MYC(N) suppression and NFκB activation, the latter triggered by both tumor cell–intrinsic and –extrinsic factors, mediate cellular persistence in high-risk neuroblastoma and reveal new therapeutic approaches to prevent relapse.
Abstract
Relapse rates in high-risk neuroblastoma remain exceedingly high. The malignant cells that are responsible for relapse have not been identified, and mechanisms of therapy resistance remain poorly understood. In this study, we used single-nucleus RNA sequencing and bulk whole-genome sequencing to identify and characterize the residual malignant persister cells that survive chemotherapy from a cohort of 20 matched diagnosis and definitive surgery tumor samples from patients treated with high-risk neuroblastoma induction chemotherapy. We show that persister cells share common mechanisms of chemotherapy escape, including suppression of MYC(N) activity and activation of NFκB signaling, and the latter is further enhanced by cell–cell communication between the malignant cells and the tumor microenvironment. Overall, our work dissects the transcriptional landscape of cellular persistence in high-risk neuroblastoma and paves the way to the development of new therapeutic strategies to prevent disease relapse.
Significance: Approximately 50% of patients with high-risk neuroblastoma die of relapsed refractory disease. We identified the malignant cells that likely contribute to relapse and discovered key signaling pathways that mediate cellular persistence. Inhibition of these pathways and their downstream effectors is postulated to eliminate persister cells and prevent relapse.
Introduction
Chemotherapy resistance and disease relapse remains the leading cause of mortality among children with cancer. In high-risk neuroblastoma, a childhood cancer derived from the developing sympathetic nervous system, relapse rates are approximately 40% to 60% even when there is an apparent complete radiographic remission, suggesting that a subset of malignant cells can persist through multimodal intensive therapy, which includes induction chemotherapy followed by definitive surgery, myeloablative autologous stem cell transplant, external beam radiation therapy, systemic radiotherapy with 131I-metaiodobenzylguanidine (131I-MIBG), differentiating agents, and immunotherapy (1).
Multiple mechanisms of chemotherapy resistance have been proposed over the years, and our knowledge about therapy resistance in children with high-risk neuroblastoma is derived primarily from in vitro and in vivo models as well as a limited number of diagnostic and relapse patient samples. These include the upregulation of drug efflux transporters (2), acquisition of mutations in drug resistance–conferring genes, in particular, genes of the RAS-MAPK pathway (3, 4), and epigenetic reprogramming from an adrenergic toward a more chemoresistant mesenchymal cell state (5–10). However, these data have yet to be leveraged therapeutically to prevent neuroblastoma recurrence, and the cells responsible for relapse have not been identified. This is in part due to the difficulty in detecting rare malignant cells that survive therapy in patient samples, which are often highly necrotic and consist primarily of cells from the tumor microenvironment (TME). We therefore hypothesized that identification and characterization of the residual persisting malignant cells that survive chemotherapy in patient samples can lead to the discovery of novel therapeutic vulnerabilities to eliminate persister cells and prevent neuroblastoma relapse and/or inform on more precise relapsed neuroblastoma therapeutic interventions.
Recent advances have allowed the use of single-cell technologies to detect rare cellular subpopulations at an unprecedented level and dissect malignant from nonmalignant cells (11). The majority of neuroblastoma single-cell/nucleus RNA sequencing (RNA-seq) studies using patient samples focused on the developmental origin and intratumoral heterogeneity of neuroblastoma (12–14) as well as the TME (15–17). In addition, several studies leveraged single-cell technologies to investigate therapy resistance in cell lines and patient derived xenografts (PDX; refs. 18, 19), but the relevance of these models in patients is yet to be determined. Importantly, although a “pure” malignant mesenchymal population has not yet been identified in patient samples and PDXs, an aggressive transitional state (20) and a subpopulation of adrenergic cells with mesenchymal features have been reported in patient tumor samples (14, 21), linking intratumoral heterogeneity with therapy resistance. Furthermore, Thirant and colleagues have also proposed CD44, a stem cell marker, as a cell-surface marker to identify this subpopulation of more chemoresistant cells in patients (21).
Although the abovementioned studies have tremendously advanced our understanding of neuroblastoma intratumoral heterogeneity, the ability to study cellular changes under the pressure of chemotherapy in patient samples requires serially paired samples. In this study, we used single-nucleus RNA-seq (snRNA-seq) coupled with bulk whole-genome sequencing (WGS) from 20 matched diagnostic neuroblastoma tumor samples paired with definitive surgery tumor samples acquired after 4 to 5 cycles of induction chemotherapy to identify and characterize chemotherapy-resistant persister cells.
Results
Chemotherapy Remodels the Cellular Composition of Neuroblastoma Tumors
To identify neuroblastoma persister cells, we studied a cohort of matched diagnosis and definitive surgery samples from 20 patients with high-risk neuroblastoma who were treated at the Children’s Hospital of Philadelphia (CHOP) from 2007 to 2022 (all patients ended their therapy by the end of 2022); the database was locked in January 2024. The key clinical characteristics of these patients are summarized in Fig. 1A, and additional clinical and genetic information is provided in Supplementary Tables S1 and S2. Of note, patients were treated on our institutional or Children’s Oncology Group (COG) frontline studies for high-risk neuroblastoma with induction chemotherapy generally consisting of alternating cycles of topotecan and cyclophosphamide, cisplatin and etoposide, and vincristine, doxorubicin, and cyclophosphamide (ANBL1531, NCT03126916; refs. 22–25). Three patients (CN5, CN11, and CN17) received 131I-MIBG therapy, and one patient (CN11) received two cycles of irinotecan, temozolomide, and the monoclonal anti-GD2 antibody, dinutuximab, prior to surgery. Patients received 1 to 2 additional cycles of induction chemotherapy after surgery and prior to response evaluation by 123I-MIBG imaging. Responses were classified into complete response, partial response, stable disease, or progressive disease, defined according to the International Neuroblastoma Response Criteria (26).
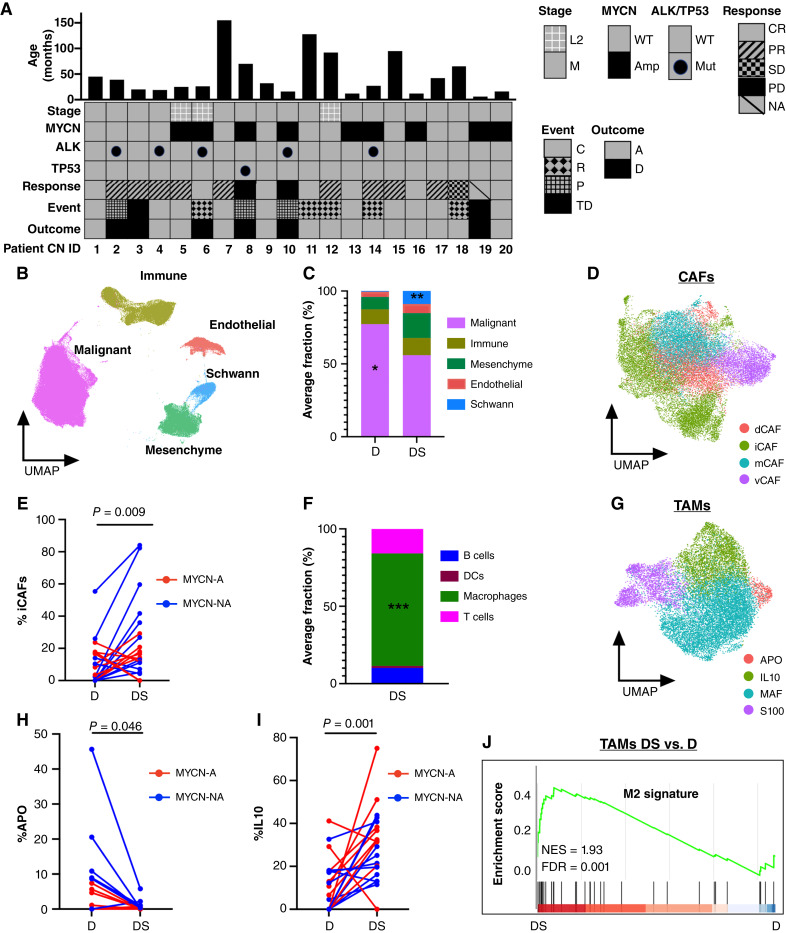
Figure 1.
Chemotherapy remodels the cellular composition of neuroblastoma tumors. A, Patients characteristics of a paired matched cohort of 20 patients with high-risk neuroblastoma at diagnosis and surgical resection. Response was determined at the end of induction therapy, typically after 1–2 additional cycles of chemotherapy following definitive surgery. B, Uniform Manifold Approximation and Projection for Dimension (UMAP) of 248,591 nuclei integrated from diagnostic and surgical specimens from 20 patients with high-risk neuroblastoma showing the main cellular components of tumors. C, Stacked bar plot of average percent of cell type at diagnosis and definitive surgery. Malignant cells showed a statistically significant decrease (P = 0.013; paired Wilcoxon test) whereas Schwann cells demonstrated a statistically significant increase (P = 0.006; paired Wilcoxon test) at definitive surgery. D, UMAP of CAFs annotated according to recently identified CAF subtypes (30). E, Connected pairs plot showing the percent of relative iCAFs at diagnosis and definitive surgery in 20 tumor pairs. Lines are colored by MYCN amplification status. F, Stacked bar plot of average percent of immune cell types at definitive surgery (P = 2.96e−05; Wilcoxon test). G, UMAP of TAMs annotated according to recently identified neuroblastoma TAM subtypes (17). H, Connected pairs plot showing the percent of relative APO TAMs (APO) at diagnosis and definitive surgery in 20 tumor pairs. I, Connected pairs plot showing the percent of relative IL10 TAMs (iL10) at diagnosis and definitive surgery in 20 tumor pairs. J, Gene set enrichment analysis (GSEA) analysis of DEGs between TAMs at definitive surgery and diagnosis shows enrichment for M2 signatures (15). *, P < 0.05; **, P < 0.01; ***, P < 0.001. A, laive; AMP, amplified (red) ; C, censor; CR, complete remission; D, death; DS, definitive surgery; D, diagnosis; M, metastatic; MUT, mutated; NES, normalized enrichment score; NA; not amplified (blue); NA, not available (patient died before end of induction response evaluation); NS, not significant; P, progression; PD, progressive disease; PR, partial response; R, relapse; SD, stable disease; TD, toxic death; WT, wildtype.
Whenever available, two regions from each tumor sample were obtained, resulting in a total of 68 samples (30 from diagnostic biopsies and 38 from surgical resection specimens), which were then subjected to snRNA-seq using 10x Genomics droplet sequencing platform. After stringent filtering, we classified nuclei into malignant and nonmalignant using previously published cell type markers and copy-number inference from RNA with InferCNV (Supplementary Fig. S1A and S1B; ref. 27). We further excluded nuclei that belonged to adjacent nonmalignant tissues, such as hepatocytes and the adrenal cortex, and integrated a total of 248,591 nuclei representing both malignant and nonmalignant cells, such as immune, mesenchymal, endothelial, and Schwann cells (Fig. 1B). Of note, using our stringent criteria for malignant cell identification, we did not identify “pure” malignant mesenchymal cells in our cohort, i.e., cells characterized by mesenchymal markers that harbor typical neuroblastoma recurrent copy-number aberrations (CNA; Supplementary Fig. S1B), in accordance with previous studies (13, 14, 21).
To examine the effect that multiagent chemotherapy exerts on the cellular composition of neuroblastoma tumors, we compared the frequency of different cell types at the initial diagnostic biopsy and subsequent definitive surgery. Overall, there was an expected statistically significant reduction in malignant nuclei percentage following induction therapy (Fig. 1C). Specifically, 16 (80%) tumor pairs demonstrated a decrease in tumor percentage across different radiographic responses, whereas 4 tumor pairs displayed an increase in malignant nuclei percentage following chemotherapy in a patient achieving complete response, a patient with stable disease, and two patients with progressive disease (Supplementary Fig. S1C). Of note, response to therapy was measured by MIBG after surgical resection of the primary tumor and thus, tumors demonstrating complete radiographic response but only slight decrease or even increase in malignant nuclei percent at the time of definitive surgery could be explained by discordance of response in primary compared with metastatic compartments and/or differentiation effect, resulting in MIBG nonavid disease.
Consistent with previous single-cell studies (12–14, 21), we observed a paucity of Schwann cells in our high-risk cohort of neuroblastomas and a statistically significant increase of Schwann cells after the time of definitive surgery (Fig. 1C). Importantly, the identified Schwann cells did not have CNAs (Supplementary Fig. S1B) and were therefore not defined as malignant cells, consistent with previous reports (28). Of note, at the time of definitive surgery, the proportion of Schwann cells displayed a statistically significant increase in patients with MYCN-non-amplified tumors compared with patients with MYCN-amplified tumors (Supplementary Fig. S1D). We validated this observation using immunohistochemistry (IHC) staining for S100A, a marker of Schwann cells, in MYCN-amplified and non-amplified surgical specimens of two patients in our cohort (Supplementary Fig. S1E). Furthermore, as the presence of Schwann cells has been associated with differentiating tumors (28, 29), we observed a statistically significant increase in expression of differentiation markers, such as synaptophysin, synaptic vesicle protein II, and neuron-specific enolase in the malignant cells of definitive surgery specimens with abundant Schwann cells (Supplementary Fig. S1F).
