Abstract
Background: Hepatocellular carcinoma (HCC) is a highly heterogeneous tumor and a primary cause of cancer-relevant deaths worldwide. The role of reactive oxygen species (ROS) in HCC development is less studied. Methods: Seurat package and CellMarker database were employed for single-cell RNA sequencing (scRNA-seq) analysis based on the GSE189175 dataset from Gene Expression Omnibus (GEO). DAVID and MsigDB database were utilized for pathway analysis. SCENIC analysis was performed to map a transcription factors (TFs) regulatory network. CellChat was used for cellular communication analysis. Results: Six major cell subpopulations were identified, among which hepatocytes accounted for the highest proportion in both cancer and adjacent tissues. The enrichment scores of the 50 hallmark gene sets showed that the ROS pathway was abnormally activated in HCC hepatocytes and positively correlated with energy metabolism-related pathways (glucose metabolism, lipid metabolism, amino acid metabolism, etc.). Then, the hepatocytes were divided into four subgroups. Noticeably, GPX4+ hepatocytes with the highest activity of the ROS pathway was related to a worse prognosis of HCC. Mechanism analysis revealed that JUND was involved in the positive regulation of the ROS pathway in GPX4+ hepatocytes. It was found that the interdependent ligand-receptor interaction between GPX4+ liver cells and immune cells facilitated the malignant development of HCC. Conclusion: ROS pathway was over-activated in the hepatocytes of HCC tissues. GPX4+ hepatocytes having the highest activity of the ROS pathway closely interacted with T cells and M2 macrophage cells. Molecular subtypes and risk score signature based on the ROS pathway and its potential target gene JUND are encouraged to be developed for improving the precision treatment of HCC.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81481-0.
Keywords: Single-cell transcriptomics, Hepatocellular carcinoma, Reactive oxygen species pathway, GPX4+ HCC hepatocytes, CellChat
Subject terms: Cancer, Oncology
Introduction
Hepatocellular carcinoma (HCC) is the most frequently detected primary liver cancer that accounts for about 90% of all liver cancer cases1,2, ranking the third highest cause for cancer-relevant deaths worldwide3. Such a high death rate is related to a late diagnosis and a high occurrence of metastasis in HCC. At present, liver transplantation, hepatectomy and ablation are the main treatments for HCC4, but 70% of patients with resectable HCC will develop intra- or extra-hepatic recurrent metastasis after surgery5,6. Moreover, a highly heterogeneous nature of HCC also significantly influences the recurrence and drug resistance in patients with HCC7. Hence, exploring the features of HCC tumor heterogeneity is instrumental for improving the clinical therapies and survival for HCC.
Reactive oxygen species (ROS) are produced by eukaryotic cells via aerobic metabolism as a by-product in mitochondria8,9. Currently, researchers have increasingly focused on the role of ROS pathways in cancer development. Tumor cells produce more ROS than normal cells10. The production of ROS is affected by a variety of carcinogenic events including activation of neoplastic genes, deprivation of tumor inhibitory function, altered mitochondrial activity, enhanced hypoxia and changes in stromal interactions11. ROS is involved in tumor-promoting processes through modulating inflammation, assisting angiogenesis and migration12 and also plays a crucial role in drug-resistant cancer cells13. Study have widely explored different chemical drugs and Chinese herbs to trigger ROS-mediated cell death in cancers14. Due to a close relationship between ROS and malignant tumors, it is imperative to analyze the potential molecular mechanisms for improving HCC treatment.
This study was the first research that examined the ROS pathway in HCC progression. It was found that hepatocytes had abnormally increased ROS pathway activity in human HCC tissues. GPX4+ liver cell subpopulation with the highest ROS pathway activity had an inhibitory effect on immune cells and was associated with a worse prognosis. Mechanism analysis revealed that JUND regulated the ROS pathway in GPX4+ hepatocytes. These findings indicated that ROS facilitated HCC development through inhibiting immune activity, and that JUND was a target for treating HCC patients. To the best of our knowledge, this was the first single-cell RNA study based on the ROS pathway for HCC management.
Materials and methods
Acquisition and pre-processing of obtained datasets
Single-cell RNA sequencing (scRNA-seq) expression dataset GSE18917515, which included 3 paired tumor and adjacent samples, were collected from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The data were pre-processed in the SangerBox website (http://sangerbox.com/home.html)16. Specifically, the Read10X function of the Seurat (v3.1.2) package17 was used to read the offline data. The proportion of mitochondrial genes within the cell was not quantified as there were no mitochondrial genes. Cells expressing genes in the range between 200 and 10,000 were retained and then the data were standardized applying the SCTransform function. After reducing the dimensionality by principal component analysis (PCA), Harmony packages18 was used to remove the batch effects (max. iter. harmony = 50, lambda = 0.5, assay. use="SCT”). Next, we performed Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction based on the top 30 PCs, followed by using FindNeighbors and FindClusters functions to cluster the cells (for all cells, resolution = 0.05; for hepatocytes, resolution = 0.1). Cell types were annotated based on the specifically high-expressed marker genes in the CellMarker database. The FindAllMarkers function was employed identify the differentially expressed genes (DEGs) among each cell type (only.pos = T, min.pct = 0.25, logfc.threshold = 0.25).
Another RNA-seq dataset of HCC with the expression value in the form of log2 (fpkm + 1) were collected from The Cancer Genome Atlas (TCGA) database. After excluding samples with a total survival time shorter than 90 days, there remained 327 samples in this dataset.
