Abstract
Objective
Our objectives were to identify correlation patterns between complement and lipid pathways using a multiomics data integration approach and to determine how these interconnections affect age-related macular degeneration (AMD).
Design
Nested case-control study.
Subjects and Controls
The analyses were performed in a subset of the Singapore Indian Eye Study. We randomly selected 155 AMD cases and age- and sex-matched them with 155 controls.
Methods
Firstly, a multiomics data integration method was used to identify correlation patterns between the omics data. Then, we tested possible interactions between the lipids and complement proteins using logistic regression models.
Main Outcome Measures
Age-related macular degeneration was determined according to the Beckman classification system. We measured in serum samples 35 complement proteins and 66 lipids, and used 9 genetic variants.
Results
Among the 155 AMD cases, 93 (60.0%) had early and 62 (40.0%) intermediate AMD. Firstly, we identified 2 clusters between complement proteins and lipids involving (1) mannan-binding lectin serine protease 1 and several different high-density lipoprotein particles, and (2) complement factor H-related protein 1, carboxypeptidase N subunit 2 and complement component C8 gamma chain, and sphingomyelin and different cholesterol. Secondly, we identified 1 interaction between complement protein 1R and sphingomyelin with an odds of AMD 2 times higher for individuals with low levels of sphingomyelin and complement protein 1R (odds ratio = 2.13 [1.09, 4.17]).
Conclusions
We report here, using a cutting-edge multiomics integration approach, the complex interconnections between genetic, metabolomics, and proteomic data. This method permitted us to obtain a holistic picture and identify multiomics signature of AMD pathophysiology. These results advocate for a personalized therapeutic approach that accounts for multiple pathways. However, these results need to be validated in larger studies with different ethnic groups.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: Age-related macular degeneration, Complement proteins, Lipids, Multiomics data integration
Age-related macular degeneration (AMD) is a leading cause of irreversible blindness in Asia and worldwide.1 It is known to be a multifactorial disease caused by a combination of lifestyle and genetic factors.2, 3, 4, 5 Several pathways are involved in the pathogenesis of AMD, such as complement system (CS) activation and lipid dysregulation.6, 7, 8, 9, 10, 11 Although progress has been made in the understanding of these pathways, treatment options to prevent AMD onset and progression remain limited because the etiology and pathogenesis of AMD remain incompletely understood.
Lipid metabolism and CS have close relationships;12, 13, 14 however, the mechanisms by which they interact to alter the risk of AMD are unknown. Firstly, multiple complement components and complement-regulatory proteins have indeed been detected in high-density lipoprotein (HDL) particles, supporting close relationships between these 2 systems.15, 16, 17 Secondly, some lipid-related metabolites have opposing effects on the risk of developing AMD, depending on the CS allele distribution.12 For example, the effect of extremely large very low density lipoproteins (VLDL) particles shift from protective for individuals with the genotype GG in the genetic variant rs116503776 [complement component 2- complement factor-ski2 like RNA helicase] (odds ratio [OR] = 0.82 [0.73, 0.91]), to increased risk for individuals with the genotype AA (OR = 3.83 [1.04, 14.18]); suggesting that the CS itself may regulate lipid metabolism. Moreover, it has been shown that changes in complement factor H (CFH) concentration can modify the anti-inflammatory properties of large HDL particles.16 However, it is still unclear how these complex interactions affect AMD pathophysiology.
Exploring the interactions between complex data corresponding to different biological systems is methodologically challenging and requires the use of advanced statistical models of systems biology. Integrating multiomics data sets may allow a holistic picture to be built up that can explain such complex pathophysiology. This approach has been, for example, used to investigate the molecular changes during the first week of human life by integrating different omics such as transcriptomic, proteomic, and metabolomics and has produced novel and robust biological insights.18 To the best of our knowledge, a systematic multiomics data integration analysis has not been conducted in the context of AMD. We propose here to integrate 3 omics layers, i.e., genetics, metabolomics and proteomics, in a data-driven approach to generate new hypotheses and to further test them using classical inferential models. Our objectives were (1) to identify correlation patterns between genetic variants, complement proteins and lipids using multiomics data integration approach and (2) to determine how these interconnections affect AMD.
Methods
Study Design and Participants
The analyses were performed in a subset of the Singapore Indian Eye Study. The Singapore Indian Eye Study is a prospective population-based study of 3400 subjects aged ≥40 years conducted in Singapore.19 Participants underwent a standardized interview and laboratory investigations. Informed written consent was obtained from participants, ethical approval was obtained from the Institutional Review Board of SingHealth, and all research adhered to the tenets of the Declaration of Helsinki.
Among the 3400 participants, we excluded individuals (1) with incomplete clinical data (n = 431), or with missing information on lipid levels (n = 175) or genetic variants (n = 862) and (2) with any retinopathy (n = 192), any cataract (n = 810), or any type of glaucoma (n = 44). Among the remaining population (n = 1213), we randomly selected 200 individuals with AMD at the baseline examination. Then, we selected our control population (individuals free of AMD) using a matching based on age (5-year categories) and sex. We found 197 pairs of cases and controls (3 cases did not have any control with the same age category and sex). Finally, we selected 155 pairs (n = 310) who had enough serum stored (if the case or the control within a pair did not have enough blood, then the whole pair was excluded).
