Abstract
Background
One of the wild fruit species with a natural distribution in Türkiye, and historically used for medicinal purposes due to its rich composition, is Berberis crataegina DC. Various parts of the plant, including its roots, bark, leaves, flowers, and fruits, have been utilized in traditional medicine, while its fruits are also consumed in various forms as food. This study aimed to characterize the morphological, biochemical, and molecular traits of B. crataegina genotypes naturally growing in the Kayseri region, located in central Türkiye.
Results
The fruit weight of the genotypes ranged from 0.047 to 0.137 g, fruit width from 3.06 to 4.64 mm, and fruit length from 5.80 to 9.05 mm. Similarly, the leaf traits of the genotypes exhibited wide variation. The total phenolic content ranged from 190.53 to 297.55 mg GAE/100 g, total flavonoid content from 82.03 to 203.89 mg QE/100 g, total anthocyanin content from 4.54 to 11.76 mg cyn-3 gluc/100 g, and total antioxidant capacity between 57.76 and 87.93%. A principal component analysis (PCA) of 11 traits identified four principal components with eigenvalues greater than 1. The first four components accounted for 71.89% of the total variation, with PCA1 explaining 23.48%, PCA2 18.68%, PCA3 16.39%, and PCA4 13.34%. ISSR molecular analysis using nine markers revealed a band count ranging from 4 to 13, polymorphic band count between 3 and 10, and polymorphism rates from 61.54 to 100%, with band lengths ranging from 200 to 1000 base pairs. According to the UPGMA dendrogram based on molecular analyses, the genetic similarity between the genotypes ranged from 0.64 to 1.00, with B2 and B13 being the most similar genotypes.
Conclusions
In conclusion, the Kayseri region is rich in B. crataegina genotypes with wide genetic variation. The genotypes identified in this region may serve as valuable genetic resources for future studies.
Keywords: Antioxidant, Berberis crataegina, Biplot, Genetic diversity, Heatmap, Molecular marker
Introduction
The importance of natural and nutrient-rich foods in healthy diets has increased interest in wild fruit species. Wild fruits are widely used as food supplements that strengthen nutrition and the immune system due to their antioxidant, antimicrobial, and anti-inflammatory properties, as well as their rich composition of beneficial components [15]. Compared to cultivated fruits, wild fruits collected from nature are richer in nutrients and bioactive compounds [26, 45].
Berberis crataegina DC., belonging to the Berberidaceae family, is one of the important wild fruit species in Türkiye, used both as food and extensively for medicinal purposes. The Berberidaceae family, generally distributed in temperate regions of the northern hemisphere, comprises approximately 500 species, with about 300 found in Eurasia and 200 in South America [37]. Four species of Berberis (B. vulgaris L., B. integerrima Bunge, B. cretica L., and B. crataegina) grow naturally in Türkiye [10]. The species B. crataegina grows naturally in Türkiye, Iran, and Turkmenistan. In Türkiye, this species is distributed across various regions, including İzmir, Kırklareli, Ankara, Kastamonu, Çankırı, Kayseri, Konya, Niğde, and Sivas [40].
B. crataegina, which is important in Türkiye, is drought- and cold-tolerant, thriving in mountainous areas where cold winters prevail [3]. This shrub, reaching about 2 m in height, typically grows in rocky areas at elevations between 800 and 1500 m. Its leaves are yellow, small, and oval-shaped, with flowers that bloom in late summer. The fruit, which initially appears red, turns dark purple to black as it ripens in the fall [24]. The fruits of B. crataegina are known by different names depending on the region in Türkiye, including Karamuk, Karamuk Diken, Diken Üzümü, Şam Püremi, and Kadın Tuzluğu [14].
B. crataegina is especially rich in phenolic compounds, particularly anthocyanins and polyphenols [16]. Due to its phenolic composition, it exhibits antifungal, anti-inflammatory, antipruritic, and diuretic effects [41, 44]. The roots, leaves, and fruits of B. crataegina plants are traditionally used in the treatment of rheumatism, as pain relievers, and in treating gynecological, circulatory, and diabetic disorders. Its fruits are used for treating hypertension, stomach and intestinal disorders, and colds, while the roots are used in treating jaundice, bronchitis, and colds, and the leaves for treating wounds and cuts in cases of intestinal disorders [15].
The identification of Berberis species is challenging due to their high genetic and geographical diversity, inter- and intra-species crossbreeding, and mutations [32]. Introgression in Berberis species often results in the formation of intermediate forms, making identification difficult [5]. Therefore, characterizing the germplasm of B. crataegina is crucial for breeding studies. Morphological traits are the simplest markers used to evaluate intra- and inter-population diversity [42]. The use of morphological traits in breeding studies is important for identifying germplasm. However, since morphology-based traits are influenced by environmental factors and may vary depending on plant development stages, they are not entirely reliable for researchers. Therefore, for accurate germplasm identification, it is necessary to use morphological, biochemical, and molecular methods that are not affected by environmental factors [38, 43, 47].
The first step in preserving plant diversity is to reveal genetic diversity by creating germplasm collections. Recent advancements in biotechnology have highlighted the importance of molecular markers in determining the genetic diversity of plant germplasm. AFLP [9], ISSR [31, 34], and SSR [25, 32] markers have been successfully used in the molecular characterization of different Berberis species. ISSR markers, which are simple, fast, low-cost, do not require prior sequence information, and offer high stability, have been successfully used in the identification of many species [8].
Previous studies have mainly focused on the medicinal properties and biochemical content of B. crataegina. However, there are limited studies on its genetic diversity. This study aimed to reveal the genetic diversity of B. crataegina germplasm using morphological, biochemical, and molecular approaches. Within the scope of the research, genetic diversity based on fruit and leaf morphology, biochemical content, and ISSR markers was examined. Identifying germplasm resources in genetic diversity studies provides important contributions for planning future breeding programs.
Materials and methods
Plant material
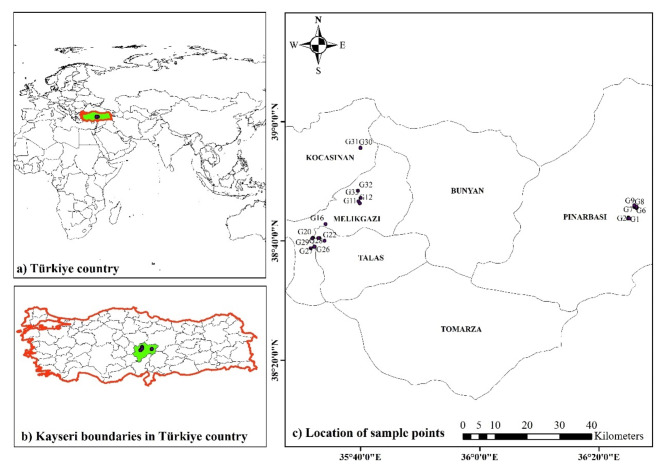
The study was conducted on 33 genotypes of B. crataegina naturally growing in Kayseri province, located in central Türkiye (Table 1; Fig. 1). The selection of genotypes was based on plant habitus and differences in Fruit length, color, shape, and yield. Fruits and leaves of the genotypes were randomly collected from different parts of the plants during the harvest maturity stage. A total of 100 fruit and 30 leaf samples were collected from each genotype and necessary fruit and leaf measurements were carried out. The samples were brought to the laboratory for analysis. For correct sampling, a proper distance of at least 200 m between the genotypes was regarded so that the clone samples are not collected. The formal identification of the specimens was performed by Assoc. Prof. Dr. Mehmet Yaman. A herbarium voucher specimen with sediment number BI-3801 has been donated to the public available herbarium of the Faculty of Agriculture of Erciyes University, Türkiye.
Table 1.