Enrichment analysis
Gene sets of interest were uploaded to the DAVID database (https://david.ncifcrf.gov/) for biological process analysis.
Enrichment score analysis
The h.all.v2023.1.Hs.symbols.gmt data file was retrieved from the MsigDB database. The keggGet function19 in the KEGGREST package was used to download the list of genes contained in the pathways such as Carbohydrate metabolism, Lipid metabolism, Amino acid metabolism, and Energy metabolism. Finally, the enrichment scores of the hallmark genes related to liver cells and energy metabolism were calculated using the AUCell package20 and single-sample gene set enrichment analysis (ssGSEA).
SCENIC analysis
The transcription status of cells is determined by the gene regulatory network (GRN) in which hub transcription factors (TFs) and cofactors regulate each other and their downstream target genes. SCENIC21 is an algorithm developed specifically for single-cell data analysis by introducing a gene co-expression network regulated by TFs to map highly reliable GRNs comprising the TFs. We used the GENIE3 method to compute potential target genes for each TF and utilized the top5perTarget to map a TF regulatory network. Finally, highly reliable relationship pairs of TF and target gene were identified, and the AUCell function was used to calculate the degree of regulon activity (TF + target genes) within each cell.
Cellular communication analysis
CellChat is a tool for quantitative inference, visualization, and analysis of intercellular communication networks. CellChat can distinguish major characteristics of intercellular communication in given scRNA-seq data and infer the optential functions of poorly understood signaling pathways. In this study, CellChat package22 was applied to calculate the possibility of ligand receptor interactions (cell-cell contact and secreted signaling) between cell subpopulations.
Statistical analysis
Statistical analysis was carried out in the R software (version 3.4.0). Survival analysis was performed based on Kaplan–Meier plot with log-rank tests. The Wilcoxon rank-sum test was used to compare two-group differences, while the Kruskal Wallis test was employed to compare the differences among three or more groups. A p-value < 0.05 was considered statically significant.
Results
Single-cell atlas of HCC
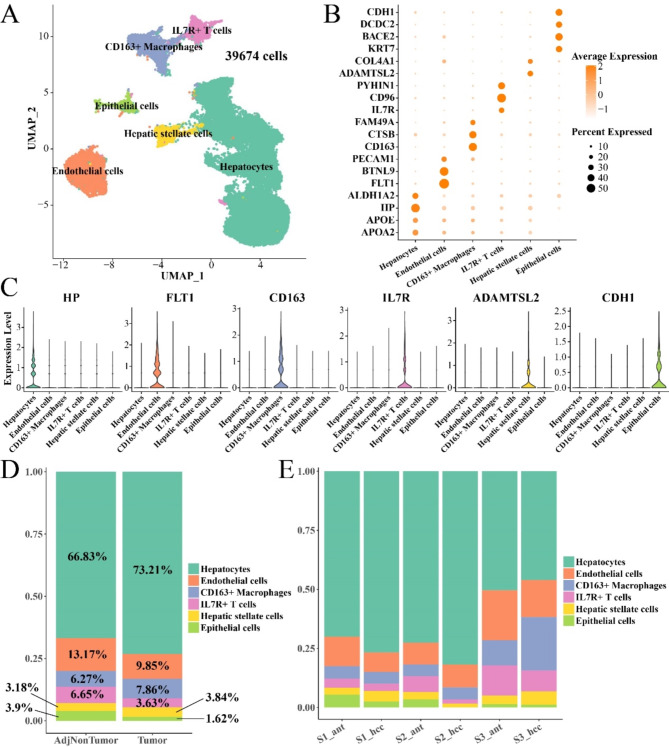
Six main cell subpopulations were obtained by conducting cell annotation analysis in Seurat package (Fig. 1A). The marker genes for hepatocytes included HP, APOE, and APOA223 and hepatocytes also high-expressed ALDH1A224, a marker gene in cancer stem cells, suggesting that hepatocytes were in a proliferative state similar to stem cells. The marker genes of endothelial cells included FLT1, BTNL9, and PECAM123,24. Macrophages high-expressed CD16325, a marker gene of M2 type macrophages, indicating a higher proportion of M2-type macrophages in HCC. T cells expressed high levels of IL7R, CD96, and PYHIN126. Hepatic stellate cells high-expressed ADAMTSL2 and COL4A127. Epithelial cells expressed high levels of KRT7, BACE2, DCDC2, and CDH128,29 (Fig. 1B, C, Supplementary Table 1). Analysis of cell subpopulations revealed a higher proportion of hepatocytes in the cancer group but lower proportions of endothelial cells, IL7R + T cells, and epithelial cells (Fig. 1D). Difference in the proportion of cell subpopulations between adjacent and cancerous areas within each patient was also similar (Fig. 1E). Detailed single-cell data of the samples in GSE189175 were shown in Supplementary Figure S1.
Fig. 1.
Detailed characterization of cell types in HCC. (A) UMAP plot of 39,674 single cells from 6 samples, showing the formation of 6 main clusters. Each dot represents a single cell and is colored based on the cell cluster. (B-C) Bubble and violin plots of marker genes in cell subpopulations. (D) The proportion of cell subpopulations between cancer and adjacent groups. (E) Proportion of cell subpopulations within the sample.
