Abstract
Chemerin, a chemotactic adipokine, plays essential roles in adipogenesis and inflammation. Serum chemerin concentration is closely associated with obesity and metabolism disorders. The mature form of chemerin (residues 21-157) acts primarily through chemerin receptor 1 (CMKLR1) for transmembrane signaling. As a result, CMKLR1 serves as a promising target for therapeutic intervention of immunometabolic diseases such as diabetes and multiple sclerosis. Here, we present a high-resolution cryo-EM structure of CMKLR1-Gi signaling complex bound to biologically active full-length chemerin. The mature chemerin shows binding features distinct from its C-terminal nonapeptide including interaction with both the extracellular loops (ECLs) and the N-terminus of CMKLR1. Combining results from functional assays, our studies demonstrate that chemerin interacts with CMKLR1 in a “two-site” mode similar to chemokine-chemokine receptor interactions, but acting as a “reverse chemokine” by inserting its C-terminus instead of the N-terminus as in the case of chemokines into the transmembrane binding pocket of CMKLR1. These structural insights are expected to help develop synthetic analogs with therapeutic potential.
Subject terms: Cryoelectron microscopy, Chemokines
A high-resolution cryo-EM structure of the CMKLR1-Gi signaling complex bound to full-length chemerin reveals its unique binding mechanism that is distinct from chemokine-chemokine receptor interactions.
Introduction
The chemotactic adipokine chemerin is a clinical serum marker of inflammation. Its receptor CMKLR1, a G protein-coupled receptor (GPCR), has also been identified as an inflammation biomarker. Chemerin-CMKLR1 axis regulates various immune-metabolic processes and diseases, such as obesity, diabetes, Alzheimer’s disease, multiple sclerosis, and cancer1–5. Therefore, it serves as an attractive target for developing drug candidates with pharmacological potential. Several candidates have already been identified and currently in different stages of clinical trials6–11.
Despite these progresses, however, the lack of structural details of chemerin and chemerin-bound CMKLR1 has long hindered the understanding of the molecular mechanism of the chemerin-CMKLR1 axis as well as the process of targeted drug development. Although the 9-amino peptide derived from the C-terminus of chemerin (C9) has been found sufficient to activate CMKLR112, C9 is a synthetic agonist that does not exist in nature. The reported physiological function of C9 as a drug candidate for inflammatory or anti-inflammatory disorders is controversial13–15. It is worth noting that the physiologically relevant agonist of CMKLR1 remains to be the full-length chemerin (a.a. 21-157), as most drug candidates reported for CMKLR1 use different sites with uncharacterized binding mode. For example, the reported antibodies use ECL3 for recognition and Resolvin E1 uses an allosteric site13,16,17. In consideration of the diverse functions of the chemerin-CMKLR1 axis in immune cells, a fine regulation of CMKLR1 functions by chemerin is necessary.
Here, we report the cryo-EM structure of CMKLR1-Gi signaling complex bound to human full-length mature chemerin, which provides a structural framework for comprehending chemerin action at CMKLR1 from a structural perspective. The high-resolution map unambiguously provides the chemerin binding pose of CMKLR1 in the Gi coupling state, which unravels a chemokine-like “two-site” model of interaction between chemerin and CMKLR1. In particular, extracellular domains of CMKLR1 including the N-terminus and ECLs, are involved in the recognition of full-length chemerin. This observation confirmed the crucial role of the extracellular domains of CMKLR1, previously found to be the recognition site of therapeutic antibodies, in binding of full-length chemerin. As reported in this study, the interaction of chemerin by CMKLR1 consists of 5 different sites. Combined with functional assays, our studies give a thorough structural framework and extend the knowledge of the mechanism underlying adipokine signaling through GPCRs. These elucidations have paved the way for the precise modulation of ligands, such as small molecules, peptides, and antibodies, for innovative drug discovery, and excite further investigation into receptor-dependent physiological functions and pathological functions as well as structure-based design of drug candidates that regulate inflammation by targeting CMKLR1.
Methods
Constructs and vector design
The constructs of full-length CMKLR1 and chemerin were designed as we reported before18,19. The full-length coding sequence of human wild-type CMKLR1 was synthesized by General Biol (Chuzhou, China). DNA sequence coding for a hemagglutinin (HA) signal peptide, a FLAG-tag, a human rhinovirus 3C (HRV 3C) protease cleavage site (LEVLFQGP), and a thermostabilized apocytochrome b562RIL (BRIL) was added to the N terminal for convenience of protein expression and purification. The CMKLR1 and DNGαi1 (dominant negative Gαi1 with G203A and A326S mutation) were cloned into a pFastbac1 vector, respectively. Human Gβ1 and Gγ2 with N-terminal 6×His-tag were cloned into a pFastBac-Dual vector. Human chemerin with a C-terminal 6 x His tag was cloned into a pFastbac1 vector. The single-chain antibody scFv16 was cloned into a pFastbac1 vector with the GP67 signal peptide sequence at the N-terminus and 6 × His tag at the C-terminus. Point mutations for functional assays and an N-terminal truncation of CMKLR1 were introduced by homologous recombination using the ClonExpress Ultra One Step Cloning Kit (C115, Vazyme Biotech). The wild type and mutant CMKLR1 cDNA were cloned into pcDNA3.1(+) vector for cellular assays.
Chemerin and scFv16 expression and purification
The chemerin and scFv16 were expressed and purified in a similar manner. The Trichoplusia ni Hi5 insect cells (B85502, Invitrogen) were cultured in suspension at 27 °C with shaking. Baculoviruses were prepared using the bac-to-bac expression system (Invitrogen). When reaching a density at 2 × 106, the cells were infected by baculoviruses of chemerin or scFv16, respectively. The supernatant was collected after 60 h of culture by centrifugation at 2000 × g for 20 mins. The pH of the medium was adjusted to 7.4 by 2 M HEPES solution (pH 8.0). The supernatant was loaded onto the Ni-NTA resin gravity column. The Ni-NTA resin was washed with 10 column volumes of buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, and 20 mM imidazole. Then, the protein was eluted by a buffer containing 20 mM HEPES (pH 7.4), 100 mM NaCl, and 250 mM imidazole. The eluted fraction was concentrated and subjected to size-exclusion chromatography on a SuperdexTM 75 Increase 10/300 column (Cytiva). Finally, the peak fractions corresponding to the protein were collected, analyzed by SDS-PAGE and Western blotting, concentrated to approximately 10 mg/mL, frozen in liquid nitrogen, and stored at -80°C until further use.
Protein complex expression and purification
The insect sf9 cells (11496015, Invitrogen) were cultured in SIM SF Expression Medium (Sino Biological) to reach a density at 2 × 106. Then the cells were co-infected with baculovirus expressing CMKLR1, DNGαi1, and Gβ1γ2 at a multiplicity of infection (MOI) ratio of 1:1:1. After an additional 48 h for protein expression, cells were harvested by centrifugation and store at −80 °C until further use.
For purification of the chemerin-CMKLR1-Gi complex, cell pellets were lysed in a hypotonic buffer containing 20 mM HEPES (pH 7.4), 50 mM of NaCl, 5 mM of CaCl2, 5 mM of MgCl2, 25 mU/mL apyrase, 2.5 μg/ml leupeptin, 0.16 mg/ml benzamidine. The purified chemerin was added during protein purification for complex formation. After 30 min incubation, the cell membrane fraction was collected by centrifugation at 18,000 g for 15 min and homogenized in buffer containing 20 mM HEPES (pH 7.4), 100 mM of NaCl, 2 mM of CaCl2, 2 mM of MgCl2, 10% glycerol, 0.8% lauryl maltose neopentyl glycol (LMNG, Anatrace), 0.08% cholesteryl hemisuccinate (CHS, Anatrace), 25 mU/mL apyrase, 2.5 μg/ml leupeptin, 0.16 mg/ml benzamidine. The purified antibody scFv16 was also added to further stabilize the protein complex. The solubilized fraction was collected by centrifugation at 18,000 g for 30 min and loaded onto an anti-FLAG affinity resin gravity column (GenScript Biotech). After washing with 10 column volume of buffer containing 20 mM HEPES (pH 7.4), 100 mM of NaCl, 0.08% LMNG, 0.008% CHS, the protein complex was eluted by buffer containing 20 mM HEPES (pH 7.4), 100 mM of NaCl, 0.08% LMNG, 0.008% CHS, 0.2 mg/ml FLAG peptide. Then, the eluted fraction was concentrated by an Amicon® Ultra-15 Centrifugal Filter Unit (Millipore) and loaded on a Superdex 200 Increase 10/300 column (GE Healthcare Life Sciences) with running buffer containing 20 mM HEPES (pH 7.4), 100 mM of NaCl, 0.0015% LMNG, and 0.0005% GDN, 0.0003% CHS. The peak fractions corresponding to the protein complex were collected, concentrated, analyzed by SDS-PAGE and Western blotting, flash frozen in liquid nitrogen, and stored at −80 °C until further use.
