Dear editor,
Enhancers are cis-regulatory sequences, along with trans-factors, that spatiotemporally control gene expressions(Jindal & Farley, 2021; Kim & Wysocka, 2023). Many active enhancers can produce transcripts, including bidirectionally transcribed enhancer RNA (eRNA) and unidirectionally transcribed long non-coding RNAs – enhancer-associated lncRNA (eaRNA). While eRNAs are generally nonpolyadenylated, unspliced and unstable, eaRNAs are mostly polyadenylated, spliced and stable(Sartorelli & Lauberth, 2020; Tan, Biasini et al., 2020). Although previous studies indicated that enhancer derived transcripts may actively promote transcription, some believe they are just byproducts of polymerase II at enhancer sites. We previously revealed the presence of an upstream enhancer for Ctnnb1, the coding gene for β-Catenin. The enhancer, named as neCtnnb1 (neocortical enhancer of Ctnnb1), was found to maintain Ctnnb1's transcription predominantly in developing cerebral cortex (neocortex) of the brain to promote neurogenesis of excitatory neurons in superficial layers(Wang, Wang et al., 2022). It is unknown whether the neCtnnb1 locus could transcribe eRNA or eaRNA, and if so, does it regulate the expression of Ctnnb1?
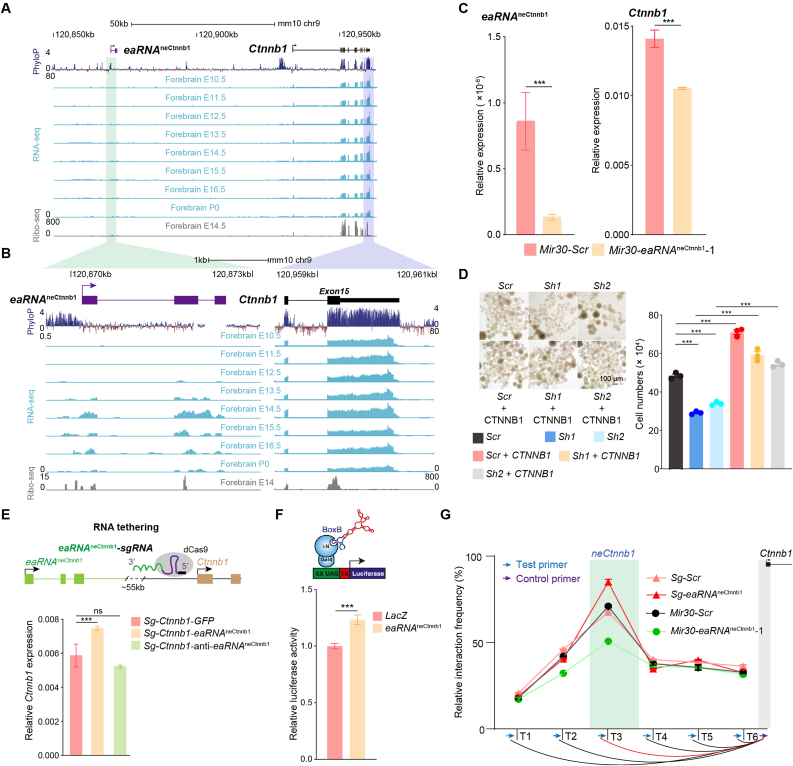
We first analyzed RNA-seq data of developing mouse forebrains deposited in public databases. Strikingly, transcripts were detected downstream of the most conserved region of neCtnnb1, starting at embryonic (E) day 12.5, with peak expression at E14.5, followed by a gradual decline through E16.5 to birth (P0) (Fig. 1A and 1B). This temporal expression pattern was further validated using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) in developing forebrain tissues (Fig. S1A). Notably, the transcription activity from neCtnnb1 coincides with, but around one day precedes with the enhancer activity of neCtnnb1. Therefore, the transcripts were named as eaRNAneCtnnb1. 5′ and 3′ RACE (Rapid Amplification of cDNA Ends) experiments revealed that eaRNAneCtnnb1 is 835 nucleotides long with three exons (Fig. 1A and 1B). And because of the 3’ polyadenylation capture method used in regular RNA-seq, eaRNAneCtnnb1 is polyadenylated. Notably, the sequence of eaRNAneCtnnb1 was predicted to have a low protein-coding potential score (Fig. S1B) and shows weak ribosome profiling signals relative to Ctnnb1 (Fig. 1B, bottom). This suggests that eaRNAneCtnnb1 is less likely to be translated into a functional protein, indicating its potential role as a regulatory RNA rather than a protein-coding RNA. Next, we carried out in situ hybridization of eaRNAneCtnnb1 on coronal sections of embryonic brains, revealing that eaRNAneCtnnb1 is predominantly enriched at the ventricular zone (VZ) and subventricular zone (SVZ), where most neural progenitor cells reside and exhibit the highest canonical Wnt/β-Catenin signaling activity (Fig. S1C).
