Abstract
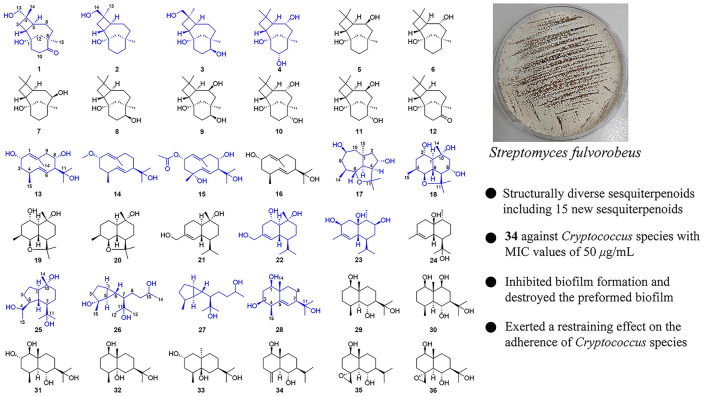
Thirty-six structurally diverse sesquiterpenoids, including caryolanes (1–12), germacranes (13–16), isodaucane (17), cadinanes (18–22), epicubenols (23, 24), oplopanane (25), pallenanes (26, 27), and eudesmanes (28–36), were isolated from the fermentation broth of Streptomyces fulvorobeus derived from Elephas maximus feces. Pallenane is a kind of rarely reported sesquiterpene with a distinctive C5/C3 bicyclic skeleton and was firstly found from microbial source. The structures of fifteen new compounds (1–4, 13–15, 17, 18, 22, 23, 25–28) were established through detailed spectroscopic data analysis, which included data from experimental and calculated ECD spectra as well as Mosher’s reagent derivative method. Compound 34 exhibited moderate antifungal activity against Cryptococcus neoformans and Cryptococcus gattii with MIC values of 50 μg/mL. It effectively inhibited biofilm formation and destroyed the preformed biofilm, as well as hindered the adhesion of Cryptococcus species. The current work would enrich the chemical diversity of sesquiterpenoid family.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s13659-024-00481-9.
Keywords: Sesquiterpenoids, Streptomyces fulvorobeus, Fermentation, Antifungal activity
Introduction
A huge amount of microorganisms colonize the guts of mammals [1, 2]. These microbes and their metabolites possess the ability to regulate intestinal epithelial cell proliferation, angiogenesis, host energy, lipid metabolism, and inflammatory immune response [3–6]. The bacterial community in fresh and unpolluted feces could largely represent the distal gut bacteria. Due to its noninvasive and convenient collection, fecal samples are commonly used for studying gut bacteria [7, 8]. In the past few years, the authors have reported on research regarding the structural diversity, antimicrobial, anti-inflammatory, and cytotoxic activities associated with animal feces [9–15]. These findings demonstrated that animal gut microorganisms could be considered as an abundant and significant microbial resource, which has prompted an investigation into the secondary metabolites produced by actinobacteria inhabiting in the animal intestinal tract.
Sesquiterpenoids undergo diverse cyclization cascades with their substrate farnesyl diphosphate (FPP), resulting in a variety of structural skeleton types and are widely distributed in plants, fungi, and red algae [16–18]. However, the discovery of these metabolites in bacteria has been limited due to difficulties in separation, low yield, and the absence of chromophores [19, 20]. With further research, the genome mining technology was applied and it has been found that terpene synthase and cyclase are also widely distributed in bacteria, especially actinomycetes [20–22]. According to the literatures, germacrane, pentalenene, zizaane, cadinane, and caryolane are the five most commonly detected type of sesquiterpenoids in bacteria [21, 23]. The exploration of a wider range of sesquiterpenoids in bacteria holds great prospect.
In an ongoing search for structurally diverse sesquiterpenes from actinomycetes associated with animal feces, we systematically investigated the secondary metabolites of Streptomyces fulvorobeus (YIM 103582), which was isolated from Elephas maximus feces. The chemical analysis of fermentation broth of S. fulvorobeus led to obtain fifteen new compounds, (1S,2R,4S,5S,8R)-9-oxocaryolane-1,13-diol (1), (1S,2R,4R,5S,8R)-caryolane-1,14-diol (2), (1S,2R,4R,5S,8R,9S)-caryolane-1,9,14-triol (3), caryolane-1,6α,10α-triol (4), (2S,4S,7S,8S)-1(10)E,5E-germacradiene-2,8,11-triol (13), (2S,4S,7R)-1(10)E,5E-germacradiene-2,11-diol 2-methyl ether (14), (2S,4R,7S,8S)-1(10)E,5E-germacradiene-2,4,8,11-tetraol 2-acetate (15), (1α,3α,4β,5α,6α,7β,9β)-6,11-epoxyisodaucane-3,9-diol (17), 8α-hydroxyganodermanol L (18), (1S,2S,6R,7S,10R)-cadinane-2,10,15-triol (22), (1R,3S,6R,7S,9S,10R)-3,9-dihydroxyepicubenol (23), oplopanane-4,10α,11-triol (25), 4-epi-pallenane-4α,10,11-triol (26), 4-epi-pallenane-10,11-diol (27), (1R,3S,4R,7R,10R)-eudesm-5-ene-1,3,11-triol (28) as well as twenty-one known analogues. The types of structural skeleton include caryolane, germacrene, isodaucane, cadinane, epicubenol, oplopanane, pallenane, and eudesmane. The structures of the new compounds were elucidated based on detailed spectroscopic data analysis. In this study, we report the fermentation, isolation, structural elucidation, and evaluation of antimicrobial activities of the isolated compounds.
Results and discussion
The S. fulvorobeus was obtained from fresh feces of E. maximus collected in the Xishuangbanna National Nature Reserve. The fermentation broth of S. fulvorobeus was clarified with a centrifuge to collected 150 L of culture supernatant. The clarified supernatant was extracted with ethyl acetate and the extract was isolated by repeated column chromatography over silica gel, Sephadex LH-20, and ODS to afford thirty-six sequiterpenoids (Fig. 1).
Fig. 1.
Chemical structures of compounds 1–36 from S. fulvorobeus
Structural elucidation of isolated new compounds
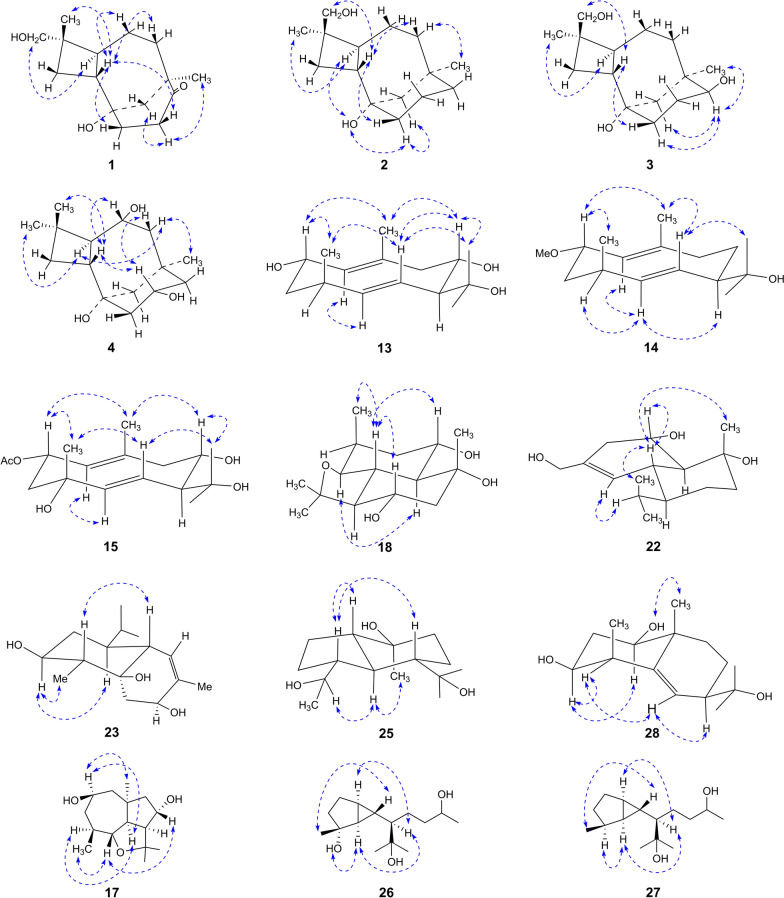
Compound 1 was isolated as a colorless oil with the molecular formula C15H24O3 based on HRESIMS and 13C NMR data. The IR spectrum of 1 showed characteristic absorption bands for hydroxy groups (3382 cm−1) and carbonyl (1699 cm−1). The 1H NMR (Table 1) of 1 displayed the presence of an oxygenated methylene (δH 3.14, 3.11), two singlet methyls (δH 1.01, 0.88), and other aliphatic residues at δH 0.98–2.82. The 13C NMR data (Table 2) of 1 showed 15 carbons signals, including one carbonyl (δC 216.9), two oxygen bearing carbons (δC 71.0, 68.1), two quaternary carbons (δC 46.5, 38.4), six methylenes (δC 49.6, 38.6, 36.9, 34.7, 30.3, 26.4), two methines (δC 43.5, 39.9), and two methyls (δC 31.1, 17.7). Analysis of the 1D and 2D NMR data indicated that 1 was consistent with a caryolane-type sesquiterpenoid and exhibited similarity to the reported bacaryolane A [19], except for the presence of an additional hydroxymethyl (δC 71.0, δH 3.14, 3.11) and the absence of a methyl. The HMBC correlations from H-13 (δH 3.14, 3.11) to C-3 (δC 30.3) and C-14 (δC 17.7), from H-3β (δH 1.78) to C-13 (δC 71.0), and from H-14 (δH 0.88) to C-13 (δC 71.0) established the hydroxymethyl was located at C-4. Moreover, the HMBC correlations from H-10 (δH 2.82, 2.17), H-11 (δH 1.84, 1.70), H-12 (δH 1.89), and H3-15 (δH 1.01) to C-9 (δC 216.9) confirmed that the carbonyl was located at C-9 (Fig. 2A). Caryolanes derived from plants and bacteria possessed varied stereochemical structures as they were biosynthesized by different cyclization from the humulyl cation [19]. The carbon skeleton of caryolanes in the current study was defined the same as those from bacteria such as bacaryolanes A-C [19]. The NOESY correlations found between H-13 (δH 3.14, 3.11) and H-5 (δH 1.82), between H3-14 (δH 0.88) and H-2 (δH 1.59) established that H-2 and H3-14 were β-orientated, whereas H-5 and H2-13 were α-orientated. In addition, NOE correlations between H3-15 (δH 1.01) and H-10α (δH 2.82), between H-10β (δH 2.17) and H-2 (δH 1.59) further confirmed the structure (Fig. 3). The absolute configuration 1S,2R,4S,5S,8R were deduced from comparison of experimental and calculated ECD spectra of 1 (Fig. 4A). According to the literature, the proton signals of the oxymethylene protons in the (S)- and (R)-α-methoxy-α-trifluoromethylphenylacetyl (MTPA) esters of primary alcohol showed a unique split pattern [24]. 1 was treated with (R)-MTPA-Cl and (S)-MTPA-Cl to afford the (S)- or (R)-MTPA ester derivatives 1a and 1b, respectively. The signals of oxymethylene protons at C-13 for the (S)-MTPA ester 1a appeared at δlow 4.26 and δhigh 4.24 (Δδ 0.02), while those for the (R)-MTPA ester 1b were observed as two separated doublet signals at δlow 4.30 and δhigh 4.22 (Δδ 0.08). The (R)-MTPA esters of primary alcohol analogue possessing 4S-configuration had relatively larger Δδ (δlow–δhigh) values. Therefore, the structure of 1 was elucidated as (1S,2R,4S,5S,8R)-9-oxocaryolane-1,13-diol.
Table 1.