Geographical location and district distribution of Berberis crataegina genotypes growing in Kayseri region
| Genotype | Location | County | Genotype | Location | County |
|---|---|---|---|---|---|
| G1 | 38°43’55"N 36°24’56"E | Pınarbaşı | G18 | 38°40’30"N 35°33’02"E | Talas |
| G2 | 38°43’50"N 36°24’54"E | Pınarbaşı | G19 | 38°40’28"N 35°32’57"E | Talas |
| G3 | 38°45’30"N 36°26’17"E | Pınarbaşı | G20 | 38°40’31"N 35°32’02"E | Talas |
| G4 | 38°45’42"N 36°26’12"E | Pınarbaşı | G21 | 38°40’26"N 35°31’56"E | Talas |
| G5 | 38°45’31"N 36°26’18"E | Pınarbaşı | G22 | 38°40’01"N 35°33’58"E | Talas |
| G6 | 38°45’41"N 36°25’58"E | Pınarbaşı | G23 | 38°39’03"N 35°32’19"E | Talas |
| G7 | 38°45’47"N 36°25’58"E | Pınarbaşı | G24 | 38°39’02"N 35°32’19"E | Talas |
| G8 | 38°45’35"N 36°26’20"E | Pınarbaşı | G25 | 38°39’01"N 35°32’15"E | Talas |
| G9 | 38°45’45"N 36°26’15"E | Pınarbaşı | G26 | 38°39’00"N 35°32’19"E | Talas |
| G10 | 38°43’47"N 36°24’51"E | Pınarbaşı | G27 | 38°39’02"N 35°32’19"E | Talas |
| G11 | 38°46’39"N 35°39’40"E | Melikgazi | G28 | 38°39’02"N 35°32’18"E | Talas |
| G12 | 38°46’19"N 35°39’53"E | Melikgazi | G29 | 38°38’46"N 35°31’40"E | Talas |
| G13 | 38°43’47"N 36°25’04"E | Pınarbaşı | G30 | 38°55’35"N 35°39’59"E | Kocasinan |
| G14 | 38°45’54"N 36°25’55"E | Pınarbaşı | G31 | 38°55’39"N 35°40’00"E | Kocasinan |
| G15 | 38°45’58"N 36°25’55"E | Pınarbaşı | G32 | 38°48’25"N 35°39’34"E | Melikgazi |
| G16 | 38°42’50"N 35°34’09"E | Talas | G33 | 38°47’12"N 35°40’02"E | Melikgazi |
| G17 | 38°40’30"N 35°33’06"E | Talas |
Fig. 1.
Geographical location map of Kayseri region
Fruit and leaf analyses
For the analysis of fruit and leaf characteristics, 30 intact fruits and 30 leaves were randomly selected. The dimensions of fruit width, fruit length, leaf width, leaf length, petiole thickness, and petiole length were measured using a digital caliper with a sensitivity of 0.01 mm. The weight of the fruit was measured using a precision balance with a sensitivity of 0.01 g.
Biochemical analyses
For biochemical analyses, the fruits collected from the genotypes were first washed with distilled water and dried. The samples were homogenized using a blender, and the homogenized fruit samples were transferred into 50 mL Falcon tubes and stored at -20 °C until analysis.
Total phenolic content
The total phenolic content was determined using the Folin-Ciocalteu reagent according to the method proposed by Singleton and Rossi [36]. To 500 µL of the fruit sample, 4.1 mL of distilled water, 100 µL of Folin-Ciocalteu reagent, and 2% Na2CO3 were added, and the mixture was incubated in the dark for 2 h. The absorbance values of the samples were measured at 760 nm using a UV-Vis spectrophotometer, and the results were expressed as mg GAE/100 g fresh weight in terms of gallic acid equivalent.
Total flavonoid content
The total flavonoid content was determined according to the method proposed by Chang et al. [6]. A mixture of 1000 µL fruit sample, 3.3 mL methanol, 0.1 mL 10% AlCl3.6H2O, and CH3COO was prepared. The absorbance values of the samples were measured at 415 nm using a UV-Vis spectrophotometer, and the results were expressed as mg QE/100 g fresh weight in terms of quercetin equivalent.
Total anthocyanin content
The total anthocyanin content was determined according to the pH differential method (pH 1.0 and pH 4.5) proposed by Giusti and Wrolstad [19]. The absorbance values of the samples were measured at 533 nm and 700 nm using a UV-Vis spectrophotometer, and the results were expressed as mg cyn-3-gluc/100 g fresh weight based on a molar extinction coefficient of 26,900.
Total antioxidant content
The total antioxidant content was determined using the DPPH (1.1-diphenyl-2-picryl-hydrazyl) method proposed by Brand-Willams et al. [4]. A mixture of 100 µL fruit sample, 2900 µL ethanol, and 1 mL of 0.26 mM DPPH solution was prepared. After 30 min of incubation in the dark, the absorbance values were measured at 527 nm using a UV-Vis spectrophotometer, and the total antioxidant content was calculated as percent (%) of inhibition.
Molecular marker analysis
DNA used for molecular analysis was isolated from approximately 100 mg of leaf samples using the CTAB (Cetyl Trimethyl Ammonium Bromide) method developed by Doyle and Doyle [12]. The concentration of the extracted DNA was determined using a NanoDrop ND100 spectrophotometer (NanoDrop Technologies Inc. Wilmington, DE, USA). DNA samples were subjected to electrophoresis on a 2% agarose gel using a 100 bp DNA ladder. For PCR amplification, 9 ISSR markers were used. The PCR components included 1.5 µL PCR buffer, 0.2 µL DNA polymerase enzyme, 0.6 µL dNTPs, 1 µL MgCl2, 1 µL ISSR marker, and 8.7 µL nuclease-free water, with a total reaction volume of 15 µL. The PCR cycle consisted of an initial denaturation step at 94 °C for 3 min, followed by 35 cycles at 94 °C for 1 min, annealing at a specific temperature of 53 °C for 50 s, extension at 72 °C for 2 min, and a final extension step at 72 °C for 7 min. PCR products were separated by electrophoresis on a 2% agarose gel prepared in 1X TAE buffer at 110 V for 4 h and stained with ethidium bromide before being visualized under UV light using a Kodak gel imaging system.
Data analyses
The data on fruit, leaf, and biochemical features were evaluated using correlation analysis, principal component analysis (PCA), and heatmap analysis. These analyses were carried out using the statistical software tool JMP Pro 17, which was used for doing the study. Differences between means were determined using Tukey test at the 5% significance level. For molecular analysis, the presence (1) or absence (0) of bands from the gel electrophoresis results was scored, and a data matrix was created. This data was analyzed using the NTSYS (Numerical Taxonomy and Multivariate Analysis System, NTSYS-pc version 2.11) software package. Similarity indices were calculated using the Dice [11] method, and a dendrogram was constructed using the UPGMA (Unweighted Pair Group Method with Arithmetic Mean) clustering method. Additionally, the average and total number of bands, polymorphic band numbers, and the percentage of polymorphism (number of polymorphic bands × 100/total number of bands) were calculated for each marker analyzed in the study. The relative discriminatory value of the ISSR marker was estimated using polymorphic information content (PIC) (30). Different marker attributes like effective multiplex ratio (EMR), marker index (MI) and resolving power (Rp) were also calculated to assess the discriminatory power and informativeness of the marker system utilized [27].
The GenAlExver 6.5 program was used to determine Observe allele number (Na), Effective allele number (Ne), Shannon index (I) and Nei index (H) (uHe). Also analysis of molecular variance (AMOVA) were calculated through GenAlExV6.5 [29].
Results and discussion
Fruit and leaf analysis
In the study, significant statistical differences were found between the averages of the genotypes in terms of fruit (fruit weight, fruit width, and fruit length) and leaf (leaf width, leaf length, petiole length, and petiole thickness) characteristics (p ≤ 0.05) (Table 2). The fruit weights of the genotypes varied between 0.047 g (G1) and 0.137 g (G17), with the highest fruit weight in G17, followed by G20 (0.109 g) and G23 (0.094 g). The fruit width ranged between 3.06 mm (G12) and 4.64 mm (G17), fruit length between 5.80 mm (G5) and 9.05 mm (G17), leaf length between 22.56 mm (G13) and 39.20 mm (G26), leaf width between 5.98 mm (G15) and 14.45 mm (G21), petiole length between 4.51 mm and 11.84 mm (G31), and petiole thickness between 0.35 mm (G20) and 0.68 mm (G12).
Table 2.