Abnormally activated ROS pathway in HCC hepatocytes
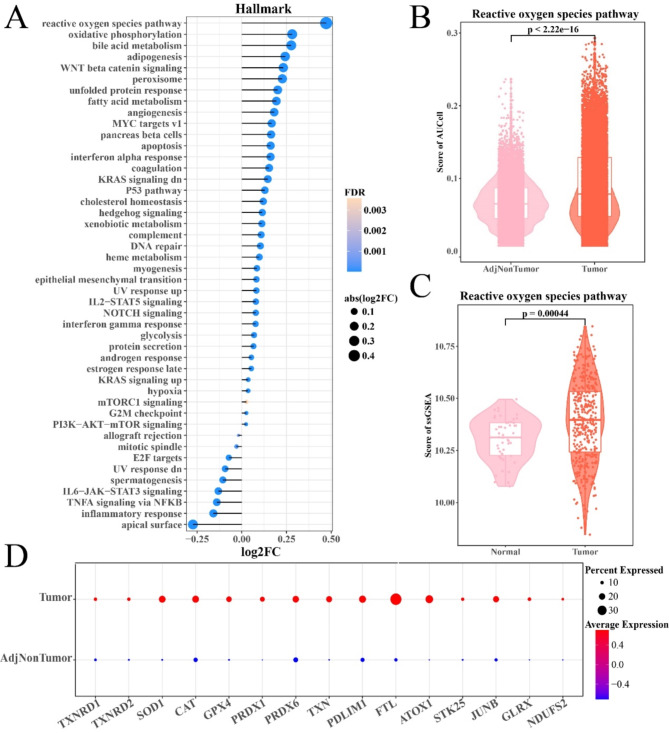
To explore the differential enrichment pathways of hepatocytes between tumors and para-cancerous tissue, we applied the AUCell algorithm to calculate the enrichment scores of 50 hallmark gene sets in each hepatocyte. It was found that hepatocytes of HCC tissues exhibited higher activities of ROS pathway, oxidative phosphorylation, bile acid metabolism, adipogenesis, and Wnt/β-catenin signaling, etc. but lower activity of inflammatory responses (Fig. 2A). Noticeably, the difference in the score of the activity of ROS pathway between tumor and normal samples was the highest (Fig. 2B), and the TCGA-LIHC cohort also showed similar results (Fig. 2C). Many gene involved in the ROS pathway genes were over-expressed in HCC tumor samples (Fig. 2D). Additionally, we calculated the Pearson correlation between the score of the activity of the ROS pathway and rest 49 hallmark gene sets. The results showed that the ROS pathway was highly positively correlated with peroxidase, bile acid metabolism, lipogenesis, oxidative phosphorylation, fatty acid metabolism, and heterobiotic metabolism (Supplementary Figure S2). These data indicated the presence of abnormally activated ROS pathways in HCC tissue, which was closely correlated with many biological pathways involved in cancer occurrence.
Fig. 2.
Abnormally activated ROS pathway in HCC hepatocytes. (A) The enrichment score of 50 hallmark gene signatures in HCC hepatocytes. (B-C) The difference in ROS pathway enrichment scores of hepatocytes between HCC and adjacent tissues in GSE189175 and TCGA-LIHC cohorts. (D) Bubble diagram describing the expression of genes within the ROS pathway of hepatocytes between HCC and adjacent tissues.
The correlation between ROS pathway and energy metabolism in HCC hepatocytes
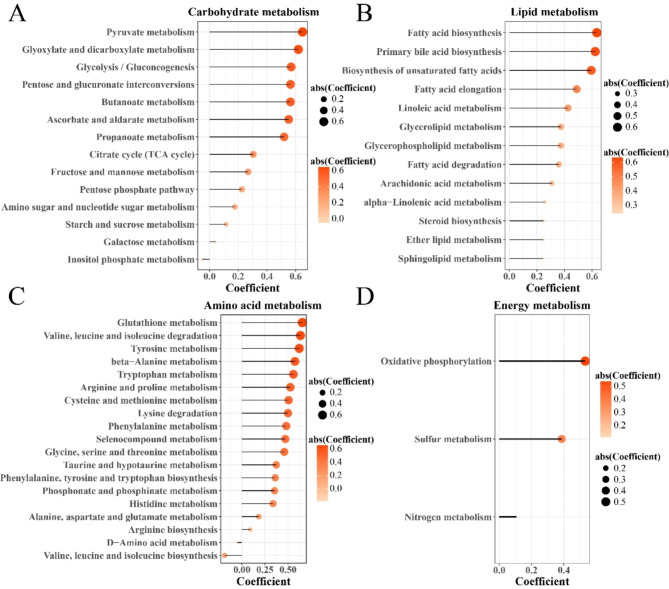
For the purpose of further investigating the impact of abnormally activated ROS pathway on energy metabolism in HCC hepatocytes, we calculated Pearson correlation enrichment scores among ROS pathway, glucose metabolism, lipid metabolism, amino acid metabolism, and energy metabolism. Specifically, the ROS pathway was positively correlated with pyruvate metabolism, glycolysis/gluconeogenesis, and TCA cycle in glucose-related metabolism (Fig. 3A). In lipid -related metabolism, the ROS pathway was positively correlated with fatty acid production, primary bile acid production, unsaturated fatty acid production, and fatty acid elongation (Fig. 3B). In amino acid-related metabolism, the ROS pathway was positively associated with glutathione metabolism, valine, leucine and isoleucine degradation, tyrosine metabolism but negatively related to D-amino acid metabolism, valine, leucine and isoleucine production (Fig. 3C). In energy -related metabolism, the ROS pathway was positively correlated with oxidative phosphorylation, sulfur metabolism, and nitrogen metabolism (Fig. 3D). These results showed that abnormally activated ROS pathway in hepatocytes increased the degree of glucose, fat, amino acid, and oxidative phosphorylation metabolism, providing sufficient energy for the infinite proliferation and invasion of tumor cells.