1H NMR (600 MHz, DMSO-d6) spectroscopic data for compounds 1–4
| No | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 2 | 1.59, q (9.6) | 2.22, brq (10.8) | 2.27, brq (11.5) | 1.86, brq (10.2) |
| 3 |
1.78, t (10.2) 1.26, t (9.6) |
1.61, m 1.40, m |
1.59, m 1.35, m |
1.50, t (10.0) 1.37, t (9.0) |
| 5 | 1.82, m | 1.85, dt (12.1, 7.6) | 1.89, ddd (12.2, 9.4, 6.3) | 1.61, dd (12.0, 8.4) |
| 6 |
1.34, m 1.11, qd (12.6, 5.4) |
1.48, m 1.38, m |
1.49, m 1.37, m |
3.51, m |
| 7 |
1.87, m 0.98, td (12.7, 5.4) |
1.50, m 1.00, m |
1.58, m 0.87, m |
1.51, m 1.15, dd (14.4, 7.8) |
| 9 |
1.31, m 0.95, m |
3.10, t (10.2) |
1.57, dd (12.0, 4.2) 0.88, t (12.0) |
|
| 10 |
2.82, ddd (16.6, 12.0, 4.8) 2.17, ddd (16.6, 9.6, 3.6) |
1.63, m 1.53, m |
1.63, m 1.58, m |
3.68, m |
| 11 |
1.84, m 1.70, m |
1.45, m 1.16, td (12.0, 5.2) |
1.43, m 1.30, m |
1.74, brd (11.4) 1.05, m |
| 12 |
1.89, brs 1.89, brs |
1.57, brd (12.8) 0.91, brd (12.8) |
1.46, m 0.89, m |
1.83, brd (12.7) 0.92, d (12.7) |
| 13 |
3.14, d (10.2) 3.11, d (10.2) |
0.98, s | 0.98, s | 1.01, s |
| 14 | 0.88, s |
3.48, dd (10.6, 4.8) 3.40, dd (10.6, 5.4) |
3.49, d (10.4) 3.42, d (10.4) |
1.03, s |
| 15 | 1.01, s | 0.81, s | 0.82, s | 0.95, s |
| 1-OH | 3.94, s | 3.97, brs | 4.08, s | |
| 6-OH | 4.08, s | |||
| 9-OH | 4.25, brs | |||
| 10-OH | 4.37, s | |||
| 14-OH | 4.28, t (5.2) | 4.33, brs |
Table 2.
13C NMR (150 MHz, DMSO-d6) spectroscopic data for compounds 1–4, 13–15
| No | 1 | 2 | 3 | 4 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|
| 1 | 68.1, C | 69.4, C | 69.2, C | 69.5, C | 136.1, CH | 131.4, CH | 129.8, CH |
| 2 | 43.5, CH | 39.6, CH | 37.5, CH | 41.5, CH | 63.8, CH | 73.8, CH | 69.3, CH |
| 3 | 30.3, CH2 | 30.6, CH2 | 30.3, CH2 | 35.5, CH2 | 40.5, CH2 | 40.0, CH2 | 45.4, CH2 |
| 4 | 38.4, C | 39.7, C | 39.9, C | 34.1, C | 32.3, CH | 32.0, CH | 70.5, C |
| 5 | 39.9, CH | 44.1, CH | 43.4, CH | 52.6, CH | 139.7, CH | 138.9, CH | 142.1, CH |
| 6 | 26.4, CH2 | 21.5, CH2 | 19.7, CH2 | 70.2, CH | 121.7, CH | 125.6, CH | 123.7, CH |
| 7 | 38.6, CH2 | 36.8, CH2 | 30.1, CH2 | 49.7, CH2 | 61.8, CH | 58.7, CH | 60.9, CH |
| 8 | 46.5, C | 34.6, C | 38.8, C | 34.4, C | 67.6, CH | 21.9, CH2 | 68.6, CH |
| 9 | 216.9, C | 37.6, CH2 | 77.1, CH | 48.6, CH2 | 52.1, CH2 | 41.4, CH2 | 51.6, CH2 |
| 10 | 34.7, CH2 | 20.7, CH2 | 30.0, CH2 | 66.1, CH | 132.9, C | 138.2, C | 137.7, C |
| 11 | 36.9, CH2 | 38.6, CH2 | 38.3, CH2 | 50.0, CH2 | 73.4, C | 70.8, C | 73.4, C |
| 12 | 49.6, CH2 | 49.3, CH2 | 48.4, CH2 | 48.2, CH2 | 30.8, CH3 | 29.8, CH3 | 30.8, CH3 |
| 13 | 71.0, CH2 | 26.6, CH3 | 26.6, CH3 | 31.8, CH3 | 24.5, CH3 | 26.0, CH3 | 24.6, CH3 |
| 14 | 17.7, CH3 | 65.3, CH2 | 65.3, CH2 | 21.5, CH3 | 18.4, CH3 | 17.5, CH3 | 18.4, CH3 |
| 15 | 31.1, CH3 | 33.6, CH3 | 29.6, CH3 | 36.9, CH3 | 16.1, CH3 | 16.0, CH3 | 24.3, CH3 |
| 2-OAc | 169.9, C | ||||||
| 21.5, CH3 | |||||||
| 2-OCH3 | 54.5, CH3 |
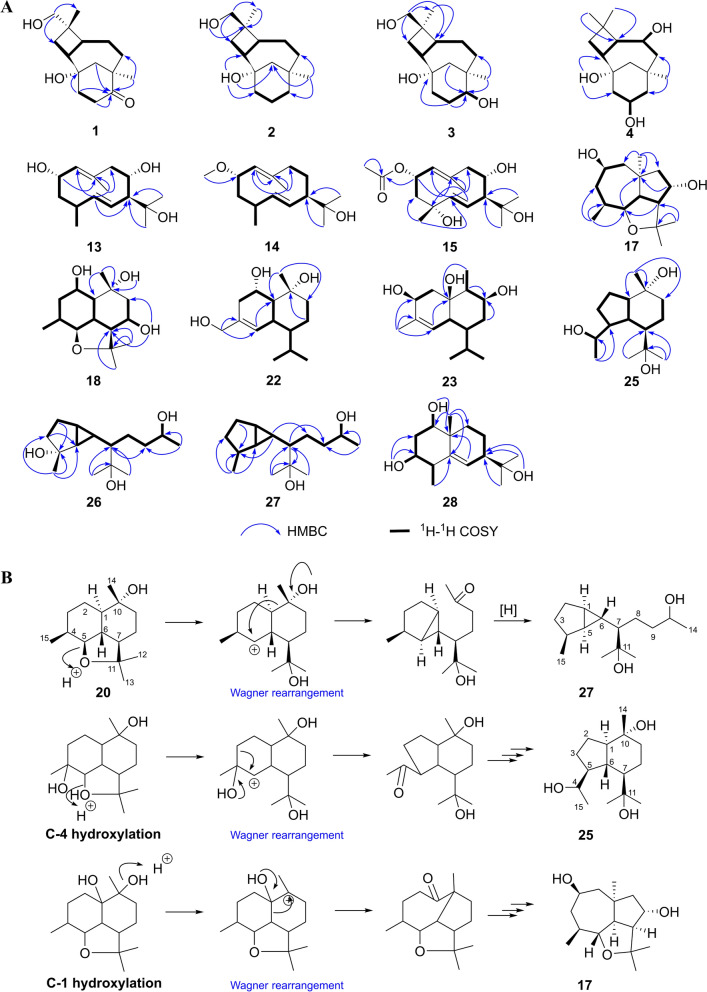
Fig. 2.
Main HMBC and COSY correlations of compounds 1–4, 13–15, 17, 18, 22, 23, 25–28 (A). proposed biosynthetic pathway of compounds 17, 25, 27 (B)
Fig. 3.
Main NOE correlations of compounds 1–4, 13–15, 17, 18, 22, 23, 25–28
Fig. 4.
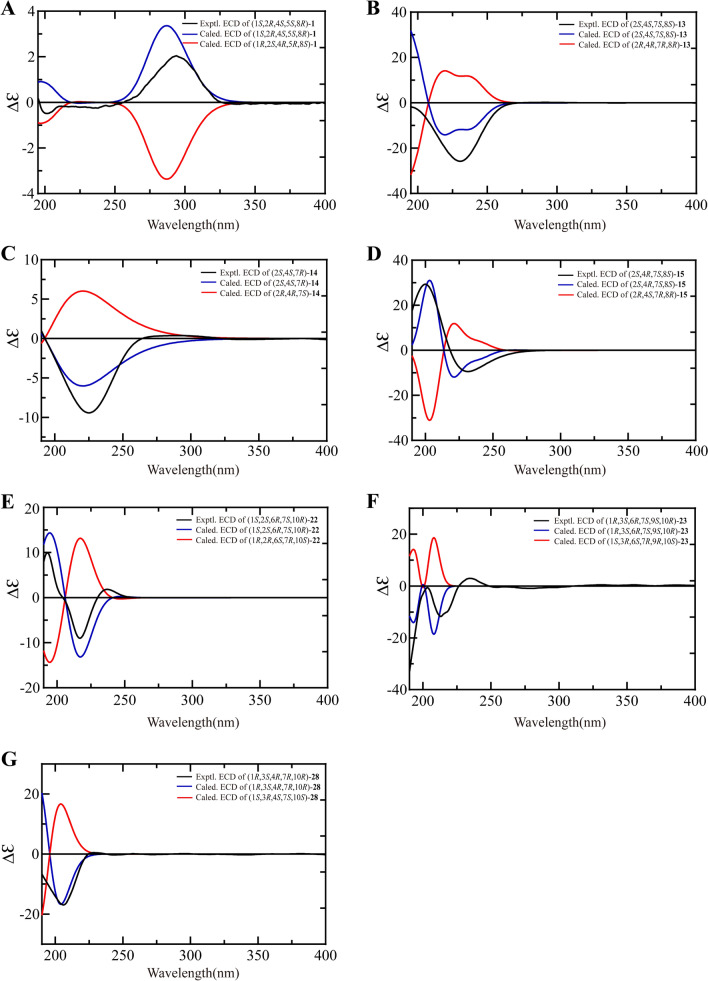
Experimental ECD spectra and calculated ECD spectra of compounds 1 (A), 13–15 (B−D), 22 (E), 23 (F), 28 (G)
Compound 2 had the molecular formula C15H26O2 by its HRESIMS and 13C NMR data. Its 1H and 13C NMR data revealed that 2 was a caryolane type sesquiterpenoid and related to the known compound caryolan-1-ol [25]. The distinct difference was that a methyl of caryolan-1-ol was replaced by a hydroxymethyl (δC 65.3, δH 3.48, 3.40) in 2. The HMBC correlations between H-14 (δH 3.48, 3.40) and C-3 (δC 30.6) and C-13 (δC 26.6), between H-3 (δH 1.61, 1.40), H-13 (δH 0.98) and C-14 (δC 65.3) determined the hydroxymethyl at C-4 (Fig. 2A). NOE correlations between H-5 (δH 1.85) and 1-OH (δH 3.94), H3-13 (δH 0.98), between H-2 (δH 2.22) and H-14 (δH 3.48, 3.40), H-7β (δH 1.50), between H-7α (δH 1.00) and H3-15 (δH 0.81) indicated that H-2, H-14 were β-orientation whereas 1-OH, H-5, H3-13, H3-15 were α-orientation (Fig. 3). The absolute configuration of C-4 was determined by Mosher’s method. Treatment of 2 with (R)-MTPA-Cl or (S)-MTPA-Cl obtained the S or R Mosher’s esters 2a and 2b. The signals of oxymethylene protons at C-14 for the (S)-MTPA ester 2a showed two separated doublet signals at δlow 4.69 and δhigh 4.55 (Δδ 0.14), while those for the (R)-MTPA ester 2b were presented at δH 4.63 as a broad singlet peak. These findings suggested the R-configuration of C-4 in 2 by comparing the Δδ values of oxymethylene protons with those of 4S-configuration analogue 1. Thus, 2 was deduced to be (1S,2R,4R,5S,8R)-caryolane-1,14-diol as the same biosynthesis pathway from bacteria.