AMD Grading
In the Singapore Indian Eye Study, color fundus photographs were graded by the Singapore National Eye Centre Ocular reading center. Features of AMD were identified and severity of AMD was determined according to the Beckman classification system.20 In brief, an individual was considered free of any AMD if pigmentary abnormalities (hyperpigmentation or depigmentation) and drusen (>63 μm) were absent in both eyes. Early AMD was defined as the presence of drusen >63 μm and ≤125 μm in ≥1 eye and without pigmentary abnormalities. Intermediate AMD was defined as the presence of large drusen (>125 μm) or the presence of pigmentary abnormalities in ≥1 eye. The analyses were performed at the individual level with the Beckman grading of the more severe eye considered for each individual.
Omics Data
Proteomics
A targeted liquid chromatography–mass spectrometry proteomics technique was used to quantify protein’s serum concentrations. This technique allows the simultaneous measurement of many proteins in a single experiment. We used 20 μL of stored serum samples at the baseline visit. A reference peptide mix standard was added to the reconstituted peptide sample according to manufacturer’s protocol (PlasmaDive Reference Peptides kit, Biognosys AG). The reconstituted peptide samples were then analyzed on an EASY-nLC 1200 system coupled to Orbitrap Exploris 480 mass spectrometer (ThermoFisher Scientific). Orbitrap Exploris 480 mass spectrometer was operated in data-independent and positive ionization mode. The resulting mass spectrometry/mass spectrometry data were processed using Spectronaut (Biognosys AG) data-independent acquisition analysis. Among the 313 proteins quantified, we kept the 35 complement proteins. Five individuals had missing complement protein values because they were below the detection threshold (4 for mannose-binding lectin 2 and 1 for CFH-related protein 5 proteins, respectively). We imputed these values by the minimum values of the remaining individuals divided by 5.21 The distributions were log transformed and standardized (centered and scaled). Finally, 13 individuals with values lower than −5 and higher than +5 standard deviation were excluded.
Metabolomics Data
A high-throughput proton nuclear magnetic resonance metabolomics platform (Nightingale Health Ltd) was used to measure metabolite’s serum concentrations at the baseline visit. Details of the methodology and applications of the nuclear magnetic resonance metabolomics platform have been described previously.22 This method provides simultaneous quantification of the following 150 blood lipid-related metabolites: total and subfractions of cholesterol, triglyceride, phospholipid, cholesterol ester, and free cholesterol in HDL, low density lipoprotein and VLDL particles, phosphoglyceride, choline, apolipoproteins, fatty acids, and small/medium/large/very large, and chylomicrons and extremely large HDL/low density lipoprotein and VLDL lipoprotein subclasses. Moreover, in each subclass, the concentrations of lipids, triglycerides, cholesterol esters, free cholesterol, and phospholipids were quantified. As the correlations among these 150 lipid-related metabolites were very high, we did not consider the levels of these subfractions in each lipoprotein subclass, thus including a total of 66 lipid-related metabolites. The distributions were log transformed and standardized (centered and scaled). Finally, 10 individuals with values lower than −5 and higher than +5 standard deviation were excluded.
Genomics Data
We considered the lead single nucleotide polymorphisms (SNPs) identified to be associated with AMD in the latest large international genome-wide association studies conducted on 16 144 patients with AMD and 17 832 controls.3 Among the 34 significant SNPs, we considered the 9 involved in the lipid (rs2043085 [hepatic lipase C], rs5817082 [cholesteryl ester transfer protein], rs429358 [apolipoprotein E], rs2740488 [ATP-binding cassette transporter A1], and rs11080055 [transmembrane protein 97-vitronectin]) or complement pathways (rs10922109 [CFH], rs10033900 [complement factor I], rs116503776 [complement component 2-complement factor-ski2 like RNA helicase], and rs2230199 [complement component 3]). For the 2 SNPs rs2230199 and rs429358, too few individuals were homozygous for the risk allele (n = 4 and 2, respectively); we thus regrouped individuals with at least a risk allele together (leading to binary variables). Otherwise, for the rest of the SNPs, the 3 categories were considered. All the SNPs were considered as continuous variables.
Statistical Analyses
The statistical analyses were conducted in 2 steps. First, we generated new hypotheses using a data-driven approach, and second, we formally tested these hypotheses using a classical statistical framework. Firstly, we used a multiomics data integration approach to determine (1) the correlation patterns between the 3 omics layer of information, i.e., genomic (genetic variants), metabolomics (lipid-related metabolites), and proteomics (complement proteins) and (2) the relative contributions of each of the variables in each omics layer. Secondly, we tested the possible interactions between the lipid-related metabolites and the complement proteins based on the results of the first step using logistic regression models (with multiple testing correction). Due to the limited sample size, early and intermediate AMD cases were pooled into a single AMD category.