Morphological properties of the leaves and fruits of Berberis crataegina genotypes
| Genotype | Fruit Weight (g) |
Fruit Width (mm) |
Fruit Length (mm) |
Leaf Length (mm) |
Leaf Width (mm) |
Petiole Length (mm) |
Petiole Thickness (mm) |
|---|---|---|---|---|---|---|---|
| 1 | 0.047 ± 0.01 m* | 3.28 ± 0.20 kl | 7.43 ± 0.30 i-m | 27.49 ± 2.97 i-o | 8.39 ± 1.08 h-m | 10.67 ± 3.26 a-d | 0.39 ± 0.04 k-n |
| 2 | 0.076 ± 0.02 d-j | 3.53 ± 0.19 g-k | 7.34 ± 0.33 j-m | 24.19 ± 3.45 no | 7.07 ± 1.05 l-n | 6.27 ± 1.25 i-n | 0.52 ± 0.05 c-j |
| 3 | 0.093 ± 0.01 b-d | 3.56 ± 0.10 g-k | 8.85 ± 0.23 a-c | 33.97 ± 2.91 a-h | 11.55 ± 1.53 b-f | 7.74 ± 1.12 f-l | 0.62 ± 0.09 a-d |
| 4 | 0.093 ± 0.01 b-d | 3.36 ± 0.19 j-l | 8.58 ± 0.40 a-d | 35.92 ± 2.59 a-f | 10.61 ± 0.58 b-h | 8.71 ± 0.79 c-i | 0.58 ± 0.02 a-f |
| 5 | 0.056 ± 0.01 lm | 3.58 ± 0.31 f-k | 5.80 ± 0.48 q | 31.73 ± 4.28 d-l | 7.66 ± 1.23 j-n | 11.31 ± 2.92 ab | 0.50 ± 0.09 d-k |
| 6 | 0.069 ± 0.01 i-l | 3.74 ± 0.23 e-j | 7.86 ± 0.21 f-i | 32.65 ± 4.30 c-k | 7.61 ± 0.93 k-n | 10.38 ± 1.84 a-e | 0.51 ± 0.04 d-k |
| 7 | 0.075 ± 0.01 e-k | 3.74 ± 0.21 e-j | 7.03 ± 0.26 m-p | 23.00 ± 1.67 o | 6.48 ± 0.13 mn | 5.86 ± 1.16 j-n | 0.50 ± 0.06 d-k |
| 8 | 0.072 ± 0.01 g-l | 3.63 ± 0.05 f-k | 7.17 ± 0.32 l-n | 31.12 ± 3.38 e-l | 8.63 ± 1.59 h-m | 6.71 ± 1.09 h-n | 0.61 ± 0.05 a-d |
| 9 | 0.080 ± 0.01 c-i | 4.59 ± 0.02 ab | 6.61 ± 0.22 o-p | 27.79 ± 2.52 i-o | 9.86 ± 0.88 e-k | 6.21 ± 0.36 i-n | 0.53 ± 0.03 b-i |
| 10 | 0.087 ± 0.01 c-g | 4.19 ± 0.26 b-d | 7.54 ± 0.32 h-l | 29.01 ± 1.64 g-n | 11.03 ± 1.09 b-g | 10.59 ± 1.54 a-d | 0.48 ± 0.08 e-l |
| 11 | 0.062 ± 0.01 j-m | 3.44 ± 0.06 i-l | 6.59 ± 0.49 p | 31.11 ± 6.64 e-l | 10.44 ± 2.51 b-h | 10.71 ± 4.33 a-c | 0.67 ± 0.14 a |
| 12 | 0.059 ± 0.01 k-m | 3.06 ± 0.53 kl | 7.68 ± 0.05 h-k | 37.01 ± 4.79 a-d | 10.03 ± 2.06 d-j | 7.67 ± 1.12 l | 0.68 ± 0.05 a |
| 13 | 0.074 ± 0.01 f-k | 3.70 ± 0.18 f-k | 7.34 ± 0.22 j-m | 22.56 ± 1.79 o | 7.14 ± 0.98 l-n | 5.28 ± 0.72 l-n | 0.41 ± 0.05 j-n |
| 14 | 0.069 ± 0.02 h-l | 3.47 ± 0.53 h-l | 8.18 ± 0.77 d-g | 29.07 ± 3.44 g-n | 9.23 ± 0.42 f-l | 5.56 ± 1.71 k-n | 0.43 ± 0.09 i-n |
| 15 | 0.071 ± 0.01 g-l | 3.88 ± 0.30 c-h | 7.69 ± 0.36 g-k | 25.11 ± 3.69 m-o | 5.98 ± 0.85 n | 6.26 ± 0.88 i-n | 0.48 ± 0.08 f-m |
| 16 | 0.082 ± 0.00 c-i | 3.30 ± 0.12 kl | 8.26 ± 0.02 d-f | 24.81 ± 0.18 m-o | 6.40 ± 1.06 mn | 5.06 ± 1.15 mn | 0.46 ± 0.07 g-n |
| 17 | 0.137 ± 0.00 a | 4.64 ± 0.35 a | 9.05 ± 0.14 a | 35.11 ± 3.17 a-f | 9.06 ± 0.56 g-l | 8.73 ± 1.40 c-i | 0.37 ± 0.03 l-n |
| 18 | 0.080 ± 0.02 c-i | 3.34 ± 0.27 j-l | 8.44 ± 0.26 c-e | 30.49 ± 1.79 f-m | 7.64 ± 0.86 k-n | 9.22 ± 1.33 b-h | 0.50 ± 0.07 d-k |
| 19 | 0.081 ± 0.02 c-i | 4.22 ± 0.20 a-d | 6.61 ± 0.27 op | 27.04 ± 2.93 k-o | 8.59 ± 1.13 h-m | 4.51 ± 0.55 n | 0.48 ± 0.08 e-m |
| 20 | 0.109 ± 0.02 b | 4.01 ± 0.40 c-f | 9.00 ± 0.26 ab | 32.91 ± 1.67 b-j | 12.67 ± 0.66 ab | 8.11 ± 0.74 d-k | 0.35 ± 0.02 n |
| 21 | 0.085 ± 0.00 c-i | 4.18 ± 0.08 b-d | 7.15 ± 0.11 l-n | 36.68 ± 4.52 a-e | 14.45 ± 2.00 a | 10.27 ± 2.80 a-f | 0.57 ± 0.03 a-g |
| 22 | 0.086 ± 0.01 c-h | 4.16 ± 0.46 b-e | 7.23 ± 0.06 k-m | 32.18 ± 0.50 c-l | 10.46 ± 1.00 b-h | 8.18 ± 0.45 c-j | 0.58 ± 0.02 a-f |
| 23 | 0.094 ± 0.00 bc | 4.18 ± 0.10 b-d | 7.78 ± 0.46 f-j | 28.94 ± 4.55 h-n | 9.90 ± 0.86 e-k | 7.09 ± 1.69 g-m | 0.57 ± 0.11 a-g |
| 24 | 0.069 ± 0.01 i-l | 3.92 ± 0.36 c-g | 7.10 ± 0.28 l-o | 27.33 ± 2.64 j-o | 7.92 ± 1.43 i-n | 6.18 ± 0.37 i-n | 0.51 ± 0.05 d-j |
| 25 | 0.077 ± 0.01 c-j | 3.69 ± 0.08 f-k | 8.40 ± 0.15 c-e | 26.67 ± 2.99 l-o | 10.12 ± 1.75 c-i | 6.32 ± 0.63 i-n | 0.45 ± 0.02 h-n |
| 26 | 0.087 ± 0.01 c-g | 4.30 ± 0.29 a-c | 7.22 ± 0.25 k-n | 39.20 ± 2.39 a | 11.57 ± 1.08 b-f | 9.27 ± 0.59 b-h | 0.64 ± 0.14 a-c |
| 27 | 0.076 ± 0.01 c-j | 3.83 ± 0.25 d-i | 7.46 ± 0.21 i-m | 32.83 ± 4.39 c-k | 12.36 ± 2.64 a-d | 9.42 ± 1.13 a-g | 0.55 ± 0.08 b-h |
| 28 | 0.074 ± 0.01 f-k | 3.51 ± 0.23 g-k | 7.99 ± 0.23 e-h | 37.58 ± 3.23 a-c | 11.86 ± 1.84 b-e | 7.61 ± 0.27 g-m | 0.36 ± 0.06 mn |
| 29 | 0.078 ± 0.02 c-j | 3.62 ± 0.25 f-k | 7.49 ± 0.37 i-m | 33.75 ± 0.45 a-h | 8.99 ± 0.13 g-l | 8.48 ± 1.17 c-i | 0.59 ± 0.04 a-e |
| 30 | 0.089 ± 0.01 c-f | 3.81 ± 0.28 d-i | 7.86 ± 0.19 f-i | 38.72 ± 4.51 ab | 12.41 ± 2.94 a-c | 7.36 ± 0.91 g-m | 0.65 ± 0.06 ab |
| 31 | 0.051 ± 0.00 m | 3.58 ± 0.20 f-k | 6.73 ± 0.31 n-p | 32.73 ± 4.49 c-k | 10.02 ± 1.20 d-j | 11.84 ± 2.41 a | 0.49 ± 0.06 e-k |
| 32 | 0.075 ± 0.02 e-k | 3.74 ± 0.21 e-j | 7.46 ± 0.06 i-m | 33.20 ± 5.11 b-i | 12.81 ± 0.74 ab | 7.83 ± 1.26 e-l | 0.43 ± 0.05 i-n |
| 33 | 0.091 ± 0.00 c-e | 3.41 ± 0.15 i-l | 8.51 ± 0.30 b-d | 34.78 ± 6.79 a-g | 11.25 ± 2.97 b-g | 8.92 ± 0.99 b-h | 0.47 ± 0.14 f-m |
* The difference between the averages indicated by different letters in the same column is significant (p ≤ 0.05)
Fruit and leaf characteristics are important indicators in differentiating plant species and revealing morphological diversity. Numerous Berberis species have undergone morphological characterization studies across various regions. Ahmed et al. [1] reported the fruit weight of B. aristata genotypes in Pakistan to range between 0.115 g and 0.122 g, fruit width between 2.65 mm and 4.34 mm, fruit length between 7.12 mm and 9.75 mm, leaf length between 66.70 mm and 96.90 mm, and leaf width between 25.80 mm and 33.10 mm. Goodarzi et al. [20] reported the fruit weight of B. vulgaris genotypes in Iran to range between 0.11 g and 0.43 g, fruit width between 4.39 mm and 8.05 mm, fruit length between 7.40 mm and 10.84 mm, leaf length between 18.89 mm and 42.90 mm, leaf width between 7.54 mm and 17.49 mm, and petiole length between 4.50 mm and 12.65 mm. Another study on B. vulgaris genotypes in Iran by Talebi et al. [39] reported fruit weights between 0.106 g and 0.285 g, fruit widths between 4.66 mm and 7.23 mm, and fruit lengths between 7.35 mm and 11.97 mm. Jannatizadeh and Khadivi-Khub [23] reported the fruit weight of B. integerrima genotypes in Iran to range between 0.10 g and 0.25 g, fruit width between 4.28 mm and 6.23 mm, fruit length between 7.70 mm and 11.31 mm, and leaf length between 22.00 mm and 38.00 mm. A study on B. vulgaris genotypes in Türkiye reported fruit weights ranging from 0.102 g to 0.342 g [17]. Observations revealed some differences, despite the fruit and leaf characteristics generally showing similarities with those in the literature. These differences may be attributed to species differences, genotypic variation, and ecological factors.
