Abstract
Background
The three-amino-acid-loop-extension (TALE) is a ubiquitous homeodomain transcription factor among plant species involved in regulating plant growth, development, and environmental responses. However, this has not been systematically analyzed or reported in sorghum.
Results
In this study, 23 SbTALE genes were identified using bioinformatics and other methods at the genome level of sorghum, classified into two families, KNOX and BEL1-like family, and localized on ten chromosomes. One pair of tandem duplicated and seven pairs of segmentally duplicated genes were found, and the conserved motifs of SbTALEs among the same subfamilies were highly conserved, with highly conserved gene structures. SbTALEs genes have the most collinear genes with monocotyledonous Zea mays and are more closely related; SbTALEs have undergone purification and diversification selection in the evolutionary process. Overall, except for SbTALE21 and SbTALE23, the expression of the other six SbTALEs was higher in the stems, whereas the expression of SbTALE21 and SbTALE23 was higher in the leaves. In sorghum grain development, the lowest relative expression of SbTALEs was observed in grains in the late stage, and the expression of SbTALE21 was higher in grains in the early stage and husks in the late stage. In addition, SbTALE14 and SbTALE21 showed higher expression in the roots and stems under the cold treatment, and SbTALE02 and SbTALE12 showed higher expression in the roots and stems under the PEG treatment. Under the four hormone treatments, the expression of eight SbTALEs was relatively low in stems, the expression of SbTALE13 was higher in leaves than in roots and stems, and the expression of SbTALE23 was higher under the MeJA and SA treatments.
Conclusion
This study lays a theoretical foundation for the study of the biological function and mechanism of SbTALE genes and is of great significance for the mining of resistance genes and trait improvement.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05735-9.
Keywords: Sorghum bicolor, TALE gene family, Genome-wide, Evolution, Gene expression
Background
A homeobox (HB) encodes a protein domain, and the homeodomain (HD) consists of a conserved 60-amino acid motif, the 60 amino acids fold into a three-helix structure, with the first and second helices forming a loop structure and the second and third helices forming a helix-turned-helix structure. Most HDs can bind DNA monomers with high affinity [1, 2], and this domain is highly conserved in different proteins, suggesting that this structure is essential for maintaining HD function and plays an important role in plant growth and development [3]. There is a special class of HD proteins that encode atypical DNA-binding structural domains consisting of 63 amino acids, with three additional amino acid residues (P-Y-P) inserted between the 1st and 2nd helical structures, known as the TALE superfamily of transcription factors [4]. In plants, the TALE family is divided into two subfamilies, KNOX and BEL1-like, according to the structural domains they contain [5].
The KNOX gene family is classified into three classes (KNOX Class I, Class II, and Class III) based on homology and gene structure in Arabidopsis thaliana [6, 7]. The expression pattern of the KNOX Class I member, KNAT1, in the leaf primordium directly affects leaf initiation and development, which ultimately determines leaf differentiation into simple or compound leaves [8, 9], and KNAT1 expression is also repressed by the phytohormone AUX [10]. For KNOX Class II, Arabidopsis KNAT 3/4/5 are involved in root development [11], and KNAT3 interacts with BLH1 to regulate the response to ABA during Arabidopsis seed germination and early seedling development. KNATM is the only KNOX Class III gene that has been shown to be involved in proximal-distal leaf patterns, serration, and compound leaf development [12].
In Arabidopsis, all 13 BEL1-like family members interact with KNOX proteins to form heterodimers [13], while BEL1-like proteins are critical for the control of meristematic organization and flower development. For example, AtBLH3 interacts with AtOFP1 to regulate the timing of the transition from nutrient availability to reproductive growth in Arabidopsis [14]. AtBEL1 mutations hinder the development of the Arabidopsis bead coat [15], and BEL1-like also has regulatory roles in plant growth, development, and stress physiology. For example, in tomato, SlBEL11 regulates the formation and development of chloroplasts and the synthesis of chlorophyll II in tomato fruits [16], and the GhBLH7-D06/GhOFP3-D13 complex negatively regulates resistance to Verticillium wilt in cotton by decreasing the JA signaling pathway and inhibiting lignin biosynthesis [17].
Sorghum bicolor (L.) Moench, a diploid (2n = 20) C4 crop of the genus Sorghum in the family Gramineae, is widely grown in subtropical, tropical, and temperate regions around the world [18]; it is the fifth largest cereal crop globally after wheat, maize, rice, and barley, and it is an important multigrain crop in China [19, 20]. Compared to other energy crops, sorghum is characterized by high stress tolerance and growth adaptability [21] and contains high levels of phenolic compounds with antioxidant properties and other bioactivities favorable to human health compared to other grain crops [22].
The TALE gene family has been identified in Arabidopsis [4], soybean [23], and wheat [24], but has not been systematically identified and analyzed in sorghum. In the present study, bioinformatics and other methods were used to identify sorghum TALE family members at the sorghum genome level, including physicochemical properties, chromosomal location, gene structure, Cis-acting elements, and evolutionary relationships, were used to identify sorghum TALE family members at the sorghum genome level. This study also investigated the tissue specificity of different subfamily TALE members during sorghum grain filling, grain developmental characteristics, and expression levels of various abiotic stresses and hormone responses during the seedling stage, with the aim of providing a theoretical basis for studying the evolutionary relationships and biological functions of SbTALE transcription factors.
Results
Genome-wide identification of the TALE gene family of Sorghum bicolor
In this study, 23 TALEs genes were identified within the whole sorghum genome, and they were named SbTALE01 to SbTALE23 according to their positions on the chromosome (Table S1) and the physicochemical properties of their encoded proteins such as molecular weight (MW/KD), isoelectric point (pI), and instability index (II). (Table 1). The amino acid content of the 23 SbTALE proteins ranged from to 294 to 770 aa, with SbTALE02 possessing the highest amino acid content (770 aa) and SbTALE13 possessing the lowest amino acid content (294 aa) (Table S1). The mean molecular weight of the 23 SbTALEs proteins was 51.46 KD. SbTALE02 was the largest, while SbTALE13 was the smallest (Table S1), thus indicating that the molecular weight of the proteins was positively correlated with their amino acid content. Overall, the isoelectric point of the 23 SbTALEs ranged from 5.34 (SbTALE19) to 7.89 (SbTALE20), but the vast majority of the SbTALE proteins (21/23) exhibited a pI < 7 (Table S1), suggesting that the SbTALE family is rich in acidic amino acids. In this study, the instability indices of 23 SbTALE proteins were analyzed, and it was observed that the instability index (II) of all 23 proteins was greater than 40, among which the instability coefficient of SbTALE06 was the highest (66.01) (Table S1). In this study, we predicted the subcellular locations of 23 SbTALE proteins and determined that all of them were located in the nucleus (Table S1).
Table 1.
