Abstract
SARS-CoV-2 is a highly transmissible virus that causes COVID-19 disease. Mechanisms of viral pathogenesis include excessive inflammation and viral-induced cell death, resulting in tissue damage. Here we show that the host E3-ubiquitin ligase TRIM7 acts as an inhibitor of apoptosis and SARS-CoV-2 replication via ubiquitination of the viral membrane (M) protein. Trim7-/- mice exhibit increased pathology and virus titers associated with epithelial apoptosis and dysregulated immune responses. Mechanistically, TRIM7 ubiquitinates M on K14, which protects cells from cell death. Longitudinal SARS-CoV-2 sequence analysis from infected patients reveal that mutations on M-K14 appeared in circulating variants during the pandemic. The relevance of these mutations was tested in a mouse model. A recombinant M-K14/K15R virus shows reduced viral replication, consistent with the role of K15 in virus assembly, and increased levels of apoptosis associated with the loss of ubiquitination on K14. TRIM7 antiviral activity requires caspase-6 inhibition, linking apoptosis with viral replication and pathology.
Subject terms: SARS-CoV-2, Viral infection
TRIM7 acts as an antiviral factor during SARS-CoV-2 infection, by ubiquitinating the M protein on K14 and inhibiting caspase-6-dependent apoptosis. The natural K14 mutations in circulating strains support the physiological role of M ubiquitination.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly transmissible positive single-stranded RNA virus of the Coronaviridae family1,2. Its RNA genome encodes four structural proteins, which include Spike (S), Nucleocapsid (N), Envelope (E), and Membrane (M) proteins3. Of the structural proteins, M is the most abundant in the virion and is essential for the sorting of structural proteins to promote the assembly and release of viral particles4.
The pathogenesis of SARS-CoV-2 in humans includes a combination of excessive inflammatory responses and viral-induced tissue damage that causes lung injury, called acute respiratory distress syndrome2,5–7. The severity of disease and respiratory failure in human-infected patients correlates with the increased presence of cytokines, including IL-1β, IL-6, and TNF-α in serum and BAL8,9. SARS-CoV-2 infects multiciliated cells of the respiratory tract and alveolar type 2 (AT2) cells expressing the ACE2 receptor and the TMPRSS2 protease5,10–12. Viral infection can increase levels of apoptosis and other forms of cell death leading to tissue damage. Together, increased cell death and enhanced inflammation can correlate with disease12–14. Multiple mechanisms have been proposed to promote cell death during infection, including cytokine-induced or intrinsic apoptosis directly triggered by viral proteins, including M15,16. The M protein can also promote apoptosis by inhibiting the activation of the PDK1-AKT pathway17, or by inducing mitochondrial intrinsic apoptosis18.
The innate immune response elicited by SARS-CoV-2 infection includes innate lymphoid cells, monocytes, macrophages, and neutrophils19,20. In COVID-19 patients, low levels of circulating lymphocytes and increased levels of neutrophils correlate with the severity of the infection21,22. However, neutrophils are not the main cell type found in the lungs of human patients with prolonged severe disease, and there is evidence suggesting that neutrophils may be protective early during infection19. It is still unclear which specific factors during SARS-CoV-2 infection contribute to this shift from protective to detrimental responses by neutrophils and monocytes. Similarly, type-I interferons (IFN-I), which are well-known antiviral cytokines, can play protective or detrimental roles during infection depending on timing23,24, however, it is also unclear what factors determine protective IFN-I induction.
The innate antiviral response against SARS-CoV-2 is mediated primarily by the cytosolic nucleic acid sensor melanoma differentiation-associated protein 5 (MDA5) that recognizes the viral RNA to activate the mitochondrial antiviral signaling protein (MAVS) leading to the production of IFNs and proinflammatory cytokines25,26. Multiple viral proteins, including M, can inhibit the IFN-I pathway by targeting cytosolic receptors or by promoting the degradation of the TANK-binding kinase (TBK-1), reducing IRF3 phosphorylation and IFN-I induction27–33.
TRIM7 belongs to a large family of E3-ubiquitin (Ub) ligases, which transfer Ub to target proteins34 and can play protective or detrimental roles during infection. TRIM7 has been reported to play antiviral roles against Enteroviruses35,36, and proviral roles during Zika virus infection (ZIKV)37. TRIM7 can also regulate immune responses by promoting the production of IFN-β, TNF-α, and IL-6 in macrophages after TLR4 stimulation38. Although there is previous evidence that TRIM7 may interact with M39 and previous reports identified another E3-ligase, RNF5, as a proviral factor by ubiquitinating M on K1540, the pathophysiological roles of TRIM7 and ubiquitination of M in vivo during SARS-CoV-2 infection remains unknown. In this work we characterize in detail the multiple roles of TRIM7 during infection in vivo. We found that TRIM7 regulates the expression of inflammatory cytokines, including the chemokine CXCL1, which promotes the recruitment of immune cells to the infection site. TRIM7 also acts as an antiviral factor during SARS-CoV-2 infection, by ubiquitinating the M protein and inhibiting caspase-6-dependent apoptosis, in an IFN-I independent manner. We also identified the presence of natural K14 mutations in circulating SARS-CoV-2 during the pandemic, supporting a physiological role for ubiquitination of M.
Results
TRIM7 ubiquitinates SARS-CoV-2 M protein on the K14 residue
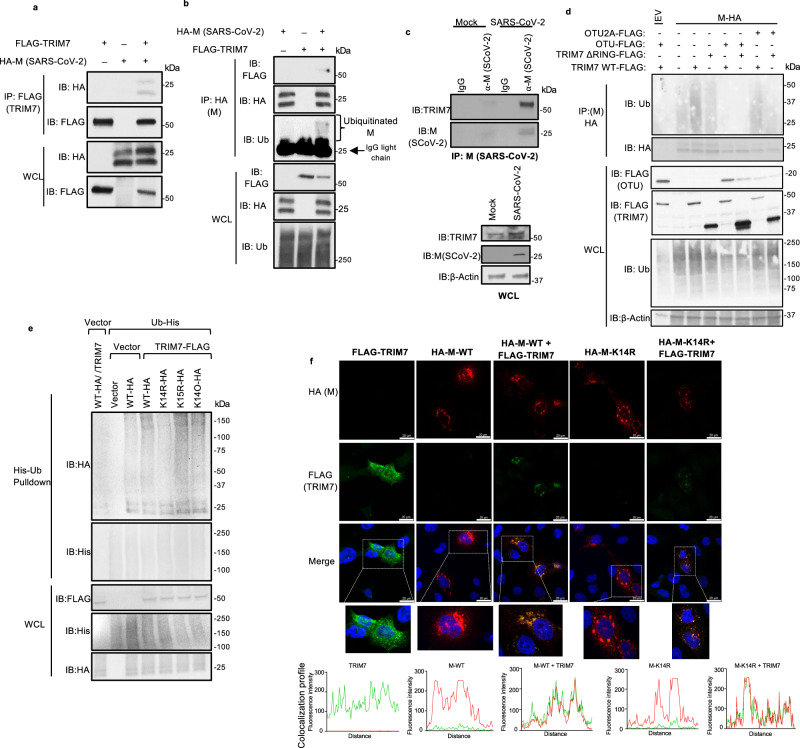
Previous mass spectrometry studies identified TRIM7 as a potential binding partner of SARS-CoV-2 M protein39, although this interaction and its functional relevance were not further investigated. We first confirmed that TRIM7 and M interact using co-immunoprecipitation assays (coIP), when ectopically expressed (Fig. 1a, b). Endogenous TRIM7 also interacted with M during SARS-CoV-2 infection in human cells and mouse lung tissue (Fig. 1c and S1a, b). This interaction does not require the RING domain of TRIM7 (TRIM7-ΔRING) (Fig. S1c), and is mediated by the C-terminal PRY-SPRY domain of TRIM7 (Fig. S1d). As previously proposed39,40, ubiquitinated M can be detected when ectopically expressed (Fig. S1e). In addition, overexpression of TRIM7 further enhanced the ubiquitination of M (Fig. 1b, d), whereas the catalytically inactive mutant TRIM7-ΔRING did not (Fig. 1d). Ectopic expression of the ovarian tumor deubiquitinase (OTU), which cleaves endogenous polyubiquitin chains (polyUb) from modified proteins37, removed all polyUb that coimmunoprecipitated with M, while a catalytically inactive mutant of OTU (OTU-2A) was used as a negative control (Fig. 1d). These results suggest that TRIM7 promotes ubiquitination of SARS-CoV-2 M protein.
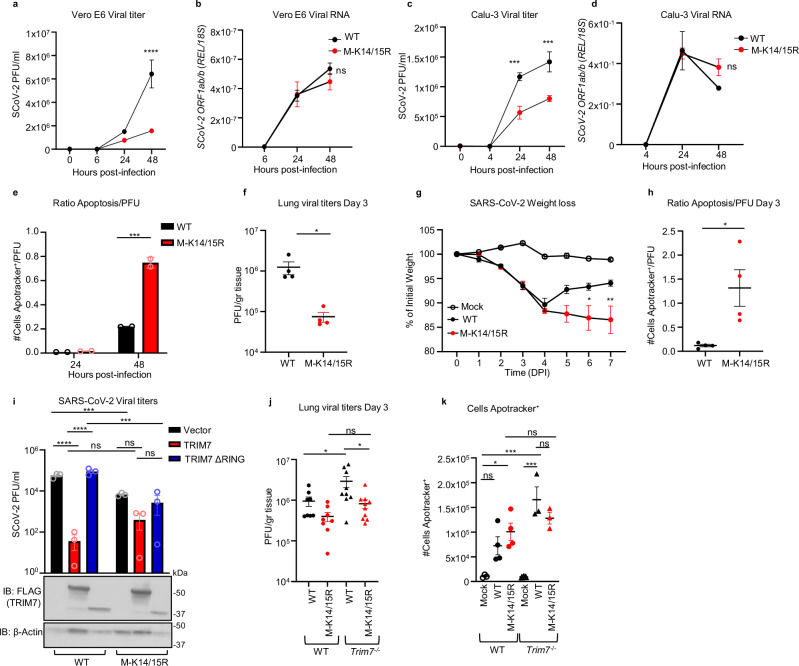
Fig. 1. TRIM7 ubiquitinates SARS-CoV-2 Membrane protein.
a, b HEK293T cells were transfected with vector, M-HA, ±TRIM7 WT-FLAG, and immunoprecipitated using anti-FLAG beads (a) or anti-HA beads (b). c Immunoprecipitation of M protein of SARS-CoV-2 (SCoV-2) from Calu-3 cells infected with SARS-CoV-2 MOI 1 for 24 h using anti-M SCoV-2 or IgG control. d HEK293T cells transfected with M-HA, of TRIM7 WT-FLAG or TRIM7-ΔRING-FLAG (TRIM7 lacking the RING domain) ±OTU-WT-FLAG or OTU-2A-FLAG followed by immunoprecipitation with anti-HA beads, e denaturing pulldown using NiNTA beads. HEK293T cells transfected with 100 ng of His-Ub, M-WT-HA, M-K14R, M-K15R, M-K14O (all Ks mutated to Rs except for K14), +/- TRIM7 WT-FLAG. f Confocal microscopy of Hela cells transfected with TRIM7-FLAG (488), M-WT-HA or M K14R-HA (555), for 24 h. Colocalization profile graphs are shown. Data are representative of 2 independent experiments with at least 4 micrographs per condition. All western blot panels are representative of at least two independent experiments. Ub Ubiquitin, OTU ovarian tumor deubiqitinase. Source data are provided in the Source data file.