Compound 3 afforded a molecular formula of C15H26O3 on the basis of HRESIMS and 13C NMR data. Analysis of its 1H and 13C NMR data (Tables 1, 2) indicated that 3 has a close structural relationship to 2. The only difference was that one methylene was absent and one oxygenated methine was present (δC 77.1, δH 3.10) in 3. The HMBC correlations from H-10 (δH 1.63, 1.58), H-12 (δH 1.46, 0.89), H3-15 (δH 0.82) to C-9 (δC 77.1) as well as 1H–1H COSY correlations between H-9 (δH 3.10) and H-10 (δH 1.63, 1.58) confirmed a hydroxy at C-9 (Fig. 2A). The NOE cross peaks observed between H3-13 (δH 0.98) and H-5 (δH 1.89), between H3-15 (δH 0.82) and H-9 (δH 3.10), between H-9 (δH 3.10) and H-11α (δH 1.30), and between H-2 (δH 2.27) and H-14 (δH 3.49, 3.42) and H-11β (δH 1.43) indicated that H-2, H-14, and 9-OH were β-oriented, while H-5, H-9, H3-13, and H3-15 were α-oriented (Fig. 3). The (S)-MTPA ester (3a) or (R)-MTPA ester (3b) were obtained by the same derivative process as those of 1 and 2. Both 9-OH and 14-OH in compound 3 were esterified by MTPA-Cl according to NMR data. Unfortunately, it was impossible to assign the absolute configuration of C-4 as the Δδ values of two separated peaks of H2-14 for 3a and 3b were very approximate (Δδ = 0.12 for 3a and Δδ = 0.10 for 3b) being influenced by two MTPA groups. As 3 presented almost identical 13C NMR data of C-4, C-13, and C-14 with those of compound 2, the R-configuration of C-4 could be determined. The configuration of C-9 could be deduced by comparing the chemical shift of H3-15 in (S)- and (R)-MTPA esters. The signal of H3-15 for (S)-MTPA ester 3a was observed at lower field (δH 0.99) compared to the signal for (R)-MTPA ester 3b (δH 0.88), which exhibited the absolute configuration of C-9 to be S. Thus, the structure of 3 was confirmed as (1S,2R,4R,5S,8R,9S)-caryolane-1,9,14-triol.
The molecular formula of 4 was deduced as C15H26O3 according to its HRESIMS and 13C NMR data. The 1H NMR and 13C NMR data (Tables 1, 2) indicated that 4 was similar to bacaryolane C (6) [19], also isolated in the current study. The obvious alteration was that a methylene in 6 was replaced by an oxygenated methine (δC 66.1, δH 3.68) in 4. The HMBC correlations from H-9 (δH 1.57, 0.88), H-11 (δH 1.74, 1.05) to C-10 (δC 66.1) confirmed that a hydroxy was located at C-10. The COSY correlations from H-10 (δH 3.68) to H-9 (δH 1.57, 0.88), H-11 (δH 1.74, 1.05), and 10-OH (δH 4.37) further supported the above inference (Fig. 2A). The NOESY cross peaks from H-2 (δH 1.86) to H-6 (δH 3.51) and H-10 (δH 3.68), from H-10 (δH 3.68) to H-7β (δH 1.51) indicated that H-2, H-6, and H-10 were on the same side. Correspondingly, the NOESY correlations from H3-15 (δH 0.95) to H-7α (δH 1.15), from H-7α (δH 1.15) to H-5 (δH 1.61) suggested that H-5, H3-15 were on the opposite side (Fig. 3). Consequently, 4 was deduced to be caryolane-1,6α,10α-triol.
Compound 13 was isolated as a colorless oil with a molecular formula C15H26O3 by HRESIMS and 13C NMR data. Its IR spectrum revealed the presence of hydroxy groups (3315 cm−1) and double bonds (1668 cm−1). The 1H NMR data (Table 3) showed three olefinic hydrogens (δH 5.41, 4.90, 4.65), two oxygenated methines (δH 4.19, 3.80), three singlet methyls (δH 1.53, 1.14, 0.93), one doublet methyl (δH 0.97), and other aliphatic hydrogens (δH 1.37–2.41). The 13C NMR data (Table 2) of 13 displayed 15 carbon signals, including four olefinic carbons (δC 139.7, 136.1, 132.9, 121.7), three oxygen-bearing carbons (δC 73.4, 67.6, 63.8), two methines (δC 61.8, 32.3), two methylenes (δC 52.1, 40.5), and four methyls (δC 30.8, 24.5, 18.4, 16.1), as determined in an HSQC experiment. The 1H and 13C NMR data of 13 were very similar to those of 1(10)E,5E-germacradiene-2α,11-diol (16) [26], a germacrane-type sesquiterpenoid. The major differences were the disappearance of a methylene in 16 and the existence of an oxygen-bearing methine (δC 67.6, δH 3.80) in 13. The 1H–1H COSY correlations from H-8 (δH 3.80) to H-7 (δH 2.28) and H-9 (δH 2.27, 2.25) as well as the HMBC correlations between H-7 (δH 2.28), H-9 (δH 2.27, 2.25) and C-8 (δC 67.6) indicated that a hydroxy was connected with C-8 (Fig. 2A). Thus, the planar structure of compound 13 was established. The NOE interactions observed between H-2 (δH 4.19) and H3-15 (δH 0.97), between H-6 (δH 4.65) and H-8 (δH 3.80), H3-15 (δH 0.97), between H3-13 (δH 1.14) and H-6 (δH 4.65), H-8 (δH 3.80) revealed that H-2, H-8, and H3-15 were on the same side, while 2-OH, 8-OH, H-4, and H-7 were located on the opposite side (Fig. 3). Furthermore, the coupling constants of H-2 (δH 4.19, td, J = 10.2, 4.2 Hz) suggested that the 2-OH presented in the equatorial position. In addition, NOE interactions observed from H3-14 (δH 1.53) to H-2 (δH 4.19) and H-8 (δH 3.80), from H-1 (δH 4.90) to H-5 (δH 5.41) as well as the large coupling constant of H-5/H-6 elucidated the E configurations of C-1/C-10 and C-5/C-6 double bonds. The experimental ECD spectrum of 13 had a consistent trend with its corresponding calculated ECD curve which determined the 2S,4S,7S,8S-configurations (Fig. 4B). Consequently, 13 was established as (2S,4S,7S,8S)-1(10)E,5E-germacradiene-2,8,11-triol.
Table 3.
1H NMR (600 MHz, DMSO-d6) spectroscopic data for compounds 13–15, 17, 18
| No | 13 | 14 | 15 | 17 | 18 |
|---|---|---|---|---|---|
| 1 | 4.90, d (9.6) | 4.80, d (10.2) | 4.82, d (10.2) | 1.22, dd (12.5, 9.7) | |
| 2 | 4.19, td (10.2, 4.2) | 3.90, td (10.2, 3.6) | 5.21, dd (11.4, 4.2) | 1.82, m | 3.67, brt (9.6) |
| 1.53, dd (13.8, 3.6) | |||||
| 3 |
1.67, dt (13.2, 4.2) 1.37, ddd (12.6, 10.8, 3.6) |
1.76, dt (13.2, 4.2) 1.52, ddd (13.8, 10.8, 3.6) |
1.76, dd (12.0, 3.6) 1.48, t (12.0) |
4.17, quint (5.6) |
1.69, ddd (13.8, 4.2, 3.0) 1.42, m |
| 4 | 2.41, m | 2.41, m | 2.30, dd (8.4, 4.8) | 2.05, m | |
| 5 | 5.41, dd (15.6, 3.6) | 5.35, dd (15.6, 3.0) | 5.20, d (15.6) | 1.94, t (9.5) | 3.46, dd (10.2, 4.8) |
| 6 |
4.65, ddd (15.6, 10.2, 2.4) |
4.84, ddd (15.6, 9.6, 1.8) |
4.72, dd (15.6, 10.2) | 3.13, t (9.9) | 1.16, m |
| 7 | 2.28, m | 2.10, brq (10.8) | 2.25, t (10.2) | 1.42, m | 1.38, m |
| 8 | 3.80, td (9.6, 3.6) |
1.71, m 1.24, brq (11.4) |
3.87, td (10.2, 4.2) |
1.82, m 1.04, q (12.0) |
3.38, m |
| 9 |
2.27, m 2.25, m |
2.27, dd (12.6, 4.2) 2.16, td (12.6, 1.8) |
2.32, dd (12.3, 9.7) 2.29, dd (12.3, 4.0) |
3.51, td (10.2, 1.8) |
1.77, dd (12.0, 3.6) 1.40, m |
| 10 | 1.62, brd (13.6) | ||||
| 1.48, dd (13.6, 10.2) | |||||
| 12 | 0.93, s | 0.99, s | 0.98, s | 1.17, s | 1.26, s |
| 13 | 1.14, s | 0.94, s | 1.14, s | 1.17, s | 1.07, s |
| 14 | 1.53, s | 1.57, s | 1.61, s | 0.87, d (6.0) | 1.17, s |
| 15 | 0.97, d (6.6) | 1.02, d (6.6) | 1.15, s | 1.16, s | 0.84, d (7.2) |
| 2-OH | 4.34, d (3.0) | ||||
| 2-OAc | 1.93, s | ||||
| 2-OCH3 | 3.06, s | ||||
| 8-OH | 4.56, d (5.4) | ||||
| 10-OH | 4.33, s | ||||
| 11-OH | 4.04, s |
The 1H and 13C NMR data (Tables 2, 3) as well as a molecular formula of C16H28O2 exhibited that 14 bore a close resemblance to the known compound 16 [26], except for the presence of an additional methoxy (δC 54.5, δH 3.06). The HMBC correlation between methoxy (δH 3.06) and C-2 (δC 73.8) determined that the methoxy was located at C-2 (Fig. 2A). In NOESY spectrum, the key cross peaks from H-2 (δH 3.90) to H3-15 (δH 1.02), from H-5 (δH 5.35) to H-4 (δH 2.41) and H-7 (δH 2.10) indicated that H-2 and H3-15 were β-orientation, while H-4 and H-7 were α-orientation (Fig. 3). Moreover, the double bonds at C-5/C-6, C-1/C-10 were elucidated as E geometry based on the NOE correlations from H3-14 (δH 1.57) to H-2 (δH 3.90) and H-6 (δH 4.84), from H-1 (δH 4.80) to H-5 (δH 5.35) as well as the large coupling constant of H-5/H-6 (J = 15.6 Hz). The absolute configurations of C-2, C-4, and C-7 were elucidated as 2S,4S,7R based on comparing the experimental and calculated ECD spectra (Fig. 4C). Therefore, the structure of 14 was confirmed as (2S,4S,7R)-1(10)E,5E-germacradiene-2,11-diol 2-methyl ether.
Compound 15 was assigned a molecular formula of C17H28O5 based on its HRESIMS and 13C NMR data. The characteristic 1H and 13C NMR data (Tables 2, 3) of 15 suggested that it was a germacrane-type sesquiterpenoid and had a close structural relationship to 13, except for the different oxygenated position and an additional acetyl (δC 169.9, 21.5). HMBC correlations from H-3 (δH 1.76, 1.48), H-6 (δH 4.72) and H3-15 (δH 1.15) to C-4 (δC 70.5) confirmed that the extra hydroxy was located at C-4. HMBC correlations from H-2 (δH 5.21) to C-16 (δC 169.9) as well as 1H–1H COSY correlations from H-2 (δH 5.21) to H-1 (δH 4.82) and H-3 (δH 1.76, 1.48) indicated that the acetyl was located at C-2. (Fig. 2A). 15 presented the same relative configuration as those of 13 and 14 on the basis of NOE correlations between H-2 (δH 5.21) and H3-15 (δH 1.15), H3-14 (δH 1.61), between H3-14 (δH 1.61) and H-8 (δH 3.87), between H3-13 (δH 1.14) and H-6 (δH 4.72), H-8 (δH 3.87) (Fig. 3). Besides, the geometry at C-5/C-6 double bonds was assigned as E according to the large coupling constants of H-5/H-6 (J = 15.6 Hz). The (+) Cotton effect at 200 nm (+ 29.25) and (−) Cotton effect at 231 nm (− 9.42) of 15 detected by CD spectrum was consistent with those of the calculated ECD curve of 2S,4R,7S,8S-15 (Fig. 4D). Thus, 15 was identified as (2S,4R,7S,8S)-1(10)E,5E-germacradiene-2,4,8,11-tetraol 2-acetate.