To dissect the heterogeneity within cancer-associated fibroblasts (CAF), we reclustered the mesenchymal compartment and identified seven clusters (Supplementary Fig. S1G). Importantly, the CAF subclusters did not harbor inferred CNAs (Supplementary Fig. S1B), confirming that the CAFs are derived from nonmalignant cells. The clusters were then annotated using recently described CAF signatures (30), resulting in four subpopulations: matrix-generating CAFs, inflammatory CAFs (iCAF), vascular CAFs, and dividing CAFs (Fig. 1D). iCAFs exhibited a statistically significant increase in relative frequency following induction chemotherapy (Fig. 1E), consistent with a recent study (17). In line with this observation, differentially expressed genes (DEG) between CAFs at definitive surgery and diagnosis were enriched in inflammatory CAF signature (Supplementary Fig. S1H).
Within the immune compartment, tumor-associated macrophages (TAM) constituted the most abundant subpopulation following induction therapy (Fig. 1F) and exhibited a statistically significant increase in both MYCN-amplified and -non-amplified tumors compared with diagnosis (Supplementary Fig. S1I). To further investigate their heterogeneity, we reclustered TAMs and identified six clusters (Supplementary Fig. S1J), which were subsequently annotated according to recently published neuroblastoma-specific TAM signatures (17). This resulted in four major TAMs subtypes: APO, IL10, MAF, and S100 (Fig. 1G), with APO TAMs demonstrating a statistically significant decrease (Fig. 1H) whereas IL10 TAMs exhibiting a statistically significant increase in relative frequency at the time of definitive surgery (Fig. 1I). In addition, DEGs between TAM definitive surgery and diagnosis were enriched in the M2 signature (Fig. 1J), supporting an immunosuppressive role of TAMs at the time of definitive surgery.
Finally, lymphocytes (T, B, and NK cells) were rare at diagnosis except from samples obtained from lymph nodes, consistent with previous reports (14, 31, 32). Moreover, we did not observe a statistically significant change in lymphocyte frequency at the time of definitive surgery (Supplementary Fig. S1K).
Pathway-Based Clustering Identifies Four Major Transcriptional Programs of Cellular Persistence
Next, we focused on the malignant cells that survived induction chemotherapy in our patient cohort, which we refer to hereon as persister cells, and sought to identify mechanisms of cellular persistence. Reclustering a total of 86,901 persister nuclei from the 38 definitive surgery samples of 19 tumors (CN19 had no malignant nuclei identified at the time of definitive surgery) resulted in a Uniform Manifold Approximation and Projection for Dimension (UMAP) in which nuclei were clustered by patient (Fig. 2A), a phenomenon that has been previously reported in single-cell RNA-seq studies of other cancer types (27). To determine whether persister cells from different patients can be grouped according to the signaling pathway that they activate, we used VISION to calculate the scores of 52 hallmark pathways for individual persister cells (33, 34) and noted that certain pathways varied in a nonrandom fashion across patients with similar clinical outcomes (Supplementary Fig. S2A). For example, MYC(N) target genes were high in three of the four patients who died from disease (Fig. 2B). We use the term MYC(N) here and throughout as the pathway classifier cannot distinguish between c-MYC (MYC) and MYCN target genes signatures. We then selected the top 9 most informative pathways (see “Methods” section for our stringent selection criteria) to cluster the persister cells, yielding a cell–cell similarity map whereby persister cells are largely distinguished by their MYC(N) target genes signature scores (Fig. 2C). On one end of the UMAP, persister cells with high MYC(N) target scores activated E2F targets, oxidative phosphorylation, and mTOR signaling, whereas persister cells with low MYC(N) target genes scores activated TNFα via NFκB signaling, IFNα signaling, and myogenesis (Supplementary Fig. S2B).
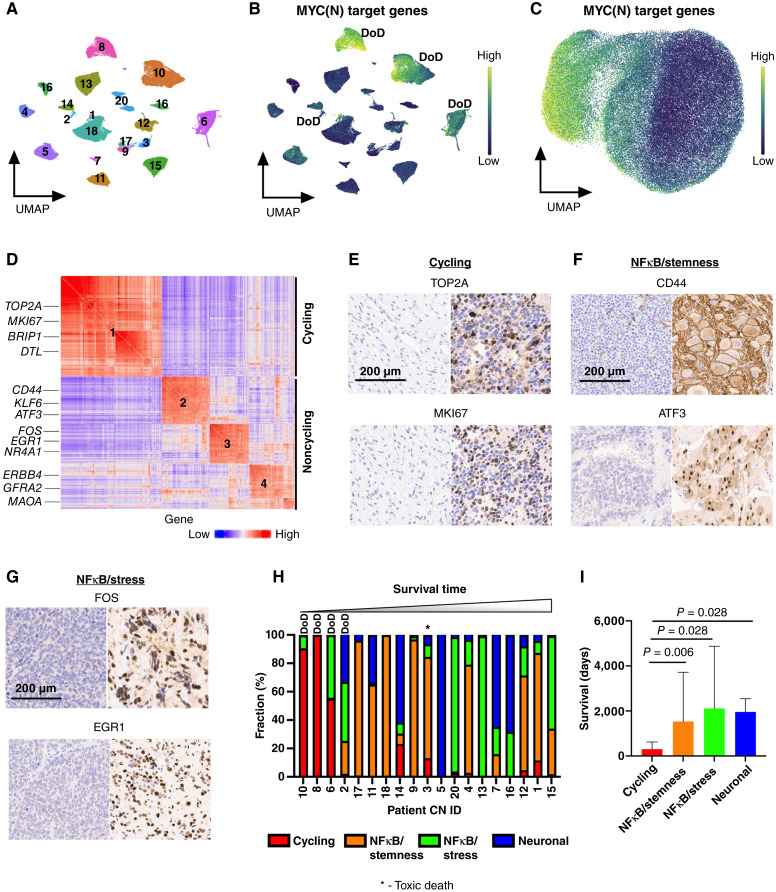
Figure 2.
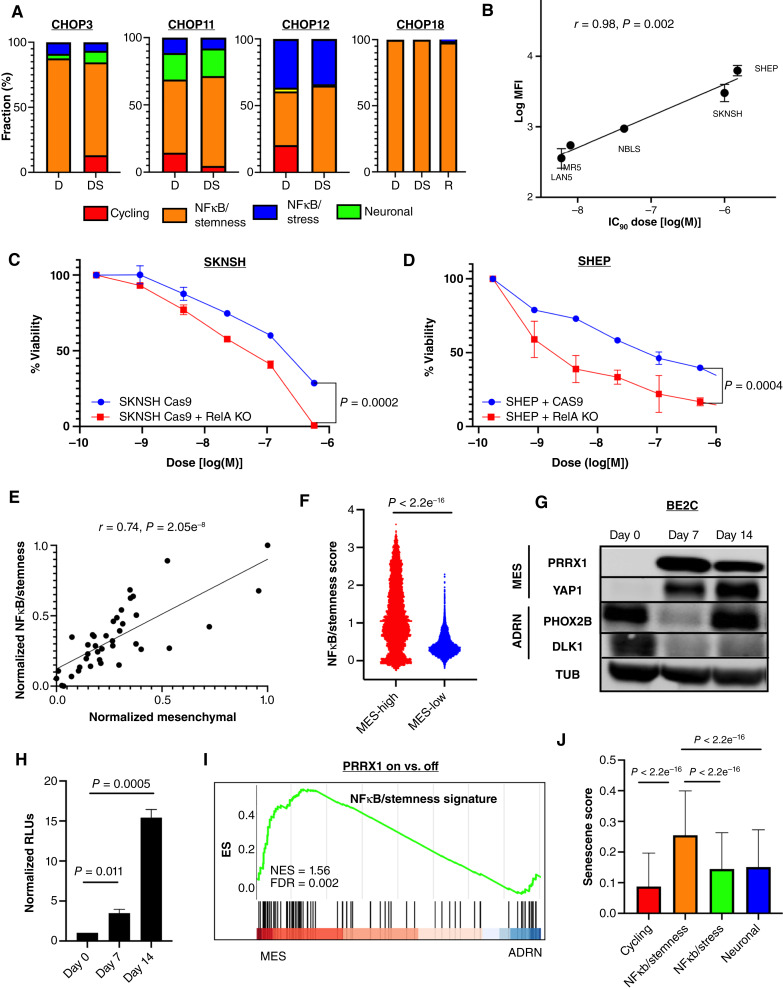
Pathway-based clustering identifies four persister cell subtypes. A, UMAP of 86,901 persister nuclei from 38 surgical resection specimens. Specimens from the same patient cluster together. Numbers correspond to patient CN sample ID. B, UMAP showing the score of MYC(N) target genes [MYC(N) activity] for each persister nucleus. Yellow, high score; dark blue, low score. Three of four patients with high MYC(N) activity at time of definitive surgery died of their disease, with two patients experiencing progressive disease (CN8 and CN10), and one experiencing relapse (CN6). DoD, dead of disease. C, UMAP showing 86,901 persister nuclei clustered according to the pathway-based clustering algorithm using the top 9 most autocorrelated pathways (C > 0.5; FDR < 0.01; see “Methods” for definition of C), here with MYC(N) target genes pathway score shown. Yellow – high score, dark blue – low score. D, Gene to gene (426 genes × 426 genes) correlation heatmap. Rows and columns corresponding to the genes in the meta-modules identified by Hotspot. Red, high correlation; blue, low correlation. Meta-modules were annotated using cell-cycle and pathway enrichment analyses and are numbered as follows: 1–cycling, 2–NFκB/stemness, 3– NFκB/stress, and 4–neuronal. Genes enriched in each meta-module are depicted on the left. E, IHC staining for TOP2A and MKI67, two markers of cycling persister meta-module (#1), of a tumor with low frequency of cycling persister cells at time of definite surgery (left) and tumor with high frequency of cycling persister cells obtained from patient CN8 (right). F, IHC staining for CD44 and ATF3, two markers of the NFκB/stemness meta-module (#2), of a tumor with high NFκB/stemness cells at time of definite surgery obtained from patient CN9. G, IHC staining for FOS and EGR1, two markers of the NFκB/stress meta-module (#3), of a tumor with high NFκB/stress cells at time of definite surgery obtained from patient CN13. H, Bar plot of persister subtypes fraction of 19 surgical specimens (CN19 had no malignant nuclei identified at the time of definitive surgery) ordered in increasing order of the corresponding patients’ survival times. I, Bar plot of patients’ survival times with corresponding tumors assigned to the most abundant persister subtypes. The malignant nuclei of definitive surgery specimens from each patient were decomposed into persister subtypes as in H, and each tumor was classified according to its most abundant persister subtype. The patients’ survival times were then assigned to the corresponding tumors.
To identify recurrent transcriptional programs in persister cells that activate similar pathways, we used Hotspot (35), a computational tool that identifies genes that act in concert, i.e., modules, within a given cell–cell similarity map. Hotspot identified four nonoverlapping transcriptional meta-modules that were largely distinguished by their cell-cycle state (Fig. 2D; Supplementary Fig. S2C and S2D). We then used pathway enrichment analysis to further annotate each meta-module and identified a cycling meta-module (meta-module #1) that was characterized by high expression of genes regulated by the E2F family and MYC(N) transcription factors as well as genes involved in oxidative phosphorylation and mTOR signaling (TOP2A, MKI67, TPX2, DTL, and BRIP1). Two noncycling meta-modules exhibited high expression of NFκB target genes, one associated with stemness (meta-module #2 - ATF3, BCL6, KLF6, CD44) and one associated with stress response (meta-module #3 - FOS, GCH1, NR4A1, EGR1). A third noncycling meta-module (meta-module #4) was characterized by genes enriched with neuronal lineage specification signature (ERBB4, GABRB1, MAOA, and GFRA2; Supplementary Fig. S2E; Supplementary Table S3). Thus, our analysis identified four transcriptional programs of persister cells: cycling, NFκB/stemness, NFκB/stress, and neuronal. We performed IHC that validated the protein expression in the relevant subcategories of key markers belonging to the first three meta-modules which are the main focus of our study: MKI67 and TOP2A (cycling), ATF3 and CD44 (NFκB/stemness), and FOS and EGR1 (NFκB/stress; Fig. 2E–G).
Persister Subtypes Are Associated with Clinical Outcome
We next classified persister cells according to their highest scoring meta-module and decomposed the malignant compartment of each definitive surgery tumor into four transcriptional persister subtypes, revealing both intertumoral and intratumoral heterogeneity (Fig. 2H). Importantly, patients whose tumors consisted of high cycling persister cells fraction had the shortest survival time and died from progressive or relapse refractory disease (Fig. 2I). Of note, we did not observe statistically significant differences in spatial distribution of persister subtypes when we compared their frequency in different samples obtained from the same tumors at definitive surgery (Supplementary Fig. S2F).
Genetic Alterations at Diagnosis Shape Persister Cell Phenotype
To determine whether the observed intratumoral heterogeneity of persister subtypes is driven by genetic events, we subjected tumor pairs to bulk WGS and compared both copy-number variations (CNV) and single-nucleotide variants (SNV) at diagnosis and definitive surgery. For diagnostic samples, CNVs and SNVs were validated using SNP array and a next-generation sequencing targeted panel, respectively, performed as part of the patients’ clinical care.
We did not observe obvious major changes in recurrent copy number changes in high-risk neuroblastoma, including MYCN amplification, partial deletions of chromosome arms 1p, 3p, 4p, and 11q, and partial gain of 2p, 7q, and 17q (36, 37), between matched diagnosis and definitive surgery samples in pairs in which reliable CNVs could be detected at both timepoints (N = 12; Fig. 3A; Supplementary Fig. S3A). Of note, the treated tumors of two patients, CN7 and CN15, harbored 11q deletion and 4p deletion, respectively, which were not present at diagnosis; however, the corresponding diagnostic samples were obtained from readily accessible metastatic lymph nodes rather than abdominal primary tumors. Thus, it is possible that the observed differences reflect spatial rather than temporal heterogeneity.