Biochemical analyses
The biochemical characteristics of the genotypes (total phenolic content, total flavonoid content, total anthocyanin content, and total antioxidant content) were found to be statistically significant on average (p ≤ 0.05) (Table 3). The total phenolic contents of the genotypes ranged from 190.53 mg GAE/100 g (G12) to 297.55 mg GAE/100 g (G7), with the G7 genotype having the highest phenolic content, followed by the G19 genotype with 295.12 mg GAE/100 g. Flavonoids are phenolic compounds with antioxidant properties. The genotypes with the highest flavonoid content were G31 (203.89 mg QE/100 g), G11 (203.14 mg QE/100 g), and G33 (202.03 mg QE/100 g), while the genotypes with the lowest flavonoid content were G30 (82.03 mg QE/100 g) and G26 (93.89 mg QE/100 g). Anthocyanins are pigments that give plants blue, purple, and red colors and possess strong antioxidant properties. The genotypes G9 (11.76 mg cyn-3-gluc/100 g) and G28 (11.52 mg cyn-3-gluc/100 g) had the highest anthocyanin content, while the G29 genotype (4.54 mg cyn-3-gluc/100 g) had the lowest. In terms of antioxidant capacity, genotypes G16 (87.93%) and G1 (87.07%) had the highest values, while genotype G23 (57.76%) had the lowest antioxidant capacity.
Table 3.
Comparison of biochemical contents in Berberis crataegina genotypes
| Genotype | Total Phenolic (mg GAE/100 g) |
Total Flavonoids (mg QE/100 g) |
Total Anthocyanin (mg cyn-3-gluc/100 g) |
Total Antioxidant (%) |
|---|---|---|---|---|
| 1 | 219.72 ± 2.00 m* | 161.66 ± 2.10 f | 6.35 ± 0.04 n | 87.07 ± 0.50 b |
| 2 | 274.31 ± 2.30 fg | 194.30 ± 1.04 c | 6.83 ± 0.04 k | 68.97 ± 0.12 n |
| 3 | 276.20 ± 2.20 f | 140.92 ± 1.40 i | 6.16 ± 0.02 pq | 84.48 ± 0.40 d |
| 4 | 279.45 ± 1.30 e | 145.00 ± 0.85 h | 6.51 ± 0.03 m | 62.93 ± 0.80 s |
| 5 | 228.09 ± 1.04 k | 139.07 ± 0.60 j | 4.76 ± 0.01 v | 67.24 ± 0.40 o |
| 6 | 194.58 ± 1.10 r | 128.70 ± 0.80 kl | 6.28 ± 0.01 no | 81.03 ± 0.10 e |
| 7 | 297.55 ± 1.00 a | 150.18 ± 0.50 g | 6.33 ± 0.03 no | 60.34 ± 0.13 v |
| 8 | 285.53 ± 0.80 c | 127.59 ± 0.63 l | 6.06 ± 0.03 qr | 64.66 ± 0.25 q |
| 9 | 279.58 ± 1.20 e | 137.59 ± 1.10 j | 11.76 ± 0.03 a | 59.48 ± 0.70 w |
| 10 | 263.91 ± 1.10 i | 114.63 ± 0.80 o | 7.38 ± 0.03 h | 62.93 ± 0.70 s |
| 11 | 283.23 ± 1.21 d | 203.14 ± 1.18 ab | 6.68 ± 0.04 l | 65.52 ± 0.40 p |
| 12 | 190.53 ± 1.03 s | 117.22 ± 0.40 n | 9.07 ± 0.06 c | 80.17 ± 0.10 f |
| 13 | 235.80 ± 1.30 j | 112.40 ± 0.30 p | 4.88 ± 0.03 u | 73.28 ± 0.38 l |
| 14 | 267.96 ± 2.00 h | 128.70 ± 1.20 kl | 6.31 ± 0.01 no | 61.21 ± 0.41 u |
| 15 | 196.07 ± 1.07 r | 112.40 ± 0.60 p | 4.58 ± 0.03 w | 61.21 ± 0.40 u |
| 16 | 212.69 ± 0.40 o | 151.66 ± 0.30 g | 8.00 ± 0.02 f | 87.93 ± 0.70 a |
| 17 | 219.99 ± 1.20 m | 124.63 ± 1.10 m | 4.96 ± 0.01 u | 74.14 ± 0.13 k |
| 18 | 274.72 ± 1.20 fg | 183.89 ± 0.80 d | 7.25 ± 0.01 i | 75.00 ± 0.37 j |
| 19 | 295.12 ± 0.90 b | 116.48 ± 1.5 2n | 8.17 ± 0.05 e | 63.79 ± 0.21 r |
| 20 | 213.09 ± 0.70 o | 115.74 ± 1.20 no | 5.48 ± 0.01 s | 76.72 ± 0.32 h |
| 21 | 282.15 ± 1.20 d | 138.43 ± 1.05 j | 6.61 ± 0.04 lm | 61.21 ± 0.43 u |
| 22 | 208.77 ± 1.20 p | 116.11 ± 0.70 no | 7.43 ± 0.05 h | 62.07 ± 0.50 t |
| 23 | 273.50 ± 3.00 g | 129.81 ± 1.01 k | 5.46 ± 0.02 s | 57.76 ± 0.50 x |
| 24 | 222.28 ± 1.10 l | 97.96 ± 1.50 q | 4.98 ± 0.03 u | 69.83 ± 0.50 m |
| 25 | 228.23 ± 1.20 k | 123.89 ± 0.60 m | 6.04 ± 0.02 r | 64.66 ± 0.30 q |
| 26 | 196.74 ± 1.04 r | 93.89 ± 0.70 r | 8.73 ± 0.01 d | 67.24 ± 0.60 o |
| 27 | 216.61 ± 1.01 n | 145.37 ± 1.10 h | 7.58 ± 0.03 g | 62.07 ± 0.60 t |
| 28 | 261.74 ± 1.70 i | 175.37 ± 1.20 e | 11.52 ± 0.02 b | 60.34 ± 0.20 v |
| 29 | 209.85 ± 0.70 p | 115.74 ± 0.78 no | 4.54 ± 0.02 w | 75.86 ± 0.60 i |
| 30 | 202.28 ± 1.10 q | 82.03 ± 0.60 s | 7.01 ± 0.01 j | 77.59 ± 0.70 g |
| 31 | 274.05 ± 1.09 fg | 203.89 ± 0.64 a | 5.34 ± 0.02 t | 85.34 ± 0.24 c |
| 32 | 272.96 ± 1.40 g | 194.26 ± 0.48 c | 6.23 ± 0.03 op | 67.24 ± 0.40 o |
| 33 | 283.36 ± 2.10 cd | 202.03 ± 1.16 b | 6.58 ± 0.04 lm | 64.66 ± 0.76 q |
* The difference between the averages indicated by different letters letter in the same column is significant (p ≤ 0.05)
Many studies have been conducted on the biochemical contents of Berberis species. Researchers have reported the total phenolic content of Berberis species as follows: Akbulut et al. [2] reported 789.32 mg GAE/100 g, Sasikumar et al. [35] reported 410 mg GAE/100 g, Hassanpour and Alizadeh [21] reported 347.52–623.07 mg GAE/100 g, Ersoy et al. [17] reported 228.10–346.20 mg GAE/100 g, Goodarzi et al. [20] reported 4.44–28.93 mg GAE/100 g, Gholizadeh-Moghadam et al. [18] reported 25.98–94.04 mg GAE/100 g, and Eroğlu et al. [16] reported 140.00–448.30 mg GAE/100 g. For total flavonoid content, Demirci [13] reported 62.58 mg QE/100 g, Charehsaz et al. [7] reported 274.20 mg QE/100 g, Hassanpour and Alizadeh [21] reported 132.66–280.00 mg QE/100 g, and Gholizadeh-Moghadam et al. [18] reported 2.96–76.70 mg QE/100 g. For total anthocyanin content, Hassanpour and Alizadeh [21] reported 16.32–91.66 mg cyn-3-gluc/100 g, Goodarzi et al. [20] reported 7.00–70.00 mg cyn-3-gluc/100 g, and Ersoy et al. [17] reported 36.00–87.40 mg cyn-3-gluc/100 g. The total antioxidant content (DPPH) was reported by Yildiz et al. [46] as 75.01 – 90.64%, Hassanpour and Alizadeh [21] as 20.47 – 74.72%, Gholizadeh-Moghadam et al. [18] as 13.20 – 56.84%, and Demirci [13] as 90.74%.