We next asked whether TRIM7 promotes ubiquitination on a specific lysine residue on M. A denaturing pulldown of ectopically expressed His-tagged Ub and M encoding K-to-R mutations, showed reduced TRIM7-mediated ubiquitination on an M-K14R mutant as compared to WT M (Fig. 1e). The K15 residue on M, which is ubiquitinated by the E3-ligase RNF540, did not appear to be an acceptor for ubiquitination by TRIM7. In further support of this, a K14-only mutant of M, in which all its K residues were mutated to R except for K14 (K14O), showed similar ubiquitination levels as compared to WT M, in the presence of overexpressed TRIM7 (Fig.1e). Together, these data suggest that TRIM7 specifically ubiquitinates the M protein on the K14 residue.
Ubiquitination of M on K15 by RNF5 is necessary to promote the efficient formation of virus-like-particles (VLPs) and virus release40. To rule out a functional role for TRIM7 in virus release, we evaluated the efficiency of VLP formation upon transfection of all viral structural proteins in A549 WT or TRIM7 knockout cells (KO), previously generated in our lab37. No apparent differences were observed in the amount of VLPs released from WT and TRIM7 KO (Fig. S1f), suggesting that TRIM7 does not affect virus release, which is consistent with previous observations that the M-K14R mutant is still able to form VLPs40.
Finally, upon ectopic expression, TRIM7 re-localized from discrete punctate cytoplasmic bodies to larger vesicle-like compartments where it colocalized with M (Fig. 1f). These vesicles could be associated with Golgi or ER-Golgi intermediate compartment (ERGIC), as M localized with the Golgi marker GM130 (Fig. S1g), as previously reported41. The M-K14R mutant still colocalized and coimmunoprecipitated with TRIM7 (Fig. 1f and S1h), indicating that the interaction did not depend on M-K14 ubiquitination.
Since TRIM7 has been proposed to bind some target proteins, including M, via a terminal-glutamine (Q) residue, termed c-degron signal42, we tested whether TRIM7 binding with M requires this degron signal. In contrast to the previously observed interactions of NSP5 and NSP842 with TRIM7, a mutant of M lacking the degron signal (M-Q222A) interacted efficiently with TRIM7 (Fig. S1i), suggesting that this Q residue is not a determinant for M-TRIM7 interaction. This is in line with the observation that TRIM7 does not appear to degrade M.
Overall, these data provide evidence that TRIM7 ubiquitinates M on its K14 residue, and this ubiquitination does not significantly degrade M or affect M’s function in the assembly and release of viral particles.
TRIM7 has antiviral activity during SARS-CoV-2 infection
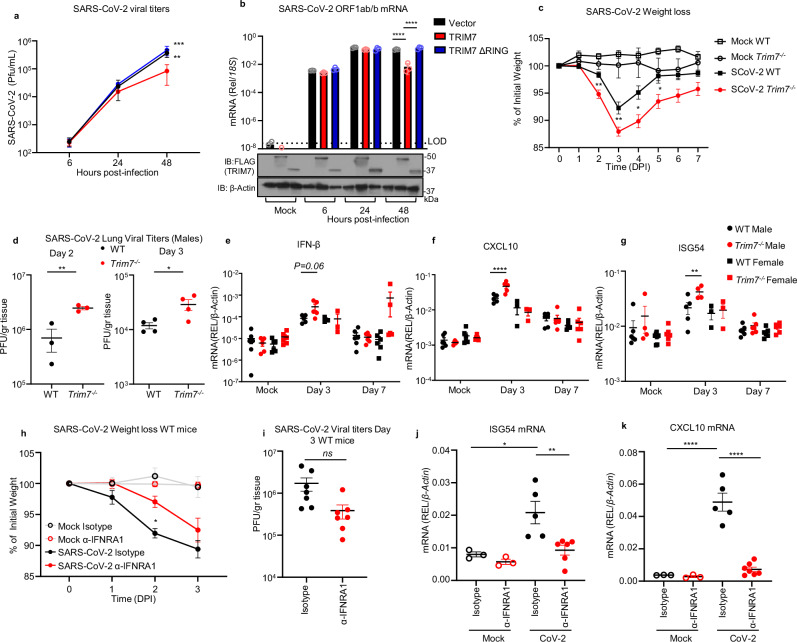
Since TRIM7 has been reported to have both proviral and antiviral roles, we next evaluated the role of TRIM7 during SARS-CoV-2 infection. Overexpression of TRIM7 in HEK293T cells stably expressing human ACE2 (293T-hACE2) significantly reduced SARS-CoV-2 titers (plaque assay) and viral RNA (qPCR) as compared to the inactive TRIM7-ΔRING, or a vector control (Fig. 2a, b).
Fig. 2. TRIM7 has antiviral activity during SARS-CoV-2 infection.
a, b HEK293T-hACE-2 cells were transfected with 200 ng of TRIM7 WT or TRIM7-ΔRING followed by infected with SARS-CoV-2 (MOI 0.1). Viral titers quantified by plaque assay (a) or viral RNA by qPCR (b). Bottom (b) shows immunoblot control for TRIM7 expression. Two-way ANOVA Tukey’s multiple comparisons (**p = 0.0038, ***p = 0.0001, ****p < 0.0001). c C57BL/6NJ WT (n = 18, 9 females, 9 males) and Trim7−/− mice (n = 23, 10 females, 13 males) infected with maSARS-CoV-2 (1 × 106 PFU). Weight loss (c) data were from three combined independent experiments and multiple T-test (**p = 0.001, p = 0.003, *p = 0.005). d Viral titers (day 2, n = 3 males and day 3, n = 4 males per group). Two-tailed T-test (**p = 0.0083, *p = 0.03). e IFN-β gene expression in lung tissue by qPCR (n = 5 male day 3, 6 male day 7, and 3 female per infected group, mock n = 3 male and 3 female). f CXCL10 (same animal number as in (e). g ISG54 (mice numbers as in e, except for day 3, n = 5 WT and 4 Trim7−/− male). Two-way ANOVA Tukey’s multiple comparisons (****p < 0.0001, *p = 0.026). h–k IFNAR1 blockade: C57BL/6J WT (n = 7/group five females and two males) were treated with anti-IFNRA1 or isotype 24 h before infection with maSARS-CoV-2 (1×106 PFU). h Weight loss, two-way ANOVA Tukey’s multiple comparisons (*p = 0.023), i viral titers in the lung. j ISG54 gene expression by qPCR (n = 5 isotype, 6 α-IFNRA1 *p = 0.01, **0.009). k CXCL10 gene expression, one-way ANOVA Tukey’s multiple comparisons (n = 5 isotype, 7 α-IFNRA1, ****p < 0.0001). DPI days post infection, α-IFNRA1 anti-IFNRA1. Data were depicted as mean + SEM. Source data are provided in the Source data file.
We then tested whether TRIM7 also has antiviral function in vivo. WT and Trim7−/− mice37 were infected with a mouse-adapted strain of SARS-CoV-2 (CMA3p20)43. Trim7−/− mice lost significantly more weight at the acute phase of infection and showed slower recovery than WT controls (mixed males and females, Fig. 2c). Trim7−/− male mice exhibited significantly higher lung viral titers (Fig. 2d) and viral RNA at day 2 and 3 p.i., while we observed smaller differences between females (Fig. S2a–c). The increase in weight loss and viral titers in Trim7−/− mice correlated with clinical scores (e.g., ruffled fur and/or hunched posture, Fig. S2d), as well as consolidation of the airway at later time points (Fig. S2e, f). TRIM7 antiviral effects were most likely independent of the IFN-I response because the levels of IFN-β mRNA were trending higher in Trim7−/− and correlated with significantly increased levels of ISG54 and CXCL10, which are well-known ISGs (Fig. 2e–g). In further support of this, IFN-I receptor (IFNAR1) blockade resulted in less weight loss without significantly affecting virus titers as compared to isotype control-treated mice (Fig. 2h, i). As expected, anti-IFNAR1 treated mice showed reduced levels of ISGs (Fig. 2j, k). These results are in line with previous reports showing that IFN-I has a pathogenic effect during SARS-CoV-2 infection, by regulating the infiltration of inflammatory cells but not affecting virus levels23,24.
Together, our data indicates that TRIM7 plays an antiviral role in cell culture and in vivo. These effects require TRIM7 E3-ubiquitin ligase activity and do not appear to be IFN mediated.
TRIM7 is a negative regulator of IFN-I induction during SARS-CoV-2 infection
Although our data suggest that IFN-I is not involved in the TRIM7-mediated antiviral response, elevated IFN induction could still affect inflammatory responses leading to disease. Therefore, we determined whether the increased levels of ISGs observed in Trim7−/− infected mice are due to a direct effect of TRIM7 in the IFN pathway. TRIM7 represses expression of IFN-I because bone marrow-derived dendritic cells (BMDCs) from Trim7−/− mice infected with SARS-CoV-2 showed increased levels of IFN-β mRNA as compared to WT BMDCs (Fig. S2g). In contrast, Trim7−/− cells expressed lower levels of IL-1β mRNA when compared with WT BMDCs (Fig. S2h). Although SARS-CoV-2 does not productively replicate in DCs44, the presence of similar levels of viral RNA in WT and KO cells indicated that the effects observed are not due to differences in virus infection (Fig. S2i).
To further examine how TRIM7 inhibits IFN-I induction, we tested interactions with the pattern recognition receptors (PRRs) RIG-I and MDA5. Results from coIP assays revealed that TRIM7 interacts with both PRRs (Fig. S2j). Since there is evidence that MDA5 is the major cytosolic receptor for SARS-CoV-225,26, we also evaluated if TRIM7 can affect MDA5’s induction of IFN-β. IFN luciferase reporter assays showed that increased concentrations of TRIM7 reduced the IFN-β promoter activity (Fig. S2k), suggesting that TRIM7 can negatively regulate IFN-β by inhibiting MDA5-mediated signaling.
Ubiquitination on M-K14 does not affect IFN-I antagonist function
M has also been shown to inhibit both the IFN-I production as well as the IFN-I signaling pathways27–30,45. Therefore, we examined if M ubiquitination can affect IFN antagonism. Ectopic expression of WT M or a mutant lacking all ubiquitination sites (M-KallR) inhibited IRF3 phosphorylation at comparable levels upon stimulation with the dsRNA mimic poly (I:C) (Fig. S2l), suggesting that ubiquitination on M does not play a role in inhibition of IFN-I production. M has also been reported to inhibit the induction of ISGs downstream of the IFN-I receptor30. M-WT, as well as the mutants M-K14R, M-K15R, and M-KallR reduced the IFN-induced ISRE luciferase reporter activity (Fig. S2m), suggesting that ubiquitination of the lysine residues is not necessary for antagonism of IFN-I signaling.
