Abstract
NIP7 encodes a conserved Saccharomyces cerevisiae nucleolar protein that is required for 60S subunit biogenesis (N. I. T. Zanchin, P. Roberts, A. DeSilva, F. Sherman, and D. S. Goldfarb, Mol. Cell. Biol. 17:5001–5015, 1997). Rrp43p and a second essential protein, Nop8p, were identified in a two-hybrid screen as Nip7p-interacting proteins. Biochemical evidence for an interaction was provided by the copurification on immunoglobulin G-Sepharose of Nip7p with protein A-tagged Rrp43p and Nop8p. Cells depleted of Nop8p contained reduced levels of free 60S ribosomes and polysomes and accumulated half-mer polysomes. Nop8p-depleted cells also accumulated 35S pre-rRNA and an aberrant 23S pre-rRNA. Nop8p-depleted cells failed to accumulate either 25S or 27S rRNA, although they did synthesize significant levels of 18S rRNA. These results indicate that 27S or 25S rRNA is degraded in Nop8p-depleted cells after the section containing 18S rRNA is removed. Nip7p-depleted cells exhibited the same defects as Nop8p-depleted cells, except that they accumulated 27S precursors. Rrp43p is a component of the exosome, a complex of 3′-to-5′ exonucleases whose subunits have been implicated in 5.8S rRNA processing and mRNA turnover. Whereas both green fluorescent protein (GFP)-Nop8p and GFP-Nip7p localized to nucleoli, GFP-Rrp43p localized throughout the nucleus and to a lesser extent in the cytoplasm. Distinct pools of Rrp43p may interact both with the exosome and with Nip7p, possibly both in the nucleus and in the cytoplasm, to catalyze analogous reactions in the multistep process of 60S ribosome biogenesis and mRNA turnover.
Ribosome biogenesis in eukaryotes occurs mostly in the nucleolus, where rRNA is transcribed, processed, and covalently modified, newly synthesized ribosomal proteins (rproteins) are delivered, and 40S and 60S subunits are assembled. While the mechanism of ribosome synthesis is complex and poorly understood, significant progress has been made toward identifying and characterizing rproteins, and the many nonribosomal factors that are required for ribosome biogenesis, in Saccharomyces cerevisiae. The completion of the yeast genome project secured the identification of 137 genes (59 of which were duplicated) that encode 32 40S and 46 60S rproteins (32). The 18S rRNA of 40S subunits and the 5.8S and 25S rRNAs of 60S subunits are transcribed as a single 35S pre-rRNA. The organization of 35S pre-rRNA, including the locations of the mature rRNAs; two internal transcribed spacers, ITS1 and ITS2; and two external transcribed spacers, 5′ ETS and 3′ ETS, is shown in Fig. 1. rRNAs are formed by ordered endo- and exonucleolytic digestion of the spacer sequences and by covalent modifications that include ribose and base methylation and conversion of uridine residues to pseudouridine (27, 40, 61).
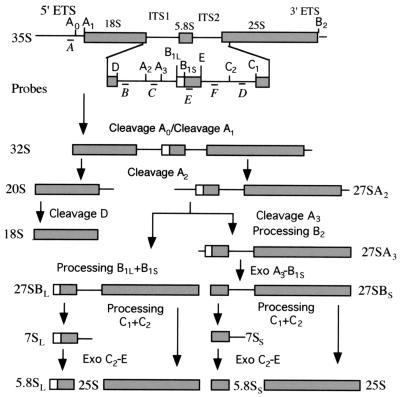
FIG. 1.
Structure and relevant processing steps of the 35S pre-rRNA in S. cerevisiae. In the 35S pre-rRNA, sequences of the mature 18S, 5.8S, and 25S rRNAs are separated by two internal transcribed spacers (ITS1 and ITS2) and flanked by 5′ and 3′ external transcribed spacers. The 5′ ETS is removed by sequential cleavages at A0 and A1 to generate 32S pre-rRNA. Processing of ITS1 and ITS2 is more complex. Cleavage of 32S pre-rRNA at A2 separates 20S pre-rRNA from 27SA2 pre-rRNA. Cytoplasmic processing of 20S pre-rRNA to mature 18S rRNA occurs by cleavage at D. 5′ processing of 27SA2 pre-rRNA follows alternative pathways. Approximately 90% of 27SA2 pre-rRNA undergoes cleavage at A3, producing 27SA3, which is subsequently processed at B1S to produce 27SBS. The remaining 10% is processed at B1L, which produces 27SBL pre-rRNA. At the stage of 27S pre-rRNA, the mature 3′ end of 25S rRNA is formed by cleaving 27S pre-rRNAs at B2. Subsequent processing of 27SBS and 27SBL precursors is identical. Cleavage of these precursors at C2 and C1 releases 25S rRNA and two forms of 7S pre-rRNA, 7SS and 7SL, respectively. The mature end of 5.8SS and 5.8SL rRNAs are produced by 3′→5′ digestion to site E of 7S pre-rRNAs. The longest form, 5.8SL rRNA, contains a 5′ 6- to 8-nucleotide extension. Letters below the 35S pre-rRNA diagram (probes A to F) indicate the positions of the oligonucleotide probes used in Northern blot analysis.
Many, but certainly not all, trans-acting factors that function in pre-rRNA processing and ribosome assembly have been identified. Four small nucleolar RNAs (snoRNAs), U3, U14, snR10, and snR30 (20, 28, 36, 51), have been implicated in pre-rRNA cleavages leading to the synthesis of 18S rRNA, but most snoRNAs function as guides for enzymes that catalyze either rRNA methylation or pseudouridinylation (48, 54). Several nucleases are responsible for endo- and exonucleolytic processing and for degradation of the spacer sequences. Cleavages at site A0 and at a site in the 3′ ETS are dependent on Rnt1p, a homologue of prokaryote RNase III (12). RNase MRP is responsible for endonucleolytic processing at A3 in ITS1 (31). Two 5′-to-3′ exonucleases, Rat1p and Xrn1p, are required for maturation of the 5.8SS rRNA 5′ end (17) and for the degradation of several excised spacer sequences, including fragments A0 to A1, A2 to A3, and D to A2 (27, 49). The five subunits of the exosome complex (35) and Rrp6p (5) are required for maturation of 5.8S rRNA. The putative RNA helicases Fal1p, Dbp4p, Rok1p, and Rrp3p (25, 30, 39, 56) are required for 40S subunit synthesis, and the putative helicases Dbp3p, Dbp6p, Dbp7p, Dob1p, Drs1p, and Sbp4p are required for 60S synthesis (7, 8, 26, 41, 43, 60).
