Abstract
Background
The progression of liver cancer is a complicated process that involves genetic and epigenetic changes. Paired box 6 (PAX6) is a critical transcription factor for embryonic development. PAX6 is abnormally methylated in human cancer. The role of the PAX6 gene in the pathogenesis of hepatocellular carcinoma (HCC) is still unclear.
Methods
Transcriptional silencing of PAX6 mediated by promoter methylation was confirmed using quantitative methylation-specific polymerase chain reaction (PCR) and reverse-transcription (RT)-PCR. Then we conducted gain-and-loss of function approaches to evaluate the function of PAX6 in HCC progression in vitro. Moreover, we designed xenograft mouse models to assess the effect of PAX6 on tumor growth and metastasis. Finally, we used RNA sequencing (RNA-seq) strategy and phenotypic rescue experiments to identify potential targets of PAX6 performing tumor-suppressive function.
Results
Constitutive expression of PAX6 suppressed anchorage-independent growth and cell invasion in vitro as well as tumor growth and metastasis in xenograft mouse models. In contrast, the inhibition of PAX6 using knockout and knockdown strategies increased tumor growth both in vitro and in vivo. Downregulation of PAX6 by doxycycline depletion partially reversed the malignant phenotypes of HCC cells induced by PAX6. Moreover, we identified E-cadherin (CDH1) and thrombospondin-1 (THBS1) as targets of PAX6. Ultimately, we demonstrated that the knockdown of CDH1 and overexpression of THBS1 in PAX6-expressing HCC cells partly reversed the tumor-suppressive effect.
Conclusion
PAX6 functions as a tumor suppressor partly through upregulation of CDH1 and downregulation of THBS1. Promoter hypermethylation-mediated suppression of PAX6 reduces the tumor suppressor function in the progression of liver cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01789-6.
Keywords: Hepatocellular carcinoma, PAX6, Tumor suppressor, CDH1, THBS1
Introduction
Hepatocellular carcinoma (HCC) is a serious disease with a poor prognosis and high mortality rate due to obstacles in early detection [1–5]. The risk factors associated with hepatocarcinogenesis include chronic hepatitis B or C viral infection, alcohol, aflatoxin B1, and others [6–8]. Inactivation of tumor suppressor genes (TSGs) through promoter hypermethylation plays an essential role in carcinogenesis [9–12]. Although many studies have investigated the development of HCC, the genetic and epigenetic alterations that result in the initiation and progression of HCC remain unknown [6–14].
Epithelial-mesenchymal transition (EMT) is a complex process involving the loss of epithelial cell features and the acquisition of a mesenchymal phenotype [15–17]. This process is important in cancer progression, drug resistance and cancer progenitor cells [16, 17]. Many transcriptional regulators, such as Twist, Snail, Slug, and ZEB1/ZEB2, can induce EMT via a decrease in epithelial markers and an increase in mesenchymal markers [16, 17]. Moreover, the Wnt/β-catenin pathway, transforming growth factor-β pathway, receptor tyrosine kinase pathway, Notch pathway, and Sonic hedgehog signaling pathway are often dysregulated and lead to EMT in cancer[16, 17].
Paired box 6 (PAX6), a member of the homeobox gene superfamily, is a critical transcription factor for embryonic development [18–21]. PAX6 plays an important role in eye development via the Wnt/β-catenin, TGF-beta, Notch, and extracellular matrix (ECM)-related signaling pathways. Abnormal PAX6 expression contributes to developmental disorders and the formation of tumors [18–22]. In human cancer, the expression and biological function of PAX6 differ depending on the cancer type. PAX6 is downregulated in glioblastoma and prostate cancer and acts as a tumor suppressor [23–25]. In contrast, PAX6 acts as an oncogene in pancreatic cancer, retinoblastoma, and breast cancer cell lines [26–28]. PAX6 was shown to promote proliferation and cell cycle progression in human non-small cell lung cancer (NSCLC) cell lines [29]. Moreover, PAX6 overexpression notably enhanced EMT and metastasis via ZEB2 and the PI3K/AKT signaling pathway in NSCLC [30].
Our previous study demonstrated that the promoter of the PAX6 gene is frequently methylated in HCC [31]. Hypermethylation of gene promoters is frequently associated with transcriptional silencing and results in tumor suppressor gene (TSG) inactivation in cancer cells [9, 11, 14]. Elucidation of the role of the PAX6 gene in the pathogenesis of HCC requires further study. In this study, we investigated the role of PAX6 in liver cancer. We confirmed that PAX6 downregulation is significantly correlated with promoter hypermethylation. Furthermore, PAX6 protein overexpression suppressed anchorage-independent growth (AIG) and invasion in HCC cells. In xenograft mouse models, PAX6 also repressed tumor growth and metastasis. Moreover, PAX6 suppresses metastasis through the upregulation of E-cadherin (CDH1) and downregulation of thrombospondin-1 (THBS1) in HCC. Collectively, our findings suggest that PAX6 functions as a tumor suppressor in HCC progression.
Materials and methods
Clinical tissue samples
A total of 60 paired samples of HCC and adjacent nontumor tissues were obtained from the Taiwan Liver Cancer Network (TLCN). TLCN is funded by the National Science Council to provide researchers in Taiwan with primary liver cancer tissue specimens and their associated clinical information. These specimens were obtained during surgery and frozen immediately in liquid nitrogen and/or at − 80 °C until DNA extraction and RNA extraction. The diagnosis of HCC was confirmed by histology. The deidentified demographic data was anonymously collected by the staff of the TLCN to protect all subjects’ privacy. The use of clinical samples in this study was approved by the TLCN User Committee and Tri-Service General Hospital (TSGH) Institutional Review Board (TSGHIRB: B-109–02.)
Bioinformatics analysis
Bioinformatics analysis was carried out by using the following websites: UALCAN (http://ualcan.path.uab.edu) [32] and DNA Methylation Interactive Visualization Database (DNMIVD, http://119.3.41.228/dnmivd/) [33]. UALCAN were used to assess the PAX6 methylation levels in datasets from The Cancer Genome Atlas (TCGA). We used DNMIVD to analyze the correlation between PAX6 gene expression and methylation.
Cell lines
Nine human liver cancer cell lines (HepG2, Hep3B, Huh6, SK-Hep1, Mahlavu, TONG, PLC/PRF5, HA22T, and Huh7) were used in this study. HepG2, Hep3B and SK-Hep1 cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). Mahlavu, TONG, PLC/PRF5, HA22T, Huh6, and Huh7 cells were obtained from Professor K.H. Lin (Chuang-Gung University, Taiwan). The culture conditions and other components used were described in the Supplementary Materials and Methods.
DNA methylation and gene expression analysis
Genomic DNA from tissues and HCC cell lines was extracted, and bisulfite was used for conversion as previously described [31, 34]. Genomic DNA was extracted from cell lines and tissue samples by using a commercial DNA extraction kit (QIAmp Tissue Kit; Qiagen, Hilden, Germany). We treated DNA with bisulfite using an EZ DNA methylation kit (Zymo Research, Orange, CA) as described previously[35–37]. Quantitative methylation-specific polymerase chain reaction (qMSP) was performed as previously described [31, 34, 35]. RNA extraction and reverse transcription polymerase chain reaction (RT-PCR) were conducted as previously described [35–37]. Supplementary Table S1showed the primer sequences. Detailed explanations are available in the Supplementary Materials and Methods.
Plasmids and shRNA clones
The full-length PAX6 open reading frame (ORF) was cloned and inserted into the pLAS2.1 constitutive expression vector (termed pLAS2.1-PAX6) or the inducible expression vector pAS4.1 (termed pAS4.1-PAX6) as previously described [38]. All-in-one CRISPR vector, pAll-Cas9.Ppuro (from the C6 RNAi core facility, Academia Sinica, Taiwan), was digested with BsmBI and ligated with annealed oligonucleotides (GGCCCCATATTCGAGCCCCG and GGCTTGGCTCTTCTCG ATAC) for expression of PAX6 sgRNAs, which could bind to the targeting regions of PAX6 gene. pLKO.1-shLacZ, CDH1-shRNAs and THBS1-shRNAs were purchased from the National RNAi Core Facility of Taiwan. The pcDNA3.1-V5-THBS1 plasmid was purchased from Thermal ViGene BioSciences. The shRNA sequences are described in Supplementary Table S1.
