Abstract
We found previously that neither a 6-kbp promoter fragment nor even a 120-kbp yeast artificial chromosome (YAC) containing the whole GATA-3 gene was sufficient to recapitulate its full transcription pattern during embryonic development in transgenic mice. In an attempt to further identify tissue-specific regulatory elements modulating the dynamic embryonic pattern of the GATA-3 gene, we have examined the expression of two much larger (540- and 625-kbp) GATA-3 YACs in transgenic animals. A lacZ reporter gene was first inserted into both large GATA-3 YACs. The transgenic YAC patterns were then compared to those of embryos bearing the identical lacZ insertion in the chromosomal GATA-3 locus (creating GATA-3/lacZ “knock-ins”). We found that most of the YAC expression sites and tissues are directly reflective of the endogenous pattern, and detailed examination of the integrated YAC transgenes allowed the general localization of a number of very distant transcriptional regulatory elements (putative central nervous system-, endocardium-, and urogenital system-specific enhancers). Remarkably, even the 625-kbp GATA-3 YAC, containing approximately 450 kbp and 150 kbp of 5′ and 3′ flanking sequences, respectively, does not contain the full transcriptional regulatory potential of the endogenous locus and is clearly missing regulatory elements that confer tissue-specific expression to GATA-3 in a subset of neural crest-derived cell lineages.
GATA-3 belongs to a family of transcription factors that bind to the consensus sequence (A/T)GATA(A/G) and share a steroid hormone receptor superfamily C4 zinc finger DNA binding motif (14, 24, 27, 41) that is also evolutionarily conserved in lower eucaryotic GATA factors. The GATA factor family is composed of six vertebrate members (1, 19, 41), and from gene ablation studies, GATA-1 through GATA-4 have been shown to be individually indispensable for embryonic development (26, 28, 32, 37, 38). GATA-1 is expressed in myeloerythroid lineage cells and Sertoli cells of the testis (13, 25, 33). GATA-2 is expressed in multipotential hematopoietic progenitors, megakaryocytes, mast cells, and endothelial cells as well as in an overlapping pattern with GATA-3 in the placenta and central nervous system (CNS) (6, 15, 29, 30, 34). GATA-3 is, like GATA-2, expressed more widely than GATA-1, and it is the only family member expressed in T lymphocytes (41, 42). Based on comparisons of cDNA sequences and intron/exon boundaries, the GATA-4, -5, and -6 factors constitute a distinct subfamily, principally implicated in cardiac and ventral/dorsal patterning (28).
Previous in situ hybridization analysis showed that GATA-3 transcription is controlled both temporally and spatially during early embryonic development (8, 31). GATA-3 mRNA is detected at high levels in the ectoplacental cone at 8.5 days postcoitus (dpc) and persists over the course of the next several gestational days. Within the embryo proper, GATA-3 is expressed first and most abundantly in the CNS and peripheral nervous system (PNS), the kidney, the adrenal gland, and the primitive thymus; these initial experiments also suggested that there might be weak expression in the heart and the skin (8).
Our initial transgenic experiments examining GATA-3 transcriptional regulation using plasmid constructs identified a number of discrete regulatory elements (namely, the genital tubercle and branchial arch elements), but these promoter proximal sequences were clearly unable to fully recapitulate the wild-type developmental expression pattern of the GATA-3 locus (20). Therefore, we isolated and characterized yeast artificial chromosomes (YACs) bearing the GATA-3 gene, in anticipation that they would provide a means of analyzing this locus as a single, large contiguous fragment of genomic DNA (17). We identified two GATA-3 YACs that together delineate approximately 1 Mbp of gene flanking sequence, with the GATA-3 structural gene located approximately in the center. We found that a 120-kbp YAC lacZ reporter transgene (called C4lacZ), which contains approximately 35 kbp of 5′ GATA-3 flanking sequence as well as 60 kbp of 3′ GATA-3 flanking sequence, could direct the expression of the reporter gene at new anatomical sites not identified previously in the smaller plasmid expression constructs. However, even this 120-kbp YAC failed to direct the expression of the reporter gene in several tissues that are known to normally express GATA-3. In accord with this observation, the 120-kbp C4 YAC was not able to rescue embryonic lethality caused by the original gene targeted mutation (17).
To localize the regulatory determinants of its expression during embryogenesis, we have significantly extended the boundaries of the murine GATA-3 locus under scrutiny. Here, we describe the expression profile of a lacZ reporter gene that was targeted to the initiation codon of the chromosomal GATA-3 gene in ES cells, generating GATA-3/lacZ “knock-in” mice (11, 44) as the reference point. We then asked whether large transgenic YACs encoding GATA-3 could reproduce this same pattern. In independent transgenic lines bearing a 625-kbp GATA-3 YAC, the lacZ transgene reflected the endogenous expression profile in all tissues except the thymus and specific neural crest-derived cells (i.e., the sympathetic chain and the adrenal gland), while the smaller, 540-kbp YAC conferred a less complete pattern.
Surprisingly, these studies indicate that the complete GATA-3 locus lies beyond the boundaries of even the largest YAC (625 kbp) examined here. Nonetheless, detailed structural analysis of these integrated transgenes led to the general localization of at least three positive regulatory elements that direct the expression of GATA-3 in distinct tissues. The element(s) required for GATA-3 expression in the endocardial cushions of the embryonic heart is located quite far from the 3′ end of the gene, between +105 and +145 kbp with respect to the GATA-3 transcription initiation site. At least two other tissue-specific element(s) are located far 5′ to the gene: these elements regulate GATA-3 expression in the developing CNS and the urogenital system, and reside between −6 to −35 kbp and −35 to −150 kbp, respectively. These studies also lead to the conclusion that the patterning elements controlling GATA-3 expression in specific cell lineages derived from the neural crest (in the sympathetic chain and in the adrenal medulla) are located beyond the boundaries of the 625-kbp YAC and therefore lie more than 450 kbp 5′, or more than 150 kbp 3′, to the GATA-3 structural gene.
MATERIALS AND METHODS
LacZ targeting of GATA-3 YACs.
The Escherichia coli lacZ gene was targeted into the initiation codon within the first coding exon of the GATA-3 gene in B124 and B125 YACs by homologous recombination in yeast (4, 40), as described previously (17). The resulting YACs therefore precisely mimic the structure of the original term line targeted mutation (32) as well as that of the lacZ gene in the knock-ins (12).
Preparation and analysis of high-molecular-weight DNA.
Yeast and mouse thymic DNA agarose plugs were prepared as described previously (17). Pulsed-field gel electrophoresis (PFGE) was performed by using 1% agarose gels in 0.5× Tris-borate-EDTA at 14°C. For resolution of DNA up to 2 Mbp in size, the electrophoresis conditions were 120 V with 10- to 200-s ramped switch time for 20 h. The gels were transferred onto nylon membranes (BioRad) and hybridized at 65°C. The blots were then washed and finally exposed for autoradiography. Probes were generated by random primer labeling (7).
Isolation of YAC DNA for microinjection.