Cryo-EM sample preparation and data collection
The cryo-EM samples were prepared by using a Vitrobot Mark IV (Thermo Fisher Scientific). For cryo-EM grid preparation, Ultrafoil 300 mesh R1.2/1.3 holy Au grids were glow discharged at Tergeo-EM plasma cleaner. Then, 3 μL of purified protein complex was loaded onto a grid at 4 °C in 100% humidity. After 3 s blotting with a blot force of 1 to remove extra liquid, the grid was quickly plunged into liquid ethane cooled by liquid nitrogen. Finally, the cryo-EM grids were stored in liquid nitrogen until further use.
The cryo-EM grids were screened, and the cryo-EM data was collected using a 300 kV Titan Krios Gi3 electron microscope (Thermo Fisher Scientific) equipped with a Gatan K3 direct electron detector. The cryo-EM data were collected at a nominal magnification of 105,000, and the pixel size is 0.85 Å. Inelastically scattered electrons were removed by a GIF Quantum energy filter (Gatan) with a slit width of 20 eV. The defocus range was set from −1.2 to −2.5 μm. A total exposure time of 2.5 s fractionated to 50 frames was set, and the dose rate was 21.3 e/pixel/s. A total of 6408 movie stacks were collected in semi-automatic data acquisition using SerialEM.
Cryo-EM data processing
CryoSPARC version v4.2.1 (Structura Biotechnology Inc.) was used for the overall datasets process of the chemerin-CMKLR1-Gi complex. The movie stacks were first subjected to motion correction and aligned with dose-weighting. Then, patch CTF estimations were applied to the aligned micrographs. The micrographs were manually inspected and obviously bad micrographs were discarded. A small dataset of particles was manually picked for the generation of initial two-dimensional (2D) templates for autopicking. A total of 2,682,313 particles were template-based picked and then subjected to 3 rounds of 2D classification. After that, the selected particles were used for ab initio reconstruction and heterogeneous refinements. Finally, 181,855 particles were used to generate the final map at a global resolution of 3.18 Å with a Fourier shell correlation of 0.143.
Model building and refinement
The model of CMKLR1-Gi complex (PDB: 7YKD) and chemerin-GPR1-Gi complex (PDB: 8XGM) was used as an initial model and docked into the corresponding EM density maps by UCSF Chimera-1.14. The structure model was manually rebuilt and adjusted by COOT-0.9.8. The chemical restraints were further refined by iterative real-space refinement in Phenix-1.18.2. The final model was validated by MolProbity, and the statistics are provided in Table 1. The illustrating graphic figures were prepared using UCSF Chimera-1.14, ChimeraX-1.274 and PyMOL-2.0.
Table 1.
Cryo-EM data collection, model refinement and validation statistics
| Data collection and processing | Chemerin-CMKLR1-Gi |
|---|---|
| Magnification | 105,000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 52.0 |
| Defocus range (μm) | –1.2 to −2.5 |
| Pixel size (Å) | 0.85 |
| Symmetry imposed | C1 |
| Initial particle projections (no.) | 2682313 |
| Final particle projections (no.) | 181855 |
| Refinement | |
| Map resolution (Å) | 3.18 |
| FSC threshold | 0.143 |
| Model composition | |
| Non-hydrogen atoms | 10123 |
| Protein residues | 1274 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.675 |
| Validation | |
| MolProbity score | 2.35 |
| Clashs core | 9.14 |
| Ramachandran plot | |
| Favored (%) | 94.65 |
| Allowed (%) | 5.35 |
| Outliers(%) | 0.00 |
G protein dissociation assay
A NanoBiT-based G protein dissociation assay was performed to test the activation of G proteins20. HEK293T cells (CRL-3216, ATCC) were seeded on a 24-well plate 24 h before transfection. A mixture of expression vectors including 100 ng pcDNA3.1-CMKLR1 (WT/mutants), 50 ng pcDNA3.1-Gαi1-LgBiT, 200 ng pcDNA3.1-Gβ1 and 200 ng pcDNA3.1-Gγ2-SmBiT per well of the 24-well plate was transfected into the cells. After 24 h transfection, the cells were collected and resuspended in HBSS with 20 mM HEPES. The cells were loaded onto a 384-well culture white plate (PerkinElmer Life Sciences, Waltham, MA) at a volume of 20 μL supplied with coelenterazine H at a final concentration of 10 μM (Yeasen Biotech, Shanghai, China). After 2 h of incubation at room temperature, the baseline signals were measured using an EnVision 2105 multimode plate reader (PerkinElmer). Then, cells were stimulated by full-length chemerin (Abcam, Cambridge, UK; ab256228) in different concentrations. The chemiluminescence signals were measured 15 mins after ligand addition. The fold changes of signals were further normalized to HBSS-treated negative control signals and the values were calculated based on three independent experiments, each with duplicate measurements.
cAMP assay
HeLa cells (CCL2, ATCC) were transiently transfected with CMKLR1 (WT/mutants) for 24 h. Cells were later collected in HBSS containing 5 mM HEPES, 0.1% BSA (w/v) and 0.5 mM 3-isobutyl-1-methylxanthine. Plated cells were then stimulated by different concentrations of chemerin and 2.5 μM forskolin for 30 mins in a cell incubator. Intracellular cAMP levels were measured with the LANCE Ultra cAMP kit (TRF0263, PerkinElmer) following the manufacturer’s instructions using an EnVision 2105 multimode plate reader (PerkinElmer).
Surface expression analysis
HEK293T cells were transfected with WT or mutant CMKLR1 expression vectors for 24 h. The cells were then harvested and washed in HBSS containing 5% BSA on ice. Then, the cells were incubated with a human chemR23 (CMKLR1) APC-conjugated antibody (FAB362A, R&D Systems, Minneapolis, MN) at a concentration of 10 μL/106 cells. The fluorescence signals associated with the antibody-receptor complex on the cell surface were measured by flow cytometry (CytoFLEX, Beckman Couter, Brea, CA).
