Abstract
Lytic reactivation of Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, from latency requires transcriptional transactivation by the viral protein RTA encoded by the ORF50 gene. Very little is known about how RTA functions and the cellular factors that may be involved in its transactivation function. Using the yeast two-hybrid system, we have identified a human cellular protein that can interact with KSHV RTA. The cellular protein, referred to as the human hypothetical protein MGC2663 by GenBank, is encoded by human chromosome 19. This protein is 554 amino acids (aa) in size and displays sequence similarity with members of the Krueppel-associated box–zinc finger proteins (KRAB-ZFPs). MGC2663 expression could be detected in all primate cell lines tested, and its expression level was neither stimulated nor inhibited by RTA. MGC2663 specifically synergizes with RTA to activate viral transcription, and overexpression of MGC2663 in the presence of RTA further enhances RTA transactivation of several viral promoters that were identified as targets for RTA. Coimmunoprecipitation and pull-down assays further demonstrated that MGC2663 interacts with RTA both in vivo and in vitro, and the N-terminal 273 aa of KSHV RTA and the potential zinc finger domain of MGC2663 are required for their interaction. Our results indicate that this novel human cellular protein, MGC2663, named K-RBP (KSHV RTA binding protein) due to its RTA binding feature, specifically interacts with the KSHV RTA protein and functions as a cellular RTA cofactor to activate viral gene expression. Though its normal cellular function needs to be further studied, K-RBP may play a significant role in mediating RTA transactivation in vivo.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, was first discovered in AIDS-associated Kaposi's sarcoma (KS) tissue in 1994 (4). Since then, KSHV DNA has been detected in all other forms of KS, including classic, endemic, and posttransplantation KS, as well as in all clinical stages of KS (5, 6, 24, 31). Moreover, detection of KSHV in peripheral blood mononuclear cells of human immunodeficiency virus (HIV)-infected individuals predicts the subsequent development of KS lesions (41). KSHV is also associated with two other neoplastic disorders, primary effusion lymphoma, also termed body cavity-associated lymphoma, and multicentric Castleman's disease (3, 12, 35). Since KS cell lines usually do not harbor viral DNA, several KSHV-infected human B-cell lines were established from primary effusion lymphomas for molecular studies (2, 28). Viral gene expression is highly restricted in these cell lines, indicating that the cells are latently infected. Certain inducing agents, such as n-butyrate and tetradecanoyl phorbol acetate (TPA), can induce lytic replication of KSHV from latency (28, 42).
The entire genomic sequence of KSHV has been reported, and the virus has been classified in the Gamma 2 herpesvirus (Rhadinovirus) subfamily (29, 31). Members of this subfamily, including herpesvirus saimiri and murine herpesvirus 68, share a common genome structure in which the encoding region of a central open reading frame (ORF) is flanked by multirepetitive high-GC-content DNA (29). KSHV reading frames are assigned the same number as their homologous versions in herpesvirus saimiri. Those genes unique to KSHV are given the prefix K (for KSHV) and numbered sequentially (25). Based on expression kinetics, herpesvirus genes can be categorized into four groups: latent, immediate-early (IE), early, and late genes. Expression of IE genes does not require any viral protein synthesis and is induced immediately after reactivation. The genes of this group usually encode regulatory proteins that up- or down-regulate the expression of various viral and cellular genes and therefore play a crucial role in the switch from latent to lytic replication.
The closest known relative of KSHV in humans is Epstein-Barr virus (EBV), whose mechanism of reactivation has been studied extensively (10, 27, 43). In this model, reactivation from latency is controlled by two IE genes, BZLF1 and BRLF1, whose products, ZEBRA and RTA, respectively, are transcription activators that augment expression of downstream viral target genes (14, 16, 21, 38). KSHV also has a homolog of the EBV BRLF1 gene, which is the ORF50 gene. Expression of KSHV ORF50 can be detected as early as 1 h after induction of viral replication and in the presence of the protein synthesis inhibitor cycloheximide (18, 26, 37, 45). The ORF50 gene has been shown to be crucial for viral replication and gene transcription (13, 18, 19, 23, 36). Due to its similarity to the EBV gene, its gene product has been referred to as the KSHV RTA. As a typical transcription activator, KSHV RTA contains an N-terminal basic domain and a C-terminal acidic activation domain located from amino acid (aa) 527 to 634. The activation domain consists of three partially overlapping hydrophobic motifs which are homologous with the transcriptional activation domains of many transcription factors found in viruses, yeast, and mammalian cells (39). It has been reported that RTA activates expression of early genes, late genes, and polyadenylated nuclear (PAN) RNA. The affected early genes include ORF57, K8, and the gene for virus-encoded interleukin 6. The late genes include the small viral capsid antigen gene (also known as ORF65) (17, 19, 20, 32, 36, 39). An RTA-responsive element (RRE) that can potentially form a palindrome has been identified on both the ORF57 and K8 gene promoters (20, 39). RTA was also found to autoregulate its own expression through a mechanism in which Oct-1 binding plays an important role (8, 30). These observations strongly suggest that RTA may interact directly or indirectly with its target sequences to stimulate transcription and that it serves as a molecular switch for KSHV reactivation. Indeed, RTA was recently found to bind directly to sequences in the PAN RNA promoter region (34) and the RRE palindromes in ORF57 and K8 promoters (20).
It has also recently been reported that both CREB-binding protein (CBP) and histone deacetylase (HDAC) regulate RTA function (15), and it has been suggested that cellular proteins are involved in RTA functions. Identification and characterization of these cellular cofactors for RTA would be important for deciphering the mechanism involved in the latent-lytic transition during viral reactivation. We performed a yeast two-hybrid screening for RTA binding proteins (RBPs) with an EBV-transformed B-cell cDNA library using an RTA C-terminal mutant ORF50c as bait. A single protein of unknown function was found to interact with RTA consistently. This protein is identical to a hypothetical cellular protein termed MGC2663 (from GenBank clone MGC:2663). The identified protein consists of 554 aa, and it appears to encode an N-terminal Krueppel-associated box (KRAB) and a C-terminal zinc finger domain. Overexpression of MGC2663 stimulated RTA transactivation of various target viral promoters in different mammalian cells. Both in vivo and in vitro interaction between RTA and MGC2663 were confirmed by coimmunoprecipitation and pull-down assays. Moreover, this association was found to be independent of the C-terminal activation domain of RTA but required the RTA N terminus and the MGC2663 zinc finger domain. Our data strongly suggests that this novel cellular protein, MGC2663, also referred to as K-RBP (for KSHV RBP), may be an important cofactor involved in KSHV RTA transactivation and viral reactivation.
MATERIALS AND METHODS
Plasmids.
All PCR-generated plasmids and fusion protein expression plasmids were sequenced to verify the cloned inserts and their reading frames. Plasmid pGBK/ORF50c, which was used as bait for yeast two-hybrid screening, was generated from an RTA clone, pcDNA-ORF50c, consisting of the N-terminal 671 aa of RTA followed by the K8.1 region from nucleotide (nt) 76433 to 76690 (encoding 86 aa). The pcDNA-ORF50c clone was generated to study the effect of K8.1 sequence on RTA transactivation function; it was cloned from a potential spliced product observed in a 3′ rapid amplification of cDNA ends study of the ORF50-K8-K8.1 region. This clone retains both RTA DNA binding and transactivation domains but is not active in transactivation (unpublished data). The insert of this clone was released with NotI and then cloned into the NcoI/SmaI sites of the pGBKT7 DNA-BD vector (Clontech, Palo Alto, Calif.). This insert was fused downstream of the N-terminal 147-aa DNA binding domain (DNABD) of Gal4 to generate plasmid pGBK/ORF50c for yeast two-hybrid screening. Plasmid pGBK/ORF50 contains the full-length 691-aa RTA coding region fused to the Gal4 DNA BD. Another plasmid that was used in the study, pGBK/ORF50AD2, has an RTA insert with a deletion of 101 aa at the C terminus. The plasmids pGBK-53 and pGAD-T encode the Gal4 DNA BD/murine p53 and Gal4 activation domain (AD)/simian virus 40 large T antigen, respectively. These two clones are known to interact in a yeast two-hybrid assay and were used together as a positive control or in combination with our Gal4 fusion constructs as negative controls.