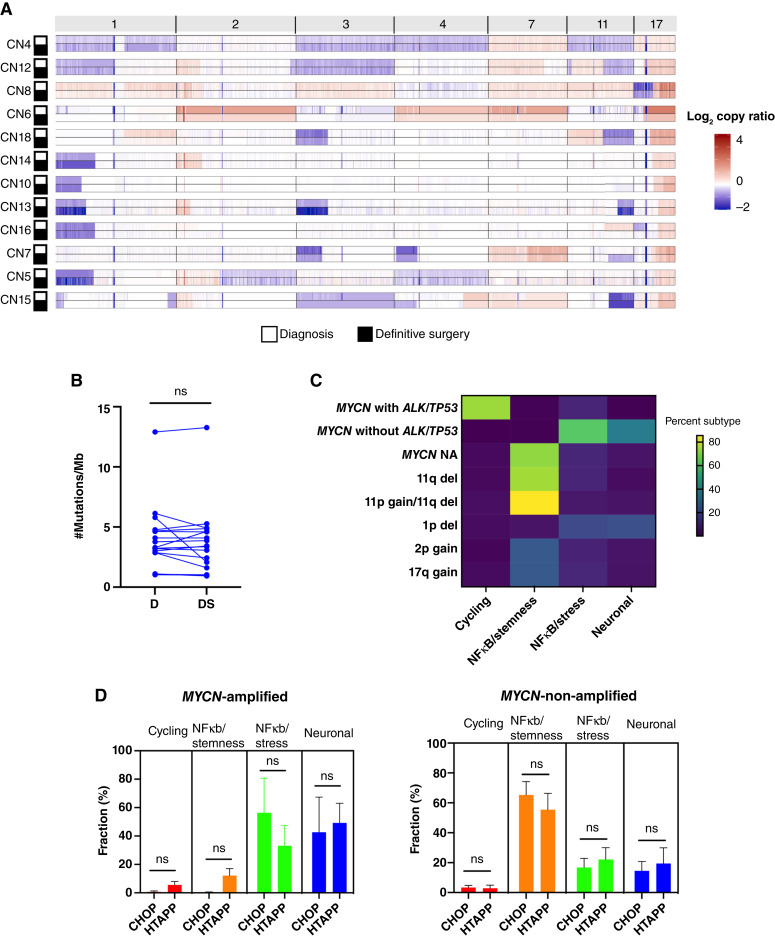
Figure 3.
Persister cell phenotype is shaped by genetic alterations present at diagnosis. A, Heatmap of log2 copy ratio of chromosomes 1, 2, 3, 4, 7, 11, and 17 for 11 matched pairs with malignant cells percentage at diagnosis and definitive surgery sufficient to identify reliable copy number. Red, gain; blue, loss. B, Connected pairs plot showing the mutational burden, measured as the number of mutations per Mb, in 17 matched pairs at diagnosis and definitive surgery. C, Heatmap of median percent persister subtype fraction as a function of genetic alterations at diagnosis. del - deletion, NA - Non-Amplified. D, Bar plots of persister subtypes percentages for the CHOP and HTAPP cohorts in MYCN-amplified tumors (left) and -non-amplified tumors (right). Plots do not include tumors with MYCN amplification and TP53/ALK (N = 3, CHOP cohort). A, Amplified; NA, non-amplified; del, deletion; D, diagnosis; DS, definitive surgery; HTAPP, Human Tumor Atlas Pilot Project; ns, not significant.
In addition, we did not observe a statistically significant difference in mutational burden from diagnosis to definitive surgery (Fig. 3B) and therefore focused our analysis on frequency changes occurring in known driver mutations of neuroblastoma (36, 37). At diagnosis, canonical somatic ALK mutations were identified in five patients (four with F1174L and one with R1275Q), and a TP53 mutation (C135F) was present in one patient tumor, whereas no mutations in other neuroblastoma-associated genes were detected in our cohort. ALK mutations in four tumors were clonal at diagnosis, whereas one diagnostic tumor had a subclonal ALK mutation [F1174L; variant allele frequency (VAF) = 1.5%] that was not originally called in clinical sequencing of tumors using a custom gene panel (38) and was subsequently enriched in the surgical specimen (VAF = 30%), indicating clonal evolution as described by our group and others (Supplementary Fig. S3B and S3C; refs. 39–41).
As our data do not support the acquisition of new genetic aberrations during induction therapy as a major driver of cellular persistence, we sought to determine the association between persister subtypes and genetic events present at diagnosis. We found that cycling persister cells were almost exclusively present in MYCN-amplified tumors also carrying either TP53 or ALK mutations. Consistent with a previous report (42), patients with tumors harboring MYCN amplification with either ALK or TP53 mutation at diagnosis had the shortest survival time. Importantly, the NFκB/stemness subtype was predominant in patients with MYCN-non-amplified tumors, especially in tumors harboring 11q deletion with distal 11p/proximal 11q gain, whereas NFκB/stress and neuronal subtypes were more prevalent in MYCN-amplified tumors without additional mutations (Fig. 3C; Supplementary Fig. S3D). Persister cells in tumors harboring 1p deletion displayed a slight bias toward NFκB/stress and neuronal subtypes, whereas persister subtypes did not differ across tumors harboring 2p gain and 17q gain.
To validate the association between MYCN amplification status at diagnosis and persister subtypes after induction chemotherapy, we used an independent dataset of post-induction chemotherapy definitive surgery samples obtained from 14 patients with high-risk neuroblastoma that were subjected to snRNA-seq as part of the Human Tumor Atlas Pilot Project (HTAPP). Using the signatures of the four persister subtypes defined above, we decomposed the malignant compartment of each tumor in the HTAPP dataset into persister subtypes and observed a similar pattern of persister subtypes distribution in MYCN-amplified and -non-amplified tumors (Fig. 3D). Collectively, our data suggests that the genetic background at diagnosis is a major determinant of response to therapy and clinical outcome.
Noncycling Persister Cells Suppress MYC(N) Activity
To determine the transcriptional changes that occur in malignant cells in response to induction chemotherapy, we decomposed the malignant compartment of diagnostic tumors into persister subtypes (Supplementary Fig. S4A) and compared the changes in their frequency between matched diagnosis and definitive surgery pairs. Cycling and neuronal malignant cells demonstrated a statistically significant decrease whereas NFκB/stemness and NFκB/stress exhibited a statistically significant increase at the time of definitive surgery (Fig. 4A; Supplementary Fig. S4B)
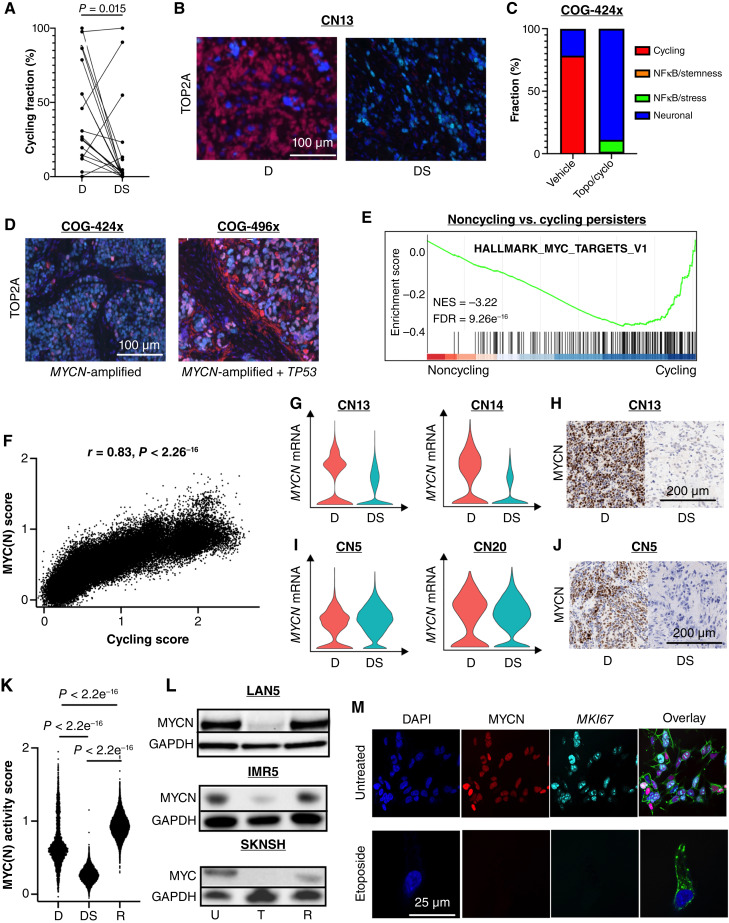
Figure 4.
Noncycling persister cells suppress MYC(N) activity. A, Connected pairs plot showing the decrease in percentage of cycling persister subtype at diagnosis and definitive surgery in 19 matched pairs. B, Dual IF staining for the cycling marker, TOP2A (red), and neuroblastoma marker, PHOX2B (turquoise), at diagnosis and definitive surgery in a MYCN-amplified tumor of patient CN13. Blue represents DAPI nuclear staining. C, Bar plot of persister subtypes decomposition from tumors obtained from a PDX model with MYCN-amplification (COG-424x) before and after treatment with two cycles of topotecan and cyclophosphamide. D, Dual IF staining for the cycling marker, TOP2A (red), and neuroblastoma marker, PHOX2B (turquoise), in two PDX models, one with MYCN amplification (COG-424x; left) and one with MYCN amplification and TP53 mutation (COG496x; right). Blue represents DAPI nuclear staining. E, Gene set enrichment analysis (GSEA) of DEGs between noncycling and cycling persister cells. F, Pearson correlation between MYC(N) activity score and cycling persister subtype scores (meta-module #1). G, Violin plots for MYCN mRNA expression in the malignant cells of MYCN-amplified tumors obtained from patients CN13 and CN14 at diagnosis and definitive surgery. H, IHC staining for MYCN protein in tumors obtained from patient CN13 at diagnosis and definitive surgery. I, Violin plots for MYCN mRNA expression in the malignant cells of MYCN-amplified tumors obtained from patients CN5 and CN20 at diagnosis and definitive surgery. J, IHC staining for MYCN protein in tumors obtained from patient CN5 at diagnosis and definitive surgery. K, Violin plots of MYC(N) activity scores of the malignant cells of a tumor obtained from a patient with an intermediate-risk neuroblastoma with high-risk features at diagnosis, definitive surgery, and relapse. L, Western blots for MYCN and MYC proteins of two MYCN-amplified cell lines (LAN5 and IMR5) and a MYCN-non-amplified, MYC overexpressing cell line (SKNSH) before treatment, at the end of treatment with etoposide, and at regrowth after 3 weeks. M, Dual IF staining for MYCN (red) and MKI67 (turqoise) of LAN5 prior to treatment (top) and at the end of treatment with etoposide (bottom). Blue represents DAPI nuclear staining. Green represents phalloidin staining for actin. D, diagnosis; DS, definitive surgery; U, untreated; T, treated with etoposide; R, regrowth.
We focused on the dynamics of cycling persister cells first and validated the decrease in protein expression from diagnosis to definitive surgery of two cycling markers, TOP2A and MKI67, in both MYCN-amplified and -non-amplified tumor pairs from our patient cohort using immunofluorescence (IF) and IHC staining (Fig. 4B; Supplementary Fig. S4C). We then sought to model the redistribution of cycling persister cells in response to chemotherapy in clinically relevant animal models. We selected two PDX murine models—one from a diagnostic MYCN-amplified tumor lacking TP53 or ALK mutation but harboring BRAF V600E mutation (COG-424x) and another obtained from a diagnostic MYCN-amplified tumor harboring TP53 R342T mutation (COG-496x). To mimic the clinic schedule, mice were treated with a combination of topotecan and cyclophosphamide for five days in two 21-day cycles, and tumors were collected prior to therapy and at the time of maximal tumor shrinkage following the second cycle of chemotherapy (Supplementary Fig. S4D). The tumors of COG-424x were subjected to snRNA-seq and malignant nuclei were classified into persister subtypes at each time point, revealing a profound decrease in cycling persister cells after two cycles of chemotherapy, with a proportionate increase in neuronal and NFκB/stress modules (Fig. 4C). Of note, although we noted an overall decrease in neuronal subtypes following chemotherapy in our patients’ cohort (Supplementary Fig. S4B), few patients’ tumors, such as CN5, showed a similar increase in neuronal subtypes at the time of definitive surgery. To validate the difference in the frequency of cycling persister cells between treated tumors with MYCN amplification and MYCN amplification + TP53 mutation, we performed IF and IHC staining for MKI67 and TOP2A of the treated tumors from our PDX models. Staining confirmed that cycling persister cells were indeed rare in the MYCN-amplified treated tumor (COG-424x) but abundant in the MYCN-amplified + TP53 mutation–treated tumor (COG-496x; Fig. 4D).
As MYCN amplification drives proliferation in neuroblastoma (43) and MYC is overexpressed in many MYCN-non-amplified tumors (44), we hypothesized that MYC(N) activity, as measured by the score of MYC(N) target genes signature, must be suppressed in noncycling persister cells. To test our hypothesis, we performed gene set enrichment analysis (GSEA) of the DEGs between noncycling and cycling persister cells and found that indeed MYC(N) target gene signature was the most negatively enriched pathway (Fig. 4E; Supplementary Table S4). The same negative enrichment was observed in separate analyses for MYCN-amplified and MYCN-non-amplified tumors (Supplementary Fig. S4E). Moreover, the scores of the cycling persister subtype signature were strongly and positively correlated with the scores of MYC(N) target genes signature for individual nuclei in MYCN-amplified tumors (Fig. 4F), further suggesting that MYC(N) activity must be suppressed in noncycling persister cells even in the presence of MYCN amplification.