Fig. 3.
Correlationship between ROS pathway and energy metabolism in HCC hepatocytes. (A) Carbohydrate metabolism. (B) Lipid metabolism. (C) Amino acid metabolism. (D) Energy metabolism.
GPX4+ HCC hepatocytes had the highest ROS pathway activity
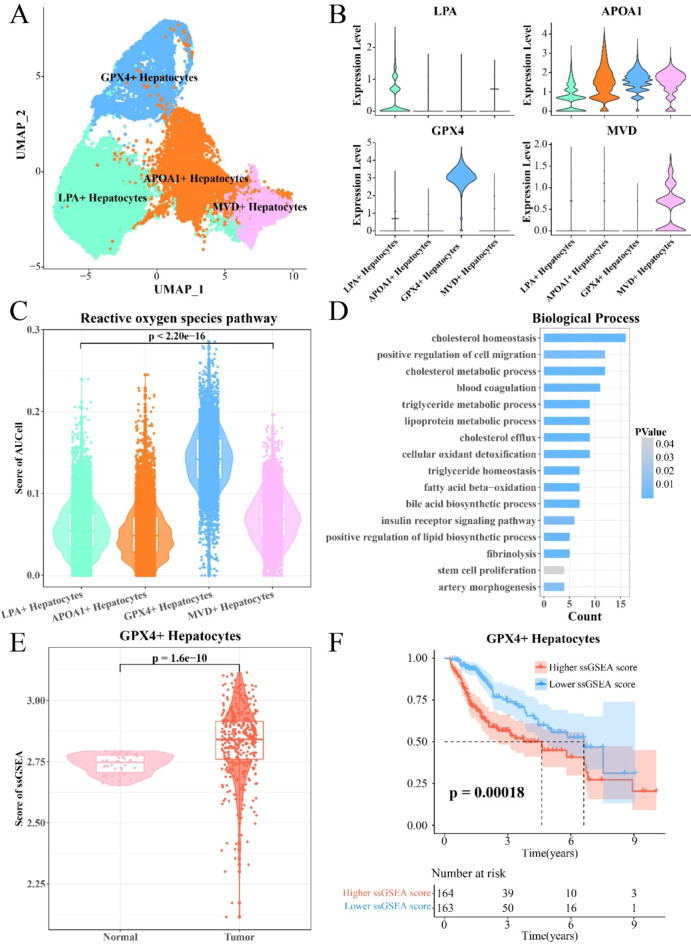
The Seurat was adopted to classify four subpopulations hepatocytes to identify the hepatocyte subpopulations with the highest ROS pathway activity (Fig. 4A). The representative high-expressed genes in each hepatocyte subpopulation were shown in Fig. 4B. It was found that the ROS pathway activity score was the highest in GPX4+ hepatocyte subpopulation (Fig. 4C). Gene Ontology (GO) analysis for biological processes revealed that the high-expressed genes in GPX4+ hepatocyte subpopulations were enriched in cholesterol homeostasis, cell migration, cholesterol metabolism process, triglyceride metabolism process, lipoprotein metabolism process, fatty acid β-oxidation, bile acid production, stem cell proliferation, and arterial morphogenesis, etc. (Fig. 4D). These results indicated that the activated ROS pathway may be closely related to the enhanced cholesterol, triglycerides, lipoprotein, and fatty acid metabolism. Also, we calculated the ssGSEA enrichment scores for the overexpressed genes in GPX4+ hepatocytes of the samples from the TCGA-LIHC cohort, and observed that these genes had a higher enrichment score in the tumor group (Fig. 4E), which indicated a shorter survival time for the patients (Fig. 4F). These results suggested that the GPX4+ hepatocyte subpopulation played an important role in the development of tumors.
Fig. 4.
The highest ROS pathway activity in GPX4+ HCC hepatocytes. (A) UMAP diagram for subdivision of HCC hepatocytes. (B) Violin diagram of representative high expression genes in hepatocyte subpopulations. (C) Violin plot describing the enrichment score of ROS pathway in four hepatocytes subpopulations. (D) Enriched biological processes of GPX4+ hepatocytes highly expressed genes. (E) Violin plot describing the enrichment score of GPX4+ hepatocytes highly expressed genes between TCGA-LIHC tumor and normal groups. (F) Survival curve of GPX4+ hepatocytes highly expressed genes enrichment scores in TCGA-LIHC samples.