Taken together, the increased IFN response observed in Trim7−/− mice is unlikely to be mediated by M ubiquitination and does not explain the increased virus replication observed in the knockout mice.
TRIM7 promotes innate immune inflammation while protecting from cell death during SARS-CoV-2 infection
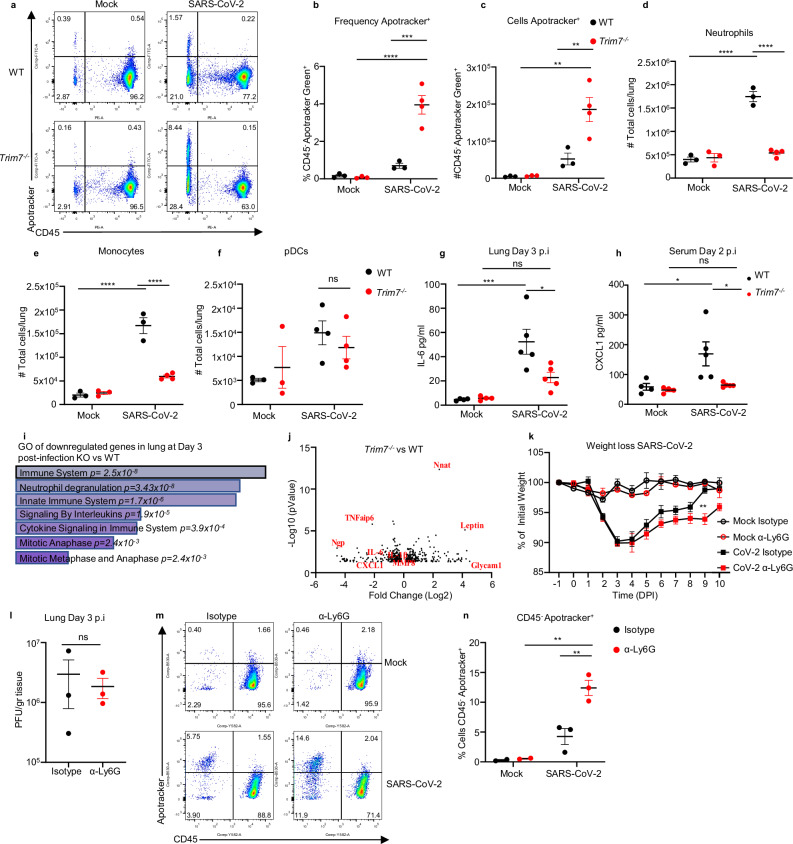
TRIM7 has been associated with the induction of genes involved in cell growth, proliferation, and survival46. Conversely, SARS-CoV-2 has been shown to induce cell death in lung cells47, and its M protein has been associated with apoptotic effects17. At day 3 post infection, we observed a significantly higher proportion of cells positive for Apotracker staining (cells undergoing apoptosis) in the lungs of Trim7−/− mice as compared to WT controls (Fig. 3a). These effects were evident in CD45- cells (Fig. 3b, c and S3a). In contrast, no differences were observed in apoptosis between WT and KO mice within the hematopoietic CD45+ compartment (Fig. S3b).
Fig. 3. Trim7−/− mice have impaired innate immune response to SARS-CoV-2 infection.
a–j Male C57BL/6NJ WT and Trim7−/− mice were infected with maSARS-CoV-2 (WT n = 3, KO n = 4). On day 3 post infection, lung cells were stained for CD45 and Apotracker Green (a representative FACS dot plots; b frequency of CD45-/Apotracker Green+ cells; c total number of CD45-/Apotracker cells per lung). d Total number of neutrophils (CD45+CD11c-CD11b+Ly6CintLy6Ghi). e Total monocytes (CD45+CD11c-CD11b+Ly6ChiLy6G-). f Total pDCs (CD45+CD11cloPDCA-1+CD11b−). Two-way ANOVA Tukey’s multiple comparisons (***p = 0.0002, ****p < 0.0001). g 23-Bioplex analysis shows IL-6 levels in lung homogenate, or h CXCL1 in serum (complete Bioplex shown in S4a, b). Two-way ANOVA Tukey’s multiple comparisons (***p = 0.0006, *p = 0.01, p = 0.02). i, j RNAseq analysis of infected lungs at day 3 post infection. i Gene ontology of genes downregulated in Trim7−/− mice. j Volcano plot of the RNAseq (n = 3 per group, Wald test). k–n Neutrophil depletion in WT mice using anti-Ly6G antibody or isotype as control followed by infected with maSARS-CoV-2 (n = 8) (**p = 0.001), weigh loss (k), viral titers (l, n = 3), two-tailed T-test analysis, representative dot blot of lung cells (m, CD45/Apotracker Green), frequency CD45−/Apotracker Green+ cells (n), two-way ANOVA Tukey’s multiple comparisons (**p = 0.001, p = 0.006). Data were depicted as Mean ± SEM. DPI days post infection, GO gene ontology, α-Ly6G anti-Ly6G. Source data are provided in the Source data file.
We evaluated whether TRIM7 antiviral effects were associated with changes in the innate immune cell composition in the lungs. Trim7-/- mice showed reduced neutrophil and monocyte infiltration as compared to WT mice at day 3 p.i. (Fig. 3d, e and S3c), whereas no differences in infiltration of plasmacytoid DCs (pDCs) were observed (Fig. 3f). Multiplex analysis of lung and serum cytokines showed reduced proinflammatory cytokines IL-6, IL-1β, and IL-1α in Trim7−/− mice (Fig. 3g and S4a, b). In line with the reduced cellular infiltration to the lung, the neutrophil chemoattractant CXCL1 in serum was lower in Trim7−/− mice as compared to controls (Fig. 3h). In further support of the role of TRIM7 in promoting immune inflammation, RNAseq and Gene Ontology analysis (GO) of infected lungs showed that downregulated genes were enriched in pathways related to the inflammatory response (neutrophil degranulation, innate immune signaling, and cytokine signaling) as well as cell division/survival (mitotic genes) (Fig. 3i). Specifically, induction of Il6, Il1b, Cxcl1, Tnfaip6, and Mmp8 was reduced in Trim7−/− mice (Fig. 3j). These results correlate at the protein level in the lung (Fig. S4a) and in the serum (Fig. S4c).
Since we found a lower number of neutrophils in the lungs of Trim7−/− mice, we asked whether neutrophil recruitment to the lung could be associated with protection from disease. To test this, C57BL/6 J WT mice were depleted of neutrophils (Fig. S4d). Anti-Ly6G-treated mice lost weight at a similar rate as isotype-treated mice until the peak of viral titers (day 3 p.i.). However, neutrophil-depleted mice recovered from infection significantly slower than control mice (Fig. 3k). These effects did not appear to be due to differences in virus replication because control and neutrophil-depleted mice showed similar virus titers (Fig. 3l). These data suggest that neutrophils are not responsible for the antiviral role mediated by TRIM7 but may be involved in tissue repair/healing during the recovery phase. In support of this, neutrophil-depleted mice had a significantly higher frequency of apoptotic cells, specifically in the CD45- compartment (Fig. 3m, n), suggesting that neutrophils are important for the removal of apoptotic cells either directly or indirectly, promoting tissue repair during the recovery phase.
Overall, our data indicate that TRIM7 is antiviral during SARS-CoV-2 infection and suggest that TRIM7 regulates inflammatory immune responses.
TRIM7 protects from SARS-CoV-2-induced apoptosis and requires an intact K14 residue on M
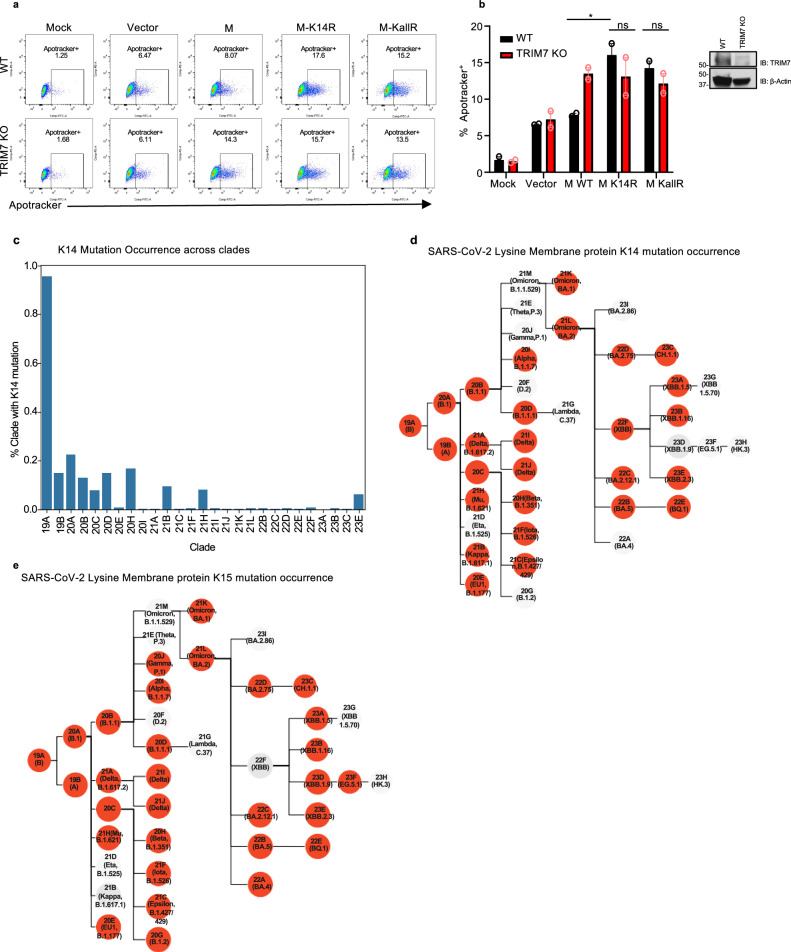
Apoptosis during viral infection is a process that can either limit virus replication or promote virus dissemination48. To evaluate the relationship between M, TRIM7, and apoptosis, WT and TRIM7 KO A549 cells were transfected with vectors expressing WT or M mutants. Upon transfection of WT M, a significantly higher frequency of apoptotic cells was observed in TRIM7 KO cells as compared to WT cells. These effects required the presence of an intact K14 residue on M because expression of an M-K14R mutant that cannot be ubiquitinated by TRIM7 induced higher frequency of cells in apoptosis in WT cells, and no further difference was observed in TRIM7 KO cells (Fig. 4a, b, controls for expression shown in Fig. S5a). TRIM7 KO cells also display reduced AKT phosphorylation upon stimulation with TNF, while no differences were observed in IKKα/β phosphorylation (Fig. S5b), providing further evidence that TRIM7 is involved in signaling pathways associated with apoptosis and cytokine signaling. Overall, these data suggest that TRIM7 protects from cell apoptosis via ubiquitination on the K14 residue and this potentially reduces virus replication.