In addition, we used our previously published neuroblastoma chromatin immunoprecipitation sequencing (ChIP-seq) dataset for MYCN and MYC (45) to validate that both MYCN and MYC bind to the promoters of the cycling persister markers MKI67 and TOP2A (Supplementary Fig. S4F). Thus, the low protein expression of MKI67 and TOP2A (Fig. 4B) could be attributed, at least in part, to the low MYC(N) activity level.
Transcriptional and Post-Transcriptional Mechanisms Regulate MYC(N) Activity in Noncycling Persister Cells
To explore the potential mechanisms of MYC(N) activity suppression in noncycling persister cells, we examined the mRNA and protein levels of MYCN and MYC in our patients’ samples. In two out of five tumors with MYCN amplification and low MYC(N) activity at definitive surgery, there was a decrease in MYCN mRNA expression, with a proportionate decrease in protein expression in response to chemotherapy (Fig. 4G and H). In contrast, three MYCN-amplified tumors displayed little to no change in MYCN mRNA expression, but MYCN protein expression was absent in the samples available for IHC staining (Fig. 4I and J; Supplementary Fig. S4G), suggesting that post-transcriptional regulation might mediate MYC(N) activity suppression in noncycling persister cells. Of note, a similar pattern of downregulation was observed in two MYCN-non-amplified tumors with MYC overexpression (Supplementary Fig. S4H and S4I).
The decrease in MYC(N) protein in the presence of high MYCN/MYC mRNA levels was further validated in PDXs and neuroblastoma cell lines treated with multiagent chemotherapy in intervals that mimic the clinical schedule (Supplementary Fig. S4J, S4K, and S4L). Importantly, we did not observe a decrease in MYCN protein levels after treatment in cell lines with MYCN amplification and ALK/TP53 mutation (e.g., KELLY; Supplementary Fig. S4M), consistent with the high MYC(N) activity observed in patients with MYCN amplification and ALK/TP53 mutation.
MYC(N) Activity Is Restored Upon Relapse
Next, we investigated the dynamics of MYC(N) activity from diagnosis through relapse. We obtained serial samples from a tumor of a patient with intermediated-risk neuroblastoma and with high-risk features (including 11q deletion and 17q gain) who suffered from local relapse. Samples from three time points—diagnosis, definitive surgery, and relapse—were subjected to snRNA-seq, and malignant cells were identified and classified into persister subtypes at each time point, as described above (Supplementary Fig. S4N). We then compared the MYC(N) activity scores at diagnosis, definitive surgery, and relapse and observed high MYC(N) activity at diagnosis followed by an expected decrease at definitive surgery and an increase in MYC(N) activity at the time of relapse (Fig. 4K).
To model the restoration of MYC(N) activity upon relapse, we treated MYCN-amplified and -non-amplified neuroblastoma cell lines with chemotherapy as outlined above and allowed the residual cells to regrow for additional 3 weeks after drug wash. Consistently, MYCN/MYC protein levels were high before treatment, low during treatment, and high 3 weeks after treatment (Fig. 4L).
The above findings raise two possible and nonmutually exclusive mechanisms for MYC(N) activity restoration upon relapse. One, by which chemotherapy primarily selects for pre-existing noncycling, low–MYC(N) activity malignant cells, while at the same time, a rare subpopulation of high–MYC(N) activity cells escapes chemotherapy and outgrows after drug wash. The other potential mechanism is one by which individual malignant cells downregulate MYCN mRNA and/or MYCN protein under the pressure of chemotherapy and upregulate its expression during drug holiday and relapse. Although lineage tracing experiments are required to definitively study these two mechanisms, we show that at least in seemingly homogenous cell lines, the latter is more likely. We performed dual IF staining for MYCN and MKI67 in LAN5, a MYCN-amplified cell line, before and after 3 days of etoposide treatment. We did not detect pre-existing noncycling, low–MYC(N) activity cells prior to therapy or cycling, high–MYC(N) activity cells during treatment (Fig. 4M), suggesting that plasticity in MYCN expression might account for the restoration of MYC(N) activity during relapse.
Constitutive NFκB Activation Mediates Cellular Persistence
We next investigated the transcriptional programs that are activated rather than suppressed in noncycling persister cells and focused on the two NFκB subtypes, namely, NFκB/stemness and NFκB/stress. The NFκB/stemness subtype demonstrated a high expression of stem cell markers, such as CD44 and KLF6, suggesting that this subtype may represent constitutive NFκB activation that is present even prior to therapy. Indeed, four patients’ tumors displayed a high percentage of the NFκB/stemness subtype at diagnosis [median 70.4%, range (40.26%, 99.88%)] which remained high during definitive surgery [median 69.1%, range (65.04%, 99.94%)] and persisted through relapse in a patient (CN18) whose relapse sample was available (Fig. 5A). We further validated the persistence of the NFκB/stemness subtype from diagnosis through relapse using NFκB /stemness markers CD44 and ATF3 (Supplementary Fig. S5A). Importantly, none of these patients achieved complete radiographic response and three patients experienced relapse/progression of disease while on therapy, implying that constitutive NFκB activation confers de novo resistance to therapy. Indeed, in vitro measurements of NFκB activity using the GFP reporter showed high correlation between basal NFκB activity and resistance to topotecan in various neuroblastoma cell lines (Fig. 5B). Furthermore, CRISPR−CAS9 knock-out of RelA (p65) in SKNSH and SHEP, two cell lines with high basal NFκB activity, resulted in a statistically significant decrease in cell viability (up to 30%) in response to chemotherapy (Fig. 5C and D), supporting the link between constitutive NFκB activation and therapy resistance.
Figure 5.
Constitutive NFκB activation mediates cellular persistence. A, Bar plots of persister subtypes percent of three tumors obtained from three patients (CN3, CN11, and CN12) at diagnosis and definitive surgery and a tumor obtained from one patient (CN18) at diagnosis, definitive surgery, and relapse. B, Scatter plot showing NFκB basal activity, measured as the log of the median fluorescence intensity (MFI), in five cell lines as a function of their corresponding IC90 dose of topotecan. C, Dose–response curve of cell viability plot as a function of etoposide dose (log scale) for SKNSH (left) with and without RelA CAS9-CRISPR knockout. D, Dose–response curve of cell viability plot as a function of etoposide dose (log scale) for SHEP with and without RelA CAS9-CRISPR knockout. E, Scatter plot showing the NFκB/stemness score as a function of the mesenchymal score in 39 neuroblastoma cell lines (46). F, Violin plots of NFκB/stemness score in persister nuclei with high mesenchymal and reduced adrenergic scores vs. persister nuclei with low mesenchymal and high adrenergic scores. G, Western blot of mesenchymal and adrenergic markers in an adrenergic cell line, BE2C, with doxycycline-inducible PRRX1 expression along the transdifferentiation trajectory. Day 0 is day of doxycycline administration, and day 14 represents the end of assay. H, NFκB luciferase activity, measured as the relative luminescence, following induction of PRRX1 expression in BE2C. I, GSEA of DEGs between PRRX1-transduced and -untransduced BE2 and KPNYN (47) J, Senescence signature (50) scores of the persister nuclei classified according to the four persister subtypes. ADRN, adrenergic; D, diagnosis; DS, definitive surgery; MES, mesenchymal; MFI, median fluorescence intensity; ns, not significant.
The Mesenchymal Cell State Is Associated with High NFκB Activity
We next investigated the association between NFκB activation and the recently described adrenergic and mesenchymal transcriptional states, of which the latter has been shown to be more chemoresistant (5–10). As these two subtypes were primarily described in cell lines, we focused our studies first on in vitro models. We calculated the NFκB/stemness signature scores and the mesenchymal transcriptional state scores of 39 neuroblastoma cell lines from our publicly available bulk RNA-seq dataset (46) and observed a strong and positive correlation between NFκB/stemness scores and the mesenchymal state scores of the cell lines (Fig. 5E). This observation was replicated in our snRNA-seq dataset and the TARGET bulk RNA-seq cohort, although perhaps not surprisingly to a lesser extent (Supplementary Fig. S5B). This is likely explained by the paucity of mesenchymal malignant cells in patients’ neuroblastoma samples in accordance with previous studies that did not identify a “pure” mesenchymal malignant subpopulation, that is, cells characterized by a mesenchymal transcriptional state that also harbored recurrent chromosomal aberrations typical of neuroblastoma (12–14, 21). Nevertheless, similar to these studies, we identified malignant nuclei in our patients’ definitive surgery samples with reduced adrenergic and increased mesenchymal signature scores. These persister nuclei displayed statistically significant higher NFκB/stemness signature scores compared with persister nuclei with low mesenchymal and high adrenergic signature scores (Fig. 5F).
To functionally establish the association between the mesenchymal state and NFκB activation, we conditionally transduced three adrenergic cell lines, BE2C, IMR5, and SY5Y, with PRRX1, a mesenchymal transcription factor, and cotransfected them with an NFκB luciferase reporter. In parallel to the transdifferentiation from the adrenergic to the mesenchymal state upon PRRX1 expression (Fig. 5G), we observed a statistically significant increase in NFκB activity, indicating a close association between the mesenchymal state and NFκB activation (Fig. 5H; Supplementary Fig. S5C and S5D). Furthermore, we used a publicly available bulk RNA-seq dataset from transdifferentiation assays of two additional adrenergic cell lines (47), BE2 and KPNYN, and performed differential gene expression analysis between PRRX1-transduced and -untransduced cells. Consistent with our observations, upregulated genes in the mesenchymal state were enriched in NFκB /stemness signature (Fig. 5I).
In addition, CD44, a marker of NFκB/stemness, has been recently proposed as a marker of the mesenchymal state in cell lines, PDXs, and patient samples (21), further supporting the association between NFκB/stemness and the mesenchymal state. We validated the surface expression of CD44 in neuroblastoma cell lines with high basal NFκB activity and noted that whereas SHEP exhibited a homogeneous CD44 surface expression, CD44 expression in SKNSH was heterogenous (Supplementary Fig. S5E). This likely reflects the presence of two subpopulations with different basal NFκB activities, namely, SY5Y with low CD44 and baseline NFκB activity (Supplementary Fig. S5F) and SHEP with high CD44 and baseline NFκB activity.
Activation of NFκB has been shown to induce senescence (48), which in turn can promote cancer stemness (49). We therefore asked whether the NFκB/Stemness subtype may be associated with senescence. Using a recently published senescence RNA-based signature (50), we compared the senescence scores of the persister cells corresponding to each persister subtype and observed that the NFκB/stemness subtype displayed a statistically significant higher senescence score than the other three persister subtypes (Fig. 5J).
TAMs and CAFs Promote NFκB Activation in Persister Cells
Lastly, we sought to identify potential activators of NFκB signaling in persister cells. The NFκB/stress subtype was predominantly present in surgical specimens (Fig. 6A) and exhibited a high expression of immediate early genes, such as EGR1, FOS, and NR4A1, suggesting that NFκB signaling could be activated in response to chemotherapy. Using clinically relevant doses of chemotherapy (IC70 and above), we treated both MYCN-amplified and MYCN-non-amplified cell lines with topotecan followed by etoposide and confirmed a statistically significant increase of NFκB activity following chemotherapy across MYCN amplification status (Supplementary Fig. S6A). Combination of chemotherapy with the specific IκB kinase β inhibitor ML120B (51) resulted in increased killing of persister cells, supporting a prosurvival role for NFκB activation (Supplementary Fig. S6B). Thus, chemotherapy alone is sufficient to trigger NFκB activation in both MYCN-amplified and -non-amplified neuroblastoma cell lines.
Figure 6.