Statistics and reproducibility
The data were analyzed using Prism 9.5.0 (GraphPad, San Diego, CA). Dose-response curves were plotted in the software using the log[agonist] vs. response equation with three parameters. For cAMP and G protein dissociation assays, data points were expressed as percentages (mean ± SEM) of the maximal response level for each sample, based on at least three independent experiments with duplicates, as detailed in the figure legends. EC50 values were derived from the dose-response curves. For cell surface expression, data were represented as percentages (mean ± SEM) of the flow cytometry fluorescence signals of WT CMKLR1. Statistical comparisons were performed using one-way Analysis of Variance (ANOVA), with a p-value of 0.05 or less considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Overall structure of the chemerin-CMKLR1-Gi complex
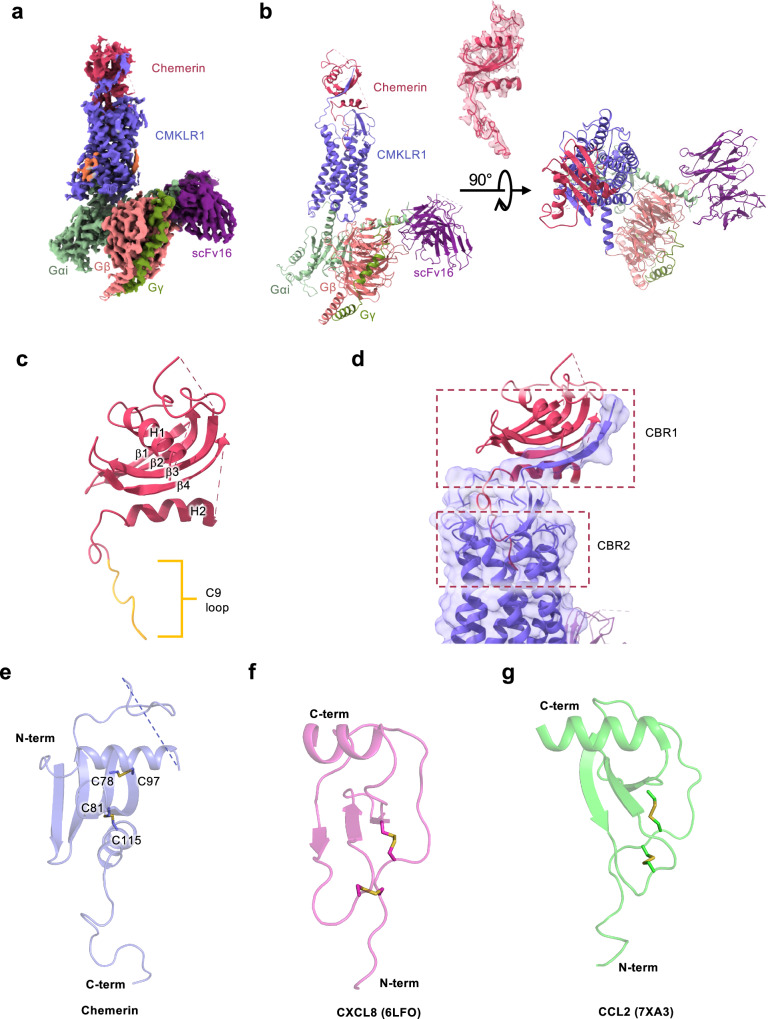
The human full-length mature chemerin (a.a. 21-157 after removal of signal peptide and the C-terminal 6 a.a.21,22; designated 1–137 in this study) was used to induce an active conformation of CMKLR1 that coupled to the Gi proteins and formed a signaling complex (Fig. S1a, S7). The chemerin-CMKLR1-Gi heterotrimer complex was purified for cryo-EM analysis (Fig. S1b–f), resulting in an EM map at a global resolution of 3.18 Å (Fig. 1a). A 3D model of the complex was built (Fig. 1b, Table 1). Most residues in the complex, including the 7 transmembrane (TM) helices, the extracellular loops and intracellular loops of CMKLR1, were unambiguously assigned with well-defined side chains (Fig. S2).
Fig. 1. Overall features of chemerin binding to the CMKLR1-Gi complex.
a Cryo-EM density of chemerin-CMKLR1-Gi complex. b Overall structure of chemerin-CMKLR1-Gi complex at 3.18 Å. c Molecular structure of the mature human chemerin. The C-terminal nonapeptide (C9) loop is highlight in yellow. d Schematic illustration of chemerin binding to CMKLR1. Chemerin is shown in magenta, and CMKLR1 is shown in purple. Chemerin binding region 1 (CBR1) and 2 (CBR2) are marked with dashed squares. e Structure of chemerin, with disulfide bonds shown as sticks. f Structure of CXCL8 (PDB ID: 6LFO), with disulfide bonds shown as sticks. g Structure of CCL2 (PDB ID: 7XA3), with disulfide bonds shown as sticks.
The structure of the CMKLR1-bound chemerin contains an overlying N-terminal α-helix (H1), a central four-stranded β-sheet body (β1-β4), a C-terminal α-helix (H2) and an unstructured C-terminal loop consisting of 17 amino acid residues (Fig. 1c). In the chemerin-CMKLR1-Gi complex, the chemerin is positioned above the orthosteric ligand-binding pocket similarly to a chemokine on top of its binding pocket of a chemokine receptor (Fig. 1d). One shared feature is that the N-terminal main body of chemerin interacts with the N-terminus and ECLs of CMKLR1 (chemerin binding region 1, CBR1, dotted box in Fig. 1d), and the unstructured C-terminal of chemerin (N-terminal instead for chemokines) interacts with the TM binding pocket forming CBR2 (lower box with dotted line, Fig. 1d). The architecture of the adipokine chemerin could be described as a “reverse” chemokine from the aspects of sequence and structure (Fig. S3). The C-terminus of chemerin instead of the N-terminus of a chemokine is deeply inserted into the orthosteric binding pocket of the receptor (Fig. 1d, S3). A slide-bolt helix is extended from the C-terminus and horizontally located above the pocket, which is a distinctive feature of chemerin not observed in the chemokine-chemokine receptor interactions (Fig. 1d, S3). The rest part of the chemerin forms a β-sheet core half-surrounding the N-terminal helix (Fig. 1c), which is topologically similar to a chemokine (Fig. S3). However, there is one more β-strand in the chemerin (β1-β4) than in a chemokine (β1-β3), which makes chemerin slightly heavier (15.9 kDa) than a typical chemokine (~12 kDa) and results in a more stable framework to contact the N-terminus of CMKLR1 with electrostatic interactions (Fig. 1c–f, S3).
The distinct interaction profile of chemerin
Similar to chemokines, chemerin has two intramolecular disulfide bonds: C78chemerin to C97chemerin and C81chemerin to C115chemerin (Fig. 1e). However, in chemerin the N-terminal 2 Cys residues (C78chemerin and C81chemerin) are separated by two amino acids, forming a CX2C sequence motif which is different from chemokines containing C-C, C-X-C and CX3C motifs23–25 (Fig. 1e–g). In chemerin, the first two Cys are located on the β3 strand, forming a disulfide bond with one cystine in the β4 strand (C97chemerin) and the other in the slide-bolt helix H2 (C115chemerin) (Fig. 1e). Considering the “reverse” topological architecture compared to chemokines and the widely accepted chemotactic function of chemerin, the structural information supports the notion that chemerin stands as an atypical chemokine.