The pcDNA-MGC2663 mammalian expression clone was generated from the clone pACT-clone7 that was selected from a pACT vector-based cDNA library. The insert of pACT-clone7 was released with BamHI/BglII and inserted into the XhoI/BamHI sites of the pcDNA3.1(−) vector (Invitrogen, Carlsbad, Calif.) to generate pcDNA-MGC2663 or into the EcoRI/BglII sites of the pCMV-HA vector (Clontech) to generate pHA-MGC2663. Another MGC2663 yeast expression clone, pGAD-clone7, was generated by releasing the same BamHI/BglII insert from pACT-clone7 and inserting it into the NcoI/BamHI sites of the pGADT7 AD vector (Clontech) following the coding sequence for the Gal4 AD from aa 768 to 881. The His-tagged mammalian fusion expression clone pcDNA4His-2663 was generated from pcDNA-MGC2663 by releasing the BstUI/XhoI insert of pcDNA-MGC2663 and inserting it into the EcoRV/XhoI sites of the pcDNA4HisMax vector (Invitrogen). The bacterial expression clone pET-MGC2663 was also generated by ligating the BstUI/XhoI insert of pcDNA-MGC2663 into the NheI/XhoI sites of the pET28a vector (Novagen, Madison, Wis.). The deletion clones pcDNA-MGC2663/242 and pHA-MGC2663/242, which encode only the N-terminal 242 aa of MGC2663, were obtained by inserting the small XhoI/NdeI fragment of pACT-clone7 into the XhoI/EcoRV sites of pcDNA3.1(−) and inserting the SfiI/NdeI fragment of pHA-MGC2663 into the SfiI/XhoI sites of pCMV-HA, respectively. Plasmid pEGFP-MGC2663, which expresses MGC2663-green fluorescent protein (GFP) fusion protein, was generated by inserting the MGC2663 XhoI/EcoRI fragment of pcDNA-MGC2663 into the corresponding sites of the pEGFP-N1 vector (Clontech).
Clones pcDNA-ORF50, which encodes the full-length RTA, and pCMV-Tag50, which encodes the Flag tag-fused RTA, were described previously (9, 39). Clone pGEX-RTA, which expresses the glutathione-S-transferase (GST)-RTA fusion protein, was generated by inserting the NotI/EcoRI fragment that contains the entire ORF50 coding region obtained from an ORF50 clone, pcDNA-ORF50R, into the EcoRI/NotI sites of pGEX-5X-3 (Amersham Pharmacia, Piscataway, N.J.). Clone pGEX-RTA/273, which expresses the GST-fused RTA N-terminal 273 aa, was derived from pGEX-RTA. It was generated by digesting pGEX-RTA with NcoI/NotI to eliminate the small fragment. Plasmid pcDNA-ORF50/548, which encodes the N-terminal 548 aa of RTA, was generated from pcDNA-ORF50 by digestion with KpnI to eliminate the 470-bp fragment.
Reporter plasmids p57Pluc1, pK8Pluc, pMIPPluc, and p50LPluc contain PCR-cloned promoter regions of ORF57 (from nt 81556 to 82008), ORF K8 (from nt 73851 to 74849), ORF K6 (vMIP-I) (from nt 27783 to 27422), and ORF50 (from nt 70011 to 71576), respectively. The promoter fragments were inserted into the NcoI/SacI sites upstream of the luciferase reporter gene of the pGL3-Basic vector (Promega, Madison, Wis.) as described earlier (9, 39). Plasmid pGL3-promoter, which was used as a control, was from Promega. Plasmid pHIVLTR-luc contains the PCR-generated HIV long terminal repeat (LTR) region upstream of the luciferase gene on the pGL3-Basic vector. Plasmid pcDNA-Tat expresses the full-length HIV Tat protein.
Yeast two-hybrid selection.
Plasmid pGBK/ORF50c was transformed into Saccharomyces cerevisiae AH109, which contains three reporter genes, his3, ade2, and lacZ, by the lithium acetate-mediated method (yeast protocols handbook; Clontech). The expression of yeast reporter genes was under the control of three completely heterologous upstream activation sites (UAS) and promoter elements—gal1, gal2, and mel1, respectively. A pACT-based EBV-transformed human B-cell cDNA library tagged with the Gal4 AD (Clontech) was then transformed into a pGBK/ORF50c yeast transformant for two-hybrid screening. After a 3- to 5-day incubation at 30°C, yeast colonies that grew on synthetic dropout (SD) plates lacking Ade, His, Leu, and Trp were streaked at least twice into a single colony on SD agar lacking Leu and Trp to allow clones that contained more than one library plasmid to segregate and then tested on SD agar lacking Ade, His, Leu, and Trp to verify the phenotype. Positive colonies were transferred to nitrocellulose filters, permeabilized on the filters by freezing-thawing, and then assayed for β-galactosidase (β-Gal) activity. Positive blue colonies were picked and grown in SD medium lacking Leu and Trp. Plasmid DNA was isolated and transformed into Escherichia coli DH5α to recover the positive library clones. These clones were analyzed by restriction enzyme digestion followed by sequencing. To further exclude the possibility of false positives, these initially positive clones were cotransformed with pGBK/ORF50c into yeast strains AH109 and Y187, which contain the lacZ reporter gene under control of the gal1 UAS. Growth of AH109 cotransformants was examined on SD plates lacking His. More than three independent β-Gal assays were performed from the liquid culture of Y187 cotransformants using o-nitrophenyl-β-d-galactopyranoside as a substrate (yeast protocols handbook; Clontech).
Northern blot analysis.
Total cellular RNA was prepared with the RNeasy kit (Qiagen, Valencia, Calif.). Twenty micrograms of RNA from each sample was analyzed on a 1.2% formaldehyde-agarose gel and then transferred to a nitrocellulose membrane. Restriction enzyme-released inserts from pcDNA-MGC2663 and pcDNA-ORF50 plasmid DNAs, as well as a PCR-generated GAPDH (glyceraldehyde-3-phosphate dehydrogenase) fragment, were 32P labeled and used as probes in Northern blotting. After hybridization and washing, the hybridized bands were detected with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Cell culture, transfection, and luciferase assays.
CV-1, 3T3, and human 293 cells were grown in Dubecco's modified Eagle medium (Gibco BRL, Rockville, Md.) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in 5% CO2. The cell lines BC-3 (EBV−/KSHV+) and BJAB (EBV−/KSHV−) were cultured in RPMI 1640 medium (Gibco BRL) supplemented with 10% fetal bovine serum and penicillin-streptomycin. To induce lytic replication of KSHV in BC-3 cells, TPA was added to complete RPMI 1640 medium at a concentration of 30 ng/ml for 24 h before the cells were harvested.
CV-1, 3T3, and 293 cells were transfected with Lipofectamine (Gibco BRL) as described previously (7). Transfections of BJAB cells were performed by mixing 107 cells with 5 to 15 μg of plasmid DNA in 0.6 ml of complete RPMI 1640 medium and then electroporating the mixture at 250 V and 960 μF using the GenePulser (Bio-Rad, Hercules, Calif.). The transfected cells were harvested 48 h posttransfection, and luciferase activities were measured (Luciferase Assay System; Promega) according to the manufacturer's procedure. Each result was obtained from an average of multiple transfections from at least three independent experiments.
Expression and purification of recombinant proteins.
E. coli BL21(DE3) cultures harboring recombinant protein expression plasmids were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. The CV-1 cells transfected with pcDNA4His-2663 or the vector control were harvested 40 h posttransfection. The bacteria or cell pellets were then resuspended in ice-cold phosphate-buffered saline (150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, pH 7.3) and sonicated. The cleared lysates were either resolved by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) or purified by affinity chromatography. For affinity purification, glutathione-Sepharose 4B beads (Amersham Pharmacia) were used for the GST fusion proteins and Ni-nitrilotriacetic acid (NTA) beads (Qiagen) were used for the His-tagged fusion proteins. Western blot analysis was performed using a rabbit antiserum raised against the His-tagged N-terminal 376 aa of RTA or mouse anti-polyhistidine (Sigma, St. Louis, Mo.) as the primary antibody, followed by alkaline phosphatase-conjugated goat anti-rabbit or anti-mouse antibody (Bio-Rad), respectively, as the secondary antibody. Signals were detected by chemiluminescence using CDP-Star (Boehringer-Mannheim, Indianapolis, Ind.) as the substrate.
Transcription-translation of proteins and in vitro binding assay.
35S-labeled proteins were synthesized in vitro using the TNT quick coupled transcription-translation system (Promega) according to the manufacturer's procedures, using pcDNA-MGC2663, pcDNA-MGC2663/242, pcDNA-ORF50, or pcDNA-ORF50/548 as a template. Recombinant proteins immobilized on beads were pretreated with 0.2 U of DNase I and 0.2 μg of RNase A per μl for 0.5 h at 25°C in pretreating buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 2.5 mM CaCl2, 100 mM NaCl, 5% glycerol, 1 mM dithiothreitol) as described previously (22). The beads were washed twice with binding buffer (20 mM Tris-Cl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM dithiothreitol) and resuspended in the same buffer before the labeled proteins were added, followed by incubation for 1 h at 4°C. The beads were then washed four times in binding buffer, heated in SDS gel loading buffer, and analyzed by SDS-PAGE. The gels were exposed to a phosphorimaging screen for 2 to 3 days and then analyzed.