JUND regulated the ROS pathway in GPX4+ hepatocytes
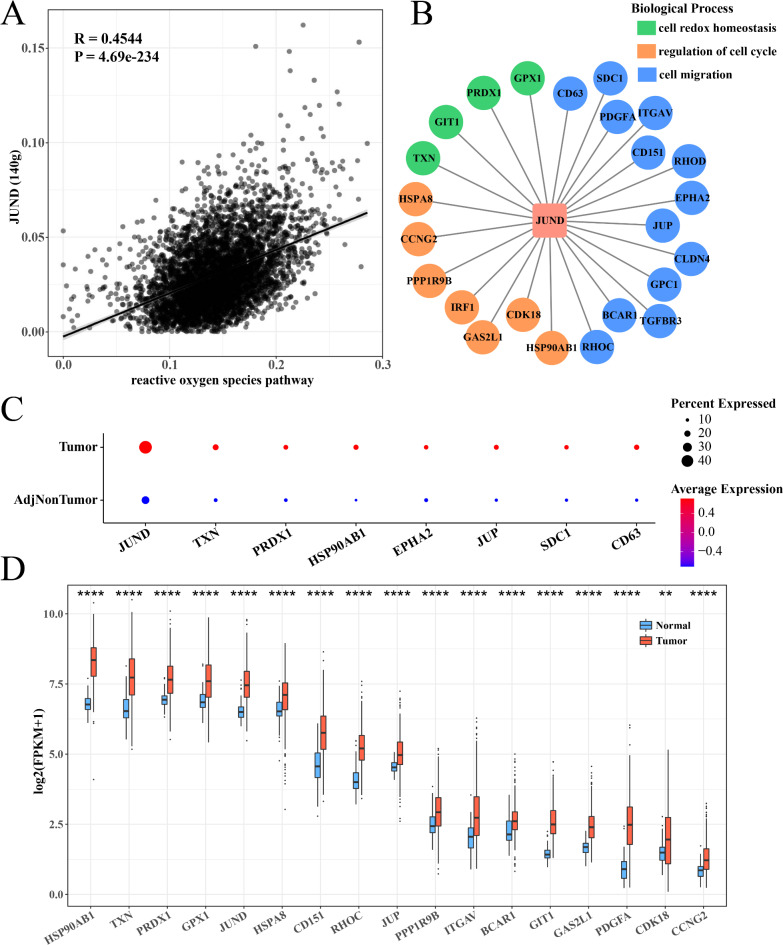
The SCENIC method was used to calculate the AUCell enrichment score for the TFs in GPX4+ hepatocytes. JUND showed the strongest correlation with ROS pathway (Fig. 5A) and played a positive regulatory role in the activation of the ROS pathway in GPX4+ hepatocytes. Furthermore, GO analysis on the biological processes involving the target genes of JUND revealed that these genes were enriched in cell redox homeostasis, cell cycle regulation, and cell migration (Fig. 5B). High-expressed JUND and its target genes were also found in the tumor group of both GSE189175 and TCGA-LIHC cohorts (Fig. 5C-D).
Fig. 5.
JUND regulated the ROS pathway in GPX4+ hepatocytes. (A) The correlation between JUND and ROS pathway enrichment score in GPX4+ hepatocytes. (B) JUND regulated target genes related to redox homeostasis, cell cycle, and cell migration. The red square represented transcription factors, while the circle represented target genes. The biological functions of the target genes were identified by different colors. (C) Bubble diagram describing the expression levels of JUND and its target genes between adjacent and cancerous regions in GSE189175 dataset. (D) Box plot of JUND expression levels and its target genes between normal and tumor tissues in TCGA-LIHC cohort. **p < 0.01 and ****p < 0.0001.
GPX4+ hepatocytes closely interacted with immune cells in HCC
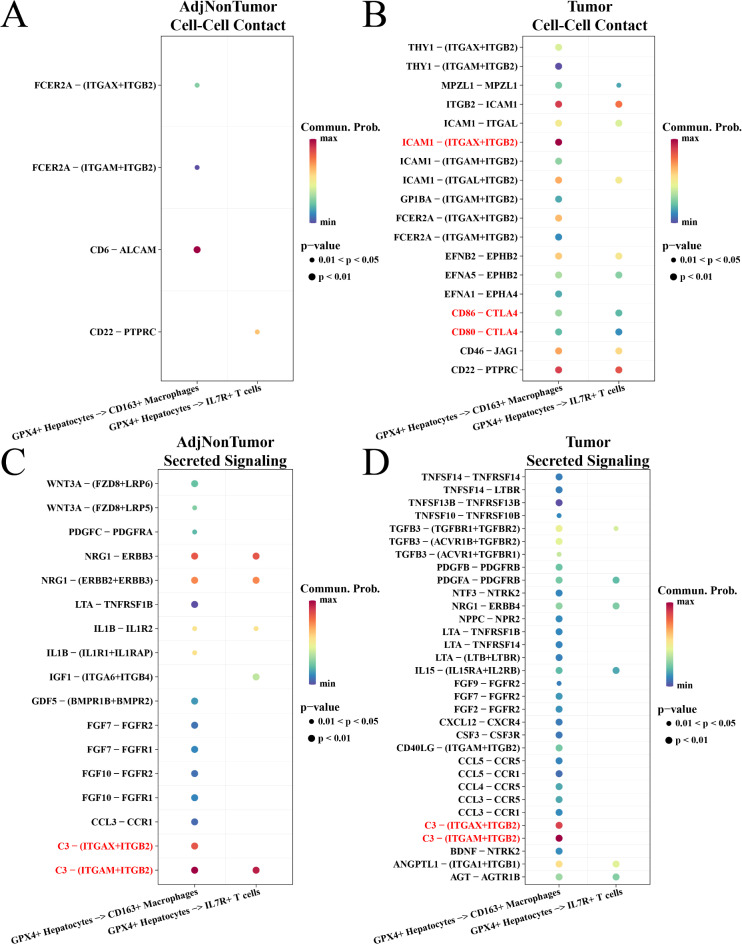
To further study the impact of the GPX4+ hepatocyte subpopulation on intracellular immune cells of HCC tissues, we analyzed the correlation between GPX4+ hepatocytes and immune cells in the para-cancerous and tumor groups. For direct cell-cell contact, we found that CD86-CTLA4 and CD80-CTLA4 with the abilities to inhibit T cell activity were only presented in the tumor group. Noticeably, GPX4+ hepatocytes were actively interacted with D163 + macrophages through ICAM1-(ITGAX + ITGB2) to promote the development of HCC (Fig. 6A-B). For secreted signaling, C3-(ITGAX + ITGB2) and C3-(ITGAM + ITGB2) activated the activity of D163 + macrophages in both the adjutant and tumor groups (Fig. 6C-D). These results indicated that GPX4+ hepatocytes were closely related to immune cells in HCC tissues.