Although our findings are, for the most part, consistent with those found in the existing body of research, it was discovered that they were greater than the findings of certain researchers and lower than those of others. Several factors, including species, genotype, climate, soil, and geographical variances, might be responsible for these disparities.
Correlation analysis
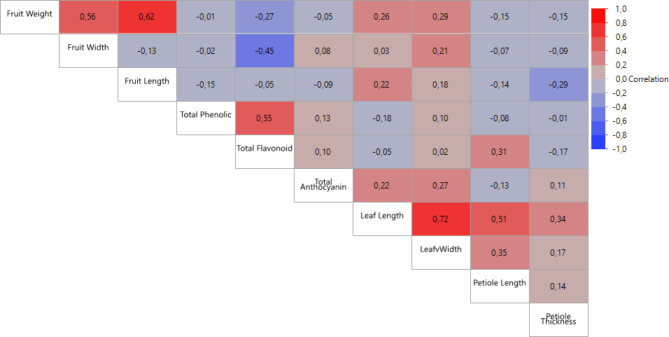
Significant correlations were detected among the traits examined in this study (Fig. 2). Leaf and fruit characteristics showed a strong positive correlation among themselves. The highest correlation among leaf traits was observed between leaf length and leaf width (r = 0.73). Similarly, the strongest correlation among fruit characteristics was found between fruit weight and fruit length (r = 0.62). In terms of the biochemical contents of Berberis species, the highest correlation was observed between total phenolic content and total flavonoid content (r = 0.94). The correlation findings of this study align with previous studies conducted on Berberis [20, 23, 33]. Correlation analysis helps breeders in parent selection by identifying relationships among the traits studied and can provide valuable information about the most important traits in the evaluation of genotypes [28].
Fig. 2.
Pearson correlation matrix of morphological and biochemical traits of Berberis crataegina genotypes (significance level p ≤ 0.05)
Principal component analysis (PCA)
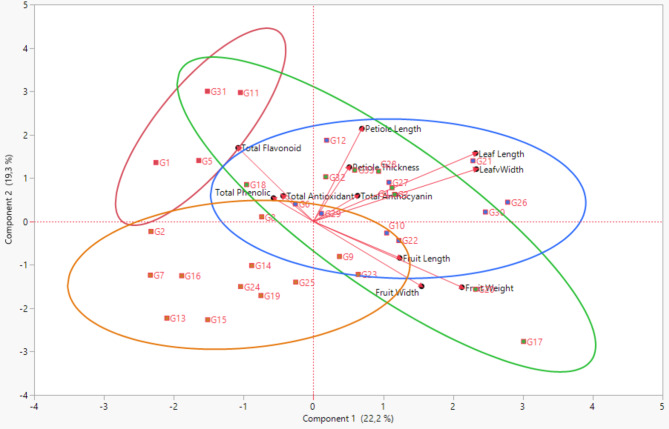
Principal component analysis (PCA) is widely used to reduce the number of traits investigated for distinguishing genotypes and to determine the degree of influence these traits have on the variation. Furthermore, the first three principal components provide significant time savings in the characterization of genotypes [22]. In this study, PCA was performed on 11 parameters related to fruit, leaf, and biochemical characteristics. To identify the traits that explain the largest proportion of variation, components with an eigenvalue greater than 1 were considered. The analysis revealed four components with eigenvalues greater than 1. These four components explained 71.89% of the total variation. However, the first three principal components accounted for 58.55% of the total variation. PCA1 explained 23.48% of the variation, PCA2 explained 18.68%, and PCA3 explained 16.39%. Our findings were similar to the results of Hassanpour and Alizadeh [21] (PCA3 explained 63.90% of the variation), but higher than the results of Jannatizadeh and Khadivi-Khub [23] and Rezaei et al. [33] (PCA3 explained 37.77% and 31.77%, respectively). Genotype numbers and types of parameters likely caused study differences. The contribution of each trait to the principal components varied. For PCA1, the traits that had the greatest impact were fruit weight and fruit width among fruit traits, and leaf length, leaf width, and petiole length among leaf traits. The traits that showed the highest effect on PCA1 showed a positive effect. When the traits that showed the highest effect on PCA2 were examined; fruit weight, fruit width and fruit length had a negative effect among fruit traits, while total flavonoids among biochemical traits and petiole length among leaf traits had a positive effect. For PCA3, fruit width and total phenolics made the largest positive contributions, while total antioxidants made the largest negative contributions (Table 4; Fig. 3).
Table 4.
Contributions of variables to principal components in PCA analysis
| Variables | PCA1 | Contribution (%) | PCA2 | Contribution (%) | PCA3 | Contribution (%) | PCA4 | Contribution (%) |
|---|---|---|---|---|---|---|---|---|
| Fruit Weight | 0.37 | 13.51 | -0.43 | 18.69 | 0.13 | 1.69 | 0.29 | 8.13 |
| Fruit Width | 0.29 | 8.19 | -0.34 | 11.80 | 0.34 | 11.75 | -0.17 | 2.86 |
| Fruit Length | 0.21 | 4.32 | -0.33 | 10.68 | -0.21 | 4.49 | 0.52 | 27.29 |
| Total Phenolic | -0.10 | 0.98 | 0.26 | 6.66 | 0.50 | 25.04 | 0.36 | 12.79 |
| Total Flavonoids | -0.15 | 2.14 | 0.44 | 19.06 | 0.09 | 0.77 | 0.53 | 28.47 |
| Total Anthocyanin | 0.11 | 1.10 | 0.16 | 2.49 | 0.31 | 9.54 | -0.06 | 0.33 |
| Total Antioxidant | -0.05 | 0.30 | 0.02 | 0.02 | -0.62 | 38.79 | 0.13 | 1.75 |
| Leaf Length | 0.54 | 29.43 | 0.21 | 4.28 | -0.13 | 1.81 | 0.00 | 0.00 |
| Leaf Width | 0.50 | 25.05 | 0.21 | 4.47 | 0.09 | 0.82 | 0.07 | 0.55 |
| Petiole Length | 0.31 | 9.73 | 0.37 | 13.47 | -0.23 | 5.20 | 0.06 | 0.37 |
| Petiole Thickness | 0.23 | 5.25 | 0.29 | 8.37 | -0.03 | 0.10 | -0.42 | 17.47 |
| Eigenvalue | 2.58 | 2.06 | 1.80 | 1.47 | ||||
| Percent | 23.48 | 18.68 | 16.39 | 13.34 | ||||
| Cumulative values | 23.48 | 42.16 | 58.55 | 71.89 |
Fig. 3.
