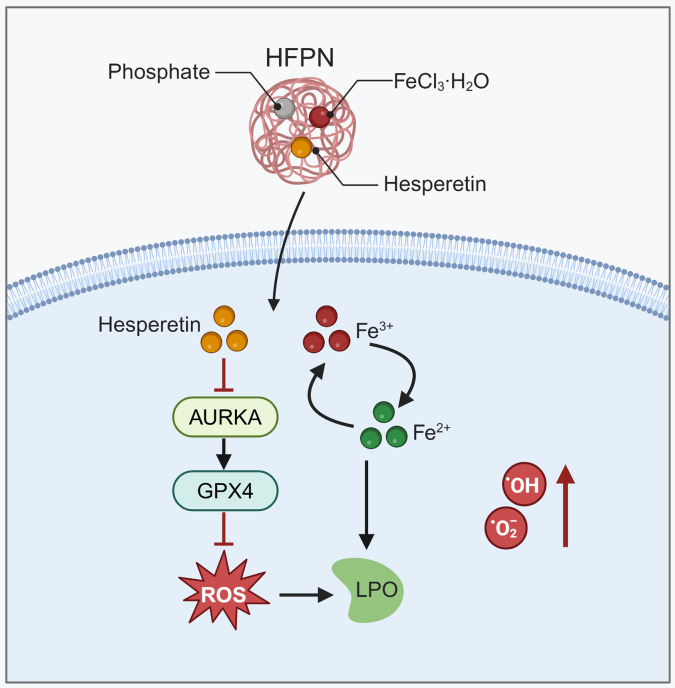
Abstract
Triple-negative breast cancer (TNBC) is an aggressive cancer type that lacks targeted treatment options. Ferroptosis, a novel therapeutic strategy, induces cell death by disrupting the oxidative-reductive balance. Hesperetin, a potential TNBC therapeutic drug, has unidentified regulatory targets. The objective of this study was to explore the potential targets of hesperetin in TNBC and investigate whether the nanocomposites carrier hesperetin-loaded ferroptosis-inducing nanocomposites (HFPN), which activates ferroptosis, can enhance the anti-tumor efficacy of hesperetin. Bioinformatics methods were employed to screen hesperetin targets in TNBC, and a molecular docking model between hesperetin and the core target aurora kinase A (AURKA) was successfully constructed. The stability and anti-tumor activity of HFPN were validated in cell and mouse models, including tumor suppression and increased radiation sensitivity. These results suggest that HFPN can regulate the core target AURKA in TNBC, disrupt tumor oxidative-reductive balance, promote ferroptosis in tumor cells, and ultimately enhance the effectiveness of radiation therapy for TNBC.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12951-024-02987-3.
Keywords: Triple-negative breast cancer, Ferroptosis, Hesperetin, AURKA, Redox homeostasis, Radiotherapy efficacy
Introduction
Breast cancer is a common malignant tumor, of which triple-negative breast cancer (TNBC) is a severe and deteriorating subtype [1–3]. Traditional radiotherapy has limited effectiveness in treating TNBC patients, hence the need to search for more effective treatment methods [4–6]. In recent years, ferroptosis, a novel type of cell death, has attracted attention and shown potential application value in anti-tumor therapy [7–9].
Hesperetin is a natural flavonoid compound found in citrus fruit peels with multiple biological activities [10–12]. Previous studies have shown that hesperetin exhibits anti-proliferation, anti-invasion, and pro-apoptosis activities in various cancers [13–15]. However, the anti-tumor effects of hesperetin in TNBC and its mechanisms of action remain unclear [16, 17].
Iron plays an important role in tumor cells, including cell proliferation, redox homeostasis, and tumor development [18, 19]. Ferroptosis is a mechanism that induces cell death by regulating intracellular iron ion metabolism and oxidative reduction processes [20, 21]. Recent studies have shown that promoting ferroptosis in tumor cells may serve as a novel tumor treatment strategy [19].
This study aims to investigate the target of hesperetin in TNBC and explore the potential molecular mechanism of ferroptosis promoter nanocomposites loaded with hesperetin in enhancing the efficacy of TNBC radiotherapy. This research is expected to provide a novel strategy for TNBC treatment by promoting ferroptosis in tumor cells, thereby improving the efficacy of radiotherapy and enhancing the survival rate and quality of life for TNBC patients.
Through bioinformatics analysis and experimental validation, this study identified the candidate target AURKA associated with hesperetin and successfully synthesized the ferroptosis promoter nanocomposites HFPN. Experimental evidence demonstrates that HFPN can promote tumor cell ferroptosis and enhance the radiotherapy sensitivity of TNBC by regulating reactive oxygen species (ROS) accumulation and disrupting the tumor’s redox homeostasis. The results of this study have significant scientific and clinical implications, providing new therapeutic options for improving the efficacy of radiotherapy for TNBC patients. Further investigation into the mechanism of action of hesperetin and ferroptosis in TNBC holds promise for providing a basis for individualized treatment of TNBC and opening up new directions for breast cancer treatment strategies.
Results and discussion
Potential targets of hesperetin for TNBC regulation
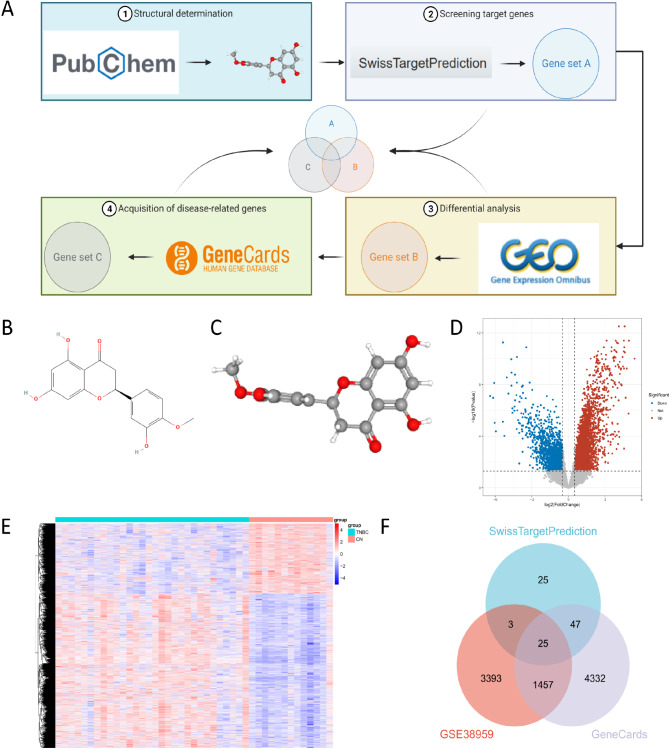
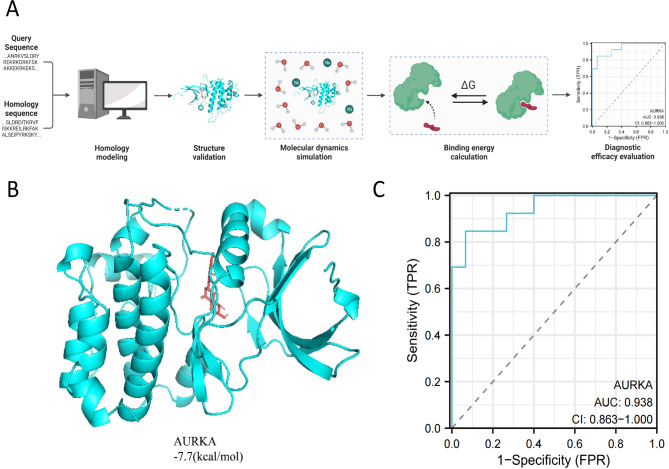
TNBC is a distinct type of breast cancer characterized by the absence of estrogen receptors (ER), progesterone receptors (PR), and human epidermal growth factor receptor 2 (HER2) expression. This implies that TNBC does not respond to hormone therapies targeting ER, PR, and HER2, such as tamoxifen or HER2-targeted drugs like trastuzumab [22]. Hesperetin, a naturally occurring flavonoid compound primarily found in citrus fruits, particularly oranges and grapefruits, has been reported to have potential as a therapeutic agent for TNBC [16]. In this study, we aim to identify potential targets of Hesperetin in TNBC and analyze its mechanism of action using molecular docking techniques. The workflow for this analysis is depicted in (Fig. 1A).
Fig. 1.
Screening for common targets between hesperetin and TNBC. Note: (A) The schematic diagram of the bioinformatics analysis workflow; (B) The 2D chemical structure of Hesperetin; (C) The 3D chemical structure of Hesperetin; (D) Volcano plot illustrating the differential gene expression in GSE38959 (upregulated genes: 3359; downregulated genes: 1519). Blue dots represent significantly downregulated genes, red dots represent significantly upregulated genes, and gray dots represent genes with non-significant differential expression; (E) Heatmap depicting the differential gene expression in GSE38959; (F) Venn diagram showing the intersection of genes retrieved from SwissTargetPrediction database, GSE38959, and GeneCards database
First, we determined the 2D and 3D chemical structures of hesperetin using the PubChem database (Fig. 1B-C). The SwissTargetPrediction server was then utilized to analyze and screen potential target genes of hesperetin, resulting in a total of 100 relevant genes. Additionally, we obtained 4,878 differentially expressed gene (DEGs) from the TNBC dataset GSE38959, including 3,359 upregulated DEGs and 1,519 downregulated DEGs (Fig. 1D-E). Through a search in the GeneCards database using TNBC as a keyword and setting a relevance score ≥ 1 as the screening criterion, we identified 5,861 genes associated with TNBC.
Using Venn analysis, we intersected the target genes from the SwissTargetPrediction server, the DEGs from the GSE38959 dataset, and the TNBC-associated genes from the GeneCards database, revealing 25 hesperetin target genes related to TNBC regulation (Fig. 1F). These genes include CA12, ABCG2, ESR1, MAOB, ABCC1, CBR1, MMP13, SRC, KLK2, CA2, AURKA, PGD, FUT7, STAT1, SQLE, PIM1, IGF1R, ODC1, PARP1, MAP4K4, BCL2, CHEK1, MMP3, KIT, and MAPKAPK2.