The protocol used was essentially as described (17) except for the following modifications. For the 540- and 625-kbp YACs, a 25- to 80-s ramped switch time was used for 20 h. The YAC DNA was excised, rotated 90° (perpendicular to the electric field), and cast in a NuSieve 4% agarose gel (FMC Corp.). The electrophoresis conditions used were 300-s ramped switch time for 15 h. The concentrated YAC DNA band was excised and equilibrated with injection buffer (10 mM Tris-HCl [pH 7.2], 0.1 mM EDTA, 70 mM spermine, and 30 mM spermidine) on ice for 2 to 24 h prior to digestion with β-agarase (4 U/100 μl) for 2 to 4 h. Finally, the YAC DNA was dialyzed against injection buffer by using a floating dialysis membrane (100-kDa exclusion limit) for 2 to 24 h. The integrity of the YAC DNA was verified by PFGE prior to microinjection.
Transgenic mice.
Transgenic mice were generated using standard protocols (20) and identified by PCR by using lacZ and YAC left and right vector arm primers, and by Southern blotting (17).
LacZ staining and tissue sectioning.
The morning that vaginal plugs were detected was designated 0.5 dpc. Embryos were isolated at gestation days from 10.5 to 18.0 and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (39). They were then frozen in O.C.T. compound (Sakura Finetek) and sectioned at 10-μm thickness prior to counterstaining with nuclear fast red.
Genomic mapping.
The generation of probes used in mapping the transgenic YAC lines has been described previously (17, 20). To illustrate the analysis that must be performed to characterize the integrity of YAC transgenes in each line, the deduction of the transgene structure of the most complicated multicopy line described in the present study, B125Z89 (see Fig. 1E), is discussed below.
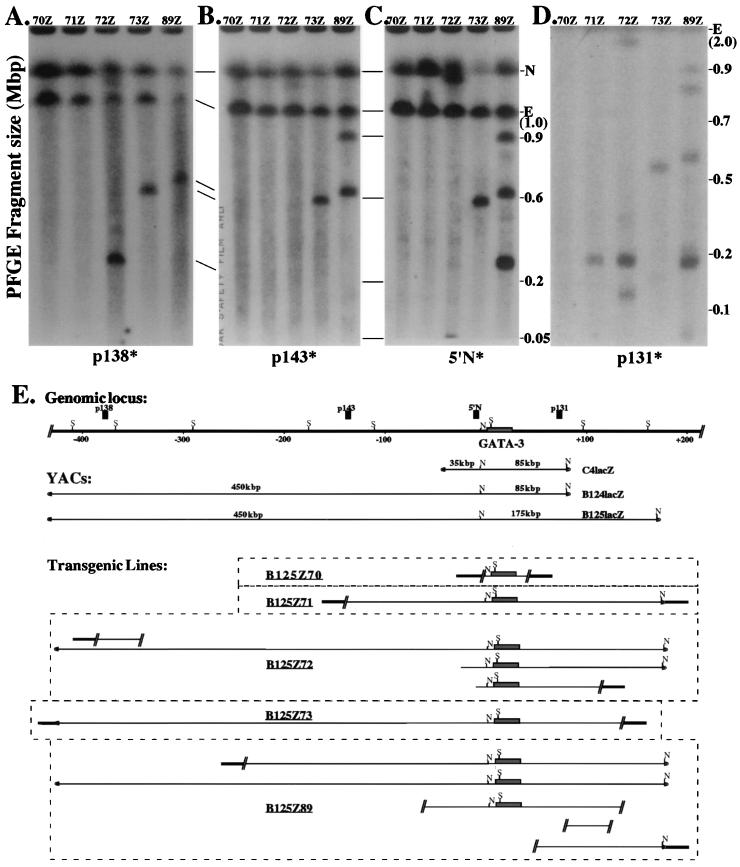
FIG. 1.
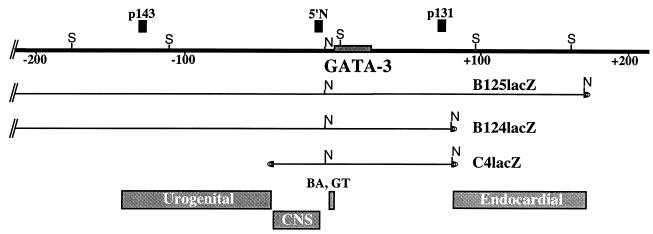
Structural integrity of the B125lacZ transgenes. Agarose plugs containing thymus DNA from the transgenic lines Z70, Z71, Z72, Z73, and Z89 were digested in situ with NotI restriction endonuclease. After PFGE electrophoresis, the DNA was transferred to a nylon membrane and independently hybridized to either p138 probe (A) or p143 probe (B) or 5′ N probe (C) or p131 probe (D). (E) Summary diagram for five of the B125lacZ YAC transgenic lines. The top line shows an abbreviated map of the GATA-3 locus (S denotes SfiI sites; data not shown) and the positions of several markers used as probes, as well as the positions of the two NotI (N) sites in the YAC at −4.5-kbp (20) in the locus and in the YAC right vector arm. The shaded line represents mouse DNA flanking the integrated transgene. The actual order of integration of multicopy transgenes (lines Z72 and Z89) is arbitrary, since they have not been determined, but all are integrated at a single site in the mouse genome. For those two multicopy lines, it is not possible to determine which fragment lying 5′ to the genomic NotI site is physically contiguous with which fragment lying 3′ to the site, and thus the 5′ and 3′ fragments in these two lines are depicted as separate and not connected. Lines Z70, Z71, and Z73 contain fragmented, single-copy transgenes. E, endogenous mGATA-3 NotI fragment; N, nonspecific hybridization.
Since the transgenes in line B125Z89 segregated together, we assumed that they represented multiple copies integrated at a single genomic site. The 5′-most probe, p138, is located at approximately −400 kbp with respect to the GATA-3 transcription start site (see Fig. 1E and also reference 17) and hybridized to one band of approximately 650 kbp in line B125Z89 (see Fig. 1A) as well as to an endogenous 1-Mbp NotI fragment and a nonspecific band (see Fig. 1A through D). The 5′ p143 probe lies at approximately −125 kbp (17) and hybridized to two bands that were 650 and 900 kbp as well as to the same endogenous and nonspecific bands as the p138 probe (see Fig. 1B). These data indicated that line B125Z89 contained one transgene copy (represented by the 650-kbp band) that was contiguous between −400 and −125 kbp, and that a second copy (900 kbp) was fragmented somewhere between these two points. The 5′ N probe is located at −4.5 kbp (17, 20) and hybridized to the same 650- and 900-kbp fragments as well as to an additional 280-kbp band (see Fig. 1C). This indicated that both larger bands (i.e., 650- and 900-kbp bands) were contiguous from −125 to −4.5 kbp, while the 280-kbp band represented a third YAC copy that was fragmented between −125 and −4.5 kbp.