Principal component analysis (PCA) biplot of Berberis crataegina genotypes based on morphological and biochemical traits
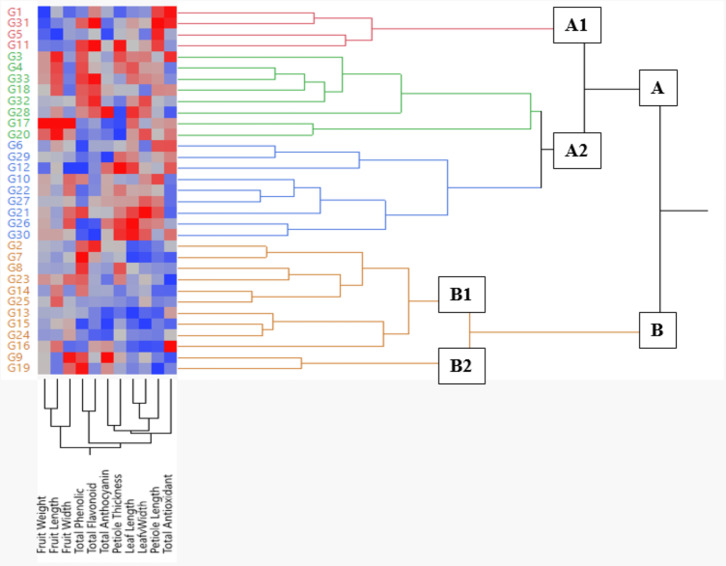
The heatmap and hierarchical clustering analysis revealed that the genotypes were divided into two main groups (Fig. 4). Each group was further subdivided into two subgroups. The A1 group, consisting of genotypes G1, G31, G5, and G11, displayed high values for traits such as petiole length, total antioxidant content, total phenolic content, and total flavonoid content. The A2 group was further divided into two subgroups. Group B was also divided into two subgroups. Genotypes G9 and G19, which showed high values for traits like fruit width, total phenolic content, and total anthocyanin content, formed the B2 subgroup. Heatmap analysis provides a classification of genotypes based on morphological traits and offers breeders collective information about the characteristics of the genotypes (Fig. 4).
Fig. 4.
Heatmap and hierarchical clustering dendrogram of Berberis crataegina genotypes based on morphological and biochemical traits
Molecular analyses
A total of 9 ISSR markers were used for the molecular characterization of the examined Berberis genotypes. The lengths of the bands generated by these markers, the total number of bands, the number of polymorphic bands, and polymorphism rates are presented in Table 5. A total of 72 scorable bands were obtained, with 58 of them being identified as polymorphic. The number of bands obtained from the markers ranged from 4.00 with the (CAC)3GC marker to 13 bands with the HVH (TCC)7 marker. The band lengths produced by the ISSR markers ranged between 190 and 2500 bp. The average number of bands per marker was found to be 8.00, with an average of 6.44 polymorphic bands and an average polymorphism rate of 82.24%. The band lengths produced by the markers ranged from 200 to 1100 bp. While the PIC of ISSR primers was between 0.27 ((AGC)6G) and 0.39 (HVH (TCC)7), the mean PIC was determined as 0.32. Among the features of ISSR primers, RP varied between 3.20 ((AGC)6G) and 5.42 (HVH (TCC)7), EMR varied between 8.42 ((GA)8YG) and 10.23 ((GT)6GG), and MI varied between 3.75 ((GT)6GG) and 5.12 (BDB (CA)7 C) (Table 5).
Table 5.
Band information and polymorphism rates of ISSR markers in Berberis crataegina genotypes
| Marker | NB | NPB | PR | BL (bp) | PIC | RP | EMR | MI |
|---|---|---|---|---|---|---|---|---|
| (GA)8YG | 9.00 | 8.00 | 88.89 | 200–1000 | 0.35 | 4.25 | 8.42 | 4.24 |
| (GAA)6 | 6.00 | 6.00 | 100.00 | 220–750 | 0.33 | 3.85 | 9.17 | 3.80 |
| (CAC)3GC | 4.00 | 3.00 | 75.00 | 300–1000 | 0.28 | 4.37 | 10.06 | 4.36 |
| BDB (CA)7C | 8.00 | 8.00 | 100.00 | 350–1000 | 0.30 | 4.12 | 8.75 | 5.12 |
| VHV (GTG)7 | 11.00 | 10.00 | 90.91 | 250–950 | 0.32 | 4.33 | 9.46 | 4.01 |
| (GT)6GG | 5.00 | 5.00 | 100.00 | 400–1000 | 0.32 | 4.74 | 10.23 | 3.75 |
| (AGC)6G | 7.00 | 4.00 | 57.14 | 250–950 | 0.27 | 3.20 | 9.16 | 4.41 |
| DBDA(CA)7 | 9.00 | 6.00 | 66.67 | 400–1000 | 0.33 | 3.98 | 9.78 | 5.01 |
| HVH (TCC)7 | 13.00 | 8.00 | 61.54 | 400–1100 | 0.39 | 5.42 | 8.47 | 4.83 |
| Mean | 8.00 | 6.44 | 82.24 | - | 0.32 | 4.25 | 9.28 | 4.39 |
| Total | 72.00 | 58.00 | - | - | 2.89 | - | - | - |
Number of Bands: NB, Number of Polymorphic Bands: NPB, Polymorphism Rate: PR, Base Length: BL, Resolving Power: RP, Polymorphic Information Content: PIC, Effective Multiplex Ratio: EMR, Marker Index: MI
There are a limited number of molecular studies on Berberis species in the literature. In their research conducted with twenty ISSR markers, Pınar et al. [30] reported that each marker had an average of 7.50 bands, with an average of 5.50 polymorphic bands, an average polymorphism rate of 74.00%, and band lengths ranging from 190 to 1400 base pairs. In another study, Safamanesh et al. [33] used 10 ISSR and 5 SSR markers. They reported an average number of 9.80 bands per ISSR marker, with 8.60 polymorphic bands, an average polymorphism rate of 79.98%, and band lengths between 300 and 1300 bp. For SSR markers, they found an average of 8.60 bands, with an identical number of polymorphic bands and an average polymorphism rate of 79.98%, with band lengths ranging from 100 to 1100 bp.
In the ISSR primers, Observe allele number (Na) ranged from 1.19 ((CAC)3GC) to 1.51 ((GT)6GG), Effective allele number (Ne) ranged from 1.04 ((GAA)8) to 1.31 ((GT)6GG), Shannon index (I) ranged from 0.20 (BDB (CA)7 C) to 0.40 ((CAC)3GC) and Nei index (H) ranged from 0.21 (BDB (CA)7 C) to 0.36 (DBDA(CA)7) (Table 6).
Table 6.
Genetic diversity data and differentiation parameters of ISSR molecular markers in Berberis crataegina genotypes in Kayseri province. Observe allele number (na), effective allele number (ne), Shannon index (I) and Nei index (H)
| Marker | Na | Ne | I | H |
|---|---|---|---|---|
| (GA)8YG | 1.37 | 1.19 | 0.24 | 0.35 |
| (GAA)6 | 1.28 | 1.04 | 0.33 | 0.31 |
| (CAC)3GC | 1.19 | 1.12 | 0.40 | 0.28 |
| BDB (CA)7C | 1.42 | 1.26 | 0.20 | 0.21 |
| VHV (GTG)7 | 1.46 | 1.06 | 0.27 | 0.27 |
| (GT)6GG | 1.51 | 1.31 | 0.34 | 0.32 |
| (AGC)6G | 1.33 | 1.14 | 0.28 | 0.30 |
| DBDA(CA)7 | 1.45 | 1.09 | 0.35 | 0.36 |
| HVH (TCC)7 | 1.23 | 1.18 | 0.21 | 0.29 |
| Mean | 1.36 | 1.15 | 0.29 | 0.30 |
Even though the findings of this study are generally comparable to those of other studies in terms of ISSR marker banding and diversity features, there were some differences, with certain parameters exhibiting larger values than others. The changes in the markers and the genetic variances between the germplasms are responsible for these discrepancies. Since these markers revealed polymorphism in B. crataegina, it is advised that future molecular research on Berberis species use ISSR markers, which were used in this work.
Analysis of molecular variance using ISSR markers showed that, the greatest variation was related to within populations. Variations within populations were 17 for ISSR markers, while the variation between populations were 83 (Fig. 5).
Fig. 5.