Hesperetin targets associated with protein kinase activity in TNBC
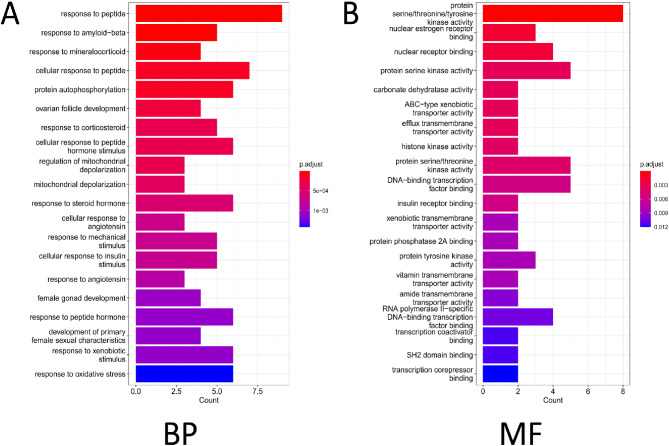
Further, gene ontology (GO) functional analysis was performed on the selected 25 candidate targets. The results of the GO functional analysis revealed that these candidates are involved in various BP, including “response to peptide,” “response to mineralocorticoid,” and “protein autophosphorylation” (Fig. 2A-B). In terms of MF, they are primarily enriched in “protein serine/threonine/tyrosine kinase activity,” “nuclear estrogen receptor binding,” and “nuclear receptor binding.” Protein kinases play a crucial role in cancer development and progression. Disruption of protein kinase activity or regulation can result in dysregulated control of cell growth, differentiation, migration, and survival, an important mechanisms underlying cancer [23]. The enrichment analysis suggests that these 25 candidate targets of hesperetin may be involved in TNBC processes through the regulation of protein kinase activity, making them potential targets for the anti-TNBC effect of hesperetin.
Fig. 2.
Enrichment analysis results for 25 genes. Note: (A) Bar chart depicting the GO functional analysis of candidate anti-TNBC targets at the BP level. The colors represent the p-values of the enrichment analysis. (B) Bar chart illustrating the GO functional analysis of candidate anti-TNBC targets at the MF level. The colors represent the P of the enrichment analysis
AURKA and FUT7: potential hesperetin targets for TNBC therapy
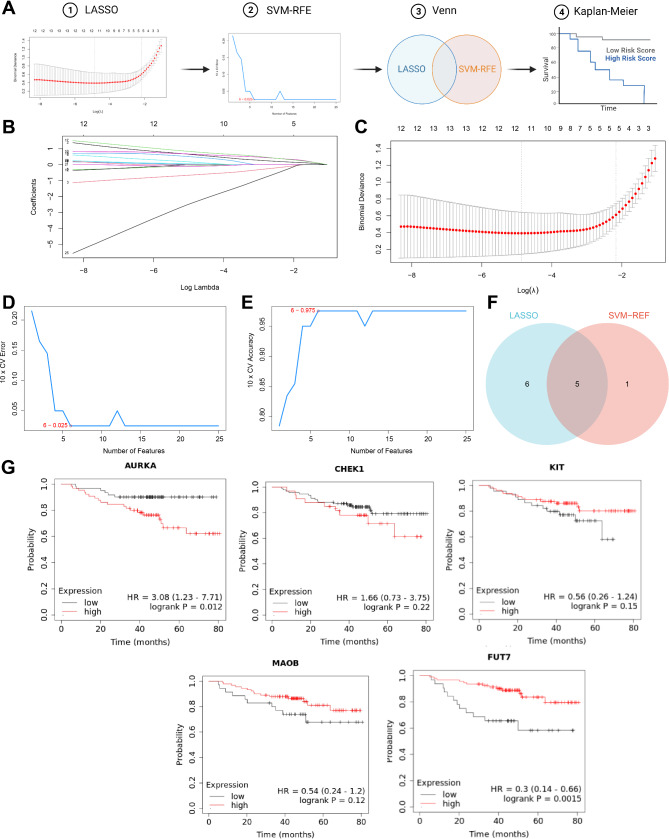
Among the 25 identified candidate targets, in order to further select more meaningful genes, a combination of machine learning and survival curve analysis was employed. The analytical process for this section can be seen in Fig. 3A. By applying the least absolute shrinkage and selection operator (LASSO) regression, 11 candidate targets were identified (Fig. 3A-B). Subsequently, the support vector machine-recursive feature elimination (SVM-RFE) algorithm was used to screen out 6 candidate targets for further analysis (Fig. 3C-E), with an intersection of 5 remaining candidate targets (Fig. 3F). The impact of the expression of these 5 candidate targets on survival in TNBC patients was analyzed using the Kaplan-Meier Plotter database. The results of the survival curve demonstrated a significant influence on patient survival time due to the expression changes in AURKA and FUT7 (Fig. 3G). The protein encoded by AURKA is a cell cycle-regulating kinase that exists in the centrosome of interphase cells and the spindle of mitotic cells. It is involved in the formation and/or stability of spindle pole microtubules during chromosome separation. This gene may play a role in the occurrence and development of tumors, and numerous studies have reported its relationship with breast cancer [24, 25]. However, there are fewer reports on FUT7 and breast cancer; thus in subsequent analysis, we will focus on exploring the potential of AURKA as a target for hesperetin therapy in TNBC.
Fig. 3.
Further selection of core targets using machine learning and survival curve analysis. Note: (A) The schematic of the bioinformatics analysis process; (B) Coefficients of different genes vary with different λ values; (C) The optimal parameter (lambda) identified through cross-validation of LASSO regression analysis; (D-E) Important features recognition of the 25 genes based on the SVM-RFE algorithm; F: 5 candidate targets obtained from the results of LASSO and SVM-RFE algorithms; G: Survival curve plots for the five candidate targets in TNBC
Stable binding and promising diagnostic performance of AURKA and hesperetin
We further carried out docking analysis of candidate proteins and Hesperetin using software such as AutoDockTools 1.5.6 and Vina 1.1.2. The analytical workflow for this section is depicted in Fig. 4A. The docking results of candidate proteins with hesperetin were visualized in 3D (Fig. 4B), providing a clear representation of the binding mode between the target protein receptor and the compound, as well as the interactions with surrounding amino acid residues.
Fig. 4.
Molecular docking of candidate targets with hesperetin and their ROC analysis. Note: (A) A schematic diagram of the bioinformatics analysis workflow; (B) Docking of AURKA with the Hesperetin molecule, where blue represents the receptor’s secondary structure, and red represents the small molecule Hesperetin structure; (C) Diagnostic performance of candidate targets
When the binding energy is < 0 kJ/mol, it indicates spontaneous binding and interaction between the protein and the molecule. Moreover, lower binding energy corresponds to a more stable molecular conformation [26]. The results demonstrate that AURKA shows a binding free energy of -7.7 kcal/mol, implying a relatively stable conformation when bound to hesperetin. Additionally, the diagnostic performance of candidate targets was further evaluated using ROC analysis, where a higher AUC value signifies better diagnostic performance. The evaluation of AURKA reveals diagnostic performance (Fig. 4C).
Nanocomposites-mediated hesperetin delivery promotes ferroptosis
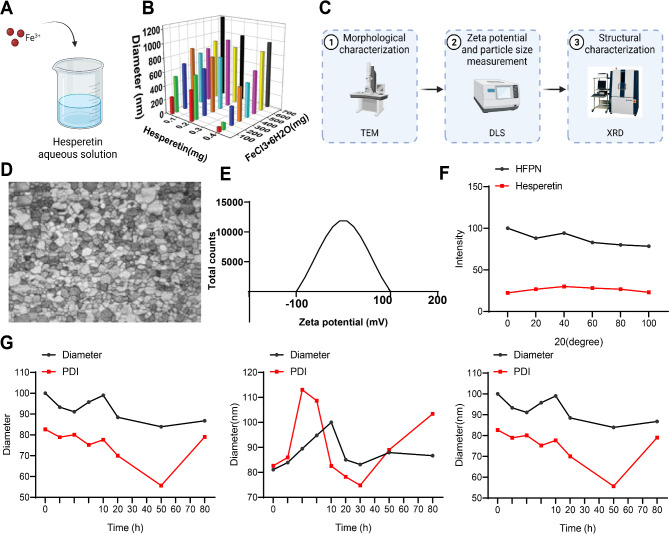
Due to their biocompatibility, nanocomposites have gained approval from the USA Food and Drug Administration (FDA) in most materials used for nanocomposites research [27]. In this study, we prepared ferroptosis-inducing nanocomposites loaded with hesperetin through a synergistic self-assembly approach. Specifically, FeCl3•6H2O was added to an aqueous solution of hesperetin at room temperature (Fig. 5A). The size of the nanocomposites was optimized using Taguchi orthogonal experiments, employing an L24 orthogonal array design with two different operational parameters: Hesperetin concentration and FeCl3•6H2O concentration. The average size of 24 samples was measured using DLS, with the nanocomposites with an average size of (68.43 ± 0.47) nm selected as the optimized formulation and named HFPN for further investigation (Fig. 5B).
Fig. 5.
Characterization of HFPN. Notes: (A) Schematic illustration of the preparation of HFPN; (B) DLS measurement of the particle size of nanocomposites with different concentrations of Hesperetin and FeCl3•6H2O; (C) Schematic diagram of the experimental procedure for the physical characterization of nanocomposites; (D) TEM observation of the morphology of HFPN (200 nm); (E) Zeta potential of HFPN; (F) Structure characterization of HFPN nanomaterials using powder X-ray diffraction spectroscopy; (G) DLS measurement of the particle size distribution and surface charge of HFPN in deionized water, PBS, and DMEM. The experiment was repeated three times
Subsequently, the nanocomposites were physically characterized (Fig. 5C). Transmission electron microscopy (TEM) revealed that the HFPN exhibited a shuttle-like shape with an average length of (66.39 ± 22.17) nm and a width of (22.84 ± 4.69) nm (Fig. 5D). HFPN exhibited a slight negative surface charge (zeta potential of -0.28 ± 0.021 mV) (Fig. 5E). HFPN displayed consistent particle size distribution and polymer dispersity index (PDI) in deionized water, phosphate-buffered saline (PBS), and DMEM, sustaining for over 72 h (Fig. 5G). The powder X-ray diffraction pattern showed no obvious diffraction peaks, indicating a well-dispersed amorphous structure of the assembled HFPN (Fig. 5F). These results validated its colloidal stability. These results demonstrate the successful preparation of ferroptosis-inducing nanocomposites loaded with Hesperetin, which exhibit favorable stability and dispersity.