The HRESIMS and 13C NMR data determined the molecular formula of 17 as C15H26O3. The NMR data (Tables 3, 4) suggested 17 was a 6,11-epoxyisodaucane type sesquiterpenoid and related to the known compound 6,11-epoxyisodaucane [27]. The difference was that two oxygenated methines replaced two methylenes in 17. 1H–1H COSY correlations from H-3 (δH 4.17) to H-4 (δH 2.30) and H-2 (δH 1.82, 1.53), from H-9 (δH 3.51) to H-10 (δH 1.62, 1.48) and H-8 (δH 1.82, 1.04) determined the two hydroxys at C-3 and C-9, respectively. The HMBC correlations between H3-15 (δH 1.16) and C-1 (δC 38.9), C-2 (δC 51.9), C-5 (δC 61.6), and C-10 (δC 51.7), between H-6 (δH 3.13) and C-1 (δC 38.9), C-8 (δC 47.7), between H-5 (δH 1.94) and C-2 (δC 51.9), C-3 (δC 73.0), C-10 (δC 51.7), and C-15 (δC 31.3), between H-12 (δH 1.17) and C-4 (δC 64.2), C-11 (δC 80.0) confirmed the planar structure (Fig. 2A). The absolute configuration of 6,11-epoxyisodaucane was determined by total synthesis method [27]. The similar coupling constants of H-5 (δH 1.94, t, J = 9.5 Hz), H-6 (δH 3.13, t, J = 9.9 Hz) indicated that 17 possessed the same 4α-H, 5α-H, 6β-H, 7β-methyl configuratinos as those of synthesized 6,11-epoxyisodaucane. The NOE interactions from H-5 (δH 1.94) to H-9 (δH 3.51), H-7 (δH 1.42), from H3-14 (δH 0.87) to H-6 (δH 3.13), from H-6 (δH 3.13) to H-3 (δH 4.17), from H3-15 (δH 1.16) to H-9 (δH 3.51) determined the configurations as in Fig. 3.
Table 4.
13C NMR (150 MHz, DMSO-d6) spectroscopic data for compounds 17, 18, 22, 23, 25–28
| No | 17 | 18 | 22 | 23 | 25 | 26 | 27 | 28 |
|---|---|---|---|---|---|---|---|---|
| 1 | 38.9, C | 52.9, CH | 53.8, CH | 74.6, C | 58.9, CH | 23.8, CH | 23.8, CH | 76.5, CH |
| 2 | 51.9, CH2 | 66.7, CH | 70.1, CH | 34.3, CH2 | 25.7, CH2 | 26.2, CH2 | 28.0, CH2 | 35.6, CH2 |
| 3 | 73.0, CH | 39.8, CH2 | 36.9, CH2 | 68.0, CH | 24.2, CH2 | 37.2, CH2 | 30.2, CH2 | 68.7, CH |
| 4 | 64.2, CH | 31.0, CH | 136.9, C | 136.4, C | 67.2, CH | 78.8, C | 35.0, CH | 45.5, CH |
| 5 | 61.6, CH | 80.8, CH | 120.7, CH | 124.2, CH | 48.1, CH | 36.3, CH | 30.1, CH | 145.9, C |
| 6 | 82.3, CH | 40.0, CH | 40.5, CH | 47.4, CH | 44.3, CH | 23.1, CH | 18.9, CH | 126.6, CH |
| 7 | 38.2, CH | 59.1, CH | 46.5, CH | 45.7, CH | 55.5, CH | 52.0, CH | 52.1, CH | 48.2, CH |
| 8 | 47.7, CH2 | 67.0, CH | 21.8, CH2 | 34.3, CH2 | 28.4, CH2 | 27.6, CH2 | 27.7, CH2 | 21.1, CH2 |
| 9 | 66.7, CH | 52.8, CH2 | 41.4, CH2 | 71.3, CH | 42.8, CH2 | 39.4, CH2 | 39.4, CH2 | 38.3, CH2 |
| 10 | 51.7, CH2 | 74.1, C | 73.0, C | 51.3, CH | 71.2, C | 67.1, CH | 67.1, CH | 39.3, C |
| 11 | 80.0, C | 83.0, C | 26.5, CH | 26.6, CH | 72.4, C | 72.9, C | 73.0, C | 71.5, C |
| 12 | 24.8, CH3 | 24.4, CH3 | 15.4, CH3 | 15.5, CH3 | 24.5, CH3 | 26.5, CH3 | 26.3, CH3 | 28.2, CH3 |
| 13 | 32.8, CH3 | 31.1, CH3 | 21.8, CH3 | 22.0, CH3 | 31.9, CH3 | 30.6, CH3 | 30.7, CH3 | 25.6, CH3 |
| 14 | 20.3, CH3 | 23.4, CH3 | 21.7, CH3 | 11.5, CH3 | 20.6, CH3 | 24.2, CH3 | 24.3, CH3 | 21.1, CH3 |
| 15 | 31.3, CH3 | 11.9, CH3 | 64.9, CH2 | 20.3, CH3 | 23.3, CH3 | 26.4, CH3 | 18.9, CH3 | 16.7, CH3 |
Analysis of 1H NMR, 13C NMR (Tables 3, 4), and HRESIMS data of 18 indicated it was a sesquiterpenoid and was related to the reported ganodermanol L (19) [28]. The only difference was one more oxygenated methine (δC 67.0, δH 3.38) in 18 replaced a methylene in 19. The HMBC correlations between 8-OH (δH 4.56) and C-7 (δC 59.1), C-8 (δC 67.0), and C-9 (δC 52.8) determined the C-8 position of extra hydroxy. Moreover, COSY correlations from H-8 (δH 3.38) to H-7 (δH 1.38), H-9 (δH 1.77, 1.40), and 8-OH (δH 4.56) further supported the conclusion (Fig. 2A). NOE cross peaks observed between H-6 (δH 1.16) and H-2 (δH 3.67), H-8 (δH 3.38), H3-15 (δH 0.84), between H-5 (δH 3.46) and H-1 (δH 1.22) demonstrated that H-2, H-6, H-8, and H3-15 were located on the same side, whereas H-1, H-5, and H-7 were located on the opposite side (Fig. 3). There were potential inaccuracies of NOE correlations due to the overlapped signals of H-6 (δH 1.16) and H3-14 (δH 1.17). The configuration of C-10 was then confirmed through comparing the 13C NMR data of C-1, C-2, C-10, and C-14 with those of ganodermanol L (19) [28], which was elucidated the structure by X-ray crystallographic analyses. In addition, the peak shapes and coupling constants of H-1 (δH 1.22, dd, J = 12.5, 9.7 Hz), H-2 (δH 3.67, brt J = 9.6 Hz), and H-5 (δH 3.46, dd, J = 10.2, 4.8 Hz) further confirmed the relative configuration.
The 1H and 13C NMR data (Tables 4, 5) as well as molecular formula, C15H26O3, of 22 showed it was a cadinene-type sesquiterpenoid and related to 15-hydroxy-α-cadinol (21) [29], except an additional oxygenated methine (δC 70.1, δH 3.82) in 22 instead of a methylene in 21. 1H–1H COSY correlations from H-2 (δH 3.82) to H-3 (δH 2.20, 1.89) and H-1 (δH 1.30) as well as HMBC correlations from H-1 (δH 1.30), H-3 (δH 2.20, 1.89) to C-2 (δC 70.1) revealed that a hydroxy was located at C-2 (Fig. 2A). The large coupling constants of H-1 (δH 1.30, brt, J = 10.6 Hz), H-6 (δH 1.73, brt, J = 10.6 Hz) indicated a trans fusion of the bicyclic system. NOE correlations from H-6 (δH 1.73) to H-2 (δH 3.82), H3-14 (δH 1.18), and H3-12 (δH 0.73) indicated that the H-2, H-6, and H3-14 were β-orientated, whereas H-1, H-7, 2-OH, and 10-OH were α-orientated (Fig. 3). Comparison of the experimental and calculated ECD spectra of 22 revealed the absolute configuration as 1S,2S,6R,7S,10R (Fig. 4E). Thus, compound 22 was elucidated as (1S,2S,6R,7S,10R)-cadinane-2,10,15-triol.
Table 5.
1H NMR (600 MHz, DMSO-d6) spectroscopic data for compounds 22, 23, 25–28
| No | 22 | 23 | 25 | 26 | 27 | 28 |
|---|---|---|---|---|---|---|
| 1 | 1.30, t (10.6) | 1.26, m | 1.04, m | 0.97, m | 3.03, dt (11.6, 4.8) | |
| 2 |
3.82, td (10.2, 6.0) |
1.80, dd (12.8, 6.8) 1.19, m |
1.53, m 1.00, m |
1.87, m 1.56, dd (12.0, 7.9) |
1.65, m 1.65, m |
1.65, brq (12.0) 1.60, m |
| 3 |
2.20, dd (16.2, 4.8) 1.89, m |
3.96, brt (8.0) |
1.66, ddd (11.5, 7.8, 3.0) 1.22, m |
1.34, dd (13.4, 8.4) 1.11, m |
1.54, m 0.70, dq (12.8, 10.0) |
3.52, m |
| 4 | 4.23, m | 2.15, m | 2.39, quint (7.0) | |||
| 5 | 5.61, s | 5.33, d (5.6) | 2.00, brt (9.0) | 1.13, dd (6.0, 3.6) | 1.12, dt (6.3, 3.5) | |
| 6 | 1.73, brt (10.6) | 1.56, dd (11.2, 4.8) | 1.37, td (10.9, 8.0) | 0.13, dt (10.3, 3.4) | 0.27, dt (10.3, 3.3) | 5.53, m |
| 7 | 1.02, m | 1.16, m |
1.18, ddd (13.3, 10.5, 3.0) |
0.57, ddd (10.0, 8.0, 3.5) |
0.56, ddd (10.6, 7.8, 3.5) |
2.02, ddd (11.0, 6.2, 2.0) |
| 8 |
1.51, m 1.04, m |
1.67, dt (11.6, 4.3) 0.91, brd (11.6) |
1.61, dt (13.3, 3.5) 0.95, m |
1.62, m 1.26, m |
1.65, m 1.27, m |
1.56, m 1.19, m |
| 9 |
1.61, dt (12.6, 3.0) 1.38, td (12.6, 3.6) |
3.00, td (10.8, 4.4) |
1.57, dt (12.6, 3.4) 1.28, m |
1.60, m 1.25, m |
1.61, m 1.25, m |
1.80, dt (12.4, 3.2) 1.06, m |
| 10 | 1.23, m | 3.50, m | 3.51, m | |||
| 11 | 2.10, m | 1.95, m | ||||
| 12 | 0.73, d (7.2) | 0.78, d (7.2) | 1.02, s | 1.05, s | 1.06, s | 1.04, s |
| 13 | 0.88, d (6.6) | 0.86, d (7.2) | 1.09, s | 1.16, s | 1.16, s | 0.97, s |
| 14 | 1.18, s | 0.96, d (6.4) | 0.98, s | 1.04, d (6.1) | 1.03, d (6.1) | 0.95, s |
| 15 |
3.77, brd (10.8) 3.75, brd (10.8) |
1.64, s | 0.94, d (6.3) | 1.22, s | 0.99, d (6.6) | 0.96, d (6.4) |
| 1-OH | 4.45, d (4.8) | |||||
| 3-OH | 4.50, d (4.0) | |||||
| 4-OH | 4.04, d (5.6) | 4.24, s | ||||
| 10-OH | 4.03, s | 4.26, d (4.4) | 4.27, brs | |||
| 11-OH | 4.08, brs | 3.95, s | 3.93, brs | 4.15, s |
The molecular formula of 23 was determined to be C15H26O3 according to HRESIMS and 13C NMR data. Analysis of its 1H and 13C NMR data (Tables 4, 5) indicated that 23 possessed a resemble structural relationship to 3β-hydroxyepicubenol [13], except for one oxygenated methine (δC 71.3, δH 3.00) instead of a methylene in 23. 1H–1H COSY correlations from H-9 (δH 3.00) to H-8 (δH 1.67, 0.91) and H-10 (δH 1.23) supported that an additional hydroxy was located at C-9. This conclusion was confirmed by HMBC correlations between H-8 (δH 1.67, 0.91), H-10 (δH 1.23) and C-9 (δC 71.3) (Fig. 2A). The relative configuration of 23 was established from detailed analysis of NOE correlations. NOE interactions from H-9 (δH 3.00) to H-7 (δH 1.16) and H3-14 (δH 0.96), from H-6 (δH 1.56) to H-10 (δH 1.23) revealed that H-7, H-9, and H3-14 were α-orientation, while H-6 and H-10 were β-orientation (Fig. 3). Although there was no obvious NOE correlation to demonstrate the configuration of 1-OH and 3-OH, comparing the 13C NMR data of C-1 (δC 74.6), C-2 (δC 34.3), C-3 (δC 68.0), C-4 (δC 136.4), C-5 (δC 124.2), and C-6 (δC 47.4) of 23 with those of the analogues indicated that 23 possessed the same configurations of C-1 and C-3 as 3β-hydroxyepicubenol [13] and muurol-4-ene-1β,3β,10β-triol [30]. The absolute configurations of the chiral carbons were determined to be 1R,3S,6R,7S,9S,10R according to the experimental ECD spectrum of 23 closely matched the calculated ECD curve (Fig. 4F).