Coimmunoprecipitation.
CV-1 cells were cotransfected with pcDNA-ORF50 and pHA-MGC2663. At 48 h posttransfection, the cells were lysed in ice-cold IP buffer (1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, and 2.5 mM phenylmethylsulfonyl fluoride in phosphate-buffered saline) and sonicated. After centrifugation, the cell lysates were precleared with 5 μl of preimmune rabbit serum and 50 μl of a 50% slurry of protein A-Sepharose beads (Amersham Pharmacia). RTA protein was precipitated with 5 μl of rabbit antiserum against RTA and protein A beads. The beads were washed four times with IP buffer and heated in sample buffer to be analyzed by SDS-PAGE. Western blot analysis was performed using polyvinylidene difluoride membranes with anti-hemagglutinin (HA) polyclonal antibody (Clontech) and alkaline phosphatase-conjugated goat anti-rabbit antibody.
Immunofluorescence assay.
The CV-1 cells grown on glass coverslips were cotransfected with pEGFP-MGC2663 and pCMV-Tag50. At 48 h posttransfection, the cells were fixed and immunostained using mouse anti-Flag (Stratagene, La Jolla, Calif.) at 1:5,000 dilution as the primary antibody, followed by cyanine 5 (Cy5)-conjugated donkey anti-mouse (Jackson ImmunoResearch, West Grove, Pa.) at 1:100 dilution as the secondary antibody. Samples were examined under a Bio-Rad MRC1024ES confocal laser-scanning microscope equipped with an argon-krypton laser. Optical images of GFP fusion proteins and Cy5 (laser lines, 488 and 640 nm, respectively; emission filters, 520 and 690 nm, respectively), as well as the corresponding phase-contrast images, were collected simultaneously using the dual-channel display mode of the Bio-Rad LaserSharp imaging program.
Nucleotide sequence accession number.
The GenBank accession number for the KSHV sequences reported in this study is KSU75698.
RESULTS
Yeast two-hybrid library screening identified a cellular protein that interacts with KSHV RTA.
Yeast two-hybrid systems provide a sensitive method for detecting relatively weak and transient protein-protein interactions. In the selection system, we used RTA as our bait and fused it downstream of the DNA BD of the yeast transcription factor Gal4. A second construct expressing the AD of Gal4 fused to sequences from a cDNA library is able to activate reporter gene expression under the control of the Gal4-responsive UAS and therefore enables the selection of positive colonies. This activation will occur if the library-encoded protein can interact with the bait protein to bring the DNA BD and AD of Gal4 into close physical proximity to the promoter region. The full-length RTA functions as a transcription activator and showed a strong autologous activation of reporter genes in the absence of the Gal4 AD when it was fused to the Gal4 DNA BD (Table 1). Therefore, the full-length RTA cannot be used as bait. To overcome this problem, we used a C-terminal mutant of RTA, ORF50c, which contains the N-terminal 671-aa segment (with a C-terminal 20-aa deletion) of RTA fused to an 86-aa segment encoded by the downstream K8.1 gene. This clone was constructed from a spliced ORF50 mRNA that was previously used to study the trans-acting function of RTA and was found to be inactive against its target ORF57 promoter (unpublished data). The clone pGBK/ORF50c, when fused to the Gal4 DNA BD, was found to be incapable of activating reporter gene expression by itself (Table 1) and was thus used as bait to screen a Gal4 AD fusion B-cell cDNA library.
TABLE 1.
Reporter gene expression by yeast transformantsa
| Plasmid with Ga14 DNA BD | Plasmid with Ga14 AD | LacZ expression in Y187b | His expression in AH109c |
|---|---|---|---|
| pGBK/ORF50 | pGAD-T control | +++ | ++ |
| pGBK/ORF50c | pGAD-T control | − | − |
| pGBK/ORF50c | pACT-clone7 | + | + |
| pGBK/ORF50c | pGAD-clone7 | ++ | + |
| pGBK/ORF50AD2 | pGAD-T control | − | − |
| pGBK/ORF50AD2 | pACT-clone7 | + | + |
| pGBK/ORF50AD2 | pGAD-clone7 | ++ | + |
| pGBK-53 control | pACT-clone7 | − | − |
| pGBK-53 control | pGAD-clone7 | − | − |
| pGBK-53 control | pGAD-T control | +++ | ++ |
The indicated yeast strains were cotransformed with the indicated Ga14 DNA BD and Ga14 AD plasmids. Cotransformants were selected using nutrition selection markers for the vectors. The phenotypes of the colonies were analyzed by liquid β-Gal assays (for yeast strain Y187) and by their growth on SD plates lacking His (for yeast strain AH109).
+++, intense color development within 1 h in the liquid β-Gal assays (about 20- and 10-fold higher, respectively, than clones designated + and ++); −, clone was unable to develop any color after incubation with ONPG for more than 24 h in the liquid β-Gal assays.
++, colonies appeared after 2 days of incubation at 30°C on SD plates lacking His; +, colonies appeared after 4 days; −, no growth.
Among ≈3.5 × 106 yeast transformants that were screened, 17 clones were capable of continuous efficient growth in high-stringency double-selection SD medium. Of the 17 clones that were selected, 7 expressed β-Gal activities and gave a positive blue color in a colony lift filter assay. The other 10 clones were found to be false positive and did not express any β-Gal upon retesting. The seven clones were then further characterized by restriction enzyme pattern and DNA sequence analyses and were subdivided into three groups. The first group of three clones contained cDNA inserts that encode a hypothetical human protein, MGC2663; the second group of two clones contained partial sequence of the human mitochondrial genome; the third group of two clones contained a cDNA insert encoding the EBV nuclear protein BMRF2, but in the reverse orientation. When the plasmids purified from these positive clones were cotransformed with the bait plasmid into yeast to verify their positive interactions with RTA, only the first group, which contained MGC2663 inserts, gave positive results in both growth selection and liquid culture β-Gal assays in different yeast strains (Table 1). These results demonstrated that the insert contained in the first group of clones encodes a product that can interact with RTA. To further analyze this interaction, one clone (clone 7) from the first group was further characterized. In addition to the positive interaction between clone 7 and ORF50c, clone 7 also interacted with an RTA deletion mutant, ORF50AD2, which retains the N-terminal 589 aa. This RTA mutant was not active in transactivation but was still able to interact with clone 7 when both were cotransformed into yeast cells. To further confirm the interaction between the clone 7 gene product and RTA, the insert of clone 7 was released from the library vector and then transferred to another Gal4 AD vector, pGADT7, which can express higher levels of the fusion protein. When pGAD-clone7 was cotransformed into yeast cells with the RTA constructs, the same positive results were observed with both ORF50c and ORF50AD2, while no interaction with the pGBK-53 negative control was observed (Table 1).
Clone 7 encodes the putative human MGC2663 protein located on chromosome 19.
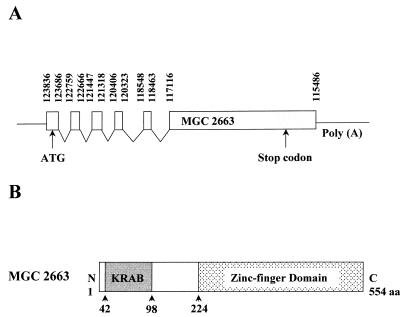
Sequence analysis showed that the insert of clone 7 is 2,062 nt in length and contains a long ORF which fuses to the Gal4 AD region of the vector at the N terminus in the same reading frame. Comparison of the GenBank database with the clone 7 sequence revealed that the ORF is the complete coding sequence of a hypothetical human protein, MGC2663, named after clone MGC:2663 (GenBank accession numbers, BC001791 for the nucleotide and AAH01791 for the protein). Further analysis of the MGC2663 gene revealed that it is located on chromosome 19 and is encoded by the human chromosome 19 clone CTC-543D15 (GenBank accession number AC008567). The function of MGC2663 is currently not known, and its amino acid coding sequence was derived entirely from the sequence of its cDNA clone. Comparison between our cDNA sequence and the corresponding genomic sequence showed that the MGC2663 protein is generated by multiple splicing. It is encoded by six spliced exons, with five small exons at the N terminus and a large one at the C terminus (Fig. 1A). Although the structure of MGC2663 is not known, amino acid sequence analysis identified a KRAB at the N terminus from aa 42 to 98 and a C-terminal zinc finger domain, starting from aa 224 (Fig. 1B). Up to 11 well-conserved C2H2-type zinc finger motifs can be found in the C-terminal domain.