Fig. 6.
Communications among GPX4+ hepatocytes, macrophages, and T cells across tumor and normal individuals. (A-B) Bubble diagram of the cell-cell contact type in adjacent cancer and tumor group. (C-D) Bubble plot of ligand receptors for secretory types within the para-cancerous and tumor groups.
Discussion
The present study analyzed the cell heterogeneity during HCC progression using large-scale single-cell transcriptomics30,31, classified four HCC hepatocyte subtypes, and analyzed the cell interactions between the main subpopulation of HCC hepatocytes and immune cells.
The results demonstrated that the HCC tissues had a variety of hepatocytes, which was consistent with the findings of Bai YM et al.32. Apart from this, Chi H et al.33 detected high proportions of macrophages, and Fu S34 discovered enriched innate-like mucosa-associated invariant T cells in HCC tissues. Such a compositional difference could be explained by different sampling locations, which also supported a high heterogeneity of HCC. Hepatocytes also high-expressed ALDH1A2, a marker gene for stem cells35,36, suggesting that hepatocytes are under a proliferative state similar to stem cells. Noticeably, the hepatic stellate cells, which play a vital role in hepatic fibrosis, accounted for a relatively high proportion in HCC tissues37,38. Study showed that hepatic stellate cells can help awaken the dormancy of cancer cells by secreting immunosuppressive CXCL12 chemokines to drive fibrosis damage, causing NK cells to enter a resting state to promote the development of cancer39.
On one hand, the production of ROS contributes to the development of cancer through destroying cell DNA to trigger the accumulation of carcinogenic changes11, on the other hand, ROS could strengthen immune defense and a high level of ROS helps induce tumor cell apoptosis40. This study found that the ROS pathway promoted the development of HCC and functioned together with multiple cancer-related pathways. Moreover, highly activated ROS pathway in GPX4+ hepatocytes had an inhibitory effect on immune cells. Therefore, our research focused on the genes related to the ROS pathway. Ferritin light chain (FTL) was overexpressed in HCC hepatocytes in comparison with adjacent tissues. This was consistent with Ke Set al.‘s finding41 as they also found that high levels of FTL were correlated with unfavorable survival of HCC patients. In acute myeloid leukemia, SOD1, an oxidative stress marker, was associated with poor overall survival42. Interestingly, SOD1 was also upregulated in the animal model of HCC43 and TCGA-LIHC tissues. ROS scavengers (SOD1 and CAT) help maintain ROS level for stabilizing the biological system. Patient with high levels of SOD1 and CAT exhibited longer survival44. Prognostic differences among patients may be related to different roles of ROS, which needs to be further investigated. Moreover, TFs that regulated the ROS pathway in GPX4+ hepatocytes were also investigated. JUND showed the strongest positive correlation with the ROS pathway. Previous study verified JUND as a crucial mediator for higher ROS levels induced by androgen in prostate cancer cells45,46, suggesting that JUND may be a therapeutic target for diseases related to the ROS pathway.
High activation of the ROS pathway in the hepatocyte subpopulation GPX4+ hepatocytes encouraged us to further investigate the role of GPX4+ hepatocytes in HCC development. As immune cells play an important role in the development of cancers, we explored the interactions between immune cells and GPX4+ hepatocytes. D163 + macrophages are M2-type macrophages with a cancer-promoting effect47. In direct cell-cell contract, we found that GPX4+ hepatocytes had a strong contact with D163 + macrophages via ICAM1-(ITGAX + ITGB2).
Intercellular adhesion molecule-1 (ICAM1) is involved in cancer metastasis such as bone metastasis48 and gallbladder cancer cell metastasis49. A recent study revealed that tumor could be induced to express ICAM1 through macrophage co-culture50, pointing to a close connection between ICAM1 and macrophages. ITGB2 is inversely correlated with the prognosis of gliomas patients51. In-depth research uncovered that this gene was closely linked to the dominance of M2-type macrophages in microenvironment infiltration52. ITGAX/CD11 is a macrophage-related gene53 that could notably facilitate the proliferation and metastasis of melanoma cells54. For secreted signaling, C3-(ITGAX + ITGB2) and C3-(ITGAM + ITGB2) receptor ligand pairs were found to be the communication bridges for GPX4+ hepatocytes and D163 + macrophages. Complement component 3 (C3) is the dominant effector molecule of the complement system55, and upregulated C3 could mediate tumor progression56,57. Accordingly, relevant inhibitors have been studied in preclinical models58. Collectively, all these discoveries supported that GPX4+ hepatocytes had a simulative effect on D163 + macrophages, which contributed to a dismal prognosis of HCC patients.