In addition to the snoRNAs, nucleases, and RNA helicases, a number of other nucleolar proteins are required for ribosome biogenesis. The best characterized of these proteins is Nop1p, an abundant protein that is required for many steps of both 40S and 60S synthesis, including rRNA methylation and processing and subunit assembly (52, 53). Nop1p is common to a large number of snoRNAs (45) and associates with four nucleolar proteins, Nop56p, Nop58p, Nop77p/Nop4p, and Sof1p (4, 13, 21, 50). Several other proteins, including Gar1p, Mpp10p, Sof1p, and Ssb1p, were shown to interact with one or more snoRNAs (6, 11, 15, 21). Genetic depletion of Gar1p, Mpp10p, and Sof1p impairs 18S synthesis and 40S subunit formation (11, 15, 21). On the other hand, Nip7p, Nop2p, Nop56p, Nop58p, and Nop77p/Nop4p are involved in 60S subunit synthesis (4, 13, 19, 50, 62). Rrp5p has been implicated in 18S and 5.8S rRNA synthesis, since its deficiency blocks cleavages in the 5′ ETS and ITS1, leading to accumulation of an unusual 24S intermediate (55). Although these nucleolar proteins are implicated in the process of ribosome biogenesis, the biochemical activities for most are unknown.
We previously characterized Nip7p in S. cerevisiae (62). Nip7p homologues have been identified in, for example, humans, Caenorhabditis elegans, and Arabidopsis thaliana. The depletion of Nip7p preferentially impairs the synthesis of 60S subunits, most probably due to a kinetic delay in 27S pre-rRNA processing. Nip7p localized to the nucleolus, but significant amounts cosedimented with free 60S subunits. The biochemical function of Nip7p, especially its role when it is associated with free 60S subunits, is unknown. In order to learn more about the function of Nip7p, we identified several putative Nip7p-interacting proteins, using a two-hybrid screen. We report here on the interaction of Nip7p with the exosome subunit Rrp43p (34) and with a previously uncharacterized nucleolar protein, Nop8p. Rrp43p shows sequence similarity to bacterial RNase PH, a 3′-to-5′ exoribonuclease, and in vivo depletion of Rrp43p impairs processing of the 5.8S 3′ end (34). The depletion of Nop8p, which is unlike other proteins, revealed that it too is required for 60S subunit biogenesis. These results suggest that Nip7p, Rrp43p, and Nop8p are subunits of a dynamic complex with essential roles in 60S subunit biogenesis.
MATERIALS AND METHODS
DNA analysis methods and plasmid construction.
DNA cloning and electrophoretic analysis were performed as described by Sambrook et al. (44). DNA sequencing was performed by the Big Dye method (Perkin-Elmer). A cDNA library fused to the GAL4 activation domain was obtained from the American Type Culture Collection (ATCC 87002). Plasmids used in this study are summarized in Table 1, and cloning strategies are briefly described below. The lexA::NIP7 fusion used in the two-hybrid screen was constructed by inserting a PCR-amplified NIP7 open reading frame (ORF) into the BamHI and SalI sites of plasmid pBTM116 (2), generating plasmid pBTM-NIP7. For construction of green fluorescent protein (GFP) fusion proteins, RRP43 and NOP8 ORFs were PCR amplified from yeast genomic DNA with primers containing suitable restriction sites (XbaI and SalI for RRP43 and HindIII and SalI for NOP8) and inserted at the C terminus of GFP into the vector pGFP-N-FUS (38), generating the vectors pGFP-RRP43 and pGFP-NOP8. pDN291 was used for the expression of GFP alone (37). Plasmids YCpGAL-RRP43 and YCpGAL-NOP8 contain a protein A (PrtA) tag fused to the N termini of Rrp43p and Nop8p, respectively, under the control of the GAL1 promoter. The vector YCpGAL-RRP43 was constructed by ligating the following four DNA fragments: the DNA of the vector YCplac33 (14) digested with EcoRI and SalI, the GAL1 promoter isolated from YCpGAL-NIP7 (62) digested with EcoRI and NdeI, the RRP43 ORF isolated from pGFP-RRP43 digested with XbaI and SalI, and a double Staphylococcus aureus PrtA immunoglobulin G (IgG)-binding domain PCR amplified from pBD20 (10) as an NdeI-XbaI fragment. YCpGAL-NOP8 was constructed by a similar strategy, except that the NOP8 ORF was isolated from pGFP-NOP8 as an EcoRI-SalI DNA fragment and the PrtA tag was an NdeI-EcoRI fragment. Plasmids pACT-RRP43 and pACT-NOP8, which bear genes encoding hybrid proteins of the Gal4p activation domain and Rrp43p or Nop8p, respectively, were isolated from L40-derivative strains that showed positive two-hybrid interaction with Nip7p. pGAD424 was used for expression of the Gal4p activation domain (2).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pBTM116 | lexA DNA-binding domain, TRP1 2μm | 2 |
| pGAD-424 | GAL4 activation domain, LEU2 2μm | 2 |
| pBTM-NIP7 | lexA DNA-binding domain::NIP7 TRP1 2μm | This study |
| pACT-RRP43 | GAL4 activation domain::RRP43 LEU2 2μm | This study |
| pACT-NOP8 | GAL4 activation domain::NOP8 LEU2 2μm | This study |
| pGFP-N-FUS | MET25::GFP URA3 CEN6 | 38 |
| pGFP-RRP43 | GFP::RRP43 URA3 CEN6 | This study |
| pGFP-NOP8 | GFP::NOP8 URA3 CEN6 | This study |
| pDN291 | GFP URA3 CEN4 | 37 |
| YCpGAL-RRP43 | GAL::PrtA::RRP43 URA3 CEN4 | This study |
| YCpGAL-NOP8 | GAL::PrtA::NOP8 URA3 CEN4 | This study |
Yeast strains, media, and genetic techniques.