The multiomics data integration method used here, called multiblock sparse projection to latent structure-discriminant analysis (sPLS-DA), is an extension of a projection to latent structure-discriminant analysis with multiple blocks of omics information.23,24 Projection to latent structure-discriminant analysis is a linear multivariate model which performs classification tasks by seeking components that best separate the sample groups. By calculating components from the original variables, this method reduces the data dimension and is thus able to process a very high number of variables. Sparse projection to latent structure-discriminant analysis is the sparse version that also selects variables using lasso penalization that best discriminate between groups. The analytical aim of multiblock sPLS-DA is to identify a signature composed of correlated features across different types of omics (a multiomics signature) which can classify a given outcome. This method is a holistic approach with the potential to find new biological insights not revealed by any single-data omics analysis.
The multiblock sPLS-DA requires in a single optimization problem to both maximize correlations between the omics blocks and classification of the outcome. To do so, it is required to provide a design matrix which indicates which blocks should be connected to maximize the covariance between components, and to what extent. This pertains to the complexity of the analytical task involved as several constraints are included in the optimization procedure. The choice of this matrix can be based on prior knowledge or data-driven. Due to the lack of knowledge on how much our 3 omics layers are correlated, we have performed a pairwise projection to latent structure analysis to examine the correlation between pairs of components associated to each block. The other arguments needed for this model were the number of components (1 single value) and the number of variables to select (1 value for each omics block and for each component). Both were tuned using repeated cross-validation (fivefold cross-validation repeated 50 times). We selected the arguments that minimized the classification error rates. For the number of components, up to 5 components were tested. For the number of variables to select, we tested the following grid values: [1, 2, 3, 4, 5, 6, 7, 8, 9] for the genetic variants (n = 9), [2, 5, 10, 15, 20, 25, 30, 35, 45, 55, 65] for the lipids (n = 66), and [2, 4, 6, 8, 10, 15, 20, 25, 30, 35] for the complement proteins (n = 35).
The second objective was to identify possible interactions between the lipids and the complement proteins in relation to AMD. Because the multiblock sPLS-DA model both optimizes correlations between the omics data sets and the disease classification, it provides an elegant way to identify interactions candidates. We chose pairs of lipids/proteins that were selected by the multiblock sPLS-DA with correlations ≥0.5. Then, all these interaction candidates were tested using logistic regression models adjusted on the following possible confounders: age, sex, hypertension, diabetes, body mass index, smoking status, and lipid-lowering medications. The lipids and complement proteins were considered as continuous variables. We corrected for multiple testing issues using the method developed by Gao et al to account for the correlations.25 As 20 components explained 99.2% of the data variability of the lipids and complement proteins, we used a corrected P value equal to 0.05/20 = 0.0025. Finally, for the significant interactions, we performed 2 additional analyses: 1 using a logistic regression model with the significant interacting terms considered as binary variables (lower and higher than the median value) and a generalized additive model (gam) to generate a 3-dimensional plot of the interaction. Both models were adjusted with the same possible confounders.
Finally, we compared the contributions of the lipids, complement proteins, and genetic variants selected by the multiblock sPLS-DA model to the ORs estimated with a logistic regression model. Each variable was tested in a separate model adjusted for the same covariates considered in the previous analyses. All the analyses were performed using R software version 4.3.1 (R Foundation for Statistical Computing). The multiomics data integration was done using the R package “mixOmics.”24
Results
Study Population
Among the 155 individuals with AMD, 93 (60.0%) had early and 62 (40.0%) intermediate AMD. The age and sex distribution were very similar, as a result of the matching between individuals with and without AMD, as well as other possible confounders such as body mass index, and diabetes status (Table 1). Projection to Latent Structure-Discriminant Analysis were performed for each omics layer to explore their discrimination abilities (Fig S1, available at www.ophthalmologyscience.org). Complement proteins visually enabled the best discrimination, followed by genetic variants and lipids.
Table 1.
Characteristics of the Study Population According to the AMD Status
| No AMD (N = 155) | AMD (N = 155) | P Value | |
|---|---|---|---|
| Age, median (IQR) | 52.4 (47.1,59) | 52.3 (47.7,59.1) | 0.694 |
| Sex, female (%) | 77 (49.7) | 77 (49.7) | 1 |
| BMI, median (IQR) | 26.2 (23.8,29) | 26.5 (23.9,28.8) | 0.776 |
| Hypertension, n (%) | 75 (48.4) | 83 (53.5) | 0.363 |
| Diabetes, n (%) | 37 (23.9) | 45 (29) | 0.303 |
| Lipid-lowering medication, n (%) | 37 (23.9) | 45 (29) | 0.303 |
| Smoking status, n (%) | 0.199 | ||
| Never smoked | 119 (76.8) | 114 (73.5) | |
| Current smoker | 26 (16.8) | 22 (14.2) | |
| Past smoker | 10 (6.5) | 19 (12.3) |
AMD = age-related macular degeneration; BMI = body mass index; IQR = interquartile range.
Multiblock sPLS-DA model tuning
The first argument to be tuned was the design matrix. The correlations between the pairs of components in projection to latent structure analyses were 0.29 (between complement proteins and lipids), 0.23 (between lipids and genetic variants), and 0.32 (between complement proteins and genetic variants). We therefore considered an overall level of correlations between the omics layers of 0.3 in the design matrix. Using fivefold cross-validation repeat 50 times, we selected 2 components (Fig S2, available at www.ophthalmologyscience.org). For the number of variables to retain, we selected 3 genetic variants (2 in the first component and 1 in the second), 32 lipids (2 in the first component and 30 in the second), and 19 complement proteins (15 in the first component and 4 in the second). Therefore, among 110 variables included in the multiblock sPLS-DA model (9 genetic variants, 66 lipids, and 35 complement proteins), a total of 54 were selected in the final model. The distributions of the lipids and complement proteins are presented Figure S3 (available at www.ophthalmologyscience.org).
