the bc-GenExMiner web tool in oncology]. Bull Cancer. 2021;108:1057-1064. doi: 10.1016/j.bulcan.2021.05.008 [DOI] [PubMed] [Google Scholar]
- 32. Ding W, Chen J, Feng G, et al. DNMIVD: DNA methylation interactive visualization database. Nucleic Acids Res. 2020;48:D856-D862. doi: 10.1093/nar/gkz830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang H, Li J, Tang Y, et al. MethHC 2.0: information repository of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2021;49:D1268-D1275. doi: 10.1093/nar/gkaa1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10:277-288. doi: 10.2217/epi-2017-0118 [DOI] [PubMed] [Google Scholar]
- 35. Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018;27:233-244. doi: 10.1002/pro.3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W514. doi: 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li K, Tan G, Zhang X, et al. EIF4G1 is a potential prognostic biomarker of breast cancer. Biomolecules. 2022;12:1756. doi: 10.3390/biom12121756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900-D908. doi: 10.1093/nar/gky939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fonseka P, Pathan M, Chitti S, Kang T, Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. 2021;433:166747. doi: 10.1016/j.jmb.2020.166747 [DOI] [PubMed] [Google Scholar]
- 40. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447-D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Hu Y, Xue J, et al. Advances in immunotherapy for triple-negative breast cancer. Mol Cancer. 2023;22:145. doi: 10.1186/s12943-023-01850-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weisenberger DJ. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J Clin Invest. 2014;124:17-23. doi: 10.1172/jci69740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Zhu J. The altered DNA methylation pattern and its implications in liver cancer. Cell Res. 2005;15:272-280. doi: 10.1038/sj.cr.7290296 [DOI] [PubMed] [Google Scholar]
- 44. Yi M, Li T, Niu M, et al. Exploiting innate immunity for cancer immunotherapy. Mol Cancer. 2023;22:187. doi: 10.1186/s12943-023-01885-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li YJ, Zhang C, Martincuks A, Herrmann A, Yu H. STAT proteins in cancer: orchestration of metabolism. Nat Rev Cancer. 2023;23:115-134. doi: 10.1038/s41568-022-00537-3 [DOI] [PubMed] [Google Scholar]
- 46. Hassani MA, Murid J, Yan J. Regulator of telomere elongation helicase 1 gene and its association with malignancy. Cancer Rep (Hoboken). 2023;6:e1735. doi: 10.1002/cnr2.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zinatizadeh MR, Miri SR, Zarandi PK, et al. The hippo tumor suppressor pathway (YAP/TAZ/TEAD/MST/LATS) and EGFR-RAS-RAF-MEK in cancer metastasis. Genes Dis. 2021;8:48-60. doi: 10.1016/j.gendis.2019.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-onc-10.1177_11795549241301341 for High Expression of ZNF208 Predicts Better Prognosis and Suppresses the Tumorigenesis of Breast Cancer by Jing Wei, Fangzheng Jiao, Xiaoya Wang, Yifan Qiao, Zihan Yuan, Fang Liu, Yan Fang and Yanfang Pan in Clinical Medicine Insights: Oncology
Supplemental material, sj-docx-2-onc-10.1177_11795549241301341 for High Expression of ZNF208 Predicts Better Prognosis and Suppresses the Tumorigenesis of Breast Cancer by Jing Wei, Fangzheng Jiao, Xiaoya Wang, Yifan Qiao, Zihan Yuan, Fang Liu, Yan Fang and Yanfang Pan in Clinical Medicine Insights: Oncology