The 3′ p131 probe, which is located at approximately +80 kbp, hybridized to a full-length NotI fragment (175 kbp) that is more intense than a single-copy band, as well as to three larger fragments. Thus, the 3′ p131 probe detected five transgene copies, three of which were fragmented between +80 kbp and the right arm of the YAC, while the two remaining copies represent intact 175-kbp 3′ end fragments. Similar mapping studies using SfiI restriction endonuclease (16) demonstrated that both the 5′ N and p131 probes (Fig. 1E) detect no aberrant bands, and thus all the transgenes that contain the mGATA-3 gene in line Z89 are contiguous within the 5′ 120-kbp or the 3′ 100-kbp SfiI fragments. In summary, the data indicate that none of the breakpoints in any of the transgene copies in this line occurred between −110 and +80 kbp, including the entire 23-kbp GATA-3 structural gene (8, 17, 20). Thus, both the sets of data for SfiI (16) and NotI restriction mapping (see Fig. 1A through D) indicate that the minimum contiguous mGATA-3 lacZ YAC present in line Z89 must be >500 kbp but leave open the equally likely possibility that this line carries one intact transgene copy.
RESULTS
Generation of GATA-3 YAC transgenic lines.
YAC B124 contains approximately 450 kbp of the 5′ end genomic information, the GATA-3 structural gene (approximately 25 kbp; see reference 8), and 65 kbp of 3′ end genomic information, while YAC B125 is identical to YAC B124 except for 85 kbp of additional sequence at the 3′ end (8, 17). Both YACs were modified by insertion of the lacZ gene at the site of the GATA-3 initiation codon by sequential homologous integration and excision in yeast (4, 17). Gel-purified B124lacZ and B125lacZ YAC DNAs were injected into fertilized ova to generate transgenic founders (4, 5).
Thirteen transgenic founders bearing the larger, B125lacZ, YAC were obtained from 133 pups. Of these, ten transmitted the transgene through the germ line, and seven of them contained the lacZ gene as well as both the left and right YAC vector arms as detected by the initial PCR screens. The copy numbers of the B125lacZ YAC lines ranged from one to five (Fig. 1, and data not shown).
Thymus nuclei recovered from each established line were digested with NotI or SfiI followed by PFGE. The DNA was transferred to nylon, and the blots were then probed with radiolabeled DNA fragments from throughout the locus (p138, p143, 5′ N and p131; Fig. 1), hence enabling us to identify the approximate positions of breakpoints in the integrated transgenes (4, 17, 23). These experiments, for which a representative description is provided in Materials and Methods, showed that four of the B125lacZ lines contained large internal segments of the GATA-3 locus, including unaltered 5′ and 3′ SfiI fragments flanking the unique NotI site at −4.5 kbp (Fig. 1E; references 8 and 20). A summary of the B125lacZ YAC maps in the five lines characterized here is diagrammed in Fig. 1E. Although only two of the five lines harbored transgenes that contained large uninterrupted blocks of contiguous B125lacZ YAC DNA, comparison of the integrated transgene structures to the expression patterns of these YAC lines turned out to be uniquely informative (see below and Discussion).
Six transgenic lines bearing the smaller, B124lacZ, YAC were obtained from 52 pups, and five were determined by PCR to contain the lacZ gene and both left and right YAC vector arms. Four of the five lines transmitted the transgene through the germ line, while one line was mosaic (multiple sites of transgene integration) and therefore was not characterized further. When the four remaining lines were analyzed for copy number and integrated YAC structure, two were found to contain intact B124lacZ transgenes, while the other two were badly fragmented and hence not examined further. The copy numbers of the four B124lacZ YAC transgenic lines ranged from one to three (16).
GATA-3 expression from the chromosomal locus.
In order to establish a reference point for endogenous GATA-3 expression, we characterized the pattern for GATA-3LacZ/+ knock-in mice and compared this pattern to that detected by in situ hybridization (8, 31). Generation of the GATA-3LacZ knock-in allele, which results in precisely the same lacZ insertion in the GATA-3 genomic locus in ES cells as that in the YACs, has been described (12). Although both the YAC and ES cell targeting events placed the reporter gene at the GATA-3 initiation codon, one difference was that the knock-in reporter gene incorporated a nuclear localization signal, thus allowing us to clearly distinguish cell morphology in expressing tissues.
Whole-mount staining for β-galactosidase of GATA-3LacZ/+ embryos showed that GATA-3 expression began in the ectoplacental cone around 8.5 dpc and was also observed in the branchial arches and cloaca or genital tubercle (16, 19). Strong staining was also noted in the midbrain, hindbrain, and spinal cord (abbreviated collectively as the CNS below; references 16 and 19–22). By 10.5 dpc, expression became more intense in the CNS and in the branchial arches and was also prominent in the otic vesicle, the developing eye, the heart, the mesonephric duct, and the cloaca (Fig. 2A and below). By 12.5 dpc, the pattern was largely unchanged, with localized expression in the CNS, in the developing eye, organs of the inner ear, the jaw, and the neck region (elaborated from the developing branchial arches) as well as in proximal regions of the developing limbs, the mesonephros, and the mesonephric ducts (Fig. 2B). Expression was also apparent in the developing sympathetic trunk (see below) and umbilical vessels. The cloaca continued to strongly express GATA-3 as it developed into the urogenital sinus and rectum (see below). All of these sites and times of expression were previously detected in the in situ hybridization studies (8).
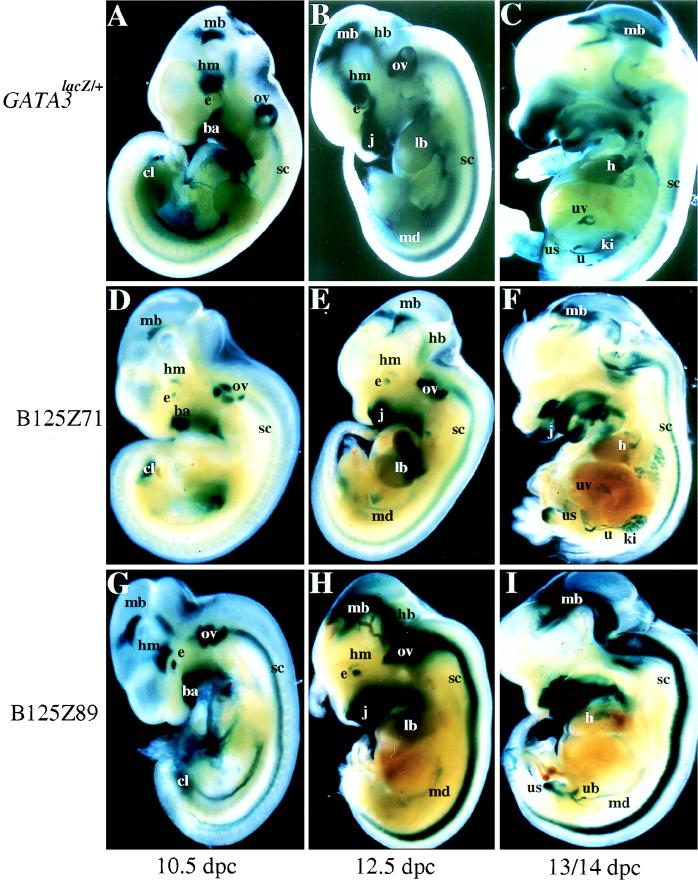
FIG. 2.