Yeast strains used in this work are listed in Table 2. Yeast strains were grown and analyzed as described by Sherman and coworkers (46, 47). Various carbon sources were added to yeast extract-peptone medium (YP) and synthetic complete medium (SC). YP-dextrose (YPD) or SCGlu and YPGal or SCGal contained either 2% glucose or 1% galactose plus 1% raffinose, respectively, as a carbon source.
TABLE 2.
Yeast strains
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| W303-1a | MATa ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 | M. Nomura |
| W303α | MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 | S. Wente |
| W303a | MATa/MATα ade2-1/ade2-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11 trp1-1/trp1-1 ura3-1/ura3-1 can1-100/can1-100 | This study |
| DG456 | MATa ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 Nop8::KAN p(URA3 GAL::PrtA::NOP8) | This study |
| DG457 | MATa/MATα NOP8/nop8::KAN ade2-1/ade2-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11 trp1-1/trp1-1 ura3-1/ura3-1 can1-100/can1-100 | This study |
| L40 | MATa his3Δ200 trp1-901 leu2-3,311 ade2 lys2-801am URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3 | 18 |
| L40-0 | L40 pBTM-NIP7, pGAD-424 | This study |
| L40-41 | L40 pBTM-NIP7, pACT-NOP8 | This study |
| L40-61 | L40 pBTM-NIP7, pACT-RRP43 | This study |
Generated by mating W303-1a and W303α.
NOP8 gene disruption was achieved by transforming strain W303-1a with a kanr marker (16), which was PCR amplified with primers containing sequences overlapping the NOP8 ORF at the 5′ and 3′ ends. The sequences of the primers used to amplify the kanr marker are 5′CTGAAGTGAGAACTAGGTAATA TACGACGATGGATAGTGTAATTCCAGCTGAAGCTTCGTACGC3′ (5′ primer) and 5′TATACTATATATGTATATATACTCACTATAGAAGAAGCCCGCTCTGCATAGGCGACTAGTGGATCTG3′ (3′ primer). kanr colonies were allowed to sporulate, and tetrads were dissected. Only two spores per tetrad were able to germinate. Both spores were geneticin sensitive (kan−). In order to construct a conditional strain for NOP8, strain DG457 (NOP8/nop8::KAN) was transformed with plasmid YCpGAL-NOP8, which contains NOP8 under the control of the GAL1 promoter, and sporulated. kanr spores carrying this plasmid were able to grow on medium containing galactose but not on medium containing glucose.
Yeast two-hybrid screen for proteins that interact with Nip7p.
The host strain for the two-hybrid screen, L40 (Table 2) (18), contains both yeast HIS3 and Escherichia coli lacZ as reporters for two-hybrid interaction integrated into the genome. Strain L40 was transformed with plasmid pBTM-NIP7, which bears the gene that encodes a hybrid protein containing the lexA DNA-binding domain and the full-length NIP7 ORF. Expression of the fusion protein was confirmed by immunoblot analysis with an antibody raised against Nip7p (data not shown). Subsequently, a large-scale transformation of L40 carrying pBTM-NIP7 was performed with a yeast cDNA library fused to the GAL4 activation domain (ATCC 87002). Approximately 6 × 104 clones were tested for auxotrophy for histidine by plating transformed cells on selective synthetic medium. Fast-growing colonies were transferred to nitrocellulose filters and tested for β-galactosidase (β-Gal) activity, as described by Vojtek and Hollenberg (58). Plasmid DNA was isolated from colonies showing both fast growth on selective plates and high β-Gal activity and submitted to sequencing analysis.
IgG affinity column and immunoblot analysis.
Two hundred optical density at 600 nm (OD600) units of exponentially growing cells was resuspended in 2 ml of ice-cold buffer A (10 mM Tris-Cl [pH 7.6], 100 mM NaCl, 5 mM MgCl2, 1 mM dithiotreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of aprotinin per ml, 1 μg of pepstatin A per ml, 1 μg of leupeptin per ml). Cells were disrupted by vortexing with 1 volume of glass beads, and extracts were cleared by centrifugation at 30,000 × g for 15 min. Seventy-five OD280 units of extract was adjusted to 1.9 ml with buffer A and incubated with 100 μl of IgG-Sepharose beads (Pharmacia) for 2 h. Subsequently, the suspension was transferred to a small column, which was washed extensively with buffer A, and eluted with buffer A containing 0.5 M KCl. A fraction of both PrtA-tagged Nop8p (PrtA-Nop8p) and PrtA-Rrp43p eluted from the IgG columns with buffer A containing 0.5 M KCl, indicating that the affinity of the PrtA-tagged proteins for IgG may be less than that of native PrtA, which normally remains bound under these conditions. All of the steps described above were performed at 4°C. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted to an Immobilon-P membrane (Millipore) as described previously (62). Membranes were blocked with 2% nonfat dry milk in TST buffer (20 mM Tris-Cl [pH 8.0] 150 mM NaCl, 0.05% [vol/vol] Tween 20] and then probed with rabbit polyclonal antiserum raised against Nip7p. Subsequently, the blots were incubated with alkaline phosphatase-conjugated anti-rabbit IgG and visualized with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium as previously described (44).
Subcellular localization of Rrp43p and Nop8p.
The subcellular localization of Rrp43p and Nop8p was analyzed by monitoring the fluorescent signal produced by green fluorescent protein (GFP) fusions to the amino termini of Rrp43p and Nop8p. GFP-Rrp43p and GFP-Nop8p reporter proteins were expressed from the vectors pGFP-RRP43 and pGFP-NOP8 (Table 1), respectively, transformed into strain W303-1a. Plasmid pDN291 (37), which bears the gene for the GFP protein, was used as a control. Images were obtained with a Leica TCS NT confocal microscope, and digital images were processed with MetaMorph software (Universal Imaging Corporation).
Growth curves and analysis of Nop8p depletion.
Growth rates of strains W303-1a (NOP8) and DG456 (GAL::NOP8) in YPGal and YPD cultures were analyzed as follows: exponentially growing YPGal cultures were divided into two fractions, and cells were harvested by centrifugation and resuspended in either YPD or YPGal. Cultures were incubated at 30°C, and the OD600 values were determined at various time points. In order to keep cultures in exponential growth, they were diluted in fresh medium whenever the OD600 reached 0.8. Samples were collected at various times from YPD cultures for the analysis of Nop58p depletion. For isolation of cell extracts, cells were harvested by centrifugation, resuspended in 200 μl of breaking buffer (20 mM HEPES-KOH [pH 7.4], 2 mM Mg diacetate, 100 mM KCl, 1 mM DTT, 0.5 mM EDTA, 1 mM PMSF), and disrupted by vortexing in the presence of 1 volume of glass beads. Cell extracts was cleared by centrifugation and submitted to immunoblot analysis as described above. Rabbit antibody against translation initiation factor eIF-2α (kindly provided by John McCarthy) was used as an internal control.