HFPN-mediated generation of ROS for ferroptosis induction
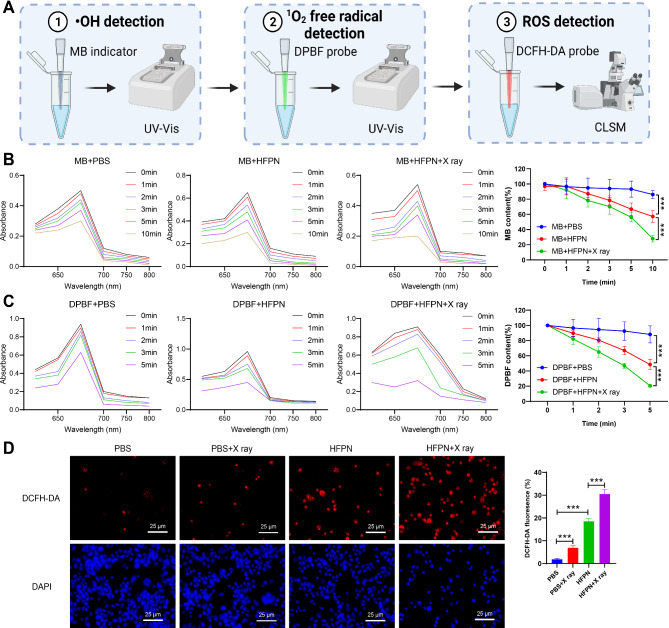
Previous studies have indicated that Fe3+ can induce Fenton-like reactions in tumor cells, leading to the generation of highly toxic •OH. To investigate the efficacy of HFPN in generating •OH, we utilized methylene blue (MB) as an indicator to detect •OH production (Fig. 6A). The results demonstrated that there was no significant decrease in absorbance in the PBS control group, while the absorbance of MB in HFPN continuously decreased, with a more pronounced decline in the HFPN + X-ray solution (Fig. 6B). This indicates that HFPN can generate •OH and X-ray irradiation can enhance the production of •OH by HFPN.
Fig. 6.
Performance evaluation of HFPN and its impact on ROS production. Note: (A) The experimental procedure; (B) Absorbance spectra of MB in different cell groups; (C) Changes in absorbance values of MB in different cell groups; (D) Visualization of ROS in MDA-MB-231 cells in different groups using CLSM, with red fluorescence (DHE) representing ROS. *** indicates a significant difference between the two groups with P < 0.001. The experiment was repeated three times
Furthermore, we employed the DPBF probe to monitor the excessive generation of 1O2 induced by the combined irradiation of HFPN and X-rays in different media. With increasing time, there was a gradual decrease in absorption at 410 nm for DPBF, indicating that HFPN can generate 1O2 radicals, and X-ray irradiation can enhance the production of 1O2 by HFPN (Fig. 6C). We further utilized the DHE probe to measure the levels of ROS within MDA-MB-231 cells. The red fluorescence intensity increased gradually in the PBS group, PBS + X-ray group, HFPN group, and HFPN + X-ray group, indicating an increase in ROS generation (Fig. 6D). Together, these results provide comprehensive evidence of the potential of HFPN in effective ferroptosis therapy, which may be mediated by the excessive generation of ROS to promote ferroptosis.
HFPN-enhanced radio sensitization in breast cancer cells
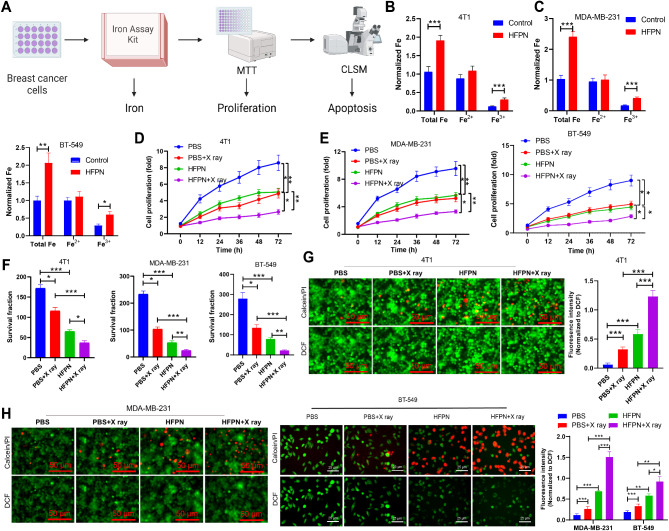
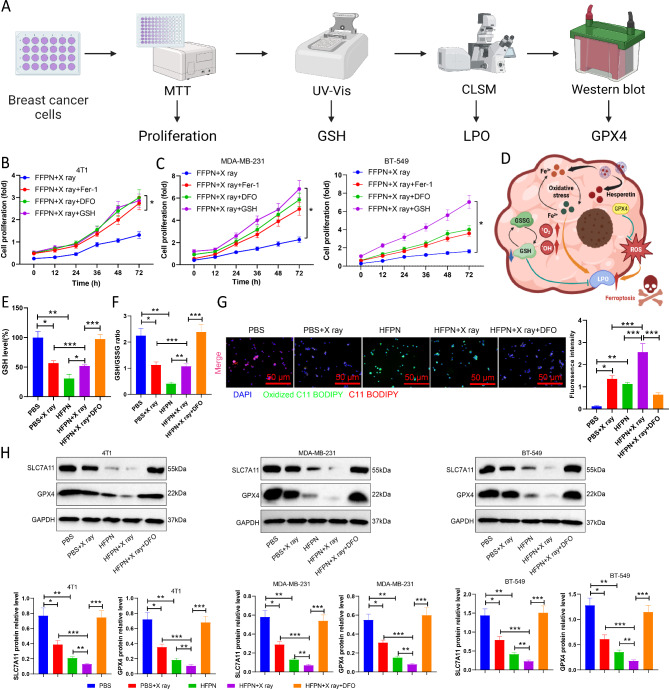
Due to its outstanding ability to generate ROS, we further evaluated the anti-cancer effect and radiosensitizing effect of HFPN in breast cancer cells (4T1 and MDA-MB-231). The analysis process for this section is illustrated in Fig. 7A. Cellular uptake was assessed by measuring intracellular iron concentration using an iron assay kit. After co-incubation with HFPN for 4 h, both 4T1, MDA-MB-231 and BT-549 cells showed increased iron accumulation compared to cells treated with PBS (Fig. 7B-C). Furthermore, cell proliferation viability was determined using the MTT assay. The results indicated a significant decrease in cell viability for both 4T1, MDA-MB-231 and BT-549 cells in the HFPN group compared to the PBS group. Additionally, the viability of 4T1, MDA-MB-231 and BT-549 cells treated with a combination of X-rays and HFPN was significantly lower than that of cells treated with HFPN or X-rays alone (Fig. 7D-E), confirming the effective radio-sensitizing property of HFPN. Colony formation experiments also revealed that post X-ray irradiation, HFPN significantly inhibited colony formation in 4T1, MDA-MB-231 and BT-549 cells compared to treatment with HFPN or X-rays alone (Fig. 7F). The results of live/dead staining showed a significant increase in red fluorescence (indicating dead cells) and decreased green fluorescence (indicating live cells) in the HFPN group compared to the PBS + X-ray group. Moreover, the HFPN + X-ray group exhibited even more pronounced red fluorescence and a decrease in green fluorescence compared to the HFPN and X-ray alone groups (Fig. 7G-H).
Fig. 7.
Impact of HFPN on the efficacy of radiation therapy in TNBC. Note: (A) Experimental flowchart illustrating the effect of HFPN on TNBC radiotherapy efficacy; (B) Measurement of intracellular iron concentration in breast cancer 4T1 cells using an iron assay kit; (C) Measurement of intracellular iron concentration in breast cancer MDA-MB-231 and BT-549 cells using an iron assay kit; (D) Assessment of cell proliferation in 4T1 cells using the MTT assay; (E) Assessment of cell proliferation in MDA-MB-231 and BT-549 cells using the MTT assay; (F) Colony formation assay to evaluate colony formation in 4T1, MDA-MB-231 and BT-549 cells; (G) CLSM observation of the distribution of live/dead cells in 4T1 cells; (H) CLSM observation of the distribution of live/dead cells in MDA-MB-2311 and BT-549 cells. * indicates a statistically significant difference between the two groups (P < 0.05), ** indicates a statistically significant difference between the two groups (P < 0.01), *** indicates a statistically significant difference between the two groups (P < 0.001). The experiments were repeated three times
Furthermore, we conducted a similar evaluation of the impact of HFPN on normal breast epithelial cells MCF 10 A. The assessment involved measuring intracellular iron levels using an iron assay kit to evaluate cellular uptake and assessing cell proliferation vitality by the MTT method. Our results indicated that after a 4-hour co-incubation of HFPN with MCF 10 A cells, there was no significant effect on intracellular iron accumulation than cells treated with PBS. Moreover, the vitality of MCF 10 A cells in the HFPN group showed no significant change compared to the PBS group (Fig. S1A-B). Additionally, colony formation experiments revealed that post X-ray irradiation, there was no change in colony formation of MCF 10 A cells treated with HFPN, in contrast to cells treated separately with HFPN and X-ray (Fig. S1C). Cell viability staining results showed no observable changes in red fluorescence in the HFPN + X-ray group compared to the HFPN + X-ray group (Fig. S1D).
HFPN-induced ferroptosis through ROS accumulation and disruption of redox homeostasis
The previous findings have confirmed the potential of HFPN in effective ferroptosis treatment. To further investigate whether cells undergo cell death through the mechanism of ferroptosis, we pre-treated the cells in HFPN combined with X-ray irradiation with the iron-regulatory protein-1 (Fer-1), iron chelator (DFO), and GSH inhibitor to inhibit ferroptosis. The relative cell viability of the different groups was then assessed (Fig. 8A). The results reveal that the cell survival rates of 4T1, MDA-MB-231, and BT-549 cells significantly increased after pretreatment with Fer-1, DFO, and GSH, as compared to the group treated with HFPN + X-ray (Fig. 8B-C), further indicating the crucial role of ferroptosis in HFPN-mediated cell death.
Fig. 8.
Impact of HFPN on redox homeostasis. Note: (A) Experimental flowchart of HFPN affecting redox homeostasis; (B) Proliferation of 4T1 cells detected by MTT assay; (C) Proliferation of MDA-MB-231 and BT-549 cells detected by MTT assay; (D) Schematic diagram of the potential mechanism by which HFPN mediates iron death; (E) Depletion of GSH in 4T1 cells in each group; (F) GSH/GSSG ratio in 4T1 cells in each group; (G) Expression of LPO in 4T1 cells in each group observed by CLSM; (H) Expression of GPX4 and SLC7A11 proteins in 4T1 cells, MDA-MB-231, and BT-549 cells detected by Western blot. * denotes a significant difference between the two groups with P < 0.05, ** denotes a significant difference with P < 0.01, *** denotes a significant difference with P < 0.001. The experiments were repeated three times
Based on previous experiments suggesting that HFPN promotes excessive ROS production, we propose that HFPN disrupts the redox homeostasis by depleting GSH and inactivating glutathione peroxidase 4 (GPX4), leading to lipid peroxidation enzyme family (LPO) accumulation and ultimately promoting cell ferroptosis (Fig. 8D). Different groups of cells were assessed for their levels of GSH, revealing a significant reduction in GSH levels in cells treated with HFPN and HFPN combined with X-ray compared to cells treated with PBS. Pre-treatment with the iron chelator DFO significantly increased the levels of cellular GSH (Fig. 8E) and the GSH/GSSG ratio (Fig. 8F). Further investigation using fluorescent probes to study the levels of LPO showed intense green fluorescence in 4T1 cells in the HFPN group and HFPN + X-ray group after 24 h of incubation; however, pre-treatment with the iron chelator DFO resulted in a significant decrease in LPO fluorescence signal (Fig. 8G). Western blot analysis revealed that GPX4 and solute carrier family 7 (SLC7A11) protein expression in 4T1, MDA-MB-231, and BT-549 cells in the HFPN and X-ray groups was significantly decreased compared to the PBS group. Moreover, the expression of GPX4 and SLC7A11 proteins in cells in the HFPN + X-ray group was significantly lower than in the HFPN and X-ray groups, while in the HFPN + X-ray + DFO group, the expression of GPX4 and SLC7A11 proteins was significantly higher than in the HFPN + X-ray group (Fig. 8H). These experimental findings indicate that HFPN can disrupt the redox homeostasis crucial for tumor survival by inducing GSH depletion and LPO accumulation through the excessive accumulation of ROS.