LacZ expression of GATA-3LacZ/+ versus GATA-3lacZ YAC transgenic embryos. The expression patterns of GATA-3LacZ/+ and two GATA-3 B125 lacZ YAC transgenic lines are shown here at three developmental stages (10.5, 12.5, and 13/14 dpc) for direct comparisons of their coincidence. The embryos were cleaved roughly along the lateral midline and then stained with X-Gal. The 10.5- and 12.5-dpc embryos are displayed as the outside halves of the embryos, while the 13/14-dpc embryos are displayed as a view with the internal organs presented en face. A, B, and C, GATA-3LacZ/+ embryos at 10.5, 12.5, and 13 dpc, respectively; D, E, and F, line B125Z71 transgenic embryos at 10.5, 12.5, and 14 dpc, respectively; and G, H, and I, line B125Z89 transgenic embryos at 10.5, 12.5, and 13 dpc, respectively. Although the intensity of staining varies, consistent patterns of expression are detected in all three cases in the CNS (labeled mb, hb, and sc, for midbrain, hindbrain, and spinal cord, respectively), the eye (e), the head mesenchyme (hm), the otic vesicle (ov), the branchial arches/jaw (ba/j), the limb buds (lb), the heart (h), the umbilical vessels (uv), the mesonephric duct (md), the ureteric bud (ub), the urogenital sinus (us), the ureters (u), and the kidneys (ki). The labeling in the lung of the single-copy Z71 embryo (panel F) was not detected in the other B125lacZ lines nor in the GATA-3LacZ/+ embryos and therefore was rejected as ectopic staining due to the transgene integration position.
By 14 dpc, β-galactosidase continued to be most predominantly expressed in the CNS (Fig. 2C) and in the sympathoadrenal system (see below). As the embryo matured further, expression diminished in the spinal cord but persisted in the midbrain. The eyes, the semicircular canals of the inner ear, the jaws (both mandibular and maxillary structures), the neck region, and the base of the tongue also continued to express lacZ (Fig. 2C). In the circulatory system, expression was confined to the base of the heart, outflow vessels, and umbilical vessels (Fig. 2C), but by days 14 to 15, when the development of the fetal circulatory system was complete, expression had vanished from these sites.
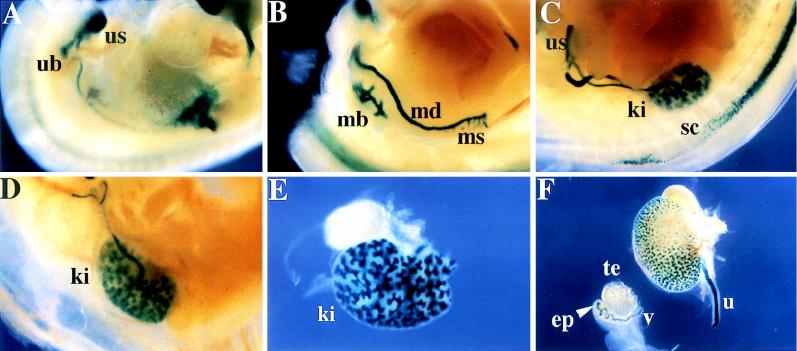
In the urogenital system, staining was strong in the mesonephros and mesonephric duct (Fig. 2C). At later stages in embryogenesis, expression persisted in structures derived from the mesonephros (the epididymis) and the mesonephric ducts (the van deferens; see below). Expression was quite prominent in the metanephric duct and ureteric bud and continued as the metanephric duct differentiated into the renal collecting tubules and the ureters (Fig. 2C and below). The primitive urinary bladder, derived from the ventral aspect of the urogenital sinus, strongly expressed the reporter gene (Fig. 2C and below), and this expression persisted until bladder development was complete. The epithelium lining the mesonephric tubules and ducts also expressed β-galactosidase (data not shown). The tubules derived from the metanephric duct, which eventually contribute to the adult kidney, also expressed the lacZ gene (e.g., Fig. 3A). The epithelium lining the bladder was also stained (16), as were neural crest descendent cells that contributed to the adrenal medulla (Fig. 3A).
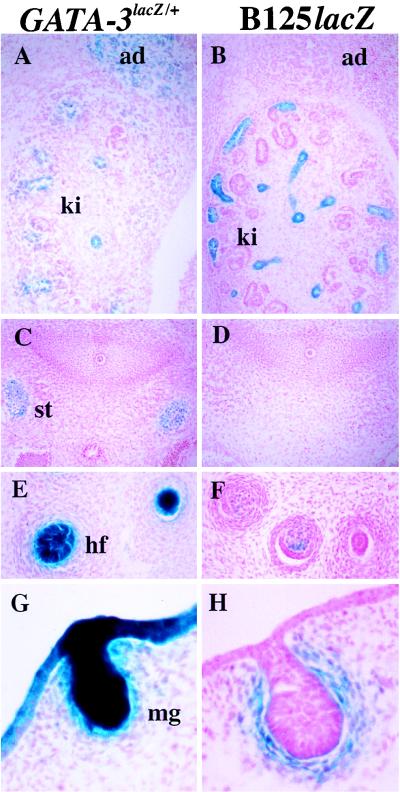
FIG. 3.
Differential expression of 14.5-dpc GATA-3LacZ/+ and B125lacZ embryos. A and B, axial sagittal sections at the level of the developing kidney and adrenal gland. The cells of the adrenal medulla (ad) stain only in the GATA-3LacZ/+ embryos, while the tubules of the kidney (ki) in both embryos express β-galactosidase. C and D, transverse sections at the axial level of the cervical sympathetic trunk (st). The sympathetic trunk is negative for β-galactosidase expression in the YAC embryo (panel C), while the GATA-3LacZ/+ allele is expressed strongly (panel D). E and F, sagittal sections of developing hair follicles (hf) in the skin, showing expression of β-galactosidase in the dermal papillae of both GATA-3LacZ/+ and B125lacZ embryos. G and H, sagittal sections showing expression in the developing mammary glands (mg) of both GATA-3LacZ/+ and B125lacZ89 embryos.
In the developing ear, expression was confined to the developing semicircular canals, saccule, and cochlea (Fig. 4A). β-galactosidase expression was also detected in the ganglion of the vestibulocochlear cranial nerve. However, we found no evidence for expression in the trigeminal and facial ganglia, as reported previously for our in situ hybridization experiments (8). Sectioning through the developing eye revealed that expression was confined to the differentiating cuboidal cells in the equatorial zone that form secondary lens fibers (Fig. 4D).