List of the identified SbTALE genes and their physicochemical properties in the study
| Gene name | Accession number/Gene ID | Chromosome | Subfamily | Encoded protein | ||||
|---|---|---|---|---|---|---|---|---|
| Length/aa | Molecular weight (average)/KD | Isoelectric point (pI) | Instability index (II) | Subcellular localization prediction | ||||
| SbTALE01 | SORBI_3001G075101 | Chr 1 | KNOX Class I | 307 | 34.18 | 6.08 | 61.20 | nuclear |
| SbTALE02 | SORBI_3001G102300 | Chr 1 | Leaf morphology | 770 | 80.77 | 6.55 | 51.37 | nuclear |
| SbTALE03 | SORBI_3001G106000 | Chr 1 | KNOX Class I | 371 | 40.47 | 6.29 | 46.06 | nuclear |
| SbTALE04 | SORBI_3001G106200 | Chr 1 | KNOX Class I | 360 | 39.86 | 6.56 | 46.23 | nuclear |
| SbTALE05 | SORBI_3001G137100 | Chr 1 | OFPs partners | 649 | 71.79 | 5.53 | 50.61 | nuclear |
| SbTALE06 | SORBI_3001G137200 | Chr 1 | OFPs partners | 356 | 38.95 | 5.88 | 66.01 | nuclear |
| SbTALE07 | SORBI_3001G140200 | Chr 1 | KNOX Class I | 334 | 36.80 | 5.53 | 56.43 | nuclear |
| SbTALE08 | SORBI_3001G314900 | Chr 1 | Ovule morphology | 623 | 65.72 | 6.53 | 47.55 | nuclear |
| SbTALE09 | SORBI_3001G494000 | Chr 1 | OFPs partners | 590 | 62.83 | 5.87 | 41.73 | nuclear |
| SbTALE10 | SORBI_3001G525900 | Chr 1 | Ovule morphology | 626 | 66.68 | 6.14 | 43.41 | nuclear |
| SbTALE11 | SORBI_3001G526200 | Chr 1 | KNOX Class II | 323 | 35.15 | 5.72 | 58.31 | nuclear |
| SbTALE12 | SORBI_3002G023900 | Chr 2 | KNOX Class I | 356 | 38.88 | 6.23 | 49.11 | nuclear |
| SbTALE13 | SORBI_3003G144200 | Chr 3 | KNOX Class I | 294 | 32.46 | 5.60 | 46.57 | nuclear |
| SbTALE14 | SORBI_3003G356200 | Chr 3 | Meristem function | 593 | 63.83 | 7.00 | 53.11 | nuclear |
| SbTALE15 | SORBI_3004G067100 | Chr 4 | KNOX Class II | 306 | 33.12 | 5.89 | 46.44 | nuclear |
| SbTALE16 | SORBI_3004G097700 | Chr 4 | OFPs partners | 564 | 58.84 | 6.47 | 55.11 | nuclear |
| SbTALE17 | SORBI_3005G045200 | Chr 5 | OFPs partners | 690 | 71.66 | 6.15 | 43.73 | nuclear |
| SbTALE18 | SORBI_3008G188900 | Chr 8 | OFPs partners | 658 | 73.13 | 5.59 | 56.97 | nuclear |
| SbTALE19 | SORBI_3009G030200 | Chr 9 | KNOX Class I | 303 | 32.96 | 5.34 | 50.11 | nuclear |
| SbTALE20 | SORBI_3009G159900 | Chr 9 | Meristem function | 570 | 61.22 | 7.89 | 53.99 | nuclear |
| SbTALE21 | SORBI_3010G006300 | Chr 10 | Ovule morphology | 524 | 57.04 | 6.88 | 53.53 | nuclear |
| SbTALE22 | SORBI_3010G169200 | Chr 10 | OFPs partners | 494 | 52.61 | 6.64 | 61.78 | nuclear |
| SbTALE23 | SORBI_3010G207500 | Chr 10 | KNOX Class II | 319 | 34.61 | 5.79 | 50.83 | nuclear |
Phylogenetic analysis and classification of TALEs of Sorghum bicolor
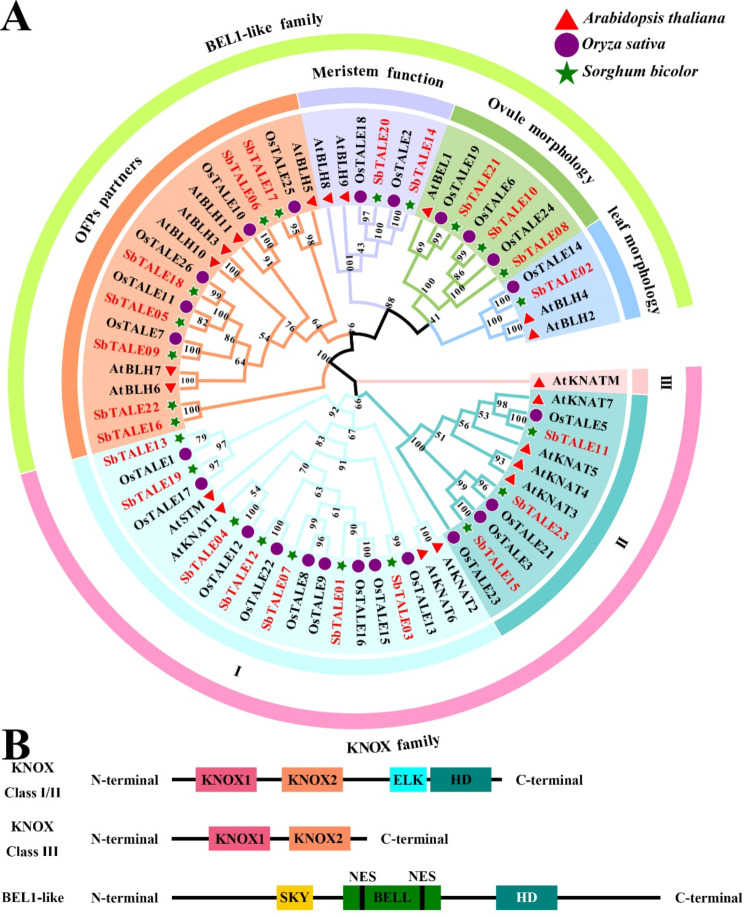
In this study, we constructed an evolutionary tree based on the identified 23 SbTALEs and the reported TALEs of Arabidopsis thaliana and Oryza sativa, and classified the 23 SbTALE proteins into two major groups according to the classification of the AtTALE family, which includes the KNOX and BEL1-like family. The KNOX family is divided into three subfamilies (Class I, II, and III) and the BEL1-like family is divided into four subfamilies (OFPs partners, meristem function, ovule morphology, and leaf morphology) (Fig. 1A). Of the two families, the BEL1-like family contains more SbTALE family members (13), all of which contain BELL structures (Fig. 1B), with the OFPs partner subfamily containing the most SbTALE members (7) (Fig. 1A and B). In the KNOX family, KNOX Class I contained the most SbTALE family members (seven), whereas KNOX Class III exhibited no distribution of SbTALE members (Fig. 1A). The structures of KNOX classes I/II and III are quite different, but both contain KNOX1 and KNOX2 domains (Fig. 1B) [4].
Fig. 1.
Classification of phylogenetic tree among the TALE proteins of Sorghum bicolor, Arabidopsis thaliana, and Oryza sativa and schematic structures of the KNOX and BEL1-like proteins. (A) Different colored outer arcs represent different families or subfamilies. The red triangle represents Arabidopsis, the purple circle represents Oryza sativa, and the green pentagram represents Sorghum bicolor. (B) Colorful modules represent different structures
Gene structures and conserved motifs analysis of TALEs family of Sorghum bicolor
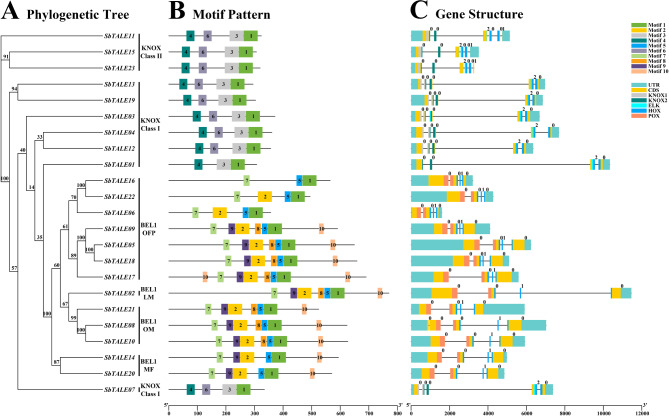
To further understand the differences among family members and structural changes in SbTALEs during the evolutionary process, we used TBtools to construct a family evolutionary tree (Fig. 2A) and gene structures (Fig. 2C) of the 23 SbTALE proteins, and analyzed the conserved motif constructs through the MEME online website motif pattern (Fig. 2B and Table S2). The conserved motifs were found to be highly conserved among the KNOX family, with all nine SbTALE proteins having the conserved motif 4-6-3-1, except for SbTALE01, which did not contain motif 6 (Fig. 2A and B). For the BEL1-like family, the vast majority (10/13) of the SbTALE proteins were highly conserved, with a conserved motif distribution of motifs 7-9-2-8-5-1-10, whereas the conserved motifs of the proteins in the OFPs partner subfamily varied to some extent (Fig. 2A and B). For example, there are only three motifs in SbTALE16 (motif 7-5-1), and the conserved motifs of SbTALE06 and SbTALE22 have more motif 2 (motif 7-2-5-1) than SbTALE16. The conserved motif of SbTALE17 had more motif 10 than most members of the BEL1-like family and was distributed at the front, that is, motif 10-7-9-2-8-5-1-10 (Fig. 2B). In general, the average number of conserved motifs in the KNOX family distribution was 3.9, which was 2.4 less than that in the BEL1-like family, but both the KNOX and BEL1-like families contained motif 1 (Fig. 2A and B).
Fig. 2.
Phylogenetic tree, motif pattern, and gene structure of TALE genes in Sorghum bicolor. (A) The phylogenetic tree is constructed by SbTALE protein sequences with 1000 replicates on each node. (B) The amino acids conserved motifs (motif 1 to 10) in SbTALEs are displayed in ten colored boxes, and black lines indicate protein length. (C) Colorful rectangles indicate UTR (untranslated region), CDS (coding sequence or exons), KNOX1, KNOX2, ELK, HOX, and POX domain, respectively. The black lines represent introns of SbTALE genes
Based on the sorghum genome database, the gene structures of the 23 SbTALEs were analyzed in this study (Fig. 2C). Overall, the vast majority of SbTALEs (22/23) contained two UTR regions, all of which were distributed at both ends of the sequence. The majority of the members of the KNOX family (8/10) contained five exon regions, and the intron regions ranged from three to five. There were also 12 BEL1-like family SbTALE members containing three exon regions, and their intron regions were between all three exon regions (Fig. 2C). In the KNOX family, the gene structure of KNOX Class II is more conserved than that of KNOX Class I, with one KNOX1 domain, two KNOX2 domains, two HOX domains, and two ELK domains. The vast majority (6/7) of KNOX Class I proteins contain two KNOX1 domains and one ELK domain, and all seven SbTALEs contain two KNOX2 domains and two HOX domains (Fig. 2C). For the BEL1-like family that possesses no KNOX1, KNOX2 and ELK domains, the 13 SbTALEs members are highly conserved, and all contain three HOX domains and two POX domains (Fig. 2C).