Fig. 4. Mutations on M lysine 14 induce apoptosis and are present in circulating stains of SARS-CoV-2.
a, b A549 WT or TRIM7 KO transfected with 500 ng of M-WT, M-K14R, or M-KallR mutants for 24 h and then stained with Apotracker Green, a representative dot blot. b Frequency of Apotracker+ cells, representative data of two independent experiments and each with two biological replicates. c–e M lysine mutations in SARS-CoV-2 sequences of circulating variants. Percentage of occurrence of M-K14 mutation across the clades (c) and membrane protein K14 mutation occurrence (d), or membrane protein K15 mutation occurrence (e). Red highlighted nodes indicate at least one mutation occurrence in the specific clade. Data were depicted as Mean ± SEM. Source data are provided in the Source data file.
SARS-CoV-2 membrane protein mutations on K14 appeared during the pandemic in COVID-19 patients
We next asked whether mutations that lead to loss of ubiquitination and cause more apoptosis can appear in circulating strains of SARS-CoV-2. Data analysis from all ~8.5 million SARS-CoV-2 genomes present in GenBank from the beginning of the pandemic to March 2024, was performed with CoV-Spectrum49. From the samples analyzed, we observed that 985 showed mutations on K14 residue representing 0.01% of the total samples. Mutations on this site were relatively more frequent early in the pandemic, with a higher occurrence (~0.95%) in samples from clade 19A (Fig. 4c). The most common mutation was a deletion of K14, followed by the K14R mutation (Fig. 4d and Table S1). The mutations on the K15 residue were more frequent, present in 0.02% of associated samples in GenBank (2433), (Fig. 4e and Table S2). Samples with K14 and K15 mutations were more infrequent, being observed 51 times in total, with all except one of these double mutations being a double deletion (Table S3). This analysis shows that these mutations can occur in nature and were overrepresented in early clades during the pandemic.
A recombinant virus with M K14/K15 mutations causes more apoptosis in mice
Since we are unable to correlate these mutations with clinical data from patients, we tested the relevance of these mutations in viral pathogenesis in a mouse model. To this end, we generated a recombinant mouse-adapted double mutant virus M-K14/K15R (CMA5 M-K14/15R), which cannot be ubiquitinated on either K14 or K15 sites. We used this double mutant to avoid generating a virus with increased replication ability due to the loss of the target site for TRIM7 ubiquitination. Since previous studies have shown that ubiquitination on M-K15 by another E3-Ub ligase, RNF5, is required for efficient virus release40, introducing the K15R mutation on the K14R mutant virus should result in an attenuated virus. This would still allow us to dissect the roles of ubiquitination on virus replication and apoptosis by both the K14 and K15 residues of M.
As predicted, this M-K14/K15 mutant virus showed reduced replication kinetics in the IFN-incompetent Vero E6 as well as in IFN-competent Calu-3 cell lines, as compared to the parental WT virus (Fig. 5a, c and S6a, c). Consistent with the role of K15 in virus particle formation and budding40, the viral RNA accumulated in the cells at similar rates between the K14/15R and the parental virus strain (Fig. 5b, d and S6b, d). Since Vero cells do not produce active IFN-I, the differences observed are likely not IFN-I dependent. Consistent with this, no difference in ISG54 mRNA levels was observed between the parental and the mutant virus in Calu-3 cells (Fig. S6e). These data contrast with the phenotype we observed of enhanced virus replication in Trim7−/− mice but can be explained by the loss of ubiquitination on M-K15 that is required for virus release. Importantly, even though the M-K14/15R mutant virus is highly attenuated, it showed a higher ratio of cells in apoptosis when normalized by PFU (Fig. 5e). Similarly, the M-K14/15R virus replicated to lower levels in the lungs of WT mice (Fig. 5f) but caused increased weight loss (Fig. 5g), and increased apoptosis as compared to the WT parental virus (Fig. 5h and S6f, g). No differences in the production of IFN-β or ISG54 were observed in the lung at day 3 p.i. (Fig. S6h, i). Together, these data and the data described above suggest that the K15 site promotes virus replication while the K14 site protects cells from apoptosis during SARS-CoV-2 infection.
Fig. 5. Recombinant virus with M K14/K15 mutations shows increased pathogenesis.
Vero E6 (a, b) or Calu-3 (c, d) infected with SARS-Cov-2 WT or M-K14/15R MOI 0.1 and 1, respectively. Viral titers (a, c) and viral RNA by qPCR (b, d). Two-way ANOVA Tukey’s multiple comparisons (****p < 0.0001, ***p = 0.001, p = 0.0007). e Ratio of cells in apoptosis Apotracker+ normalized by viral titers (Apotracker+/PFU) in Calu-3 cells. Data were representative of two independent experiments, each with two biological replicates. Multiple T-test comparison (***p = 0.003). f–h WT C57BL/6J male mice infected with SARS-CoV-2 WT (n = 8) or M-K14/15R mutant viruses (n = 10) and mock (n = 5). f viral titers in lung (n = 4), two-tailed t-test analysis (*p = 0.036). g weight loss curve. Two-way ANOVA Tukey’s multiple comparisons (*p = 0.02, **p = 0.001). h Ratio of cells in apoptosis in the lung (Apotracker+/PFU, n = 4). Two-tailed t-test analysis (*p = 0.02). i Viral titers of HEK293T-hACE-2 cells transfected with 200 ng of TRIM7 WT or TRIM7 ΔRING and infected with SARS-CoV-2 WT or M K14/15R at MOI 0.1 (bottom panel shows immunoblot for TRIM7). Two-way ANOVA Tukey’s multiple comparisons (****p < 0.0001, ***p = 0.0007). WT (n = 8, 6 males, 2 females) and Trim7−/− (n = 9 and 11, 7 or 9 males and 2 females) infected with SARS-CoV-2 WT or M-K14/15R and euthanized at day 3. j Lung viral titers are combined data from two independent experiments (*p = 0.038, 0.016). k Total number of cells in apoptosis (CD45 +Apotracker+) are representative of 1 of the 2 independent experiments shown in (j). Mock n = 3/group, SARS-CoV-2 WT n = 4 in WT and 3 in Trim7−/− mice. M K14/15R virus n = 4 in WT and 3 in Trim7−/−. One-way ANOVA Tukey’s multiple comparisons (*p = 0.014, ***p = 0.0002). Data were depicted as mean ± SEM. DPI days post infection. Source data are provided in the Source data file.
Next, we examined whether TRIM7 can still inhibit virus replication in the absence of the K14/K15 ubiquitination sites. As expected, overexpression of TRIM7 in 293T-hACE2 cells reduced replication of the parental WT virus (Fig. 5i). In contrast, overexpression of TRIM7 did not significantly reduce replication of the K14/15 R virus as compared to the empty vector control (Fig. 5i). The mutant virus showed reduced replication as compared to the parental virus, confirming that this mutant virus is attenuated. Overexpression of the inactive TRIM7-ΔRING did not affect the replication of either virus and served as an additional control (Fig. 5i). In line with these results, while the parental WT virus replicated to higher levels in Trim7−/− compared to WT mice, no significant difference was observed when comparing M-K14/15R titers between WT and Trim7−/− mice (Fig. 5j). The loss of the K14 ubiquitination site, which is mediated by TRIM7, would explain the lost difference between WT and KO mice. The K14/15R virus still replicated to a lower titer than the WT virus in Trim7−/− mice, and this can likely be explained by the loss of the K15 ubiquitination site, which is dependent on RNF5 and not TRIM7. The loss of ubiquitination on K14/15 resulted in a slightly increased number of cells in the lung undergoing apoptosis as compared to the parental virus in WT mice. As expected, the parental virus promoted greater levels of apoptosis in Trim7−/− mice, however, the M-K14/15R mutant virus did not (Fig. 5k). These data suggest that TRIM7 restricts apoptosis in the lung during SARS-CoV-2 infection and this requires intact K14/15 residues on the M protein.
TRIM7 mediates its antiviral effects by inhibiting caspase-6 activation
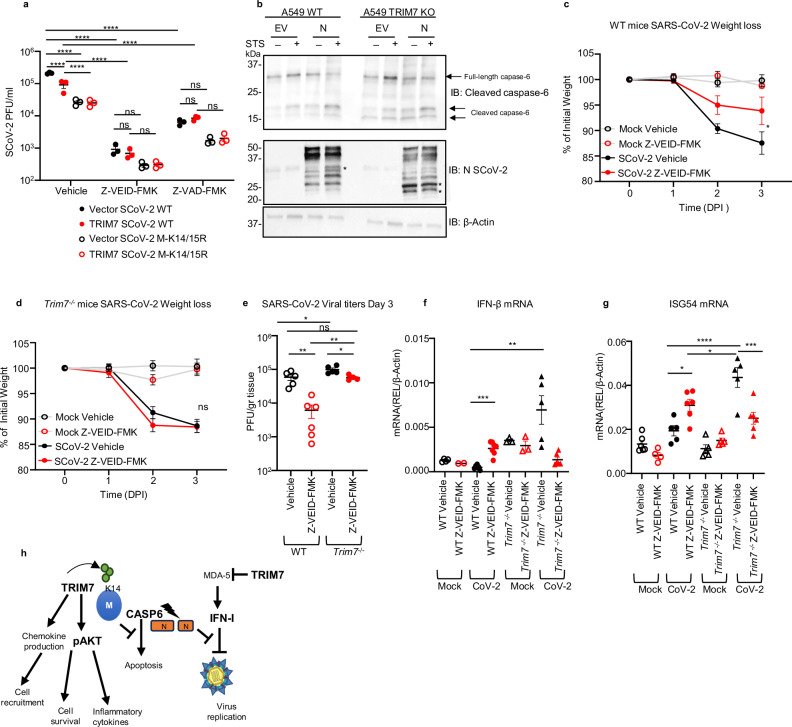
Apoptosis during viral infection is known to play an important role in limiting virus replication50,51. Intriguingly, coronaviruses can take advantage of the apoptosis machinery to promote their replication52,53. Therefore, we evaluated if TRIM7’s antiviral mechanism depends on its ability to inhibit apoptosis. To test this, we used inhibitors of apoptosis, Z-VAD-FMK (a pan caspase inhibitor), and Z-VEID-FMK, (which targets caspase-6 and has been shown to inhibit SARS-CoV-2 replication52). Consistent with this previous study, treatment with caspase-6 inhibitor (Z-VEID-FMK) strongly reduced replication of both the parental and the M-K14/15R viruses in 293T-hACE2 cells (Fig. 6a) and completely inhibited apoptosis (Fig. S6j). However, while overexpression of TRIM7 reduced viral titers in DMSO-treated cells, TRIM7 lost its ability to further reduce SARS-CoV-2 replication as compared to vector control in cells treated with Z-VEID-FMK or Z-VAD-FMK (Fig. 6a and expression controls in Fig. S6k). This suggests that TRIM7 requires, at least in part, an active caspase-6 pathway to exert its antiviral activity. Since it has been shown that caspase-6 can cleave the N protein of coronaviruses53–55 and cleaved N inhibits the IFN-I response leading to increased virus replication, we evaluated if TRIM7 is involved in the cleavage of N. Treatment with staurosporine (STS), which activates apoptotic pathways, enhanced cleavage of N as compared to vehicle control in WT A549 cells. These effects were further increased in TRIM7 KO cells, in which additional products of N cleavage were evident (Fig. 6b). These effects correlated with slightly enhanced cleavage of caspase-6 in TRIM7 KO cells (Fig. 6b and S6l). In support of these results, WT mice treated with Z-VEID-FMK show less weight loss (Fig. 6c) and significantly reduced viral titers in the lungs as compared to DMSO-treated mice (Fig. 6e). In contrast, Z-VEID-FMK treatment of Trim7−/− mice did not prevent weight loss (Fig. 6d), indicating that TRIM7 deficient mice are more resistant than WT mice to the protective effects of the inhibitor. Importantly, treatment with Z-VEID-FMK reduced virus titers in the Trim7−/− mice to the levels observed in the DMSO-treated WT animals (Fig. 6e), suggesting that the antiviral activity of TRIM7 is, in part, mediated by inhibition of caspase-6 activity. Notably, TRIM7 also shows antiviral activity in vivo that is independent of caspase-6, because Trim7−/− mice treated with the inhibitor have significantly higher viral titers compared to WT-treated mice (Fig. 6e).