Main AMD Contributors
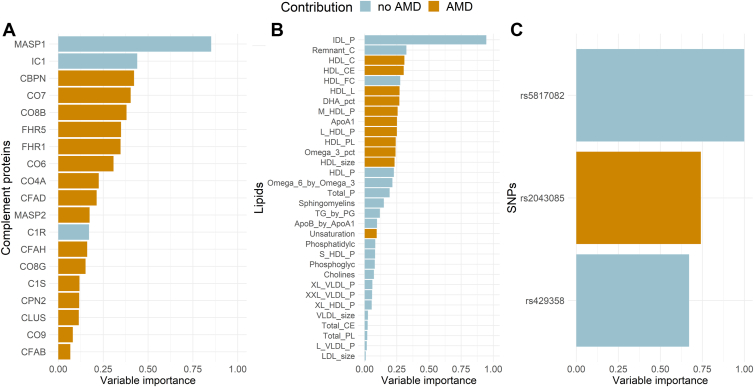
The Figure 4 presents the contribution to AMD of each genetic variant, lipid, and complement protein selected by the multiblock sPLS-DA model. The 3 stronger contributors were rs5817082 (variable importance [VI] = 1.00), rs2043085 (VI = 0.74), and rs429358 (VI = 0.67) for the genetic variants; intermediate-density lipoprotein (IDL) particle (VI = 0.95), remnant cholesterol (VI = 0.32), and HDL cholesterol (VI = 0.31) for the lipids; and mannan-binding lectin serine protease 1 (MASP1) (VI = 0.85), inhibitor of complement (VI = 0.44), and complement binding protein N (VI = 0.42) for the complement proteins (Fig 4).
Figure 4.
Contributions to AMD, expressed as variable importance, of the complement proteins (n = 19) (A), lipids (n = 32) (B), and genetic variants (n = 3) (C) selected by the multiblock sPLS-DA model. AMD = age-related macular degeneration; SNP = single nucleotide polymorphism; sPLS-DA = sparse projection to latent structure-discriminant analysis.
The contributions estimated using the multiblock sPLS-DA model were similar to the estimates obtained a logistic regression model (Fig S5, available at www.ophthalmologyscience.org). The 4 complement proteins significantly associated with AMD using the logistic regression model were all among the top 6 proteins with the highest contributions with the multiblock sPLS-DA model. Regarding the lipids, the only significant variable identified with the logistic model was the level of IDL particles, which was ranked as the most important lipid in the multiblock sPLS-DA model. Finally, for the genetic variants, among the 3 identified by the multiblock sPLS-DA model, 2 were significant using the logistic model.
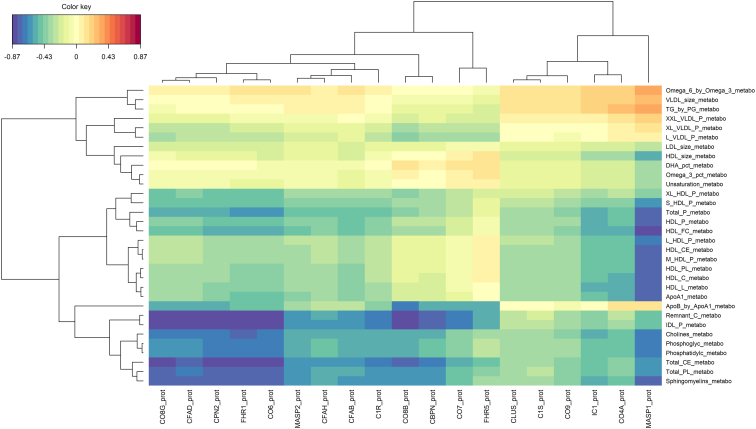
Correlation Patterns between Complement Proteins and Lipids
Figure 6 shows the correlations between the complement proteins and the lipids selected by the multiblock sPLS-DA model. There were 2 main clusters of negative correlations between the following complement proteins: complement component 8 gamma (CO8G), complement factor A domain, carboxypeptidase N subunit 2 (CPN2), CFH-related protein 1 (FHR1), complement component 6, complement component 8 beta; and the following lipids: remnant cholesterol, IDL particle, choline, phosphoglyceride, phosphatidylcholine, total cholesterol ester, total phospholipid, and sphingomyelin and between the complement protein MASP1 and several lipids such as concentration of HDL particles, free cholesterol, and cholesterol ester in HDL particle (Fig 6). Furthermore, there was one cluster of positive correlations (with weaker absolute values) between MASP1 and ratio omega6/omega3, VLDL particle size, and ratio triglyceride/phosphoglyceride. Figure S7 (available at www.ophthalmologyscience.org) and Figure 8 present these correlations in network graphs, with the subsets of correlations ≥0.50 and 0.75, respectively, to simplify the visualization. Additional graphical outputs showing these complex correlation patterns are presented in the supplementary materials (Figs S9, S10, available at www.ophthalmologyscience.org).