Promoter Cis-elements analysis of TALEs of Sorghum bicolor
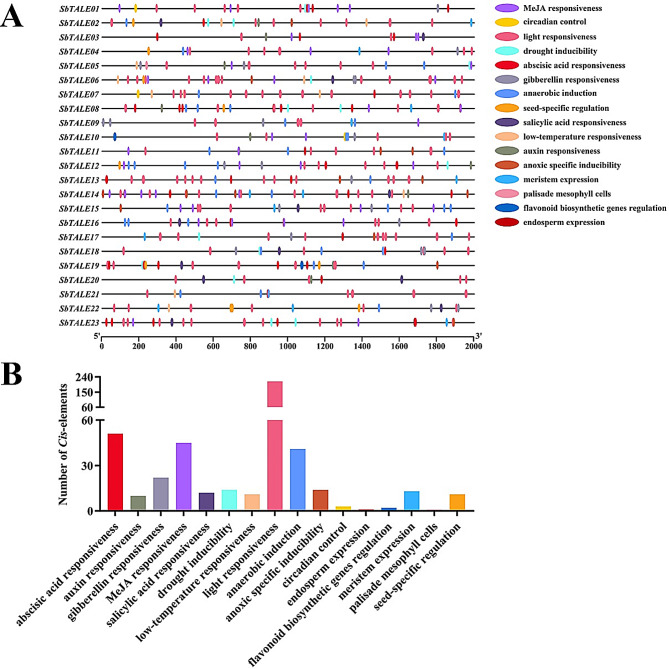
In this study, we selected 2000 bp upstream of the transcription start site as promoter sequences and used PlantCARE to predict their Cis-acting elements, and found that the 23 SbTALE promoter sequences mainly contained light responsiveness, hormone responsiveness, abiotic stress responsiveness, and growth and development elements (Fig. 3A). Among them, light responsiveness and hormone responsiveness elements were distributed in all promoter sequences. The number of light responsiveness elements was the highest (214), followed by abscisic acid responsiveness (51), whereas the two types of abiotic stress responsiveness elements (drought inducibility and low-temperature responsiveness) were less abundant (Fig. 3A and B). In this study, we found that SbTALE02, SbTALE05, and SbTALE12 contained most of the hormone- and abiotic-responsive elements, and SbTALE05 contained the majority (13/16) of Cis-acting elements that were hormone- and abiotic-responsive (Fig. 3A and B).
Fig. 3.
Distribution of Cis-acting elements and their number statistics of SbTALE promoters. (A) The promotor Cis-element of the promoter region (upstream 2000 bp) of 23 TALE genes in Sorghum bicolor. (B) Statistics of Cis-elements in the SbTALE promoters
Chromosomal location, duplication, and collinearity analysis of TALEs from Sorghum bicolor
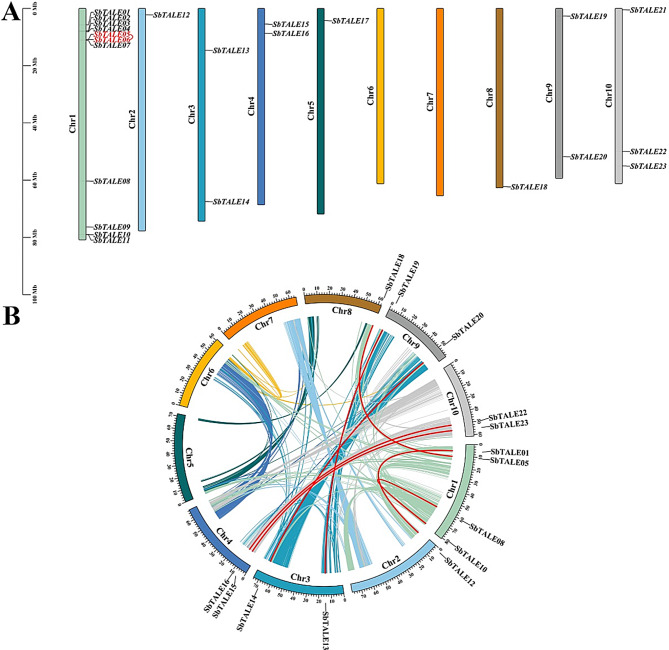
In this study, 23 SbTALE genes were located on 10 chromosomes (Chr 1 to 10) based on genomic analysis of sorghum, and these genes exhibited an uneven distribution (Fig. 4A). It was observed that the largest number of genes was distributed on Chr 1 (47.83%, 11/23), and this was followed by Chr 10 containing three genes (SbTALE21, SbTALE22, and SbTALE23, 12.5%), whereas Chr 6 and Chr 7 did not possess SbTALE genes (Fig. 4A). In this study, we also determined that only one pair of SbTALE genes (SbTALE05 and SbTALE06) possessed tandem duplications and were distributed on chromosome 1. Both of these genes belong to OFPs partners in the BEL1-like family (Fig. 4A and Table S4).
Fig. 4.
Chromosomal location and collinearity of TALE genes in Sorghum bicolor. (A) The colorful rectangular bars represent the different chromosomes (Chr 1 to 10) of Sorghum bicolor; red fonts represent gene tandem duplications, and the 0 to 100 Mb scale represents chromosome length. (B) Colored lines indicate the collinear blocks in the genomes of Sorghum bicolor; the chromosome number is shown inside each chromosome, and red lines indicate collinear TALE gene pairs
To further analyze the characteristics of SbTALE genes on sorghum chromosomes, we analyzed 23 SbTALE genes for gene duplication events occurring within the sorghum genome (Fig. 4B and Table S5). It was found that a total of 14 homoeologous loci and 7 pairs of segmental duplication events of SbTALEs were identified on 10 sorghum chromosomes, respectively SbTALE01/SbTALE12, SbTALE05/SbTALE18, SbTALE08/SbTALE10, SbTALE13/ SbTALE19, SbTALE14/SbTALE20, SbTALE15/ SbTALE23, and SbTALE16/SbTALE22, of which, 3 and 4 pairs of SbTALE genes belonged to the KNOX family or the BEL1-like family, respectively, and both the KNOX Class I and OFPs partners were distributed with two pairs of SbTALEs duplication events (Fig. 4B and Table S5). In this study, it was observed that SbTALE genes were unevenly distributed in 10 linked regions (LG) of sorghum, of which LG1 possessed the most SbTALEs (28.57%, 4/14), and LG5, LG6, LG7, and LG8 did not possess SbTALE genes (Fig. 4B and Table S5).
Synteny of TALEs of Sorghum bicolor and Ka/Ks analysis
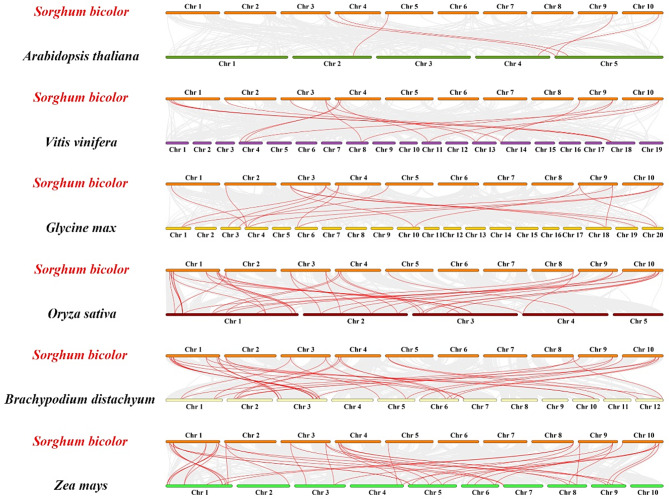
To further investigate the evolutionary mechanism of SbTALE, interspecific synteny maps were constructed between sorghum and three monocotyledons (Arabidopsis thaliana, Vitis vinifera, and Glycine max) and three dicotyledons (Oryza sativa, Brachypodium distachyon, and Zea mays) (Fig. 5). Sorghum TALE genes were observed to possess more synteny with monocotyledons than with dicotyledons, with Sorghum bicolor sharing the most synteny genes with Zea mays (42 pairs) followed by Brachypodium distachyon (37 pairs). They shared the least amount of synteny genes (5 pairs) with dicotyledons, whereas Arabidopsis thaliana possessed the least number of synteny genes (5 pairs) (Fig. 5 and Table S6). We observed that all 20 SbTALEs, with the exceptions of SbTALE03, SbTALE04 and SbTALE06, exhibited several synteny relationships with three dicotyledons, whereas all four genes, SbTALE14, SbTALE15, SbTALE20 and SbTALE23, possessed homologous genes with six plants, thus indicating that these genes were formed before monocotyledonous and dicotyledonous differentiation (Table S6).
Fig. 5.