FIG. 1.
Schematic representations of MGC2663 (clone 7) splicing pattern and its potential functional domains. (A) Putative genomic organization and splicing pattern of the MGC2663 gene. The indicated nucleotide numbers of the potential splicing sites reflect the numbers that were assigned to the GenBank human chromosome 19 clone CTC-543D15 (GenBank AC008567). Conserved splicing donor (GT) and acceptor (AG) sites are found at the ends of each exon. (B) Schematic representation of MGC2663 protein. The putative KRAB and the zinc finger domain are indicated.
Northern blot analysis of MGC2663 transcription.
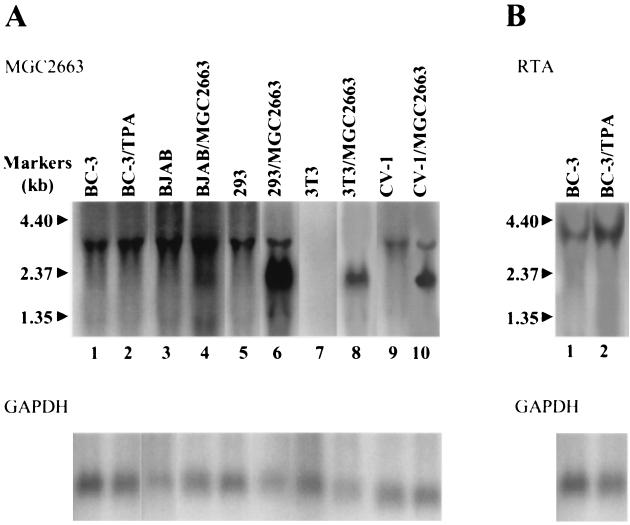
To determine the size of the MGC2663 transcript and the possible effect of KSHV reactivation on its expression, Northern blot analyses were performed using various mammalian cell lines. Total RNA was extracted from BC-3, BJAB, 293, 3T3, and CV-1 cells and analyzed with a 32P-labeled MGC2663 probe. A band of approximately 3.0 kb was detected from all human cells and CV-1 cells tested but not from 3T3 cells. Since our cDNA insert sequence would predict only a 2.1-kb transcript, the detecting of a 3.0-kb novel cellular transcript suggests that the MGC2663 transcript contain an untranslated sequence of about 0.9 kb at the 5′ end. Interestingly, TPA treatment of BC-3 cells, which stimulates RTA expression (Fig. 2B), did not seem to enhance MGC2663 expression (Fig. 2A, lanes 1 and 2), suggesting that MGC2663 may not be induced upon lytic KSHV replication. In the MGC2663-transfected cells, in addition to the 3.0-kb cellular MGC2663 transcript, a 2.3-kb transcript encoded by the MGC2663 expression clone was also observed. The transcript of 2.3 kb was expected from the transfected MGC2663 expression plasmid because of the presence of an extra 200 bp of plasmid-encoded flanking sequences in addition to the 2.1-kb MGC2663 insert.
FIG. 2.
Northern blot analysis of the expression pattern of MGC2663 gene in various mammalian cell lines. (A) MGC2663 transcript was detected in all human cells tested and in CV-1 cells. Total cellular RNAs were extracted from BC-3 cells (untreated [BC-3] or TPA treated for 24 h [BC-3/TPA]) and BJAB cells (transfected with vector control [BJAB] or pcDNA-MGC2663 [BJAB/MGC2663]), as well as transfected (/MGC2663) or control 293 cells, 3T3 cells, and CV-1 cells. Northern blotting was performed using a 32P-labeled MGC2663-specific probe and analyzed by a PhosphorImager after a 2-day exposure. The RNA sizes according to the molecular size markers are indicated. The same membrane was then rehybridized with a 32P-labeled GAPDH probe to ensure similar amounts of RNA were loaded onto each lane. (B) Northern blot analysis of the RTA RNA in BC-3 cells. The same membrane used for panel A was stripped and reprobed with a 32P-labeled ORF50 probe. A 3.6-kb RTA band was detected in both control and TPA-stimulated BC-3 cells.
MGC2663 synergizes with RTA to activate transcription.
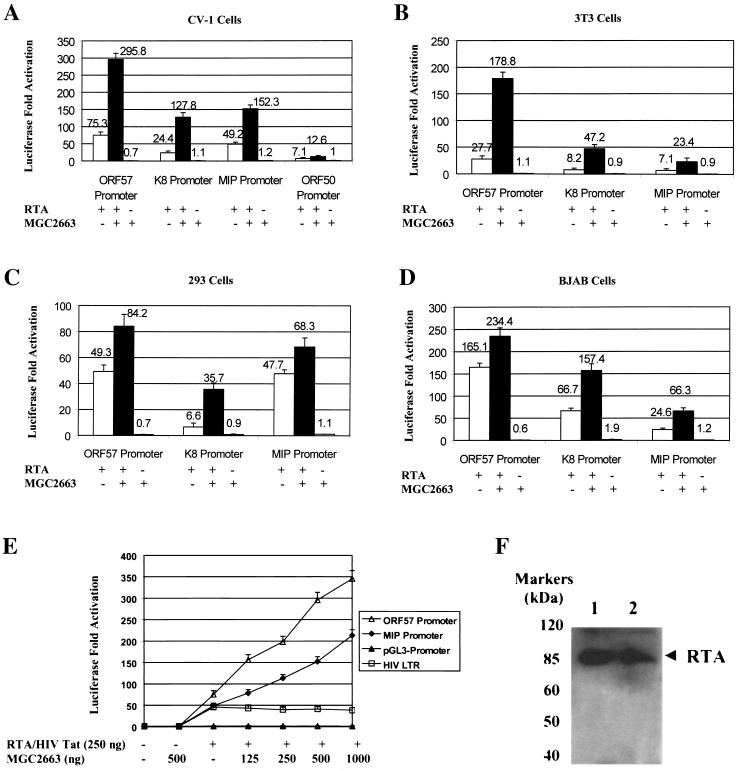
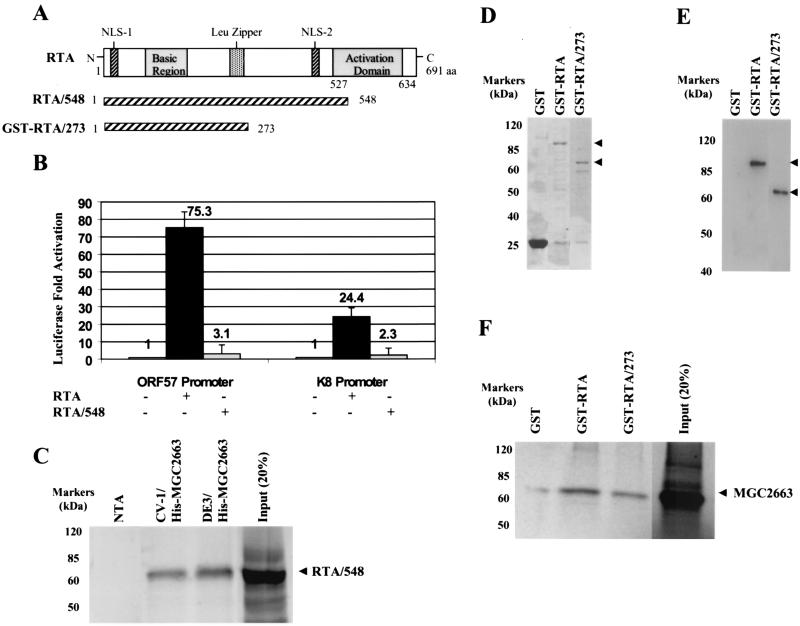
Based on the observed interaction between MGC2663 and RTA in the yeast cells, we predicted that MGC2663 may be involved in RTA transactivation and may be required for optimal activation of those viral promoters that are regulated by RTA. To study this possibility, we tested several viral promoters, such as the ORF57, K8, K6 (which encodes a homolog of macrophage inflammatory protein vMIP-I), and ORF50 promoters, which have been previously shown to be activated in vitro by RTA (8, 9, 17, 32, 36, 39), and examined their RTA responsiveness in the absence or presence of overexpressed MGC2663 in various cell lines. As expected, the ORF57 promoter was strongly activated by RTA in all the cell types tested, ranging from 27-fold in 3T3 cells to 165-fold in BJAB cells. Lower levels of activation were observed for K8, MIP, and ORF50 promoters. Cotransfection of an MGC2663-expressing plasmid further enhanced RTA activation of these viral promoters in all four cell lines tested, while overexpression of MGC2663 by itself did not seem to have any effect on the activities of the promoters (Fig. 3). The enhancement effect of MGC2663 varied among different promoters in different cell lines. In general, higher levels of activation of the promoters by RTA alone resulted in lower levels of enhancement in the presence of MGC2663, such as in the activation of the ORF57 promoter in BJAB cells (Fig. 3D). The highest level of synergy between RTA and MGC2663 was observed with the K8 promoter in all cell types tested. When an increasing amount of MGC2663 was cotransfected into CV-1 cells with an equal amount of RTA plasmid DNA, a parallel increase in the levels of MGC2663 stimulation was observed on the ORF57 and MIP promoters tested (Fig. 3E). The effect of MGC2663 on RTA transactivation appears to be specific for RTA and for the promoters that are responsive to RTA, and it was found to have no effect on the HIV transactivator Tat. Cotransfection of MGC2663 with Tat and the HIV LTR did not show any synergistic effect on Tat transactivation of the HIV promoter (Fig. 3E). Activity of minimal simian virus 40 promoter was not affected by either MGC2663 alone or the RTA/MGC2663 combination, suggesting that MGC2663 has no detectable effect on basal transcription activities (Fig. 3E). Our transfection studies suggest that MGC2663 synergizes with RTA and plays an accessory role in enhancing RTA transactivation. To further demonstrate that the RTA expression level was not altered in the MGC2663-cotransfected CV-1 cells, Western blot analysis using rabbit anti-RTA antibody was carried out, and similar levels of RTA protein were detected in both RTA-transfected and RTA/MGC2663-cotransfected cells (Fig. 3F). Therefore, the enhanced RTA transactivation in the presence of MGC2663 was not due to the stimulation of RTA expression at the transcriptional or translational level.