Figure 6.
Correlations between complement proteins (n = 19) and lipids (n = 32) selected by the multiblock sPLS-DA model. sPLS-DA = sparse projection to latent structure-discriminant analysis.
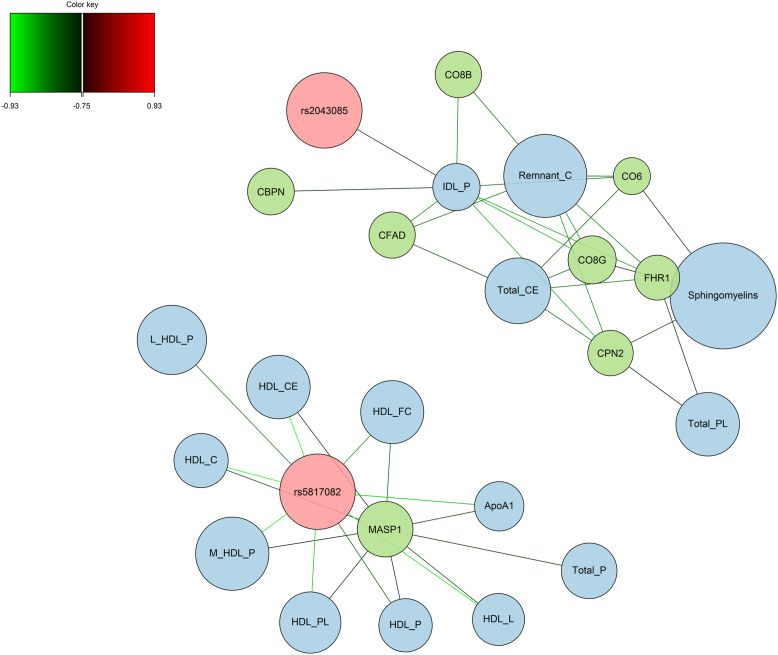
Figure 8.
Correlations between complement proteins (n = 8), lipids (n = 15), and genetic variants (n = 2) selected by the multiblock sPLS-DA model (only correlations ≥0.75 are considered in this graph to simplify the reading). sPLS-DA = sparse projection to latent structure-discriminant analysis.
Correlation Patterns between Genetic Variants with Complement Proteins and Lipids
The 3 genetic variants selected by the multiblock sPLS-DA model were all located in genes involved in lipid metabolism, respectively hepatic lipase C (rs2043085), cholesteryl ester transfer protein (rs5817082), and apolipoprotein E (rs429358). The strongest correlations were found between rs2043085 and the concentration of IDL particles and between rs5817082 and apolipoprotein A1 and several HDL subfractions such as cholesterol ester, free cholesterol, and phospholipids (Fig 8, Fig S11, available at www.ophthalmologyscience.org). Interestingly, these genetic variants also show high correlations (although <0.75) with complement proteins. The genetic variant rs5817082 was positively correlated with MASP1; rs2043085 was positively correlated with FHR1, CO8G, and CPN2; and rs429358 was negatively correlated with CO8G and CPN2 and complement component 7 (Fig S12, available at www.ophthalmologyscience.org).
Interaction between Complement Proteins and Lipids
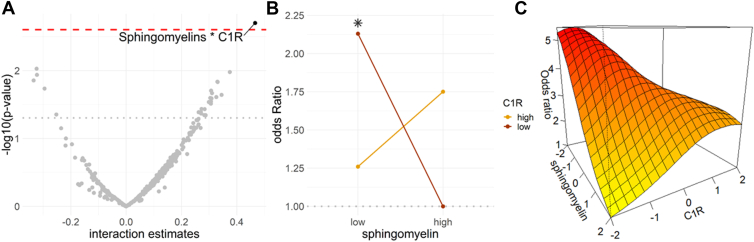
We tested the interactions between all the pairs of complement proteins and lipids selected by the multiblock sPLS-DA model at a correlation cutoff ≥0.50. To do so, we tested each interaction separately in a logistic regression model adjusted for confounders (with lipids and complement proteins considered as continuous variables). After correcting for multiple testing, only 1 interaction was significant (P = 0.002) between sphingomyelin and complement protein 1R (C1R) (Fig 13A). The odds of AMD were 2 times higher for individuals with low levels of sphingomyelin and C1R (lower than the median values) (OR = 2.13 [1.09, 4.17], P = 0.028), compared with individuals with high levels of sphingomyelin and low level of C1R (Table 2, Fig 13B, C).
Figure 13.
A, Interaction estimates corresponding to all the combinations between the complement proteins (n = 16) and lipids (n = 23) selected by multiblock sPLS-DA model (at a cutoff 0.50), i.e., the interaction candidates. Estimates above the dotted red line corresponded to the significant interactions after correction for multiple testing. B and C increased in the odds of AMD (expressed in odds ratio) according to sphingomyelin and C1R using a logistic regression adjusted on age, sex, hypertension, diabetes, body mass index, smoking status, and lipid-lowering medications (with sphingomyelin and C1R considered as binary variables with low and high values corresponding to values lower and higher than the median) (B); and using a generalized additive model adjusted on the same variables (with sphingomyelin and C1R considered as continuous variables) (C). The asterisk in panel B corresponds to a significant effect. AMD = age-related macular degeneration; C1R = complement protein 1R; sPLS-DA = sparse projection to latent structure-discriminant analysis.