Fig. 6. TRIM7 mediates its antiviral effects by inhibiting caspase-6 activation.
a Viral titers of HEK293T-hACE-2 cells overexpressing TRIM7 WT and infected with SARS-CoV-2 WT or M-K14/15R at MOI 0.1 and treated with vehicle (DMSO) or 50 μM of Z-VEID-FMK 24 h post infection. Two-way ANOVA Tukey’s multiple comparisons (****p < 0.0001). b Western blot analysis of A549 WT and TRIM7 KO cells transfected with SARS-CoV-2 N protein or empty vector and treated with Staurosporine (STS) *indicates a cleaved form of N (blot is representative of two independent experiments). c Weight loss of WT female mice infected intranasally with SARS-CoV-2 WT, treated intraperitoneally with caspase-6 inhibitor or vehicle. Mock (n = 6 each) vehicle (n = 5) or Z-VEID-FMK (n = 6). Two-way ANOVA Tukey’s multiple comparisons (*p = 0.03) (d) weight loss Trim7−/− female mice infected as in (c), vehicle mocks (n = 5 each) infected vehicle (n = 4) or Z-VEID-FMK (n = 6). e Viral titers in lung (WT n = 5 and 6, Trim7−/− n = 5 and 4, respectively). One-way ANOVA Tukey’s multiple comparisons (**p = 0.002, p = 0.003, *p = 0.02). f IFN-β mRNA multiple T-test comparison (***p = 0.0003, **p = 0.001). g ISG54 mRNA expression levels in the lung. One-way ANOVA Tukey’s multiple comparisons (*p = 0.03, p = 0.01, ***p = 0.0002, ****p < 0.0001). h Scheme of the multiple functions of TRIM7 during SARS-CoV-2 infection. Data were depicted as mean ± SEM. DPI days post infection, CoV-2 SARS-CoV-2. Source data are provided in the Source data file.
In line with the proposed role of N cleavage in IFN antagonism, WT mice treated with Z-VEID-FMK showed higher levels of IFN-β and ISG54 mRNA in infected lungs, suggesting that the caspase-6 inhibition indeed results in increased IFN responses that could potentially inhibit virus replication, in WT mice. Surprisingly, these effects did not recapitulate in Trim7−/− mice. Consistent with our data described above, vehicle control-treated Trim7−/− mice showed higher IFN responses than WT mice, however caspase inhibition in Trim7-/- did not increase but rather reduced IFN/ISGs (Fig. 6f, g). These data suggest that TRIM7 limits virus replication via a mechanism that partially requires inhibiting caspase-6 activity, but it is mostly independent of IFN-I.
Discussion
In this study, we show that TRIM7 has antiviral activity against SARS-CoV-2 by ubiquitinating the K14 residue on M, and these effects are associated with reduced apoptosis during infection. Our experiments using caspase inhibitors suggest that the antiviral effects of TRIM7 require an active caspase-6 pathway, linking apoptosis to virus replication and pathology. While our study is in line with a previous report that coronaviruses use apoptosis to replicate52, in our study the effects do not seem to be dependent on IFN-I. Although TRIM7 depletion does result in increased cleavage of the viral protein N as well as increased IFN-I induction, these effects do not lead to reduced virus replication. Furthermore, blocking IFN-I signaling did not change virus titers, further suggesting that in this model, IFN-I does not play an antiviral role. In previous studies in mice, IFN-I has been associated with pathology24. Our data agree with these studies, in which IFN-I seems pathogenic and not a major antiviral mechanism. Higher levels of IFN produced by the Trim7-/- mice do not reduce virus titers to the levels observed in WT mice. Intriguingly, blocking IFN-I signaling in the Trim7−/− mice, which induces higher IFN responses, does not affect weight loss (Fig. S6m), although it does result in increased virus titer as compared to isotype-treated Trim7−/− mice (Fig. S6n). This suggests that there is a threshold for IFN-I to have antiviral effects, but without affecting pathology. Therefore, together these results suggest that the increased disease phenotype observed in Trim7-/- mice is IFN-I independent.
At the moment, the connection between the cleavage of N and the increased virus replication observed in Trim7−/− mice remains unclear. However, it is clear that TRIM7 and M-K14 are associated with inhibiting the caspase-6 pathway, which also correlates with a reduction in virus replication.
Our data also indicate that ubiquitination on M-K14 leads to opposite effects from those of the ubiquitination mediated by RNF5 on M-K15, which has a proviral activity40. We further confirmed the previously proposed proviral role of K15, using a recombinant mutant virus.
Although we cannot completely rule out that ubiquitination on K15 can also contribute to effects on apoptosis, our data suggest that ubiquitination on both residues is not mutually exclusive. Using structures of the M protein in its long and short form, we modeled ubiquitinated forms of M with ubiquitin covalently attached to either K14 and/or K15. Our structural modeling analysis suggests that ubiquitination of both lysine residues is energetically possible, either with covalent ubiquitination to one lysine in each M protein monomer, or even with ubiquitination happening in neighboring residues of the same chain (Fig. S7a–d). Our calculations indicate that there is a small energetic advantage for the long form of M, suggesting that ubiquitination may drive the population ensemble of M towards the long form.
According to our data (Fig. S1g) and previous reports41, M can localize in the Golgi membrane or ERGIC vesicles during infection. An important unresolved question is how TRIM7 ubiquitinates the K14 site on M, which is predicted to face the lumen of the Golgi. Since TRIM7 localizes in cytoplasmic structures, how does TRIM7 reach the lumen of the Golgi? Although our technical approaches do not have the resolution to demonstrate where and how TRIM7 ubiquitinates M-K14, it is still possible that TRIM7 may translocate to the lumen. A previous study proposed that TRIM7, may localize in the Golgi56. In addition, we observed that ectopically expressed TRIM7 can colocalize with a truncated N-terminal form of M containing only the transmembrane domains and the ectodomain, which faces the lumen (Fig. S7e). Furthermore, our data suggest that TRIM7 does not act via the degron sequence, which is predicted to face the cytoplasmic side. Although it is still unclear when ubiquitination of M occurs during the viral replication cycle, other examples of viral proteins ubiquitinated on regions predicted to face the luminal side of the Golgi have been reported37,40. Our experiments and previous reports are not yet sufficient to conclude whether TRIM7 can re-localize to the lumen or ubiquitinate viral ectodomains, and future experiments (e.g., cryo-electron tomography) are needed to resolve these questions.
The advantage of using this double mutant virus is that we can avoid any compensatory ubiquitination on one residue if the neighboring one is missing. We also show that these mutations do not affect the IFN-I response, and it is unlikely that the effects on virus replication are IFN-I mediated.
We observed a dysregulation in the inflammatory response in the Trim7−/− mice with a reduced number of infiltrating neutrophils and monocytes in the lung. Inflammatory monocytes responsible for producing inflammatory cytokines such as IL-6, TNF-α, and IL-1β are recruited to the lung in patients with COVID-19. These cytokines have been associated with detrimental inflammation but can also have protective roles57–59. In this mouse model, the decreased levels of proinflammatory cytokines in Trim7−/− mice may correlate with dysfunctional activation of the inflammatory responses associated with severe COVID-19 patients58,60–62. Furthermore, a decrease in monocytes in Trim7−/− correlated with an increase in viral load, consistent with the finding that reduction of monocyte recruitment in ccr2−/− mice increases virus in the lungs and also increases IFN-I RNA during infection63.
In addition, neutrophils have been associated with pathology through the induction of neutrophil extracellular traps (NETs)64–66. Our data suggest that the reduction of neutrophils in Trim7−/− mice is not the reason for the increased virus titers but could contribute to the increased apoptosis. Neutrophils appear to play an important protective role in the recovery phase and could be associated with healing and protecting from apoptosis. This is in line with studies showing that neutrophils can be involved in tissue repair by MMP-9, which can regulate activation of PRRs and promote angiogenesis67–71, and is relevant given the degree of damage to blood vessels/endothelialitis in COVID-1972. These effects could be mediated by a specific subpopulation of neutrophils that needs further characterization, which could also potentially be important for the removal of apoptotic bodies or possibly indirectly by recruiting other cells responsible for this clearance.
Our study suggests that TRIM7 protects the host against SARS-CoV-2 via three independent mechanisms; (1) TRIM7-mediated ubiquitination of M limits a cell autonomous/intrinsic caspase-6-dependent pathway resulting in less tissue damage and less virus replication, (2) by promoting recruitment of innate immune cells that protects against SARS-CoV-2 and may promote tissue repair, and (3) TRIM7 negatively regulates MDA5 signaling, which may help control the detrimental inflammatory effects of IFN-I73 (Fig. S8). The intrinsic apoptotic pathway is unlikely to be dependent on the previously proposed stabilization of B-cell lymphoma 2 (BCL-2) ovarian killer (BOK)18, because the effects on BOK required the endodomain of M18, while the K14 residue required for the effects observed in our study are in the ectodomain of M.
Finally, we identified that mutations on residues K14 and K15 can occur in the circulating strains of SARS-CoV-2. Although the presence of these mutations is relatively low and does not correlate with a specific variant of concern (VOC), the presence of these mutations could indicate that these strains are potentially more pathogenic. Therefore, we propose that monitoring mutations on M in infected individuals might predict disease severity if the effects can be correlated with clinical profiles in infected patients.