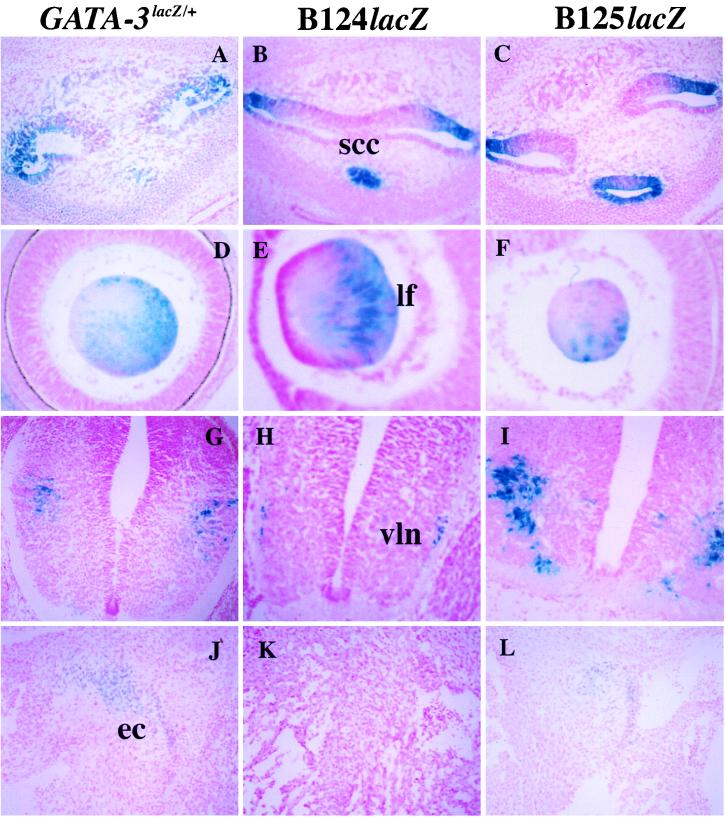
FIG. 4.
Expression in the ear, eye, nervous system, and heart of GATA-3LacZ/+ and B124lacZ and B125lacZ embryos. A through C, transverse sections at the level of the developing ear showing expression of β-galactosidase in saccules of the semicircular canals (scc) in the inner ear of 12.5-dpc embryos. D through F, sagittal sections at the level of the developing eye of 12.5-dpc embryos showing expression of β-galactosidase in the posterior lens fibers (lf). G through I, transverse sections at the level of the thoracic spinal cord. Neurons expressing β-galactosidase are located in the ventrolateral (vln) areas of the spinal cord in these 13.5- to 14.5-dpc embryos. J through L, transverse sections at the level of the developing heart. Note that the endocardial cushions (ec) express β-galactosidase in both the GATA-3LacZ/+ and B125lacZ 13.5-dpc embryos, but not in the B124lacZ embryo at the same stage.
In the heart, where only weak expression was detected in our previous in situ experiments (8), sectioning of the lacZ gene-targeted embryos showed that expression was confined to cells that contribute to the endocardial cushions at the atrioventricular junction as well as in the outflow track (Fig. 4J) and was temporally visualized only between 10.5 and 14 dpc. Caudal cervical cross-sections revealed that β-galactosidase was expressed in the developing thyroid gland (data not shown), which was also not evident from in situ hybridization. Other sites of expression included the mammary gland (Fig. 3G) and the hair bulb and dermal papillae of hair follicles (Fig. 3E). Since expression in the mammary gland and hair follicles was also observed for a 6-kbp GATA-3 promoter reporter transgene (20), these data indicate that these two sites were simply overlooked in the earlier in situ hybridization analysis (8).
GATA-3 expression from 540- and 625-kbp YAC reporter transgenes.
The expression patterns of the GATA-3LacZ/+ mice revealed by whole-mount staining (Fig. 2 and 5) and tissue sectioning (Fig. 3 and 4) were compared to the patterns reflected in two B125lacZ YAC transgenic lines as well as in two B124lacZ transgenic lines at multiple stages during embryogenesis. Only the most informative sections are reproduced here to highlight specific tissue or temporal expression differences among the three different kinds of mice examined (GATA-3LacZ/+ knock-in mice and B124LacZ or B125LacZ YAC transgenic embryos).
FIG. 5.
Expression of YAC B125lacZ during urogenital and renal development. (A) A 11.5-dpc B125Z71 embryo displaying lacZ expression in the mesonephric duct and the ureteric bud (ub), which gives rise to the adult kidney. The ureteric bud is beginning to expand into the metanephric blastema (mb). The urogenital sinus (us) also stains. (B) A 12.5-dpc embryo displaying expression in the metanephric blastema (mb) that has differentiated from the ureteric bud to form the primitive renal pelvis. This embryo also displays transgene expression in remnants of the mesonephros (ms) and the mesonephric duct (md). (C, D, and E) The same region, or the isolated organs, from 14.5-, 15.5-, and 16.5-dpc embryos, respectively. The mesonephros has regressed, while the metanephros (the definitive kidney; ki) continues to differentiate, enlarge, and gradually ascend rostrally from the us site of origin. The metanephric (collecting) tubule continues to divide to form the collecting system and expresses intense β-galactosidase activity (D, E, and F). (F) A dissected kidney attached to its ureter (u), and the testis (te), epididymis (ep), and vas deferens (v) (the ep and v are derived from the mesonephric duct) of an embryo at 17.5 dpc; strong β-galactosidase activity is detected throughout the collecting system and in the ep.
For the B125LacZ YAC studies, transgenic line Z71 was chosen for further analysis because it bears only one transgene copy. NotI and SfiI mapping of this line showed not only that the 5′ breakpoint in the Z71 transgene lies between 125 and 150 kbp from the GATA-3 transcription start site (Fig. 1B and C) but also that the entire 3′ end (175 kbp) is intact (Fig. 1D). Line Z89 was also selected for further analysis because, in addition to providing independent confirmation for genuine sites of B125 YAC expression, it bears at least one substantially intact copy of the entire 625-kbp YAC (Fig. 1E). Both the Z71 and Z89 transgenic YAC expression patterns did not significantly differ from one another (other than ectopic staining in the lungs of Z71 embryos; Table 1 and Fig. 2D to I), leading us to conclude that many of the regulatory elements controlling GATA-3 transcription (see below) must lie within an approximately 300-kbp radius surrounding the gene described by transgene B125Z71 (containing, at most, 150 kbp of 5′ end as well as an intact 175-kbp 3′ end). The B124LacZ YAC contains 85 kbp less 3′-end genomic information than the B125lacZ YAC, and thus comparison of the two expression patterns allowed us to further delimit the position of 3′ GATA-3 regulatory sequences (Table 1 and Fig. 3 and 4).
TABLE 1.
Expression of GATA-3a
| Expression site | Branchial arches | Genital tubercle/ cloaca | Mammary gland | Hair follicles | Vestibulo-cochlear ganglia | CNSe | Eye | Ectoplacental cone/placenta | Thyroid gland | MT/Kf | Semicircular canals | Endocardial cushions | Sympathetic trunk | Adrenal medulla | Thymus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| In situb | + | + | − | ? | + | + | + | + | − | + | + | ? | + | + | + |
| GATA-3LacZ/+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| GATA-3 promoterc | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − |
| C4lacZd | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − |
| B124lacZ | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − |
| B125lacZ | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
Sites of expression are labeled +, sites where expression was not detected are labeled −, and sites where expression was ambiguous in previous studies are labeled with a ?.
−4500/+1012 GATA-3 promoter fragment (20).
YAC C4lacZ transgenic mice (17).
Midbrain, hindbrain, and spinal cord.
MT/K, mesonephric and metanephric tubules and kidney.