Table 2.
Conjoint Effect of Sphingomyelin and C1R on the Odds of AMD
| Sphingomyelin | C1R | n | OR | P |
|---|---|---|---|---|
| Continuous | Continuous | 310 | 1.59 [1.18, 2.14] | 0.002 |
| High | Low | 88 | 1 | |
| High | High | 67 | 1.75 [0.90, 3.39] | 0.097 |
| Low | High | 88 | 1.26 [0.67, 2.37] | 0.482 |
| Low | Low | 67 | 2.13 [1.09, 4.17] | 0.028 |
AMD = age-related macular degeneration; C1R = complement protein 1R; high = higher than the median value; low = lower than the median value; OR = odds ratio.
Median value for sphingomyelin = 0.14; median value for C1R = 0.11.
Discussion
In this study, we used a cutting-edge statistical method to integrate multiple omics data in order to better understand the complex pathophysiology of AMD. More specifically, we used this approach to investigate the interplay between genetic variants, complement proteins, and lipids and determine how it can affect AMD. First, we showed that complement proteins and lipids were strongly interconnected within 2 main clusters. The first one was composed of MASP1 and different HDL particles and HDL subfractions. The second one was composed of several complement proteins (such as FHR1, CPN2, and CO8G) and different types of lipids (such as sphingomyelin, remnant cholesterol, and IDL particles). Second, and more importantly, we identified one interaction between sphingomyelin and C1R, with 2 times higher odds of AMD for individuals with low levels of sphingomyelin and C1R.
Integrating omics data sets offer the unprecedented opportunity to assess interactions between multiple omics data and provide a more comprehensive understanding of diseases' biological pathways. The method chosen here allowed the identification of molecular biomarkers across different omics data that are correlated and associated with a phenotype of interest. The classical reductionist approaches which identify key features within each omics individually would have failed to provide a comprehensive picture due to the complexity of the relationships between the genetic variants, the lipids, and the complement proteins. On the contrary, integrating different omics data sets allowed us to explore these complex multiomics interplays and determine how they affect AMD. To make it possible, the method relies both on dimension reduction and feature selection, and maximizes both correlations between the omics blocks, and classification of the outcome.
Because this multiomics integration model is complex and more difficult to interpret compared to tradition inferential models, we have compared the contributions of the lipids, complement proteins, and the genetic variants using this model to the estimates obtained using a logistic regression model (Fig S5, available at www.ophthalmologyscience.org). The results were very similar except for the protein MASP1 which was not identified using the logistic model. This discrepancy arises from the high correlation of MASP1 with several lipids, as illustrated in Figure 6, Figure 8. The multiblock sPLS-DA model optimizes both the correlations between the omics blocks and the contributions of each block to AMD. Therefore, this model gave this protein a higher importance due to the overall correlation structure of the data which could not be accounted for by a simple logistic model. This difference highlights the relevance of the multiomics model that integrates the different omics blocks in a single model to obtain a multiomics signature of AMD.
The presence of complement proteins in lipoprotein particles has been reported.15,16 For example, it has been shown that CFH levels could be greatly increased in large HDL and may explain the ability of HDL to prevent the organization of complement membrane attack complex and to suppress inflammation.16 However, most studies were conducted only in HDL particles. Here, in this study, we included many different types of lipids. We found 2 clusters between complement proteins and lipids: the first one involving different HDL particles and subfractions with MASP1 and the second one involving different cholesterol (remnant cholesterol and total cholesterol ester) and lipoprotein particles such as IDL, with different complement proteins such as complement component 6, FHR1, CPN2, and complement factor A domain. A similar negative correlation between HDL particles and MASP1 has been reported in a study conducted among 255 patients with AMD cases and 221 controls selected from the Dutch and German European Genetic Database (EUGENDA-Nijmegen and EUGENDA-Cologne).26 The associations in the second cluster further extend our understanding of the interconnections between lipids and complement proteins. Interestingly, we also found high correlations between genetic variants located in genes involved in lipid metabolism with complement proteins such as MASP1, CO8G, and CPN2. The understanding of the interplay between complement proteins and lipids is crucial as the complement proteins and lipids can regulate each other (e.g., modification of the lipid inflammatory properties by complement proteins and regulation of the complement activation by lipid components), which may have important implications in AMD pathophysiology.