Synteny analysis of the SbTALE genes between Sorghum bicolor and six plants (Arabidopsis thaliana, Vitis vinifera, Glycine max, Oryza sativa, Brachypodium distachyon, Zea mays). The gray lines represent synteny blocks in wide genomes of Sorghum bicolor and other plants, while red lines highlight syntenic TALE gene pairs
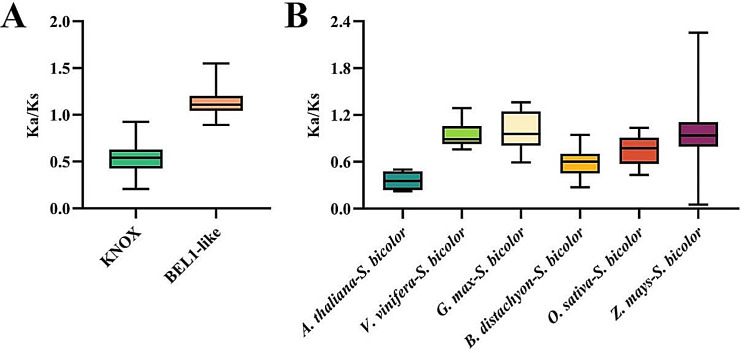
To understand the dynamics and selective pressure of SbTALE genes during the evolutionary process, in this study, two families and syntenic TALE gene pairs of interspecies were subjected to non-synonymous (Ka) to synonymous (Ks) ratios (Ka/Ks) (Fig. 6, Table S7, and Table S8). The results show that the average Ka/Ks ratio of the KNOX family is smaller than that of the BEL1-like family and is less than 1, indicating that the KNOX family has undergone purification selection in the evolutionary process. However, most BEL1-like families were greater than 1, suggesting that these genes played a key role in evolution (Fig. 6A and Table S7). For the Ka/Ks calculation of homologous gene pairs between Sorghum bicolor and the six species, we determined that the Ka/Ks ratio of homologous gene pairs belonging to Sorghum bicolor, Vitis vinifera, Glycine max and Zea mays was close to 1. The Ka/Ks ratio was the lowest between Sorghum bicolor and Arabidopsis thaliana at 0.357 (Fig. 6B and Table S8).
Fig. 6.
The ratio of TALE genes nonsynonymous and synonymous substitution (Ka/Ks). (A) Ka/Ks values of different families of SbTALEs. (B) Ka/Ks values of syntenic TALE gene pairs of interspecies
Evolutionary analysis of TALEs among Sorghum bicolor and different plants
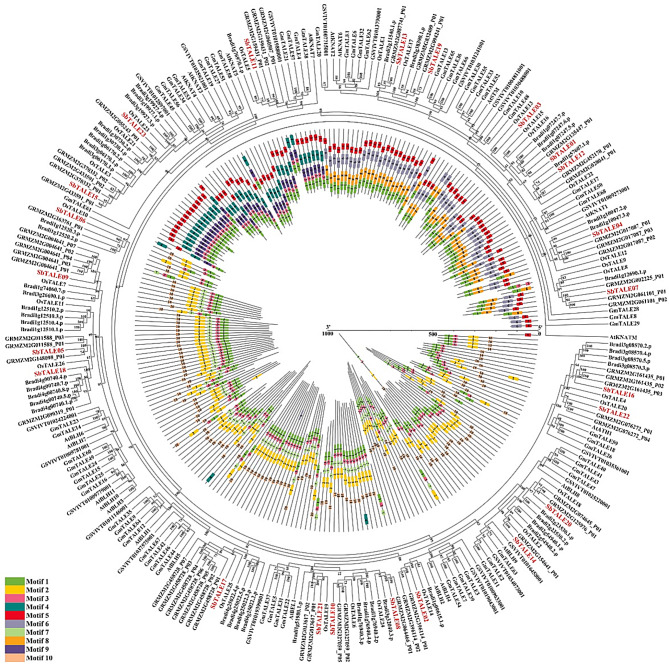
For further assessment of the evolutionary relationship between SbTALE proteins and different plants, an interspecies evolutionary tree was constructed between the 23 sorghum TALE proteins and six plant TALEs proteins with predicted conserved motifs (motif 1 to 10) (Fig. 7, Table S9, and Table S10). Sorghum TALEs clustered closest to Zea mays followed by Brachypodium distachyon, thus indicating that sorghum TALEs are more closely related to monocotyledons (Fig. 7). By predicting their conserved motifs, this study determined that all members contained motif 1 and that all SbTALE proteins possessed motifs 3 − 1 with the exception of KNOX Class I. In the same subfamily, the distribution of conserved motifs is relatively close. For example, the three SbTALEs (SbTALE11, SbTALE15, and SbTALE23) in KNOX Class II all possess the conserved motif 5-4-9-3-1, and the proteins of the SbTALEs in the BEL1-like family all contain the motif 10-2-3-1 (Fig. 7 and Table S10), suggesting that members of proteins in the same subfamily are structurally similar and may also possess similar functions. The conserved motifs of certain SbTALE proteins in the same subfamily also exhibit some differences. For example, the conserved motifs of SbTALE06, SbTALE16, and SbTALE22 contained less motif 7 than did the other four members in OFPs partners, and the conserved motifs of SbTALE01 contained less motif 6 than did the other six proteins in KNOX Class I (Fig. 7 and Table S10).
Fig. 7.
Phylogenetic relationship and motif patterns of TALE proteins among Sorghum bicolor and different plants (Arabidopsis thaliana, Vitis vinifera, Glycine max, Oryza sativa, Brachypodium distachyon, Zea mays). The colored legends represent the conserved motifs (motif 1 to 10), the outer circle represents the phylogenetic tree of TALE proteins of seven plants, and the inner circle represents protein length and conserved motifs. The red fonts represent 23 SbTALEs
Expression patterns of TALEs of Sorghum bicolor in tissue-specificity
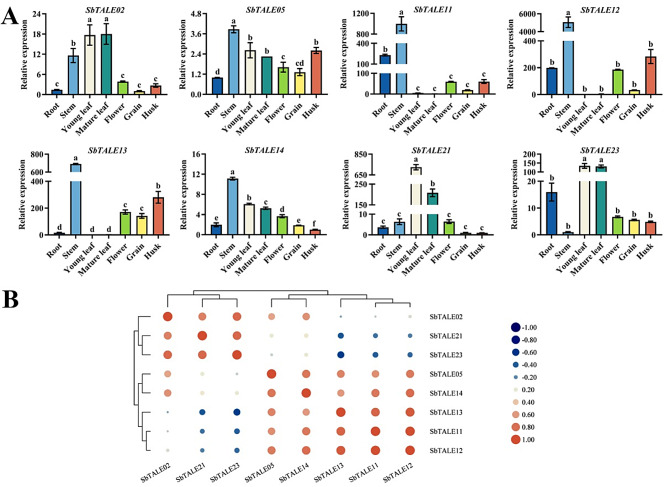
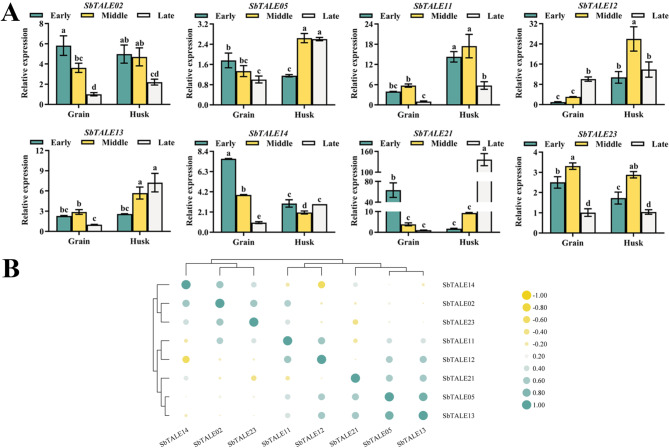
TALE is a class of transcription factors that play an important regulator role in plant growth and development, and to explore the tissue specificity of TALE transcription factors in sorghum, the tissue specificity of different subfamilies of SbTALEs in different tissues was investigated in this paper (Fig. 8A). Overall, with the exceptions of SbTALE21 and SbTALE23, the expression of the other six SbTALEs was higher in the stems, especially that of SbTALE11, SbTALE12, and SbTALE13 that belong to the same KNOX family, whereas the expression of SbTALE21 and SbTALE23 was higher in the leaves (Fig. 8A). SbTALE02, SbTALE11, SbTALE12, SbTALE21, and SbTALE23 did not show significant differences among flowers, grains, and husks (Fig. 8A). This study also revealed that the expression patterns of the three groups of genes, SbTALE05/SbTALE14, SbTALE11/SbTALE12, SbTALE21/SbTALE23, were similar, with the highest correlation coefficients (r = 0.982) observed for SbTALE11 and SbTALE12. However, there was a negative correlation between SbTALE21 and SbTALE23 and between SbTALE11, SbTALE12, and SbTALE13 (r < 0) (Fig. 8A and B).
Fig. 8.
The tissue-specific expression levels and correlation analysis of eight SbTALEs (SbTALE02, SbTALE05, SbTALE11, SbTALE12, SbTALE13, SbTALE14, SbTALE21, and SbTALE23). (A) Expression levels of eight SbTALEs at the mid-grain filling stage in roots, stems, young leaf, mature leaf, flower, grain, and husk. Values of the column chart are expressed as mean ± SD, the lowercase letters represent significant differences (p < 0.05, Duncan). (B) Correlation hierarchical cluster analysis between their expression in different tissues. Positive number represents positive correlation and negative number indicates negative correlation
Expression levels of TALEs of Sorghum bicolor in grain development
In this study, seed-specific regulatory elements were found to be distributed in multiple SbTALE promoters when analyzed by Cis-acting element analysis. Therefore, the relative expression of SbTALEs in grains and husks was further analyzed (Fig. 9A). The lowest relative expression of SbTALEs was observed in grains during the late stages of sorghum grain development. The expression trends of SbTALE02, SbTALE05, SbTALE21 genes and SbTALE11, SbTALE13, SbTALE23 genes in grains were similar. These two groups of genes belonged to the BEL1-like and KNOX families, respectively (Fig. 9A). According to the analysis shown in Fig. 3, seed-specific regulatory elements are distributed in the SbTALE21 promoter sequence, and this study found that the expression of SbTALE21 was higher in grains in the early stage and husks in the late stage (Fig. 9A). In addition, the expression patterns of SbTALE12 and SbTALE14 were the opposite in grains and husks, and the correlation between these two genes was significantly negative (r = − 0.627) (Fig. 9A and B). In addition, we observed that both SbTALE21 and SbTALE23 were negatively correlated (r < 0) with SbTALE12, which was similar to the results shown in Fig. 8 (Fig. 9B).