Compound 25 had a molecular formula of C15H28O3 from its HRESIMS and 13C NMR data. The characteristic 1H and 13C NMR data (Tables 4, 5) of 25 demonstrated an oplopanane sequiterpenoid and related to the known oplopanane-4,10α-diol [31]. The difference was a 2-hydroxypropan-2-yl (δC 72.4, 31.9, 24.5, δH 1.09, s, δH 1.02, s) in 25 at C-7 replaced an isopropyl, which was confirmed by HMBC correlations between H3-12 (δH 1.02) and C-7 (δC 55.5), C-11 (δC 72.4), between H3-13 (δH 1.09) and C-7 (δC 55.5), C-11 (δC 72.4). The large coupling constants of H-5 (δH 2.00, brt, J = 9.0 Hz), H-6 (δH 1.37, td, J = 10.9, 8.0 Hz), and H-7 (δH 1.18, ddd, J = 13.3, 10.5, 3.0 Hz) indicated the both trans configurations of H-5/H-6, H-6/H-7. In addition, the trans configuration of H-1/H-6 also could be deduced from the peak shape and coupling constants of H-6 (δH 1.37, td, J = 10.9, 8.0 Hz). The relative configuration of 25 was further determined by NOE correlations observed from H-5 (δH 2.00) to H-7 (δH 1.18) and H-1 (δH 1.26), from H-6 (δH 1.37) to H3-14 (δH 0.98) and H-4 (δH 4.23) (Fig. 3). Furthermore, comparing the 13C NMR data of C-14 (δC 20.6) with that of oplopanone (δC 20.3) or 10-epi-oplopanone (δC 28.2) further confirmed above inference [31, 32]. Thus, the structure of 25 was identified as 4,10α,11-oplopananetriol and the configuration of C-4 undetermined as the less amount.
The molecular formula of compound 26 was determined as C15H28O3 based on HRESIMS and 13C NMR data. The 1H NMR data (Table 5) presented one oxygenated methine (δH 3.50), three singlet methyls (δH 1.22, 1.16, 1.05), one doublet methyl (δH 1.04), and three active hydrogens (δH 4.26, 4.24, 3.95). The 13C NMR data (Table 4) of 26 displayed 15 carbon signals, which were assigned to three oxygen-bearing carbons (δC 78.8, 72.9, 67.1), four methines (δC 52.0, 36.3, 23.8, 23.1), four methylenes (δC 39.4, 37.2, 27.6, 26.2), and four methyls (δC 30.6, 26.5, 26.4, 24.2) with the aid of HSQC experiment. The obviously upfield of H-6 (δH 0.13, dt, J = 10.3, 3.4 Hz) and H-7 (δH 0.57, ddd, J = 10.0, 8.0, 3.5 Hz) hinted 26 was a pallenane and related to 3β,4β-dihydroxypallenone [33, 34]. The C5/C3 bicyclic skeleton of 26 was established by 1H–1H COSY correlations between H-1/H-2/H-3, between H-1/H-6, H-1/H-5, and H-5/H-6. Furthermore, the 1H–1H COSY correlations from H-6/H-7/H-8/H-9/H-10/H3-14 and H-10/10-OH revealed a 4-hydroxypentyl were connected with C-6. HMBC correlations from H-12 (δH 1.05) to C-7 (δC 52.0), C-11 (δC 72.9), from H-13 (δH 1.16) to C-7 (δC 52.0), C-11 (δC 72.9) elucidated a 2-hydroxypropan-2-yl was connected with C-7. Moreover, HMBC correlations from H-3 (δH 1.34) to C-4 (δC 78.8), C-5 (δC 36.3), from H-6 (δH 0.13) to C-2 (δC 26.2) and C-4 (δC 78.8), from H-7 (δH 0.57) to C-6 (δC 23.1), C-8 (δC 27.6), C-9 (δC 39.4), and C-11 (δC 72.9), from H3-15 (δH 1.22) to C-3 (δC 37.2), C-4 (δC 78.8), C-5 (δC 36.3) further confirmed the planar structure (Fig. 2A). Comparing the 1H NMR data of H-5 (δH 1.13, dd, J = 6.0, 3.6 Hz), H-6 (δH 0.13, dt, J = 10.3, 3.4 Hz), and H-7 (δH 0.57, ddd, J = 10.0, 8.0, 3.5 Hz) with those of 3β,4β-dihydroxypallenone [33, 34] indicated 26 possessed the same H-1/H-5 cis, H-1/H-6 trans, H-5/H-6 trans configurations. This conclusion was confirmed by NOE correlations observed from H-7 (δH 0.57) to H-1 (δH 1.04) and H-5 (δH 1.13) (Fig. 3). The methyl at C-4 was determined as β deduced from NOE correlations observed from H-6 (δH 0.13) to H3-15 (δH 1.22), from H-5 (δH 1.13) to 4-OH (δH 4.24). As there were potential inaccuracies of NOE correlations as the overlapped signals of H-1 and H3-14. 1D and 2D NMR spectra of 26 were remeasured in pyridine-d5 to get the distinct signals of H-1, H-5, H-6 and analysis of NOE correlations (in pyridine-d5) further confirmed the configurations (Table S1, Additional file 1). Due to the configuration of methyl at C-4 differed from the congeners, 26 was named as 4-epi-pallenane-4α,10,11-triol.
The characteristic 1H and 13C NMR data (Tables 4, 5) of 27 showed it was also a pallenane sesquiterpenoid. The NMR data of 27 were similar to those of 26, except for the presence of one methine (δC 35.0) instead of one oxygenated quaternary carbon as well as a doublet methyl replaced a singlet methyl. 1H–1H COSY correlations from H-4 (δH 2.15) to H3-15 (δH 0.99) combining with HMBC correlations between H3-15 (δH 0.99) and C-3 (δC 30.2), C-4 (δC 35.0), C-5 (δC 30.1) confirmed the methyl was located at C-4. (Fig. 2A). The similar 1H NMR data of H-5 (δH 1.12, dt, J = 6.3, 3.5 Hz), H-6 (δH 0.27, dt, J = 10.3, 3.3 Hz), and H-7 (δH 0.56, ddd, J = 10.6, 7.8, 3.5 Hz) with those of 26 suggested they possessed the identical configurations. In NOESY spectrum, the key cross peaks from H-6 (δH 0.27) to H3-15 (δH 0.99), from H-4 (δH 2.15) to H-5 (δH 1.12), from H-7 (δH 0.56) to H-1 (δH 0.97) and H-5 (δH 1.12) further indicated that H-1, H-4, H-5, and H-7 were α-oriented, while H-6, and H3-15 were β-oriented (Fig. 3). Finally, the structure of compound 27 was confirmed. Pallenane is a kind of rarely reported sesquiterpene with a distinctive C5/C3 bicyclic skeleton. Till now, only two pallenane congeners (3β,4β-dihydroxypallenone and 3β-acetoxy-4β-hydroxypallenone) were found from the plant Pallenis spinosa [33, 34]. The current compounds 26 and 27 were firstly obtained from streptomycete. According to the literature [33, 34], the isodaucane (17), oplopanane (25), and pallenane (26, 27) skeleton metabolites were derived from the pinacol-type cadinane glycols through Wagner rearrangement in organisms (Fig. 2B). The 2-hydroxypropan-2-yl groups at C-7 in 26 and 27 were suggested as β-orientation according to the biogenetic way.
The characteristic 1H and 13C NMR signals implied that 28 was an eudesmane-type sesquiterpene and related to eudesmane-1β,6α,11-triol (29) [10]. The alterations were one more oxygenated methine replaced a methylene and a double bond replaced two methines. The HMBC correlations from H-4 (δH 2.39), H-7 (δH 2.02) to C-5 (δC 145.9) and H-4 (δH 2.39), H-7 (δH 2.02) to C-6 (δC 126.6) indicated double bond was located at C-5 and C-6, from 3-OH (δH 4.50) to C-2 (δC 35.6) and C-3 (δC 68.7) demonstrated an extra hydroxy was connected with C-3 (Fig. 2A). The relative configuration was determined by analysis of coupling constants and NOE correlations. The large coupling constants of H-1 (δH 3.03, dt, J = 11.6, 4.8 Hz), H-3 (δH 3.52, m) and H-7 (δH 2.02, ddd, J = 11.0, 6.2, 2.0 Hz) suggested that H-1, H-3, H-7 were axial orientation. While the peak shape and coupling constants of H-4 (δH 2.39, quint, J = 7.0 Hz) suggested H-4 was equatorial orientation. NOE interactions from H-3 (δH 3.52) to H-4 (δH 2.39), H-1 (δH 3.03), from H-6 (δH 5.53) to H-4 (δH 2.39), H-7 (δH 2.02), from H3-14 (δH 0.95) to 1-OH (δH 4.45) demonstrated that H-1, H-3, H-4, and H-7 were α-oriented, while 1-OH, 3-OH, H3-14, and H3-15 were β-oriented (Fig. 3). The configurations of chiral carbons of 28 were identified as 1R,3S,4R,7R,10R by comparing the experimental ECD spectrum of 28 with calculated ECD spectrum (Fig. 4G). Hence, the structure of 28 was determined as (1R,3S,4R,7R,10R)-eudesm-5-ene-1,3,11-triol.
The twenty-one known compounds were identified as bacaryolane B (5) [19], bacaryolane C (6) [19], caryolane-1,7α-diol (7) [35], caryolane-1,9α-diol (8) [36], 6α,9α-dihydroxy-β-caryolanol (9) [37], 6β,9β-dihydroxy-β-caryolanol (10) [37], caryolane-1,6β,9α-triol (11) [10], bacaryolane A (12) [19], 1(10)E,5E-germacradiene-2,11-diol (16) [26], ganodermanol L (19) [28], (1α,4β,5β,6β,7β,10α)-5,11-epoxy-10-cadinanol (20) [10], 15-hydroxy-α-cadinol (21) [29], pubinernoid C (24) [38], eudesmane-1β,6α,11-triol (29) [10], eudesmane-1β,6α,9β,11-tetrol (30) [10], ganodermanol J (31) [39], eudesmane-1β,5α,11-triol (32) [10], (2α,4β,5β,7β,10α)-2,5,11-eudesmanetriol (33) [10], ent-4(15)-eudesmene-1β,6α-diol (34) [40], 1β,6α-dihydroxy-4β(15)-epoxyeudesmane (35) [41] and 4α,15-epoxyeudesmane-1β,6α,11-triol (36) [28] by comparing their spectroscopic data with those reported in the literatures.
Evaluation of antimicrobial activity in vitro
The isolated sesquiterpenoids which amounted to more than 3 mg were chosen to evaluate their antimicrobial activities against five bacteria and four fungi. As presented in Table S2, Additional file 1, compounds 5–7, 9, 12, 31–34 inhibited the growth against C. albicans and C. parapsilosis with the MIC80 values ranged from 100 to 400 μg/mL. Furthermore, 34 exhibited moderate antifungal activity against C. neoformans and C. gattii with MIC values of 50 μg/mL. The caryolane sesquiterpenoids 5–7 exhibited weak antibacterial activity against B. subtilis, E. faecium, and E. coli with MIC values ranged from 100–200 μg/mL, respectively (Table S3, Additional file 1). Meanwhile, the other tested compounds (10, 11, 13, 16, 19–21, 23, 29, 30, 35, 36) didn’t exhibit antimicrobial activity against test microorganisms at a concentration of 400 μg/mL.
34 inhibited Cryptococcus species in vitro
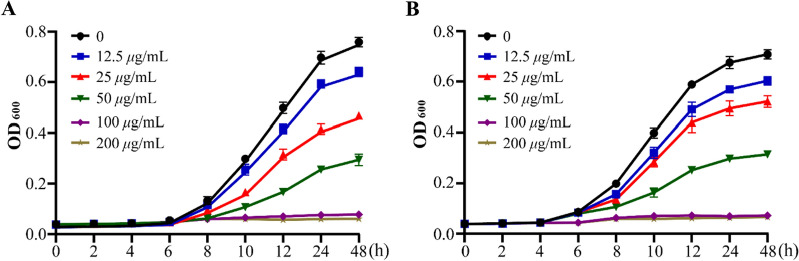
The optimal antifungal activity of 34 prompted us to further investigation its effect on Cryptococcus species. As shown in Fig. 5A, B, 34 consistently suppressed the growth of Cryptococcus species cells at all tested time points. Notably, at a concentration of 100 μg/mL, 34 could effectively eliminate almost all Cryptococcus species fungi. The above data demonstrated that 34 significantly inhibited the growth of Cryptococcus species in a time- and dose-dependent manner.