Among all the possible interactions between complement proteins and lipids, we found a significant one between sphingomyelin and C1R. The odds of AMD were 2 times higher for individuals with low levels of sphingomyelin and C1R. Sphingolipids are known to be involved in the regulation of many physiological and pathophysiological processes in the retina.27 Their dysregulation is an important element in the onset and progression of retinal diseases, including AMD.27 Moreover, homeostasis of different sphingolipids plays a role in the maintenance, progression, and regulation of the immune response.28 Regarding C1R, studies have shown increased expression of mRNAs of C1R gene in some AMD lines compared with controls.29,30 Interestingly, it has been shown that the NOD-like receptor family pyrin domain containing 3 inflammasome, that is responsible for a chronic self-perpetuating inflammatory process which has a prominent role in AMD,31 is promoted by several factors including lipid accumulation and complement activation.32 It is therefore possible that the interaction we found here between sphingomyelin and C1R represents a molecular signature of inflammasome activation in AMD pathophysiology. It is worth noting that a similar synergy between the complement (C5a and its C5aR1 receptor) and glycosphingolipid systems results in a massive generation of proinflammatory cytokines, chemokines, and growth factors and shown to promote immune dysregulation and tissue damage in coronavirus disease 2019 and Gaucher’s disease.33 Similarly, sphingolipid metabolism and complement activation products have essential roles in promoting tumor survival.34 Although promising, our findings need further research to elucidate the exact mechanisms at play between lipids and complement components.
Several clinical trials targeting different components of the CS have completed or are currently ongoing. So far, most of the candidate therapeutic complement compounds tested have shown limited success.35 The mechanism by which the different risk factors interact and converge toward AMD are not fully understood and therefore drug discovery is challenging. Our findings suggest that to fully understand the role of the complement pathway and to be able to use it as a therapeutic candidate, other pathways should be accounted for, notably those pertaining to lipid metabolism and complement dysregulation. Our findings also indicate that eventually a personalized medicine approach which takes into account lipid metabolism and complement regulation will be needed in managing AMD.
The principal strength of this study is its unique combination of multiomics data sets, i.e., genetic, metabolomics, and proteomic, to explore their complex interconnections and how these affect the risk of AMD. We have used cutting-edge analytical techniques that have allowed us to combine information from different evaluations into a single analysis instead of considering data from each omics separately. Our study also suffers from limitations. First, we only included individuals of Indian ancestry. Therefore, further studies are crucial, especially with larger sample sizes and including other ethnicities to investigate the generalizability and reproducibility of our results in different populations and geographical locations where cultural factors influence diet and consequently lipid metabolism. Second, the associations between genetic variants and the other omics, i.e., complement proteins and lipids, were performed using correlations. While this may not be the most appropriate metric to use here, the methods implemented are not yet able to include categorical variables and thus still need to be improved. Finally, because of the limited sample size, early and intermediate AMD were pooled in a single category. We also did not include individuals with late AMD since early stages of the disease are more relevant for translational purposes. Further studies thus need to be conducted based on AMD stages.
To summarize, we report here, using a cutting-edge multiomics integration approach, the complex interconnections between genetic, metabolomics, and proteomic data, and their role in AMD pathophysiology. This method permitted us to integrate the outputs from different omics data to obtain a holistic picture and identify multiomics signature of AMD pathophysiology. We confirm the existence of connectivity between complement proteins and lipids and identify among the high number of lipids and complement proteins a specific interaction between sphingomyelin and C1R that might be responsible for driving an inflammatory process, thus increasing the risk of AMD. These findings have clinical implications in terms of understanding AMD pathophysiology and advocate the development of a personalized therapeutic approach that could account for the multiple etiological pathways. However, these findings need to be validated in larger studies with different ethnic groups.
Manuscript no. XOPS-D-24-00085.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s):
This research is supported by A∗STARBMRC (08/1/35/19/550; C.-Y.C.) and Lee Foundation grant (S.N.).
HUMAN SUBJECTS: Human subjects were included in this study. Ethical approval was obtained from the Institutional Review Board of SingHealth and all research adhered to the tenets of the Declaration of Helsinki. Informed, written consent was obtained from participants.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Nusinovici, Chakravarthy, Cheng
Data collection: Nusinovici, Zhou
Analysis and interpretation: Nusinovici, Zhou, Raghavan, Tham, Li, D. Cheung, Wang, C.M.C. Cheung, Wong, Chakravarthy, Cheng
Obtained funding: N/A
Overall responsibility: Nusinovici, Zhou, Raghavan, Tham, Li, D. Cheung, Wang, C.M.C. Cheung, Wong, Chakravarthy, Cheng
Supplementary Data
References