Inhibition of AURKA expression by HFPN disrupts redox homeostasis
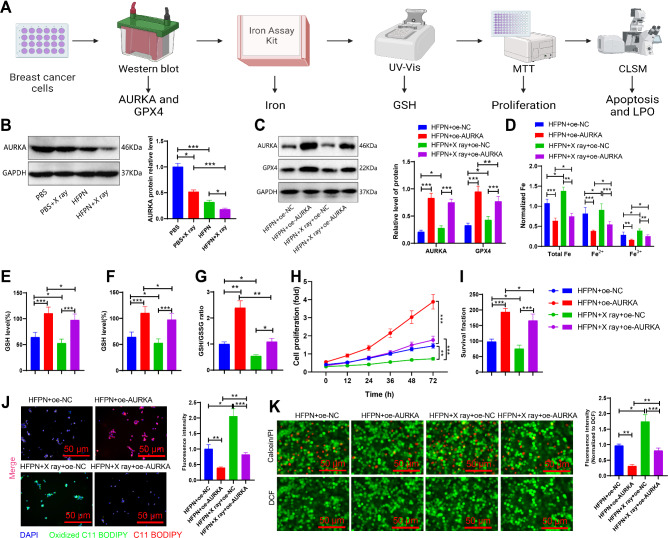
We hypothesize that HFPN may suppress the expression of the AURKA factor, thereby affecting redox homeostasis. To further validate this hypothesis, we performed the analysis as outlined in Fig. 9A. Western blot analysis revealed a significant decrease in AURKA protein expression in the cells treated with HFPN compared to the PBS group (Fig. 9B). To confirm that HFPN affects redox homeostasis by inhibiting AURKA expression, we subjected 4T1 cells to AURKA overexpression pretreatment and assessed the expression of AURKA and GPX4 proteins. Cells with AURKA overexpression pretreatment demonstrated a significant increase in AURKA and GPX4 protein expression (Fig. 9C). Intracellular iron concentration was determined using the iron assay kit. The results showed that cells treated with HFPN + oe-AURKA exhibited reduced iron accumulation compared to the HFPN + oe-NC group; a similar reduction was observed in the cells treated with HFPN + X ray + oe-AURKA compared to the HFPN + X ray + oe-NC group (Fig. 9D). The results of the measurements of the levels of GSH, GSH/GSSG ratio, and GSH-PX activity in different cell groups indicate that cells pre-treated with the combination of AURKA overexpression and HFPN exhibit significantly higher levels of GSH and GSH/GSSG ratio compared to cells treated with HFPN alone. In addition, the cells in the HFPN + X-ray + oe-AURKA group show markedly higher levels of GSH and GSH/GSSG ratio than the cells in the HFPN + X-ray + oe-NC group, while the GSH-PX activity shows an opposite trend (Fig. 9E-G). Methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay results also demonstrated that cells subjected to AURKA overexpression pretreatment exhibited higher cell viability compared to cells without such pretreatment (Fig. 9H). Colony formation assay revealed that cells in the HFPN + oe-AURKA group exhibited a significant increase in colony formation compared to the HFPN + oe-NC group; a similar increase was observed in the cells treated with HFPN + X ray + oe-AURKA compared to the HFPN + X ray + oe-NC group (Fig. 9I). Cell viability staining results showed a decrease in red fluorescence and an increase in green fluorescence in the HFPN + oe-AURKA group compared to the HFPN + oe-NC group; a similar trend was observed in the cells treated with HFPN + X ray + oe-AURKA compared to the HFPN + X ray + oe-NC group (Fig. 9J). The LPO probe assay showed a significant reduction in LPO fluorescence signal in the cells treated with HFPN + oe-AURKA compared to the HFPN + oe-NC group; a similar reduction was observed in the cells treated with HFPN + X ray + oe-AURKA compared to the HFPN + X ray + oe-NC group (Fig. 9K).
Fig. 9.
Regulation of AURKA factor expression by HFPN. Note: (A) Experimental flowchart illustrating the effect of HFPN on AURKA expression and disruption of redox homeostasis; (B) Western blot analysis of AURKA protein expression in different groups of 4T1 cells; (C) Western blot analysis of AURKA and GPX4 protein expression in different groups of 4T1 cells; (D) Iron concentration measurement in MDA-MB-2311 breast cancer cells using the iron assay kit; (E) Depletion of GSH in 4T1 cells of each group; (F) GSH/GSSG ratio in 4T1 cells of each group; (G) GSH-PX activity in 4T1 cells of each group; (H) Proliferation of 4T1 cells detected by MTT assay; (I) Colony formation experiment assessing colony formation of 4T1 cells; (J) CLSM observation of the distribution of live/dead cells in 4T1 cells; (K) CLSM observation of LPO expression in 4T1 cells of each group. * indicate statistical significance at P < 0.05, ** at P < 0.01, and *** at P < 0.001 when comparing between two groups. The experiments were repeated three times
Consistent results were obtained in MDA-MB-2311 cells as in 4T1 cells (Fig. S2). Taken together, these findings suggest that HFPN effectively inhibits the expression of AURKA and affects the redox homeostasis.
HFPN exhibits targeted anticancer efficacy and enhances radiotherapy in breast cancer models
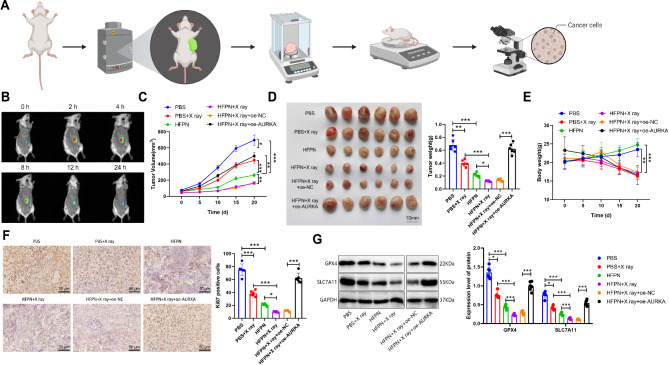
To investigate the in vivo anti-tumor efficacy and targeting ability of HFPN, we established a subcutaneous tumor model in mice using 4T1 cells. The analytical procedure for this section is shown in Fig. 10A. We utilized the in vivo imaging system and observed a strong fluorescence signal at the tumor site 2 h after intravenous injection of HFPN, as depicted in Fig. 10B. This suggests that HFPN can rapidly accumulate at the tumor site.
Fig. 10.
Effects of HFPN in the breast cancer model mice. Note: (A) Experimental flowchart illustrating the targeted anti-cancer effects of HFPN and enhancement of radiotherapy efficacy; (B) In vivo distribution of HFPN in mice observed using the Maestro in vivo fluorescence imaging system at 0, 2, 4, 8, 12, and 24 h after injection, with a total of 6 mice; (C) Tumor volume changes over a 20-day period in each group of mice; (D) Tumor dissection images and statistical analysis of tumor weight in each group of mice; (E) Body weight changes over a 20-day period in each group of mice; (F) Immunohistochemical detection of positive expression of the proliferation-related protein Ki67 in tumor tissues; (G) Western blot analysis of the expression of ferroptosis-related proteins in tumor tissues of each group of mice. * indicates a significant difference between the two groups with P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001. Each group in B-D consisted of 6 mice
Furthermore, we further evaluated the in vivo anti-tumor efficacy of HFPN by assessing tumor volume and weight. Compared to the PBS group, both the PBS + X-ray and HFPN groups displayed significant reductions in tumor volume and weight. Additionally, compared to the PBS + X-ray group, the HFPN + X-ray group also exhibited a significant decrease in tumor volume and weight. Conversely, the HFPN + X-ray + oe-NC group showed a significant increase in tumor volume and weight compared to the HFPN + X-ray + oe-AURKA group, as shown in Fig. 10C-D. Changes in mouse body weight were also monitored, revealing a decrease in the PBS + X-ray group and no significant impact on the body weight of mice in the HFPN group, as presented in Fig. 10E.
Immunohistochemical (IHC) analysis was performed to assess the expression of the proliferation marker Ki67 in tumor tissues from each group of mice. The results demonstrated a significant decrease in the number of Ki67-positive cells in the tumor tissues of both the PBS + X-ray and HFPN groups compared to the PBS group. Additionally, the HFPN + X-ray group exhibited a significant decrease in Ki67-positive cells compared to the PBS + X-ray group. Notably, the HFPN + X-ray + oe-AURKA group displayed a significant increase in the number of Ki67-positive cells compared to the HFPN + X-ray + oe-NC group, as depicted in Fig. 10F.
Western blot analysis was conducted to evaluate the expression of the ferroptosis-related proteins GPX4 and SLC7A11 in the tumor tissues of each group of mice. Compared to the PBS group, the X-ray, HFPN, and HFPN + X-ray groups displayed a significant decrease in the expression of GPX4 and SLC7A11 proteins. The HFPN + X-ray group exhibited lower expression of GPX4 and SLC7A11 proteins compared to the HFPN group, while the HFPN + X-ray + oe-AURKA group showed even lower expression compared to the HFPN + X-ray + oe-NC group, as shown in Fig. 10G. These results collectively demonstrate that HFPN exhibits good anti-tumor efficacy and targeting ability, and through AURKA inhibition, promotes ferroptosis, thereby effectively enhancing the radiation therapy efficacy in a mouse model of breast cancer.
Conclusion
TNBC is a highly aggressive and malignant subtype of breast cancer, and current radiotherapy methods have certain limitations in treatment [28–30]. Therefore, it is of great significance to identify new therapeutic targets [31–33]. Hesperetin, a natural compound, has the potential to play an important role in the treatment of TNBC [34–36].