Methods
Cell culture
HEK293T (cat. CRL-11268) and A549 (cat. CCL-185) cell lines were purchased from ATCC. Calu-3 2B4 cells were kindly provided by Vineet D. Menachery (The University of Texas Medical Branch at Galveston)74 Vero E6 cells were kindly provided by Pei-Yong Shi (The University of Texas Medical Branch at Galveston). HEK293T-hACE2 cells were kindly provided by Benhur Lee (Mount Sinai)75. A549 TRIM7 KO cells were generated as described by ref. 37. All cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (GIBCO) supplemented with 10% v/v fetal bovine serum (FBS) (HyClone) and 1% v/v penicillin-streptomycin (Corning) in a humidified 5% CO2 incubator at 37 ˚C.
Viruses
Viruses used in this study were handled under biosafety level 3 (BSL-3) conditions at UTMB facilities in accordance with institutional biosafety approvals. SARS-CoV-2 (USA-WA1/2020) was kindly provided by The World Reference Center of Emerging Viruses and Arboviruses (WRCEVA) (The University of Texas Medical Branch at Galveston), SARS-CoV-2 (CMA3p20) was kindly provided by Vineet D. Menachery (The University of Texas Medical Branch at Galveston) and grown in Vero E6 cells as described by ref. 43. SARS-CoV-2 USA-WA1/2020 + D614G was provided by Dr. Pei-Yong Shi (The University of Texas Medical Branch at Galveston).
The infectious cDNA clone icSARS-CoV-2 M-K14/15R was constructed through mutagenesis of a mouse-adapted USA-WA1/2020 SARS-CoV-2 (CMA5 strain) used for in vivo studies76,77. To generate the CMA5 strain, an adaptive mutation (Spike_Q493H) was identified and engineered into the backbone of the CMA3p20 strain43. The full-length cDNA was assembled via in vitro ligation and used as a template for T7 in vitro transcription. The full-length viral RNA was electroporated into Vero E6 cells. Forty-eight hours post electroporation, the original P0 virus was harvested and used to infect another flask of Vero E6 cells to produce the P1 virus. The titer of the P1 virus was determined by plaque assay on Vero E6 cells. The viral RNA of P1 virus was extracted and sequenced to confirm the designed mutations using the primers: M-K14R/K15R-F-ACCGTTGAAGAGCTTCGCCGCCTCCTTGAACAATGG and M-K14R/K15R-R CCATTGTTCAAGGAGGCGGCGAAGCTCTTCAACGGT. The P1 virus was used for all the experiments performed in this study. All work following electroporation was performed in a BSL3 laboratory.
Plasmids
The M-WT, M-K14R, M-K15R, M-KallR, and M-K14O, M-Q22A, and M-NTD were cloned into pXJ-HA plasmid, Flag-TRIM7 constructs Variant 1 and 2 were purchased from Origene (Rockville, MD), TRIM7-PRY-SPRY domain was PCR amplified from the full-length TRIM7 plasmid cloned in pCMV6-Entry Vector (Origin Cat. No. RC224754) using the primer F: CGCGATCGCCATGCTGAAGAAGTTCAAAGAG and R: GCGTACGCGTAGGCCAGATTCGCAAGTAGG and cloned into pCMV6-FLAG plasmid. the Flag-OTU and -OTU-2A were kindly provided by Adolfo Garcia-Sastre (Mount Sinai), the Ub plasmids have been described before78.
Transfections
Transient transfections of DNA were performed with TransIT-LT1 (Mirus Bio) for HEK293T cells, and Lipofectamine 3000 (Invitrogen) for A549 cells according to the manufacturer’s guidelines. For lipofectamine transfection media was exchanged 6–8 h. All transfections were performed in DMEM 10% v/v FBS without penicillin-streptomycin.
Cell lysis and co-immunoprecipitation
Cells were harvested in RIPA lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% (v/v) IGEPAL CA-630, 0.5% (w/v) sodium deoxycholate, 0.1% (v/v) SDS, protease inhibitor cocktail79, 5 mM N-ethylmaleimide (Sigma), and 5 mM iodoacetamide (Sigma) as deubiquitinase inhibitors. Cell lysates were clarified by centrifugation at 21,000×g for 20 min at 4 ˚C. About 10% of the clarified lysate was added to 2X SDS-PAGE loading buffer containing 2-mercaptoethanol, heated for 30 min at 37 ˚C, and stored at −20 ˚C as a whole-cell lysate (WCL). The remaining lysate was subjected to immunoprecipitation with anti-FLAG M2 or anti-HA, EZview Red agarose beads (Sigma) overnight at 4 ˚C on a rotating platform. Beads were washed seven times in RIPA buffer (150 or 550 mM NaCl) and the bound proteins were eluted using FLAG or HA peptide respectively, elution was reduced in 2X SDS-PAGE loading buffer containing 2- Mercaptoethanol and incubated for 30 min at 37 ˚C.
Denaturing pull-down
Cell lysis and WCL collection were performed as above. Lysates were subjected to pull down using nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) overnight at 4 ˚C on a rotation platform. Beads were washed seven times using denaturing buffer containing 50 mM Tris-HCl pH 8.0 (Sigma), 6 M urea, 350 mM NaCl, 0.5%(v/v) IGEPAL CA-630 (Sigma), and 40 mM imidazole (Sigma). The proteins were eluted at 4 ˚C for 30 min, using elution buffer containing 50 mM Tris-HCl pH 8.0 and 300 mM imidazole. Eluted proteins were treated with 2X SDS-PAGE loading buffer containing 2-mercaptoethanol and incubated for 30 min at 37 ˚C.
Confocal immunofluorescence
HeLa cells were seeded into six-well plates. After 16 h, the cells were transfected with 1 µg of M-WT or M-K14R-HA tagged with the co-expression of TRIM7-FLAG tagged for 24 h. The cells were washed with DPBS 1X, fixed with 4% paraformaldehyde 20’, permeabilized with 0.1% Triton X100 (v/v) in DPBS 1X for 5 min, and blocked with 0.5% pork skin gelatin (w/v) in DPBS for 1 h. The staining was performed with rabbit anti-HA (Milipore Sigma H6908, 1:100 dilution), anti-FLAG (Sigma-Aldrich F1804, 1:100 dilution in P) overnight at 4 °C. The next day, cells were washed with DPBS 1X and incubated with the secondary antibodies anti-mouse Ig Alexa Fluor 488 (Invitrogen A21202) and anti-rabbit Ig Alexa Fluor 555 (Invitrogen A31572) at 1:200 dilution each in DPBS 1X) and washed with DPBS 1X after 2 h incubation at RT. DAPI staining (BioLegend) working solution (1 µg/mL in PBS) was added for 5 min at RT and washed with PBS before mounting with Merck FluorSaveTM reagent. Micrographs were taken with the Leica Stellaris 8 tau-STED Microscope (Leica Microsystems). Microscope parameters and LAS-X software post-processing were set constant for each experiment. Fluorescence intensity values were obtained with ImageJ software (National Institute of Health), and curves were graphed with Graphpad Prism 10 (Graphpad Software, Inc.).
Virus-like particles (VLPs) generation
VLPs were generated by transfection of the plasmids for expression of S-HA, M-HA, N-FLAG, and E-FLAG, into A549 WT and TRIM7 knockout, briefly; 2 × 105 cells were seeded into a six-well plate and transfected using a total of 2 µg of plasmid using Lipofectamine 3000 (Invitrogen, USA) as per the manufacturer’s instruction. The molar ratio for the S, E, M, and N plasmid was 8:8:6:3, as described by ref. 80. Seventy hours after transfection, the supernatant was collected, and the cells were harvested in RIPA buffer for immunoblotting. The supernatant was clarified by centrifugation at 4000 rpm for 10 min, then the supernatant was filtered through a 0.45 μM mesh to remove the debris, subsequently, the supernatant was layered over in a 20% sucrose gradient and ultra-centrifugated at 74,000×g for 3 h at 4 ˚C to pellet down the VLPs and subsequently loaded on to discontinuous, 20–60% sucrose solution and centrifuged at 74,000×g for 3 h at 4 ˚C. The opaque band containing the VLPs were collected and analyzed by western blot.
Western blot
Cell lysates were resolved on 7.5 or 4–15% Mini-PROTEAN and Criterion TGX SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes using a Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked with 5% (w/v) non-fat dry milk in TBST (TBS with 0.1% (v/v) Tween-20) for 1 h and then probed with the indicated primary antibody in 3% (w/v) BSA in TBST at 4 ˚C overnight. Following overnight incubation, membranes were probed with secondary antibodies in 5% (w/v) non-fat dry milk in TBST for 1 h at room temperature in a rocking platform: anti-rabbit or anti-mouse IgG-HRP conjugated antibody from sheep (both 1:10,000 NA934 and NA931 GE Healthcare). Proteins were visualized using ECL or SuperSignal West Femto chemiluminescence reagents (Pierce) and detected by autoradiography. Protein band intensity was quantified using ImageJ software (version 1.53 m NIH).
M Polyclonal antibody generation
M SARS-CoV-2 antibody was generated by ProSci using the antigen sequence of the N-terminal end of SARS-CoV-2 Membrane protein MADSNGTITVEELKKLLEQ to immunize rabbits, then the serum was purified by affinity purification to obtain the polyclonal antibody.
Mice
All animal experiments were carried out following Institutional Animal Care and Use Committee (IACUC) guidelines and have been approved by the IACUC of the University of Texas Medical Branch at Galveston protocol numbers: 2103023 and 1904044, and Rutgers University protocol number PROTO202200007. Our studies utilized males and females of 20- to 25-week-old C57BL/6NJ WT mice (The Jackson Laboratory) that match the Trim7−/− mice generated as described by ref. 37 and male 25-week-old C57BL/6J WT mice (The Jackson Laboratory). Mice were maintained under specific pathogen-free conditions in the Animal Resource Center (ARC) facility at UTMB or Rutgers University in a climate control room with access to food and water at libitum and 12 h light/dark cycles, at a temperature of 18–23 °C and humidity of 40–60%. Animal experiments involving infectious viruses were performed under animal biosafety level 3 (ABSL-3) conditions at UTMB and Rutgers University in accordance with institutional biosafety approvals.
In vitro virus infection
HEK293t-ACE-2 overexpressing TRIM7 or TRIM7ΔRING domain were seeded onto 24-well plates at a confluency of 100,000 cells/well and infected with SARS-CoV-2 USA/WA1 D614G strain MOI 0.1 for 1 h, cells were washed once with DPBS 1X and incubated with 6, 24, and 48 h after infection. supernatant, RNA, and protein were collected to measure virus titers, gene, and protein expression respectively.