Fig. 5.
The growth curve of 34 against C. neoformans (A) and C. gattii (B). Data are presented as mean ± SD of three independent determinations
Effect of 34 on biofilm
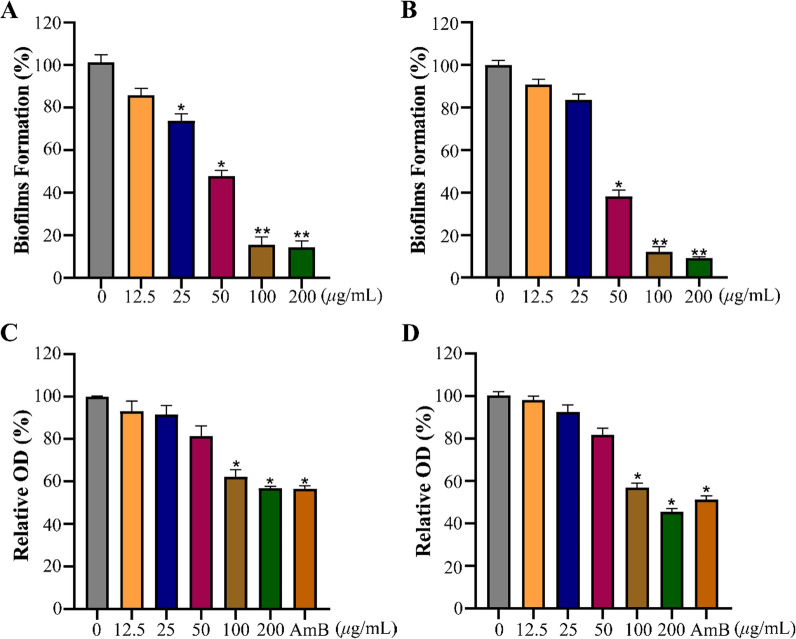
The majority of fungal infections are attributed to biofilm, so inhibiting biofilm formation is crucial for effective antifungal treatment [42]. Thereafter, the effect of 34 was further investigated on biofilm formation and preformed biofilm of Cryptococcus species. As displayed in Fig. 6A, B, the formation rate of biofilms for C. neoformans and C. gattii were decreased to 47.8% and 38.3%, respectively, at a dose of 50 μg/mL. As shown in Fig. 6C, D, 34 effectively destroyed the preformed biofilms of Cryptococcus species at a concentration of 100 μg/mL. These results indicated that 34 could not only inhibit biofilm formation, but also destroy preformed biofilms, which has the potential source for discovering novel therapeutic agents for treatment of fungal diseases.
Fig. 6.
The effect of 34 on the biofilm of Cryptococcus species. Effect of 34 on biofilm formation of C. neoformans (A) and C. gattii (B). Effect of 34 on the preformed biofilms of C. neoformans (C) and C. gattii (D). 2 μg/mL AMB was used as positive control. Data are presented as mean ± SD of three independent determinations. *P < 0.05, **P < 0.01 vs. control group
34 inhibited the adhesion of Cryptococcus species
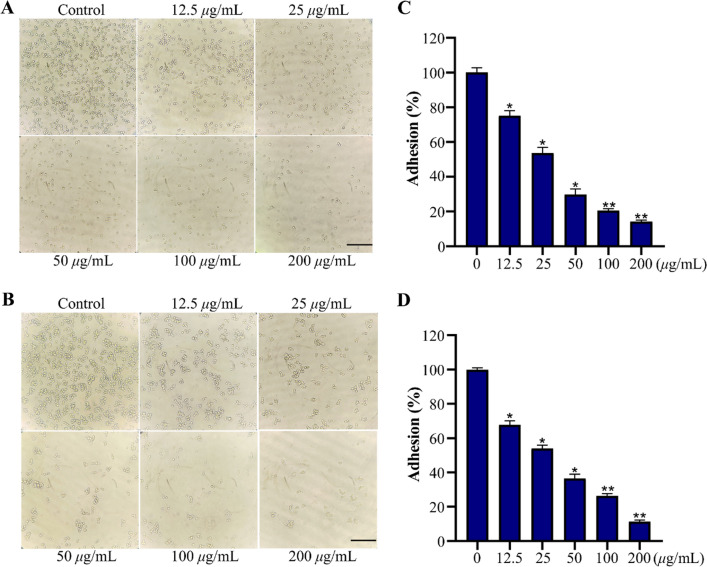
Adhesion is the initial step in host colonization and dissemination [43]. 34 was further evaluated for its ability to inhibit the adhesion of Cryptococcus species. Following treatment with various concentrations of 34, the number of adherent Cryptococcus sp. cells exhibited a dose-dependent reduction trend compared to the untreated group (Fig. 7A, B). The above observation was further confirmed by the results of the XTT reduction assay. As shown in Fig. 7C, D, the adhesion rates for C. neoformans and C. gattii significantly decreased from 75.2% to 14.1% and from 67.7% to 11.3%, respectively after treated with 34 (12.5 to 200 μg/mL).
Fig. 7.
The effect of 34 on Cryptococcus species adhesion. The photographs of C. neoformans cells (A) and C. gattii cells (B) treated with 34 for 4 h at 37 °C; The adhesion rates of C. neoformans cells (C) and C. gattii cells (D) was measured by XTT reduction assay. The bar in panel (A, B) indicates 40 µm. Data are presented as mean ± SD of three independent determinations. *P < 0.05, **P < 0.01 vs. control group
Experimental procedures
General
Optical rotation was determined using an Anton Paar MCP200 automatic polarimeter (Graz, Austria). IR spectra were recorded with a Bruker Tensor 27 FT-IR spectrometer. A biologic MOS-450 spectra polarimeter (Biologic Science, Claix, France) was used to measured ECD spectra. NMR spectra were recorded on a Bruker Advance III-600 MHz spectrometer (Bruker, Rheinstetten, Germany). ESIMS were recorded on an Agilent 1290–6420 Triple Quadrupole LC-MS spectrometer (Santa Clara, CA, USA). HRESIMS experiments were conducted using a Bruker Micro TOF-Q mass spectrometer (Bruker Daltonics, Billerica, MA). Silica gel (100–200 mesh, 200–300 mesh, Qingdao Marine Chemical, Ltd., Qingdao, China), Sephadex LH-20 (GE Healthcare Biosciences AB, Uppsala, Sweden), YMC*GEL ODS-A (S-50 μm, 12 nm) (YMC Co., Ltd., Kyoto, Japan) were used for column chromatography. Mosher’s reagents R-MTPA-Cl and S-MTPA-Cl were purchased from Sigma Aldrich (Shanghai) Trading Co., Ltd. XTT and antimicrobial assays were analyzed using a microplate reader (BioTek Synergy H1, BioTek Instruments, Inc., Vermont, USA). The images of cells were observed directly with a microscope (Olympus IX71, Olympus, Tokyo, Japan).
Microbial material
The producing organism was derived from fresh fecal samples excreted by healthy adult E. maximus living in Xishuangbanna National Nature Reserve, Xishuangbanna, Yunnan Province, China. The strain was identified to be S. fulvorobeus by Dr. Yi Jiang based on morphological characteristics and 16S rRNA gene sequences. The BLAST result showed that the sequence was most similar (99.89%) to the sequence of S. fulvorobeus (strain: NBRC 15897, GenBank accession no. AB184711). The strain (No. YIM 103582) was deposited at the Yunnan Institute of Microbiology, Yunnan University, China.
Fermentation, extraction, and isolation
The strain was inoculated to 100 mL seed medium consisting of 4 g/L yeast extract, 4 g/L glucose, 5 g/L malt extract, 1.0 mL of multiple vitamin solution, and 1.0 mL of trace element solution at a pH of 7.2 without adjustment. The flasks were cultured for 2 days at 28 °C on a rotary shaker at 160 rpm, followed by inoculation to fermentation medium (24 g/L soluble starch, 3 g/L beef extract, 1 g/L glucose, 3 g/ L peptone, 5 g/L yeast extract, 4 g/L CaCO3, pH 7.0) with a 10% volume. The fermentation was incubated at 28 °C for 7 days on a rotary shaker at 160 rpm.
The completed fermentation culture (150 L) was centrifuged (4000 rpm, 5 min) to separate into supernatant and mycelium, and the supernatant was extracted with ethyl acetate three times and evaporated to yield a crude extract 60 g. The dried extract was subjected to a silica gel column chromatography eluting with a CH2Cl2-MeOH solvent system (from 100:1 to 30:1, 10:1, and finally 1:1) to yield five fractions Fr.1–5. Fraction 1 was subjected to Sephadex LH-20 chromatography (MeOH) to produce three subfractions Fr.1.1-Fr.1.3. Fr.1.3 was subjected to silica gel column chromatography (P. ether-EtOAc 10:1) to yield three subfractions Fr.1.3.1-Fr.1.3.9. Fr.1.3.3 was isolated through ODS column chromatography MeOH-H2O (60:40) to yield compounds 5 (4.6 mg), 6 (3.0 mg), and 7 (14.3 mg). Fr.1.3.5 was purified by silica gel column chromatography (P. ether-EtOAc 7:1) to afford compound 12 (8.8 mg), 20 (26.7 mg), 25 (2.0 mg), and 35 (5.0 mg). Fraction 2 was subjected to Sephadex LH-20 chromatography (MeOH) to obtain three subfractions Fr.2.1-Fr.2.3. Fr.2.3 was separated by silica gel column chromatoraphy (P. ether-EtOAc 4:1), followed by eluting with MeOH-H2O (40:60) using ODS column chromatography to afford compounds 8 (2.2 mg), 16 (4.0 mg), 34 (16.6 mg), and 36 (6.2 mg). Fraction 3 was subjected to silica gel column chromatography (CH2Cl2-MeOH 50:1) to yield seven subfractions Fr. 3.1-Fr. 3.7. Fr. 3.1was put on an ODS column and eluted with MeOH-H2O (40:60) to yield three subfractions Fr. 3.1.1-Fr. 3.1.3. Compounds 2 (2.5 mg), 18 (2.3 mg), 19 (57.8 mg), 29 (23.3 mg), and compound 33 (16.0 mg) were obtained from Fr.3.1.1 through a silica gel column chromatography (P. ether-EtOAc 6:1). Fr.3.3 was subjected to ODS column chromatography, eluting with MeOH-H2O (45:55) to yield nine subfractions Fr.3.3.1-Fr.3.3.9. Fr.3.3.3 was fractionated by silica gel column chromatography (CH2Cl2-MeOH 50:1) to give compounds 9 (4.0 mg), 11 (15.0 mg), and 26 (1.6 mg). Fr.3.3.4 was separated by silica gel column chromatography (P. ether-EtOAc 2:1), then further purified by silica gel column chromatography (CH2Cl2-MeOH 50:1) to give compounds 1 (2.8 mg), 15 (2.3 mg), 17 (2.6 mg), 30 (23.7 mg), and 31 (3.2 mg). Compounds 21 (6.2 mg) and 32 (5.8 mg) were obtained from Fr.3.3.5 through a silica gel column chromatography (CH2Cl2-MeOH 60:1). Similarly, compounds 13 (5.2 mg) and 14 (2.6 mg) were isolated from Fr. 3.3.7 through a silica gel column chromatography (CH2Cl2-MeOH 60:1). Fr. 3.3.9 was purified by silica gel column chromatography (P. ether-EtOAc 2:1) to afford compounds 22 (2.3 mg) and 24 (2.0 mg). Fr.3.5 was separated by ODS column chromatography and eluted with MeOH-H2O (30:70) to yield six fractions Fr.3.5.1–3.5.6. Fr.3.5.2 was purified by silica gel column chromatography (EtOAc–MeOH 40:1) to afford compounds 3 (2.0 mg) and 4 (2.6 mg). Fr.3.5.4 was purified by silica gel column chromatography (CH2Cl2-MeOH 25:1) to afford compounds 10 (3.8 mg) and 27 (1.5 mg). Fr.3.5.5 was firstly subjected to silica gel column chromatography (CH2Cl2-MeOH 25:1), then purified by silica gel column chromatography (P. ether-EtOAc 2:1) to give compounds 23 (3.2 mg) and 28 (2.0 mg).