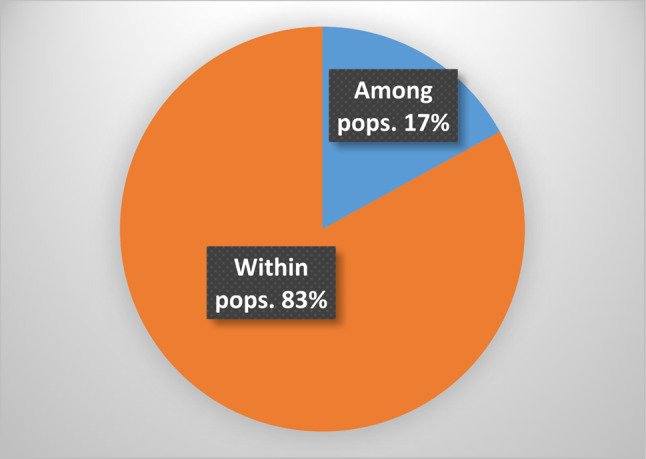
The percentage of molecular variance between and within populations of 33 Berberis crataegina genotypes investigated using ISSR primers
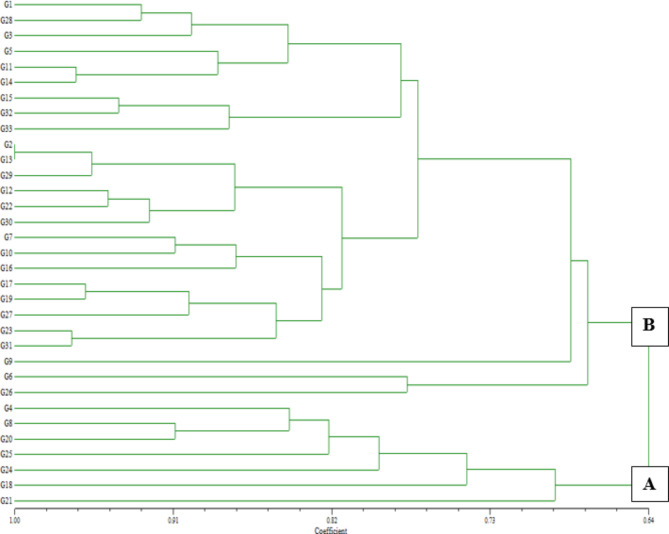
A UPGMA dendrogram was created based on the scoring results obtained from the ISSR markers to investigate the genetic relationship between the Berberis genotypes. According to the UPGMA method, the similarity index of the genotypes in the dendrogram ranged between 0.64 and 1.00 (Fig. 6). Two distinct main groups were formed in the dendrogram. Group A was further divided into two subgroups. The G21 genotype, with a similarity index of 0.70, was the most genetically distant genotype from the others within group (A) Other genotypes in group A included G4, G8, G18, G20, G24, and G25. The remaining genotypes formed group (B) Among the genotypes, G2 and G13 were the closest, sharing a 1.00 similarity index.
Fig. 6.
UPGMA dendrogram showing genetic relationships among Berberis crataegina genotypes based on ISSR marker data
The similarity indices obtained in this study are in line with previous molecular analyses of Berberis species. For ISSR markers, Pınar et al. [31] reported similarity indices ranging from 0.84 to 1.00, while Safamanesh et al. [33] found values between 0.27 and 0.70. Using AFLP markers, Cote and Leduc [9] reported similarity indices between 0.30 and 1.00, and for SSR markers, Rezaei et al. [33] observed a range of 0.54 to 0.96, while Safamanesh et al. [34] recorded values from 0.18 to 0.57. The differences in similarity ratios observed between Berberis species are mainly attributed to the variations in genetic material sources used in these studies.
Conclusion
This study identified the genetic diversity of 33 genotypes of B. crataegina, naturally growing in Kayseri province, located in the central part of Anatolia, through fruit, leaf, biochemical, and molecular analyses. Significant variations were detected among the genotypes in terms of fruit, leaf, and biochemical characteristics. The G17 genotype was found to be superior to the other genotypes in terms of fruit weight. While the G7 genotype was superior in terms of total phenolics, the G16 genotype was determined to be superior in terms of total antioxidants. The genotypes were observed to possess rich biochemical content. The integration of morphological and biochemical data with ISSR markers has shown that combining these factors provides more reliable results for distinguishing genotypes. The findings of this study are expected to guide future breeding studies for the conservation and improvement of this species.
Acknowledgements
This study was supported by Erciyes University Scientific Research Projects Coordination Unit under the project number FYL-2024-13697.
Author contributions
This study was conceived by MY and AS. Plant materials were collected by MS and MY. MY, AS, MS, and KUY conceptualized and established the methodology, while AS and YT performed the analyses. AS, MY, FD, and YT wrote the manuscript. AS, FD, YT, and AK revised and edited the manuscript. All authors read and approved the final version of the manuscript.
Funding
Not applicable.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Research involving human participants and, or animals
Not applicable.
Clinical Trial Study
Not applicable.
Clinical trial number
Not applicable.
Informed consent
Not applicable.
Statement specifying permissions
For this study, we acquired permission to study Berberis crataegina issued by the Agriculture and Forestry Ministry of the Republic of Türkiye.
Statement on experimental research and field studies on plants
The either cultivated or wild-growing plants sampled comply with relevant institutional, national, and international guidelines and domestic legislation of Türkiye.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mehmet Yaman, Email: mehmetyaman@erciyes.edu.tr.
Ali Khadivi, Email: a-khadivi@araku.ac.ir.
References
- 1.Ahmed M, Anjum MA, Naz RMM, Khan MR, Hussain S. Characterization of indigenous barberry germplasm in Pakistan: variability in morphological characteristics and nutritional composition. Fruits. 2013;68:409–22. 10.1051/fruits/2013085. [Google Scholar]
- 2.Akbulut M, Çalışır S, Marakoğlu T, Coklar H. Some physicomechanical and nutritional properties of barberry (Berberis vulgaris L.) fruits. J Food Proces Eng. 2009;32:497–511. 10.1111/j.1745-4530.2007.00229.x. [Google Scholar]
- 3.Alemardan A, Asadi W, Rezaei M, Tabrizi L, Mohammadi S. Cultivation of Iranian seedless barberry (Berberis integerrima ‘Bidaneh’): a medicinal shrub. Indus Crop Produc. 2013;50:276–87. 10.1016/j.indcrop.2013.07.061. [Google Scholar]
- 4.Brand–Williams W, Cuvelier ME, Berset CLWT. Use of a free radical method to evaluate antioxidant activity. Food Sci Tech. 1995;28:25–30. 10.1016/S0023-6438(95)80008-5. [Google Scholar]
- 5.Bottini MCJ, De Bustos A, Sanso AM, Jouve N, Poggio L. Relationships in Patagonian species of Berberis (Berberidaceae) based on the characterization of rDNA internal transcribed spacer sequences. Botan J Linn Societ. 2007;153:321–8. 10.1111/j.1095-8339.2007.00586.x. [Google Scholar]
- 6.Chang CC, Yang MH, Wen HM, Chern JC. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J Food Drug Analy. 2002;10:178–82. 10.38212/2224-6614.2748. [Google Scholar]
- 7.Charehsaz M, Sipahi H, Celep E, Üstündağ A, Cemiloğlu Ülker Ö, Duydu Y, Aydın A, Yesilada E. The fruit extract of Berberis crataegina DC: exerts potent antioxidant activity and protects DNA integrity. Daru J Pharmaceut Sci. 2015;23:1–7. 10.1186/s40199-015-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary RS, Zagade VS, Khalakar GD. ISSR based genotypic differentiation of grape (Vitis vinifera L). Bioscan. 2014;9:823–8. 10.1007/s10722-024-01861-3. [Google Scholar]
- 9.Cote MJ, Leduc L. Molecular identification of Japanese barberry (Berberis thunbergii) cultivars using amplified fragment length polymorphism. HortScience. 2007;42:478–82. 10.21273/HORTSCI.42.3.478. [Google Scholar]
- 10.Davis PH. (1965–1988) Flora of Türkiye and the East Aegean Islands. Edinburgh University Press. Edinburgh, pp 1–10.
- 11.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. 10.2307/1932409. [Google Scholar]
- 12.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:39–40. [Google Scholar]
- 13.Demirci ŞY. (2022) Investigation of the thermal stability and bioavailability of barberry (Berberis crataegina DC.) fruit anthocyanins. İstanbul Sabahattin Zaim University. Master Thesis. (in Turkish).