Fig. 1.
eaRNAneCtnnb1maintains the transcription ofCtnnb1and facilitates the interaction of neCtnnb1 andpCtnnb1. (A–B) Transcripts derived from neCtnnb1 and Ctnnb1 in forebrains at indicated time points are shown. (B) indicates enlarged regions of neCtnnb1 (green shaded) and the Ctnnb1 (blue shaded). Please note that eaRNAneCtnnb1 consists of three exons and is transcribed at the same direction as Ctnnb1. The sequence homology among vertebrates is shown at the top, while the Ribo-seq signal of E14.5 forebrains is displayed at the bottom. RNA-seq data were obtained from ENCODE (The Encyclopedia of DNA Elements) and the Ribo-seq data were retrieved from the GEO database (GSE169457). (C) qRT-PCR showing relative mRNA levels of eaRNAneCtnnb1 and Ctnnb1 in Neuro-2a cells transfected with shRNAs with scrambled (Scr) sequence or against eaRNAneCtnnb1 for 72 hours. n = 3 independent experiments. (D) Neocortical neurospheres were transfected with lentiviruses expressing indicated shRNAs and human CTNNB1 for seven days followed by cell counting. n = 3 independent experiments. (E) In the RNA-tethering experiments, indicated transcripts were attached with gRNA to target the promoter of Ctnnb1. 72 hours after transfection, the expression levels of Ctnnb1 in Neuro-2a cells were measured by qRT-PCR. n = 3 independent experiments. (F) Relative luciferase activity of Neuro-2a cells transfected with plasmids expressing BoxB-tagged LacZ or eaRNAneCtnnb1 along with Gal4-kN and UAS-TK-Luciferase for 24 hours. n = 3 independent experiments. (G) In the 3C experiments, the relative association strength between indicated sites with the promoter of Ctnnb1 were measured in Neuro-2a cells. The test primer T3 is located within the neCtnnb1 region. Quantification data are shown as means ± SEM, statistical significance was determined using one-way ANOVA analysis (D–E) and an unpaired two-tailed Student's t-test (C and F). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. ns, not significant.
We next asked whether eaRNAneCtnnb1 is functional. We devised short hairpin RNAs (shRNAs) against eaRNAneCtnnb1, which could efficiently downregulate its expression in Neuro-2a neuroblastoma cells. The expression levels of Ctnnb1 were decreased by ∼30% upon eaRNAneCtnnb1 shRNA treatment, indicating eaRNAneCtnnb1 positively regulates Ctnnb1 transcription (Fig. 1C). Importantly, numbers of cultured neocortical progenitor cells were decreased by ∼40% on loss of eaRNAneCtnnb1, which could be completely reversed by overexpressing shRNA-resisting human CTNNB1 (Fig. 1D). Therefore, eaRNAneCtnnb1 maintains self-renewal of neural progenitors in a Ctnnb1-dependent manner. We next explored whether eaRNAneCtnnb1 has a trans-activating role. To this end, the transcript of eaRNAneCtnnb1 was attached with the guide RNA to target the promoter of Ctnnb1 (pCtnnb1), with the transcript of Gfp and antisense eaRNAneCtnnb1 as controls. Data showed that eaRNAneCtnnb1 could significantly enhances the transcription of Ctnnb1 in Neuro-2a cells, whereas the antisense eaRNAneCtnnb1 was unable to do so (Fig. 1E). Consistently, the Gal4-λN/BoxB reporter assay revealed that eaRNAneCtnnb1 could boost the luciferase reporter activity (Fig. 1F). Together, eaRNAneCtnnb1 bears intrinsic activity to promote transcription.
Because neCtnnb1 physically contacts with the pCtnnb1, we then examined whether eaRNAneCtnnb1 could facilitate the association. Neuro-2a cells were transfected with the shRNA against eaRNAneCtnnb1. The chromosome conformation capture (3C) assay revealed that downregulating eaRNAneCtnnb1 could significantly compromise the association of neCtnnb1 with pCtnnb1. Moreover, CRISPR/dCas9-mediated activation (CRISPRa) of the promoter of eaRNAneCtnnb1 greatly increased the transcription of eaRNAneCtnnb1 and Ctnnb1 (Fig. S1D), which simultaneously enhanced the association of neCtnnb1 and pCtnnb1 (Fig. 1G). Thus, eaRNAneCtnnb1 mediates the enhancer-promoter (E-P) contact.