Previous studies have shown that hesperetin has anti-tumor effects in other types of cancer, but its specific mechanisms in TNBC are still unclear [37, 38]. Further investigation into the mechanisms of action between hesperetin and TNBC could provide a better understanding of its role [17, 39].
In this study, we employed bioinformatics methods to identify potential targets associated with TNBC regulation. Compared to previous methods, we utilized the SwissTargetPrediction and GeneCards databases to improve the accuracy and quantity of targets. These potential targets will provide important clues for further research on the mechanism of action of hesperetin.
Through machine learning and survival curve analysis, we identified AURKA as a key target of Hesperetin. Our findings show certain discrepancies compared to previous studies. Research indicates that AURKA is associated with various tumors. AURKA acts as an inhibitor of erastin-induced iron death in meningiomas [40]; AURKA inhibition significantly induces apoptosis and iron death in Ewing’s sarcoma (ES) cells and reduces tumor formation in vivo [41]. Furthermore, loss of Aurka in intestinal epithelial cells leads to dysbiosis and elevated levels of SCFAs (particularly acetates) in the gut, resulting in AKT activation and regulation of lipid metabolism genes, thereby promoting age-induced obesity [42]. Signaling pathway analysis reveals that AURKA overexpression impedes oxidative stress and subsequent lipid peroxidation both in vitro and in vivo, with alterations in lipid peroxidation biomarkers (lipid ROS, GPX4, SLC7A11, ALOX5, and ASLC4) suggesting an interaction between diabetes-associated limb ischemia-induced iron death and AUKRA [43]. Iron death is an iron-dependent regulated cell death characterized by uncontrolled lipid peroxidation and membrane damage. Inhibiting iron death involves two main surveillance mechanisms: one mediated by GPX4, which catalyzes the reduction of phospholipid peroxides; GPX4 plays a major role in blocking iron death by eliminating phospholipid hydroperoxides [44]. The other mechanism is mediated by enzymes such as FSP1, generating metabolites with free radical scavenging antioxidant activity [45, 46]. Further validation of this regulatory relationship is needed, along with an exploration of its clinical potential in the treatment of TNBC.
We successfully synthesized a nanocomplex called HFPN and found that it has outstanding characteristics and advantages. Compared to other nanocomplexes, HFPN has a more pronounced effect on TNBC cells. This provides strong support for further studying the combination application of hesperetin and HFPN.
Our experimental results demonstrate that treating TNBC cells with HFPN significantly reduces cell viability, induces excessive ROS accumulation, and inhibits cell uptake. Compared to other studies, we found that HFPN has better effects in inhibiting TNBC cell growth. This provides stronger evidence for the application of HFPN in TNBC treatment.
In mouse tumor experiments, we observed that HFPN accumulates well at the tumor site, significantly suppresses tumor proliferation, and regulates the expression of ferroptosis-related proteins. In conclusion, HFPN is a new type of nanocomplex that combines hesperetin and ferroptosis promoter, exhibiting good colloidal stability, safety, and targeting ability. Compared to other radiosensitizers, HFPN has similar or even better effects in enhancing radiotherapy efficacy.
The scientific value of this study lies in demonstrating the potential therapeutic effect of the ferroptosis promoter nanocomplex loaded with hesperetin in TNBC through bioinformatics, cell experiments, and animal experiments. Firstly, we screened target genes of hesperetin, a flavonoid compound, using bioinformatics methods combined with the intersection of TNBC-related DEGs and TNBC-related genes to determine candidate targets associated with TNBC regulation. Then, using machine learning algorithms and survival curve construction, we confirmed the potential of hesperetin in regulating AURKA in TNBC processes. Experimental testing and animal experiments demonstrated that the synthesized ferroptosis promoter nanocomplex HFPN exhibited good stability and dispersion, promoted excessive ROS accumulation, inhibited AURKA, disrupted tumor redox homeostasis, and improved radiotherapy sensitivity. Finally, through mouse tumor experiments, we demonstrated that HFPN rapidly accumulates at the tumor site, inhibits tumor proliferation, and suppresses the expression of ferroptosis-related GPX4 protein, thereby improving radiotherapy efficacy.
However, this study still has some limitations. Firstly, the use of bioinformatics methods to screen target genes of hesperetin is a preliminary screening approach that requires further experimental validation to confirm the accuracy and feasibility of these candidate targets. Secondly, this study only involved in vitro and mouse experiments, so more preclinical studies and clinical trials are needed to verify its effectiveness and safety in humans. Moreover, this study only focused on the radiotherapy efficacy of TNBC and further research is needed to explore its application in other cancer treatments. This study provides clues for further exploring the anti-tumor mechanism of hesperetin as a ferroptosis promoter nanocomplex. Future research can investigate the combined application of hesperetin with other compounds to explore more effective treatment strategies. In addition, researchers can further optimize the preparation methods of the ferroptosis promoter nanocomplex to improve its stability and dispersion for better application in clinical practice. Additionally, combination applications with other treatment methods, such as chemotherapy and immunotherapy, could be considered to improve treatment outcomes. In conclusion, this study provides new ideas and methods for the treatment of TNBC, which is of great significance for improving the efficacy of radiotherapy and has potential clinical application prospects.
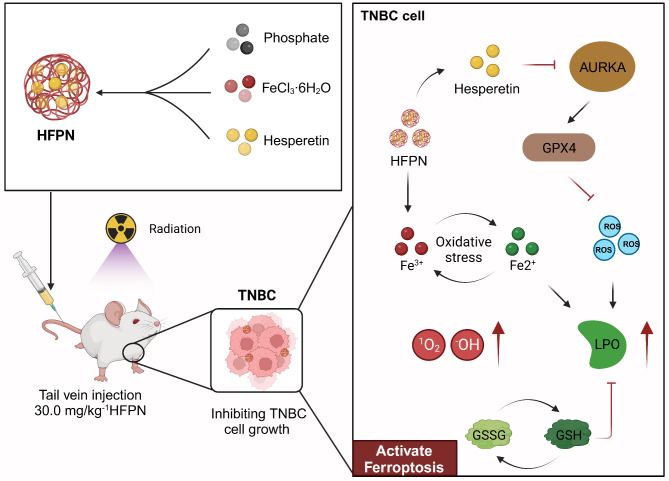
In summary, this study discovered the regulatory relationship between hesperetin and AURKA, as well as the anti-tumor effects of HFPN on TNBC and its role in enhancing radiotherapy efficacy. These findings have important scientific and clinical value for the treatment of TNBC (Fig. 11). However, this study still has limitations, such as the incomplete understanding of the mechanisms of hesperetin and the challenges faced by HFPN in clinical applications. Future research can further explore the regulatory mechanisms of hesperetin, optimize the design of HFPN, and conduct more preclinical studies to expand its clinical potential.
Fig. 11.
Molecular mechanism diagram illustrating the inhibition of AURKA factor and promotion of tumor cell ferroptosis by ferroptosis-inducing nanocomplex loaded with hesperetin to improve the therapeutic efficacy of radiation treatment in TNBC
Experimental section
Hesperetin target gene selection
The 2D and 3D chemical structures of hesperetin (compound CID: 72281) were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The 3D chemical structure was saved in “SDF” format and uploaded to the SwissTargetPrediction database (http://www.swisstargetprediction.ch/). “Homo sapiens” was selected as the species to obtain potential targets for hesperetin.
GEO chip data analysis
The TNBC expression profile dataset, GSE38959, was acquired from the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds). This dataset includes data from 30 TNBC cells and 13 normal mammary duct cells. Differential analysis was performed using the “limma” package in the R language [47], with normal mammary duct cells as the control. The probe with the highest expression level was selected, and log2 was transformed. The criteria for selecting differential genes were set as |log2(FoldChange)| > 0.5 and adj.P < 0.05.
Candidate target selection
TNBC-related target genes were obtained from the GeneCards database (https://www.genecards.org/). By inputting the search term “triple negative breast cancer,” a list of relevant genes was obtained. Subsequently, a Venn analysis was performed to integrate the potential targets of hesperetin, the differential gene analysis results of GSE38959, and the search results from the GeneCards database to identify candidate targets [48, 49].
Functional enrichment analysis of candidate targets
The “ClusterProfiler” package in the R language was used for GO enrichment analysis of the candidate targets, including biological processes (BP) and molecular functions (MF) [50]. Furthermore, the major impact of potential targets on cellular functions and signaling pathways was examined.
Machine learning
LASSO and SVM-RFE were employed as machine learning algorithms for further target selection. LASSO is a feature selection and regularization method applied in supervised learning tasks such as linear regression and logistic regression. It utilizes L1 regularization to compress model parameters to zero, thereby achieving sparsity in feature selection. In this way, essential features can be effectively identified and retained [51]. On the other hand, SVM-RFE combines the principles of support vector machines (SVM) and recursive feature elimination (RFE). It is a feature selection technique used to reduce the dimensionality of high-dimensional data. By preserving the most relevant features, SVM-RFE aims to enhance the efficiency and performance of machine learning models in dealing with complex data [52].
Survival curve analysis
Survival curves for candidate targets were constructed among TNBC patients using the Kaplan-Meier Plotter database (https://kmplot.com/analysis/) [53]. In the analysis, ER, PGR, and HER2 status were set to negative to analyze TNBC patients specifically, and the names of the candidate targets were inputted to generate the Kaplan-Meier plot.
Molecular docking
The crystal structure of the core protein AURKA (PDB ID: 1MQ4) was downloaded from the Protein Data Bank (https://www.rcsb.org). The structure of hesperetin was downloaded from the PubChem database, and the compound structure was converted into a three-dimensional structure using Chem3DUltra 14.0 software, followed by energy minimization using the MM2 algorithm. Subsequently, the target protein receptor was processed for dehydration and removal of organic molecules using PyMOL software. AutoDockTools 1.5.6 was used to add hydrogen atoms, calculate charges, and convert the compound and target protein receptor to “pdbqt” files, with appropriate settings for the center and grid parameters of the docking box. Finally, molecular docking evaluation was performed using Vina 1.1.2 to calculate the docking energy values [26].
Synthesis of HFPN
HFPN was synthesized using the conventional nanoprecipitation method. Hesperetin with a concentration of 0.1 mg/mL (abs42078166, Absin, China) and FeCl3•6H2O with a concentration of 1 mg/mL (10025-77-1, Shanghai Kelaiman Reagent Co., Ltd., China) were dissolved in a phosphate buffer solution. After vigorous stirring, purification was performed by dialysis using deionized water in a 3000 Da molecular weight cutoff ultrafiltration tube (10.1002/adfm.202303899).