Western blot, cell viability, AIG, and invasion assays
Western blot, MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, Inner Salt] cell viability, AIG and invasion assays were conducted as described previously [34, 39, 40]. Detailed descriptions are available in the Supplementary Materials and Methods. The following antibodies were used for western blotting: anti-PAX6 (Cell Signaling Technology), anti-THBS1 (Cell Signaling Technology), anti-V5 (Cell Signaling Technology), anti-CDH1 (BD Biosciences), anti-CDH2 (BD Biosciences), anti-Vimentin (Santa Cruz Biotechnology), anti-Twist (GeneTex), anti-Snail (GeneTex), and anti-beta-actin (GeneTex).
Immunofluorescence staining
Immunofluorescence staining was described as reported previously [34, 40].
In vivo tumor xenograft and metastasis model
We used Six-week-old nonobese diabetic severe-combined immunodeficiency (NOD-SCID) female mice to perform tumorigenicity and metastasis analyses. Animal studies were approved by the Institutional Animal Care and Use Committee of the National Defense Medical Center (Taipei, Taiwan). Details regarding xenograft mouse experiments were performed as described previously [34, 39, 40].
RNA sequencing (RNA-seq) analysis
RNA-seq data generation and normalization were performed on an Illumina NovaSeq 500 platform. The data analysis was performed as described in our previous report [41]. Next, Gene Ontology (GO) analysis and KEGG pathway analysis were performed with DAVID (https://david.ncifcrf.gov/). [42] Gene set enrichment analysis (GSEA) was performed with GSEA software download form website (https://www.gsea-msigdb.org/gsea/index.jsp) [43, 44].
Statistical analysis
We used GraphPad Prism software (version 5; GraphPad Software, La Jolla, CA) and SPSS software (IBM SPSS Statistics 21; Asia Analytics Taiwan, Taipei, Taiwan) to perform statistical analyses. All values are expressed as the mean ± SE. We used the Wilcoxon signed rank test to determine differences in disease status and gene methylation levels (MI). The correlation between PAX6 mRNA and promoter methylation was analyzed by Spearman’s rho analysis. Mann‒Whitney U test and the Student's t test were used to compare AIG number, cell invasion and relative gene expression between the different stable clones. We used unpaired two-tailed t test to analyze the in vivo data. In all cases, p < 0.05 was defined to indicate statistical significance.
Results
Promoter hypermethylation of PAX6 contributes to PAX6 silencing/or downregulation in liver cancer.
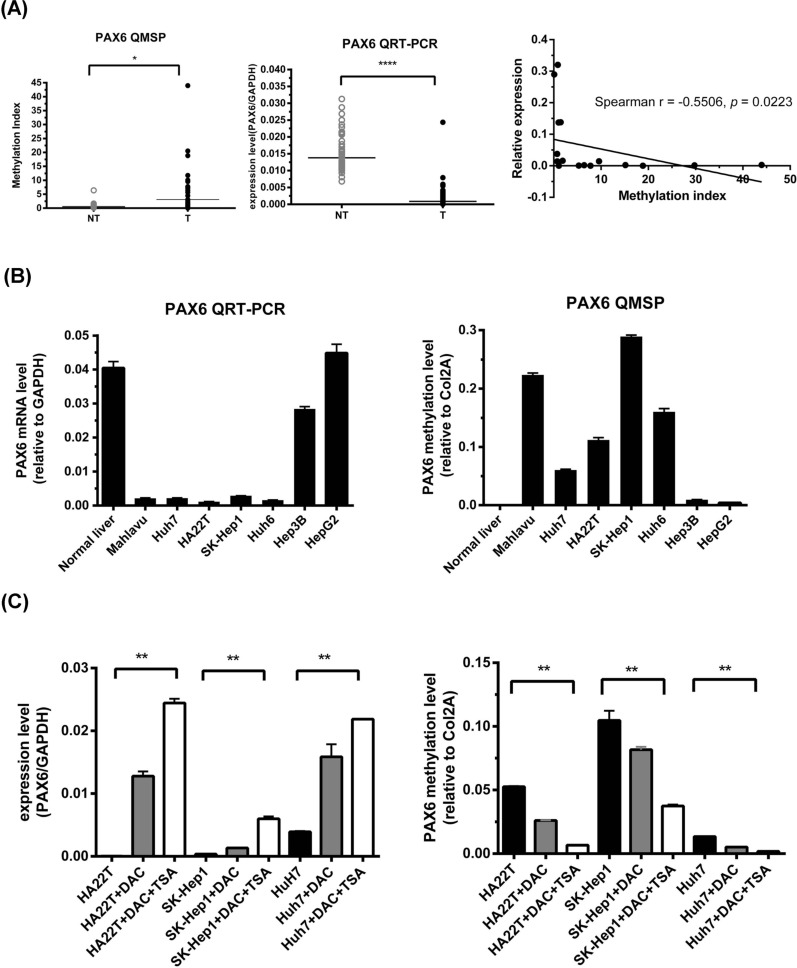
First, we performed qMSP to investigate the methylation levels of PAX6 in 60 pairs of HCC samples from the Taiwan Liver Cancer Network (TLCN) biobank and found that PAX6 methylation was significantly greater in the tumors than in the corresponding nontumor samples (Fig. 1A). Next, we confirmed the PAX6 methylation level in 377 liver hepatocellular carcinoma (LIHC) samples from TCGA using the UALCAN website (http://ualcan.path.uab.edu). The average beta value was greater in the LIHC group than in the normal control group (p < 0.0001) (Supplementary Figure S1). Using quantitative reverse transcription polymerase chain reaction (qRT‒PCR), we confirmed that the PAX6 mRNA (NM_000280.4) level in the 60 pairs of HCC tissues was significantly lower than that in their nontumor counterparts (Fig. 1A). Furthermore, we identified an inverse correlation between PAX6 mRNA expression and methylation in seventeen HCC tissues, which showed hypermethylation at the PAX6 promoter (Fig. 1A). Based on these data, promoter hypermethylation might contribute to the downregulation of PAX6 in HCC. Then, we analyzed the mRNA expression of PAX6 and its methylation level in HCC cell lines using qRT‒PCR and qMSP (Fig. 1B). The results confirmed that PAX6 was poorly methylated in normal liver tissues and HepG2 and Hep3B cells and highly methylated in Mahlavu, SK-Hep1, HA22T, Huh6, and Huh7 cells. Moreover, three HCC cell lines (HA22T, SK-Hep1, and Huh7) with low PAX6 expression were treated with 5-aza-2’-deoxycytidine (DAC) to confirm that promoter methylation is involved in the regulation of PAX6. We found a decreased methylation status of PAX6 and re-expression of PAX6 mRNA in all cell lines examined (Fig. 1C), further revealing that the transcriptional silencing of PAX6 was mediated by promoter methylation.
Fig. 1.
Promoter hypermethylation of PAX6 contributes to PAX6 downregulation in HCC A The promoter methylation and mRNA expression levels of PAX6 were determined in 60 paired HCC tissues and adjacent nontumor tissues (NT) by qMSP and qRT‒PCR. The results are presented as the relative expression of PAX6 mRNA and the difference in the MI. The black lines indicate the mean PAX6 mRNA and MI levels. The P value for methylation levels among the groups was determined by the Wilcoxon signed rank test. The correlation between PAX6 mRNA and promoter methylation was analyzed in HCC tissues with PAX6 promoter hypermethylation by Spearman’s rho analysis. (B) The gene expression levels of PAX6 and the internal reference gene GAPDH were analyzed by qRT‒PCR. The DNA methylation levels of PAX6 in normal liver tissue and seven HCC cell lines were determined by qMSP. (C) Gene expression levels of PAX6 and the internal reference gene GAPDH in HA22T, SK-Hep1, and Huh7 cells treated with 0.5 µM DAC or 0.5 µM DAC combined with 0.3 µM trichostatin A (TSA) were analyzed by qRT‒PCR. At the same time, the DNA methylation levels of PAX6 were determined by qMSP. The results are represented as differences in the MI.** p < 0.01, *** p < 0.001, and **** p < 0.0001 (Student’s t test)
PAX6 overexpression inhibits liver cancer cell growth and invasion.