Polysome profile analysis.
For polysome profile analysis cell extracts were isolated from 300-ml cultures grown to mid-exponential phase in YPGal or shifted to YPD for 12 h. Following addition of 3 ml of cycloheximide (10 mg/ml) to the cultures, cells were harvested by centrifugation and resuspended in 0.5 ml of breaking buffer (20 mM HEPES-KOH [pH 7.4], 2 mM Mg diacetate, 100 mM KCl, 1 mM DTT, 1 mM PMSF, 100 μg of cycloheximide per ml). Cell extracts and sucrose gradients were prepared as described previously (62). Polysomes were separated by centrifugation at 40,000 rpm for 4 h at 4°C with a Beckman SW41 rotor. Gradients were fractionated with a Buchler Auto-densiflow IIC fractionator and monitored at 254 nm with a UA-5 absorbance-fluorescence monitor (ISCO).
Pulse-chase labeling, rRNA electrophoresis, and Northern blot analysis.
Metabolic labeling of rRNA was performed as described previously (59, 62). Exponentially growing cultures of W303-1a (NOP8) and DG456 (GAL::NOP8) were shifted from SCGal to SCGlu lacking methionine and incubated at 30°C for 18 h. Subsequently, cells were pulse-chase labeled with 100 μCi of [methyl-3H]methionine (DuPont-NEN) per ml for 2 min and chased with 100 μg of unlabeled methionine per ml. At various times, samples were taken and quickly frozen in a dry ice-ethanol bath. Total RNA was isolated from yeast cells by the hot-phenol method (24). For analysis of pre-rRNA steady-state levels, RNAs were isolated from strains W303-1a and DG456 incubated in YPGal or shifted to YPD. Samples were collected for RNA extraction at time zero and 28 h after the shift to YPD for W303-1a and at time zero and 6, 12, 20, and 28 h after the shift to YPD for DG456. RNAs were separated by electrophoresis on 1.2% agarose–6% formaldehyde gels and transferred by Northern blotting to Hybond nylon membranes (Amersham) as described previously (44). Membranes were probed with 32P-labeled oligonucleotides complementary to specific regions of the 35S pre-rRNA under the hybridization conditions described previously (51) and submitted to autoradiography. The oligonucleotide probes used (see also Fig. 1) were 5′GGTCTCTCTGCTGCCGGAAATG3′ (probe A); 5′GCTCTCATGCTCTTGCCAAAAC3′ (probe B); 5′TGTTACCTCTGGGCCCCG3′ (probe C); 5′GTTCGCCTAGACGCTCTCTTC3′ (probe D); 5′CGTATCGCATTTCGCTGCGTTC3′ (probe E); and 5′GGCCAGCAATTTCAAGTTAAC3′ (probe F).
RESULTS
Rrp43p and Nop8p interact with Nip7p.
We previously described the function of Nip7p, an evolutionarily conserved protein that is required for pre-rRNA processing and the accumulation of 60S subunits in S. cerevisiae (62). A two-hybrid screen to identify Nip7p-interacting proteins was performed by using the complete NIP7 ORF fused to the E. coli lexA DNA-binding domain as the bait. This bait was screened against a yeast cDNA library fused to the Gal4p activation domain in L40 cells (18). His+ clones with relatively high β-Gal activities were further characterized by DNA sequence analysis. Three of the positive clones contained cDNA for the exosome complex subunit Rrp43p (34), and another three clones contained an uncharacterized ORF (YOL144W in the Saccharomyces Genome Database). YOL144W contains a gene that encodes a 484-amino-acid (57-kDa) polypeptide that is dissimilar to known proteins and lacks any telling sequence motifs. Based on the subcellular localization of this gene’s product (shown below), we named the gene NOP8 for nucleolar protein.
Comparative two-hybrid analysis was performed with the control strain (L40-0) and with strains carrying the Nip7p bait together with prey plasmids carrying cDNAs for Rrp43p (L40-61) and Nop8p (L40-41) (Table 1). The β-Gal activities of extracts isolated from strains L40-61 and L40-41 were, respectively, 21- and 65-fold higher than those of extracts from L40 cells (data not shown). The stringency of the selection for histidine auxotrophy was enhanced by adding 3-aminotriazole (3-AT), an inhibitor of imidazoleglycerol phosphase-dehydratase (23). Whereas the three strains grew at similar rates on media selective for the vector auxotrophy markers, only L40-61 and L40-41 cells grew on 3-AT-supplemented plates (Fig. 2A). L40-41 (Nop8p) cells grew faster than L40-61 (Rrp43p) cells on 3-AT plates, which is consistent with their higher β-Gal content.
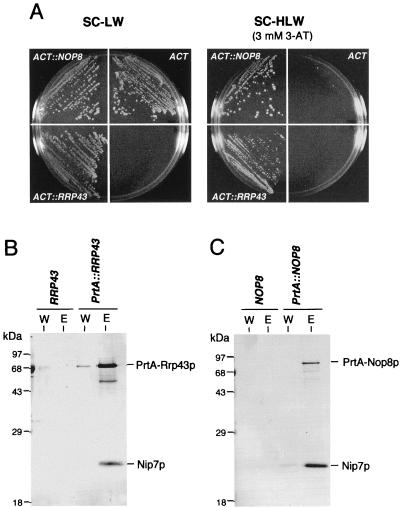
FIG. 2.