Fig. 9.
The grain development expression levels and correlation analysis of eight SbTALEs (SbTALE02, SbTALE05, SbTALE11, SbTALE12, SbTALE13, SbTALE14, SbTALE21, and SbTALE23). (A) Expression levels of eight SbTALEs of grain and husk in the early, middle, and late stages of grain filling. Values of the column chart are expressed as mean ± SD, the lowercase letters represent significant differences (p < 0.05, Duncan). (B) Correlation hierarchical cluster analysis between their expression in different tissues. Positive number represents positive correlation and negative number indicates negative correlation
Expression levels of TALEs of Sorghum bicolor in response to abiotic stresses
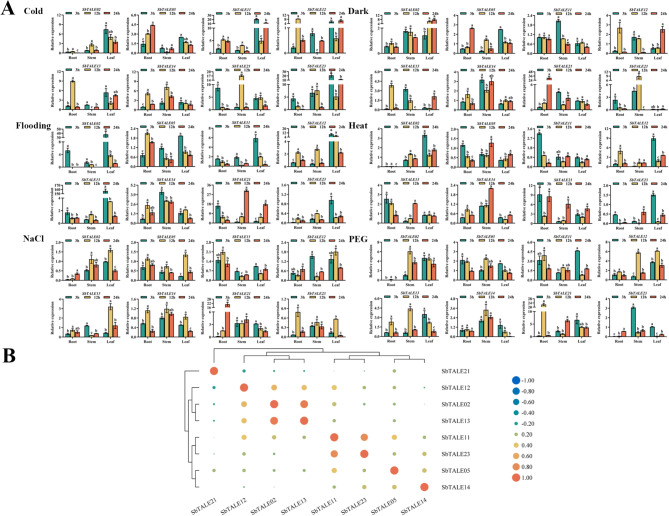
To investigate the regulatory role of TALEs in adversity stress, the expression patterns of SbTALEs under different abiotic stress conditions were explored (Fig. 10A). In general, there were certain differences in the expression patterns of the eight SbTALEs under six abiotic stresses. Among that relative expression of SbTALE02 was low in the roots (Fig. 10A). SbTALE05 and SbTALE14, which belong to the same BEL1-like family, exhibited similar expression trends under the four treatments that included flooding, heat, NaCl, and PEG. For example, the relative expression of the two genes in the roots, stems, and leaves was highest under NaCl treatment for 12 h, and the expression of the two genes in leaves gradually decreased with the extension of PEG treatment time (Fig. 10A). Meanwhile, under cold, flooding, heat, and PEG treatments, SbTALE13 and SbTALE23, both belonging to the KNOX family, showed similar expression patterns in the leaves, with their expression being the highest at 3 h of treatment (Fig. 10A). The analysis of Cis-acting elements (Fig. 3) showed that there were several low-temperature responsiveness and drought inducibility elements distributed in the promoter sequences of SbTALEs. In this study, we found that the expression of SbTALE14 and SbTALE21 was higher in the roots and stems under cold treatment, whereas SbTALE02 and SbTALE12 had higher expression levels in roots and stems under PEG treatment, and both had the highest expression in the 12 h stem (Fig. 10A); at the same time, the correlation between SbTALE02 and SbTALE12 was high (r = 0.520) (Fig. 10B). In addition, this study found a high correlation between SbTALE02 and SbTALE13 and between SbTALE11 and SbTALE23 with correlation coefficients of 0.917 and 0.800, respectively (Fig. 10B).
Fig. 10.
The spatiotemporal expression levels and correlation analysis of eight SbTALEs (SbTALE02, SbTALE05, SbTALE11, SbTALE12, SbTALE13, SbTALE14, SbTALE21, and SbTALE23) under six abiotic stresses (cold, dark, flooding, heat, NaCl and PEG) at the seedling stage. (A) Expression level of eight SbTALEs at 3 h, 12 h, and 24 h in root, stem, and leaf. Values of column chart are expressed as mean ± SD, the lowercase letters represent significant differences (p < 0.05, Duncan). (B) Correlation hierarchical cluster analysis between their expression in different tissues. Positive number represents positive correlation and negative number indicates negative correlation
Expression levels of TALEs of Sorghum bicolor in response to hormones
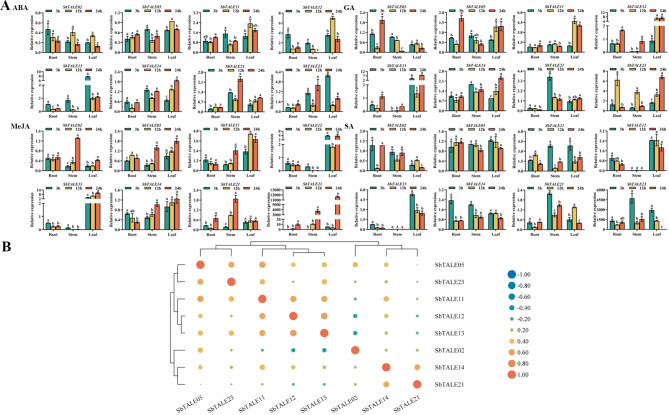
A previous analysis of the Cis-acting elements of the SbTALE promoters revealed that the hormone-responsive elements were unevenly distributed within each promoter (Fig. 3). Therefore, this study was conducted to explore the regulatory roles of SbTALE genes under different hormone treatments (Fig. 11A). Overall, the expression of SbTALEs in stems was relatively low under ABA, GA, MeJA, and SA treatments, the expression of SbTALE13 was higher in leaves than in roots and stems, and the expression of SbTALE23 was higher under MeJA and SA treatments (Fig. 11A). Cis-acting element analysis also revealed that SbTALE23 was distributed with MeJA responsiveness and salicylic acid responsiveness elements (Fig. 3). In this study, we found that the expression trends of SbTALE05 and SbTALE14 were similar under the four hormone treatments, and both belonged to the BEL1-like family. The trends of SbTALE12 and SbTALE13 were similar and both belonged to the KNOX family, and their correlation was high (r = 0.633) (Fig. 11A and B).
Fig. 11.
The spatiotemporal expression levels and correlation analysis of eight SbTALEs (SbTALE02, SbTALE05, SbTALE11, SbTALE12, SbTALE13, SbTALE14, SbTALE21, and SbTALE23) under four hormonal treatments (ABA, GA, MeJA, and SA) at the seedling stage. (A) Expression levels of eight SbTALEs at 3 h, 12 h, and 24 h in root, stem and leaf. Values of the column chart are expressed as mean ± SD, the lowercase letters represent significant differences (p < 0.05, Duncan). (B) Correlation hierarchical cluster analysis between their expression in different tissues. Positive number represents positive correlation and negative number indicates negative correlation
Discussion
Identification and characteristics of TALE genes in Sorghum bicolor
TALE regulates plant growth and development, controls meristematic tissue formation, and maintains organ morphogenesis [25]. TALE family members have been identified in various crops, including 35 TALE family members in poplar [26] and 22 TALE family members in apples [27]. In the present study, the physicochemical properties, conserved structures, evolutionary relationships, and spatiotemporal expression of SbTALEs were systematically analyzed using various bioinformatic methods. Through searching and screening within the whole sorghum genome, 23 SbTALEs (Table S1) were identified in this study, with amino acid content ranging from 294 (SbTALE02) to 770 (SbTALE13) aa and molecular weights (MW) ranging from 32.46 (SbTALE13) to 80.77 (SbTALE02) KD (Table S1). The large differences in amino acid content and molecular weights between the 23 SbTALEs indicate that sorghum has been subjected to changes in protein structure due to changes in its environment over a long period during the evolutionary selection process. The amino acid content of the 23 SbTALEs were positively correlated with their molecular weights [28]. Interestingly, the vast majority of SbTALE proteins (21/23, 91.30%) had pI < 7 (Table 1), indicating that the SbTALEs family tends to be enriched in acidic amino acids. At the same time, by predicting their subcellular location, it was found that the SbTALEs were all located in the nucleus (Table 1), indicating that the TALEs family mainly plays certain regulatory functions and roles in the nucleus, which is consistent with the findings of Zhao et al. and Ezura et al., all of which indicate that TALE genes are relatively conserved during the evolutionary process and their degree of differentiation is small [26, 29].
In order to investigate the structural differences among sorghum TALEs, this study classified SbTALEs into two major groups (KNOX family and BEL1-like family) with reference to the classification of the Arabidopsis TALE family, whereas the KNOX family was divided into three subfamilies (Class I, II, and III) and the BEL1 like family was divided into four subfamilies (OFPs partners, meristem function, ovule morphology, and leaf morphology) (Fig. 1A). Among them, the BEL1-like family contained more family members (13, 56.52%) (Fig. 1A), which is similar to the findings of other TALE families [24, 30], suggesting that the TALE family has been relatively stable during evolution, its sequences are well conserved, and the same branch may have similar biological functions. In this study, we found that KNOX Class III has only AtKNATM and no distribution of SbTALE members (Fig. 1A), and there are only two domains of KNOX Class III, KNOX1, and KNOX2 (Fig. 1B), which suggests that KNATM has evolved in a more complex manner and has a different regulatory role on plant growth and development than other KNOX members [12], and it is possible that KNATM was lost during sorghum evolution.