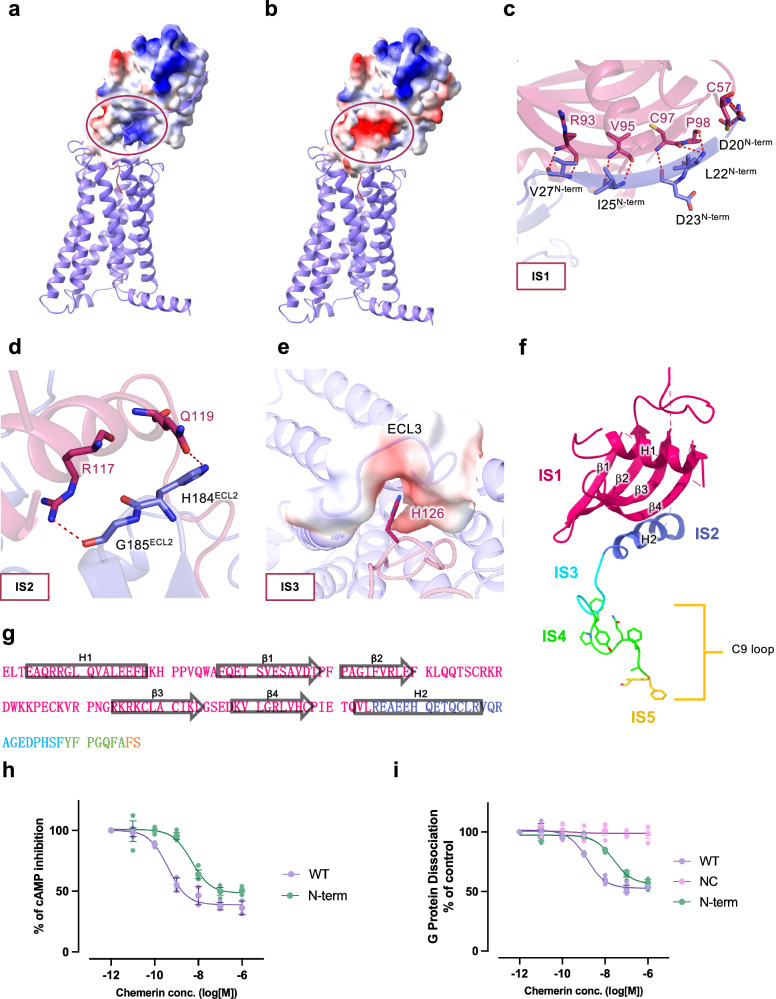
The two pairs of intramolecular disulfide binds play an important role in maintaining the overall structure of chemerin including the orientation of the C-terminal residues that is critical to receptor activation. In addition, C97chemerin forms a hydrogen bond with the side chain of D23N-term, that strengthens the interaction between full-length chemerin and CMKLR1 (Fig. 2c). This interaction is defined as “interaction site 1” or IS1, which involves positively charged groove (β4 strand in the main body of chemerin) for docking the negatively charged N-terminus of CMKLR1 (Fig. 2a, b). The IS1 gives rise to a β-sheet architecture accommodating the N-terminal β-strand of CMKLR1 through an anti-parallel β-sheet hydrogen bond network. Specifically, D20N-term forms 2 hydrogen bonds with C57chemerin, and L22N-term has 2 polar contacts with P98chemerin. D23N-term uses its backbone oxygen to form a hydrogen bond with the backbone nitrogen of C97chemerin, and I25N-term forms 2 hydrogen bonds with V95chemerin. The backbone of V27N-term has 2 hydrogen bonds with the backbone R93chemerin (Fig. 2d). This extensive hydrogen bond network between the two anti-parallel β-strands serves as an important dock for chemerin recruitment and stabilization by CMKLR1. However, as this interaction is mostly reserved by electrostatic charge and spatial shape fitness, it is expected that the N-terminus of CMKLR1 slide on the β4 strand of chemerin to precisely control the distance between chemerin and CMKLR1 during initial contact.
Fig. 2. Molecular interactions between the main body of chemerin and CMKLR1.
Chemerin is shown in magenta and CMKLR1 is shown in purple. a, b Electrostatic potential surface of (a) chemerin and (b) the N-terminus of CMKLR1. The circles mark the site where the N-terminal β-strand of CMKLR1 interacts with the β4-strand of chemerin (IS1). c Polar interactions at IS1. Hydrogen bonds between residues of chemerin and the N-terminal β-strand of CMKLR1 are shown in red dashes. d Polar interactions at IS2 of chemerin (magenta) with ECL2 of CMKLR1 (purple). Hydrogen bonds are shown in red dashes. e Electrostatic interactions at IS3 of chemerin with ECL3 of CMKLR1. The electrostatic potential surface of ECL3 and H126 of chemerin are shown. f, g Illustrations of chemerin interaction sites (f), and amino acid sequence of chemerin with IS1-5 highlighted in different colours (g). h cAMP inhibition assay in cells expressing wild-type and N-terminus truncated CMKLR1. i G protein dissociation assay in cells expressing wild-type and N-terminus truncated CMKLR1. Data are shown as mean ± SEM from n = 3 independent experiments.
Interaction site 2 (IS2) is the slide-bolt helix H2 of chemerin which is held up by the tip of the β-hairpin in ECL2 of CMKLR1 (Fig. 2d). The H2 helix of chemerin is located right above the orthosteric pocket of CMKLR1, forcing the C-terminus of chemerin to turn and insert into the TM binding pocket of CMKLR1. This interaction is highly important for the proper interaction of the C-terminal amino acids of chemerin with residues critical to receptor activation. The key residues in IS2 include R117chemerin, which interacts with G185ECL2, and Q119chemerin, which forms a hydrogen bond with the amide group on H184ECL2 (Fig. 2d). The allocation of IS2 further helps to define IS3, which involves an electrostatic interaction of H126chemerin with a crevice formed by ECL3 of CMKLR1 (Fig. 2e). The bulky residues F128chemerin, Y129chemerin, F130chemerin, and the turns made of P131chemerin and P125chemerin below and above H126chemerin further stabilize the lock to accommodate the C-terminus of chemerin in the orthosteric pocket (CBR2). Together, IS1-IS3 (Fig. 2f, g) are critical interaction sites of CBR1 and play a unique role in the interaction of full-length chemerin with CMKLR1. Next, to experimentally characterize the interaction between chemerin and the CBR1 of CMKLR1, we removed the first 27 amino acid residues from the N-terminus of CMKLR1. The truncated CMKLR1 mutant was then subjected to cAMP inhibition assay and NanoBiT G protein dissociation assay. Compared to the wild-type CMKLR1, the truncated receptor exhibited slightly reduced potency (~1 order of magnitude) upon chemerin stimulation in both assays (Fig. 2h, i). Altogether, our findings suggest that the CBR1 interactions between CMKLR1 and chemerin is responsible for stabilizing the chemerin-CMKLR1 complex, which allows the efficient insertion of the C-terminus of chemerin into the orthosteric binding pocket of CMKLR1 for receptor activation and downstream G protein signaling.
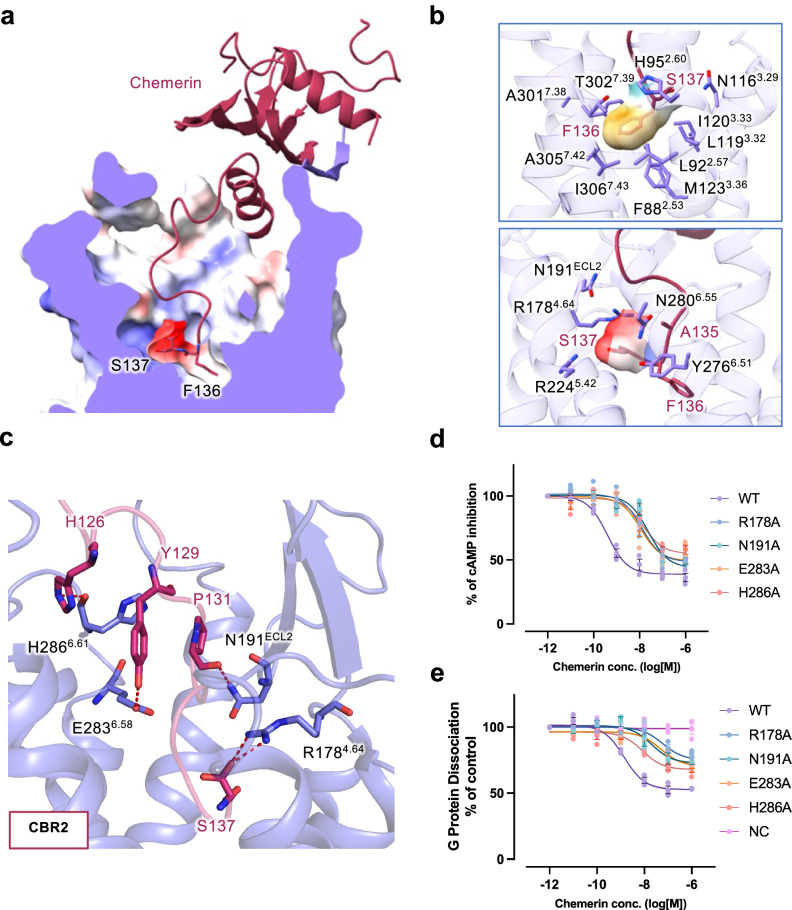
The C-terminal end of chemerin is both necessary and sufficient for receptor activation. Since the last two amino acids at the C-terminal end of chemerin contribute significantly to the agonistic activities of chemerin, we designate the conserved part from Y129chemerin to A135chemerin as IS4 and the last two amino acids, S137chemerin and F136chemerin as IS5 (Fig. 2f, g). The negatively charged S137chemerin loosely fills in a positively charged subpocket, and the hydrophobic bulky F136chemerin fits in the hydrophobic subpocket at the bottom of the orthosteric binding pocket of CMKLR1 (Fig. 3a, b). Interestingly, an extra density is found in the positively charged subpocket and a water molecule is placed for a hydrogen bond network between S137chemerin and the receptor (Fig. S3).