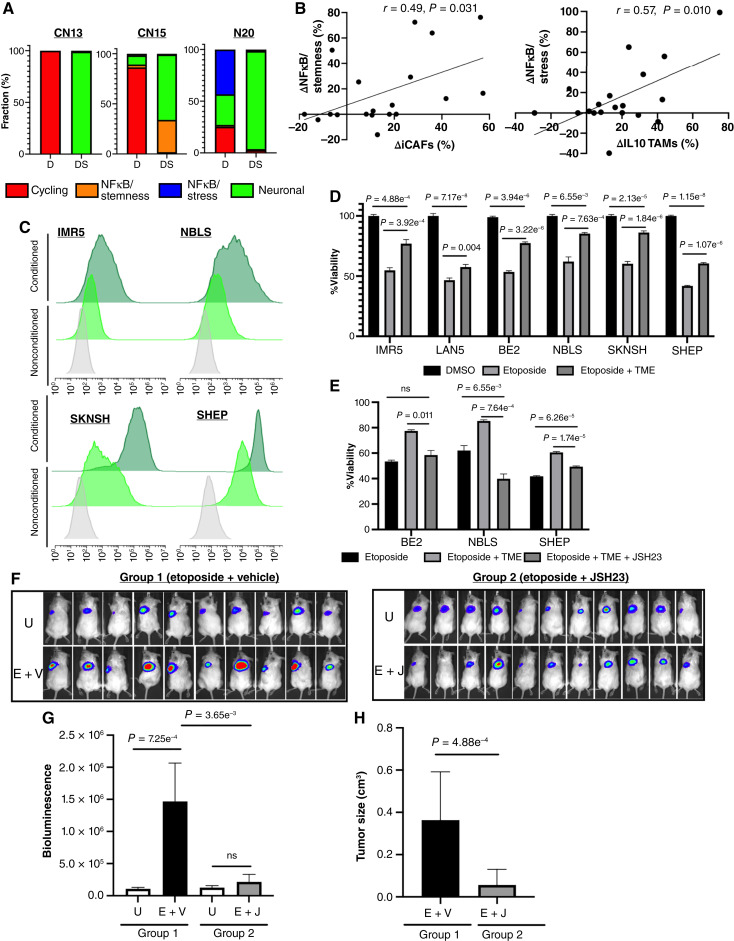
Multiple signals trigger NFκB activation in persister cells. A, Bar plots of persister subtypes percent of three tumors obtained from patients (CN13, CN15, and CN20) at diagnosis and definitive surgery. B, Scatter plots showing the relative changes in percentage of NFκB/stemness from diagnosis to definitive surgery (left) and relative changes in percentage of NFκB/stress (right) from diagnosis to definitive surgery as a function of the relative changes in iCAFs and IL10 TAMs percentages from diagnosis to definitive surgery, respectively. C, Flow cytometry histograms for the GFP (FITC) reporter under NFκB consensus sequence in MYCN-amplified or overexpressing neuroblastoma cell lines (IMR5 and NBLS; top) and MYCN-non-amplified neuroblastoma cell lines (SKNSH and SHEP; bottom) in standard medium and conditioned medium (CM). Grey, untransduced cell line, standard medium; light green, GFP-transduced cell line, standard medium; dark green, GFP-transduced cell line, CM. D, Bar plots of cell viability of six neuroblastoma cell lines, three MYCN-amplified (IMR5, LAN5, and BE2) and three MYCN-non-amplified (NBLS, SKNSH, and SHEP,) treated with etoposide (100 nmol/L for IMR5, LAN5, and BE2 and 1 µmol/L for NBLS, 5 µmol/L for SKNSH, and 10 µmol/L for SHEP) for 3 days in standard medium and in the presence of CM from neuroblastoma, TAMs, and CAFs triple cocultures. E, Bar plots of cell viability of three neuroblastoma cell lines, BE2, NBLS, and SHEP, treated with etoposide for 3 days as above in standard medium and in the presence of CM from triple cocultures with and without the NFκB nuclear translocation inhibitor JSH-23 (1 µmol/L for BE2, 35 µmol/L for NBLS, and 100 µmol/L for SHEP). F, Bioluminescence images of NOD/SCID gamma mice arms which were subcutaneously implanted with BE2-LUC neuroblastoma cells in the presence of human TAMs and CAFs (TME) treated with etoposide + vehicle (E + V; group 1) or etoposide + JSH-23 (E + J; group 2). Top, untreated in each group. G, Bar plots of bioluminescence levels of untreated mice in each group and treated mice (E + V; group 1 and E + J; group 2). H, Bar plots of tumor size in of treated mice (E + V; group 1 and E + J; group 2). D, diagnosis; DS, definitive surgery; U, untreated.
We next asked whether certain cellular components of the TME may contribute to NFκB activation in persister cells. We focused on CAFs and TAMs, which constituted the majority of TME cells during induction chemotherapy and noted that the observed increase in iCAFs frequency correlated positively with the increase in frequency of NFκB/stemness persister nuclei at definitive surgery, whereas the frequency increases of IL10 TAMs and NFκB/stress demonstrated a positive correlation with the increased frequency of NFκB/stress persister nuclei at definitive surgery Fig. 6B). We did not observe statistically significant correlations between other types of TAMs or CAFs with NFκB persister subtypes (Supplementary Fig. S6C). In light of their known protumorigenic role in neuroblastoma (52, 53), we hypothesized that CAFs and TAMs could promote resistance to chemotherapy via activation of NFκB signaling in persister cells. To test our hypothesis, we cultured neuroblastoma cell lines in the presence of conditioned medium (CM) obtained from triple cocultures of neuroblastoma cell lines, TAMs, and CAFs, as we previously described (54). We observed a robust increase in NFκB activity in cell lines cultured in CM compared with those cultured in standard medium (Fig. 6C). In addition, there was a statistically significant increase in the viability of neuroblastoma cells that were treated with etoposide in the presence of CM compared with cells that were cultured in standard medium (Fig. 6D), supporting the hypothesis that TAMs and CAFs may trigger NFκB activation in persister cells that lead to therapy resistance.
To validate that NFκB mediates therapy resistance in the presence of TAMs and CAFs, we treated MYCN-amplified and -non-amplified cell lines with a combination of etoposide and JSH-23, a specific NFκB inhibitor, which inhibits NFκB nuclear translocation without affecting IκBα degradation (55), in the presence and absence of CM for 3 days. Inhibition of NFκB nuclear translocation resulted in a statistically significant decrease of cell viability in response to etoposide in the presence of CM and was comparable to the cellular viability in response to etoposide in standard medium (Fig. 6E). To establish the role of NFκB signaling inhibition in eradicating the persister cells in vivo, we used our recently developed murine model (Fig. 6F). Briefly, NOD/SCID gamma (NSG) mice were subcutaneously injected with the luciferase transduced human neuroblastoma cell line BE2-LUC together with human TAMs and CAFs obtained as previously described (54). The mice were then treated with etoposide and vehicle (normal saline; group 1) or with a combination of etoposide and JSH-23 (group 2), and tumor size was quantified by bioluminescence as well as manually measured. Following 7 days of treatment, tumors from mice treated with etoposide alone progressed on therapy, whereas tumors from mice treated with etoposide and JSH-23 did not grow (Fig. 6G and H).
Altogether, our human snRNA-seq data coupled with in vitro and in vivo functional validations nominate NFκB signaling as a potential therapeutic target to eradicate persister cells.
Discussion
In this study, we dissected the cellular composition of high-risk neuroblastoma tumors at diagnosis and after several cycles of induction chemotherapy using snRNA-seq of approximately a quarter of a million malignant and nonmalignant cells from a cohort of 20 paired tumors from untreated and treated specimens. We identified the malignant persister cells and defined four transcriptional subtypes that are associated with cell-cycle state, genetic alterations, certain TAM and CAF subpopulations, and clinical outcome.
Leveraging these subtypes, we show that chemotherapy remodels the malignant and TME compartments of neuroblastomas, resulting in both intertumoral and intratumoral heterogeneity. Our data suggest that the malignant compartment remodeling is driven by nongenetic mechanisms, at least initially, as we did not identify acquisition of new CNVs and SNVs during therapy in the majority of our patients. This stands in contrast to the previously reported increase in resistance-conferring mutations at the time of relapse (3, 4). The role of nongenetic mechanisms in mediating cellular persistence is becoming increasingly recognized, and studies supporting such mechanisms have recently been proposed in various types of cancers (56, 57). Therefore, a model whereby malignant cells initially escape chemotherapy via nongenetic mechanisms and subsequently acquire resistance-conferring mutations upon additional genotoxic therapy is likely in high-risk neuroblastoma and warrants further investigation.
Our findings demonstrate that a large subset of persister cells enter a noncycling cell state that is characterized by suppressed MYC(N) activity and high NFκB signaling activity. Suppression of MYC(N) activity has been described in the context of development and cancer as a means of cellular adaptation in response to stress (58); however, the exact mechanism remains elusive. Our in vitro studies demonstrate that MYCN and MYC could be transcriptionally and/or post-transcriptionally repressed even in tumors with MYCN amplification or MYC overexpression, and it is, therefore, possible that epitranscriptomic modifications govern this suppression (59). Indeed, recent studies demonstrated the role of N-6 methyl-adenosine (m6A) in mediating MYCN suppression during developmental pausing (60) and in the stabilization of the MYCN transcript in neuroblastoma (61). Future studies are planned to decipher the exact mechanisms of MYCN/MYC post-transcriptional modulation in response to chemotherapy and other cellular stresses and track MYC(N) activity at the single-cell level. In addition, using tumor samples obtained at three different time points from one patient and several cell lines, we have shown that although MYC(N) activity is suppressed during therapy, it is restored upon relapse, suggesting that chemotherapy may select for low–MYC(N) activity malignant cells that pre-exist at diagnosis and/or that MYC(N) activity may be regulated in response to chemotherapy. Lineage tracing experiments have the potential to address these important questions, and additional relapse samples, acquired via national and international collaborations, are required to study the dynamics of MYC(N) activity from diagnosis to relapse at the single-cell level, although such samples are rare.
The role of NFκB signaling in tumorigenesis and therapy resistance has been well established in multiple types of cancer (62), including several pediatric tumors, such as ependymoma (63) and lymphoma (64). In neuroblastoma, activation of NFκB has been studied primarily in in vitro models in which it has been implicated in differentiation (65), migration (66), MHC I expression (67), and response to genotoxic agents, including chemotherapy and radiation (68, 69). In this study, using patients’ samples, we identified two NFκB transcriptional programs, NFκB/stemness and NFκB/stress. The former represents a constitutive mode of NFκB activation, whereas the latter represents an acquired mode of NFκB activation.
The NFκB/stemness subtype is characterized by the stem cell marker, CD44, and is also associated with the mesenchymal transcriptional cell state. These findings are consistent with a recent work of ours that links the mesenchymal state to an inflammatory state in neuroblastoma cells (9) as well as previous studies reporting NFκB activation in S-type cell lines (70) and during an adrenergic to mesenchymal transition induced by TNFα (71). Our findings also highlight the importance of CD44 surface expression as a marker of NFκB activating cells with mesenchymal features in patient samples, in agreement with a recent study that reported its role in identifying in vitro and in vivo malignant cells with mesenchymal features (21). In addition, our WGS data revealed that all tumors with a high frequency of the NFκB/stemness subtype harbored distal 11q deletion and gain of the 11p distal–11q proximal region. The genes encoding CD44 and RelA (p65) are located on 11p13 and 11q13.1, respectively, and it is therefore possible that these recurrent segmental chromosomal aberrations drive constitutive NFκB activation.
The NFκB/stress subtype represents an acquired mode of NFκB activation which can be triggered by both chemotherapy and TAMs and CAFs, of which the latter two play a protumorigenic role in neuroblastoma. Recently, our group showed that TAMs and CAFs cooperate via TGFβ signaling to promote the survival of neuroblastoma cells (54). Here, we show that the frequency of both TAMs and CAFs with immunosuppressive phenotypes are increased following induction chemotherapy in both MYCN-amplified and -non-amplified tumors and validate both in vitro and in vivo that TAMs and CAFs can stimulate NFκB activation in persister cells and increase their resistance to therapy.
Targeting NFκB pathways has been challenging due to their critical role in normal cells, primarily immune cells, lack of specific inhibitors, and toxicity. Nevertheless, in a complementary study conducted by our group (72), we have identified TGFβ as a trigger for NFκB signaling in neuroblastoma cells by TAMs and CAFs and demonstrated that inhibition of TGFβ leads to partial inhibition of NFκB signal transduction and restoration of neuroblastoma chemosensitivity.
In addition, targeting downstream effectors of NFκB, which are overexpressed in persister cells and are required for their sustenance, could potentially eradicate the persister cells and prevent relapse, a strategy that is currently under investigation by our team. Finally, we have recently showed the feasibility of targeting intracellular oncoproteins with peptide-centric chimeric antigen receptor–engineered T cells (73) and plan to define the immunopeptidome of persister cells as an orthogonal therapeutic strategy.
Methods
Patient Samples and Ethics
The de-identified tissues of 20 patients with high-risk neuroblastoma treated at CHOP between 2009 and 2022 were obtained from the Pathology Department of CHOP. Written informed consents were obtained from all patients according to the CHOP biobank protocol approved by the CHOP Institutional Review Board and was conducted in accordance with recognized ethical guidelines. The clinical data were collated from CHOP clinical databases by an honest broker. Whenever available, two regions from each tumor sample were obtained, and samples were embedded in optimal cutting temperature compounds. The samples were then reviewed by a pediatric pathologist (LFS) to assess for tumor viability at diagnosis and definitive surgery. All samples contained microscopically viable malignant cells at the time of definitive surgery expect the tumor from CN19 in which no malignant cells were visualized.
Sample Dissociation and Nuclei Isolation
Samples were received from pathology either cryosectioned from frozen tissues or embedded in optimal cutting temperature blocks. A measure of 500 µL of Nuclear Extraction Buffer (20 mmol/L Tris at pH 8, 320 mmol/L sucrose, 5 mmol/L CaCl2, 3 mmol/L MgAc2, 0.1 mmol/L Ethylenediaminetetraacetic acid (EDTA), 0.01% digitonin, 0.1% Triton X-100, and ddH2O) was added to dissociate the tissues. The samples were then transferred to a clean, prechilled 2-mL Dounce homogenizer (Millipore Sigma, D8938), and additional 500 µL of Nuclear Extraction Buffer was added to a total volume of 1 mL. Five to -ten slow strokes with pestle A were applied on ice until the pestle could easily go through the grinder. Additional 5 to 10 slow strokes with pestle B were applied on ice until the pestle could easily go through the grinder. Nuclei were examined by light microscopy to determine whether additional stroking was required. The samples were transferred from the homogenizer to a 1.7-mL low-binding tube with filtration using a 40-µm Falcon cell strainer (Falcon, 352340). Nuclei suspensions were then centrifuged at 500 × g for 5 minutes at 4°C, and the supernatant was removed without disturbing the pellet. A measure of 1.5 mL of Nuclei Wash and Resuspension Buffer (0.2 U/µL RNase inhibitor and 1% BSA) was added to each sample, and the samples were allowed to incubate for 2 minutes, after which we gently and slowly resuspended the nuclei. The samples were then centrifuged at 500 × g for 5 minutes at 4°C, and the supernatant was removed without disturbing the pellet. The last two steps were repeated three times. A measure of 150 µL of the final resuspension buffer was then added, and samples were incubated on ice for at least 2 minutes, after which we gently and slowly resuspended the nuclei. Nuclei were filtered using a 40-µm Flowmi cell strainer with a 1-mL tip to a new 1.7-mL low-binding tube. Nuclei concentration was determined using a Countess machine.
snRNA-seq Library Preparation and Sequencing
snRNA-seq libraries were prepared using 10× Genomics Chromium Single Cell 3′ Kit (version 3 and 3.1) according to the 10x Genomics protocol. The libraries were sequenced using the IIlumina Novaseq 6000 sequencing platform. Sequencing reads were aligned to the human genome reference hg38 using CellRanger v.6.0 (10x Genomics).