- 14.Erarslan ZB, Kültür Ş. Ethnoveterinary medicine in Turkey: a comprehensive review. Turkish J Vet Anim Sci. 2019;43:55–582. 10.3906/vet-1904-8. [Google Scholar]
- 15.Ercan L. Bioactive components, antioxidant capacity, and antimicrobial activity of Berberis crataegina fruit. Pharmacolog Res Nat Produc. 2024;2:100020. 10.1016/j.prenap.2024.100020. [Google Scholar]
- 16.Eroğlu AY, Çakır Ö, Sağdıç M, Dertli E. Bioactive characteristics of wild Berberis vulgaris and Berberis crataegina fruits. J Chem. 2020;2020:8908301. 10.1155/2020/8908301. [Google Scholar]
- 17.Ersoy N, Kupe M, Sagbas HI, Ercisli S. Physicochemical diversity among barberry (Berberis vulgaris L.) fruits from eastern Anatolia. Not Botani Horti Agrobot Cluj Napoca. 2018;46:336–42. 10.15835/nbha46211111. [Google Scholar]
- 18.Gholizadeh-Moghadam N, Hosseini B, Alirezalu A. Classification of barberry genotypes by multivariate analysis of biochemical constituents and HPLC profiles. Phytochem Analy. 2019;30:385–94. 10.1002/pca.2821. [DOI] [PubMed] [Google Scholar]
- 19.Giusti MM, Wrolstad RE, Wrolstad RE. Ed; John Wiley and Sons Inc: New York, NY, USA: pp 12–13. 10.1002/0471142913.faf0102s00
- 20.Goodarzi S, Khadivi A, Abbasifar A, Akramian M. Phenotypic, pomological and chemical variations of the seedless barberry (Berberis vulgaris L. var. Asperma). Scientia Horti. 2018;238:38–50. 10.1016/j.scienta.2018.04.040. [Google Scholar]
- 21.Hassanpour H, Alizadeh S. Evaluation of phenolic compound, antioxidant activities and antioxidant enzymes of barberry genotypes in Iran. Scientia Horti. 2016;200:125–30. 10.1016/j.scienta.2016.01.015. [Google Scholar]
- 22.Iezzoni AF, Pritts MP. Applications of principal component analysis to horticultural research. HortScience. 1991;26:334–8. [Google Scholar]
- 23.Jannatizadeh A, Khadivi-Khub A. Morphological variability of Berberis integerrima from Iran. Erwerbs Obstbau. 2016;58:247–252. 10.1007/s10341-016-0285-7. [Google Scholar]
- 24.Kılıç AB, Yusufbeyoğlu S. Berberis crataegina DC. Novel drug targets with Traditional Herbal Medicines: scientific and clinical evidence. Cham: Springer International Publishing; 2022. pp. 37–47. 10.1007/978-3-031-07753-1_3. [Google Scholar]
- 25.Lambert KA, Obae SG. Molecular characterisation of Berberis thunbergii cultivars using microsatellite markers. J Horti Sci Biotechnol. 2016;91:156–60. 10.1080/14620316.2015.1123406. [Google Scholar]
- 26.Milivojevic J, Slatnar A, Mikulic-Petkovsek M, Stampar F, Nikolic M, Veberic R. The influence of early yield on the accumulation of major taste and health-related compounds in black and red currant cultivars (Ribes Spp). J Agri Food Chem. 2012;60:2682–91. 10.1021/jf204627m. [DOI] [PubMed] [Google Scholar]
- 27.Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, Waugh R. Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato. Mol Breeding. 1997;3:127–36. [Google Scholar]
- 28.Norman PE, Tongoona P, Shanahan PE. Determination of interrelationships among agr-morphological traits of yams (Discorea spp.) using correlation and factor analyses. J Appl Biosci. 2011;45:3059–70. [Google Scholar]
- 29.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysisin Excel. Population genetic software for teaching andresearch—an update. Bioinformatics. 2012;28(19):2537–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinar H, Yahya HN, Ercişli S, Coskun OF, Yaman M, Turgunbaev K, Uzun A. Molecular characterization of barberry genotypes from Turkey and Kyrgyzstan. Erwerbs Obstbau. 2021;63:403–7. 10.1007/s10341-021-00599-x. [Google Scholar]
- 31.Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breeding. 1996;2:225–38. [Google Scholar]
- 32.Rezaei M, Ebadi A, Reim S, Fatahi R, Balandary A, Farrokhi N, Hanke MV. Molecular analysis of Iranian seedless barberries via SSR. Scientia Horti. 2011;129:702–9. 10.1016/j.scienta.2011.05.021. [Google Scholar]
- 33.Rezaei M, Sarkhosh A, Balandari A. Characterization of valuable indigenous barberry (Berberis sp.) germplasm by using multivariate analysis. Int J Fruit Sci. 2020;20:1–19. 10.1080/15538362.2018.1555508. [Google Scholar]
- 34.Safamanesh B, Esmaeilzadeh Bahabadi S, Izanloo A. Investigation of genetic variation in Berberis vulgaris using ISSR and SSR molecular markers. J Cell Mol Res. 2017;9:23–34. 10.22067/jcmr.v9i1.62712. [Google Scholar]
- 35.Sasikumar JM, Maheshu V, Smilin AG, Gincy MM, Joji C. Antioxidant and antihemolytic activities of common Nilgiri barberry (Berberis Tinctoria Lesch.) From south India. Int Food Res J. 2012;19:1601–7. [Google Scholar]
- 36.Singleton VL, Rossi JA. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am J Enol Viticult. 1965;16:144–58. 10.5344/ajev.1965.16.3.144. [Google Scholar]
- 37.Sodagar N, Bahrami AR, Memariani F, Ejtehadi H, Vaezi J, Khosravi AR. Biosystematic study of the genus Berberis L. (Berberidaceae) in Khorassan, NE Iran. Plant Syst Evol. 2012;298:193–203. 10.1007/s00606-011-0537-9. [Google Scholar]
- 38.Sümbül A, Yildiz E, Sabir A, Nadeem MA. Investigation of genetic diversity among autochthonous grape cultivars grown in Türkiye using molecular primers. Genet Res Crop Evol. 2024;71:3507–20. 10.1007/s10722-024-01861-3. [Google Scholar]
- 39.Talebi S, Alizadeh M, Ghasem Nejhad A, Ramezanpour SS. Characterization of some wild Berberis sp. genotypes distributed in the northeast of Iran. J Plant Physiol Breed. 2022;12:135–52. [Google Scholar]
- 40.TUBIVES. (2024). Turkish Plants Data Service. Available online: http://www.tubives.com (28 June 2024).
- 41.Ullah N, Khan S, Khan A, Ahmad W, Shah Y, Ahmad L, Ullah I. A prospective pharmacological review of medicinal herbs, Cucumis melo and Berberis vulgaris, commonly used in the treatment of renal diseases in Pakistan. Acta Pol Pharm. 2015;72:651–4. [PubMed] [Google Scholar]
- 42.Weising K, Nybom H, Wolff K, Kahl G. DNA fingerprinting in plants: principles, methods, and applications. New York: CRC; 2005. 10.1201/9781420040043. [Google Scholar]
- 43.Yaman M, Uzun A. Morphological and molecular identification of hybrid individuals obtained by interspecies hybridization (Prunus armeniaca× Prunus salicina). Int J Agr Natur Sci. 2021;14:7–15. [Google Scholar]
- 44.Yeşilada E, Küpeli E. Berberis crataegina DC. root exhibits potent anti-inflammatory, analgesic and febrifuge effects in mice and rats. J Ethnopharmacol. 2002;79:237–48. 10.1016/S0378-8741(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 45.Yildiz H, Sengul M, Celik F, Hegedus A, Ercisli S, Tosun M. Some phytochemical and antioxidant characteristics of wild and cultivated blackberry (Rubus caucasicus) fruits. J Food Agric Environ. 2010;8:156–9. [Google Scholar]
- 46.Yildiz H, Ercisli S, Sengul M, Topdas EF, Beyhan O, Cakir O, Narmanlıoğlu HK, Orhan E. Some physicochemical characteristics, bioactive content and antioxidant characteristics of non-sprayed barberry (Berberis vulgaris L.) fruits from Turkey. Erwerbs Obstbau. 2014;56:123–9. 10.1007/s10341-014-0216-4. [Google Scholar]
- 47.Yildiz E, Pinar H, Uzun A, Yaman M, Sumbul A, Ercisli S. Identification of genetic diversity among Juglans regia L. genotypes using molecular, morphological, and fatty acid data. Genet Res Crop Evol. 2021;68:1425–37. 10.1007/s10722-020-01072-6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.