Nip7p interacts with Rrp43p and Nop8p. (A) Two-hybrid-assay interaction of Nip7p with Rrp43p and Nop8p. Strains L40-41 (ACT::NOP8) and L40-61 (ACT::RRP43) grow on medium supplemented with 3-AT that is sufficient to prevent growth of control strain L40-0 (ACT). (B and C) Copurification of Nip7p with PrtA-Rrp43p and PrtA-Nop8p. Whole-cell extracts were isolated from cells expressing PrtA-Rrp43 and PrtA-Nop8p. PrtA-tagged fusions were purified by IgG-Sepharose chromatography. Control extracts were prepared from W303-1a cells, designated RRP43 in panel B and NOP8 in panel C. Samples from the last wash (lanes W) and from the 0.5 M KCl eluate (lanes E) were submitted to immunoblot analysis with anti-Nip7p serum. See Materials and Methods for details. LW, minus leucine and tryptophan; HLW, minus histidine, leucine, and tryptophan.
The two-hybrid results suggest that Nip7p associates in vivo with Rrp43p and Nop8p. Evidence for their association in a complex was obtained by the copurification of Nip7p with PrtA-tagged Rrp43p (PrtA-Rrp43p) and Nop8p (PrtA-Nop8p). Nip7p was present in IgG-Sepharose column eluates from whole-cell extracts containing either PrtA-Rrp43p or PrtA-Nop8p but not from control extract (Fig. 2B and C). With the same starting amounts of extract, more Nip7p reproducibly copurified with PrtA-Nop8p than with PrtA-Rrp43p. This result is consistent with results of the two-hybrid analysis, where L40-41 cells (Nop8p) showed both higher β-Gal activity and faster growth on 3-AT medium than L40-61 cells (Rrp43p).
Subcellular localization of Rrp43p and Nop8p.
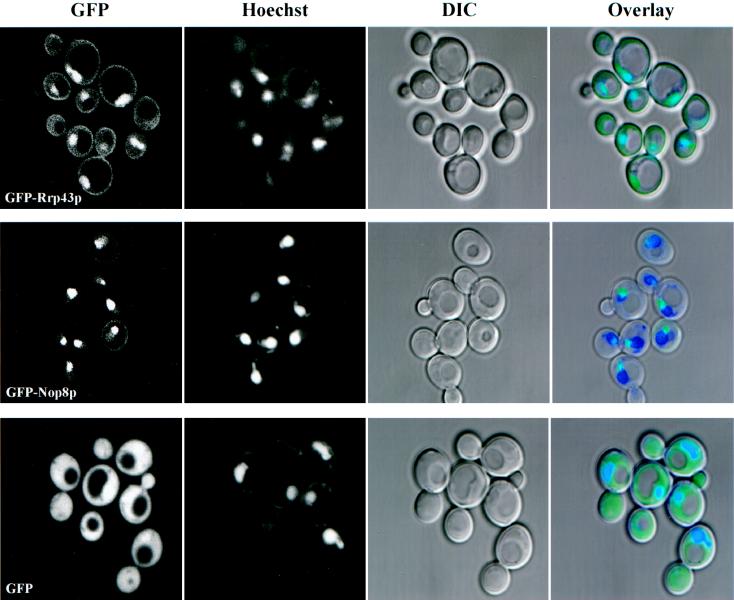
Proteins involved in pre-rRNA processing and ribosome biogenesis are usually located in the nucleolus, although there is precedent for certain nucleases and the RNA helicase Dob1p/Mtr4p to also be localized throughout the nucleoplasm and in the cytoplasm (8, 22, 29, 34). GFP-Nip7p was previously shown to be predominately nucleolar; however, native Nip7p cosedimented with free 60S ribosomes and thus Nip7p may function both in the nucleus and in the cytoplasm (62). Since Rrp43p and Nop8p interact with Nip7p in the two-hybrid system and Nip7p copurifies with PrtA-Rrp43p and PrtA-Nop8p, it was important to determine if these two proteins colocalized with Nip7p. GFP-Rrp43p and GFP-Nop8p were localized by direct fluorescence in exponentially growing cultures (Fig. 3). Mitochondrial and nuclear DNAs were stained with Hoechst 3323 (Materials and Methods). When expressed alone, GFP was distributed about equally across the nuclear envelope and was excluded from vacuoles (Fig. 3, bottom images). GFP-Nop8p fluorescence, when GFP-Nop8p was superimposed over the Hoechst 3323-stained nucleus, appeared as a cap-like extension of the nucleus, which is characteristic of the nucleolus (Fig. 3, middle images). GFP-Rrp43p appeared to localize to the nucleoplasm, nucleolus, and, to a lesser extent, cytoplasm (Fig. 3, top images). Therefore, most of GFP-Nip7p and GFP-Nop8p and a portion of GFP-Rrp43p localized to the nucleolus. The colocalization of these three proteins is consistent with the in vivo association of Nip7p with Nop8p and Rrp43p, although not necessarily in a single complex. By immunoblot analysis, PrtA-Nop8p did not appear to cosediment with 60S ribosomal subunits (not shown).
FIG. 3.
Subcellular localization of GFP, GFP-Rrp43p, and GFP-Nop8p in logarithmically growing cells. Positions of nuclei were determined by Hoechst staining. Overlay images show the superimposition of the GFP (green), Hoechst (blue), and DIC (black and white). See Materials and Methods for details.
Gene disruption analysis and construction of a conditional lethal NOP8 mutant.
A complete gene replacement of NOP8 was produced by transforming diploid W303 with a PCR-amplified KAN gene targeted for insertion into NOP8 by the method described by Güldener et al. (Materials and Methods) (16). Sporulation of a kanr transformant, followed by tetrad dissection analysis revealed that only two spores per tetrad germinated and grew to form visible colonies (data not shown). Because geneticin sensitivity (Kan−) segregated with spore viability and PCR analysis indicated that the KAN gene had inserted into the NOP8 gene (not shown), we conclude that NOP8 is an essential, single-copy gene.