Fig. 3. Molecular interactions in the transmembrane pocket of CMKLR1.
a Sliced view of the transmembrane binding pocket of CMKLR1. The C-terminal two residues of chemerin (S137 and F136) are shown in sticks. Electrostatic potential surface is shown in the binding pocket. b Hydrophobic and negatively-charged pocket for the C-terminal two amino acids of chemerin. In the upper panel, F136 is surrounded by a hydrophobic binding pocket. In the lower panel, S137 fits in a negatively-charged binding pocket. c Polar interactions at the chemerin binding region 2 (CBR2) in the transmembrane binding pocket of CMKLR1. Hydrogen bonds are shown in red dashes. d cAMP inhibition assay in cells expressing wild-type and mutant CMKLR1. e NanoBiT-based G protein dissociation assay in cells expressing wild-type and mutant CMKLR1. Data are shown as mean ± SEM from n = 3 independent experiments.
The key residues for chemerin recognition by CMKLR1
In the CBR2 of CMKLR1, R1784.64 is critical for chemerin recognition by forming extensive hydrogen bonds with IS4 and IS5 of chemerin (Fig. 3c). Specifically, R1784.64 forms hydrogen bonds with S137chemerin. Additionally, N191ECL2 interacts with P131chemerin with a polar contact in IS4 of chemerin. A hydrogen bond is also observed between E2836.58 and Y129chemerin (Fig. 3c). These interactions are further transformed into receptor conformational changes towards activation.
Above the transmembrane binding pocket, the interactions between chemerin and CMKLR1 at CBR1 are maintained by anti-parallel β-sheet hydrogen bonds between the β4 strand of chemerin and the N-terminal β-strand of CMKLR1 (Fig. 2c). Specifically, D20N-term forms 2 hydrogen bonds with C57chemerin, and L22N-term has 2 polar contacts with P98chemerin. D23N-term uses its backbone oxygen to form a hydrogen bond with the backbone nitrogen of C97chemerin, and I25N-term forms 2 hydrogen bonds with V95chemerin. The backbone of V27N-term has 2 hydrogen bonds with the backbone R93chemerin (Fig. 2c). This extensive hydrogen bond network between the two anti-parallel β-strands serves as an important dock for chemerin recruitment and stabilization by CMKLR1. At IS2, the core body of chemerin is supported by ECL2 of CMKLR1. H184ECL2 at the tip of the hairpin poses polar interactions with Q119chemerin, and the adjacent G185ECL2 forms a hydrogen bond with R117chemerin (Fig. 2d). At IS3, ECL3 of CMKLR1 forms a negatively charged dock for H126chemerin that further stabilizes chemerin binding (Fig. 2e). This binding is strengthened by a polar contact between H2866.61 and H126chemerin (Fig. 3c).
To examine the interactions between chemerin and CMKLR1 in the TM binding pocket, we conducted site-directed mutagenesis experiments together with NanoBiT-based G protein dissociation assay and cAMP inhibition assay. Alanine substitution of receptor residues constituting the CBR2 results in an overall decrease in potency and efficacy compared with the wild-type CMKLR1 (Fig. 3d, e). Cell surface expression levels of the mutants are comparable to that of wild type CMKLR1 (Fig. S4). As evidenced by the dose-response curve from the cAMP inhibition assay (Fig. 3d), R178A, N191A, E283A and H286A mutants reduce the potency by around 2 orders of magnitudes. In the G protein dissociation assay (Fig. 3e), the mutants produced a decrease in potency of around 2 orders, and a maximum response of around 50% with respect to the wild-type control. Altogether, structural analysis and functional assays support our model of binding between chemerin and CMKLR1.
Mechanism of activation and Gi coupling of CMKLR1
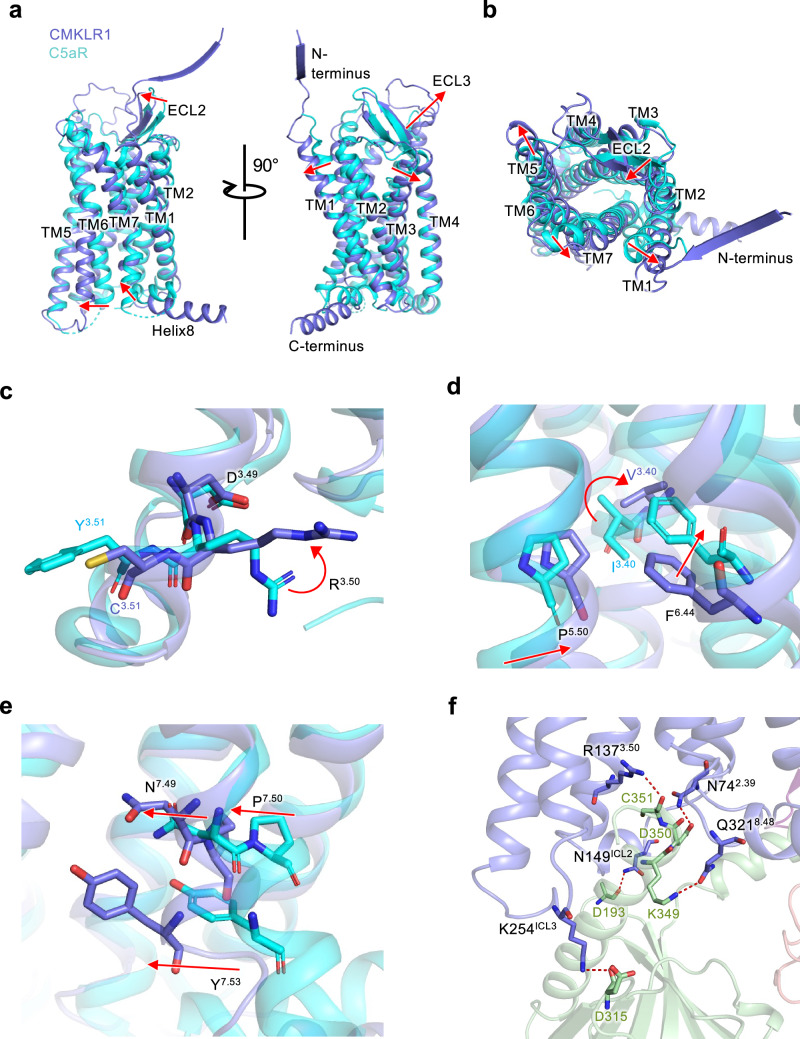
To investigate the mechanism of CMKLR1 activation, the C5aR1 structure in inactive state (PDB ID: 6C1R) is used as a reference (Fig. 4)26. From the superimposed structure, an outward movement of TM1, TM3, TM6, an inward movement of TM7, as well as an inward movement of ECL2 and an outward movement of ECL3 are observed (Fig. 4a). These changes fit the canonical conformational changes towards class A GPCR activation. To accommodate the binding of chemerin, the extracellular ends of TM1, TM5 and TM7 show an outward displacement (Fig. 4b).
Fig. 4. The activation mechanism of CMKLR1.
a Superimposed structures of active CMKLR1 (purple) and inactive C5aR1 (cyan, PDB ID 6C1R). Red arrows show the conformational changes of CMKLR1 in active state. b Extracellular view of (a). c Conformational changes at the DRY motif (DRC for CMKLR1). d Rotamer conformation changes at the P-I/V-F motif. e Side view of the conformational changes at the NPxxY motif. f The CMKLR1-Gi interface. Gαi is shown in green, and polar interactions are highlighted in red dashes.