eRNA and eaRNA can help enhancers to find their cognate promoters. Recent research has revealed that repeating sequence within eRNAs and promoter upstream transcripts (PROMPTs) facilitate E-P interactions(Liang, Cao et al., 2023). Here we showed that eaRNAneCtnnb1, the RNA transcript derived from the neocortical enhancer of Ctnnb1, positively regulates the transcription of Ctnnb1. eaRNAneCtnnb1 achieves this possibly by promoting the E-P contact. The trans-factor ASH2L has been found to associate with neCtnnb1 and pCtnnb1, sustaining Ctnnb1 transcription(Wang, Wang et al., 2022). It would be interesting to investigate whether eaRNAneCtnnb1 binds to ASH2L and whether its intrinsic ability to promote transcription depends on this interaction. Intriguingly, a larger proportion of eaRNAneCtnnb1 is localized in the cytosol than in the nucleus (Fig. S1E), a phenomenon that deserves further investigation. For example, it raises the question of whether eaRNAneCtnnb1 could regulate the Wnt/β-Catenin signaling by interacting with the post-translational machinery in the cytosol(Lin, Luo et al., 2022). The sequence of neCtnnb1 enhancer is evolutionarily conserved among amniotes with neocortical structures. The possible presence of eaRNAneCtnnb1 in other species, especially those with complex neocortical structures, might fine-tune the strength of Wnt/β-Catenin signaling, thereby contributing to the expansion of the neocortex during evolution. Recently, we identified another enhancer of Ctnnb1, ieCtnnb1 (intestinal enhancer of Ctnnb1), which plays a critical role in regulating homeostasis and tumorigenesis of intestinal epithelia(Hua, Zhao et al., 2024). The presence and role of ieCtnnb1-associated RNA also warrant further exploration.
CRediT authorship contribution statement
Chen Zhao: Formal analysis, Data curation, Conceptualization, Investigation, Validation. Liang Wang: Formal analysis, Data curation, Conceptualization. Junbao Wang: Project administration. Kuan Tian: Software, Data curation. Xiaojiao Hua: Visualization. Fangyu Wang: Funding acquisition, Conceptualization, Project administration, Supervision. Yan Zhou: Writing – review & editing, Writing – original draft, Visualization, Validation, Resources, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
None.
Acknowledgement
We thank the Core Facility and the Animal Facility of Medical Research Institute of Wuhan University for technical support. Y. Zhou was supported by grants from National Key R&D Program of China (2022YFA0806603), National Natural Science Foundation of China (32270876), and the Fundamental Research Funds for the Central Universities (2042022dx0003 and 2042023kf0234). F. Wang was supported by National Natural Science Foundation of China (32300660).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellin.2024.100212.
Contributor Information
Fangyu Wang, Email: fangyu.wang@whu.edu.cn.
Yan Zhou, Email: yan.zhou@whu.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Hua X., Zhao C., Tian J., Wang J., Miao X., Zheng G., Wu M., Ye M., Liu Y., Zhou Y. A Ctnnb1 enhancer transcriptionally regulates Wnt signaling dosage to balance homeostasis and tumorigenesis of intestinal epithelia. Elife. 2024;13 doi: 10.7554/eLife.98238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal G.A., Farley E.K. Enhancer grammar in development, evolution, and disease: Dependencies and interplay. Developmental Cell. 2021;56(5):575–587. doi: 10.1016/j.devcel.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Wysocka J. Deciphering the multi-scale, quantitative cis-regulatory code. Molecular Cell. 2023;83(3):373–392. doi: 10.1016/j.molcel.2022.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Cao C., Ji L., Cai Z., Wang D., Ye R., Chen J., Yu X., Zhou J., Bai Z., Wang R., Yang X., Zhu P., Xue Y. Complementary Alu sequences mediate enhancer–promoter selectivity. Nature. 2023;619(7971):868–875. doi: 10.1038/s41586-023-06323-x. [DOI] [PubMed] [Google Scholar]
- Lin X., Luo M.-L., Song E. Long non-coding RNA and non-coding nucleic acids: Signaling players in the networks of the tumor ecosystem. Cell Insight. 2022;1(1) doi: 10.1016/j.cellin.2022.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Lauberth S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nature Structural & Molecular Biology. 2020;27(6):521–528. doi: 10.1038/s41594-020-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.Y., Biasini A., Young R.S., Marques A.C. Splicing of enhancer-associated lincRNAs contributes to enhancer activity. Life Science Alliance. 2020;3(4) doi: 10.26508/lsa.202000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang A., Tian K., Hua X., Zhang B., Zheng Y., Kong X., Li W., Xu L., Wang J., Li Z., Liu Y., Zhou Y. A Ctnnb1 enhancer regulates neocortical neurogenesis by controlling the abundance of intermediate progenitors. Cell Discovery. 2022;8(1):74. doi: 10.1038/s41421-022-00421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.