Spectroscopic data of compounds
(1S,2R,4S,5S,8R)-9-Oxocaryolane-1,13-diol (1)
Colorless oil; + 60.0 (c 0.20, MeOH); IR (film) νmax 3727, 3382, 2938, 2866, 1699, 1455, 1054, 1033, 1013 cm−1; CD (0.5 mg/mL, MeOH) λmax (Δε) 202 (− 0.48), 294 (+ 2.04) nm; 1H and 13C NMR see Tables 1 and 2; HRESIMS m/z 275.1623 [M+Na]+ (calcd for C15H24NaO3+, 275.1618).
(1S,2R,4R,5S,8R)-Caryolane-1,14-diol (2)
Colorless oil; − 53.0 (c 0.30, MeOH); IR (film) νmax 3726, 3347, 2926, 2867, 1456, 1053, 1033 cm−1; 1H and 13C NMR see Tables 1 and 2; HRESIMS m/z 239.2021 [M+H]+ (calcd for C15H27O2+, 239.2006).
(1S,2R,4R,5S,8R,9S)-Caryolane-1,9,14-triol (3)
Colorless oil; − 40.5 (c 0.20, MeOH); IR (film) νmax 3355, 2940, 2865, 1457, 1055, 1033, 1018 cm−1; 1H and 13C NMR see Tables 1 and 2; HRESIMS m/z 277.1780 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
Caryolane-1,6α,10α-triol (4)
Colorless solid; − 54.0 (c 0.37, MeOH); IR (film) νmax 3348, 2928, 2866, 1461, 1363, 1109, 1040, 1004 cm−1; 1H and 13C NMR see Tables 1 and 2; HRESIMS m/z 277.1778 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
(2S,4S,7S,8S)-1(10)E,5E-Germacradiene-2,8,11-triol (13)
Colorless oil; − 80.5 (c 0.20, MeOH); IR (film) νmax 3315, 2968, 2930, 1668, 1382, 1046, 1010 cm−1; CD (0.5 mg/mL, MeOH) λmax (Δ ε) 231 (− 25.77) nm; 1H and 13C NMR see Tables 2 and 3; HRESIMS m/z 277.1780 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
(2S,4S,7R)-1(10)E,5E-Germacradiene-2,11-diol 2-methyl ether (14)
Colorless oil; − 112.2 (c 0.50, MeOH); IR (film) νmax 3728, 2970, 2930, 1447, 1381, 1088, 934 cm−1; CD (0.5 mg/mL, MeOH) λmax (Δ ε) 225 (− 9.42), 290 (+ 0.38) nm; 1H and 13C NMR see Tables 2 and 3; HRESIMS m/z 275.1986 [M+Na]+ (calcd for C16H28NaO2+, 275.1982).
(2S,4R,7S,8S)-1(10)E,5E-Germacradiene-2,4,8,11-tetraol 2-acetate (15)
White powder; − 120.5 (c 0.20, MeOH); IR (film) νmax 3348, 2973, 2929, 1730, 1373, 1244, 1022 cm−1; CD (0.5 mg/mL, MeOH) λmax (Δ ε) 200 (+ 29.35), 231 (− 9.42) nm; 1H and 13C NMR see Tables 2 and 3; HRESIMS m/z 335.1833 [M+Na]+ (calcd for C17H28NaO5+, 335.1829).
(1α,3α,4β,5α,6α,7β,9β)-6,11-Epoxyisodaucane-3,9-diol (17)
White powder; + 13.3 (c 0.30, MeOH); IR (film) νmax 3360, 2927, 2869, 1460, 1366, 1236, 1049, 1028 cm−1; 1H and 13C NMR see Tables 3 and 4; HRESIMS m/z 277.1788 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
8α-Hydroxyganodermanol L (18)
White powder; − 40.6 (c 0.20, MeOH); IR (film) νmax 3355, 2966, 2927, 1461, 1379, 1172, 1046 cm−1; 1H and 13C NMR see Tables 3 and 4; HRESIMS m/z 293.1729 [M+Na]+ (calcd for C15H26NaO4+, 293.1723).
(1S,2S,6R,7S,10R)-Cadinane-2,10,15-triol (22)
Yellow oil; − 40.2 (c 0.30, MeOH); IR (film) νmax 3312, 2959, 2932, 2871, 1462, 1378, 1125, 1069, 1016 cm−1; CD (0.25 mg/mL, MeOH) λmax (Δ ε) 194 (+ 9.63), 217 (− 9.01), 237 (+ 1.85) nm; 1H and 13C NMR see Tables 4 and 5; HRESIMS m/z 277.1777 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
(1R,3S,6R,7S,9S,10R)-3,9-Dihydroxyepicubenol (23)
Yellow oil; − 16.0 (c 0.50, MeOH); IR (film) νmax 3354, 2960, 2877, 1450, 1659, 1369, 1060, 1030, 1007 cm−1; CD (0.25 mg/mL, MeOH) λmax (Δ ε) 191 (− 29.60), 213 (− 11.77), 234 (+ 2.98) nm; 1H and 13C NMR see Tables 4 and 5; HRESIMS m/z 277.1775 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
Oplopanane-4,10α,11-triol (25)
White amorphous power; − 50.0 (c 0.24, MeOH); IR (film) νmax 3331, 2925, 2862, 1597, 1454, 1261, 1069, 1014 cm−1; 1H and 13C NMR see Tables 4 and 5; HRESIMS m/z 279.1937 [M+Na]+ (calcd for C15H28NaO3+, 279.1931).
4-epi-Pallenane-4α,10,11-triol (26)
Colorless oil; − 202.0 (c 0.10, MeOH); IR (film) νmax 3358, 2965, 2867, 1600, 1458, 1372, 1185, 1124, 1003 cm−1; 1H and 13C NMR see Tables 4 and 5; HRESIMS m/z 279.1936 [M+Na]+ (calcd for C15H28NaO3+, 279.1931).
4-epi-Pallenane-10,11-diol (27)
Colorless oil; − 80.6 (c 0.20, MeOH); IR (film) νmax 3349, 2927, 2865, 1602, 1457, 1372, 1126, 1082 cm−1; 1H and 13C NMR see Tables 4 and 5; HRESIMS m/z 223.2039 [M−H2O+H]+ (calcd for C15H27O+, 223.2056).
(1R,3S,4R,7R,10R)-Eudesm-5-ene-1,3,11-triol (28)
Colorless oil; − 40.4 (c 0.20, MeOH); IR (film) νmax 3350, 2971, 2936, 1659, 1468, 1376, 1145, 1066, 1007 cm−1; CD (0.5 mg/mL, MeOH) λmax (Δε) 206 (− 16.93), 229 (+ 0.46) nm; 1H and 13C NMR see Tables 4 and 5; HRESIMS m/z 277.1779 [M+Na]+ (calcd for C15H26NaO3+, 277.1774).
Esterification using Mosher′s reagent
The 1 mg sample of compound 1 was dissolved in 0.5 mL of pyridine-d5 and subsequently transferred into a pristine NMR tube. Then, the solution was subjected to treatment with (R)-MTPA-Cl (5 μL) for 12 h to obtained (S)-MTPA ester of 1 (1a). The (R)-MTPA ester of 1 (1b) was prepared by the same process. Subsequently, the tubes were directly employed for 1H-NMR measurements. The (S)-MTPA esters (2a and 3a) as well as (R)-MTPA esters (2b and 3b) of compounds 2 and 3 were prepared in a completely analog manner.
ECD calculations
The ECD calculations of compounds 1, 13–15, 22, 23, and 28 were performed with Gaussian 09 [44]. The CONFLEX software was used to perform a stable conformational analysis of all enantiomers, employing the MMFF94S molecular force field and an energy cut off of 3 kcal/mol. The selected stable conformers were further optimized using the Gaussian 09 program package at the B3LYP/6-31G+(d) level of theory. Then, the ECD was theoretically calculated at the B3LYP/6-311 g++ (2d, p) level in a methanol solution using the PCM model. SpecDis 1.51 software was used to generate the global theoretical ECD curve based on the Boltzmann weights of each conformer.
Antimicrobial assay
The antimicrobial activities were evaluated using a microbroth dilution method to determine the minimum inhibitory concentrations (MICs) [9]. The tested strains included five bacteria (Bacillus subtilis ATCC 6633, Staphylococcus aureus ATCC 25923, Enterococcus faecium ATCC19434, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC27853), as well as four fungi (Candida albicans ATCC MYA-2867, C. parapsilosis ATCC 22019, and Cryptococcus neoformans ATCC 208821, C. gattii CGMCC 2.3159). Antibacterial and antifungal tests were performed in Luria–Bertani and RPMI-1640 broth, respectively. Test compounds were dissolved in DMSO and twofold serially diluted to six different concentrations (40.0–1.25 mg/mL). Each well of the 96-well microtiter plates was added with 1 μL of test sample solutions and 100 μL of suspensions, which contained a concentration of 1 × 106 cfu/mL for bacteria (2 × 103 cfu/mL for fungi). The plates were subsequently incubated for 24 h at 28 °C for bacteria, 48 h at 28 °C for fungi. The XTT reduction assay was performed as described in the literature [45] and the absorbance at 490 nm was measured using a microplate reader for each well. The MIC was defined as the minimum concentration of the antimicrobial agent that completely inhibited visual growth of a microorganism, while the MIC80 was defined as the minimum concentration of the antimicrobial agent that inhibited 80% of the visible growth of a microorganism. Ciprofloxacin and amphotericin B were used as positive controls against bacteria and fungi, respectively.
Growth curve assay
The growth curve of Cryptococcus species was conducted according to the previously described method [45]. Briefly, the fungal suspensions were normalized to in the YPD liquid medium. Then, 100 μL fungal suspensions (1 × 106 cfu/mL) and different concentrations of compound 34 were inoculated into 96-well plate. The final concentrations of 34 were 12.5, 25, 50, 100 or 200 μg/mL. After incubating for 2, 4, 6, 8, 10, 12 and 24 h at 37 °C, the absorbance at 600 nm was determined by microplate reader (BioTek Synergy H1, BioTek Instruments, Inc., Vermont, USA).
Biofilm formation assay
The strains C. neoformans and C. gattii were selected to investigate the anti-biofilm activity of compound 34. Biofilm formation was assessed by XTT reduction assay in 96-well plate [45]. Briefly, 100 μL of fungal suspensions (1 × 106 cfu/mL) were added to 96-well plates with varying different concentrations of 34 (12.5, 25, 50, 100 or 200 μg/mL). After incubation for 24 h at 30 °C, the plate was gently washed twice with sterile phosphate-buffered saline (PBS) to remove free-floating fungal cells. The biofilm of Cryptococcus species was assayed by XTT reduction assay.
Preformed biofilms assay
To evaluate the potential ability of compound 34 to disrupt preformed biofilms was assessed according to previously reported method [45]. 100 μL of fungal suspensions (1 × 106 cfu/mL) were dispensed into 96-well plate and cultured for 24 h at 37 °C. Then, the resulting preformed biofilm were washed twice with sterile PBS and treated with various concentrations of 34 (12.5, 25, 50, 100 or 200 μg/mL) or 2 μg/mL AMB. The plates were further incubated for 24 h, and then XTT reduction assay was conducted as described previously.
Adhesion assay
The impact of 34 on Cryptococcus species adhesion on 96-well plate was examined by XTT reduction assay according to a previously described method [45]. 100 μL suspensions of C. neoformans and C. gattii (1 × 106 cfu/mL) were respectively inoculated into 96-well plate, along with 1 μL of compound 34 at different concentrations (12.5, 25, 50, 100 or 200 μg/mL) and incubated at 37 °C for 4 h without shaking. Then, the cell supernatants were discarded and the plate was gently washed three times with sterile phosphate-buffered saline (PBS) to remove non-adherent cells. Then, the adherent C. neoformans and C. gattii were directly observed under a microscope in bright field mode and detected by XTT reduction assay.
Conclusion
In summary, we report the discovery of thirty-six structurally diverse sesquiterpenoids, including fifteen new compounds (1–4, 13–15, 17, 18, 22, 23, 25–28), along with twenty-one known analogues. These compounds featured eight distinctive carbon skeletons: caryolane, germacrane, cadinane, epicubenol, isaodaucane, oplopanane, pallenane, and eudesmane. It has been proven that Streptomyces possesses unparalleled ability to produce structurally diverse and novel secondary metabolites. Notably, parenane is a rare sesquiterpene with a unique C5/C3 bicyclic skeleton, which was first discovered in microorganisms. To our knowledge, S. fulvorobeus is the first actinomycete to produce such a substantial quantity of sesquiterpenoids. Additionally, the isolated sesquiterpenoid 34 exhibited optimal antifungal activity against C. neoformans and C. gattii with MIC values of 50 μg/mL. Further experiments showed that 34 significantly inhibited biofilm formation, destroyed the preformed biofilm of fungi, and prevented adhesion of Cryptococcus species. This work not only enriches the structural diversity of bacterial terpenoids but also provides support for the genetic capacity of actinomycetes to synthesize a diverse array of terpenoids.