To precisely define sites of β-galactosidase expression at the cellular level, we sectioned GATA-3LacZ/+, B124lacZ, and B125lacZ transgenic embryos at 12.5 and 14.0 dpc. These observations showed that the CNS patterns of the YAC transgenic animals were coincident with that in the germ-line mutant lacZ-targeted animals (Fig. 2). At 12.5 dpc, expression in the brain was confined to the mantle layers of the diencephalon, the mesencephalon, and the pontine region of the metencephalon. Expression was also found in the myelencephalon, which forms the medulla oblongata (Fig. 2, and data not shown), and in ventrolateral neurons within the spinal cord (Fig. 4G). However, in the cervical region of the spinal cord, neither YAC was expressed in neurons of the sympathetic trunk (e.g., Fig. 3D).
Comparisons among the GATA-3LacZ/+ knock-in and B124lacZ and B125lacZ YAC transgenic embryos demonstrated that most of the sites of expression are coincident (Table 1 and Fig. 2 to 4). The tissues in which neither YAC transgene is expressed are the thymus (see the Discussion) and specific differentiated lineages contributing to the sympathetic nervous system (e.g., in the adrenal gland [Fig. 3B] and the sympathetic trunk [Fig. 3D]). Therefore, regulatory elements controlling expression in those cell types are not present in either YAC.
In summary, analysis of the expression patterns of the lacZ transgenes and the GATA-3LacZ/+ knock-in mice allows further refinement of the position of cis-regulatory elements directing GATA-3 expression. When we compared the profile to that previously detected by in situ hybridization (8), a promoter GATA-3 transgene (20), or a smaller, 120-kbp YAC reporter transgene (C4lacZ; reference 17), we found that a number of previously unresolved cis-regulatory elements for GATA-3 could be roughly positioned within the locus (Table 1 and Fig. 6). Taken together, detailed comparisons to earlier data reveal that the elements controlling the expression of GATA-3 in the ectoplacental cone, the CNS, the eye, and the thyroid gland all lie within the smallest (120 kbp) C4 YAC (17) but outside the boundaries described by the GATA-3 promoter transgene, which is expressed in the branchial arches, genital tubercle/cloaca, mammary glands, and whisker follicles (20).
FIG. 6.
Summary of localization of GATA-3 regulatory elements. The top of the diagram depicts the genomic GATA-3 locus (bold line), while the bottom shows the positions of several regionally defined regulatory elements in the locus. The 5′-most element identified here contains the urogenital element(s) and was defined by the 5′ breakpoint in the B125Z71 transgene at approximately −150 kbp and at its 3′ boundary by the 5′ end of YAC C4Z (17) at −35 kbp. The CNS element(s) is defined by the 5′ end of the C4Z transgene at −35 kbp and the NotI site at −4.6 kbp (17, 20). The branchial arch (BA) and genital tubercle (GT) elements were defined previously (20). The element conferring GATA-3 expression in the endocardial cushions was originally defined by the differences in the 3′ boundaries of the B124 (+85 kbp) and B125 (+175 kbp) lacZ transgenes (above), and more recent studies have localized the element to within +105 and +145 kbp 3′ to the gene (see Discussion and reference 10).
Both B125lacZ and B124lacZ YAC transgenes reproducibly express β-galactosidase in the umbilical vessels (data not shown) and in the developing urogenital system, particularly in the mesonephric duct as well as in the metanephric duct and ureteric bud (Fig. 5). Therefore, the elements controlling GATA-3 expression in the umbilical vessels and in the mesonephric and the metanephric ducts lie beyond the boundaries of the 120-kbp C4 YAC but inside the boundaries described by both the B124lacZ and B125lacZ YACs (Fig. 1E). Since the C4 and B124 YACs share the same 3′ genomic boundary, this deductively narrows the positions of these elements to between −35 and −450 kbp 5′ to the gene.
Neither of the B124lacZ transgenic lines expresses β-galactosidase in the endocardial cushion tissues in the heart (Fig. 4K), while GATA-3LacZ/+ and both B125lacZ lines express β-galactosidase there (Fig. 4J and L), demonstrating that the element(s) directing GATA-3 expression in the endocardial cushion lie in very distant 3′ flanking sequences. These results delimit the position of a positive endocardium-specific regulatory element for the GATA-3 gene to between +85 and +175 kbp 3′ to the GATA-3 gene (also see the Discussion). Finally, these experiments demonstrate deductively that the cis-regulatory elements controlling GATA-3 in a specific subset of PNS derivative lineages (the sympathetic trunk and the adrenal medulla) lie beyond either 450 kbp 5′ to or 150 kbp 3′ to the GATA-3 structural gene.
DISCUSSION
During the last decade, numerous technical advances have allowed us to dissect and decipher the effects of specific genetic perturbations introduced into the mouse genome by homologous gene targeting. Homozygous loss of GATA-3 represents one typical category of germ line-targeted mutants: embryos missing GATA-3 activity survive until midgestation but suffer multiple phenotypic abnormalities at the time of death, including partially penetrant or incompletely expressive malformation of the spinal cord, brain, and jaw (32). The homozygous null mutant embryos also display other fully penetrant defects (e.g., in either blood vessel formation or vascular connection to internal organs) at the time of demise in utero. Additionally, from other studies we know that defects in GATA-3 function profoundly affect the differentiation of cell lineages which mature later in development (e.g., during T-cell differentiation; reference 43), and these same defects would likely be manifested in vivo were the mutant embryos to survive long enough to initiate thymic organogenesis.
While gene ablation studies can sometimes provide a definitive answer to the question of the functional significance of a particular gene of interest, early embryonic lethal mutations can also serve as a significant barrier to further analysis of the gene in lineages that develop after the time of death. In order to circumvent this, various strategies, such as conditional knockouts, have been developed recently. Other alternate avenues include tissue- or lineage-specific gene ablation and the analysis of hypomorphic alleles.
An alternative strategy distinct from those mentioned above would be to define the entire genetic locus by isolation of completely complementing transgenes, thereby delimiting the boundaries for all chromatin and transcriptional elements necessary to specify the complete expression pattern of the gene of interest. With these genomic sequences in hand, one could then methodically refine the positions of individual tissue-specific regulatory regions and then examine the in vivo consequences of deleting specific control elements (and therefore ablation of specific cell lineages) from the fully complementing transgene in a null mutant background. Since the developmental expression profile of GATA-3 is dynamic in both space and time, and since previous attempts to define the boundaries of the locus by using either plasmid expression or small YAC constructs have failed, we have examined even larger GATA-3 YACs in this study. The experiments presented here underscore a major concern with this strategy of gene rescue: even the largest (625 kbp) GATA-3 transgene lacks the patterning element(s) that are critical for its expression in certain sympathetic ganglia and the adrenal gland.
The studies presented here provide direct evidence that an upper limit for the mouse GATA-3 locus has not yet been defined. However, we explicitly note that there are several caveats to the overall conclusions, since analysis of transgenes of this size presents several unique challenges. First, it is clearly impossible to exhaustively characterize every line with respect to the detailed internal structure of the integrated transgenes. Thus, a small deletion or inversion (note that even 1% of the B124 or B125 YAC transgenes would comprise 5 or 6 kbp) would probably be undetected in the PFGE assays. Second, and along the same lines as the first complication, regardless of the detail applied to both conventional and PFGE mapping, we cannot ever completely eliminate the possibility that the original YACs differ slightly in structure from the genomic locus. While one can surmount objections regarding possible position-of-integration effects by examining multiple transgenic lines, as we have done here, a third caveat regarding the general utility of this strategy is that one can position regulatory elements by mapping breakpoints in the transgenes only if a breakpoint happens to occur at an informative position. In other words, the breakpoint mapping strategy for positioning of regulatory elements within the locus depends on fortuitous opportunity rather than on intentional, directed mutagenesis.