As far as conserved motifs were concerned, motif 1 of the TALE conserved motif was distributed in all 23 SbTALEs, and the number of conserved motifs in the KNOX family was lower than that in the BEL1-like family. The conserved motifs within the same family were similar. For example, the conserved motif of most SbTALE proteins in the KNOX family was motif 4-6-3-1 (Fig. 2B). The conserved motif of SbTALE16 in the OFPs partner subfamily was motif 7-5-1, which was less than that of SbTALE06 and SbTALE22 (motif 7-2-5-1) (Fig. 2B). This study determined that the KNOX family possessed more introns and exons regions, especially the KNOX II subfamily members with five intron or exon regions (Fig. 2C). This indicates that the number of intron and exon regions is positively correlated, and this is contrary to the results of soybean [23] but consistent with the results of wheat [24], indicating that the evolutionary differentiation process between monocotyledons and dicotyledons caused great differences in gene structure. Different intron distributions among TALE family members may affect the evolution of new family members, and introns are crucial for the evolution and production of genes in gene families [31]. The greater the number of introns, the higher the frequency of recombination between genes, and genes without introns cannot be spliced or separated [32].
Transcriptional regulation is an important means of regulating gene expression; Cis-acting elements play key roles in the regulatory process and can bind to trans-acting factors to regulate the activity of target genes [33]. Cis-acting element analysis revealed a large number of light-responsive elements distributed in all SbTALE promoters, and different hormone-responsive elements were distributed in most SbTALE promoters, suggesting that SbTALE genes may respond to a variety of hormone signals, with abscisic acid responsive elements being the most numerous (51) (Fig. 3A and B). Notably, four hormone response elements and drought inducibility elements were present in both SbTALE02 and SbTALE12 promoters, suggesting that this gene pair may be involved in drought stress via the ABA or MeJA pathways (Fig. 3A).
Gene duplication and evolutionary relationship of TALE genes in Sorghum bicolor
Differences in the number of TALE family members in different species reflect differences in genome size and ploidy levels, as well as genetic recombination and replication during the natural evolution of species [34]. In this study, 23 SbTALEs identified were located on 10 chromosomes of sorghum, of which the highest number of genes was distributed on Chr 1 (47.83%, 11/23) and no SbTALE genes were distributed on Chr 6 or Chr 7 (Fig. 4A). The occurrence of tandem duplications is an important reason for the amplification of several genes [35]. In this study, a pair of tandemly duplicated genes (SbTALE05 and SbTALE06) was found on chromosome 1 and belonged to the same OFPs partners in the BEL1-like family (Fig. 4A and Table S4), suggesting that these genes may have been highly retained in their copy number after duplication during the evolutionary divergence of sorghum, allowing them to co-regulate relevant biological roles in growth and development. Fragment duplication events are another reason for the increase in the number of gene families [36]. Seven pairs of fragment duplication events occurred in the sorghum TALE family, and three and four pairs of SbTALE genes belonged to the KNOX family or BEL1-like family, respectively (Fig. 4B and Table S5), indicating that genes retain a large amount of information about gene structure and function during the duplication process. However, some bias also occurs in the process, which makes their gene structure, function regulation, and environmental response different.
In addition, we analyzed the gene synteny of sorghum TALEs with three dicotyledons and three monocotyledons and found that sorghum TALEs genes were more highly synonymous with monocotyledons, especially Zea mays (42 pairs), and the fewest genes were in synteny with dicotyledons Arabidopsis thaliana (5 pairs) (Fig. 5 and Table S6). Sorghum TALEs clustered closest to Zea mays in the interspecific evolutionary tree (Fig. 7), suggesting that Sorghum bicolor and Zea mays are more closely related and that the monocotyledons and dicotyledons differ in evolutionary direction. In this paper, we also found that four genes, SbTALE14, SbTALE15, SbTALE20, and SbTALE23, shared common syntenic homologous genes with all six plants (Table S6), suggesting that these genes were formed before monocotyledonous and dicotyledonous divergence, and also suggesting that due to the different evolutionary pressures of microenvironments in which the duplicated genes are found on the chromosomes, these genes diverged, lost function, or were chromosomally rearranged to form new genes or were lost, and the lost TALE genes may have been replaced by functionally similar genes [37]. Ka/Ks is the basis for analyzing selection pressure in gene duplication events. In this study, Ka/Ks was calculated for two families and syntenic TALE gene pairs of interspecies, and it was found that the Ka/Ks of the KNOX family was < 1, whereas the Ka/Ks of the BEL1-like family was > 1 (Fig. 6A, Table S7), implying that the two families differed in evolutionary selection, with the former undergoing purifying selection (negative selection) and being relatively conserved in evolution, whereas most of the genes in the latter underwent diversifying selection (positive selection). The Ka/Ks ratios of the homologous gene pairs between Sorghum bicolor and Vitis vinifera, Glycine max, Zea mays were close to 1, suggesting that they underwent neutral selection during evolution. The smallest Ka/Ks ratio was found between Sorghum bicolor and Arabidopsis thaliana (0.357) (Fig. 6B and Table S8), indicating that they were subjected to rapid evolution and strong natural selection.
Spatiotemporal expression levels of TALE genes in Sorghum bicolor
It has been shown that TALE transcription factors are expressed in different plant tissues, such as inflorescences and stems, and the KNOX2 gene was found to promote secondary cell wall biosynthesis in xylem conduits [38]. The KNOX1 gene has been associated with the maintenance of tissue proliferation and meristematic potential in flowering plants and moss sporophytes, and the modulation of KNOX1 activity is associated with leaf shape diversity in flowering plants [39]. Abiotic stressors such as high temperatures, drought, and salinity can reduce crop productivity and cause significant losses in crop yield. It has been found that cotton GhKNOX4-A and GhKNOX22-D may promote drought response by regulating stomatal opening and oxidative stress [40]. PagKNAT2/6b directly inhibits gibberellin (GA) synthesis to alter plant architecture and enhance drought tolerance in poplar plants under short- and long-term drought stress [41]. These studies indicate that TALE plays an essential role in the regulation of crop growth and development, environmental responses, and signal transduction.
In this study, TALE members from different subfamilies were analyzed for their different expression patterns. We found that the expression levels of the eight SbTALE genes varied significantly among different tissues and between grains and husks at different grain-filling stages (Figs. 8A and 9A). Expression of SbTALE21 and SbTALE23 was higher in the leaves, whereas the other six SbTALEs were expressed in the stems. The expression patterns of the three groups (SbTALE05/SbTALE14, SbTALE11/SbTALE12, SbTALE21/SbTALE23) of genes were similar and showed high correlation coefficients (Fig. 8A and B), indicating that the SbTALE family is tissue-specific. The lowest relative expression of SbTALEs was in grain in the late stage of development, suggesting that SbTALE genes are positively regulated in sorghum before and during the middle stage of grain-filling. In the present study, we also found that SbTALE21 with seed-specific regulatory elements distributed in the promoter, had higher expression in grains in the early stage and husks in the late stage (Fig. 9A). In this study, we found that SbTALE14 and SbTALE21, which have low-temperature responsiveness elements in their promoter sequences, showed higher expression in roots and stems under cold treatment, and SbTALE02 and SbTALE12, which contain drought inducibility elements in their promoter sequences, had higher expression in roots and stems under PEG treatment (Figs. 3 and 10A), indicating that the above genes regulate the environmental response more clearly in roots and stems and have stronger stress tolerance. In the hormonal response, the expression of eight SbTALEs in the stems was relatively low under ABA, GA, MeJA, and SA treatments (Fig. 11A). We found that some SbTALEs showed similar expression trends under different hormone treatments, such as SbTALE05 and SbTALE14, which belong to the same BEL1-like family, under all four hormone treatments, and SbTALE12 and SbTALE13, which belong to the same KNOX family (Fig. 11A). MeJA- and SA-responsive elements were predicted to be distributed in SbTALE23, and their expression was higher under both MeJA and SA treatments, especially at 24 h of MeJA and 3 h of SA treatment (Figs. 3 and 11A). The above results indicate that SbTALE genes have significantly different expression patterns in specific tissues and organs under different treatments and suggest that SbTALEs play important roles in maintaining the establishment and development of specific tissues and organs in plants.
Conclusions
In this study, 23 SbTALE genes were identified for the first time in the whole sorghum genome, which were classified into two families, KNOX and BEL1-like, and were located on 10 Sorghum bicolor chromosomes; one pair of tandem duplications and seven pairs of segment duplications were also found. The conserved motifs and gene structures of SbTALEs were highly conserved among the same subfamilies. SbTALE genes have the most collinear genes with monocotyledonous plant Zea mays, which is more closely related, and SbTALEs has undergone purification and diversification selection in the evolutionary process. In addition, SbTALEs has tissue-specific transcriptional regulatory and hormone-induced roles in sorghum growth and development. This study lays a theoretical foundation for the study of the biological functions and mechanisms of SbTALE genes, and is of great significance for the mining of resistance genes and trait improvement.