FIG. 3.
Enhancement of RTA transactivation of various KSHV promoters by MGC2663. (A) Transfection of CV-1 cells was carried out with 50 ng of viral promoter reporter construct and 250 ng of pcDNA-ORF50 (RTA) and/or 500 ng of pcDNA-MGC2663 (MGC2663) (+) as indicated. The total DNA amount used in each transfection was normalized by adding pcDNA3.1(−) vector. Luciferase activities were measured 48 h posttransfection. The error bars indicate standard deviations. (B and C) Transfections of 3T3 and 293 cells were carried out as described for panel A, using the same amounts of DNA for transfection. (D) Transfection of BJAB cells. In each transfection, 107 BJAB cells were mixed with 2 μg of reporter plasmid, 5 μg of RTA expression plasmid, and/or 5 μg of MGC2663 expression plasmid. Transfection was carried out using electroporation as described in Materials and Methods. (E) MGC2663 stimulation of RTA activation is dose dependent and is specific for RTA and its target promoters. CV-1 cells were transfected with 50 ng of KSHV promoter reporter construct, pGL3-promoter, or pHIVLTR-luc, as well as the indicated amount of pcDNA-ORF50, pcDNA-Tat, and/or pcDNA-MGC2663. Each result represents an average of at least three independent experiments. The standard deviations are shown as error bars. Transfection efficiency for each experiment was normalized using a β-Gal expression plasmid as an internal control. Fold activation was calculated based on the transfection of the reporter plasmid and the vector control, which was normalized to 1. (F) Western blot analysis of RTA expression in transfected CV-1 cells in the presence or absence of MGC2663. Lysates of cells transfected with either pcDNA-ORF50 alone (lane 1) or pcDNA-ORF50 and pcDNA-MGC2663 (lane 2) were analyzed using RTA antiserum. The protein molecular mass markers are indicated.
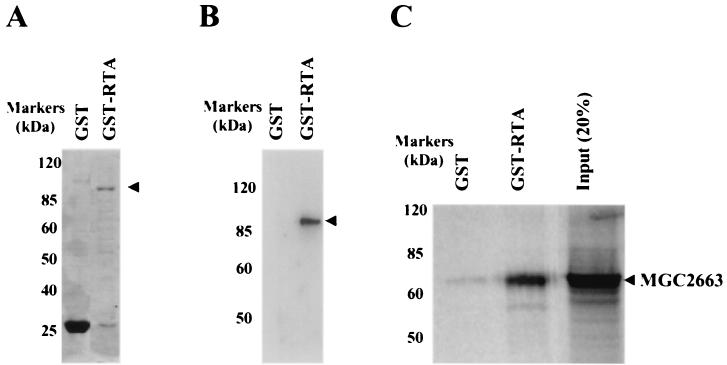
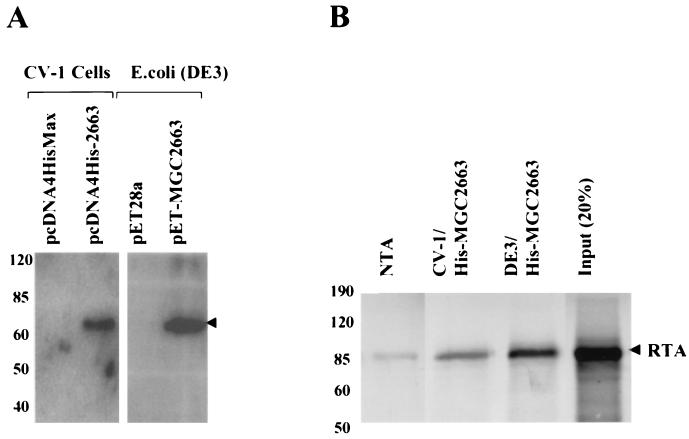
In vitro binding assay of MGC2663 and RTA.
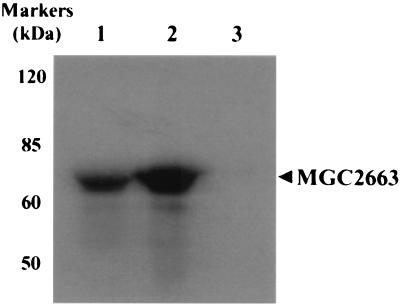
In order to further explore the direct association between MGC2663 and RTA, in vitro GST pull-down assays were performed using a 35S-labeled in vitro-translated (IVT) MGC2663 product and a recombinant GST-fused RTA protein. The GST-RTA fusion protein was first expressed in bacteria. A protein of about 95 kDa was detected by SDS-PAGE (Fig. 4A) and was confirmed by Western blot analysis using RTA antiserum (Fig. 4B). To study interaction between MGC2663 and RTA, the recombinant GST-RTA was immobilized on glutathione-Sepharose 4B beads, and 35S-labeled MGC2663 was allowed to bind to the GST-RTA. The IVT MGC2663 was found to bind strongly in the presence of GST-RTA, while only a very small amount of IVT MGC2663 was found to bind nonspecifically (Fig. 4C). To confirm that MGC2663 and RTA interact specifically, the reverse pull-down assay was carried out using the recombinant MGC2663 protein to pull down IVT RTA. The recombinant MGC2663 was expressed either in mammalian cells or in bacteria as a His-tagged fusion protein. The protein was expressed at a higher level in bacteria, which can be detected by SDS-PAGE and confirmed by Western blot analysis (Fig. 5A), whereas in CV-1 cells the MGC2663 was expressed at a much lower level, and the expressed protein could only be detected by Western blotting (Fig. 5A). Both the mammalian-cell- and bacterial-cell-expressed His-tagged MGC2663 proteins, after being immobilized on Ni-NTA beads, were found to be capable of binding IVT RTA (Fig. 5B). Only a very small amount of IVT RTA was found to bind nonspecifically to the NTA beads in the absence of MGC2663. The level of this nonspecific binding was highly variable, depending on the amount of labeled protein added, and was consistently weaker than the specific signals in the presence of MGC2663. These results are consistent with those of the GST pull-down assay (Fig. 4) and further demonstrate that RTA and MGC2663 interact directly.
FIG. 4.
Pull-down assays indicating that MGC2663 binds to GST-fused RTA in vitro. (A) Coomassie blue-stained gel of glutathione-Sepharose 4B bead-purified GST-fused RTA used in GST pull-down assay. (B) Western blot analysis of the GST-RTA using RTA antiserum to confirm the expression of GST-fused RTA. (C) GST-fused RTA protein binds to IVT MGC2663 protein. In vitro-transcribed and -translated 35S-labeled MGC2663 was added to either the immobilized GST-RTA or GST control. A control lane indicates the size of the labeled IVT MGC2663 using input counts per minute about 20% of that used in the pull-down assay with GST-RTA. The protein molecular mass markers are indicated on the side of each panel.
FIG. 5.