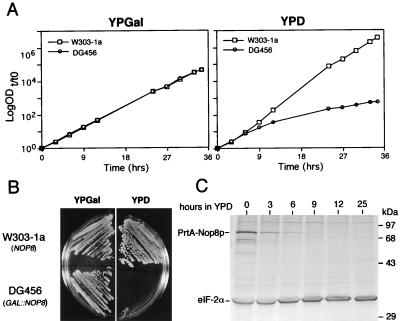
The diploid nop8::KAN disruption strain (DG457) was used to construct a NOP8 conditional strain in order to study the physiological effects of Nop8p depletion. Plasmid YCpGAL-NOP8, in which NOP8 is under the GAL1-inducible promoter, was transformed into DG457 cells (NOP8/nop8p::KAN). Transformants were sporulated, and haploid cells containing the nop8::KAN disruption and the YCpGAL-NOP8 plasmid were identified based on geneticin resistance (kanr) and conditional growth on galactose. One of these spores (DG456) was further characterized. DG456 and control NOP8 W303-1a cells grew similarly on galactose (Fig. 4A and B); however, DG456 did not grow on glucose plates (Fig. 4B). The growth of DG456 cells after a shift from galactose to glucose liquid medium slowed beginning at about 6 h and was severely impeded by 10 to 12 h (Fig. 4A). The reduction in the growth rate of DG456 cells on glucose correlated with the decrease in cellular levels of PrtA-Nop8p, which were barely detectable after 6 h (Fig. 4C). In contrast to the effect of glucose on PrtA-Nop8p levels, the levels of an internal control protein, eIF-2α, should not and did not decrease during the same time course (Fig. 4C).
FIG. 4.
In vivo depletion of Nop8p causes growth arrest. (A) Growth curves of W303-1a (NOP8) and DG456 (GAL::NOP8) cultured at 30°C in YPGal and shifted to YPD at time zero. OD600 values are plotted as log ODt/t0 units, where t0 is the initial OD600 and t is time in hours after transfer to YPD. (B) Growth at 30°C of W303-1a and DG456 on YPGal and YPD plates. (C) Immunoblot analysis showing levels of PrtA-Nop8p and endogenous eIF-2α in DG456 at various times after a shift from YPGal to YPD. The same amount of total protein was loaded in each lane. See Materials and Methods for details.
Depletion of Nop8p reduces cellular levels of 60S ribosomes.
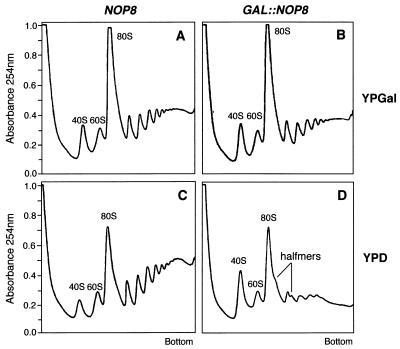
Polysome analysis was a key to gaining insight into the role of Nip7p in ribosome biogenesis (62). Therefore, the polysome profile of Nop8p-depleted cells was investigated by sucrose density gradient analysis. Extracts were prepared from DG456 (GAL::NOP8) and control W303-1a (NOP8) cells maintained in galactose or shifted to glucose for 12 h. As shown in Fig. 5A and B, the polysome profiles from galactose-grown DG456 and W303-1a cells were virtually indistinguishable. In contrast, the polysome profile of glucose-grown DG456 cells was strikingly different from that of W303-1a cells (Fig. 5C and D). Specifically, the polysome profile of extracts from Nop8p-depleted cells displayed a reduction in the level of free 60S subunits, the appearance of half-mer polysomes, and a decrease in the total amount of polysomes (Fig. 5D). Because Nip7p-depleted cells exhibited these same defects (62), these results support the hypothesis that Nip7p and Nop8p function together during the same stage(s) of ribosome biogenesis.
FIG. 5.
Nop8p-depleted cells show defects associated with decreasing 60S subunit levels. Polysome profiles were analyzed from strains W303-1a (NOP8) and DG456 (GAL::NOP8) by sedimentation through 15 to 50% sucrose gradients. Cultures were either grown in YPGal (A and B) or shifted to YPD for 12 h (C and D).
Pre-rRNA processing is defective in cells depleted of Nop8p.
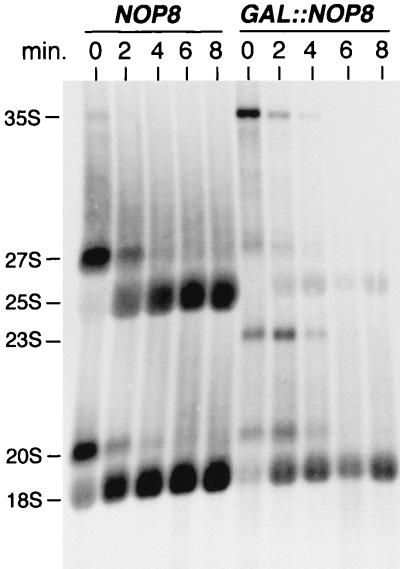
The polysome analyses shown above indicated that Nop8p is required for maintenance of normal 60S ribosome levels. For this reason, and because Nip7p-depleted cells were defective in rRNA processing, we performed [methyl-3H]methionine pulse-chase labeling and Northern blot analysis of rRNA synthesis in normal and Nop8p-depleted cells. The kinetics of rRNA processing were analyzed by pulse-chase labeling with [methyl-3H]methionine and cells shifted from galactose to glucose medium for 18 h (Fig. 6). In control cells, methyl-3H-labeled precursors were quickly chased to mature rRNAs, whereas cells depleted of Nop8p showed a drastic reduction in 25S rRNA labeling and a relatively moderate decrease in 18S rRNA labeling. In comparison to normal cells, Nop8p-depleted cells also transiently labeled 35S pre-rRNA and an aberrant 23S pre-rRNA (Fig. 6). Because there was no accumulation of unprocessed 27S pre-rRNA that could account for the defect of 25S synthesis, these data suggest that Nop8p depletion may trigger the premature degradation of 27S pre-rRNA or 25S rRNA. Consistent with this, ethidium bromide staining showed that Nop8p depletion resulted in the simultaneous decline of 5.8S and 25S rRNAs (data not shown).
FIG. 6.
Pulse-chase labeling of rRNA synthesis of Nop8p-depleted cells. Pulse-chase labeling with [methyl-3H]methionine was performed with W303-1a (NOP8) and DG456 (GAL::NOP8) shifted from SCGal to SCGlu lacking methionine for 18 h. RNA samples were collected every 2 min. Total RNA (OD = 0.9) was loaded in each lane. See Materials and Methods for details.