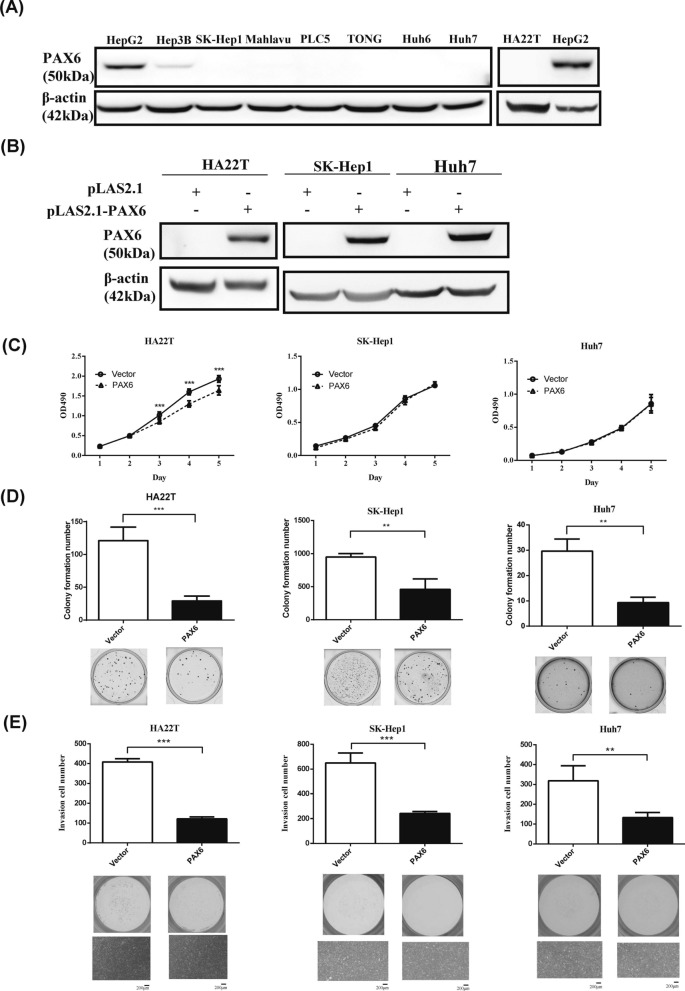
To explore the role of PAX6 in liver cancer, we evaluated endogenous PAX6 expression in these cell lines, namely, HepG2, Hep3B, SK-Hep1, Mahlavu, PLC/PRF5, TONG, Huh6, Huh7, and HA22T. PAX6 protein expression was undetectable in all liver cancer cell lines, except for HepG2 and Hep3B, and normal liver tissues (Fig. 2A). To further confirm the tumor-suppressive function of PAX6, we examined the effect of PAX6 overexpression on HAT22T, Huh7 and SK-Hep1 cancer cells. Stable expression of pLAS2.1-PAX6 in HAT22T, Huh7 and SK-Hep1 cells was verified by western blot analysis (Fig. 2B). Ectopic expression of PAX6 in HA22T cells significantly decreased cancer cell growth (Fig. 2C). As shown in Fig. 2D and E, restoration of PAX6 expression in HAT22T, Huh7 and SK-Hep1 cells significantly suppressed AIG and cancer cell invasion.
Fig. 2.
Ectopic expression of PAX6 inhibits AIG and invasion in a constitutive expression system in liver cancer cells. A The endogenous protein expression of PAX6 in liver cancer cell lines was determined by western blotting using an anti-PAX6 antibody. B Stable overexpression of pLAS2.1-PAX6 was established in different liver cancer cell lines and confirmed by western blotting. β-Actin was used as an internal control. C Cell proliferation (MTS) assays D and AIG assays were performed in HAT22T, SK-Hep1 and Huh7 cells expressing PAX6. E Matrigel invasion assays were performed in HAT22T, SK-Hep1 and Huh7 cells expressing PAX6. The data are expressed as the mean ± SE. Significant differences were determined using the Mann‒Whitney U test. ** p < 0.01, *** p < 0.001
Inhibition of PAX6 notably increases AIG
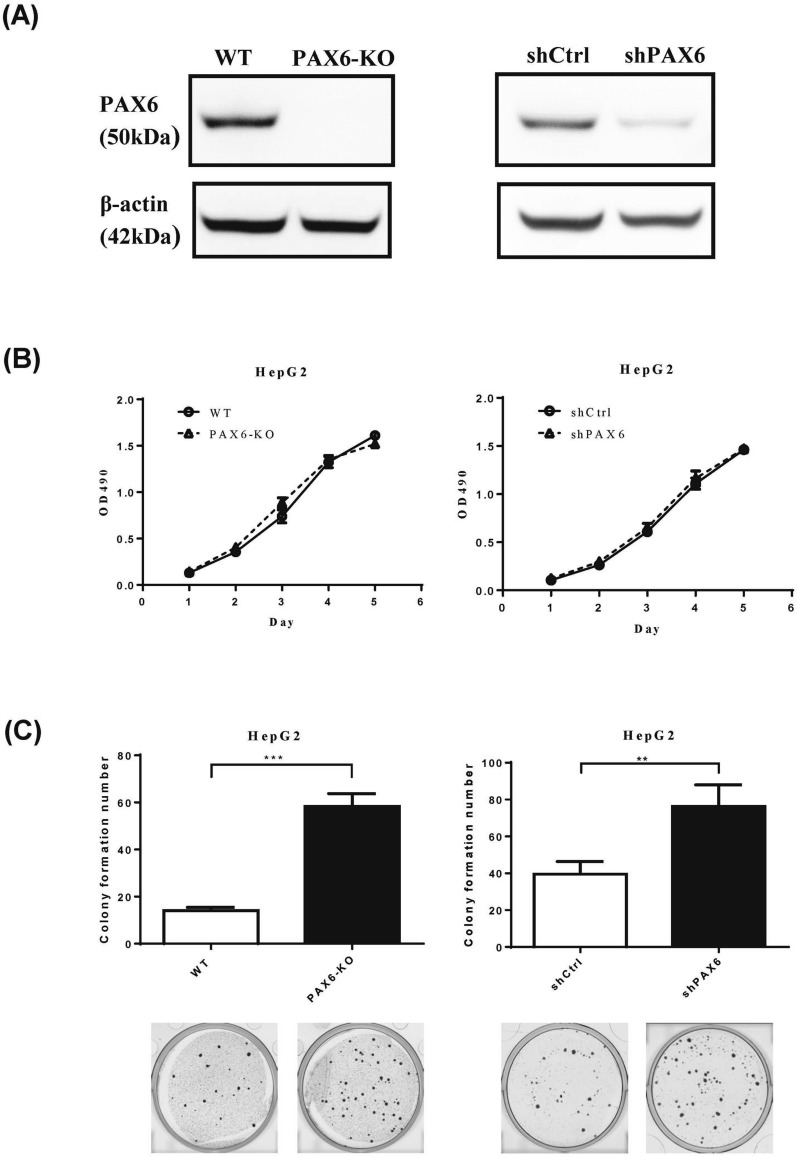
To further verify the effect of PAX6 on tumor progression, we reduced PAX6 expression in HepG2 cells using CRISPR/Cas9 knockout and shRNA knockdown strategies. Silencing/downregulation of PAX6 was confirmed by western blotting (Fig. 3A). The reduction in PAX6 expression in HepG2 cells did not affect cancer cell growth (Fig. 3B) but significantly increased AIG (Fig. 3C).
Fig. 3.