Pull-down assays indicating that RTA binds to His-tagged MGC2663 in vitro. (A) Western blot analysis of the His-tagged MGC2663 used in the pull-down assay, expressed either in CV-1 cells or in E. coli. The Western blotting was carried out using antibody against His tag. The arrowhead indicates the expressed His-tagged MGC2663. (B) His-tagged MGC2663 protein binds to RTA. In vitro-transcribed and -translated 35S-labeled RTA was added to immobilized His-tagged MGC2663 that was expressed either in CV-1 cells or in E. coli. NTA beads were used as a control for the pull-down assay. A control lane indicates the size of the labeled IVT RTA using input counts per minute about 20% of that used in the pull-down assay with His-tagged MGC2663. The positions of protein markers are indicated.
Coimmunoprecipitation of RTA and MGC2663.
The synergistic effect of MGC2663 on RTA transactivation and the binding of MGC2663 with RTA in vitro suggest that an in vivo protein-protein interaction between RTA and MGC2663 should also occur. To confirm this possibility, a coimmunoprecipitation assay in transfected CV-1 cells was carried out (Fig. 6). CV-1 cells were cotransfected with RTA and HA-tagged MGC2663 expression plasmids, and the expression of HA-tagged MGC2663 in the transfected cell lysates was first confirmed by Western blot analysis (lane 1). Immunoprecipitation of RTA from the cell lysates was then carried out using anti-RTA rabbit serum. Precipitation of RTA by antiserum led to the coprecipitation of MGC2663, which was detected by Western blotting with anti-HA tag antibody (lane 2). In control cells that were transfected with HA-tagged MGC2663 alone, RTA antiserum was unable to precipitate MGC2663 in the absence of RTA (lane 3). This result indicates that MGC2663 indeed binds to RTA in vivo.
FIG. 6.
Immunoprecipitation of transfected CV-1 cells to demonstrate that MGC2663 associates with RTA in vivo. CV-1 cells were cotransfected with pcDNA-ORF50 and pHA-MGC2663 (lanes 1 and 2) or with pHA-MGC2663 alone (lane 3). The cell lysates were precipitated with RTA antiserum (lanes 2 and 3), and the coimmunoprecipitation of MGC2663 was detected using anti-HA antibody. Expression of HA-tagged MGC2663 in the total lysates was also indicated in the same Western blot as a control (lane 1).
MGC2663 binds to the N terminus of RTA.
To further characterize the interaction between MGC2663 and RTA, an RTA AD deletion clone, pcDNA-ORF50/548, was studied (Fig. 7A). This clone contains the N-terminal 548 aa of RTA, and almost the entire AD from aa 527 to 634 was deleted. The RTA/548 deletion mutant lost its transactivation function; it was not able to activate either the ORF57 promoter- or the K8 promoter-luciferase construct in CV-1 cells (Fig. 7B). To determine if RTA with the deletion can still interact with MGC2663, the RTA/548 clone was then translated in vitro and used in the His tag pull-down assay as described earlier. Even though the deletion of the AD in RTA abolished its ability to transactivate (Fig. 7B), this did not affect its ability to bind MGC2663. IVT RTA/548 protein was able to interact with either the CV-1-expressed or bacterially expressed His-tagged MGC2663 protein (Fig. 7C), and the levels of binding were comparable to those of the intact RTA protein (Fig. 5B).
FIG. 7.
The RTA N-terminal domain is responsible for interaction with MGC2663. (A) Schematic representations of RTA and the RTA AD deletion clones. The basic region, AD, Leu zipper, and nuclear localizing signals (NLS) of RTA are indicated. The deletion clones RTA/548 and GST-RTA/273, in which the C-terminal activation domain of RTA was deleted, are also shown. (B) The RTA/548 clone failed to activate both ORF57 and K8 promoters in the transfection assays. CV-1 cells were transfected under the conditions described for Fig. 3A. Transfection was carried out using 50 ng of reporter plasmid and 250 ng of pcDNA-ORF50 (RTA) or pcDNA-ORF50/548 (RTA/548). Fold activation was calculated based on the transfection of the reporter plasmid and the vector control, which was normalized to 1. The error bars indicate standard deviations. +, present; −, absent. (C) The RTA/548 protein retained its ability to bind to the His-tagged MGC2663. The His tag pull-down assay was performed as described for Fig. 5B, using 35S-labeled IVT RTA/548. A control lane indicates the size of the labeled IVT RTA/548 using input counts per minute about 20% of that used in the pull-down assay with His-tagged MGC2663. (D) The RTA deletion clone GST-RTA/273 was expressed as a GST fusion protein and used in the pull-down assay with MGC2663. The Coomassie blue stain of the purified GST-RTA/273 indicates the correct deleted RTA protein was expressed. The intact GST-RTA protein was also purified and run on the same gel as a control. (E) Western blot analysis using RTA antiserum to confirm that the GST-RTA/273 fusion protein of the correct size was expressed. The GST-RTA was also run in parallel as a control. The arrowheads in panels D and E indicate the expressed proteins. (F) GST-RTA/273 retained its ability to bind to MGC2663, but less MGC2663 protein seemed to bind than with the intact GST-RTA. The GST pull-down assay was performed as described for Fig. 4C. A control lane indicates the size of the labeled IVT MGC2663 using input counts per minute about 20% of that used in the pull-down assay with GST-RTA. The positions of protein markers are indicated.
In our previous study, another RTA deletion clone which contains only the first 273 aa was found to have lost its transactivation activity (39), and we were interested in determining if it could still interact with MGC2663. Therefore, to further map the domain of RTA that may be involved in the interaction with MGC2663, a second RTA deletion clone that contained only the N-terminal 273-aa putative RTA DNA BD was constructed. This clone, pGEX-RTA/273, was expressed as a GST fusion protein. To determine whether this RTA deletion mutant binds to MGC2663, it was expressed in bacteria and used in the pull-down assay with IVT MGC2663. The RTA N-terminal 273-aa GST fusion protein was well expressed in bacteria and could be detected by SDS-PAGE at the expected size and confirmed by Western blotting (Fig. 7D and E). The amount of protein expressed was comparable to that of the full-length GST-RTA fusion protein. When GST-RTA/273 was immobilized on glutathione beads, it was able to pull down IVT MGC2663 but seemed to be binding less efficiently: less MGC2663 appeared to bind to GST-RTA/273 than to the intact RTA (Fig. 7F). These results indicate that the AD of KSHV RTA is dispensable for association with MGC2663 while the N-terminal 273 aa confers its ability to bind.
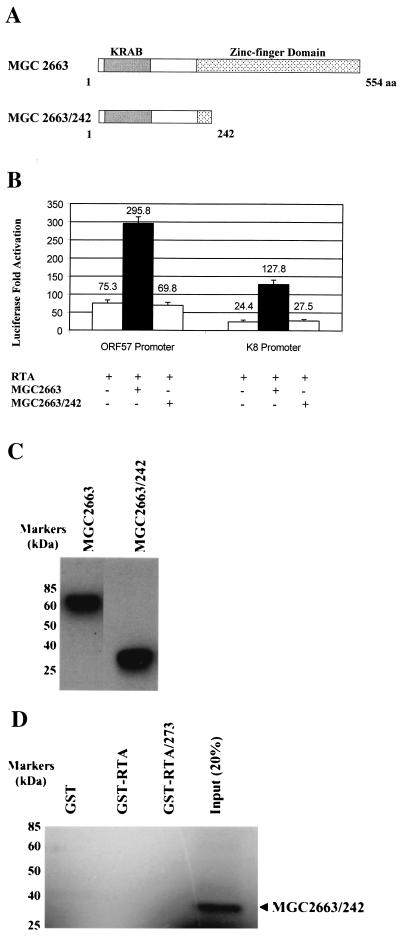
The zinc finger domain of MGC2663 is required for association with RTA.
To investigate the domain of the MGC2663 protein that may be involved in the interaction with RTA, we constructed an MGC2663 deletion mutant, MGC2663/242, that retains the N-terminal 242 aa, including the entire KRAB, but with most of the C-terminal zinc-finger domain deleted (Fig. 8A). When the MGC2663 deletion clone was cotransfected into CV-1 cells with RTA and either the ORF57 or K8 promoter, it was unable to synergize with RTA in transactivation of these viral promoters (Fig. 8B). To eliminate the possibility that the inability of MGC2663/242 to synergize with RTA is due to problems with expression and stability of the truncated protein, Western blot analysis using the transfected cell lysates was carried out, using anti-HA antibody to detect the expression of the HA-MGC2663/242 fusion protein (Fig. 8C). As expected, the approximately 28-kDa truncated HA-MGC2663/242 was detected in the transfected cells, suggesting that the protein was made but was unable to synergize with RTA. These results indicate that the deleted region containing the zinc finger domain of MGC2663 is necessary for the enhancement of RTA transactivation. We then further examined the in vitro interaction between RTA and MGC2663/242, using the immobilized GST-RTA and IVT MGC2663/242 in the GST pull-down assay. As shown in Fig. 8D, neither GST-RTA nor GST-RTA/273 pulled down the mutant protein MGC2663/242 as they did the full-length MGC2663 (Fig. 7F), suggesting that the failure of MGC2663/242 to stimulate RTA transactivation is due to its inability to interact with RTA. Our results demonstrate that the MGC2663 C-terminal zinc finger domain is required for both interaction and synergy with RTA in transcriptional activation.