Methods
Identification of TALE genes of Sorghum bicolor
In this paper, we downloaded the sorghum whole genome annotation file via the Phytozome website (https://phytozome-next.jgi.doe.gov/), as well as the TALE amino acid sequences of Arabidopsis (https://www.Arabidopsis.org/) and rice (http://Rice.plantbiology.msu.edu/), and obtained the Hidden Markov (HMM) information of the TALE structural domain (PF00046) from the Pfam database [42]. The TALE amino acid sequences were compared with those of Arabidopsis and rice using BLASTp (score value ≥ 100, e-value ≤ 1e-10) in the sorghum genome to screen all possible SbTALE proteins from Arabidopsis TALE amino acid sequences [43]. Further, SMART (http://smart.embl-heidelberg.de/) and Conserved Domains (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) were used for the SbTALE structural domain search screening to identify all members of the SbTALE family of transcription factors [44, 45]. Finally, the physicochemical properties of the identified SbTALE proteins (https://www.expasy.org/), including molecular weight (MW), theoretical isoelectric point (pI), instability index (II), and predicted SbTALE protein subcellular location information (https://wolfpsort.hgc.jp/) were determined.
Analysis of phylogenetic evolution, estimation of nonsynonymous and synonymous substitutions (Ka/Ks)
TBtools-ll (Toolbox for Biologists) v2.057 [46] was used to compare the Muscle Wrapper model among the TALE (Table S7) and SbTALA protein sequences, and the IQ-Tree Wrapper program (bootstrap number set to 1000, other default parameters) was used to construct the evolutionary tree [4, 23, 47–49]. The identified SbTALE proteins were grouped, classified, and analyzed according to the classification of the model plant Arabidopsis in the TALE family. In addition, Ka/Ks ratios were calculated using the Ka/Ks calculator, which was used to assess selection pressure on homologous genes.
Gene structure, conserved motifs, and Cis-acting elements
Multiple Em for Motif Elicitation (MEME) (https://meme-suite.org/meme/tools/meme) was used to predict the conserved motifs of SbTALEs (maximum conserved motif search value was set to 10, other default parameters) [50], and a composite map of the gene structure and conserved motifs of SbTALEs was constructed using TBtools-ll v2.057 [46]. PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict Cis-acting elements of the SbTALE promoter sequence (upstream 2000 bp) [51].
Chromosomal location, duplication, collinearity and synteny analysis
Based on the sorghum genomic location information, SbTALEs were located on 10 chromosomes of sorghum using Gene Location Visualize, while tandem repeats and segmental repeats of SbTALEs were analyzed by One Step Multiple Collinearity Scan toolkit X (MCScanX, parameters default) [52], and Dual Synteny Plot was used to analyze sorghum and six representative species (Arabidopsis thaliana, Vitis vinifera, Glycine max, Oryza sativa, Brachypodium distachyon, Zea mays) for the synteny between the genes of TALEs [46].
Plant materials, growth, and treatments in Sorghum bicolor
In this experiment, we used the Sorghum bicolor cv. Hongyingzi, a typical cultivated variety with good resistance in Guizhou province, China, which was planted in an artificial climate incubator (60% humidity, 16 h light/25℃ and 8 h dark/20℃). When the sorghum seedlings developed three leaves and one heart, the seedlings with the same growth condition were subjected to six abiotic stresses (Cold: 4℃, Dark: complete shading, Flooding: whole plant immerse, Heat: 40℃, NaCl: 150 mmol·L− 1, PEG: 30%) and four hormone treatments (ABA: 100 µmol·L− 1, GA: 100 µmol·L− 1, MeJA: 100 µmol·L− 1, SA: 100 µmol·L− 1), with three replicates for each treatment, and three tissues were taken from the roots, stems, and leaves at 0 h, 3 h, 12 h, and 24 h. The samples were stored in an ultra-low temperature refrigerator at − 80℃ [28, 53]. In addition, seven tissues of sorghum were taken from the root, stem, young leaf, mature leaf, flower, fruit, and husk during the grain-filling stage, whereas the grain and husk were taken at the early, middle, and late grain-filling stage [54].
Total RNA extraction, cDNA synthesis, and qRT-PCR analysis
Total RNA was extracted from sorghum samples (0.1 g) using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek, Inc., USA), and RNA concentration and purity were detected using an ultra-micro spectrophotometer (Beijing Kaiao Technology Development Co., Ltd., China). The first cDNA strand was synthesized according to the HiScript II Q RT SuperMix (R223, Vazyme Biotech Co., Ltd, China) for qPCR Kit instructions, and the reaction system was 20 µL. Primers specific for the qPCR of SbTALEs were designed using Primer Premier 5.0 (Premier, Canada), and SbUBQ10 was used as the internal reference gene (Table S9) [55]. Referring to the Taq Pro Universal SYBR qPCR Master Mix instructions (Q712, Vazyme Biotech Co., Ltd, China), amplification was performed using a qTOWER3/qTOWER3G Real-Time PCR Thermal Cycler instrument (Jena Analytical Instruments (Beijing) Co., Ltd, China), and the relative gene expression was calculated using the 2−ΔΔCt formula [56], and three biological and three technical replicates were set up for this experiment.
Statistical analysis
IBM SPSS Statistics 26.0 (International Business Machines Co., Ltd., USA) was used for analysis of variance (p < 0.05) and multiple comparisons (Duncan). Pearson’s correlation analysis (Pearson) was performed using OriginPro2019b software (OriginLab, USA). GraphPad Prism10.0 (GraphPad software Co., Ltd., USA) was used to draw the bar charts and box plots, and TBtools-ll v2.057 was used to draw the correlation heat maps.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Additional file 1: Table S1. SbTALE protein sequences. Table S2. Distribution and analysis of conserved motifs in SbTALE proteins. Table S3.Cis-acting elements in the promoter region of SbTALEs. Table S4. One pair of tandem duplicates in SbTALE genes. Table S5. Seven pairs of segmental duplicates in SbTALE. Table S6. One-to-one orthologous relationships between Sorghum bicolor and the six plants. Table S7. Ka/Ks ratio distributions in different subfamilies. Table S8. Distribution of Ka/Ks ratios of homologous gene pairs. Table S9. Phylogenetic analysis of TALE protein sequences from six representative plants. Table S10. Analysis and distribution of conserved motifs in TALE proteins of seven species. Table S11. Primer sequences used for qPCR
Acknowledgements
We acknowledge the College of Agronomy, Guizhou University, Guiyang, China, for providing the experimental facilities and other materials necessary for this study. We thank our colleagues for useful discussions and technical assistance.
Abbreviations
- TALE
Three-amino-acid-loop-extension
- HB
Homeobox
- HD
Homeodomain
- aa
Amino acid
- MW
Molecular weight
- pI
Isoelectric point
- II
Instability index
- UTR
Untranslated region
- CDS
Coding sequence or exons
- LG
Linked region
- Ka
Nonsynonymous site
- Ks
Synonymous site
- ABA
Abscisic acid
- GA
Gibberellin
- MeJA
Methyl jasmonate
- SA
Salicylic acid
- MEME
Multiple Em for Motif Elicitation
- MCScanX
One Step Multiple Collinearity Scan toolkit X
Author contributions
XY, SWY, DLL, YF, and JJR conceived and designed the study. XY, WFW, and WJW performed the experiments. XY, DLL, YF, and CM performed data analysis and wrote the manuscript. JPC, MLZ, and JJR edited and drafted the manuscript. All the authors contributed to the manuscript and approved the submitted version. All authors reviewed the manuscript.
Funding
This work was supported by Construction of Scientific and Technological Innovation Talent Team for High-Quality and High-Efficiency Mechanisation of Speciality Mixed Grains in Guizhou Province, Grant/Award Number: [Qiankehe Platform Talent-BQW (2024) 009], Research and Integrated Application of Key Technologies for Green and High Yield in Mountainous Characteristic Agriculture, Grant/Award Number: [Guida Lingjun Hezi (2023) 07], and the Natural Science Foundation of China, Grant/Award Number: (32160669, 32161143005, 32312051).
Data availability
The Sorghum bicolor genome sequence information was obtained from the Phytozome website (https://phytozome-next.jgi.doe.gov/). The Sorghum bicolor material (Hongyingzi) used in the experiment was supplied by JC and JR at Guizhou University. All the data is provided within the manuscript or supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xin Yao and Sanwei Yang contributed equally to this work.
Contributor Information
Meiliang Zhou, Email: zhoumeiliang@caas.cn.
Jingjun Ruan, Email: jjruan@gzu.edu.cn.