Despite these multiple complications, this strategy has allowed us to position several regulatory elements that confer proper patterning and temporal expression to a target gene without reference to surrogate methods, and indeed the overall strategy appears well suited for defining very distant constituents of a genetic locus. For example, we demonstrated previously that a 120-kbp YAC transgene (17) confers expression in the CNS, whereas a 6-kbp mGATA-3 promoter construct contained regulatory elements that were able to confer expression in other sites where the gene is normally detected (20).
Given that the 120-kbp C4 YAC did not confer expression at a number of sites where GATA-3 is known to be transcribed (17), we examined the expression patterns of two much larger YACs and compared them with the pattern obtained from targeting lacZ into the endogenous GATA-3 gene locus (GATA-3LacZ/+). The GATA-3LacZ/+ knock-in embryos showed expression in all of the tissues where GATA-3 is synthesized, including those where previous studies had indicated either ambiguous or weak expression (Table 1), with the exception of the thymus. Neither the heterozygous germ line mutant nor the YAC transgenic animals display expression in the thymus, one of the most prominent sites of midembryonic GATA-3 expression (41–43), indicating a general failure of lacZ expression in T lymphocytes. While lacZ could not be detected in thymocytes of any of these animals by normal X-Gal staining protocols (Materials and Methods), its detection was made possible by using a far more sensitive reagent, FACS-Gal (12). We do not know, at the present time, why detection of lacZ expression in the thymus is fraught with such complications.
Transgenic animals bearing the 540-kbp B124lacZ and 625-kbp B125lacZ YACs showed several sites of normal GATA-3 embryonic expression in addition to those found in mice bearing smaller transgenes; these sites include the heart, umbilical vessels, and mesonephric and metanephric ducts (Table 1). While the position(s) of regulatory elements within the B125 YAC that are required for the generation of this expression pattern are not yet finely localized, they must lie beyond the limits of C4 YAC but within the boundaries described by B125lacZ transgene Z71, which is broken at the 5′ end. The evidence indicates that the elements directing the expression of GATA-3 in the umbilical vessels, the inner ear, and the mesonephros and metanephros reside between −35 kbp (the 5′ end of the C series of YACs, including the C4lacZ; reference 17) and −150 kbp (the approximate 5′ breakpoint in B125lacZ line 71) 5′ to the GATA-3 structural gene (Fig. 1E and Fig. 6). Expression in the endocardial cushions of the heart is regulated by an element within the 85-kbp sequences that differ between the B124 and B125 YAC 3′ ends. More recent studies have refined the position of this element to an approximately 45-kbp sequence within this interval (Fig. 6; reference 10), and similarly, the CNS element has now been refined to a single 1-kbp fragment lying within the −6 to −35 kbp interval (22). Continued analysis should allow us to resolve the precise position of the urogenital and heart element(s) as well as the identity of putative upstream developmental effectors (10, 22). In this manner, we hope to determine the epistatic relationships in the regulatory cascades that lead to GATA-3 function in specific organs.
Recapitulating a complex embryonic gene expression pattern that parallels normal GATA-3 expression can be achieved by simply extending the limits of DNA surrounding the gene. This observation conceals several implications. First, the data suggest that expression of the transgene in these tissues is principally controlled by positive transcriptional regulatory elements, since extending the sequences under scrutiny from those lying very close to the gene (20) to ones lying substantially further away adds to the pattern and consistency of expression established by smaller constructs. Second, the positive regulatory elements appear to be discrete, since addition of new segments of DNA to those analyzed previously confers reproducible β-galactosidase accumulation at sites where GATA-3 is normally expressed.
To place the present analysis in perspective, it might be instructive to compare these data to other better characterized genetic regulatory models. The present data indicate that the full extent of the GATA-3 locus includes at least 150 kbp of 3′ and 150 kbp of 5′ flanking sequence information (Fig. 6). If we assume that the regulatory elements controlling the (presently unaccounted for) expression in the sympathetic chain and adrenal medulla lie immediately outside of the B125 YAC, the locus must be minimally 325 kbp in size, including the 25-kbp structural gene (8). Thus, the GATA-3 locus is at least four times the size of the human β-globin gene locus (35), at least three times larger than the known extent of any of the murine hox gene clusters (e.g., see reference 9), and even somewhat larger than the entire mating type locus complex on yeast chromosome III (e.g., see reference 36). Although other genes have been inferred to have even more distant regulatory sequences (from mutant mapping, breakpoint inversion, and similar genomic mapping studies [references 2 and 3 and references therein]), the mouse GATA-3 locus at present defines the most distant regulatory elements contributing to a single gene locus that has been characterized by direct physical isolation.
Many investigations have not shown that transcriptional control elements linked to reporter genes can confer the wild-type expression pattern to at least a subset of the appropriate tissues where and when that gene is normally expressed. Nonetheless, despite a geometric increase during the last decade in documenting regulatory elements that are required for control of metazoan transcription, phenotypic rescue of recessive loss-of-function mutants in the mouse has been achieved in only surprisingly few cases. The studies presented here show that the boundaries of mammalian loci which display complex embryonic expression patterns may extend much further than has previously been assumed and thus may represent one rather daunting hurdle that could be encountered in attempts to rescue other developmental regulatory genes.
ACKNOWLEDGMENTS
Ganesh Lakshmanan and Ken Lieuw contributed equally to this work, and both should be considered first authors.
We thank Jie Fan for outstanding technical assistance, members of the Engel lab, in particular Jorg Bungert, Ko Onodera, and Yinghui Zhou, for insightful discussions and help, and Gauri Tilak for assistance with transgenic analysis. We also thank Rick Gaber, Rob Nakamura, and Hong Liang for advice about yeast and for reagents.
This work was supported by an MSTP grant to Northwestern University (NIH T32 GM08152; to K.H.L.), a Scanlon fellowship from Evanston Hospital (to G. L.), research grants from the NWO (The Netherlands; to F.G. and A.K.) and the National Institutes of Health (GM28896; to J.D.E.).