Quality Control and Preprocessing
The count matrix of each sample was processed using the following pipeline. First, nuclei with high ambient RNA as detected by DeContX [celda, R pacakge (v1.14.2; ref. 74)] were removed. Doublets were detected and filtered with DoubletFinder R package [v2.0.3 (75)] with default settings. We then used Seurat R package (v4.3.0) to remove low-quality cells and perform normalization, feature selection, dimensionality reduction, and clustering. Specifically, we removed nuclei with less than 1,000 counts, 200 genes, or greater than 3% mitochondrial genes, which likely represented apoptotic low-quality cells (76). The resulting count matrix was scaled and normalized using the ScaleData and NormalizedData functions. The most highly variable genes were selected using the FindVariableFeatures function with methods = “vst,” and principal component analysis was performed using these genes and the RunPCA function. The top 30 principal components were used as input to RunUMAP and FindNeighbors functions. Clusters were identified with the FindClusters function.
Integration and Cell-Type Annotation
We used the Harmony R package (v1.0) with the RunHarmony function to integrate the diagnostic and definitive surgery samples from 20 patients. Following integration, cell types were annotated using two complementary approaches to assign cell types to individual nuclei. First, we used the FindAllMarker function to identify gene markers for each cluster obtained by Harmony. The following gene markers were identified: PHOX2B, PHOX2A, DBH, ALK, TH, and ISL1 for neuroendocrine cells, MPZ, PLP1, and CDH19 for Schwann cells, CD2, CD3D, and CD247 for T cells, CD22, CD79A, PAX5, and BANK1 for B cells, GZMB for NK cells, CD14 and CD68 for M1 macrophages, MRC1 and CD163 for M2 macrophages, C7, COL1A1, LAMA2, and COL3A1 for fibroblasts, VWF, CALCRL, and FLT1 for endothelial cells, ALB for hepatocytes, and CYP11A1 for the adrenal cortex.
We then used SingleR R package (v2.0.0) to annotate individual nuclei. Nuclei with a cell type assignment by SingleR that was disconcordant from their cluster assignment were excluded from downstream analysis.
Identification of Malignant Cells
We used InferCNV (v 1.14.0) to infer the CNVs from single nuclei RNA with a cutoff value of 0.1, denoise = TRUE, HMM = FALSE, and the GRCH38 reference gene positions. To avoid potential batch effects, InferCNV was run for individual samples with immune cells and endothelial cells as reference cells.
To increase our confidence in the classification of malignant cells using RNA inference only, we applied a stringent approach and excluded nuclei with inferred copy numbers that belonged to nonmalignant clusters. Specifically, nuclei were identified as malignant if they (i) belonged to a cluster with inferred recurrent segmental chromosomal aberrations, such as 1p, 3p, 4p, or 11q deletions and as 2p or 17q gain by InferCNV or (ii) belonged to a cluster expressing marker genes of neuroblastoma, such as PHOX2B, PHOX2A, ALK, DBH, and ISL1.
Pathway-Based Clustering
To identify shared mechanisms among persister nuclei across different patients, we took a pathway-based clustering approach. The pathway-based clustering algorithm consists of three sequential steps: (i) calculation of 52 signature scores for each nucleus, (ii) selection of the topmost informative signatures among all nuclei, and (iii) clustering of nuclei in the latent space represented by the selected signatures.
For the first step, we calculated the scores of 52 Hallmark pathways signatures (34) for each nucleus using VISION (33). The output of the first step is a matrix of 86,901 × 52 elements, in which the element i,j corresponds to the score of the jth Hallmark pathway in the ith nucleus. To select the most informative pathways, we used the notion of autocorrelation, denoted by C, to measure how randomly a pathway score is distributed across nuclei within the manifold. We calculated the autocorrelation scores of the 52 Hallmark pathways and selected the ones with C > 0.5 and FDR < 0.01, resulting in N = 9 pathways: MYC_TARGETS_V1 (C = 0.788), OXIDATIVE_PHOPHORYLATION (C = 0.69), G2M_CHECKPOINT (C = 0.66), E2F_TARGETS (C = 0.63), UV_RESPONSE (C = 0.58), MITOTIC_SPINDLE (C = 0.57), UV_REPSONSE (C = 0.56), MTORC1_SIGNALING (C = 0.55), and TNFA_SIGNALING_VIA_NFKB (C = 0.54). The output of this step is a matrix of 86,901 × 9 elements, in which the element i,j corresponds to the score of the jth Hallmark pathway in the ith nucleus. We normalized this matrix using z-scores of the columns to account for different scales of the signatures scores. In the step 3, we used Louvain clustering of the persister nuclei with the normalized matrix as the latent space.
Identifications of Gene Module and Persister Subtype Assignment
To identify genes that act in concert (modules) in persister nuclei that activate similar pathways, we used Hotspot with the cell–cell similarity map that is defined by the latent space of the pathway-based clustering algorithm, which in our case are derived from the topmost informative pathways. Thus, persister cells that activate similar pathways are considered similar in the cell–cell similarity map. Hotspot was run with the above latent space matrix and following parameters: model = “normal,” min_gene_thershold = 100, and n_neighbors = 295 (square root of the number of persister nuclei). To ensure that modules were robust with respect to the cell–cell similarity map, we first ran Hotspot using increasing numbers of the topmost informative pathways, and hence different cell–cell similarity maps derived from different Geary cutoffs (we used Geary scores of 0.3, 0.4, and 0.5 as there was no difference in the number of informative signatures below a Geary score of 0.3) and a FDR of less than 0.01. The Jaccard similarity between the modules was used to cluster modules from different Hotspot runs, resulting in five meta-modules. Only genes recurring in all modules with in a meta-module were retained. We calculated the module scores of persister nuclei using VISION and obtained the Spearman correlation coefficients between module scores, revealing that meta-modules #2 and #3 are highly correlated (r = 0.6; FDR < 0.0001). As meta-module #2 genes were enriched with housekeeping genes, we omitted meta-module #2 from further analyses and retained only modules #1, 3, 4, and 5.
We then calculated the scores of the four meta-modules for each nucleus using VISION and assigned the module with the highest score to each persister nucleus.
Cell-Cycle and Pathway Enrichment Analysis
The signatures of G2M and S from were used to determine the cell-cycle scores of individual persister cells using VISION. Pathway enrichment analysis was performed using the enrichPathway function in clusterProfiler (v4.6.0) R package with P value cutoff = 0.1 and Hallmark and Reactome signatures obtained from msigdbr (v7.5.1) R package. Gene set enrichment analysis was performed using the GSEA function in clusterProfiler with P value cutoff = 0.1 and Hallmark and Reactome signatures obtained from msigdbr.
Analysis of the Bulk RNA-seq Dataset
We used VISION to calculate the persister subtype scores of tumors in the bulk RNA-seq TARGET dataset. We excluded patients with low and intermediate risks, resulting in 151 patients. Tumor phenotype was determined by the maximum persister subtype score. Raw counts of the transdifferentiation experiments were obtained (GSE180516; ref. 47) and normalized and analyzed using the DESq2 pipeline (77).
Analysis ChIP-Sequencing Dataset
ChIP-seq data were obtained from three datasets (45): MYCN-amplified cell lines (GSE94782), MYCN-non-amplified cell lines (GSE138295), and histones (GSE138314). We used MACS3 to perform peak calling for two datasets, GSE138295 and GSE138314. Specifically, we used the bigwigCompare function on the IP and corresponding input (control) BigWig files to generate Bedgraph files containing the log2 ratio between the IP and input. We then used the bdgpeakcall function with cutoff of 0.5 for GSE138295 and 2 for GSE138314, corresponding to the enrichment level. For dataset GSE94782, peaks were already called by ref. 45. Integrative Genomics Viewer was used to visualize the peaks for the selected genes.
WGS
Genomic DNA was extracted from nuclei isolated from human pediatric neuroblastoma samples using Quick-DNA Microprep Plus Kit (Zymo Research, D4074). Illumina DNA Prep, (M) Tagmentation Kit (Illumina, 20018705) was used to prepare WGS libraries according to the manufacturer’s instructions. Tagmentation of gDNA and addition of adapter sequences was carried out at 55°C for 15 minutes using bead-linked transposomes. The reaction was terminated by the addition of tagmentation stop buffer, which was followed by post-tagmentation cleanup to separate adapter tagged, tagmented DNA from beads. Index adapters, i7 and i5 from Nextera DNA CD Indexes Kit (Illumina, 20018707) were used to amplify tagmented DNA, followed by double-sided size selection to obtain the fragments of interest. WGS libraries were paired-end sequenced on Novaseq 6000 as per the user guide. WGS reads were aligned to hg38 with BWA, and base recalibration was performed with GATK (v4.1.9).
Copy Number Detection in WGS Data
CNVkit (v0.9.9) software was used to compare CNA profiles using the aligned reads from WGS of pre- and post-treatment pairs of tumor samples. Sequencing coverage of genome’s sequencing-accessible regions of matched normals was used as reference for pre- and post-treatment tumor samples. For tumors without matched normals, a flat reference with a neutral copy-number profile was used. The circular binary segmentation algorithm was used to infer copy-number segments. Copy number segments were annotated to genes, and regions bearing a log2 ratios were rescaled to account for the difference in tumor purities observed across tumor samples. Log2 ratios of at least ±0.2 were indicative of shallow amplifications or deletions. Segments with log2 less than −1.1 were indicative of deep deletions, and those with log2 higher than 0.7 were identified as amplifications. Germline variants were used to calculate average B-allele frequencies for segments containing the variants. Log2 copy ratios for every pair were visualized using the cnSpec() function from GeneVisR (v1.29.3) R package, and karyoploteR (v1.20.3) R package was used to visualize B-allele frequencies across pre- and post-treated pairs of tumors.
SNV Calling in WGS Data
Somatic variant calling was run on the tumor normal pairs using Mutect (v2.2) and Strelka2 (v2.9.9). The calls were annotated using SnpEff v5.1 and Annovar (v20200608). High/moderate effect variants with Ref and Alt allele depth greater or equal to 10 were kept. Variants with gnomAD (v3.1) allele frequency greater than 0.01 were removed.
Clonal Evolution Inference
We used PyClone-Vi (Python v3.8.5) to infer the clonal population structure at diagnosis and definitive surgery in patients with either ALK or TP53 mutation. For each pair, the union set of all of SNVs called at both time points was taken. The allele counts for ALK and TP53 were extracted using Integrative Genomics Viewer (v2.14.1) to include mutations with VAFs below the level of detection of the variant caller. Tumor purity was estimated from the snRNA-seq data of the corresponding samples. Fishplot package was drawn using the fishplot (v0.5.1) R package.
Cell Lines
Human-derived neuroblastoma cell lines were obtained from the CHOP cell line bank and cultured in RPMI containing 10% FBS, 2 mmol/L L-glutamine, and 1% streptomycin/penicillin (RPMI complete) at 37°C under 5% CO2. The cell lines were routinely genotyped (GenePrint 24 System; Promega) and Mycoplasma-tested, both routinely before cells were placed into the cell bank. The cells were typically utilized for the described experiments <5 passages from thawing, genotyping, and Mycoplasma testing.
Drug Studies
We performed serial dilutions of the stock concentration and determined the cell viability after 5 days for topotecan (Teva, stock 1 mg/mL) and 3 days for etoposide (Teva, stock 20 mg/mL). Specifically, cells were plated in 96-well plates at a density of 7,000 and 10,000 cells per well for topotecan and etoposide, respectively. We examined 10 different drug concentrations corresponding to 10 columns, and four technical replicates were used for each concentration. Each well contained 90 µL of RPMI medium, and wells at the edge contained 200 µL of PBS. The day after plating, 10 µL of drug from each concentration was added to the corresponding well. Cell viability was determined using CellTiter-Glo Luminescent Cell Viability Assay (G7570, Promega). The IC50 doses for each drug and cell lines were obtained using nonlinear regression analysis in Prism (v9.1.1). IC70 and IC90 doses were calculated using the Hill equation. For MYCN/MYC RNA and protein studies, we treated cell lines with 5 days of topotecan at inhibitory concentrations of 90% (IC90), and the residual cells were allowed to regrow for 2 to 3 weeks. We then retreated the growing cells with etoposide for 3 days at the IC90 concentration, thus mimicking the clinical schedule in which multiagent chemotherapy is delivered in 3-week intervals. Live residual cells were then selected by flow sorting, and RNA and protein levels of MYCN and MYC were subsequently measured.
Similar experiments were performed to determine the viability of neuroblastoma cells in the presence of NFκB inhibition with either ML120B or JSH-23. Specifically, ML120B (Tocris, #4899) was added to reach a final concentration of 40 µmol/L in 100 µL of medium per well, and JSH-23 (MedChemExpress, 10 mmol/L, #13982) was added to reach a final concentration of 1 µmol/L (BE2), 35 µmol/L (NBLS), and 100 µmol/L (SHEP) in 100 µL of medium.