Characterization of HFPN
HFPN was observed and imaged using TEM. TEM images were captured using Talos F200X (FEI Ltd, USA). The particle size and zeta potential of HFPN were measured using dynamic light scattering (DLS) with NanoBrook 90Plus Zeta (Brookhaven Instruments Corporation, USA). Powder X-ray diffraction (XRD) measurements were carried out on a Rigaku Ultima IV X-ray diffractometer (ULTIMA IV, Rigaku Corporation, Japan). X-ray photoelectron spectroscopy was performed using the K-Alpha X-ray photoelectron spectroscopy instrument (Thermo Scientific, USA) (10.1002/adfm.202303899).
Detection of OH and 1O2 free radicals
The generation of ·OH was detected using methylene blue (MB) as an indicator (HY-B1359A, MCE, USA). A 2.7 mM MB aqueous solution (10 µL) was prepared and mixed with equal amounts of PBS, HFPN, and HFPN + X-ray treatment. The absorbance at 665 nm was measured and recorded using a UV-Vis spectrophotometer (UV1800PC, Jinghua, China) at 0, 1, 2, 3, 5, and 10 min after mixing.
The generation of 1O2 was detected using 1,3-diphenylisobenzofuran (DPBF) as a probe (HY-W011664, MCE, USA). Freshly prepared 3.7 mM DPBF solution (100 µL) was added to the solution with the same dosage. Under 450 nm (100 mw cm− 2) laser irradiation, the absorbance of DPBF at 410 nm was measured using a UV-Vis spectrophotometer (UV1800PC, Jinghua, China) (10.1002/adfm.202303899)[54].
GSH level detection
The glutathione (GSH) depletion capacity of 4T1 cells and mouse tumor tissues was determined using a UV-Vis spectrophotometer. A 1 mM GSH solution (500 µL) was mixed with the solution of equal dosage and added to 2 mL of PBS and 50 µL of 2.5 mM DTNB solution (HY-15915, MCE, USA). After centrifugation, the supernatant of the mixed solution was collected, and its absorbance was measured using a UV-Vis spectrophotometer (UV1800PC, Jinghua, China) (10.1002/adfm.202303899)[55].
Measurement of GSH-PX activity
The level of GSH-PX activity in the supernatant of 4T1 cells was assessed using the GSH-PX activity assay kit (A005-1-2, Beyotime, China), following the manufacturer’s instructions [55].
Measurement of GSH/Oxidized glutathione (GSSG) ratio
H9c2 cells (5 × 104 cells/well) were seeded into a 96-well clear flat-bottom plate. Following treatment with Hesperetin (0, 0.2, 0.5, 1, and 10 µM) at 37 °C for 12, 24, and 48 h, the GSH/GSSG ratio was measured using the GSH/GSSG ratio detection kit (catalog number ab138881; Abcam) according to the manufacturer’s protocol [56].
Cell culture and group treatment
Mouse breast cancer cells 4T1, human breast cancer cells MDA-MB-231 and BT-549, and human normal breast epithelial cells MCF-10 A were obtained from the ATCC cell repository (USA). The cells were cultured in a 5% CO2 incubator at 37 °C using Dulbecco’s Modified Eagle’s Medium (30030, Thermofisher, USA) supplemented with 10% fetal bovine serum (12484028, Thermofisher, USA), 100 µg/mL of streptomycin (HY-B1906, MCE, USA), and 100 U/mL of penicillin (HY-B0522, MCE, USA) [54, 57].
The cell groups were divided into the following: PBS group (treated with PBS), PBS + X-ray group (treated with PBS and X-ray), HFPN group (treated with HFPN), HFPN + X-ray group (treated with HFPN and X-ray), HFPN + X-ray + Fer-1 group (treated with HFPN, X-ray, and Fer-1), HFPN + X-ray + DFO group (treated with HFPN, X-ray, and DFO), HFPN + X-ray + GSH group (treated with HFPN, X-ray, and GSH), HFPN + oe-NC group (treated with HFPN and overexpressing control), HFPN + oe-AURKA group (treated with HFPN and overexpressing AURKA), HFPN + X-ray + oe-NC group (treated with HFPN, X-ray, and overexpressing control), HFPN + X-ray + oe-AURKA group (treated with HFPN, X-ray, and overexpressing AURKA).
Cells in the PBS group were incubated with PBS. Cells in the HFPN group and HFPN + X-ray group were incubated with HFPN at a concentration of 5 µM. Cells in the HFPN + X-ray group were irradiated with a dose of 4 Gy of X-ray for 24 h. Cells in the PBS + X-ray group were incubated with PBS and simultaneously exposed to a dose of 4 Gy of X-ray (ULTIMA IV, Rigaku Corporation, Japan) for 24 h. HFPN + X-ray + Fer-1, HFPN + X-ray + DFO and HFPN + X-ray + GSH groups were pre-treated with 1 µM Fer-1 (RM02804, ABclonal Technology, China), 100 µM DFO (RM02807, ABclonal Technology, China), and 2 mM GSH (RM02881, ABclonal Technology, China), respectively, for 4 h on top of the treatment protocol used in the HFPN + X-ray group.
Based on the known PRDX1 sequence in the NCBI database, overexpression constructs oe-NC and oe-AURKA were designed and constructed by Shanghai Gene Pharma Co., Ltd (Shanghai, China). The construction of the overexpression vectors was performed using the pCMV plasmid as the vector. Passage 3 of the 4T1 cells and MDA-MB-231 cells were digested with trypsin and seeded at a density of 4 × 105 cells per well in a 6-well plate. The cells were cultured until they formed a monolayer. The culture medium was then removed, and transfection was performed according to the instructions of Lipofectamine 2000 (11668-019, Invitrogen, New York, USA). The concentration of the overexpression vectors used was referred to the instructions provided by the reagent. After transfection, the cells were incubated at 37 °C with 5% CO2 for 6–8 h, then the complete culture medium was replaced, and the cells were further cultured for 48 h before protein extraction for subsequent experiments [58].
Intracellular ROS detection
Intracellular ROS levels were measured using the DCFH-DA probe (HY-D0940, MCE, USA). Intracellular lipid peroxidation levels were assessed using the C11-BODIPY581/591 probe (RM02821, ABclonal Technology, China). 4T1 cells were seeded in a 6-well plate containing 2.0 mL of DMEM medium at a density of 1.0 × 105 cells per well and incubated for 24 h. After three washes with PBS, the cells were incubated for 6 h and then washed again three times with PBS. Fluorescent microscope images were captured using CLSM (Leica TCS-SP8 SR, Leica, Germany) (10.1002/adfm.202303899) to observe the fluorescence intensity, which represented intracellular ROS levels [59].
Iron content determination
The iron content was measured using the iron content determination kit (MERCK, Germany, MAK025). 4T1 cells co-cultured with HFPN were collected and lysed with NP-40 according to the kit instructions. Finally, the absorbance of the supernatant was recorded using a microplate reader (Tecan Infinite M NANO, Tecan, Switzerland) at the specified wavelength (10.1002/adfm.202303899).
MTT assay for breast cancer cell proliferation
Healthy 4T1 and MDA-MB-231 cells in the growth phase were seeded in a 96-well plate at a density of 8 × 103 cells per well and incubated in a cell incubator for 24 h. Subsequently, the cells were treated individually with PBS, HFPN, and X-ray for 48 h, as well as with a combination of HFPN and X-ray for 48 h. After incubation, 10µL of MTT solution (Sigma-Aldrich, USA, 96992) was added to each well. Following a 4-hour incubation at 37 °C in a humidified culture chamber, the absorbance of each sample was measured at 450 nm using a spectrophotometer (Jinghua, China, UV1800PC) [60, 61]. Each group was replicated three times, with 6 replicates per group.
Experimental setup for colony formation assay
For colony formation assay, transfected cells were seeded at a density of 400 cells per well in a 6-well plate containing DMEM medium with 10% FBS. After two weeks, cells were fixed with methanol and stained with 0.1% crystal violet for colony visualization and counting. This experiment was performed in triplicate [54, 57].
Immunofluorescence staining
4T1 and MDA-MB-23 cells were seeded in a 96-well plate at a density of 1 × 104 cells per well. Once cells reached full confluence, Calcein-AM/PI dual staining reagent kit (CA1630, Solarbio, China) was used to assess cell viability. Subsequently, 4T1 and MDA-MB-23 cells were seeded in a 6-well plate and stained with PI (4.5 μm) and Calcein-AM (2 μm), followed by observation using a fluorescence microscope (IX73, OLYMPUS, TOKYO, JAPAN) [54, 57].
Western blot
Breast cancer 4T1 cells and mouse tumor tissues were lysed in an ice-cold RIPA lysis buffer containing 1% PMSF (P0013B, Biyuntian, Shanghai, China) for 30 min. The lysates were then centrifuged at 14,000 g and 4℃, and the supernatants were collected. Protein concentration in the lysates was determined using the BCA method (P0012S, Biyuntian, Shanghai, China). The lysates were mixed with an appropriate amount of 5× loading buffer and boiled at 100℃ for 10 min to denature the proteins. Protein samples (50 µg) were separated by SDS-PAGE using separating and concentrating gels. After electrophoresis, the proteins were transferred onto a PVDF membrane. The membrane was then blocked in 5% skim milk at room temperature for 1 h. Following overnight incubation at 4℃, the following primary antibodies were used: rabbit anti-GPX4 (1:10,000, ab125066, Abcam, USA), rabbit anti-AURKA (1:10,000, ab108353, Abcam, USA), rabbit anti-SLC7A11 (1:1,000, ab216876, Abcam, USA), and rabbit anti-GAPDH (1:1,000, #2118, Cell Signaling, USA), with GAPDH used as the loading control. After washing with PBST at room temperature, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:10,000, BA1056, BoDuo, Wuhan, China) at room temperature for 1 h. The membrane was washed six times with PBST for 5 min each. Finally, the membrane was treated with ECL substrate (AR1172, BoDuo, Wuhan, China) and exposed using an imaging system (Amersham Imager 600, GE, USA) for signal detection [62, 63]. ImageJ 1.48 software (National Institutes of Health) was used for protein quantification analysis. The quantification was based on the ratio of the grayscale values of each protein to the reference protein GAPDH, with the experiment repeated three times.
Mouse subcutaneous xenograft experiment
Male BALB/c mice (4–5 weeks old, weighing 18–22 g) were purchased from Beijing Vitonlihua Experimental Animal Technology Co., Ltd. (213, Beijing, China). They were housed in a specific pathogen-free (SPF) facility with constant humidity (45-50%) and temperature (25–27℃) for one week, with a 12-hour light-dark cycle each day, to acclimatize to the experimental environment. The mice were fasted for 12 h prior to drug administration and were allowed ad libitum access to food and water at all other times [60, 64]. All experiments involving mice were approved by the Animal Ethics Committee of The First Hospital of China Medical University (No. CMUXN2021734).