Knockout and knockdown of PAX6 notably reversed the AIG of HepG2 cancer cells. A For generation of PAX6 knockout cells, HepG2 cancer cells were transfected with the All-in-one CRISPR vector pAll-Cas9.Ppuro with PAX6 sgRNAs. Moreover, HepG2 cells were transduced with lentiviruses harboring control shRNA (shCtrl) or PAX6 shRNA. PAX6 expression was analyzed by western blot analysis. β-Actin was used as an internal control. B Cell proliferation (MTS) assays and C AIG assays were performed in PAX6 knockout and PAX6 knockdown HepG2 cells. The data are expressed as the mean ± SE. Significant differences were determined using the Mann‒Whitney U test. ** p < 0.01, *** p < 0.001
PAX6 overexpression inhibits colony formation and invasion of HA22T cancer cells in an inducible expression system
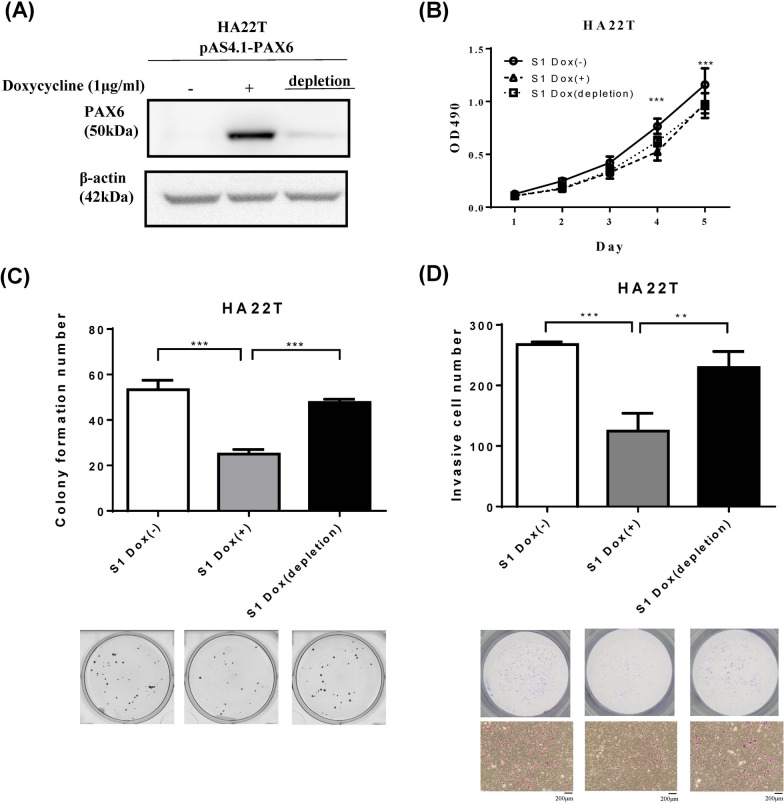
Then, we used an inducible expression system in HA22T cancer cells to confirm the suppressive effect of PAX6. We treated the cells with 1 µg/mL doxycycline (Dox) to induce PAX6 expression and performed western blotting to confirm that PAX6 protein expression was induced after Dox treatment (Fig. 4A). We evaluated the effect of PAX6 restoration on cell growth, AIG, and cell invasion after Dox induction for seven days and Dox depletion for seven days. The viability of the PAX6-overexpressing cells significantly differed from that of the controls (Fig. 4B). PAX6 restoration decreased AIG (Fig. 4C) and invasion (Fig. 4D). These results were accordant with the data acquired in PAX6 overexpression in a constitutive system. Moreover, the inhibitory effects of PAX6 on colony formation and cell invasion were reversed after PAX6 expression was decreased by withdrawal of Dox. In summary, these data supported that PAX6 represses cell transformation and the invasive phenotype in liver cancer.
Fig. 4.
The restoration of PAX6 inhibited the colony formation and invasion of HA22T cancer cells in an inducible expression system. A Dox (1 µg/mL)-induced PAX6 expression was established in HA22T cells, and PAX6 expression after treatment with Dox for 7 days or Dox depletion for another 7 days was determined by western blot analysis. Then, MTS and AIG assays were performed on schedule. B MTS assays were used to examine the effect of PAX6 on the proliferation of HA22T cells. C A colony formation assay was used to determine the effect of PAX6 on cell growth. D Matrigel invasion assays were performed on HA22T cells treated with Dox (1 µg/mL) or with Dox depleted. The data are expressed as the mean ± SE. Significant differences were determined using the Mann‒Whitney U test. ** p < 0.01, *** p < 0.001
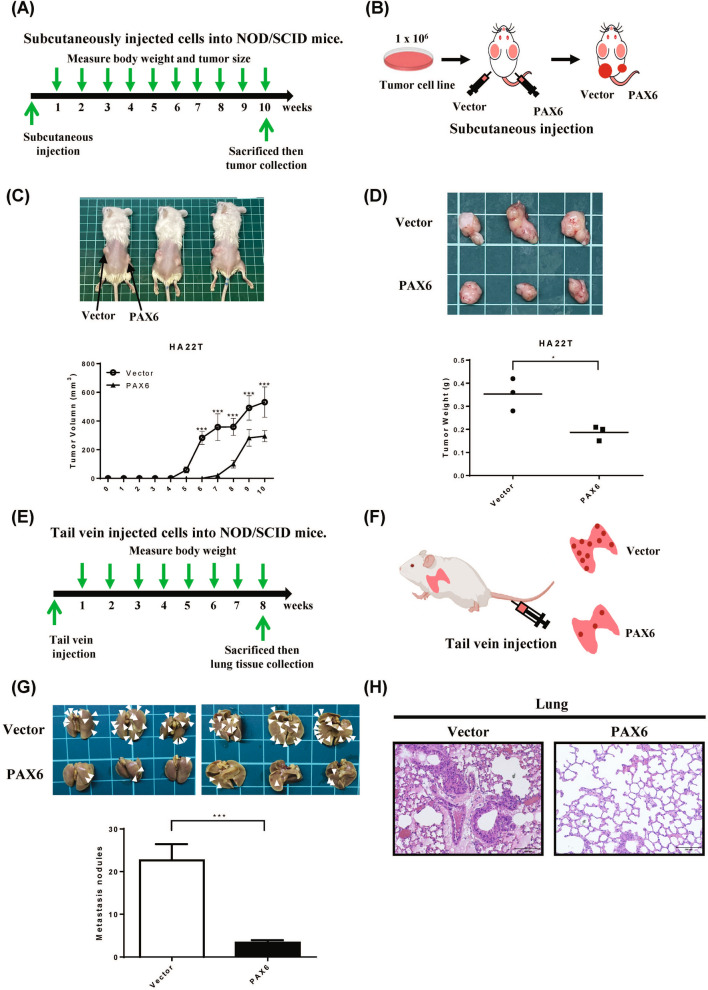
Restoration of PAX6 expression inhibits tumor growth and metastasis in NOD/SCID mice
To assess the influence of PAX6 expression on tumor development, we subcutaneously injected HA22T cells with PAX6 expression into NOD/SCID mice (Fig. 5A. B). The subcutaneous tumor in the PAX6 group grew slower than in the empty vector (Fig. 5C). The mean tumor volume in the PAX6-overexpressing group was significantly smaller than in the vector group (Fig. 5C). After ten weeks, the tumors were cut off and weighed. The mean tumor weight in the PAX6-overexpressing group was significantly lower than in the vector control group (Fig. 5D). To further validate the effect of PAX6 on metastasis in vivo, we injected HA22T cells with PAX6 expression or HA22T cells with control vector into mice through the tail vein (Fig. 5E, F ). Eight weeks after injection, the metastatic lung nodules in the PAX6 group were fewer than in the vector group (Fig. 5G, H). Our data further proved that the overexpression of PAX6 suppresses tumor growth and tumor metastasis in xenograft mouse models.
Fig. 5.
PAX6 overexpression inhibited tumor growth and metastasis in vivo. A A graph demonstrating the time schedule of the xenograft experiments. B The detailed operations of xenograft mice are illustrated. PAX6-HA22T cells or vector-HA22T cells were subcutaneously injected into the left and right flanks of NOD/SCI) mice. C The tumor growth curves of PAX6-expressing cells were compared with those of vector-only cells. D The tumor weight from the PAX6 group was compared to those of vector-alone group. E F The time schedule of the in vivo assay and the detailed procedures are illustrated in the diagrams. PAX6-HA22T cells or control vector-HA22T cells were injected into mice via the tail vein. G Eight weeks after injection, lung tissues were excised from the mice; the arrows indicate lung nodules. The data are expressed as the mean ± SE. Significant differences were determined using the unpaired two-tailed t test. * p < 0.05 and *** p < 0.001. (H) Representative images of hematoxylin and eosin (H&E)-stained lungs (original magnification, × 200) from the mice
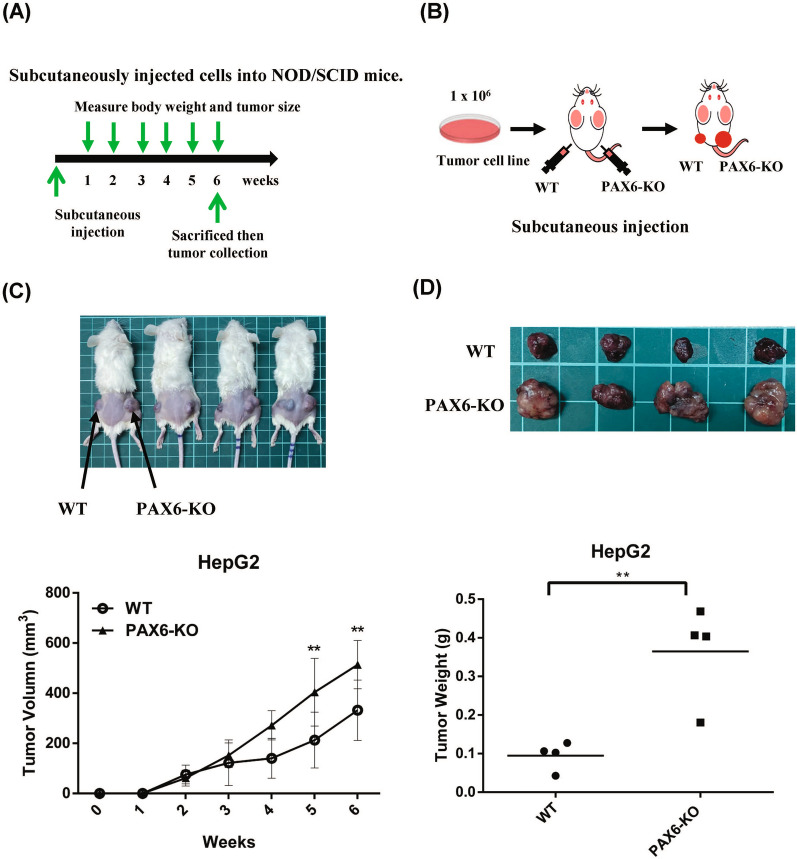
Inhibition of PAX6 in HepG2 cells increases tumor growth in a xenograft mouse model
To further confirm the effect of PAX6 expression on tumor growth, we subcutaneously injected PAX6 knockout HepG2 cells into NOD/SCID mice (Fig. 6A, B). The subcutaneous tumor growth of HepG2 cells with the empty vector or PAX6 knockout in NOD/SCID mice was shown in Fig. 6C. The mean tumor volume was significantly greater in the PAX6 knockout group than in the vector group (p < 0.01) (Fig. 6C). After six weeks, the tumors were removed and weighed. The mean tumor weight was significantly greater in the PAX6 knockout group than in the vector group (p < 0.01) (Fig. 6D).