PDX
Mice, ages 6 to 12 weeks old, were subcutaneously implanted with tumor material for tumor growth and randomized into control and treatment groups (six mice per arm). Enrollment size was reached when the xenografted tumor reached approximately 0.2 cm3. Following enrollment, control mice were administered normal saline and treated mice were given topotecan at 0.5 mg/kg and cyclophosphamide at 20 mg/kg via i.p. route for five consecutive days, and tumors were allowed to regrow. Body weights and implanted tumor volumes were measured bi-weekly. A 2 cm3 tumor volume was considered the study endpoint for a mouse after it was euthanized. Tumors were collected at enrollment and maximal shrinkage following the second administration of chemotherapy.
Western Blotting
Whole-cell protein lysates were prepared with cell lysis buffer [Cell Signaling Technology (CST), #9803], PMSF (Sigma-Aldrich), and phosphatase inhibitor cocktails 2 (Sigma-Aldrich, #P5726) and 3 (Sigma-Aldrich, P0044), briefly sonicated, rotated for 15 minutes at 4°C, and centrifuged for 10 minutes. The supernatant was removed, and protein concentration was quantified by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, #23225). Samples were prepared from lysates with NuPage LDS sample buffer (#NP0007, Thermo Fisher Scientific) and β-mercaptoethanol. Then the samples were separated on a 4% to 12% Bis-Tris gels (Life Technologies), transferred to a PVDF membrane, blocked in 5% non-fat milk in Tris-buffered saline and Tween-20 (TBS-T), and blotted using standard protocols. Membranes were typically incubated at 4°C overnight in primary antibodies, washed x three with TBS-T, then incubated in 1:2,000 diluted a horseradish peroxidase–labeled secondary antibody at room temperature for 1 hour, washed additional × 3 with TBS-T, and then developed with a chemiluminescent reagent (SuperSignal West Femto, Thermo Fisher Scientific). The following primary antibodies were used: β-actin (1:5,000; CST, #4967), GAPDH (1:5,000; CST, #2118), c-MYC (1:1,000; CST, #18583), N-MYC (1:5,000; CST, #9405), p65 (1:5,000; CST, #4767), PRRX1 (1:2,000; OriGene TA803116), YAP (1:1,000; CST, #14074), PHOX2B (1:200, sc-376997), DLK1 (1:1,000; CST, #2069), and TUB (1:10,000; Calbiochem CP06).
RT-PCR Analysis
Total RNA was isolated utilizing Direct-zol RNA MicroPrep Kit (R2062, Zymo Research), and mRNAs were converted to cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, 4368814). Taqman gene expression assays were used to quantitate MYCN (#Hs00232074), MYC (#HS00153408), and GAPDH (#HS999905). RT-PCR analysis was performed on Applied Biosystems 7900HT Sequence Detection System using standard cycling conditions, and relative expression values were calculate using the ∆∆ Ct method.
NFκB GFP Reporter Assay
NFκB GFP lentivirus was purchased form BPS Bioscience (#79926), with the response element sequence that drives eGFP reporter expression being GGGAATTTCC. Virus was added to the cells in the presence of 8 µg/mL polybrene (Sigma-Aldrich). Media was changed the next day, and puromycin (Sigma-Aldrich) was added for selection for 48 hours prior to further experiments. Cell lines with and without the NFκB GFP reporter were plated in 35-mm plates (400,000 per plate) and treated with IC90 topotecan for 5 days, allowed to regrow, and, upon reaching 80% confluency, were treated with IC90 etoposide for 3 days.
CRISPR–CAS9 Knockout
CAS9 plasmids were obtained from Ophir Shalem Laboratory and used to generate CAS9 viruses with Lipofectamine 3000 and p3000, PMD2.G (Addgene, #12259), and psPAX2 (Addgene, #12260) plasmids in 293T cells incubated in optiMem medium. Similarly, RelA plasmids (Addgene, #83944) were used to generate RelA viruses. Then SKNSH and SHEP were stably transduced with CAS9 virus and selected with 1 µg/µL of puromycin. RelA virus was then used to stably transduce SKNSH and SHEP. Knockout cells were validated by Western blotting for RelA.
Transdifferentiation Assays
BE2C, IMR5, and SY5Y cells were infected with lentivirus prepared from PRRX1 in the pLVX-TetOne vector (Takara, #631847) and selected with puromycin, as previously described (9). The cells were then transfected with the NFκB pNifty2-Luc reporter plasmid (Invivogen) using Lipofectamine 3000 transfection reagent (L3000008) followed by Zeocin selection. PRRX1 expression was induced with doxycline (1 μg/mL, Sigma-Aldrich, #D9891) and luminescence-detected using Bright-Glo Luciferase Assay System (Promega, #E2610).
IF Confocal Microscopy
Cell lines were plated in 3-mm plates in a density of 300,000 cells per plate and incubated in 2 mL of RPMI medium. The medium was removed, and cells were fixed with Fixation (4× formaldehyde buffer) at room temperature for 10 minutes. The plates were washed with PBS 3 times for 5 minutes. The cells were then permeabilized in 0.1% Triton (100×) for 10 minutes at room temperature, and the plates were washed in PBS three times. The plates were then incubated in detection buffer (1% BSA and 0.1% Tween-20) for 15 minutes at room temperature. A measure of 70 to 130 µL of primary antibody mix (CD44, CST# 3579, 1:400) was added to the plate and allowed to incubate for 2 hours at room temperature. Following incubating, the plates were washed with TBS-T (91414-10TAB, Sigma-Aldrich) for 15 minutes 3 times. A secondary anti-mouse antibody (Alexa Fluor, Invitrogen, #A32744) was then added to detection buffer at 1:500 ratio, and plates were incubated for 1 hour, after which they were washed with TBST for 15 minutes 3 times. 4’,6-diamidino-2-phenylindole (DAPI) stain was added to the detection buffer at 1:10,000 for 5 minutes of incubation. The plates were imaged by confocal microscopy (TCS SP8, Leica).
Coculture Experiments
Coculture experiments were performed in transwell (0.4 μm pore membranes, Corning). Triple coculture was used a 4:4:1 ratio for NB:MN: MSC, as previously described (54). NB cells [SK-N-BE(2)] and mesenchymal stem cells (MSC) were seeded separately the first day, and fresh monocyte (MN) was added after 24 hours in the presence of Iscove’s Modified Dulbecco’s Medium and 2% FBS. Cultures were maintained for 72 hours in the presence or absence of JSH-23 (NFκB nuclear translocation inhibitor, 1–100 µmol/L, Selleckchem). The CM used is the combination of technical triplicate, and biological duplicate was performed.
Drug Response of the Xenotransplanted Model
Animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care Utilization Committee at The Saban Research Institute of Children’s Hospital Los Angeles (Protocol #41-23 approved on October 25, 2023). A drug–response protocol was developed (54). NOD/SCID gamma mice, ages 8 to 12 weeks old, both males and females, were divided into two groups of 10 to 11 mice each and subcutaneously implanted with SK-N-BE(2)-LUC cells (1.2 × 106 cells) alone or mixed with MN and MSC (TME; ratio 4:4:1 for NB:MN:MSC) in a total volume of 100 µL. The mice treated with etoposide (5 mg/kg) received i.p. injections every 2 days on days 6, 8, and 10. The mice treated with JSH-23 received i.p. injections with 5 mg/kg of JSH-23 suspended in DMSO (Sigma-Aldrich) during 7 days (days 5–11). Following tumor implantation, the mice were monitored before and after treatment by bioluminescence imaging (Xenogen IVIS 100, Caliper Life Science) to measure luciferase intensity and tumor growth, and tumor volume was measured after treatment using the following formula: Tv = width × 2 × length × .
Statistical Analysis
For comparisons between medians, precents, and proportions, we used the Wilcoxon rank test (paired or unpaired, as appropriate). Correlations between scores of individual nuclei or cells were calculated using the Pearson method in which the test statistic is based on Pearson’s product moment correlation coefficient and follows a t-distribution. For cell viability assays, we used two-tailed, unpaired, Student t test to compare between control and treatment groups or CAS9 empty vector versus CAS9 knockout cells.
Data Availability
The snRNA-seq and WGS data are available at https://humantumoratlas.org/explore under HTAN-CHOP for the discovery dataset and HTAN-HTTAP for the validation cohort.
Supplementary Material
Supplementary Table S1 shows the patient clinical characteristics and genetic alterations at diagnosis
Supplementary Table S2 links the CN number to the HTAN biospecimen numbers
Supplementary Table S3 provides the genes of each persister subtype signature
Supplementary Table S4 shows the most negatively enriched pathways in non-cycling persister cells by GSEA
Supplementary Figure S1 shows cell type markers and inferred copy number of tumors as well as characterization of malignant, Schwann cells, tumor associated macrophages and cancer associated fibroblasts at time of definitive surgery
Supplementary Figure S2 shows the details of the pathway-based clustering algorithm including the autocorrelation pathway scores, cell cycle annotation, pathway enrichment analysis and region analysis.
Supplementary Figure S3 shows the WGS analysis of the diagnostic and definitive surgery including clonal evolution of ALK mutation from diagnosis to definitive surgery in one tumor.
Supplementary Figure S4 differences in persister cell composition between diagnosis and definitive surgery as well as decrease in MYC/N protein level in human samples, PDX and cell lines following chemotherapy
Supplementary Figure S5 shows the connection between NF-kB/stemness subtype and mesenchymal transcriptional state.
Supplementary Figure S6 shows the increase in NF-kB activity following chemotherapy and its role in survival which is enhanced by specific TAMs and CAFs
Acknowledgments
This work was supported by the Human Tumor Atlas Network U2C CA233285 (K. Tan) and NCI grants R35 CA220500 (J.M. Maris), P01 CA217959 (J.M. Maris and Y.P. Mosse), and K08 CA266914 (A.J. Wolpaw). This work was also supported by the Sheba Medical Center Physician-Scientist Program (L.D. Grossmann), the Giulio D’Angio Endowed Chair (J.M. Maris), the Richard and Sheila Sanford Endowed Chair (K. Tan), the Patricia Brophy Endowed Chair (Y.P. Mosse), and Saban Research Institute (Children’s Hospital Los Angeles) under the Research Career Development Fellowship Grant (K. Louault). The authors would like to thank the study participants and the Children’s Hospital of Philadelphia Pathology Core for their technical expertise and assistance in handling human tissue sections, the Research Information Services for providing computing support, and the Children’s Hospital Los Angeles Animal Imaging Core. The authors would also like to thank Anand Patel and Michael Dyer for sharing the St. Jude Human Tumor Atlas Pilot Project data and Nir Yosef and Matthew Jones for their help with VISION and Hotspot.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
No disclosures were reported by the authors.