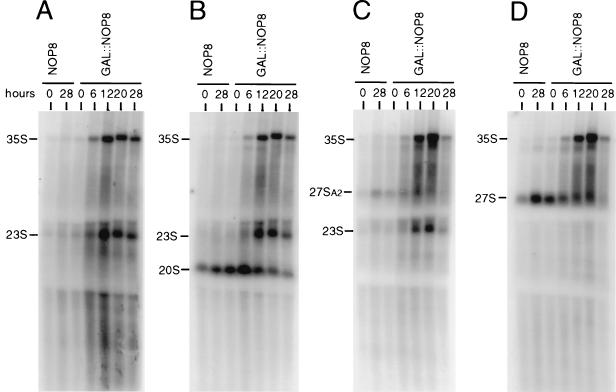
Steady-state pre-rRNA levels were analyzed from control and Nop8p-depleted cells after the shift to glucose medium for up to 28 h. Total RNAs were isolated and submitted to Northern hybridization with oligonucleotide probes specific for the 5′ ETS, ITS1, 5.8S rRNA, and ITS2 (Fig. 1). Consistent with the pulse-chase labeling results, Nop8p-depleted cells accumulated both 35S and 23S pre-rRNAs (Fig. 7). The 23S pre-rRNA incorporates the 5′ ETS, 18S rRNA, and the ITS1 region upstream of site A3, since it can be detected by probes A, B, and C (Fig. 7A to C). The decrease in the level of 20S pre-rRNA observed in Nop8p-depleted cells appears to be a consequence of the accumulation of upstream precursors, such as 35S and 23S pre-rRNAs. Hybridization with probe D, which detects all forms of 27S pre-rRNA (Fig. 7D) and probes E and F (Fig. 1 and data not shown) indicated that the 27S pre-rRNA levels decreased in accordance with the decline of the Nop8p levels. It is unclear from these data whether the decrease in 27S pre-rRNA levels is due to destabilization caused by Nop8p depletion, because at least some reduction of 27S pre-rRNA levels can be attributed to accumulation of 35S pre-rRNA. Long periods of Nop8p depletion, such as 28 h (Fig. 7), led to a reduction in the total amount of all pre-rRNAs. Finally, Northern hybridization with probe E (Fig. 1) did not detect any aberrant precursors associated with 5.8S rRNA synthesis (data not shown).
FIG. 7.
Analysis of pre-rRNA steady-state levels in Nop8p-depleted cells. RNA was analyzed from strain W303-1a (NOP8) at time zero and 28 h after transfer to YPD and from strain DG456 (GAL::NOP8) at time zero and 6, 12, 20, and 28 h after transfer to YPD. Equivalent amounts of total RNA were loaded in each lane. (A) Probe A, complementary to sequences upstream of site A0 in the 5′ ETS; (B) probe B, complementary to a region downstream of the 18S rRNA 3′ end and upstream of site A2 in ITS1; (C) probe C, complementary to sequences between sites A2 and A3 in ITS1; (D) probe D, complementary to sequences between sites C1 and C2 in ITS2. See Materials and Methods and Fig. 1 for details.
DISCUSSION
Nip7p is an evolutionarily conserved protein that is required for pre-rRNA processing and 60S ribosome biogenesis in S. cerevisiae (62). We have characterized the interactions of Nip7p with Rrp43p and Nop8p, all three of which are also required for 60S ribosome biogenesis. Two-hybrid and biochemical analyses provided genetic and physical evidence for the existence of Nip7p-Rrp43p and Nip7p-Nop8p complexes. Although both the two-hybrid and the copurification experiments suggest the existence of Nip7p-Rrp43p and Nip7p-Nop8p interactions, the mechanism of interaction remains to be determined since in both cases it may occur via a linker or adapter factor(s) in a multisubunit complex. We have not, for example, ruled out the possibility that these proteins associate with a snoRNA. We also do not know if Nip7p interacts simultaneously and/or exclusively with Nop8p and Rrp43p. These issues will have to be resolved by a structural analysis of Nip7p-containing complexes. The functional interaction between Nip7p and Nop8p is strongly supported by their colocalization in nucleoli and by their similar defects in pre-rRNA processing and 60S ribosome biogenesis caused by their depletion.
Evidence for a functional interaction between Nip7p and Rrp43p is also compelling, but this situation is more complex owing to the less restricted localization of Rrp43p to the nucleoplasm, cytoplasm, and nucleolus and its association with the exosome. The exosome contains a minimum of five putative 3′-to-5′ exonucleases that are each required for maturation of 5.8S rRNA (34). Whereas exosome subunits Rrp4p, Rrp41p or Ski6p, and Rrp44p exhibit in vitro 3′-to-5′ exoribonuclease activities, such an activity for Rrp43p is implied only from its sequence similarity to bacterial RNase PH (34). The function of certain exosomal subunits is not restricted to 5.8S rRNA processing. Benard et al. (3) have reported that the ski6-2 allele of Rrp41p/Ski6p accumulated a 38S particle derived from the 60S subunit containing a truncated part of the 25S rRNA and lacking the 5.8S rRNA. In addition, Anderson and Parker (1) have shown that Rrp4p and Rrp41p/Ski6p also participate in mRNA degradation, and there is evidence that a fraction of Rrp4p is present in the cytoplasm (34). Our results with GFP-Rrp43p suggest that it too may be partially localized in the cytoplasm.
In addition to Nip7p, other polypeptides associate with exosome subunits. Dob1p interacts genetically with Rrp4p (8), and the in vivo depletion of Dob1p and Rrp4p produce the same pre-rRNA processing defects (8, 33). With Nip7p and Rrp43p, however, in vivo depletion resulted in the accumulation of distinct precursors along the pathway leading to the synthesis of the 60S subunit rRNAs (34, 62). In Nip7p-depleted cells, the reduced synthesis of 60S subunits is associated with an accumulation of 27S pre-rRNA (62). The exosome complex, which includes Rrp43p, catalyzes the exonucleolytic digestion of the part of ITS2 that is left on the 7S pre-rRNA after cleavage of the 27S pre-rRNA at C2 (34). Although 3′-to-5′ processing of the 7S pre-rRNA 3′ end occurs subsequently to the cleavage at C2, Nip7p and Rrp43p may interact for the removal of ITS2 during the process of 5.8S and 25S rRNA maturation.