In vivo virus infection
Male and female WT and Trim7−/− mice were anesthetized with 5% isoflurane and infected intranasal with SARS-CoV-2 1 × 106 PFU of CMA3p20 strain, mice were weighed every day for 7 days. Euthanasia was performed at days 2, 3, or 7 post infection using isoflurane overdose, lungs and serum were collected for downstream analysis. For neutrophil depletion experiments, male WT C57BL/6J mice were injected intraperitoneally81 with 100 μg/mouse of anti-Ly6G (BioXCell, cat. BP0075-1, lot 695418J) or isotype (BioXCell cat. BP0089, lot 71671801) 1 day before and one after the infection, mice were infected with SARS-CoV-2 1 × 106 PFU of CMA3p20 and weighed every day for 10 days. On Day 3 post infection, a group of mice was euthanized to perform flow cytometry of lung or peripheral blood to confirm neutrophil depletion. For caspase-6 inhibition experiments, male and female C57BL/6NJ WT and Trim7−/− mice were treated with Z-VEID-FMK (APExBIO), dose: 12.5 mg/kg diluted in PBS or DMSO in PBS as the vehicle through IP injection at day 0, 1, and 2 post infection. For IFN-I blocking experiments, mice were IP injected with IFNAR1(Leinco Technologies cat. I-401, lot 1123L460) or Isotype IgG control (Leinco Technologies cat. I-536, lot 0923L495) antibody 2 mg/per mice at day 0.
Lung single-cell suspension and flow cytometry
Lungs isolated from infected mice were collected in RPMI 10% v/v FBS 1% v/v penicillin-streptomycin, lungs were rinsed with DPBS, cut into small pieces, and digested in digestion media containing 0.7 mg/ml of collagenase D and 30 µg/ml of DNase I in serum-free RPMI for 30 min in a humidified 5% CO2 incubator at 37 ˚C. FBS was added to the digestion media to inactivate the enzymes. Lungs were then passed through A 70-μm cell strainer to obtain single-cell suspension. Red blood cells were lysed using RBC lysing buffer Hybri-Max (Sigma), cells were counted, and 1 × 106 cells were stained using the following antibodies. Anti-CD45-PE (Biolegend, cat. 103105, lot B299555), anti-podoplanin PE-DAZZLE594 (Biolegend, cat.127419, lotB298016), anti-CD24-BUV395 (BD Biosciences, cat. 744471, lot 2091087) anti-CD31-BV510 (BD Biosciences cat. 740114, lot 2091082), anti- CD326-BV711 (Biolegend, Biolegend, cat. 118233), anti- MHC-II-BV605 (Biolegend, cat. 107639, lot B321059), or anti- PDCA-1-APC (Biologend, cat. 127016, lot B337341), anti- CD11b-AF700 (Biolegend, cat. 101222, lot B336447), anti- Ly6G-BV780 (Biolegend, cat. 127645, lot B313888), anti-CD11c Percp-Cy5.5 (Biolegend, cat.117328, lot B332774), anti- Ly6C-FITC (Biolegend, cat. 128005, lot B368467). To measure cell death and apoptosis, cells were stained with Ghost dye-Red780 (Tonbo Biosciences, cat. 13-0865-T100) or Fixable viability dye-eFluor506 (eBiosciences, cat. 64-0866-14, lot 2290921) and Apotracker Green (Biolegend, cat. 427402, lot B34791). After staining, samples were fixed using 4% ultrapure formaldehyde diluted in DPBS from 16% methanol-free ultrapure formaldehyde (Thermo Scientific) for 48 h. Samples were acquired using LSR II Fortessa and analyzed using FlowJo.
Plaque assay
The supernatant of infected cells or lung homogenate was used to measure viral titers. Briefly, confluent monolayers of Vero E6 cells plated in a 12-well plate were infected with virus diluted using DMEM 2% v/v FBS without penicillin-streptomycin, incubated at 37 ˚C for 1 h rocking the plate every 15 min. Infectious were removed and media was replaced with MEM containing 0.6% v/v tragacanth (Sigma), 5% v/v FBS and 1% v/v penicillin-streptomycin, plates were incubated at 37 ˚C for 2 days in humidified 5% CO2 incubator. Plates were fixed and stained using 10% buffered formalin containing 0.5% (w/v) crystal violet for 30 min.
Histology
The right inferior lobe of the lung was fixed in 10% neutral buffered formalin (HT501128, Sigma, MI) for 7 days. Tissues were cut, paraffinized and H&E stained by the Anatomic Pathology Laboratory of the Pathology Department of the University of Texas Medical Branch. The inflammatory score was calculated by analyzing the presence of peribronchiolar infiltrates (Yes = 1, No = 0) plus 1–2 Foci of inflammation (1), 2–3 foci of inflammation (2), and 3+ foci of inflammation82.
IFN-β or ISRE luciferase reporter assay
HEK293T cells were seeded into 24-well plates (50,000 cells/well) and were transfected with 30 ng of IFN-β or 180 ng of ISRE reporter plasmid together with 10 ng of Renilla luciferase plasmid. For IFN-β reporter assay, cells were co-transfected with 5 ng of MDA5 and increasing concentrations of TRIM7 20, 40, or 80 ng for 24 h. For ISRE assays, cells were co-transfected with 100 ng of M-WT or K-R mutants plasmids for 24 h and stimulated with 1000IU/ml of IFN-β for 16 h. Cells were lysed and luciferase activity was measured using the dual-luciferase reporter assay system (Promega) on a Cytation 5 Multi-Mode Reader (BioTek) according to the manufacturer’s instructions. Values were normalized to Renilla.
Quantitative reverse-transcription-PCR (qRT-PCR)
Total RNA was isolated using the Direct-zol RNA Miniprep Kit (Zymo Research) following the manufacturer’s instructions. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time qPCR was performed in 384-well plates using iTaq Universal SYBR Green Supermix and a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Gene expression was normalized to either human 18S or murine β-actin by the comparative CT method (DDCT). Primers: h18S rRNA: F-GTAACCCGTTGAACCCCATT, R-CCATCCAATCGGTAGTAGCG, hIFN-β: F-TCTGGCACAACAGGTAGTAGGC, R-GAGAAGCACAACAGGAGAGCAA, hISG54: F-ATGTGCAACCTACTGGCCTAT, R-TGAGAGTCGGCCCATGTGATA, mβ-Actin: F-CGGTTCCGATGCCCTGAGGCTCTT, R-CGTCACACTTCATGATGGAATTGA, mIFN-β F-CAGCTCCAAGAAAGGACGAAC, R-GGCAGTGTAACTCTTCTGCAT, mCXCL10: F-ATCATCCCTGCGAGCCTATCCT, R-GACCTTTTTTGGCTAAACGCTTTC, mISG54: F-CTGAGAGGGGAGTGGACTCTG, R-GCACCTGCTTCATCCAAAGAT, mIL-1β: F-TGGACCTTCCAGGATGAGGACA, R-GTTCATCTCGGAGCCTGTAGTG, SCoV-2 ORF1a/ab: F-AGTTACGGCGCCGATCTAAAGTCAT, R-TAGCCATCAGGGC CACAGAAGTT, SCoV-2 N: F-GCAATCCTGCTAACAATGCTGCA, R-ACGAGAAGAGGCTTGACTGC.
Cytokine quantification
Cytokines from serum and lung homogenate were quantified using BioPlex mouse cytokine 23-plex assay (Bio-Rad) following the manufacturer’s instructions. Serum was diluted 1:3. Samples were analyzed in a Bio-Plex200 Multiplex system (Bio-Rad).
RNAseq data processing
RNA isolated from lung homogenates of C57BL/6NJ infected as described above as isolated at day 3 post infection, was quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). The poly(A) + RNA was enriched from total RNA (1 mg) using oligo(dT)-attached magnetic beads. First- and second-strand synthesis, adapter ligation, and amplification were performed following the SMART-3Seq protocol83. The quality of the library was evaluated using an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. The library DNA templates were quantified using qRT-PCR and a known-size reference standard. The libraries were multiplexed and sequenced on Illumina NextSeq 550 using the SE75 High Output kit. NGS sequence reads were aligned using the Spliced Transcript Alignment (STAR v2.5.1b) to mouse reference genome mm10, using default parameters84.
Global analysis
Global membrane protein K14/K15 mutation occurrence was analyzed using the CoV-Spectrum49 (https://open.cov-spectrum.org) dashboard. The analysis covered all samples in the “Open Data” version of CoV-Spectrum (GenBank deposited samples) and the period 2020-01-06 to 2024-01-31. The following queries were used to determine the occurrence of mutations to K14/K15 and K14 + K15 (using Nextstrain Clade 21 A as an example):
21 A (Nextstrain clade) & M:K14
21 A (Nextstrain clade) & M:K15
21 A (Nextstrain clade) & M:K14 & M:K15
For each query, the total number of samples belonging to the underlying clade was obtained, and the percentage of samples with the particular mutation was determined using the “Substitutions and deletions” section of the resulting CoV-Spectrum reports. Data were visualized using an adaption of the nCoV Clades Schema (https://github.com/nextstrain/ncov-clades-schema)85 using Miro.
Methods for M protein + ubiquitin modeling
We utilized structures of the M protein in its long form (PDB 7VGR) and short form (PDB 7VGS). To model the membrane, we used a lipid composition reminiscent of the biological ER-Golgi intermediate compartment (ERGIC)86 and employed the CHARMM-GUI membrane builder87–89. Subsequently, we generated four models of covalent interactions with a ubiquitin structure (PDB 2JF5), each representing interactions with K14 and K15 from either the same M protein monomer or different monomers, for both conformational states of M. These models were then subjected to minimization using the Yasara web server90, followed by the calculation of the total energy of the systems utilizing adapted Surfaces functions91.
Statistical analysis
All data were presented as means ± SEM as indicated and analyzed using GraphPad PRISM software (version 9 GraphPad Software). Data were representative of 2–3 independent experiments, for in vivo experiments mice number is indicated in the figure legends, in vitro experiments include three biological replicates, unless otherwise specified in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by the US National Institute of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) grants. R01AI166668, R01AI155466, and P01AI150585 awarded to R.R. R01AI134907 and Building Interdisciplinary Research Careers in Women’s Health Program (BIRCWH) K12HD052023 awarded to M.I.G. and R01AI146081 awarded to S.M.
Author contributions
M.G.-O. performed all aspects of this study. H.X. generated mutant virus. H-c.T. and A.O. performed in vivo experiments. A.H., P.B., K.A., M.I.G., M.H., L.P.-C., Y.P.-T., S.v.T., and J.M.-C. performed in vitro experiments. M.C. and M.B. performed a global analysis of sequences. N.T. and R.J.N. performed the computational modeling analysis. T.R.S. performed histopathological analysis. K.T. and S.M. generated an M polyclonal antibody. R.S. V.M., M.E., J.E., P.-Y.S., X.X., and A.N.F. provided critical reagents and technical advice. R.R. designed, directed, contributed to data analysis, and obtained funding. M.G.-O. and R.R. organized the study and prepared the manuscript. All authors read the manuscript and provided comments.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Transcriptomic data generated in this study has been deposited with the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE268640 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE268640. Plasmids generated in this study are available upon request to the corresponding author, appropriate MTA will be required. The remaining data in this study are available within the supplementary data in the source data file provided in this manuscript. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54762-5.