We further examined the structural changes of the signaling motifs for CMKLR1 activation. D3.49-R3.50-Y3.51 motif is an “ionic lock” for GPCR activation. In CMKLR1, position 3.51 is substituted by a cysteine. R3.50 shows an upward movement compared with the inactive structure of C5aR1 (Fig. 4c). P5.50-I/V3.40-F6.44 is another important motif for receptor activation. Conformational changes are well-observed at these residues (Fig. 4d). In line with the inward displacement of TM5, P5.50 of CMKLR1 moves inwards. V3.40 shows an upward rotameric change, and F6.44 moves downwards, initiating the TM6 outward movement. In TM7, an inward displacement is observed, as is the N7.49P7.50xxY7.53 motif (Fig. 4e). These structural motifs support the active state conformation of CMKLR1.
Furthermore, we observed the interactions at the receptor-G protein interface. Multiple polar contacts secure the binding of Gαi to CMKLR1 (Fig. 4f). R1373.50 of the DRY motif interacts with C351 in the α5 helix of Gαi protein. N742.39 forms a hydrogen bond with D350 of Gαi protein. Q3218.48 on the Helix8 of CMKLR1 has a polar contact with K349 of Gαi protein. Apart from the α5 helix of the Gαi protein, which inserts into the receptor’s intracellular pocket, the Gαi protein actively engages the receptor’s intracellular loops. N149ICL2 interacts with D193 of Gαi protein, and K254ICL3 forms a hydrogen bond with D315 of Gαi protein. These interactions hold tight to the Gαi protein in favor of receptor activation.
Differences between the C9 and chemerin in binding to CMKLR1
To clarify the mechanism for signaling difference in CMKLR1 by different peptide derivatives of chemerin, we compared the structures of the chemerin-bound and C9-bound CMKLR1-Gi complexes18. Although the overall conformations are similar, differences lie in molecular details in the ligand binding mode (Fig. 5a).
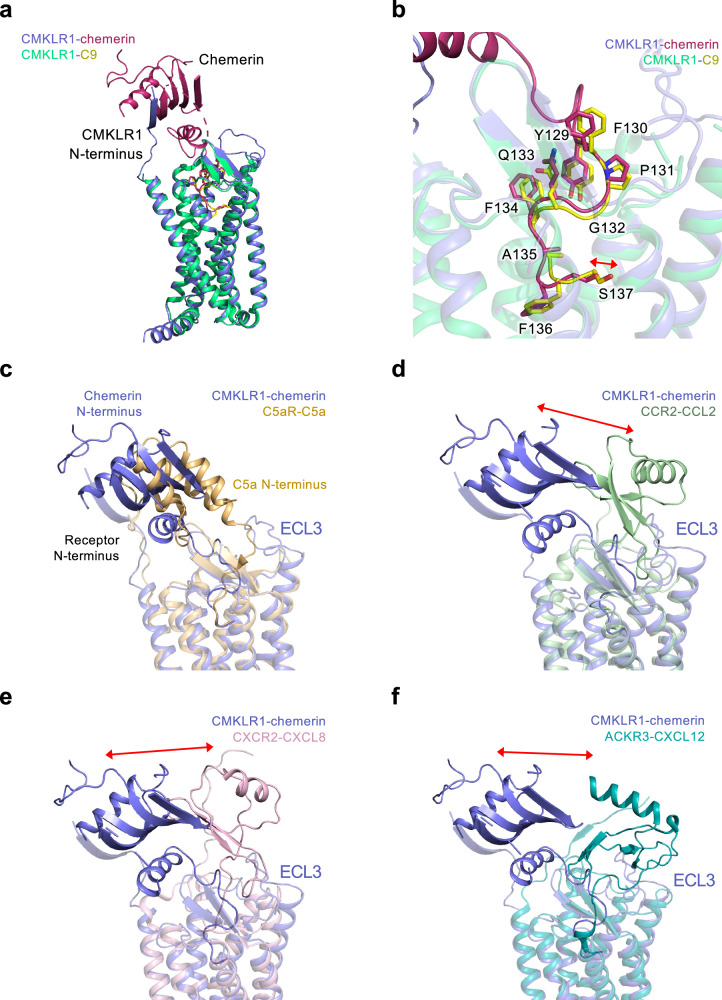
Fig. 5. Comparison of the binding modes of chemerin.
a Superimposed structure of CMKLR1-chemerin and CMKLR1-C9 complexes. b Binding poses of chemerin (magenta) and C9 (yellow) in the transmembrane binding pocket of CMKLR1. c Superimposed structure of CMKLR1-chemerin (purple) and C5aR1-C5a (wheat yellow). d Superimposed structure of CMKLR1-chemerin (purple) and CCR2-CCL2 (light green). Red arrow indicates the displacement between chemerin and the chemokine. e Superimposed structure of CMKLR1-chemerin (purple) and CXCR2-CXCL8 (light pink). Red arrow indicates the displacement between chemerin and the chemokine. f Superimposed structure of CMKLR1-chemerin (purple) and ACKR3-CXCL12 (dark cyan). Red arrow indicates the displacement between chemerin and the chemokine.
In CBR2, the conformations of chemerin and C9 are conserved in IS4, but different in IS5. A subtle shift is observed with the C-terminal two residues of chemerin and C9. Chemerin occludes the space closer to TM7 in the binding pocket. As a result, F136chemerin in chemerin-bound CMKLR1 structure inserts deeper into the local hydrophobic pocket, and S137chemerin is pointing upwards the aforementioned polar subpocket. A water-like density is found in the larger charged local pocket for S137chemerin, and this water molecule mediates the polar interaction between S137chemerin and the receptor. The differences observed in CBR2 have probably resulted from the interaction between chemerin and CMKLR1 in CBR1, which are absent in the C9-bound complex.
In the Gi protein interface, the binding poses of the α5 helix of Gαi protein coupled to CMKLR1 in the intracellular binding pocket between chemerin-bound and C9-bound CMKLR1 complex are generally similar, but with slight differences in the interaction (Fig. S5). Notably, the αN helix of Gαi bends downwards by ~10°. This downward movement into the intracellular side further reduces the chance of interactions between Gαi and the intracellular loop 2 of CMKLR1. Therefore, the chemerin-bound CMKLR1 shows a loose coupling to G protein compared with the C9-bound CMKLR1, which suggests that the chemerin-bound complex is more prone to activation and downstream signal transduction.
It was found that different chemerin C-terminal peptides could activate CMKLR1 and induce distinct signaling outcomes, which affects the progress of inflammation27,28. The C15 peptide (from A121 to A135), which lacks the last two amino acids of chemerin, is different from C9 in that it has potent anti-inflammatory effects through CMKLR129,30. The conserved “S”-shaped IS4 of chemerin may serve as a “hook” to anchor to CMKLR1 (Fig. 5b), and the molecular interaction between CBR1 and IS5 in CBR2 may retain the potency and regulate CMKLR1-mediated biased signaling towards pro-inflammatory or anti-inflammatory response.
Features of the “reverse chemokine” binding of chemerin
As a “reverse chemokine” using its C-terminal to insert into the transmembrane binding pocket of CMKLR1, chemerin shares a certain level of distinct features with chemokines binding to chemokine receptors. Therefore, we juxtaposed the chemerin-CMKLR1 binding with the interactions of its homologous receptor C5aR1 and C5a (PDB ID: 6C1R)26, C-C chemokine receptor CCR2 and CCL2 (PDB ID: 7XA3)31, CXC chemokine receptor CXCR2 and CXCL8 (PDB ID: 6LFO)32 as well as the atypical chemokine receptor ACKR3 and CXCL12 (PDB ID: 7SK3)33, respectively (Fig. 5c–f).