Fig. 6.
PAX6 knockout in HepG2 cells promoted tumor growth in xenograft NOD/SCID mice. A A diagram demonstrating the time schedule of the xenograft experiments. B The detailed operations of xenograft mice are illustrated. PAX6 knockout HepG2 cells or vector-HepG2 cells were subcutaneously injected into the left and right flanks of NOD/SCID mice. C The tumor growth curves of PAX6 knockout cells were compared with those of vector-only cells. D The weights of the PAX6 knockout tumors were compared to those of the vector-treated tumors. The data are expressed as the mean ± SE. Significant differences were determined using the unpaired two-tailed t test. ** p < 0.01
PAX6 functions as a tumor suppressor partly through CDH1 and THBS1
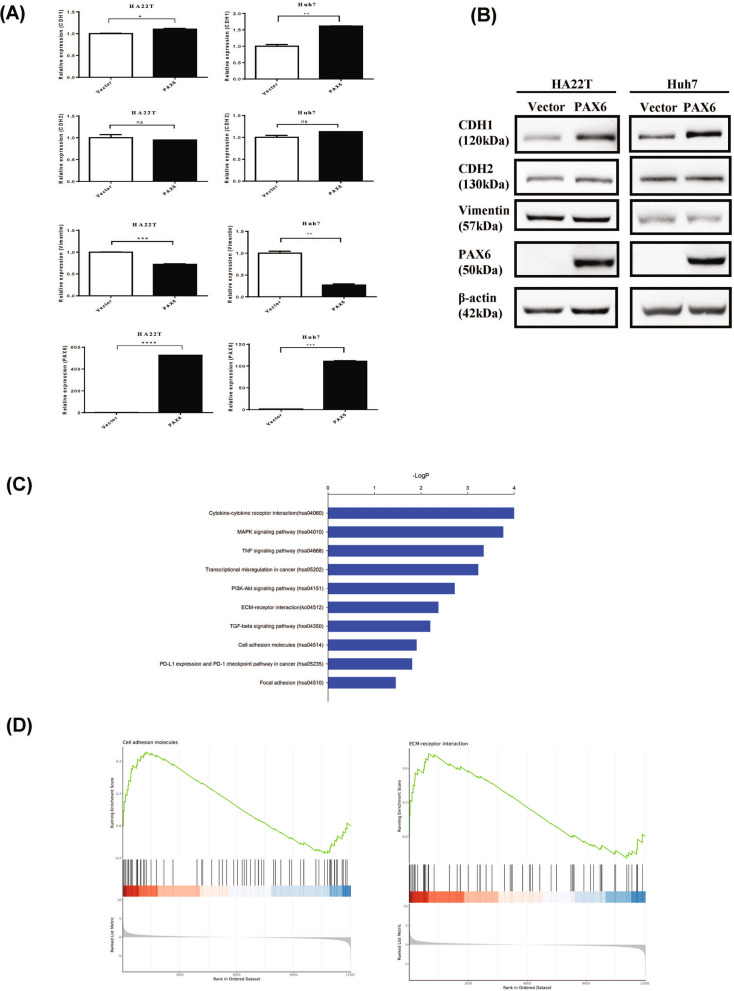
During the early stage of metastasis, a subset of cancer cells undergoes EMT, disseminate from their primary sites and travel to distant organs [17]. We first investigated whether the effect of PAX6 on invasion is associated with regulation of EMT. PAX6 overexpression did not significantly change the expression of the EMT-related transcription factors Snail, Twist1, or Slug but did change the expression of CDH1 and vimentin at the mRNA level, as indicated in Fig. 7A, B, and Supplementary Figure S2. Furthermore, CDH1 protein expression was concordant with CDH1 mRNA expression in the HCC cell lines, HA22T and Huh7.
Fig. 7.
Identification of potential target genes regulated by PAX6 overexpression in HCC. A The mRNA expression levels of genes related to EMT, such as CDH1, CDH2, and vimentin, were analyzed by qRT‒PCR. The data were normalized to the housekeeping gene GAPDH and are shown relative to the vector control. The data are expressed as the mean ± SE. Statistical significance was determined using the Mann–Whitney U test. * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001, ns: not significant. B Protein levels were determined by western blotting. C KEGG analysis of the most representative pathways controlled by PAX6. The pathway analysis was accomplished with DAVID (https://david.ncifcrf.gov/). D Gene set enrichment analysis (GSEA) was performed to search for key signaling pathways related to PAX6 TSG function, and the results revealed that ECM receptor interactions (P = 0.021) and cell adhesion molecules (P = 0.017) are key pathways
To further explore the possible downstream signaling pathways regulated by PAX6 that mediate the tumor suppressor effects, we performed RNA-seq analysis of PAX6-expressing HA22T cells. Consistent with the tumor suppressor function of PAX6, Gene Ontology (GO) analysis and KEGG pathway analysis using DAVID software (https://david.ncifcrf.gov/) revealed enrichment in pathways such as transcriptional misregulation in cancer, PI3K-Akt, ECM receptor interaction, cell adhesion molecules, the TGF-β pathway, and focal adhesion (Fig. 7C, Supplementary Figure S3, Supplementary Table S2). Gene set enrichment analysis (GSEA) revealed that ECM receptor interactions and cell adhesion molecules are key pathways (Fig. 7D). Among these genes, we identified and confirmed one putative target gene, THBS1, that plays a role in the ECM receptor interactions and cell adhesion molecules. THBS1 was inhibited in PAX6-expressing HA22T cells at the mRNA and protein levels (Supplementary Figure S4, S5).