A total of 36 mice were randomly divided into 6 groups: PBS, PBS + X-ray, HFPN, HFPN + X-ray, HFPN + X-ray + oe-NC, and HFPN + X-ray + oe-AURKA, with 6 mice in each group. The mice in the PBS, PBS + X-ray, HFPN, HFPN + X-ray + oe-NC, and HFPN + X-ray + oe-AURKA groups were injected daily with the same dosage of PBS and HFPN. The mice in the PBS + X-ray, HFPN + X-ray, HFPN + X-ray + oe-NC, and HFPN + X-ray + oe-AURKA groups received a radiation dose of 4 Gy 6 hours after each injection. The mice in the HFPN + X-ray + oe-NC and HFPN + X-ray + oe-AURKA groups were further subcutaneously injected with the same dosage of oe-NC and oe-AURKA lentivirus. 4T1 cells (5 × 106/0.2 mL) were injected subcutaneously into the back of BALB/c mice. The width (W) and length (L) of the tumors in each group were measured weekly using a caliper to monitor tumor growth, and the tumor volume (V) was calculated using the formula V = (W2×L)/2. When the tumor volume reached approximately 50 mm3, the mice were euthanized after receiving a tail intravenous injection of either PBS or HFPN. The dosage of HFPN was 30.0 mg(kg− 1), administered once every three days for a total of 21 days (10.1002/adfm.202303899) [54, 57]. The tumor tissues from each group of mice were collected, embedded, and sectioned, followed by hematoxylin and eosin staining. Immunohistochemistry staining and analysis were performed using the primary rabbit anti-ki67 antibody (1:100, ab197547, Abcam, USA).
In vivo distribution experiment of HFPN
The tissue distribution of HFPN was evaluated through in vitro fluorescence imaging of major visceral organs. Six male BALB/c mice were selected and injected with HFPN at a dose of 30.0 mg/kg-1 via the tail vein. The time points following injection were 0, 2, 4, 8, 12, and 24 h. The distribution of HFPN was observed using the Maestro in vivo fluorescence imaging system (Cambridge Research & Instrumentation Inc., Woburn, USA) [54].
Statistical analysis
All data were processed using GraphPad Prism 9.0 software. Quantitative data were presented as mean ± standard deviation (Mean ± SD). Non-paired t-tests were used for comparisons between two groups, while one-way analysis of variance (ANOVA) was employed for comparisons among multiple groups. Variances were tested for homogeneity using Levene’s test, and if homogeneity was achieved, Dunnett’s t-test and LSD-t test were employed for pairwise comparisons. If heterogeneity was observed, Dunnett’s T3 test was used. P < 0.05 indicated statistical significance for between-group comparisons [54].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
None.
Abbreviations
- TNBC
Triple-Negative Breast Cancer
- AURKA
Aurora Kinase A
- ROS
Reactive Oxygen Species
- HFPN
Hesperetin-Loaded Ferroptosis-Inducing Nanocomposites
- GPX4
Glutathione Peroxidase 4
- SLC7A11
Solute Carrier Family 7 Member 11
- GEO
Gene Expression Omnibus
- DEG
Differentially Expressed Gene
- GO
Gene Ontology
- BP
Biological Processes
- MF
Molecular Functions
- LASSO
Least Absolute Shrinkage and Selection Operator
- SVM
RFE-Support Vector Machine-Recursive Feature Elimination
- PBS
Phosphate-Buffered Saline
- TEM
Transmission Electron Microscopy
- DLS
Dynamic Light Scattering
- MB
Methylene Blue
- DPBF
1,3-Diphenylisobenzofuran
- GSH
Glutathione
- GSSG
Oxidized Glutathione
- GPX
Glutathione Peroxidase
- MTT
Methylthiazolyldiphenyl-Tetrazolium Bromide
Author contributions
Yang Guo: Conceptualization, Methodology, Formal Analysis, Investigation, Writing - Original Draft, Visualization.Huan Wang: Methodology, Validation, Formal Analysis, Investigation, Data Curation.Xinlei Wang: Software, Validation, Data Curation, Writing - Review & Editing, Visualization.Keyan Chen: Resources, Supervision, Project Administration, Funding Acquisition, Writing - Review & Editing.Liang Feng: Conceptualization, Resources, Supervision, Project Administration, Funding Acquisition, Writing - Review & Editing.
Funding
This study was supported by the Natural Science Foundation of Liaoning Province (2021-MS-202).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Guo, Huan Wang and Xinlei Wang contributed equally to this work.
Contributor Information
Keyan Chen, Email: kychen@cmu.edu.cn.
Liang Feng, Email: fengliang@cmu.edu.cn.
References
- 1.Nassif AB, Talib MA, Nasir Q, Afadar Y, Elgendy O. Breast cancer detection using artificial intelligence techniques: a systematic literature review. Artif Intell Med. 2022;127:102276. 10.1016/j.artmed.2022.102276. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo-Crespo JJ, Armiñán A, Charbonnier D, et al. Characterization of triple-negative breast cancer preclinical models provides functional evidence of metastatic progression. Int J Cancer. 2019;145(8):2267–81. 10.1002/ijc.32270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alhenc-Gelas M, Cabel L, Berger F, et al. Characteristics and outcome of breast cancer-related microangiopathic haemolytic anaemia: a multicentre study. Breast Cancer Res. 2021;23(1):9. 10.1186/s13058-021-01386-y. Published 2021 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D, Wang H, Liu H, Xu R. Liposomal ATM siRNA delivery for enhancing triple-negaitive breast cancer immune checkpoint blockade therapy. J Biomater Appl. 2023;37(10):1835–46. 10.1177/08853282231162111. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Chen A, Liu C, Zhang B. Triple-negative breast Cancer with Dermatomyositis: a Case Report and Literature Review. Cancer Manag Res. 2022;14:569–76. 10.2147/CMAR.S349400. Published 2022 Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, Wang X, Liu X, et al. Analysis of clinical characteristics, treatment, and prognostic factors of 106 breast Cancer patients with Solitary Pulmonary nodules. Front Surg. 2022;9:843913. 10.3389/fsurg.2022.843913. Published 2022 Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Chen L, Liu W. Matrix stiffness-dependent STEAP3 coordinated with PD-L2 identify tumor responding to sorafenib treatment in hepatocellular carcinoma. Cancer Cell Int. 2022;22(1):318. 10.1186/s12935-022-02634-7. Published 2022 Oct 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Huang R, Wang Y, et al. SIRT6 drives sensitivity to ferroptosis in anaplastic thyroid cancer through NCOA4-dependent autophagy. Am J Cancer Res. 2023;13(2):464–74. Published 2023 Feb 15. [PMC free article] [PubMed] [Google Scholar]
- 9.Cosialls E, El Hage R, Dos Santos L, Gong C, Mehrpour M, Hamaï A. Ferroptosis: Cancer Stem cells rely on Iron until to die for it. Cells. 2021;10(11):2981. 10.3390/cells10112981. Published 2021 Nov 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shehata MG, Awad TS, Asker D, El Sohaimy SA, Abd El-Aziz NM, Youssef MM. Antioxidant and antimicrobial activities and UPLC-ESI-MS/MS polyphenolic profile of sweet orange peel extracts. Curr Res Food Sci. 2021;4:326–35. 10.1016/j.crfs.2021.05.001. Published 2021 May 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roohbakhsh A, Parhiz H, Soltani F, Rezaee R, Iranshahi M. Molecular mechanisms behind the biological effects of hesperidin and hesperetin for the prevention of cancer and cardiovascular diseases. Life Sci. 2015;124:64–74. 10.1016/j.lfs.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 12.Vabeiryureilai M, Lalrinzuali K, Jagetia GC. NF-κB and COX-2 repression with topical application of hesperidin and naringin hydrogels augments repair and regeneration of deep dermal wounds. Burns. 2022;48(1):132–45. 10.1016/j.burns.2021.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Krishnan G, Subramaniyan J, Chengalvarayan Subramani P, Muralidharan B, Thiruvengadam D. Hesperetin conjugated PEGylated gold nanoparticles exploring the potential role in anti-inflammation and anti-proliferation during diethylnitrosamine-induced hepatocarcinogenesis in rats. Asian J Pharm Sci. 2017;12(5):442–55. 10.1016/j.ajps.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Q, Mao L, Huang H, et al. Hesperetin ameliorates glioblastoma by inhibiting proliferation, inducing apoptosis, and suppressing metastasis. Transl Cancer Res. 2022;11(6):1781–94. 10.21037/tcr-22-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poomipark N, Chaisin T, Kaulpiboon J. Anti-proliferative, anti-migration, and anti-invasion activity of novel hesperidin glycosides in non-small cell lung cancer A549 cells. Res Pharm Sci. 2023;18(5):478–88. 10.4103/1735-5362.383704. Published 2023 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap KM, Sekar M, Wu YS, et al. Hesperidin and its aglycone hesperetin in breast cancer therapy: a review of recent developments and future prospects. Saudi J Biol Sci. 2021;28(12):6730–47. 10.1016/j.sjbs.2021.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Q, Lai Y, Zhang H, et al. Hesperetin inhibits TGF-β1-Induced Migration and Invasion of Triple negative breast Cancer MDA-MB-231 cells via suppressing Fyn/Paxillin/RhoA pathway. Integr Cancer Ther. 2022;21:15347354221086900. 10.1177/15347354221086900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying JF, Lu ZB, Fu LQ, et al. The role of iron homeostasis and iron-mediated ROS in cancer. Am J Cancer Res. 2021;11(5):1895–912. Published 2021 May 15. [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JI, Ahn TG, Choi JH. Effects of Iron on Efficacy of Photodynamic Therapy using Photolon in a mouse model of CT26 Colon cancer. J Nippon Med Sch. 2023;90(1):41–9. 10.1272/jnms.JNMS.2023_90-108. [DOI] [PubMed] [Google Scholar]
- 20.Atiya HI, Frisbie L, Goldfeld E, et al. Endometriosis-Associated Mesenchymal stem cells support ovarian clear cell carcinoma through Iron Regulation. Cancer Res. 2022;82(24):4680–93. 10.1158/0008-5472.CAN-22-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapora-Kurel A, Malyszko J, NOVEL, IRON BIOMARKERS IN CHRONIC KIDNEY DISEASE. Wiad Lek. 2021;74(12):3230–3. [PubMed] [Google Scholar]
- 22.Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22(1):61. 10.1186/s13058-020-01296-5. Published 2020 Jun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigiracciolo DC, Nohata N, Lappano R et al. Focal Adhesion Kinase (FAK)-Hippo/YAP transduction signaling mediates the stimulatory effects exerted by S100A8/A9-RAGE system in triple-negative breast cancer (TNBC). J Exp Clin Cancer Res. 2022;41(1):193. Published 2022 Jun 3. 10.1186/s13046-022-02396-0 [DOI] [PMC free article] [PubMed]