References
- 1.Lamers, M. M. & Haagmans, B. L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol.20, 270–284 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Martines, R. B. et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg. Infect. Dis.26, 2005–2015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan, W., Zheng, Y., Zeng, X., He, B. & Cheng, W. Structural biology of SARS-CoV-2: open the door for novel therapies. Signal Transduct. Target Ther. 7, 26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, Z. et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat. Commun.13, 4399 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou, Y. J. et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell182, 429–446 e414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsatsakis, A. et al. SARS-CoV-2 pathophysiology and its clinical implications: an integrative overview of the pharmacotherapeutic management of COVID-19. Food Chem. Toxicol.146, 111769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delorey, T. M. et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature595, 107–113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sapir, T. et al. COVID-19 and the immune response: a multi-phasic approach to the treatment of COVID-19. Int. J. Mol. Sci.10.3390/ijms23158606 (2022). [DOI] [PMC free article] [PubMed]
- 9.Yang, L. et al. COVID-19: immunopathogenesis and Immunotherapeutics. Signal. Transduct Target. Ther.5, 128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn, J. H. et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Invest.10.1172/JCI148517 (2021). [DOI] [PMC free article] [PubMed]
- 11.Sungnak, W. et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med.26, 681–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou, X. et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med.14, 185–192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnamachary, B. et al. Extracellular vesicle-mediated endothelial apoptosis and EV-associated proteins correlate with COVID-19 disease severity. J. Extracell. Vesicles10, e12117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, Y. et al. Cell-type apoptosis in lung during SARS-CoV-2 infection. Pathogens10, 509 (2021). [DOI] [PMC free article] [PubMed]
- 15.Li, X., Zhang, Z., Wang, Z., Gutierrez-Castrellon, P. & Shi, H. Cell deaths: involvement in the pathogenesis and intervention therapy of COVID-19. Signal Transduct. Target Ther.7, 186 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riegler, A. N., Benson, P., Long, K. & Leal, S. M. Jr Differential activation of programmed cell death in patients with severe SARS-CoV-2 infection. Cell Death Discov.9, 420 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren, Y. et al. SARS-CoV-2 membrane glycoprotein M triggers apoptosis with the assistance of nucleocapsid protein N in cells. Front. Cell Infect. Microbiol.11, 706252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, Y. et al. SARS-CoV-2 membrane protein causes the mitochondrial apoptosis and pulmonary edema via targeting BOK. Cell Death Differ.29, 1395–1408 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou, Z. et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe27, 883–890 e882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, Q., Xu, Y., Wang, T. & Xie, F. Innate and adaptive immune response in SARS-CoV-2 infection-current perspectives. Front. Immunol.13, 1053437 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., Zhang, K., Zhang, Y., Gu, Z. & Huang, C. Neutrophils in COVID-19: recent insights and advances. Virol. J.20, 169 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo, Y. et al. Neutrophil extracellular traps in COVID-19. JCI Insight10.1172/jci.insight.138999 (2020). [DOI] [PMC free article] [PubMed]
- 23.Channappanavar, R. et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe19, 181–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Israelow, B. et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med.10.1084/jem.20201241 (2020). [DOI] [PMC free article] [PubMed]
- 25.Yin, X. et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep.34, 108628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampaio, N. G. et al. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep.11, 13638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu, Y. Z. et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol. Immunol.18, 613–620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sui, L. et al. SARS-CoV-2 membrane protein inhibits type I interferon production through ubiquitin-mediated degradation of TBK1. Front. Immunol.12, 662989 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng, Y. et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target Ther.5, 299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia, H. et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep.33, 108234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, K. et al. SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-mediated IFN-beta production. Viruses10.3390/v13010047 (2020). [DOI] [PMC free article] [PubMed]
- 32.Wu, J. et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep.34, 108761 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei, X. et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun.11, 3810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valerdi, K. M., Hage, A., van Tol, S., Rajsbaum, R. & Giraldo, M. I. The role of the host ubiquitin system in promoting replication of emergent viruses. Viruses10.3390/v13030369 (2021). [DOI] [PMC free article] [PubMed]
- 35.Xia, Y., Zhao, J. & Yang, C. Identification of key genes and pathways for melanoma in the TRIM family. Cancer Med.9, 8989–9005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan, W. et al. TRIM7 inhibits enterovirus replication and promotes emergence of a viral variant with increased pathogenicity. Cell184, 3410–3425 e3417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giraldo, M. I. et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature585, 414–419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, M. et al. E3 ubiquitin ligase tripartite motif 7 positively regulates the TLR4-mediated immune response via its E3 ligase domain in macrophages. Mol. Immunol.109, 126–133 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Stukalov, A. et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature594, 246–252 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Yuan, Z. et al. The E3 ubiquitin ligase RNF5 facilitates SARS-CoV-2 membrane protein-mediated virion release. mBio10.1128/mbio.03168-21 (2022). [DOI] [PMC free article] [PubMed]
- 41.Zhang, J. et al. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct. Target Ther.5, 269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang, X. et al. A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures. Nat. Chem. Biol. 10.1038/s41589-022-01128-x (2022). [DOI] [PubMed]
- 43.Muruato, A. et al. Mouse-adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. PLoS Biol.19, e3001284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bermejo-Jambrina, M. et al. Infection and transmission of SARS-CoV-2 depend on heparan sulfate proteoglycans. EMBO J.40, e106765 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Q. et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) and Spike (S) proteins antagonize host type I interferon response. Front. Cell Infect. Microbiol.11, 766922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraborty, A., Diefenbacher, M. E., Mylona, A., Kassel, O. & Behrens, A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signalling. Nat. Commun.6, 6782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, S. et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target Ther.5, 235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maharjan, S. et al. Apoptosis enhances the replication of human coronavirus OC43. Viruses10.3390/v13112199 (2021). [DOI] [PMC free article] [PubMed]
- 49.Chen, C. et al. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics38, 1735–1737 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danthi, P. Viruses and the diversity of cell death. Annu. Rev. Virol.3, 533–553 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Barber, G. N. Host defense, viruses and apoptosis. Cell Death Differ.8, 113–126 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Chu, H. et al. Coronaviruses exploit a host cysteine-aspartic protease for replication. Nature609, 785–792 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Chu, H. et al. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci. Adv.10.1126/sciadv.abf8577 (2021). [DOI] [PMC free article] [PubMed]
- 54.Diemer, C. et al. Cell type-specific cleavage of nucleocapsid protein by effector caspases during SARS coronavirus infection. J. Mol. Biol.376, 23–34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eleouet, J. F. et al. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol.74, 3975–3983 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montori-Grau, M. et al. GNIP1 E3 ubiquitin ligase is a novel player in regulating glycogen metabolism in skeletal muscle. Metabolism83, 177–187 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Hsu, R. J. et al. The role of cytokines and chemokines in severe acute respiratory syndrome coronavirus 2 infections. Front. Immunol.13, 832394 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schett, G., Sticherling, M. & Neurath, M. F. COVID-19: risk for cytokine targeting in chronic inflammatory diseases? Nat. Rev. Immunol.20, 271–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, G. et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest.130, 2620–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tay, M. Z., Poh, C. M., Renia, L., MacAry, P. A. & Ng, L. F. P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol.20, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong, R. S. Y. Inflammation in COVID-19: from pathogenesis to treatment. Int. J. Clin. Exp. Pathol.14, 831–844 (2021). [PMC free article] [PubMed] [Google Scholar]
- 63.Vanderheiden, A. et al. CCR2 signaling restricts SARS-CoV-2 infection. mBio12, e0274921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, Y. et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci. Rev.7, 998–1002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chevrier, S. et al. A distinct innate immune signature marks progression from mild to severe COVID-19. Cell Rep. Med.2, 100166 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veras, F. P. et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med.10.1084/jem.20201129 (2020). [DOI] [PMC free article] [PubMed]
- 67.Ardi, V. C., Kupriyanova, T. A., Deryugina, E. I. & Quigley, J. P. Human neutrophils uniquely release TIMP-free MMP-9 to provide a potent catalytic stimulator of angiogenesis. Proc. Natl Acad. Sci. USA104, 20262–20267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christoffersson, G. et al. Clinical and experimental pancreatic islet transplantation to striated muscle: establishment of a vascular system similar to that in native islets. Diabetes59, 2569–2578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishio, N., Okawa, Y., Sakurai, H. & Isobe, K. Neutrophil depletion delays wound repair in aged mice. Age30, 11–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hingorani, D. V. et al. Impact of MMP-2 and MMP-9 enzyme activity on wound healing, tumor growth and RACPP cleavage. PLoS ONE13, e0198464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mohan, R. et al. Matrix metalloproteinase gelatinase B (MMP-9) coordinates and effects epithelial regeneration. J. Biol. Chem.277, 2065–2072 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Vrints, C. J. M. et al. Endothelialitis plays a central role in the pathophysiology of severe COVID-19 and its cardiovascular complications. Acta Cardiol.76, 109–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, B. et al. Negative regulation of RNF90 on RNA virus-triggered antiviral immune responses targeting MAVS. Front. Immunol.12, 730483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lokugamage, K. G. et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 10.1128/JVI.01410-20 (2020). [DOI] [PMC free article] [PubMed]
- 75.Oguntuyo, K. Y. et al. Quantifying absolute neutralization titers against SARS-CoV-2 by a standardized virus neutralization assay allows for cross-cohort comparisons of COVID-19 Sera. mBio10.1128/mBio.02492-20 (2021). [DOI] [PMC free article] [PubMed]
- 76.Xie, X. et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc.16, 1761–1784 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie, X. et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe27, 841–848 e843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajsbaum, R. et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity40, 880–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hadjadj, J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science369, 718–724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu, R., Shi, M., Li, J., Song, P. & Li, N. Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol.8, 862 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcia, M. et al. Innate lymphoid cell composition associates with COVID-19 disease severity. Clin. Transl. Immunol.9, e1224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klopfleisch, R. Multiparametric and semiquantitative scoring systems for the evaluation of mouse model histopathology-a systematic review. BMC Vet. Res.9, 123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Foley, J. W. et al. Gene expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. Genome Res.29, 1816–1825 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ivan Aksamentov, C. R. et al. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw.10.21105/joss.03773 (2021).
- 86.Collins, L. T. et al. Elucidation of SARS-Cov-2 budding mechanisms through molecular dynamics simulations of M and E protein complexes. J. Phys. Chem. Lett.12, 12249–12255 (2021). [DOI] [PubMed] [Google Scholar]
- 87.Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem.29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Lee, J. et al. CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J. Chem. Theory Comput.15, 775–786 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Wu, E. L. et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem.35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krieger, E. et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins77, 114–122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teruel, N., Borges, V. M. & Najmanovich, R. Surfaces: a software to quantify and visualize interactions within and between proteins and ligands. Bioinformatics10.1093/bioinformatics/btad608 (2023). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic data generated in this study has been deposited with the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE268640 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE268640. Plasmids generated in this study are available upon request to the corresponding author, appropriate MTA will be required. The remaining data in this study are available within the supplementary data in the source data file provided in this manuscript. Source data are provided with this paper.