REFERENCES
- 1.Arceci R J, King A A J, Simon M C, Orkin S H, Wilson D B. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedell M A, Brannan C I, Evans E P, Copeland N G, Jenkins N A, Donovan P J. DNA rearrangements located over 100 kb 5′ of the Steel (Sl)-coding region in Steel-panda and Steel-contrasted mice deregulate Sl expression and cause female sterility by disrupting ovarian follicle development. Genes Dev. 1995;9:455–470. doi: 10.1101/gad.9.4.455. [DOI] [PubMed] [Google Scholar]
- 3.Bedell M A, Jenkins N A, Copeland N G. Good genes in bad neighborhoods. Nat Genet. 1996;12:229–232. doi: 10.1038/ng0396-229. [DOI] [PubMed] [Google Scholar]
- 4.Bungert J, Dave U, Lim K C, Lieuw K H, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human beta-globin gene switching by locus control region elements HS2 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 5.Dillon N, Grosveld F. Gene transcription: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. [Google Scholar]
- 6.Dorfman D M, Wilson D B, Bruns G A P, Orkin S H. Human transcription factor GATA-2: evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 7.Feinberg A P, Vogelstein B. A technique for radiolabeling RNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 8.George K M, Leonard M W, Roth M E, Lieuw K H, Kioussis D, Grosveld F, Engel J D. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 9.Giampaolo A, Acampora D, Zappavigna V, Pannese M, D’Esposito M, Care A, Faiella A, Stornaiuolo A, Russo G, Simeone A, et al. Differential expression of human HOX-2 genes along the anterior-posterior axis in embryonic central nervous system. Differentiation. 1989;40:191–197. doi: 10.1111/j.1432-0436.1989.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 10.Gu, Y., and J. D. Engel. Unpublished data.
- 11.Hanks M, Wurst W, Anson-Cartwright L, Auerbach A, Joyner A L. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 12.Hendriks, R. W., M. C. Nawijn, J. D. Engel, F. Grosveld, and A. Karis. Transcription factor GATA-3 is involved in two distinct maturation steps in early T cell development. Submitted for publication.
- 13.Ito E, Toki T, Ishihara H, Ohtani H, Gu L, Yokoyama M, Engel J D, Yamamoto M. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature. 1993;362:466–469. doi: 10.1038/362466a0. [DOI] [PubMed] [Google Scholar]
- 14.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornhauser J M, Leonard M W, Yamamoto M, LaVail J H, Mayo K E, Engel J D. Temporal and spatial changes in GATA transcription factor expression are coincident with development of the chicken optic tectum. Mol Brain Res. 1994;23:100–110. doi: 10.1016/0169-328x(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 16.Lakshmanan, G., and J. D. Engel. Unpublished data.
- 17.Lakshmanan, G., K. H. Lieuw, F. Grosveld, and J. D. Engel. Partial rescue of GATA-3 using yeast artificial chromosome transgenes. Dev. Biol., in press. [DOI] [PubMed]
- 18.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T. GATA-4, -5, and -6 constitute a new subfamily of transcription factors in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 19.Lieuw, K. H., and J. D. Engel. Unpublished data.
- 20.Lieuw K H, Li G, Zhou Y, Grosveld F, Engel J D. Temporal and spatial control of murine GATA-3 transcription by promoter-proximal regulatory elements. Dev Biol. 1997;188:1–16. doi: 10.1006/dbio.1997.8575. [DOI] [PubMed] [Google Scholar]
- 21.Lieuw K H, Roth M E, Dzierzak E, George K M, Karis A, Leonard M W, Lim K-C, Pandolfi P P, Grosveld F, Engel J D. Expression and regulation of GATA-2 and GATA-3 in hematopoietic and other cell lineages. In: Stamatoyannopoulos G, editor. Biology of Hematopoiesis and Stem Cell Gene Transfer. Andover, United Kingdom: Intercept Press; 1995. pp. 15–35. [Google Scholar]
- 22.Lim, K.-C., and J. D. Engel. Unpublished data.
- 23.Liu Q, Bungert B, Engel J D. Mutation of gene-proximal regulatory elements disrupts human epislon-, gamma-, and beta-globin expression in yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA. 1997;94:169–174. doi: 10.1073/pnas.94.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin D I K, Orkin S H. Transcriptional activation and DNA-binding by the erythroid factor [GATA-1] Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 25.Martin D I K, Zon L I, Mutter G, Orkin S H. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- 26.McDevitt M A, Shivdasani R A, Fujiwara Y, Yang H, Orkin S H. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc Natl Acad Sci USA. 1997;94:6781–6785. doi: 10.1073/pnas.94.13.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molkentin J D, Lin Q, Duncan S A, Olson E N. Requirement of the transcription factor GATA-4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 29.Nagai T, Harigae H, Ishihara H, Motohashi H, Minegishi N, Hayashi N, Gu L, Andres B, Engel J D, Yamamoto M. Transcription factor GATA-2 is expressed in erythroid, early myeloid and CD34+ human leukemia-derived cell lines. Blood. 1994;84:1074–1084. [PubMed] [Google Scholar]
- 30.Ng A, George K M, Engel J D, Linzer D I H. A GATA factor is necessary and sufficient for transcriptional regulation of the murine placental lactogen I gene promoter. Development. 1994;120:3257–3266. doi: 10.1242/dev.120.11.3257. [DOI] [PubMed] [Google Scholar]
- 31.Oosterwegel M, Timmerman J, Leiden J, Clevers H. Expression of GATA-3 during lymphocyte differentiation and mouse embryogenesis. Dev Immunol. 1992;3:1–11. doi: 10.1155/1992/27903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandolfi P P, Roth M E, Karis A, Leonard M W, Dzierzak E, Grosveld F G, Engel J D, Lindenbaum M H. Targeted disruption of the GATA-3 gene causes severe abnormalities in the nervous system and in fetal liver haemotopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 33.Romeo P-H, Prandini M-H, Joulin V, Mignotte V, Prenant M, Vainchenker W, Marguerie G, Uzan G. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990;344:447–449. doi: 10.1038/344447a0. [DOI] [PubMed] [Google Scholar]
- 34.Steger D J, Hecht J H, Mellon P L. GATA-binding proteins regulate the human gonadotropin α-subunit gene in the placenta and pituitary gland. Mol Cell Biol. 1994;14:5592–5602. doi: 10.1128/mcb.14.8.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strouboulis J, Dillon N, Grosveld F. Developmental regulation of a complete 70-kb human beta-globin locus in transgenic mice. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 36.Szeto L, Fafalios M K, Zhong H, Vershon A K, Broach J R. Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev. 1997;11:1899–1911. doi: 10.1101/gad.11.15.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi S, Onodera K, Motohashi H, Suwabe N, Hayashi N, Yanai N, Nabesima Y, Yamamoto M. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J Biol Chem. 1997;272:12611–12615. doi: 10.1074/jbc.272.19.12611. [DOI] [PubMed] [Google Scholar]
- 38.Tsai F-Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 39.Whiting J, Marshall H, Cook M, Krumlauf R, Rigby P W J, Stott D, Allemann R K. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 1991;5:2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]
- 40.Winston J F, Chumley F, Fink G R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto M, Ko L J, Leonard M W, Beug H, Orkin S H, Engel J D. Activity and tissue-specific expression of the transcription factor NF-E1 [GATA] multigene family. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Gu L, Romeo P-H, Bories D, Motohashi H, Yamamoto M, Engel J D. Human GATA-3 trans-activation, DNA binding, and nuclear localization activities are organized into distinct structural domains. Mol Cell Biol. 1994;14:2201–2212. doi: 10.1128/mcb.14.3.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng W, Flavell R A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Q Y, Palmiter R D. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell. 1995;83:1197–1209. doi: 10.1016/0092-8674(95)90145-0. [DOI] [PubMed] [Google Scholar]