- 24.Maciel-Cruz EJ, Figuera-Villanueva LE, Garibaldi-Ríos AF, et al. AURKA Gene variants rs1047972, and rs8173 are Associated with breast Cancer. J Breast Cancer. 2023;26(4):378–90. 10.4048/jbc.2023.26.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Qi Y, Yu J, et al. Aurora kinase a regulates cancer-associated RNA aberrant splicing in breast cancer. Heliyon. 2023;9(7):e17386. 10.1016/j.heliyon.2023.e17386. Published 2023 Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilipović A, Mitrović D, Obradović S, Poša M. Docking-based analysis and modeling of the activity of bile acids and their synthetic analogues on large conductance Ca2 + activated K channels in smooth muscle cells. Eur Rev Med Pharmacol Sci. 2021;25(23):7501–7. 10.26355/eurrev_202112_27449. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Li B, Cao Q, Liu T, Li J. Targeting cancer stem cells with polymer nanoparticles for gastrointestinal cancer treatment. Stem Cell Res Ther. 2022;13(1):489. 10.1186/s13287-022-03180-9. Published 2022 Oct 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grasset EM, Dunworth M, Sharma G, et al. Triple-negative breast cancer metastasis involves complex epithelial-mesenchymal transition dynamics and requires vimentin. Sci Transl Med. 2022;14(656):eabn7571. 10.1126/scitranslmed.abn7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Ende NS, Nguyen AH, Jager A, Kok M, Debets R, van Deurzen CHM. Triple-negative breast Cancer and predictive markers of response to Neoadjuvant Chemotherapy: a systematic review. Int J Mol Sci. 2023;24(3):2969. 10.3390/ijms24032969. Published 2023 Feb 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao L, Zhang YL, Deng L, et al. Protein phosphatase 1 subunit PPP1R14B stabilizes STMN1 to promote progression and Paclitaxel Resistance in Triple-negative breast Cancer. Cancer Res. 2023;83(3):471–84. 10.1158/0008-5472.CAN-22-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen DL, Cai JH, Wang CCN. Identification of key prognostic genes of Triple negative breast Cancer by LASSO-Based Machine Learning and Bioinformatics Analysis. Genes (Basel). 2022;13(5):902. 10.3390/genes13050902. Published 2022 May 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raiter A, Zlotnik O, Lipovetsky J, et al. A novel role for an old target: CD45 for breast cancer immunotherapy. Oncoimmunology. 2021;10(1):1929725. 10.1080/2162402X.2021.1929725. Published 2021 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benhamou RI, Suresh BM, Tong Y, et al. DNA-encoded library versus RNA-encoded library selection enables design of an oncogenic noncoding RNA inhibitor. Proc Natl Acad Sci U S A. 2022;119(6):e2114971119. 10.1073/pnas.2114971119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong G, Shen J, Chen Z, et al. Antioxidant and Antitumor activities of newly synthesized hesperetin derivatives. Molecules. 2022;27(3):879. 10.3390/molecules27030879. Published 2022 Jan 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Song Y, Yu L, et al. Co-delivery of Hesperetin and Cisplatin via Hyaluronic Acid-Modified Liposome for targeted inhibition of Aggression and Metastasis of Triple-negative breast Cancer. ACS Appl Mater Interfaces. 2023;15(29):34360–77. 10.1021/acsami.3c03233. [DOI] [PubMed] [Google Scholar]
- 36.Farghadani R, Naidu R. The anticancer mechanism of action of selected polyphenols in triple-negative breast cancer (TNBC). Biomed Pharmacother. 2023;165:115170. 10.1016/j.biopha.2023.115170. [DOI] [PubMed] [Google Scholar]
- 37.Gao S, Chen X, Yu Z, et al. Progress of research on the role of active ingredients of Citri Reticulatae Pericarpium in liver injury. Phytomedicine. 2023;115:154836. 10.1016/j.phymed.2023.154836. [DOI] [PubMed] [Google Scholar]
- 38.Shakiba E, Bazi A, Ghasemi H, et al. Hesperidin suppressed metastasis, angiogenesis and tumour growth in Balb/c mice model of breast cancer. J Cell Mol Med. 2023;27(18):2756–69. 10.1111/jcmm.17902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmo F, Silva C, Martel F. Inhibition of Glutamine Cellular Uptake contributes to the cytotoxic effect of Xanthohumol in Triple-negative breast Cancer cells. Nutr Cancer. 2022;74(9):3413–30. 10.1080/01635581.2022.2076889. [DOI] [PubMed] [Google Scholar]
- 40.Ye Y, Xu L, Zhang L, et al. Meningioma achieves malignancy and erastin-induced ferroptosis resistance through FOXM1-AURKA-NRF2 axis. Redox Biol. 2024;72:103137. 10.1016/j.redox.2024.103137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Hu J, Xiong X et al. AURKA inhibition induces Ewing’s sarcoma apoptosis and ferroptosis through NPM1/YAP1 axis. Cell Death Dis. 2024;15(1):99. Published 2024 Jan 29. 10.1038/s41419-024-06485-0 [DOI] [PMC free article] [PubMed]
- 42.Sun N, Meng F, Zhao J, et al. Aurka deficiency in the intestinal epithelium promotes age-induced obesity via propionate-mediated AKT activation. Int J Biol Sci. 2021;17(5):1302–14. 10.7150/ijbs.56477. Published 2021 Mar 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai T, Li M, Liu Y et al. The promotion action of AURKA on post-ischemic angiogenesis in diabetes-related limb ischemia. Mol Med. 2023;29(1):39. Published 2023 Mar 28. 10.1186/s10020-023-00635-4 [DOI] [PMC free article] [PubMed]
- 44.Xue Q, Yan D, Chen X, et al. Copper-dependent autophagic degradation of GPX4 drives ferroptosis. Autophagy. 2023;19(7):1982–96. 10.1080/15548627.2023.2165323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang D, Feng Y, Zandkarimi F, et al. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. 2023;186(13):2748–e276422. 10.1016/j.cell.2023.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–92. 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng YJ, Ren EH, Yuan WH, Zhang GZ, Wu ZL, Xie QQ. GRB10 and E2F3 as Diagnostic Markers of Osteoarthritis and Their Correlation with Immune Infiltration. Diagnostics (Basel). 2020;10(3):171. Published 2020 Mar 22. 10.3390/diagnostics10030171 [DOI] [PMC free article] [PubMed]
- 49.Peng XY, Wang Y, Hu H, Zhang XJ, Li Q. Identification of the molecular subgroups in coronary artery disease by gene expression profiles. J Cell Physiol. 2019;234(9):16540–8. 10.1002/jcp.28324. [DOI] [PubMed] [Google Scholar]
- 50.Fu C, Shuang Q, Liu Y, Zeng L, Su W. Baihe extracts reduce the activation and apoptosis of Microglia in the Hippocampus of mice with Depression-like behaviors by Downregulating MYC. ACS Chem Neurosci. 2022;13(5):587–98. 10.1021/acschemneuro.1c00439. [DOI] [PubMed] [Google Scholar]
- 51.Ding Y, Chu L, Cao Q, Lei H, Li X, Zhuang Q. A meta-validated immune infiltration-related gene model predicts prognosis and immunotherapy sensitivity in HNSCC. BMC Cancer. 2023;23(1):45. 10.1186/s12885-023-10532-y. Published 2023 Jan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao Y, Zhao J, Zhou X, Hu J, Wang Y. Potential role of a three-gene signature in predicting diagnosis in patients with myocardial infarction. Bioengineered. 2021;12(1):2734–49. 10.1080/21655979.2021.1938498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Huang Y, Zhou R, Li Z, Yan Q. Clinicopathological and prognostic significance of SMAD4 in non-small cell lung cancer: a meta-analysis and database validation. Med (Baltim). 2023;102(29):e34312. 10.1097/MD.0000000000034312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng L, Ding S, Cao Y, et al. A MOF-Based potent ferroptosis inducer for enhanced Radiotherapy of Triple negative breast Cancer. ACS Nano. 2023;17(14):13195–210. 10.1021/acsnano.3c00048. [DOI] [PubMed] [Google Scholar]
- 55.Dang R, Wang M, Li X, et al. Edaravone ameliorates depressive and anxiety-like behaviors via Sirt1/Nrf2/HO-1/Gpx4 pathway. J Neuroinflammation. 2022;19(1):41. 10.1186/s12974-022-02400-6. Published 2022 Feb 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L, Wang H, Yu S, Zhang L, Jiang J, Zhou Q. Herceptin induces ferroptosis and mitochondrial dysfunction in H9c2 cells. Int J Mol Med. 2022;49(2):17. 10.3892/ijmm.2021.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang W, Shi S, Lv H, Ju Z, Liu Q, Chen T. Tellurium-driven maple leaf-shaped manganese nanotherapeutics reshape tumor microenvironment via chemical transition in situ to achieve highly efficient radioimmunotherapy of triple negative breast cancer. Bioact Mater. 2023;27:560–73. 10.1016/j.bioactmat.2023.04.010. Published 2023 May 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahauad-Fernandez WD, Okeoma CM. Cysteine-linked dimerization of BST-2 confers anoikis resistance to breast cancer cells by negating proapoptotic activities to promote tumor cell survival and growth. Cell Death Dis. 2017;8(3):e2687. 10.1038/cddis.2017.68. Published 2017 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng P, Ding B, Jiang Z, et al. Ultrasound-augmented mitochondrial calcium Ion overload by Calcium Nanomodulator to induce immunogenic cell death. Nano Lett. 2021;21(5):2088–93. 10.1021/acs.nanolett.0c04778. [DOI] [PubMed] [Google Scholar]
- 60.Pan X, Hong X, Li S, Meng P, Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med. 2021;53(1):91–102. 10.1038/s12276-020-00510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S, Cheng M, Zheng X, et al. Interactions between lncRNA TUG1 and mir-9-5p modulate the resistance of breast Cancer cells to Doxorubicin by regulating eIF5A2. Onco Targets Ther. 2020;13:13159–70. 10.2147/OTT.S255113. Published 2020 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salem M, Shan Y, Bernaudo S, Peng C. Mir-590-3p targets cyclin G2 and FOXO3 to promote Ovarian Cancer Cell Proliferation, Invasion, and spheroid formation. Int J Mol Sci. 2019;20(8):1810. 10.3390/ijms20081810. Published 2019 Apr 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shu M, Zheng X, Wu S, et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol Cancer. 2011;10:59. 10.1186/1476-4598-10-59. Published 2011 May 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu R, Guo H, Lu S. MiR-335-5p restores cisplatin sensitivity in ovarian cancer cells through targeting BCL2L2. Cancer Med. 2018;7(9):4598–609. 10.1002/cam4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.