References
- 1. Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers 2016;2:16078. [DOI] [PubMed] [Google Scholar]
- 2. Norris MD, Bordow SB, Marshall GM, Haber PS, Cohn SL, Haber M. Expression of the gene for multidrug-resistance-associated protein and outcome in patients with neuroblastoma. N Engl J Med 1996;334:231–8. [DOI] [PubMed] [Google Scholar]
- 3. Eleveld TF, Oldridge DA, Bernard V, Koster J, Colmet Daage L, Diskin SJ, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 2015;47:864–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schramm A, Köster J, Assenov Y, Althoff K, Peifer M, Mahlow E, et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet 2015;47:872–7. [DOI] [PubMed] [Google Scholar]
- 5. Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugène C, Raynal V, et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet 2017;49:1408–13. [DOI] [PubMed] [Google Scholar]
- 6. van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet 2017;49:1261–6. [DOI] [PubMed] [Google Scholar]
- 7. Wang L, Tan TK, Durbin AD, Zimmerman MW, Abraham BJ, Tan SH, et al. ASCL1 is a MYCN- and LMO1-dependent member of the adrenergic neuroblastoma core regulatory circuitry. Nat Commun 2019;10:5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Groningen T, Akogul N, Westerhout EM, Chan A, Hasselt NE, Zwijnenburg DA, et al. A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat Commun 2019;10:1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wolpaw AJ, Grossmann LD, Dessau JL, Dong MM, Aaron BJ, Brafford PA, et al. Epigenetic state determines inflammatory sensing in neuroblastoma. Proc Natl Acad Sci U S A 2022;119:e2102358119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sengupta S, Das S, Crespo AC, Cornel AM, Patel AG, Mahadevan NR, et al. Mesenchymal and adrenergic cell lineage states in neuroblastoma possess distinct immunogenic phenotypes. Nat Cancer 2022;3:1228–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suvà ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell 2019;75:7–12. [DOI] [PubMed] [Google Scholar]
- 12. Dong R, Yang R, Zhan Y, Lai HD, Ye CJ, Yao XY, et al. Single-cell characterization of malignant phenotypes and developmental trajectories of adrenal neuroblastoma. Cancer Cell 2020;38:716–33.e6. [DOI] [PubMed] [Google Scholar]
- 13. Kildisiute G, Kholosy WM, Young MD, Roberts K, Elmentaite R, van Hooff SR, et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Sci Adv 2021;7:eabd3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jansky S, Sharma AK, Körber V, Quintero A, Toprak UH, Wecht EM, et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat Genet 2021;53:683–93. [DOI] [PubMed] [Google Scholar]
- 15. Verhoeven BM, Mei S, Olsen TK, Gustafsson K, Valind A, Lindström A, et al. The immune cell atlas of human neuroblastoma. Cell Rep Med 2022;3:100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costa A, Thirant C, Kramdi A, Pierre-Eugène C, Louis-Brennetot C, Blanchard O, et al. Single-cell transcriptomics reveals shared immunosuppressive landscapes of mouse and human neuroblastoma. J Immunother Cancer 2022;10:e004807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wienke J, Visser LL, Kholosy WM, Keller KM, Barisa M, Poon E, et al. Integrative analysis of neuroblastoma by single-cell RNA sequencing identifies the NECTIN2-TIGIT axis as a target for immunotherapy. Cancer Cell 2024;42:283–300.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avitabile M, Bonfiglio F, Aievola V, Cantalupo S, Maiorino T, Lasorsa VA, et al. Single-cell transcriptomics of neuroblastoma identifies chemoresistance-associated genes and pathways. Comput Struct Biotechnol J 2022;20:4437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hastings JF, Latham SL, Kamili A, Wheatley MS, Han JZR, Wong-Erasmus M, et al. Memory of stochastic single-cell apoptotic signaling promotes chemoresistance in neuroblastoma. Sci Adv 2023;9:eabp8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yuan X, Seneviratne JA, Du S, Xu Y, Chen Y, Jin Q, et al. Single-cell profiling of peripheral neuroblastic tumors identifies an aggressive transitional state that bridges an adrenergic-mesenchymal trajectory. Cell Rep 2022;41:111455. [DOI] [PubMed] [Google Scholar]
- 21. Thirant C, Peltier A, Durand S, Kramdi A, Louis-Brennetot C, Pierre-Eugène C, et al. Reversible transitions between noradrenergic and mesenchymal tumor identities define cell plasticity in neuroblastoma. Nat Commun 2023;14:2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Granger M, Grupp SA, Kletzel M, Kretschmar C, Naranjo A, London WB, et al. Feasibility of a tandem autologous peripheral blood stem cell transplant regimen for high risk neuroblastoma in a cooperative group setting: a Pediatric Oncology Group study: a report from the Children’s Oncology Group. Pediatr Blood Cancer 2012;59:902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park JR, Kreissman SG, London WB, Naranjo A, Cohn SL, Hogarty MD, et al. Effect of tandem autologous stem cell transplant vs single transplant on event-free survival in patients with high-risk neuroblastoma: a randomized clinical trial. JAMA 2019;322:746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weiss BD, Yanik G, Naranjo A, Zhang FF, Fitzgerald W, Shulkin BL, et al. A safety and feasibility trial of (131) I-MIBG in newly diagnosed high-risk neuroblastoma: a Children’s Oncology Group study. Pediatr Blood Cancer 2021;68:e29117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Granger MM, Naranjo A, Bagatell R, DuBois SG, McCune JS, Tenney SC, et al. Myeloablative busulfan/melphalan consolidation following induction chemotherapy for patients with newly diagnosed high-risk neuroblastoma: Children’s Oncology Group trial ANBL12P1. Transplant Cell Ther 2021;27:490.e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park JR, Bagatell R, Cohn SL, Pearson AD, Villablanca JG, Berthold F, et al. Revisions to the international neuroblastoma response criteria: a consensus statement from the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol 2017;35:2580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ambros IM, Zellner A, Roald B, Amann G, Ladenstein R, Printz D, et al. Role of ploidy, chromosome 1p, and Schwann cells in the maturation of neuroblastoma. N Engl J Med 1996;334:1505–11. [DOI] [PubMed] [Google Scholar]
- 29. Weiss T, Taschner-Mandl S, Janker L, Bileck A, Rifatbegovic F, Kromp F, et al. Schwann cell plasticity regulates neuroblastic tumor cell differentiation via epidermal growth factor-like protein 8. Nat Commun 2021;12:1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cords L, Tietscher S, Anzeneder T, Langwieder C, Rees M, de Souza N, et al. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat Commun 2023;14:4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coughlin CM, Fleming MD, Carroll RG, Pawel BR, Hogarty MD, Shan X, et al. Immunosurveillance and survivin-specific T-cell immunity in children with high-risk neuroblastoma. J Clin Oncol 2006;24:5725–34. [DOI] [PubMed] [Google Scholar]
- 32. Zhang P, Wu X, Basu M, Dong C, Zheng P, Liu Y, et al. MYCN amplification is associated with repressed cellular immunity in neuroblastoma: an in silico immunological analysis of TARGET database. Front Immunol 2017;8:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeTomaso D, Jones MG, Subramaniam M, Ashuach T, Ye CJ, Yosef N. Functional interpretation of single cell similarity maps. Nat Commun 2019;10:4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeTomaso D, Yosef N. Hotspot identifies informative gene modules across modalities of single-cell genomics. Cell Syst 2021;12:446–56.e9. [DOI] [PubMed] [Google Scholar]
- 36. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brady SW, Liu Y, Ma X, Gout AM, Hagiwara K, Zhou X, et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nat Commun 2020;11:5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Surrey LF, MacFarland SP, Chang F, Cao K, Rathi KS, Akgumus GT, et al. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med 2019;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schleiermacher G, Javanmardi N, Bernard V, Leroy Q, Cappo J, Rio Frio T, et al. Emergence of new ALK mutations at relapse of neuroblastoma. J Clin Oncol 2014;32:2727–34. [DOI] [PubMed] [Google Scholar]
- 40. Bosse KR, Giudice AM, Lane MV, McIntyre B, Schürch PM, Pascual-Pasto G, et al. Serial profiling of circulating tumor DNA identifies dynamic evolution of clinically actionable genomic alterations in high-risk neuroblastoma. Cancer Discov 2022;12:2800–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berko ER, Witek GM, Matkar S, Petrova ZO, Wu MA, Smith CM, et al. Circulating tumor DNA reveals mechanisms of lorlatinib resistance in patients with relapsed/refractory ALK-driven neuroblastoma. Nat Commun 2023;14:2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ackermann S, Cartolano M, Hero B, Welte A, Kahlert Y, Roderwieser A, et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018;362:1165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 2013;3:a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zimmerman MW, Liu Y, He S, Durbin AD, Abraham BJ, Easton J, et al. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov 2018;8:320–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Upton K, Modi A, Patel K, Kendsersky NM, Conkrite KL, Sussman RT, et al. Epigenomic profiling of neuroblastoma cell lines. Sci Data 2020;7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harenza JL, Diamond MA, Adams RN, Song MM, Davidson HL, Hart LS, et al. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data 2017;4:170033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mabe NW, Huang M, Dalton GN, Alexe G, Schaefer DA, Geraghty AC, et al. Transition to a mesenchymal state in neuroblastoma confers resistance to anti-GD2 antibody via reduced expression of ST8SIA1. Nat Cancer 2022;3:976–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rovillain E, Mansfield L, Caetano C, Alvarez-Fernandez M, Caballero OL, Medema RH, et al. Activation of nuclear factor-kappa B signalling promotes cellular senescence. Oncogene 2011;30:2356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Milanovic M, Fan DNY, Belenki D, Däbritz JHM, Zhao Z, Yu Y, et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018;553:96–100. [DOI] [PubMed] [Google Scholar]
- 50. Saul D, Kosinsky RL, Atkinson EJ, Doolittle ML, Zhang X, LeBrasseur NK, et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun 2022;13:4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hideshima T, Neri P, Tassone P, Yasui H, Ishitsuka K, Raje N, et al. MLN120B, a novel IkappaB kinase beta inhibitor, blocks multiple myeloma cell growth in vitro and in vivo. Clin Cancer Res 2006;12:5887–94. [DOI] [PubMed] [Google Scholar]
- 52. Asgharzadeh S, Salo JA, Ji L, Oberthuer A, Fischer M, Berthold F, et al. Clinical significance of tumor-associated inflammatory cells in metastatic neuroblastoma. J Clin Oncol 2012;30:3525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Borriello L, Nakata R, Sheard MA, Fernandez GE, Sposto R, Malvar J, et al. Cancer-associated fibroblasts share characteristics and protumorigenic activity with mesenchymal stromal cells. Cancer Res 2017;77:5142–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Louault K, Porras T, Lee MH, Muthugounder S, Kennedy RJ, Blavier L, et al. Fibroblasts and macrophages cooperate to create a pro-tumorigenic and immune resistant environment via activation of TGF-β/IL-6 pathway in neuroblastoma. Oncoimmunology 2022;11:2146860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kumar A, Negi G, Sharma SS. JSH-23 targets nuclear factor-kappa B and reverses various deficits in experimental diabetic neuropathy: effect on neuroinflammation and antioxidant defence. Diabetes Obes Metab 2011;13:750–8. [DOI] [PubMed] [Google Scholar]
- 56. Li B, Brady SW, Ma X, Shen S, Zhang Y, Li Y, et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020;135:41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oren Y, Tsabar M, Cuoco MS, Amir-Zilberstein L, Cabanos HF, Hütter JC, et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 2021;596:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dhimolea E, de Matos Simoes R, Kansara D, Al’Khafaji A, Bouyssou J, Weng X, et al. An embryonic diapause-like adaptation with suppressed myc activity enables tumor treatment persistence. Cancer Cell 2021;39:240–56.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201–6. [DOI] [PubMed] [Google Scholar]
- 60. Collignon E, Cho B, Furlan G, Fothergill-Robinson J, Martin SB, McClymont SA, et al. m6A RNA methylation orchestrates transcriptional dormancy during paused pluripotency. Nat Cell Biol 2023;25:1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu K, Gao T, Wang Z, Zhang L, Tan K, Lv Z. RNA N6-methyladenosine reader IGF2BP3 interacts with MYCN and facilitates neuroblastoma cell proliferation. Cell Death Discov 2023;9:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 2006;441:431–6. [DOI] [PubMed] [Google Scholar]
- 63. Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature 2014;506:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moon CS, Reglero C, Cortes JR, Quinn SA, Alvarez S, Zhao J, et al. FYN-TRAF3IP2 induces NF-κB signaling-driven peripheral T cell lymphoma. Nat Cancer 2021;2:98–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Feng Z, Porter AG. NF-kappaB/Rel proteins are required for neuronal differentiation of SH-SY5Y neuroblastoma cells. J Biol Chem 1999;274:30341–4. [DOI] [PubMed] [Google Scholar]
- 66. Zhi Y, Duan Y, Zhou X, Yin X, Guan G, Zhang H, et al. NF-κB signaling pathway confers neuroblastoma cells migration and invasion ability via the regulation of CXCR4. Med Sci Monit 2014;20:2746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lorenzi S, Forloni M, Cifaldi L, Antonucci C, Citti A, Boldrini R, et al. IRF1 and NF-kB restore MHC class I-restricted tumor antigen processing and presentation to cytotoxic T cells in aggressive neuroblastoma. PLoS One 2012;7:e46928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tsang PS, Cheuk AT, Chen QR, Song YK, Badgett TC, Wei JS, et al. Synthetic lethal screen identifies NF-κB as a target for combination therapy with topotecan for patients with neuroblastoma. BMC Cancer 2012;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Veeraraghavan J, Natarajan M, Aravindan S, Herman TS, Aravindan N. Radiation-triggered tumor necrosis factor (TNF) alpha-NFkappaB cross-signaling favors survival advantage in human neuroblastoma cells. J Biol Chem 2011;286:21588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bian X, Opipari AW Jr, Ratanaproeksa AB, Boitano AE, Lucas PC, Castle VP. Constitutively active NFkappa B is required for the survival of S-type neuroblastoma. J Biol Chem 2002;277:42144–50. [DOI] [PubMed] [Google Scholar]
- 71. Huang Y, Tsubota S, Nishio N, Takahashi Y, Kadomatsu K. Combination of tumor necrosis factor-α and epidermal growth factor induces the adrenergic-to-mesenchymal transdifferentiation in SH-SY5Y neuroblastoma cells. Cancer Sci 2021;112:715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Louault K, Blavier L, Lee MH, Kennedy RJ, Fernandez GE, Pawel BR, et al. Nuclear factor-κB activation by transforming growth factor-β1 drives tumour microenvironment-mediated drug resistance in neuroblastoma. Br J Cancer 2024;131:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yarmarkovich M, Marshall QF, Warrington JM, Premaratne R, Farrel A, Groff D, et al. Targeting of intracellular oncoproteins with peptide-centric CARs. Nature 2023;623:820–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang S, Corbett SE, Koga Y, Wang Z, Johnson WE, Yajima M, et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol 2020;21:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 2019;8:329–37.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Osorio D, Cai JJ. Systematic determination of the mitochondrial proportion in human and mice tissues for single-cell RNA-sequencing data quality control. Bioinformatics 2021;37:963–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 shows the patient clinical characteristics and genetic alterations at diagnosis
Supplementary Table S2 links the CN number to the HTAN biospecimen numbers
Supplementary Table S3 provides the genes of each persister subtype signature
Supplementary Table S4 shows the most negatively enriched pathways in non-cycling persister cells by GSEA
Supplementary Figure S1 shows cell type markers and inferred copy number of tumors as well as characterization of malignant, Schwann cells, tumor associated macrophages and cancer associated fibroblasts at time of definitive surgery
Supplementary Figure S2 shows the details of the pathway-based clustering algorithm including the autocorrelation pathway scores, cell cycle annotation, pathway enrichment analysis and region analysis.
Supplementary Figure S3 shows the WGS analysis of the diagnostic and definitive surgery including clonal evolution of ALK mutation from diagnosis to definitive surgery in one tumor.
Supplementary Figure S4 differences in persister cell composition between diagnosis and definitive surgery as well as decrease in MYC/N protein level in human samples, PDX and cell lines following chemotherapy
Supplementary Figure S5 shows the connection between NF-kB/stemness subtype and mesenchymal transcriptional state.
Supplementary Figure S6 shows the increase in NF-kB activity following chemotherapy and its role in survival which is enhanced by specific TAMs and CAFs
Data Availability Statement
The snRNA-seq and WGS data are available at https://humantumoratlas.org/explore under HTAN-CHOP for the discovery dataset and HTAN-HTTAP for the validation cohort.