When compared among these receptors, the 7TM and ECL2 β-hairpin of CMKLR1 align well for a conserved architecture of chemotactic GPCR. The coupling of chemerin to CMKLR1 involves the N-terminus and all 3 ECLs of CMKLR1 (Fig. S6). Interestingly, while chemerin and C5a have similar orientations of hydrophobic cores when bound to their corresponding receptors, chemerin and chemokines dock their hydrophobic cores to opposite positions (Fig. 5c–f). The hydrophobic core of chemerin favors the N-terminus/TM1 side of the receptor, and the hydrophobic cores of chemokines favor the ECL3/TM5 side of the receptor. Extensive β-sheet interaction between the N-terminus of CMKLR1 and the β4 strand of chemerin is responsible for this opposite orientation. Consequently, the chemerin is close to ECL2 of CMKLR1 with the upright ECL2 fitting in the slide-bolt helix of chemerin like a trigger (Fig. 2d). In addition, CMKLR1 has a long ECL2 that forms an additional loop from L195ECL2 to M209ECL2, which is not observed in other chemotactic GPCRs like C5aR, FPR2, and chemokine receptors. The ECL2 conformation of CMKLR1 is consistent with the up-moved chemerin but has steric conflicts with the position of CXCL12 in the AKCR3-CXCL12 structure. Chemerin and C5a both use their C-termini to activate the receptor. The binding poses of chemerin and C5a are similar in the TM pockets of their receptors, with their C-termini pointing towards TM4 and TM5 of the receptor (Fig. 5c). Similar binding pose in the TM pocket is also observed for CCR2-CCL2 interaction and ACKR3-CXCL12 interaction (Fig. 5d, f). However, the insertion of the N-terminus of CXCL8 is shallower, and pointing towards TM1 (Fig. 5e). Taken together, chemerin, as a distinct “reverse chemokine”, docks its hydrophobic core to the N-terminus/TM1 side of CMKLR1, rather than the opposite side of ECL3/TM5 in chemokine receptors. The special coupling feature of chemerin to CMKLR1 facilitates the regulatory role of chemerin in the function of CMKLR1, which would be critical for the collaborative regulation of inflammation by the chemerin-CMKLR1 signal axis.
Discussion
The adipokine chemerin and its receptor CMKLR1 are attractive targets for the regulation of inflammation, and successful therapeutic interventions would benefit from a deep understanding of the molecular mechanism of the chemerin-CMKLR1 axis. Under physiological conditions, only the C-terminus of chemerin is cleaved by different proteases that yield a variety of chemerin derivatives. It is therefore speculated that the conserved main body of chemerin in the N-terminus has a critical physiological role through its interaction with CBR1 of CMKLR1 in our structural model. Interestingly, most reported anti-inflammatory ligands do not use the IS5 for binding to CMKLR114,15,27,30,34. Furthermore, although the phenotype of inflammation induced by C9-CMKLR1 is controversial, the chemically modified C9 which reaches a longer physical half-life could be developed as a CMKLR1-targeting tracer in the clinic for its steady binding with CMKLR15. It is speculated that, in a full-length chemerin, the main body of chemerin is indispensable for the C-terminal nonapeptide to elicit an effective physiological function, although C9 has already been developed as an efficient experimental tool for CMKLR15,28.
The C-terminal peptide C15, which lacks the last two amino acids (IS5), shows potent anti-inflammatory effects. It was speculated that C15 blocks the transmembrane pocket of the receptor and may act as an antagonist30,35. Additionally, several other antagonists of CMKLR1 show anti-inflammatory effects36,37. However, it is confusing that agonists of CMKLR1 which do not use IS5 also hold potent anti-informatory effects, such as Aβ42 and antibodies against CMKLR117,38. As for C15, it indeed transduces signals through CMKLR1 the anti-inflammatory prophagocytic activities of C15 through CMKLR1 could be completely abrogated by pharmacological inhibition of Syk activity35. Therefore, it is more reasonable to propose that CMKLR1 activate different downstream signaling pathways when induced by different ligands and produce various biological consequences. Our structural model supports this hypothesis with an obvious signaling bias between C9-CMKLR1 and chemerin-CMKLR1. As chemerin is the only identified native ligand for CMKLR1, a thorough structural framework is informative for further research and therapeutic development.
In addition to C9 and C15, peptides that activate CMKLR1 includes the β-amyloid Aβ42 which selectively activate signaling pathways downstream of CMKLR138. Aβ42 induces chemotaxis of transfected cells expressing CMKLR1, but fails to activate signaling pathways leading to calcium mobilization38. These observations indicate that Aβ42 is a biased agonist of CMKLR1 which lacks the full agonistic activity of CMKLR1. Of note, Aβ42 is able to induce β-arrestin membrane translocation39,40, an observation that further supports its role as a biased agonist of CMKLR1. Although the protein structure of an Aβ42-bound CMKLR1 is not available currently, we predict based on our full-length mature chemerin-bound CMKLR1 structure that Aβ42 does not use the CBR2 for receptor activation. Proper interaction of CMKLR1 agonists at CBR2 is, therefore, a determinant for full agonism. Previous studies22,29,30,34,35,41,42 examined the potency of chemerin-derived peptides in activating CMKLR1, without special attention to biased agonism of these peptides. It is therefore worthwhile to re-examine these peptides for this function, especially peptides with modified C-terminal ends. In evaluating the biological consequence of chemerin binding, a number of functional assays are at our disposal. Proper use of these tools will allow us to distinguish between G protein activation and β-arrestin signaling. For G protein activation, the divergent signaling mechanism through Gα and Gβγ subunits can also be determined.
In our structural model, chemerin binds to CMKLR1 in a chemokine-like two-site model. Therefore, it is possible to consider the chemotactic adipokine chemerin as an atypical chemokine given the presence of intramolecular disulfide bonds, the structural fold, and the binding mode to the receptor43. However, chemerin interacts with CMKLR1 in a “reverse” topology, using its C-terminus to activate the receptor as opposed to the N-terminus of chemokines. In this regard, chemerin-CMKLR1 interaction also has similarities to C5a-C5aR1 interaction44, although the ligands are generated through different processes and the biological consequences also vary. From an evolutionary point of view, chemokines and chemokine receptors are in a league of their own, but at its edge there are atypical chemokine receptors that also bind chemokines but fail to signal or only activate partially. CCRL2 is an atypical chemokine receptor that binds chemerin but does not generate transmembrane signals4,27,42. Future understanding of the structural basis of CCRL2 binding to chemerin will help to elucidate the relationship between chemerin and chemokines in terms of receptor binding.
In summary, the present study provides a structural basis for CMKLR1 binding of full-length mature chemerin, which represents an active state of the receptor with full agonism. Our structural analysis indicates the presence of multiple chemerin-binding regions and critical interaction sites. These interaction sites have similarities and differences when compared with chemokine interaction with chemokine receptors. Understand the molecular details of chemerin-CMKLR1 interaction is expected to help future development of modulatory ligands that explore the biased signaling properties of CMKLR1.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the National Natural Science Foundation of China 32070950 (R.D.Y.) 82402140 (A.L.) and 82302448 (J.W.), Shenzhen Science and Technology Program Grant No. RCBS20221008093330067 (A.L.), China Postdoctoral Science Foundation 2022M713049 (A.L.) and start-up funds from the Dongguan Songshan Lake Central Hospital (A.L.). This work was also supported in part by grants from the Science, Technology and Innovation Commission of Shenzhen Municipality GXWD20201231105722002-20200831175432002 (R.D.Y.), the Ganghong Young Scholar Development Fund (R.D.Y.), the fund from Kobilka Institute of Innovative Drug Discovery at The Chinese University of Hong Kong, Shenzhen (R.D.Y.), and the fund from Shenzhen-Hong Kong Cooperation Zone for Technology and Innovation (HZQB-KCZYB-2020056).
Author contributions
A.L. and R.D.Y. designed and supervised the research; A.L. Y.L., and J.W. performed the experiment and analysis the data; A.L., Y.L., and R.D.Y. discussed the results and wrote the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Janesh Kumar and Laura Rodríguez Pérez.