However, some limitations in this study should be equally recognized. First, our study is based on a single dataset only, and in the future, we will add HCC datasets from different regions and populations and validate them with multicenter data, thus enhancing the robustness and generalizability of the results. Second, the study used Seurat v3.1.2 for data analysis, which has certain performance limitations when dealing with large-scale single-cell data. Therefore, further studies will utilize the latest version of the software in order to take advantage of its more efficient data processing and analysis capabilities. Finally, this study lacked wet experiments to verify the specific mechanism of action of specific hepatocyte subtypes in HCC progression.
Conclusion
To conclude, a single-cell atlas of HCC was constructed, in which the ROS pathway was found to be abnormally activated in HCC hepatocytes. The GPX4+ hepatocyte subpopulation with the highest ROS pathway activity had an inhibitory effect on immune cells and was associated with a worse prognosis. JUND may be a gene target for the activated ROS pathway in HCC. The findings of present research contributed to the current understanding on the role of ROS pathway in HCC, providing a theoretical basis for personalized therapy for HCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GRN
Gene regulatory network
- HCC
Hepatocellular carcinoma
- PCA
Principal Component Analysis
- ROS
Reactive oxygen species
- scRNA-seq
Single-cell RNA sequencing
- ssGSEA
Single sample gene set enrichment analysis
- TCGA
The Cancer Genome Atlas
- TFs
Transcription factors
- UMAP
Uniform Manifold Approximation and Projection
Author contributions
All authors contributed to this present work: [XPW] & [HCJ] designed the study, [ZZX], [HCJ] and [PHL] acquired and analyzed the data. [HWX] and [XPW] drafted the manuscript, [XPW], [PHL] and [ZZX] revised the manuscript. I certify that all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Henan Medical Science and Technology Joint Building Program (LHGJ20240211), and the Henan Sunshine Medical Health Development Foundation iGanDan Project (HKP2023006).
Data availability
The datasets generated and/or analyzed during the current study are available in the [GSE189175] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc= GSE189175].
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoping Wang, Email: wangxp19731230@163.com.
Huiwu Xing, Email: xhwzzu@163.com.
References
- 1.Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers7, 6 (2021). [DOI] [PubMed]
- 2.Su, Q. et al. Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in hepatocellular carcinoma. Curr. Pharm. Biotechnol. 24, 1035–1058 (2023). [DOI] [PubMed]
- 3.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Yang, J. D. et al. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol.16, 589–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erridge, S. et al. Meta-analysis of determinants of survival following treatment of recurrent hepatocellular carcinoma. Br. J. Surg.104, 1433–1442 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Yoh, T. et al. Surgery for recurrent hepatocellular carcinoma: Achieving long-term survival. Ann. Surg.273, 792–799 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Zhang, Q., Lou, Y., Bai, X. L. & Liang, T. B. Intratumoral heterogeneity of hepatocellular carcinoma: From single-cell to population-based studies. World J. Gastroenterol.26, 3720–3736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, J. et al. Anticancer effect of dihydroartemisinin via dual control of ROS-induced apoptosis and protective autophagy in prostate cancer 22Rv1 cells. Curr. Pharm. Biotechnol. 25, 1321–1332 (2024). [DOI] [PubMed]
- 9.Roy, Z., Bansal, R., Siddiqui, L. & Chaudhary, N. Understanding the role of free radicals and antioxidant enzymes in human diseases. Curr. Pharm. Biotechnol. 24, 1265–1276 (2023). [DOI] [PubMed]
- 10.Szatrowski, T. P. & Nathan, C. F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res.51, 794–798 (1991). [PubMed] [Google Scholar]
- 11.Cheung, E. C. & Vousden, K. H. The role of ROS in tumour development and progression. Nat. Rev. Cancer. 22, 280–297 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Zhao, W. et al. Double-edged sword effect of reactive oxygen species (ROS) in tumor development and carcinogenesis. Physiol. Res.72, 301–307 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou, X. et al. Molecular mechanisms of ROS-modulated cancer chemoresistance and therapeutic strategies. Biomed. Pharmacother. 165, 115036 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Tuli, H. S. et al. Molecular mechanisms behind ROS regulation in cancer: A balancing act between augmented tumorigenesis and cell apoptosis. Arch. Toxicol.97, 103–120 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Alvarez, M. et al. Human liver single nucleus and single cell RNA sequencing identify a hepatocellular carcinoma-associated cell-type affecting survival. Genome Med.14, 50 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen, W. et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta1, e36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart, T. et al. Compr. Integr. Single-Cell Data Cell.177, 1888–1902e1821 (2019). [Google Scholar]
- 18.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods. 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, Y. et al. Tissue-based metabolomics reveals metabolic signatures and major metabolic pathways of gastric cancer with help of transcriptomic data from TCGA. Biosci. Rep.41 (2021). [DOI] [PMC free article] [PubMed]