FIG. 8.
Deletion of the zinc finger domain of MGC2663 abolishes its binding to RTA. (A) Schematic representation of MGC2663/242 deletion clone that was expressed and tested for its ability to interact with RTA. A schematic representation of MGC2663 is shown for comparison. (B) Transfection of the MGC2663/242 deletion clone to determine its ability to enhance RTA activation of the ORF57 and K8 promoters. CV-1 cells were transfected under the conditions described for Fig. 3A. The plasmid pcDNA-MGC2663/242 was added and compared to the intact MGC2663 clone pcDNA-MGC2663 in the transfection studies as indicated. Fold activation was calculated based on the transfection of the reporter plasmid and the vector control, which was normalized to 1. The error bars indicate standard deviations. +, present; −, absent. (C) Western blot analysis for the expression of MGC2663 and MGC2663/242 in transfected cells. CV-1 cells were transfected with 500 ng of pHA-MGC2663 or pHA-MGC2663/242, and the cell lysates were analyzed using anti-HA antibody. (D) Pull-down assay using purified GST-RTA or RTA deletion GST-RTA/273 proteins, with 35S-labeled IVT MGC2663/242 deletion protein. MGC2663/242 did not bind to either GST-RTA or GST-RTA/273. The GST pull-down assay was performed under the conditions described for Fig. 4C but using 35S-labeled IVT MGC2663/242. A control lane indicates the size of the labeled IVT MGC2663/242 using input counts per minute about 20% of that used in the pull-down assay with GST-RTA. The positions of protein markers are indicated.
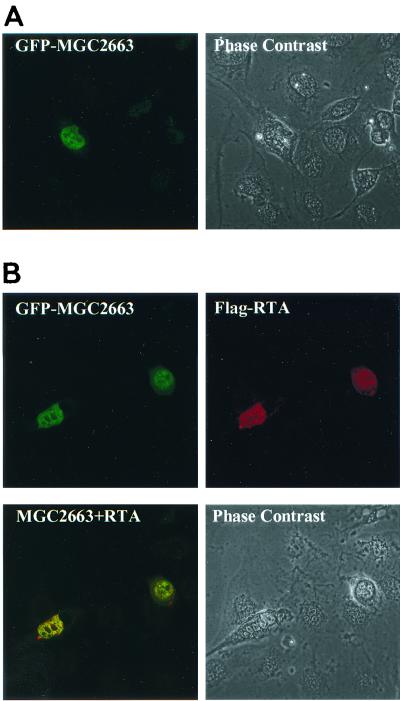
MGC2663 colocalizes with RTA in vivo.
Since MGC2663 may be involved in transcriptional regulation, it was important to determine the subcellular localization of this protein and whether the presence of RTA alters its localization. For this purpose, a plasmid expressing the GFP-MGC2663 fusion protein was constructed and then transfected into CV-1 cells. The transfected cells displayed fluorescence in the nucleus (Fig. 9A), suggesting that MGC2663 may be a nuclear protein. Since RTA has been previously demonstrated to be a nuclear protein (15), it was interesting to determine if RTA colocalizes in the nucleus with MGC2663. The CV-1 cells were cotransfected with pEGFP-MGC2663 and pCMV-Tag50, and the RTA protein was detected by immunofluorescence assay using mouse anti-Flag antibody followed by Cy5-conjugated anti-mouse antibody. The transfected RTA and MGC2663 were found to colocalize in the nucleus, and the presence of RTA did not seem to alter the localization pattern of MGC2663 (Fig. 9B). These results further support those of our pull-down assays and indicate that RTA and MGC2663 colocalize in the nucleus and can interact in vivo to transactivate viral gene expression.
FIG. 9.
Colocalization of MGC2663 and RTA proteins in the nuclei of transfected cells. (A) Cellular localization of MGC2663. CV-1 cells grown on coverslips were transfected with pEGFP-MGC2663, and the transfected cells were observed under a confocal microscope 48 h posttransfection. (B) MGC2663 colocalizes with RTA. CV-1 cells cotransfected with the plasmids pEGFP-MGC2663 and pCMV-Tag50 were observed 48 h posttransfection. The top left panel represents the localization of the GFP-MGC2663 observed using a 488- and 520-nm filter to detect green fluorescence. The top right panel represents the localization of Flag-RTA protein using mouse anti-Flag antibody followed by Cy5-conjugated anti-mouse antibody. The 640- and 690-nm filter was used to detect the red Cy5 staining. The lower left panel represents the dual detection of both GFP-MGC2663 and Flag-RTA proteins. Phase contrast represents the cells observed under bright field.
DISCUSSION
In this study, we have demonstrated that a novel cellular protein named MGC2663 can interact with the KSHV RTA directly and enhance RTA transactivation of several viral promoters. This protein has been named K-RBP due to its RTA binding feature. The KSHV RTA protein is an interesting protein which is a homolog of the EBV IE gene product RTA. KSHV RTA has been shown to be necessary for the induction of lytic viral replication and activates different early and late viral genes (13, 18, 19, 23). Several target genes for RTA activation have been identified; they include ORF57, PAN RNA, K8, and TK. The RTA protein was found to up-regulate its own gene expression (8). Several studies, including ours, have mapped the RREs for several viral promoters (9, 20, 34, 39). We found a common 16-bp element for the ORF57 and K8 promoters, but there seem to be no consensus RREs among all of the other RTA-responsive promoters. At the moment, very little is known about the mechanism of RTA transactivation, and it is possible that RTA can bind either directly or indirectly to the elements located in the target promoters to enhance transcription. It was reported recently that RTA can bind directly to the PAN RNA promoter sequences (34), suggesting that RTA-DNA binding could be involved in the transcriptional activation of PAN RNA expression. However, binding of RTA to the ORF57/K8 RRE seems to be weaker than to the PAN RNA promoter (20, 34, 39), and it is possible that RTA may be interacting with the RREs of these other target genes via cellular factors. The K-RBP protein that is identified in the present study may be one of these factors involved in the sequestering of RTA in the vicinity of the RREs of both the K8 and ORF57 promoters to activate gene expression. However, it is not clear at the moment whether K-RBP can directly and specifically interact with DNA, and whether it can bind to the RRE in the ORF57 promoter needs to be determined. Indeed, we have previously observed (unpublished data) that labeled K8 and ORF57 RRE probes consistently complexed with a cellular protein(s) in a mobility shift assay using either RTA-transfected or untransfected normal cellular nuclear extracts; whether K-RBP is part of the DNA-protein complex remains to be determined.
Cellular cofactors are often found to be involved in transcriptional activation by viral transactivators. One of the best-characterized viral transactivators is HIV Tat; it was shown to interact with cellular factors such as cyclin and CDK9 (40). The K-RBP protein that we have found is the first RTA-interacting protein identified by the yeast two-hybrid screening using RTA as bait. Since RTA itself is a potent transactivator giving high levels of background transcriptional activities in the absence of any other cellular cofactors, it cannot be used directly as bait; therefore, a nonfunctional RTA clone had to be used in our study. Interestingly, the cDNA expression library that we used for screening with RTA was from an EBV-transformed lymphoma cell line, and only K-RBP was found to consistently bind to RTA. This suggests that either there was no EBV-encoded gene product that could interact with RTA or the specific EBV gene was not represented in the lymphoma cell cDNA library. In addition to the K-RBP that we have identified in this study, it was reported recently that the cellular proteins CBP and HDAC can also interact with RTA, with CBP activating and HDAC repressing RTA-mediated viral transcription (15). Binding of CBP to RTA requires the C-terminal activation domain of RTA, while HDAC binds to a central proline-rich segment in RTA. With regard to other cellular factors, it was found that RTA transactivation of KSHV thymidine kinase promoter requires the SP-1 element, suggesting SP-1 may be involved (44). However, it is unlikely that SP-1 is involved with all RTA-responsive genes, because the SP-1 site was not observed with either the ORF57 or the K8 promoter that was found to be responsive to transactivation (39).