Data availability
The atomic coordinates and associated EM map for the chemerin-CMKLR1-Gi complex have been deposited in the Protein Data Bank and Electron Microscopy Data Bank with accession codes 8ZJG and EMD-60144, respectively. All data needed to evaluate the conclusions in the paper are included in the article and/or Supplementary Data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Aijun Liu, Yezhou Liu.
Contributor Information
Aijun Liu, Email: liuaijun@cuhk.edu.cn.
Richard D. Ye, Email: richardye@cuhk.edu.cn
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07228-9.
References
- 1.Klose, R. et al. Targeting VEGF-A in myeloid cells enhances natural killer cell responses to chemotherapy and ameliorates cachexia. Nat. Commun.7, 12528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin, Y. et al. The chemerin-CMKLR1 axis limits thermogenesis by controlling a beige adipocyte/IL-33/type 2 innate immunity circuit. Sci Immunol6, eabg9698 (2021). [DOI] [PubMed]
- 3.Ge, X. et al. Prochemerin cleavage by factor XIa links coagulation and inflammation. Blood131, 353–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy, A. J. & Davenport, A. P. International union of basic and clinical pharmacology CIII: chemerin receptors CMKLR1 (Chemerin1) and GPR1 (Chemerin2) nomenclature, pharmacology, and function. Pharm. Rev.70, 174–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mannes, P. Z. et al. Molecular imaging of chemokine-like receptor 1 (CMKLR1) in experimental acute lung injury. Proc. Natl Acad. Sci. USA120, e2216458120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rennier, K., Shin, W. J., Krug, E., Virdi, G. & Pachynski, R. K. Chemerin reactivates PTEN and suppresses PD-L1 in tumor cells via modulation of a novel CMKLR1-mediated signaling cascade. Clin. Cancer Res26, 5019–5035 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Tan, S. K. et al. Obesity-dependent adipokine chemerin suppresses fatty acid oxidation to confer ferroptosis resistance. Cancer Discov.11, 2072–2093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reznik, E., Jiang, H. & Hakimi, A. A. Chemerin tips the scales in ccRCC to evade ferroptosis. Cancer Discov.11, 1879–1880 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Fasshauer, M., Bluher, M. & Stumvoll, M. Adipokines in gestational diabetes. Lancet Diabetes Endocrinol.2, 488–499 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Eichelmann, F. et al. Chemerin as a biomarker linking inflammation and cardiovascular diseases. J. Am. Coll. Cardiol.73, 378–379 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Erdmann, S. et al. CMKLR1-targeting peptide tracers for PET/MR imaging of breast cancer. Theranostics9, 6719–6733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnea, G. et al. The genetic design of signaling cascades to record receptor activation. Proc. Natl Acad. Sci. USA105, 64–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, X. et al. Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist. PLoS Biol.21, e3002188 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su, X., Cheng, Y., Zhang, G. & Wang, B. Chemerin in inflammatory diseases. Clin. Chim. Acta517, 41–47 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Mariani, F. & Roncucci, L. Chemerin/chemR23 axis in inflammation onset and resolution. Inflamm. Res64, 85–95 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Arita, M. et al. Resolvin E1 selectively interacts with leukotriene B4 receptor BLT1 and ChemR23 to regulate inflammation. J. Immunol.178, 3912–3917 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Trilleaud, C. et al. Agonist anti-ChemR23 mAb reduces tissue neutrophil accumulation and triggers chronic inflammation resolution. Sci. Adv .7, eabd1453 (2021). [DOI] [PMC free article] [PubMed]
- 18.Wang, J. et al. Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin. Proc. Natl Acad. Sci. USA120, e2214324120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, A., Liu, Y., Zhang, W. & Ye, R. D. Structural insights into ligand recognition and activation of the succinate receptor SUCNR1. Cell Rep.43, 114381 (2024). [DOI] [PubMed] [Google Scholar]
- 20.Inoue, A. et al. Illuminating G-protein-coupling selectivity of GPCRs. Cell177, 1933–1947 e1925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittamer, V. et al. Specific recruitment of antigen-presenting cells by chemerin, a novel processed ligand from human inflammatory fluids. J. Exp. Med198, 977–985 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittamer, V. et al. The C-terminal nonapeptide of mature chemerin activates the chemerin receptor with low nanomolar potency. J. Biol. Chem.279, 9956–9962 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Wedemeyer, M. J. et al. The chemokine X-factor: Structure-function analysis of the CXC motif at CXCR4 and ACKR3. J. Biol. Chem.295, 13927–13939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, C. E. & Nibbs, R. J. B. A guide to chemokines and their receptors. FEBS J.285, 2944–2971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, M. C. & Mayo, K. H. Chemokines from a Structural Perspective. Int. J. Mol. Sci.18, 2088 (2017). [DOI] [PMC free article] [PubMed]
- 26.Liu, H. et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol.25, 472–481 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Bondue, B., Wittamer, V. & Parmentier, M. Chemerin and its receptors in leukocyte trafficking, inflammation and metabolism. Cytokine Growth Factor Rev.22, 331–338 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Zhou, J. X. et al. Chemerin C9 peptide induces receptor internalization through a clathrin-independent pathway. Acta Pharm. Sin.35, 653–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimamura, K. et al. Identification of a stable chemerin analog with potent activity toward ChemR23. Peptides30, 1529–1538 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Cash, J. L. et al. Synthetic chemerin-derived peptides suppress inflammation through ChemR23. J. Exp. Med.205, 767–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao, Z. et al. Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3. Cell Discov.8, 44 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, K. et al. Structural basis of CXC chemokine receptor 2 activation and signalling. Nature585, 135–140 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Yen, Y. C. et al. Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias. Sci. Adv.8, eabn8063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue, G. et al. The role of Chemerin in human diseases. Cytokine162, 156089 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Cash, J. L., Christian, A. R. & Greaves, D. R. Chemerin peptides promote phagocytosis in a ChemR23- and Syk-dependent manner. J. Immunol.184, 5315–5324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy, A. J. et al. Chemerin elicits potent constrictor actions via chemokine-like receptor 1 (CMKLR1), not G-protein-coupled receptor 1 (GPR1), in human and rat vasculature. J. Am. Heart Assoc.5, e004421 (2016). [DOI] [PMC free article] [PubMed]
- 37.Graham, K. L. et al. A novel CMKLR1 small molecule antagonist suppresses CNS autoimmune inflammatory disease. PLoS ONE9, e112925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng, L. et al. The chemerin receptor CMKLR1 is a functional receptor for amyloid-beta peptide. J. Alzheimers Dis.43, 227–242 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Yu, Y. & Ye, R. D. Microglial Abeta receptors in Alzheimer’s disease. Cell Mol. Neurobiol.35, 71–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haque, M. E., Kim, I. S., Jakaria, M., Akther, M. & Choi, D. K. Importance of GPCR-mediated microglial activation in Alzheimer’s disease. Front. Cell Neurosci.12, 258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, Y., Du, X. Y., Zhao, L., Morser, J. & Leung, L. L. Proteolytic cleavage of chemerin protein is necessary for activation to the active form, Chem157S, which functions as a signaling molecule in glioblastoma. J. Biol. Chem.286, 39510–39519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Henau, O. et al. Signaling properties of chemerin receptors CMKLR1, GPR1 and CCRL2. PLoS ONE11, e0164179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urvas, L. & Kellenberger, E. Structural insights into molecular recognition and receptor activation in chemokine-chemokine receptor complexes. J. Med Chem.66, 7070–7085 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Feng, Y. et al. Mechanism of activation and biased signaling in complement receptor C5aR1. Cell Res.33, 312–324 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The atomic coordinates and associated EM map for the chemerin-CMKLR1-Gi complex have been deposited in the Protein Data Bank and Electron Microscopy Data Bank with accession codes 8ZJG and EMD-60144, respectively. All data needed to evaluate the conclusions in the paper are included in the article and/or Supplementary Data.