- 1.Wong W.L., Su X., Li X., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 3.Fritsche L.G., Igl W., Bailey J.N.C., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunier V., Merle B.M.J., Delyfer M.N., et al. Incidence of and risk factors associated with age-related macular degeneration: four-year follow-up from the ALIENOR study. JAMA Ophthalmol. 2018;136:473. doi: 10.1001/jamaophthalmol.2018.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Bedell M., Zhang K. Age-related macular degeneration: genetic and environmental factors of disease. Mol Interv. 2010;10:271–281. doi: 10.1124/mi.10.5.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan P.L., Bowes Rickman C., Katsanis N. AMD and the alternative complement pathway: genetics and functional implications. Hum Genomics. 2016;10:23. doi: 10.1186/s40246-016-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan L.X., Germer C.J., La Cunza N., Lakkaraju A. Complement activation, lipid metabolism, and mitochondrial injury: converging pathways in age-related macular degeneration. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geerlings M.J., De Jong E.K., Den Hollander A.I. The complement system in age-related macular degeneration: a review of rare genetic variants and implications for personalized treatment. Mol Immunol. 2017;84:65–76. doi: 10.1016/j.molimm.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khandhadia S., Cipriani V., Yates J.R.W., Lotery A.J. Age-related macular degeneration and the complement system. Immunobiology. 2012;217:127–146. doi: 10.1016/j.imbio.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M., Jiang N., Chu Y., et al. Dysregulated metabolic pathways in age-related macular degeneration. Sci Rep. 2020;10:2464. doi: 10.1038/s41598-020-59244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nusinovici S., Zhou L., Wang X., et al. Contributions of Lipid-Related Metabolites and Complement Proteins to Early and Intermediate Age-Related Macular Degeneration. Ophthalmol Sci. 2024;4:100538. doi: 10.1016/j.xops.2024.100538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sim R.Z.H., Tham Y.C., Betzler B.K., et al. Relationships between lipid-related metabolites and age-related macular degeneration vary with complement genotype. Ophthalmol Sci. 2022;2 doi: 10.1016/j.xops.2022.100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acar İ.E., Lores-Motta L., Colijn J.M., et al. Integrating metabolomics, genomics, and disease pathways in age-related macular degeneration. Ophthalmology. 2020;127:1693–1709. doi: 10.1016/j.ophtha.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen E.M., Emri E., Merle B.M.J., et al. A new perspective on lipid research in age-related macular degeneration. Prog Retin Eye Res. 2018;67:56–86. doi: 10.1016/j.preteyeres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Vaisar T., Pennathur S., Green P.S., et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Gordon S.M., Xi H., et al. HDL subclass proteomic analysis and functional implication of protein dynamic change during HDL maturation. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbu A., Hamad O.A., Lind L., et al. The role of complement factor C3 in lipid metabolism. Mol Immunol. 2015;67:101–107. doi: 10.1016/j.molimm.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 18.Lee A.H., Shannon C.P., Amenyogbe N., et al. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat Commun. 2019;10:1092. doi: 10.1038/s41467-019-08794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan C.W., Wong T.Y., Lavanya R., et al. Prevalence and risk factors for refractive errors in Indians: the Singapore Indian eye study (SINDI) Invest Ophthalmol Vis Sci. 2011;52:3166. doi: 10.1167/iovs.10-6210. [DOI] [PubMed] [Google Scholar]
- 20.Ferris F.L., Wilkinson C.P., Bird A., et al. Clinical classification of age-related macular degeneration. Ophthalmology. 2013;120:844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Y., Malitsky S. MetaboReport: from metabolomics data analysis to comprehensive reporting. Bioinformatics. 2024;40 doi: 10.1093/bioinformatics/btae373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soininen P., Kangas A.J., Würtz P., et al. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8:192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- 23.Singh A., Shannon C.P., Gautier B., et al. DIABLO: from multi-omics assays to biomarker discovery, an integrative approach. Bioinformatics. 2016;35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohart F., Gautier B., Singh A., Lê Cao K.A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X., Starmer J., Martin E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 26.Acar I.E., Willems E., Kersten E., et al. Semi-quantitative multiplex profiling of the complement system identifies associations of complement proteins with genetic variants and metabolites in age-related macular degeneration. J Phys Math. 2021;11:1256. doi: 10.3390/jpm11121256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon M.V., Basu S.K., Qaladize B., et al. Sphingolipids as critical players in retinal physiology and pathology. J Lipid Res. 2021;62 doi: 10.1194/jlr.TR120000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee M., Lee S.Y., Bae Y.S. Functional roles of sphingolipids in immunity and their implication in disease. Exp Mol Med. 2023;55:1110–1130. doi: 10.1038/s12276-023-01018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saini J.S., Corneo B., Miller J.D., et al. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age-related macular degeneration. Cell Stem Cell. 2017;20:635–647.e7. doi: 10.1016/j.stem.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong J., Cai H., Noggle S., et al. Stem cell-derived retinal pigment epithelium from patients with age-related macular degeneration exhibit reduced metabolism and matrix interactions. Stem Cells Transl Medicine. 2020;9:364–376. doi: 10.1002/sctm.19-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marneros A.G. Role of inflammasome activation in neovascular age-related macular degeneration. FEBS J. 2023;290:28–36. doi: 10.1111/febs.16278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maran J.J., Adesina M.M., Green C.R., et al. The central role of the NLRP3 inflammasome pathway in the pathogenesis of age-related diseases in the eye and the brain. Ageing Res Rev. 2023;88 doi: 10.1016/j.arr.2023.101954. [DOI] [PubMed] [Google Scholar]
- 33.Trivedi V.S., Magnusen A.F., Rani R., et al. Targeting the complement–sphingolipid system in COVID-19 and gaucher diseases: evidence for a new treatment strategy. IJMS. 2022;23 doi: 10.3390/ijms232214340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janneh A.H., Atkinson C., Tomlinson S., Ogretmen B. Sphingolipid metabolism and complement signaling in cancer progression. Trends Cancer. 2023;9:782–787. doi: 10.1016/j.trecan.2023.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar-Singh R. The role of complement membrane attack complex in dry and wet AMD - from hypothesis to clinical trials. Exp Eye Res. 2019;184:266–277. doi: 10.1016/j.exer.2019.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.