We have found that K-RBP was expressed in different primate cells tested but not in mouse 3T3 cells, and a single K-RBP RNA species of about 3.0 kb was detected. This suggests that either the K-RBP gene is unique to primate cells or the mouse homolog of this gene is so divergent from that of the primates that it could not be detected by our human K-RBP probe. Whether K-RBP is a member of a gene family and whether similar genes can be found in other animal species need to be further investigated. Based on the genomic sequence, the K-RBP mRNA is a spliced product with six exons spanning a genomic fragment of over 8.3 kb. Interestingly, based on Northern analysis, the K-RBP mRNA seems to have a long 5′ untranslated region of about 0.9 kb. The implications for the presence of a long 5′-untranslated region also need to be further investigated.
The K-RBP protein displays a typical structure of a member of the KRAB-zinc finger protein (ZFP) family. It contains an N-terminal KRAB domain and a C-terminal zinc-finger domain composed of 11 C2H2 motifs, which are commonly found in most KRAB-ZFPs. Mammalian genomes are known to contain a large number of KRAB-zinc finger genes, and many of them encode 10 or more C2H2-type zinc finger motifs. Members of this large gene family have diverged in both sequence and expression patterns (33) and therefore may yield families of proteins with distinct, yet related, functions. In contrast to our findings here that K-RBP has no remarkable effect on general transcription but acts as a cofactor for RTA to stimulate RTA-mediated viral gene transactivation, other KRAB-containing ZFPs were found to repress both basal and VP16-activated transcription when targeted to DNA in vitro (1). It was proposed that members of the KRAB-ZFP family function as transcription factors that bind DNA through the zinc finger domain and repress gene expression via the KRAB domain (11). In this study, we have demonstrated that the zinc finger domain of K-RBP is involved in the interaction with RTA and stimulates RTA-mediated transcription activation, but it is not clear whether K-RBP and its zinc finger domain are also involved in DNA binding. Our results nevertheless suggest that human KRAB-ZFP-like K-RBP protein interacts with RTA and stimulates RTA-mediated viral gene expression through a mechanism that could be different from that involved in the transcription repression by other KRAB-ZFP family members.
In this study, we have found that four different KSHV promoters, those of ORF57, K8, MIP, and ORF50, regardless of whether they are expressed early or late after viral infection, were all further stimulated by K-RBP in the presence of RTA. Among these viral promoters, only those of ORF57 and K8 share a common RRE. It is possible that RTA may not be binding directly to the other two promoters but could be tethered to these targets via common cellular factors, such as K-RBP. The colocalization of both RTA and K-RBP in the nucleus strongly supports the notion that they complex in vivo to mediate transcriptional activation, but we could not determine whether it is RTA or yet another cellular factor that may be tethering K-RBP to the viral promoter targets in the nucleus. Our sequence analysis of K-RBP did not reveal any typical nuclear localizing signal, but further analysis needs to be carried out to determine how K-RBP can be targeted to the nucleus. Even though K-RBP appears to be localized in the nucleus, whether it plays a role in transcription regulation in the absence of RTA is unclear. Further studies are also needed in order to determine the role of K-RBP in KSHV lytic viral replication and to decipher the mechanism involved in RTA-mediated viral gene transcription.
ACKNOWLEDGMENTS
We thank Y. Zhou and M. Mathiesen of the Microscopy Core Facility for expert technical assistance and R. Weldon for helpful discussion.
This work was supported in part by PHS grants CA75903, CA76958, and RR15635 and Fogarty International Training Grant TW01429 to C.W.
REFERENCES
- 1.Abrink M, Ortiz J A, Mark C, Sanchez C, Looman C, Hellman L, Chambon P, Losson R. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 β. Proc Natl Acad Sci USA. 2001;98:1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/KSHV) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D W. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Ziegler J, Wabinga H, Katangole-Mbidda E, Boshoff C, Schulz T F, Whitby D, Maddalena D, Jaffe H W, Weiss R A, Moore P S. Kaposi's sarcoma-associated herpesvirus DNA sequences are present in African endemic and AIDS-associated Kaposi's sarcoma. Arch Intern Med. 1996;156:202–204. [PubMed] [Google Scholar]
- 6.Chang Y, Moore P S. Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5:215–222. [PubMed] [Google Scholar]
- 7.Chen H, Wilcox G, Kertayadnya G, Wood C. Characterization of the Jembrana disease virus tat gene and the cis- and trans-regulatory elements in its long terminal repeats. J Virol. 1999;73:658–666. doi: 10.1128/jvi.73.1.658-666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng H, Young A, Sun R. Auto-activation of the rtagene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J Gen Virol. 2000;81:3043–3048. doi: 10.1099/0022-1317-81-12-3043. [DOI] [PubMed] [Google Scholar]
- 9.Duan W, Wang S, Liu S, Wood C. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch Virol. 2001;146:403–413. doi: 10.1007/s007050170185. [DOI] [PubMed] [Google Scholar]
- 10.Flemington E K. Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol. 2001;75:4475–4481. doi: 10.1128/JVI.75.10.4475-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger Kruppel-associated box protein. Mol Cell Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gessain A, Sudaka A, Briere J, Fouchard N, Nicola M A, Rio B, Arborio M, Troussard X, Audouin J, Diebold J, de The G. Kaposi sarcoma-associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood. 1996;87:414–416. [PubMed] [Google Scholar]
- 13.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 1990;18:6835–6843. doi: 10.1093/nar/18.23.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gwack Y, Byun H, Hwang S, Lim C, Choe J. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J Virol. 2001;75:1909–1917. doi: 10.1128/JVI.75.4.1909-1917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S F, Robinson D R, Miller G, Kung H J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 19.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukac D M, Garibyan L, Kirshner J R, Palmeri D, Ganem D. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J Virol. 2001;75:6786–6799. doi: 10.1128/JVI.75.15.6786-6799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manet E, Rigolet A, Gruffat H, Giot J, Sergeant A. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 1991;19:2661–2667. doi: 10.1093/nar/19.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcello A, Massimi P, Banks L, Giacca M. Adeno-associated virus type 2 Rep protein inhibits human papillomavirus type 16 E2 recruitment of the transcriptional coactivator p300. J Virol. 2000;74:9090–9098. doi: 10.1128/jvi.74.19.9090-9098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore P S, Chang Y. Detection of herpesvirus-like DNA in Kaposi's sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 25.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulose-Murphy M, Ha N-K, Xiang C, Chen Y, Gillim L, Yarchoan R, Meltzer P, Bittner M, Trent J, Zeichner S. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) J Virol. 2001;75:4842–4853. doi: 10.1128/JVI.75.10.4843-4853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragoczy T, Miller G. Role of the Epstein-Barr virus Rta protein in activation of distinct classes of viral lytic cycle genes. J Virol. 1999;73:9858–9866. doi: 10.1128/jvi.73.12.9858-9866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 29.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakakibara S, Ueda K, Chen J, Okuno T, Yamanishi K. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J Virol. 2001;75:6894–6900. doi: 10.1128/JVI.75.15.6894-6900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 32.Seaman W T, Ye D, Wang R X, Hale E E, Weisse M, Quinlivan E B. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology. 1999;263:436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 33.Shannon M, Kim J, Ashworth L, Branscomb E, Stubbs L. Tandem zinc-finger gene families in mammals: insights and unanswered questions. DNA Seq. 1998;8:303–315. doi: 10.3109/10425179809034075. [DOI] [PubMed] [Google Scholar]
- 34.Song M J, Brown H J, Wu T-T, Sun R. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:3129–3140. doi: 10.1128/JVI.75.7.3129-3140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 36.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takada K, Shimuzu N, Sakuma S, Keating A. transactivation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Liu S, Wu M, Geng Y, Wood C. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch Virol. 2001;146:1415–1426. doi: 10.1007/s007050170102. [DOI] [PubMed] [Google Scholar]
- 40.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 41.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, Miller R F, Weller I V D, Weiss R A, Schulz T F. Detection of Kaposi sarcoma-associated herpesvirus (KSHV) in peripheral blood of HIV-infected individuals predicts progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 42.Yu Y, Black J B, Goldsmith C S, Browning P J, Bhalla K, Offermann M K. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J Gen Virol. 1999;80:83–90. doi: 10.1099/0022-1317-80-1-83. [DOI] [PubMed] [Google Scholar]
- 43.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Chiu J, Lin J C. Activation of human herpesvirus 8 (KSHV) thymidine kinase (TK) TATAA-less promoter by KSHV ORF50 gene product is SP1 dependent. DNA Cell Biol. 1998;17:735–742. doi: 10.1089/dna.1998.17.735. [DOI] [PubMed] [Google Scholar]
- 45.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]