Nop8p function was investigated in Nop8p-depleted cells. Nop8p depletion led to a decrease in free 60S subunit levels and the appearance of half-mer polysomes. Similar defects have been described for mutants defective in specific rproteins of the 60S subunit (9, 35, 42, 57) and for deficiencies in other factors required for pre-rRNA processing and 60S subunit assembly (4, 7, 8, 13, 19, 26, 41, 43, 50, 60), including Nip7p (62). We conclude that Nop8p is not an rprotein because it did not cosediment with cytoplasmic ribosomes on sucrose gradients (not shown) and it localized exclusively to nucleoli. The effects of Nop8p-depletion on pre-rRNA processing are consistent with a role for Nop8p in 60S subunit biogenesis. Synthesis of 25S rRNA was drastically reduced. In addition, Northern blot analysis revealed a reduction in the steady-state levels of 27S and 20S pre-rRNAs. These data are, however, insufficient to determine whether the decrease in 25S rRNA formation is due to destabilization of 27S precursors or is a consequence of premature degradation of the 60S subunit. Both pulse-chase labeling and Northern analysis of Nop8p-depleted cells detected the accumulation of the 35S pre-rRNA and the appearance of an unusual 23S precursor, indicating that multiple steps of pre-rRNA processing are directly or indirectly blocked or delayed. The accumulation of 35S and 23S pre-rRNAs has been described for deficiencies in other factors that are required for 60S subunit biogenesis, including Nip7p, Dbp3p, Dbp7p, Nop2p, Nop77p/Nop4p, and Nop56p (4, 7, 13, 19, 50, 60, 62).
Nip7p, Rrp43p, and Nop8p are all required for processing of 60S subunit rRNAs. Pre-rRNA processing analyses suggest, however, that they are required for distinct steps during 60S rRNA maturation. In vivo depletion of Rrp43p affects processing of the 5.8S rRNA 3′ end leading to accumulation of 7S pre-rRNA (34), which is generated by cleavage of the 27S pre-rRNA at C2. A deficiency of Nip7p, however, causes the accumulation of aberrant precursors containing 5.8S sequence (62). In Nop8p-depleted cells, no accumulation of the 27S precursor was observed and the defect in 60S subunit formation in these cells may be due to 27S pre-rRNA or 25S rRNA degradation.
Non-rprotein mutants with deficiencies in 60S subunit synthesis fall into two main groups. Deficiencies in Nip7p, Dob1p, Dbp3p, Drs1p, Nop2p, and Nop56p (8, 13, 19, 41, 60, 62) arrest 25S and 5.8S rRNA synthesis at the level of 27S pre-rRNA and accumulate 27S precursors. Deficiencies in Nop8p, Dbp6p, Dbp7p, and Nop77p/Nop4p (4, 7, 26, 50) also arrest 25S and 5.8S synthesis but do not accumulate 27S pre-rRNA, which presumably is targeted for degradation. In both cases, the final result is a deficit of 60S subunits. Many of these nucleolar proteins, including Nip7p and Nop8p, lack known enzymatic activities. While the identification of factors such as Nip7p and Nop8p is a prerequisite to understanding ribosome biogenesis, the elucidation of their mechanistic role in ribosome biogenesis may have to wait for the development of in vitro rRNA processing and ribosome assembly assays.
ACKNOWLEDGMENTS
We are grateful to John McCarthy for anti-eIF-2α antibody, to Bernd Dichtl and David Tollervey for the PrtA epitope tag, and to Xiaozhou Pan for assistance with confocal microscopy and image analysis.
This project was supported by American Cancer Society grant B-104C (to D.S.G.).
REFERENCES
- 1.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel P L, Chien C-T, Sternglanz R, Fields S. Analyzing protein-protein interactions using two-hybrid system. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 3.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergès T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies Nop77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs M W, Burkard K T D, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 6.Clark M W, Yip M L, Campbell J, Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nucleolar RNAs. J Cell Biol. 1993;111:1741–1751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugeron M C, Linder P. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA Publ RNA Soc. 1998;4:566–581. doi: 10.1017/s1355838298980190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshmukh M, Tsay Y-F, Paulovich A G, Woolford J L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elela S A, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 13.Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient disruption cassette for repeated use in gene deletion in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry Y, Wood H, Morrisey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix protein with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong B, Brockenbrough J S, Wu P, Aris J P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes J M X, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson A. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishore U, Shah A. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627–633. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- 24.Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 25.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kressler D, de la Cruz J, Rojo M, Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafontaine D, Tollervey D. Trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem Cell Biol. 1995;73:803–812. doi: 10.1139/o95-088. [DOI] [PubMed] [Google Scholar]
- 28.Li H V, Zagorski J, Fournier M J. Depletion of U14 small nuclear RNA snR128 disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang S, Hitomi M, Hu Y-H, Liu Y, Tartakoff A M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang W-Q, Clark J A, Fournier M J. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol Cell Biol. 1997;17:4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic precursor RNA by ribonuclease MRP in vitro. Science. 1994;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 32.Mager W H, Planta R J, Ballesta J P G, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 35.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng D T W, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nidenthal R K, Riles L, Johnston M, Hegemann A J. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 39.O’Day C L, Chavanikamannil F, Abelson J. 18S rRNA processing requires the RNA helicase-like protein Rrp3p. Nucleic Acids Res. 1996;24:3201–3207. doi: 10.1093/nar/24.16.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raué H A, Planta R J. The pathway to maturity: processing of ribosomal RNA in Saccharomyces cerevisiae. Gene Expr. 1995;5:71–77. [PMC free article] [PubMed] [Google Scholar]
- 41.Ripmaster T L, Vaughn G P, Woolford J L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rotenberg M O, Moritz M, Woolford J L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 43.Sachs A, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 47.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 48.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 49.Stevens A, Hsu C L, Isham K R, Larimer F W. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J Bacteriol. 1991;173:7024–7028. doi: 10.1128/jb.173.21.7024-7028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun C, Woolford J L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein Nop1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 54.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 55.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 56.Venema J, Bousquet-Antonelli C, Gelugne J P, Caizergues-Ferrer M, Tollervey D. Rok1p is a putative RNA helicase required for rRNA processing. Mol Cell Biol. 1997;17:3398–3407. doi: 10.1128/mcb.17.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilardell J, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vojtek A B, Hollenberg S M. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;225:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- 59.Warner J R. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 60.Weaver P L, Sun C, Chang T-H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol Cell Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woolford J L, Jr, Warner J R. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 62.Zanchin N I T, Roberts P, DeSilva A, Sherman F, Goldfarb D S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]