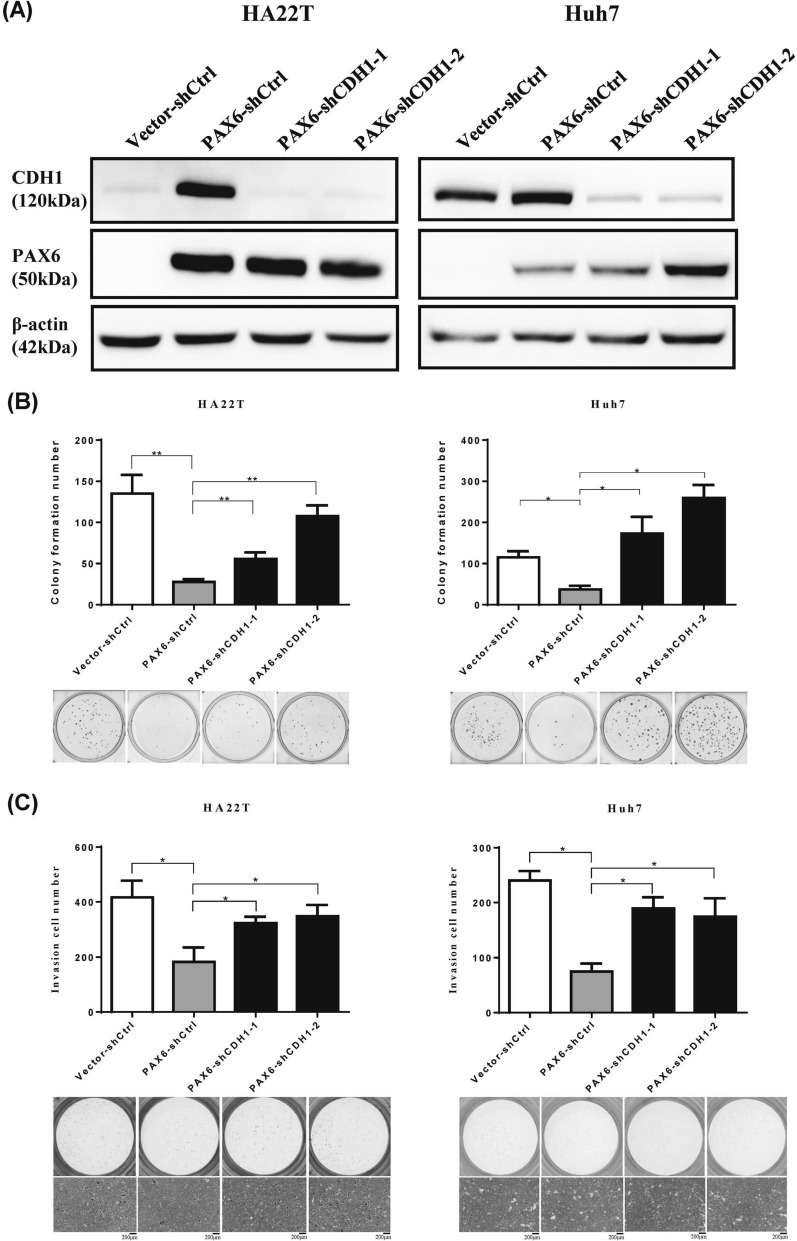
To investigate whether CDH1 is related to the tumor-suppressive function of PAX6, we knocked down CDH1 in PAX6-overexpressing cells (HA22T and Huh7). Figure 8A showed that CDH1 was successfully downregulated using CDH1-shRNA. Then, we performed colony formation and invasion assays in PAX6-overexpressing HCC cells (HA22T and Huh7 cells). Knockdown of CDH1 in PAX6-overexpressing HCC cells significantly reversed the malignant phenotypes (Fig. 8B, C).
Fig. 8.
Knockdown of CDH1 in PAX6-expressing HCC cells repressed the tumor suppressive function of PAX6. The expression of CDH1 in PAX6-expressing HCC cells (HA22T and Huh7) transfected with shCtrl or CDH1 shRNAs (shRNA CDH1-1 and shRNA CDH1-2) was analyzed by western blotting analysis. β-Actin was used as an internal control. B Colony formation and C Matrigel invasion assays were used to analyze the malignant phenotypes. The data are presented as the mean ± SE. Statistical significance was calculated with the Mann‒Whitney U test. * p < 0.05 and ** p < 0.01
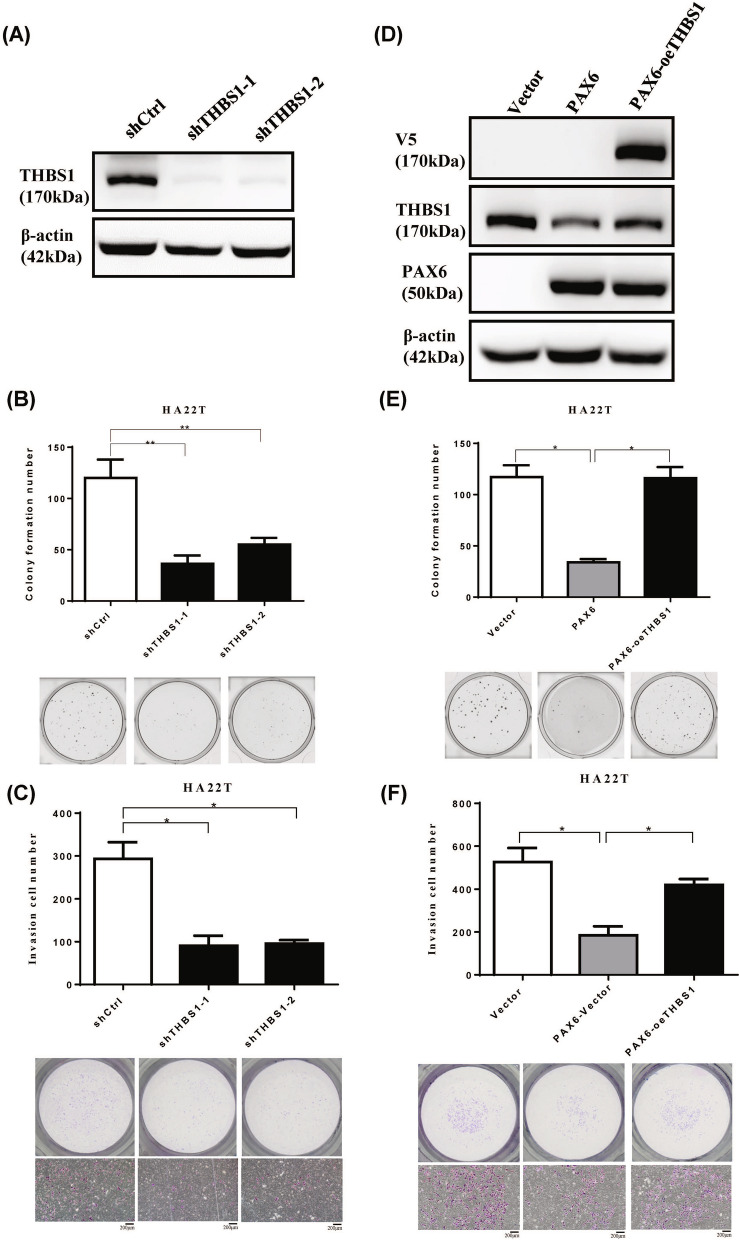
To further elucidate the role of THBS1 expression in HCC progression, THBS1 was knocked down using THBS1-shRNA. Then we performed colony formation and invasion assays in HA22T cells (Fig. 9A–C). Our data further suggest that PAX6 inhibits AIG and cell invasion by partly regulating the expression of THBS1. To test this hypothesis, we conducted phenotypic rescue experiments. Compared with that in the control cells, the overexpression of THBS1-V5 in the PAX6-expressing HA22T cells significantly reversed the antitumor effect on PAX6 expression (Fig. 9D–F). Taken together, these data suggest that CDH1 and THBS1 contribute to the tumor-suppressive function of PAX6 in HCC.
Fig. 9.
Overexpression of THBS1 in PAX6-expressing HCC cells inhibited the antitumor function of PAX6. A We check the expression of THBS1 in HA22T cells transfected with THBS1 shRNAs (shTHBS1-1 and shTHBS1-2) or shCtrl via western blotting analysis. B Colony formation assays and C invasion assays were performed in HA22T cells. D We analyze the protein level of HA22T cells transfected with the specific combination of vectors, control vector, PAX6, or THBS1-V5. E AIG assays and F Matrigel invasion assays were employed to analyze the malignant phenotypes. The data are shown as the mean ± SE. We used the Mann‒Whitney U test to perform statistical analysis. * p < 0.05 and ** p < 0.01
Discussion
We investigated the function of the transcription factor PAX6 in the progression of HCC, and the results suggested that PAX6 functions as a tumor suppressor. We provide in vitro and in vivo data demonstrating that PAX6 represses tumorigenicity and cancer metastasis. Interestingly, CDH1 upregulation and THBS1 downregulation contributed to the inhibitory effect of PAX6 on tumor growth and metastasis in HCC.
The hypermethylation of PAX6 was inversely correlated with its mRNA expression (NM_000280.4) in HCC tissue samples from TLCN. However, due to the limited clinical information and sample numbers, we did not analyze the correlation between PAX6 mRNA expression and clinical parameters. Furthermore, we confirmed the significant PAX6 promoter methylation in the LIHC groups in the TCGA database. However, there was no significant correlation between patient survival and PAX6 mRNA expression in the TCGA cohort using the DNMIVD website (http://119.3.41.228/dnmivd/) [33]. This difference might be due to the presence of many splicing variants of PAX6.
HepG2, Hep3B, and Huh6 cells were characterized as hepatoblastoma cells in the Cellosaurus database (a knowledge resource on cell lines, https://www.expasy.org/resources/cellosaurus) [45]. HepG2 was originally thought to be an HCC cell line but was recently shown to be derived from hepatoblastoma. Owing to the low invasive ability of HepG2 cells, we did not perform an invasion assay. Knockdown and knockout of PAX6 in HepG2 cells partially restored AIG. Thus, we used an inducible system to mimic the knockdown of PAX6 via the depletion of Dox in the HCC cell line HA22T. Induction of PAX6 using Dox treatment suppressed the malignant phenotype in an inducible system, whereas PAX6 downregulation via depletion of Dox significantly restored AIG and invasion in HA22T HCC cells.