References
- 1.Gehring WJ, Müller M, Affolter M, Percival-Smith A, Billeter M, Qian YQ, Otting G, Wüthrich K. The structure of the homeodomain and its functional implications. Trends Genet. 1990;6(10):323–9. [DOI] [PubMed] [Google Scholar]
- 2.Billeter M, Qian YQ, Otting G, Müller M, Gehring W, Wüthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993;234(4):1084–93. [DOI] [PubMed] [Google Scholar]
- 3.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12(9):419–26. [DOI] [PubMed] [Google Scholar]
- 4.Hamant O, Pautot V. Plant development: a TALE story. C R Biol. 2010;333(4):371–81. [DOI] [PubMed] [Google Scholar]
- 5.Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25(21):4173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerstetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell. 1994;6(12):1877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reiser L, Sánchez-Baracaldo P, Hake S. Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol. 2000;42(1):151–66. [PubMed] [Google Scholar]
- 8.Wang J, Wang HY, Zhao PM, Han LB, Jiao GL, Zheng YY, Huang SJ, Xia GX. Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol. 2010;51(8):1276–90. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Sun Y, Han L, Su L, Xia G, Wang H. Overexpression of GhPFN2 enhances protection against Verticillium Dahliae invasion in cotton. Sci China Life Sci. 2017;60(8):861–7. [DOI] [PubMed] [Google Scholar]
- 10.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96(4):587–97. [DOI] [PubMed] [Google Scholar]
- 11.Truernit E, Siemering KR, Hodge S, Grbic V, Haseloff J. A map of KNAT gene expression in the Arabidopsis root. Plant Mol Biol. 2006;60(1):1–20. [DOI] [PubMed] [Google Scholar]
- 12.Magnani E, Hake S. KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell. 2008;20(4):875–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell. 2007;19(9):2719–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Zhang X, Ju H, Chen J, Wang S, Wang H, Zhao Y, Chang Y. Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem Biophys Res Commun. 2016;470(3):492–7. [DOI] [PubMed] [Google Scholar]
- 15.Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, Kater MM, Colombo L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell. 2007;19(8):2544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng L, Fan Z, Zhang Q, Wang C, Gao Y, Deng Y, Zhu B, Zhu H, Chen J, Shan W, et al. BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J. 2018;94(6):1126–40. [DOI] [PubMed] [Google Scholar]
- 17.Ma Q, Wang N, Ma L, Lu J, Wang H, Wang C, Yu S, Wei H. The cotton BEL1-Like transcription factor GhBLH7-D06 negatively regulates the Defense response against Verticillium Dahliae. Int J Mol Sci. 2020;21(19):7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Althwab S, Carr TP, Weller CL, Dweikat IM, Schlegel V. Advances in grain sorghum and its co-products as a human health promoting dietary system. Food Res Int. 2015;77:349–59. [Google Scholar]
- 19.Shrestha K, Pant S, Huang Y. Genome-wide identification and classification of lipoxygenase gene family and their roles in sorghum-aphid interaction. Plant Mol Biol. 2021;105(4–5):527–41. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Wang W, Zhao Y. Phenolic compounds in Whole Grain Sorghum and their health benefits. Foods. 20219;10(8):1921. [DOI] [PMC free article] [PubMed]
- 21.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–6. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Johnson SK, Bornman JF, Bennett SJ, Fang Z. Changes in whole grain polyphenols and antioxidant activity of six sorghum genotypes under different irrigation treatments. Food Chem. 2017;214:199–207. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Yang X, Gao Y, Yang S. Genome-wide identification and characterization of TALE superfamily genes in soybean (Glycine max L). Int J Mol Sci. 2021;22(8):4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Y, Zhang L, Yan L, Xiong X, Wang W, Zhang XH, Min DH. Genome-wide analysis of TALE superfamily in Triticum aestivum reveals TaKNOX11-A is involved in abiotic stress response. BMC Genomics. 2022;23(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joo S, Wang MH, Lui G, Lee J, Barnas A, Kim E, Sudek S, Worden AZ, Lee JH. Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida. BMC Biol. 2018;16(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao K, Zhang X, Cheng Z, Yao W, Li R, Jiang T, Zhou B. Comprehensive analysis of the three-amino-acid-loop-extension gene family and its tissue-differential expression in response to salt stress in poplar. Plant Physiol Biochem. 2019;136:1–12. [DOI] [PubMed] [Google Scholar]
- 27.Jia P, Zhang C, Xing L, Li Y, Shah K, Zuo X, Zhang D, An N, Han M, Ren X. Genome-wide identification of the MdKNOX Gene Family and characterization of its transcriptional regulation in Malus domestica. Front Plant Sci. 2020;11:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao X, Zhou M, Ruan J, He A, Ma C, Wu W, Lai D, Fan Y, Gao A, Weng W, Cheng J. Genome-wide identification, evolution, and expression pattern analysis of the GATA Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int J Mol Sci. 2022;23(20):12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezura K, Nakamura A, Mitsuda N. Genome-wide characterization of the TALE homeodomain family and the KNOX-BLH interaction network in tomato. Plant Mol Biol. 2022;109(6):799–821. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee K, Bürglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65(2):137–53. [DOI] [PubMed] [Google Scholar]
- 31.Roy SW, Penny D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol Biol Evol. 2007;24(7):1447–57. [DOI] [PubMed] [Google Scholar]
- 32.Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol. 2010;27(8):1745–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Patra B, Pattanaik S, Wang Y, Yuan L. GATA and phytochrome interacting factor transcription factors regulate light-Induced Vindoline Biosynthesis in Catharanthus roseus. Plant Physiol. 2019;180(3):1336–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–7. [DOI] [PubMed] [Google Scholar]
- 35.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. Plant genetics. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–3. [DOI] [PubMed] [Google Scholar]
- 36.Storz JF. Genome evolution: gene duplication and the resolution of adaptive conflict. Heredity (Edinb). 2009;102(2):99–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 38.Furumizu C, Alvarez JP, Sakakibara K, Bowman JL. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015;11(2):e1004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Yamaguchi M, Grienenberger E, Martone PT, Samuels AL, Mansfield SD. The class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 2020;101(2):293–309. [DOI] [PubMed] [Google Scholar]
- 40.Sun R, Qin T, Wall SB, Wang Y, Guo X, Sun J, Liu Y, Wang Q, Zhang B. Genome-wide identification of KNOX transcription factors in cotton and the role of GhKNOX4-A and GhKNOX22-D in response to salt and drought stress. Int J Biol Macromol. 2023;226:1248–60. [DOI] [PubMed] [Google Scholar]
- 41.Song X, Zhao Y, Wang J, Lu MZ. The transcription factor KNAT2/6b mediates changes in plant architecture in response to drought via down-regulating GA20ox1 in Populus alba × P. Glandulosa. J Exp Bot. 2021;72(15):5625–37. [DOI] [PubMed] [Google Scholar]
- 42.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue): W29–37. [DOI] [PMC free article] [PubMed]
- 43.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M, Derbyshire MK, Yamashita RA, Marchler-Bauer A. NCBI’s conserved domain database and Tools for Protein Domain Analysis. Curr Protoc Bioinform. 2020;69(1):e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, Xia R. TBtools-II: a one for all, all for one bioinformatics platform for biological big-data mining. Mol Plant. 2023;16(11):1733–42. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Zhao Y, Yan M, Zhao H, Zhang X, Yuan Z. Genome-wide identification and expression analysis of TALE Gene Family in Pomegranate (Punica granatum L). Agronomy. 2020;10(6):829. [Google Scholar]
- 48.Tian F, Yang DC, Meng YQ, Jin J, Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48(D1):D1104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng W, Yang Y, Xu J, Peng E, Dai S, Dai L, Wang Y, Yi T, Wang B, Li D, Song N. TALE transcription factors in Sweet Orange (Citrus sinensis): genome-wide identification, characterization, and expression in response to biotic and abiotic stresses. Front Plant Sci. 2022;12:814252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue): W202–8. [DOI] [PMC free article] [PubMed]
- 51.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao X, Lai D, Zhou M, Ruan J, Ma C, Wu W, Weng W, Fan Y, Cheng J. Genome-wide identification, evolution and expression pattern analysis of the GATA gene family in Sorghum bicolor. Front Plant Sci. 2023;14:1163357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai D, Yao X, Yan J, Gao A, Yang H, Xiang D, Ruan J, Fan Y, Cheng J. Genome–wide identification, phylogenetic and expression pattern analysis of GATA family genes in foxtail millet (Setaria italica). BMC Genomics. 2022;23(1):549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodríguez MV, Mendiondo GM, Maskin L, Gudesblat GE, Iusem ND, Benech-Arnold RL. Expression of ABA signalling genes and ABI5 protein levels in imbibed Sorghum bicolor caryopses with contrasting dormancy and at different developmental stages. Ann Botany. 2009;104(5):975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Additional file 1: Table S1. SbTALE protein sequences. Table S2. Distribution and analysis of conserved motifs in SbTALE proteins. Table S3.Cis-acting elements in the promoter region of SbTALEs. Table S4. One pair of tandem duplicates in SbTALE genes. Table S5. Seven pairs of segmental duplicates in SbTALE. Table S6. One-to-one orthologous relationships between Sorghum bicolor and the six plants. Table S7. Ka/Ks ratio distributions in different subfamilies. Table S8. Distribution of Ka/Ks ratios of homologous gene pairs. Table S9. Phylogenetic analysis of TALE protein sequences from six representative plants. Table S10. Analysis and distribution of conserved motifs in TALE proteins of seven species. Table S11. Primer sequences used for qPCR
Data Availability Statement
The Sorghum bicolor genome sequence information was obtained from the Phytozome website (https://phytozome-next.jgi.doe.gov/). The Sorghum bicolor material (Hongyingzi) used in the experiment was supplied by JC and JR at Guizhou University. All the data is provided within the manuscript or supplementary information files.