Supplementary Information
Additional file 1: The antimicrobial activity of some compounds, HRESI-MS, 1D and 2D NMR spectra of new compounds 1–4, 13–15, 17, 18, 22, 23, 25–28, experimental ECD spectra and calculated ECD spectra of compound 12.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 32060001), the Fundamental Research Funds for the Central Universities, China (No. N2320001) and the Construction Project of Liaoning Provincial Key Laboratory, China (2022JH13/10200026).
Author contributions
Lu Cao: investigation, writing of original draft. Jun-Feng Tan: conceptualization, methodology. Zeng-Guang Zhang and Jun-Wei Yang: methodology, data curation. Yu Mu and Zhi-Long Zhao: Investigation, data analysis. Yi Jiang: resources, and funding acquisition. Xue-Shi Huang and Li Han: revised the manuscript, supervision, and funding acquisition. All authors read and approved the final manuscript.
Availability of data and materials
Data will be made available on request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yi Jiang, Email: jiangyi@ynu.edu.cn.
Xue-Shi Huang, Email: huangxs@mail.neu.edu.cn.
Li Han, Email: hanli@mail.neu.edu.cn.
References
- 1.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–9. [DOI] [PubMed] [Google Scholar]
- 2.Bartolini I, Risaliti M, Ringressi MN, Melli F, Nannini G, Amedei A, Muiesan P, Taddei A. Role of gut microbiota-immunity axis in patients undergoing surgery for colorectal cancer: focus on short and long-term outcomes. World J Gastroenterol. 2020;26(20):2498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J, Piao X, Mahfuz S, Long S, Wang J. The interaction among gut microbes, the intestinal barrier and short chain fatty acids. Anim Nutr. 2022;9:159–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–41. [DOI] [PubMed] [Google Scholar]
- 5.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12(12):562–8. [DOI] [PubMed] [Google Scholar]
- 7.Wu C, Chen T, Xu W, Zhang T, Pei Y, Yang Y, Zhang F, Guo H, Wang Q, Wang L, Zhao B. The maintenance of microbial community in human fecal samples by a cost effective preservation buffer. Sci Rep. 2021;11(1):13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Li QY, Li GD, Xu FJ, Han L, Jiang Y, Huang XS, Jiang CL. The distal gut bacterial community of some primates and carnivora. Curr Microbiol. 2018;75(2):213–22. [DOI] [PubMed] [Google Scholar]
- 9.Ding N, Jiang Y, Han L, Chen X, Ma J, Qu X, Mu Y, Liu J, Li L, Jiang C, Huang X. Bafilomycins and odoriferous sesquiterpenoids from Streptomyces albolongus isolated from Elephas maximus feces. J Nat Prod. 2016;79(4):799–805. [DOI] [PubMed] [Google Scholar]
- 10.Ding N, Han L, Jiang Y, Li G, Liu J, Mu Y, Huang X. Sesquiterpenoids from Streptomyces anulatus isolated from Giraffa camelopardalis feces. Magn Reson Chem. 2018;56(5):352–9. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Jiang Y, Cao Y, Liu J, Zheng D, Chen X, Han L, Jiang C, Huang X. Violapyrones A-G, α-pyrone derivatives from Streptomyces violascens isolated from Hylobates hoolock feces. J Nat Prod. 2013;76(11):2126–30. [DOI] [PubMed] [Google Scholar]
- 12.Ma J, Lei H, Chen X, Bi X, Jiang Y, Han L, Huang X. New anti-inflammatory metabolites produced by Streptomyces violaceoruber isolated from Equus burchelli feces. J Antibiot. 2017;70(10):991–4. [DOI] [PubMed] [Google Scholar]
- 13.Mu Y, Yu X, Zheng Z, Liu W, Li G, Liu J, Jiang Y, Han L, Huang X. New metabolites produced by Streptomyces badius isolated from Giraffa camelopardalis feces. Magn Reson Chem. 2019;57(12):1150–7. [DOI] [PubMed] [Google Scholar]
- 14.Zheng D, Han L, Qu X, Chen X, Zhong J, Bi X, Liu J, Jiang Y, Jiang C, Huang X. Cytotoxic fusicoccane-type diterpenoids from Streptomyces violascens isolated from Ailuropoda melanoleuca feces. J Nat Prod. 2017;80(4):837–44. [DOI] [PubMed] [Google Scholar]
- 15.Zheng D, Ding N, Jiang Y, Zhang J, Ma J, Chen X, Liu J, Han L, Huang X. Albaflavenoid, a new tricyclic sesquiterpenoid from Streptomyces violascens. J Antibiot. 2016;69(10):773–5. [DOI] [PubMed] [Google Scholar]
- 16.Dickschat JS. Bacterial terpene cyclases. Nat Prod Rep. 2016;33(1):87–110. [DOI] [PubMed] [Google Scholar]
- 17.Wu QX, Shi YP, Jia ZJ. Eudesmane sesquiterpenoids from the Asteraceae family. Nat Prod Rep. 2006;23(5):699–734. [DOI] [PubMed] [Google Scholar]
- 18.Rinkel J, Rabe P, Garbeva P, Dickschat JS. Lessons from 1,3-hydride shifts in sesquiterpene cyclizations. Angew Chem Int Ed Engl. 2016;55(43):13593–6. [DOI] [PubMed] [Google Scholar]
- 19.Ding L, Goerls H, Dornblut K, Lin W, Maier A, Fiebig HH, Hertweck C. Bacaryolanes A–-C, rare bacterial caryolanes from a mangrove endophyte. J Nat Prod. 2015;78(12):2963–7. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Yang Q, Zhang X, Wang Z, Xu HM, Dong LB. Discovery of diverse sesquiterpenoids from Crossiella cryophila through genome mining and NMR tracking. J Nat Prod. 2024;87(2):195–206. [DOI] [PubMed] [Google Scholar]
- 21.Yamada Y, Kuzuyama T, Komatsu M, Shin-Ya K, Omura S, Cane DE, Ikeda H. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci USA. 2015;112(3):857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citron CA, Gleitzmann J, Laurenzano G, Pukall R, Dickschat JS. Terpenoids are widespread in actinomycetes: a correlation of secondary metabolism and genome data. ChemBioChem. 2012;13(2):202–14. [DOI] [PubMed] [Google Scholar]
- 23.Rudolf JD, Alsup TA, Xu B, Li Z. Bacterial terpenome. Nat Prod Rep. 2021;38(5):905–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuda M, Toriyabe Y, Endo T, Kobayashi J. Application of modified mosher’s method for primary alcohols with a methyl group at C2 position. Chem Pharm Bull. 2003;51(4):448–51. [DOI] [PubMed] [Google Scholar]
- 25.Nakano C, Horinouchi S, Ohnishi Y. Characterization of a novel sesquiterpene cyclase involved in (+)-caryolan-1-ol biosynthesis in Streptomyces griseus. J Biol Chem. 2011;286(32):27980–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan S, Grabley S, Groth I, Lin W, Christner A, Guo D, Sattler I. Structure determination of germacrane-type sesquiterpene alcohols from an endophyte Streptomyces griseus subsp. Magn Reson Chem. 2005;43(12):1028–31. [DOI] [PubMed] [Google Scholar]
- 27.Kato R, Saito H, Ikeuchi K, Suzuki T, Tanino K. Total synthesis and structural revision of the 6,11-epoxyisodaucane natural sesquiterpene using an anionic 8π electrocyclic reaction. Org Lett. 2022;24(43):7939–43. [DOI] [PubMed] [Google Scholar]
- 28.Lu S, Hu J, Xie X, Huang R, He J. Sesquiterpenoids isolated from feces-residing Streptomyces sp. inhibit the cellular entry of influenza a viruses. Nat Prod Res. 2022;36(24):6286–96. [DOI] [PubMed] [Google Scholar]
- 29.Kuo YH, Chen CH, Chien SC, Lin YL. Five new cadinane-type sesquiterpenes from the heartwood of Chamaecyparis obtusa var. formosana. J Nat Prod. 2002;65(1):25–8. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Jiang K, Zhai YM, Tan JJ, Meng DL, Guo SL, Qu SJ, Tan CH. Sesquiterpenoids and their glycosides from Gynura procumbens. Helv Chim Acta. 2014;97(3):369–74. [Google Scholar]
- 31.Yang JL, Zhao YM, Shi YP. Sesquiterpenoids from the rhizomes of Homalomena occulta. Nat Prod Bioprospect. 2016;6(4):211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piers E, Gavai AV. A (Z)-ethylidenecyclopentane annulation method. Total syntheses of (±)-anhydrooplopanone, (±)-oplopanone, and (±)-8-epi-oplopanone. J Org Chem. 1990;55(8):2380–90. [Google Scholar]
- 33.Ahmed AA, Jakupovic J, Bohlmann F. Dihydroxypallenone, a sesquiterpene with a new carbon skeletonerom pallenisspinosa. Phytochemistry. 1990;29(10):3355–8. [Google Scholar]
- 34.Appendino G, Jakupovic J, Jakupovic S. Sesquiterpenoids from Pallenis spinosa. Phytochemistry. 1997;46(6):1039–43. [Google Scholar]
- 35.Yang Z, Yang Y, Yang X, Zhang Y, Zhao L, Xu L, Ding Z. Sesquiterpenes from the secondary metabolites of Streptomyces sp. (YIM 56130). Chem Pharm Bull. 2011;59(11):1430–3. [DOI] [PubMed] [Google Scholar]
- 36.Heymann H, Tezuka Y, Kikuchi T, Supriyatna S. Constituents of Sindora sumatrana MIQ. I. Isolation and NMR spectral analysis of sesquiterpenes from the dried pods. Chem Pharm Bull. 1994;42(1):138–46. [Google Scholar]
- 37.Chen QJ, Ouyang MA, Tan QW, Zhang ZK, Wu ZJ, Lin QY. Constituents from the seeds of Brucea javanica with inhibitory activity of Tobacco mosaic virus. J Asian Nat Prod Res. 2009;11(6):539–47. [DOI] [PubMed] [Google Scholar]
- 38.Ding L, Görls H, Hertweck C. Plant-like cadinane sesquiterpenes from an actinobacterial mangrove endophyte. Magn Reson Chem. 2021;59(1):34–42. [DOI] [PubMed] [Google Scholar]
- 39.Tan Z, Zhao J, Liu J, Zhang M, Chen R, Xie K, Dai J. Sesquiterpenoids from the cultured mycelia of Ganoderma capense. Fitoterapia. 2017;118:73–9. [DOI] [PubMed] [Google Scholar]
- 40.Nagashima F, Kishi K, Hamada Y, Takaoka S, Asakawa Y. ent-Verticillane-type diterpenoids from the Japanese liverwort Jackiella javanica. Phytochemistry. 2005;66(14):1662–70. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Yang M, Han YF, Gao K. New sesquiterpenes from Erigeron annus. Planta Med. 2005;71(3):268–72. [DOI] [PubMed] [Google Scholar]
- 42.Wu S, Wang Y, Liu N, Dong G, Sheng C. Tackling fungal resistance by biofilm inhibitors. J Med Chem. 2017;60(6):2193–211. [DOI] [PubMed] [Google Scholar]
- 43.Forsyth CB, Mathews HL. Lymphocyte adhesion to Candida albicans. Infect Immun. 2002;70(2):517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, Tan J, Mu Y, Zheng D, Huang X, Li L. New antimicrobial terpenoids and phloroglucinol glucosides from Syzygium szemaoense. Bioorg Chem. 2020;103:104242. [DOI] [PubMed] [Google Scholar]
- 45.Tan J, Zhang Z, Zheng D, Mu Y, Cao B, Yang J, Han L, Huang X. Structure–activity relationship and biofilm formation-related gene targets of oleanolic acid-type saponins from Pulsatilla chinensis against Candida albicans. Bioorg Chem. 2024;146:107311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: The antimicrobial activity of some compounds, HRESI-MS, 1D and 2D NMR spectra of new compounds 1–4, 13–15, 17, 18, 22, 23, 25–28, experimental ECD spectra and calculated ECD spectra of compound 12.
Data Availability Statement
Data will be made available on request.