- 20.Lu, Y., Li, K., Hu, Y. & Wang, X. Expression of immune related genes and possible regulatory mechanisms in Alzheimer’s disease. Front. Immunol.12, 768966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc.15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature546, 533–538 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Winterhoff, B. J. et al. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol. Oncol.144, 598–606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner, M. et al. All-In-One: Advanced preparation of human parenchymal and non-parenchymal liver cells. PLoS One. 10, e0138655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young, M. D. et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Wettere, A. J., Law, J. M., Hinton, D. E. & Kullman, S. W. Anchoring hepatic gene expression with development of fibrosis and neoplasia in a toxicant-induced fish model of liver injury. Toxicol. Pathol.41, 744–760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao, S. et al. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat. Cell. Biol.20, 721–734 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Seeberger, K. L., Eshpeter, A., Rajotte, R. V. & Korbutt, G. S. Epithelial cells within the human pancreas do not coexpress mesenchymal antigens: Epithelial-mesenchymal transition is an artifact of cell culture. Lab. Invest.89, 110–121 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Shahrajabian, M. H. & Sun, W. Survey on multi-omics, and multi-omics data analysis. Integr. Appl. Curr. Pharm. Anal.19, 267–281 (2023). [Google Scholar]
- 31.Jovic, D. et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl Med.12, e694 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai, Y. M. et al. Single-cell transcriptomic dissection of the cellular and molecular events underlying the triclosan-induced liver fibrosis in mice. Mil Med. Res.10, 7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi, H. et al. T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing. Front. Immunol.14, 1137025 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu, S. et al. Regulatory mucosa-associated invariant T cells controlled by β1 adrenergic receptor signaling contribute to hepatocellular carcinoma progression. Hepatology78, 72–87 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Tomita, H., Tanaka, K., Tanaka, T. & Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget7, 11018–11032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wazir, U., Orakzai, M., Martin, T. A., Jiang, W. G. & Mokbel, K. Correlation of TERT and stem cell markers in the context of human breast cancer. Cancer Genomics Proteom.16, 121–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He, W. et al. Single-cell transcriptomics of hepatic stellate cells uncover crucial pathways and key regulators involved in non-alcoholic steatohepatitis. Endocr. Connect.12 (2023). [DOI] [PMC free article] [PubMed]
- 38.Larsen, F. T. et al. Stellate cell expression of SPARC-related modular calcium-binding protein 2 is associated with human non-alcoholic fatty liver disease severity. JHEP Rep.5, 100615 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Correia, A. L. et al. Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature594, 566–571 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Yang, S. & Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell. Biochem.467, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ke, S., Wang, C., Su, Z., Lin, S. & Wu, G. Integrated analysis reveals critical ferroptosis regulators and FTL contribute to cancer progression in hepatocellular carcinoma. Front. Genet.13, 897683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, X. et al. High expression of LOC541471, GDAP1, SOD1, and STK25 is associated with poor overall survival of patients with acute myeloid leukemia. Cancer Med.12, 9055–9067 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghufran, H., Azam, M., Mehmood, A., Butt, H. & Riazuddin, S. Standardization of diethylnitrosamine-induced hepatocellular carcinoma rat model with time based molecular assessment. Exp. Mol. Pathol.123, 104715 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Alves, A. F. et al. Gene expression evaluation of antioxidant enzymes in patients with hepatocellular carcinoma: RT-qPCR and bioinformatic analyses. Genet. Mol. Biol.44, e20190373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehraein-Ghomi, F. et al. JunD mediates androgen-induced oxidative stress in androgen dependent LNCaP human prostate cancer cells. Prostate68, 924–934 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Mehraein-Ghomi, F. et al. Targeting androgen receptor and JunD interaction for prevention of prostate cancer progression. Prostate74, 792–803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak, M. & Klink, M. The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells 9 (2020). [DOI] [PMC free article] [PubMed]
- 48.Chen, M., Wu, C., Fu, Z. & Liu, S. ICAM1 promotes bone metastasis via integrin-mediated TGF-β/EMT signaling in triple-negative breast cancer. Cancer Sci.113, 3751–3765 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang, C. et al. Fibrinogen promotes gallbladder cancer cell metastasis and extravasation by inducing ICAM1 expression. Med. Oncol.40, 10 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Hsieh, C. Y. et al. Macrophage secretory IL-1β promotes docetaxel resistance in head and neck squamous carcinoma via SOD2/CAT-ICAM1 signaling. JCI Insight7 (2022). [DOI] [PMC free article] [PubMed]
- 51.Xu, H. et al. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas. Cancer Immunol. Immunother. 71, 645–660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, C. et al. Identifying ITGB2 as a potential prognostic biomarker in ovarian cancer. Diagnostics (Basel)13 (2023). [DOI] [PMC free article] [PubMed]
- 53.Edsfeldt, A. et al. Interferon regulatory factor-5-dependent CD11c + macrophages contribute to the formation of rupture-prone atherosclerotic plaques. Eur. Heart J.43, 1864–1877 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ni, L., Li, P., Li, M., Huang, S. & Dang, N. SERPINB8 and furin regulate ITGAX expression and affect the proliferation and invasion of melanoma cells. Exp. Dermatol.32, 24–29 (2023). [DOI] [PubMed] [Google Scholar]
- 55.Zarantonello, A., Revel, M., Grunenwald, A. & Roumenina, L. T. C3-dependent effector functions of complement. Immunol. Rev.313, 120–138 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleczko, E. K. et al. Upregulation of complement proteins in lung cancer cells mediates tumor progression. Front. Oncol.12, 1045690 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boire, A. et al. Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell168, 1101–1113e1113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwak, J. W. et al. Complement activation via a C3a receptor pathway alters CD4(+) T lymphocytes and mediates lung cancer progression. Cancer Res.78, 143–156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the [GSE189175] repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc= GSE189175].