EMT has been shown to promote cancer progression [15–17]. During the EMT process, an upregulation in mesenchymal markers and downregulation of epithelial markers are usually identified [15–17]. Several EMT-related transcription factors, including TWIST, SNAIL, SLUG, and ZEB1 can directly or indirectly increase mesenchymal markers and decrease E-cadherin expression [15–17, 46]. PAX6 did not affect EMT-related transcription factors but increased CDH1 expression at the mRNA and protein levels in HCC. However, the protein levels of mesenchymal markers CDH2 and VIM were not significantly affected. To date, increasing evidence has indicated the presence of a transitional status between epithelial and mesenchymal phenotypes, e.g., the “hybrid epithelial–mesenchymal (hybrid E/M)” state [47–49], providing a possible explanation for these incompatible results. Furthermore, CDH1 knockdown promoted the invasive phenotype in PAX6-overexpressing HCC cells. PAX6 exerts its TSG function partly through the upregulation of CDH1. Using the Eukaryotic Promoter Database (EPD) (http://epd.vital-it.ch) [50], we identified three putative Pax6 binding sites in the CDH1 promoter, which are located -1148, -365, and -292 bp from the transcriptional start site (TSS). These data confirm that CDH1 is a potential target of PAX6.
RNA-seq data revealed that the ECM might be important for downstream signaling regulated by PAX6. ECM has been shown to play a crucial role in tumor growth and metastasis [51, 52]. THSB1 is a component of the ECM that is involved in regulating cancer development and the tumor vasculature. Moreover, high THBS1 expression was associated with tumor invasiveness and progression in HCC. THBS1 was upregulated in high-grade gliomas and was associated with poor prognosis [53]. Thomas Daubon et al. showed that TGFβ1 increased THBS1 expression via direct transcriptional activation through SMAD3 in glioblastoma (GBM) [54]. In this study, compared with the control, the overexpression of THBS1-V5 in PAX6-expressing HA22T cells significantly reversed the antitumor effect of PAX6 expression in phenotypic rescue experiments (Fig. 9D–F). Using the EPD (http://epd.vital-it.ch), we identified three putative homeobox binding sites in the distal promoter of THBS1 at approximately 4 kb. The data confirms that THBS1 is also a potential target of PAX6.
However, our study has limitations. First, we did not perform an orthotopic xenograft mouse model to prove the tumor suppressor function of PAX6 in vivo. Second, we failed to find the optimum PAX6 antibody for chromatin immunoprecipitation-PCR experiments. Whether PAX6 directly or indirectly controls the expression of CDH1 and THBS1 requires further investigation.
In summary, these results demonstrated that PAX6 is frequently downregulated by promoter hypermethylation in HCC, which may lead to aberrant expression of CDH1 and THBS1. PAX6 overexpression reduced the malignant phenotype of HCC cells. These findings suggest that PAX6 may function as an important TSG by regulating CDH1 and THBS1 during the progression of HCC.
Supplementary Information
Acknowledgements
The study was conducted using resources from Taiwan Liver Cancer Network (TLCN). RNA interference reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, supported by the National Research Program for Genomic Medicine Grants of NSC (NSC 97-3112-B-001-016). We thank the National RNAi Core Facility at Academia Sinica in Taiwan for providing shRNA reagents, CRISPR/Cas reagents, and related services. We thank the Instrument Center of National Defense Medical Center for technical support. We thank Yu-Ching Chou for statistical analysis and Chia-Hsin Lin and Shin-Ping Lin for technical assistance.
Abbreviations
- PAX6
Paired box 6
- THBS1
Thrombospondin-1
- HCC
Hepatocellular carcinoma
- LIHC
Liver hepatocellular carcinoma
- RT-PCR
Reverse transcription-polymerase chain reaction
- MSP
Methylation-specific PCR
- CDH1
E-cadherin
- CDH2
N-cadherin
- EMT
Epithelial-mesenchymal transition
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- Q-MSP
Quantitative methylation-specific PCR
- DAC
5-Aza-2ʹ-deoxycytidine
- TSA
Trichostatin A
Author contributions
Conceptualization, C.-H.Y., Y.-L.S and Y.-W.L.; Formal analysis, C.-H.Y., R.-Y. C., S.-Y.C., T.-H.W., Y.-L.S. and Y.-W.L.; Methodology, C.-H.Y., R.-Y. C., Y.-L.S. and Y.-W.L.; Investigation, C.-H.Y., R.-Y. C., S.-Y.C., T.-H.W., Y.-L.S., and Y.-W.L.; Resources,, T.-Y. H., Y.-L.S. and Y.-W.L.; Supervision, T.-Y. H., Y.-L.S and Y.-W.L.; Writing—original draft, C.-H.Y. and Y.-W.L.; Writing—review & editing, Y.-L.S and Y.-W.L.; All authors reviewed the the manuscript.
Funding
This work was supported in part by the following grants: MOST 113–2314-B-016–040, MOST 110–2314-B-016–041, and MOST 107–2314-B-016–012 from the Ministry of Science and Technology, Taiwan, Republic of China; MND-MAB-110–009, MND-MAB-110–010, MND-MAB-110–011, MND-MAB-C01-111001, MND-MAB-C01-111002, MND-MAB-C01-111003, MND-MAB-C11-112044 and MND-MAB-C11-112043 from the Ministry of National Defense, Taiwan, Republic of China; TSGH-E-110191, TSGH-PH_E_111010, TSGH-PH_E_112010, TSGH-D-112103 and TSGH-E-112237 from Tri-Service General Hospital, Taiwan, Republic of China; The Liver Disease Prevention and Treatment Research Foundation, Taiwan, Republic of China.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Conflicts of interest
The authors declare no competing interests.
Informed Consent Statement
Not applicable. Patient consent was waived due to the human specimen being obtained from Taiwan Liver Cancer Network (TLCN) and approved by the TLCN User Committee and Tri-Service General Hospital (TSGH) Institutional Review Board (TSGHIRB: B-109–02).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ti Hui Wu and Shan Yueh Chang These two authors equally contributed to this work.
Contributor Information
Yu-Lueng Shih, Email: albreb@ms28.hinet.net.
Ya-Wen Lin, Email: ndmc.yawen@msa.hinet.net.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. 10.1016/s1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 3.Choi C, Choi GH, Kim TH, Tanaka M, Meng MB, Seong J. Multimodality management for barcelona clinic liver cancer stage C hepatocellular carcinoma. Liver Cancer. 2014;3:405–16. 10.1159/000343861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong ZV, Tanabe KK. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120:2824–38. 10.1002/cncr.28730. [DOI] [PubMed] [Google Scholar]
- 5.Keating GM. Sorafenib: a review in hepatocellular carcinoma. Target Oncol. 2017;12:243–53. 10.1007/s11523-017-0484-7. [DOI] [PubMed] [Google Scholar]
- 6.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 7.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–87. 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 8.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84-s101. 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Braghini MR, Lo Re O, Romito I, Fernandez-Barrena MG, Barbaro B, Pomella S, Rota R, Vinciguerra M, Avila MA, Alisi A. Epigenetic remodelling in human hepatocellular carcinoma. J Experimen Clinic Cancer Res CR. 2022;41:107. 10.1186/s13046-022-02297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feitelson MA. Parallel epigenetic and genetic changes in the pathogenesis of hepatitis virus-associated hepatocellular carcinoma. Cancer Lett. 2006;239:10–20. 10.1016/j.canlet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Meza G, von Felden J, Gonzalez-Kozlova EE, Garcia-Lezana T, Peix J, Portela A, Craig AJ, Sayols S, Schwartz M, Losic B, Mazzaferro V. DNA Methylation Profiling of Human Hepatocarcinogenesis. Hepatology. 2021;74(1):183–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Pinyol R, Kelley RK, El-Khoueiry A, Reeves HL, Wang XW, Gores GJ, Villanueva A. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nature cancer. 2022;3:386–401. 10.1038/s43018-022-00357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Lezana T, Lopez-Canovas JL, Villanueva A. Signaling pathways in hepatocellular carcinoma. Adv Cancer Res. 2021;149:63–101. 10.1016/bs.acr.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Wolinska E, Skrzypczak M. Epigenetic changes affecting the development of hepatocellular carcinoma. Cancers. 2021;13(16):4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Investig. 2009;119:1420–8. 10.1172/jci39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–86. 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Mishra R, Gorlov IP, Chao LY, Singh S, Saunders GF. PAX6, paired domain influences sequence recognition by the homeodomain. J Biol Chem. 2002;277:49488–94. 10.1074/jbc.M206478200. [DOI] [PubMed] [Google Scholar]
- 19.Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 1997;11:1122–31. [DOI] [PubMed] [Google Scholar]
- 20.Asami M, Pilz GA, Ninkovic J, Godinho L, Schroeder T, Huttner WB, Götz M. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Developm Cambridge England. 2011;138:5067–78. 10.1242/dev.074591. [DOI] [PubMed] [Google Scholar]
- 21.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5: e1000511. 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cvekl A, Callaerts P. PAX6: 25th anniversary and more to learn. Exp Eye Res. 2017;156:10–21. 10.1016/j.exer.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Shyr CR, Tsai MY, Yeh S, Kang HY, Chang YC, Wong PL, Huang CC, Huang KE, Chang C. Tumor suppressor PAX6 functions as androgen receptor co-repressor to inhibit prostate cancer growth. Prostate. 2010;70:190–9. 10.1002/pros.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayes DA, Hu Y, Teng Y, Siegel E, Wu X, Panda K, Tan F, Yung WK, Zhou YH. PAX6 suppresses the invasiveness of glioblastoma cells and the expression of the matrix metalloproteinase-2 gene. Can Res. 2006;66:9809–17. 10.1158/0008-5472.Can-05-3877. [DOI] [PubMed] [Google Scholar]
- 25.Zhou YH, Wu X, Tan F, Shi YX, Glass T, Liu TJ, Wathen K, Hess KR, Gumin J, Lang F, et al. PAX6 suppresses growth of human glioblastoma cells. J Neurooncol. 2005;71:223–9. 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]
- 26.Zong X, Yang H, Yu Y, Zou D, Ling Z, He X, Meng X. Possible role of Pax-6 in promoting breast cancer cell proliferation and tumorigenesis. BMB Rep. 2011;44:595–600. [DOI] [PubMed] [Google Scholar]
- 27.Mascarenhas JB, Young KP, Littlejohn EL, Yoo BK, Salgia R, Lang D. PAX6 is expressed in pancreatic cancer and actively participates in cancer progression through activation of the MET tyrosine kinase receptor gene. J Biol Chem. 2009;284:27524–32. 10.1074/jbc.M109.047209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai SW, Li B, Zhang H, Jonas JB, Zhao BW, Shen L, Wang YC. Pax6 regulates proliferation and apoptosis of human retinoblastoma cells. Invest Ophthalmol Vis Sci. 2011;52:4560–70. 10.1167/iovs.10-5487. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, Yue W, Zhang L, Ma L, Jia W, Qian Z, Zhang C, Wang Y. Downregulation of PAX6 by shRNA inhibits proliferation and cell cycle progression of human non-small cell lung cancer cell lines. PLoS ONE. 2014;9: e85738. 10.1371/journal.pone.0085738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu DM, Zhang T, Liu YB, Deng SH, Han R, Liu T, Li J, Xu Y. The PAX6-ZEB2 axis promotes metastasis and cisplatin resistance in non-small cell lung cancer through PI3K/AKT signaling. Cell Death Dis. 2019;10:349. 10.1038/s41419-019-1591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih YL, Kuo CC, Yan MD, Lin YW, Hsieh CB, Hsieh TY. Quantitative methylation analysis reveals distinct association between PAX6 methylation and clinical characteristics with different viral infections in hepatocellular carcinoma. Clin Epigenetics. 2016;8:41. 10.1186/s13148-016-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding W, Chen J, Feng G, Chen G, Wu J, Guo Y, Ni X, Shi T. DNMIVD: DNA methylation interactive visualization database. Nucleic Acids Res. 2020;48:D856-d862. 10.1093/nar/gkz830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsao CM, Yan MD, Shih YL, Yu PN, Kuo CC, Lin WC, Li HJ, Lin YW. SOX1 functions as a tumor suppressor by antagonizing the WNT/β-catenin signaling pathway in hepatocellular carcinoma. Hepatology. 2012;56(6):2277–87. [DOI] [PubMed] [Google Scholar]
- 35.Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, Liu J, Chan MW, Chu TY, Sun CA, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123:161–7. 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- 36.Lin YW, Tsao CM, Yu PN, Shih YL, Lin CH, Yan MD. SOX1 suppresses cell growth and invasion in cervical cancer. Gynecol Oncol. 2013;131:174–81. 10.1016/j.ygyno.2013.07.111. [DOI] [PubMed] [Google Scholar]
- 37.Tsao CM, Yan MD, Shih YL, Yu PN, Kuo CC, Lin WC, Li HJ, Lin YW. SOX1 functions as a tumor suppressor by antagonizing the WNT/β-catenin signaling pathway in hepatocellular carcinoma. Hepatology. 2012;56(6):2277–87. [DOI] [PubMed] [Google Scholar]
- 38.Chang SY, Kuo CC, Wu CC, Hsiao CW, Hu JM, Hsu CH, Chou YC, Shih YL. Lin YW.NKX6.1 hypermethylation predicts the outcome of stage II colorectal cancer patients undergoing chemotherapy. Genes, Chromosom Cancer. 2018;57(5):268–77. [DOI] [PubMed] [Google Scholar]
- 39.Liu CY, Chao TK, Su PH, Lee HY, Shih YL, Su HY, Chu TY, Yu MH, Lin YW, Lai HC. Characterization of LMX-1A as a metastasis suppressor in cervical cancer. J Pathol. 2009;219:222–31. 10.1002/path.2589. [DOI] [PubMed] [Google Scholar]
- 40.Li HJ, Yu PN, Huang KY, Su HY, Hsiao TH, Chang CP, Yu MH. Lin YW.NKX6.1 functions as a metastatic suppressor through epigenetic regulation of the epithelial–mesenchymal transition. Oncogene. 2016;35(17):2266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung HH, Lee CT, Hu JM, Chou YC, Lin YW. Shih YL.NKX6.1 represses tumorigenesis, metastasis, and chemoresistance in colorectal cancer. Int J Molecul Sci. 2020;21(14):5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216-221. 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 45.Bairoch A. The Cellosaurus, a Cell-Line Knowledge Resource. J Biomolecul Tech: JBT. 2018;29:25–38. 10.7171/jbt.18-2902-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 47.Hiew MSY, Cheng HP, Huang CJ, Chong KY, Cheong SK, Choo KB, Kamarul T. Incomplete cellular reprogramming of colorectal cancer cells elicits an epithelial/mesenchymal hybrid phenotype. J Biomed Sci. 2018;25:57. 10.1186/s12929-018-0461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umbreit C, Flanjak J, Weiss C, Erben P, Aderhold C, Faber A, Stern-Straeter J, Hoermann K, Schultz JD. Incomplete epithelial-mesenchymal transition in p16-positive squamous cell carcinoma cells correlates with β-catenin expression. Anticancer Res. 2014;34:7061–9. [PubMed] [Google Scholar]
- 49.Topel H, Bagirsakci E, Comez D, Bagci G, Cakan-Akdogan G, Atabey N. lncRNA HOTAIR overexpression induced downregulation of c-Met signaling promotes hybrid epithelial/mesenchymal phenotype in hepatocellular carcinoma cells. Cell Commun Signal. 2020;18:110. 10.1186/s12964-020-00602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Périer RC, Praz V, Junier T, Bonnard C, Bucher P. The eukaryotic promoter database (EPD). Nucleic Acids Res. 2000;28:302–3. 10.1093/nar/28.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giussani M, Triulzi T, Sozzi G, Tagliabue E. Tumor extracellular matrix remodeling: new perspectives as a circulating tool in the diagnosis and prognosis of solid tumors. Cells. 2019;8(2):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker C, Mojares E, del Río HA. Role of extracellular matrix in development and cancer progression. Int J Molecul Sci. 2018;19(10):3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poon RT, Chung KK, Cheung ST, Lau CP, Tong SW, Leung KL, Yu WC, Tuszynski GP, Fan ST. Clinical significance of thrombospondin 1 expression in hepatocellular carcinoma. Clinic Cancer Res Off J Am Assoc Cancer Res. 2004;10:4150–7. 10.1158/1078-0432.Ccr-03-0435. [DOI] [PubMed] [Google Scholar]
- 54.Daubon T, Léon C, Clarke K, Andrique L, Salabert L, Darbo E, Pineau R, Guérit S, Maitre M, Dedieu S, et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat Commun. 2019;10:1146. 10.1038/s41467-019-08480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.