Abstract
Anthropogenic activities can significantly impact wildlife in natural water bodies, affecting not only the host's physiology but also its microbiome. This study aimed to analyze the gut microbiome and antimicrobial resistance gene profile (i.e., the resistome) of yellow perch living in lakes subjected to different levels of anthropogenic pressure: wastewater effluent-impacted lakes and undeveloped lakes. Total DNA and RNA from gut content samples were extracted and sequenced for analysis. Results indicate that the gut resistome and microbiome of yellow perch differ between lakes, perhaps due to varying anthropogenic pressure. The resistome was predominated by macrolide resistance genes, particularly the MLS23S group, making up 53 % of resistome sequences from effluent-impacted lakes and 73 % from undeveloped lakes. The colistin resistance gene group (mcr) was detected in numerous samples, including variants associated with Aeromonas and the family Enterobacteriaceae. The gut microbiome across all samples was dominated by the phyla Proteobacteria, Firmicutes, and Actinobacteria, with the opportunistic pathogens Plesiomonas shigelloides and Aeromonas veronii more abundant in effluent-impacted lakes. Metagenomic analysis of wild fish samples offers valuable insights into the effects of anthropogenic pressures on microbial communities, including antimicrobial resistance genes, in water bodies.
Keywords: Anthropogenic pressure, Antimicrobial resistance, Metagenomics, Transcriptomics, Yellow perch
1. Introduction
Antimicrobial resistance (AMR) is a complex global health threat, affecting humans, animals, and the environment [1]. AMR, mediated by antimicrobial resistance genes (ARGs), occurs naturally [2], but sources of anthropogenic origin can potentially increase AMR in the environment. Effluent from wastewater treatment plants, healthcare facilities, antibiotic manufacturing plants, and animal husbandry can contribute to the antimicrobials and ARGs found in aquatic environments like rivers and lakes [3,4]. Other compounds such as heavy metals [5], pesticides [6] and non-antimicrobial pharmaceuticals [7] can also contribute to the emergence of antimicrobial resistance and ARGs in environmental ecosystems. Wildlife in these areas are exposed to these compounds, potentially altering their resistomes and microbiomes compared to unexposed wildlife [8].
Wild fish are of particular interest, as they are able to bioaccumulate compounds like antimicrobials [9,10], and microbial communities within the fish, such as the gut microbiome, have been found to harbor AMR microbes [8,11]. As such, wild fish have been proposed as sentinels and disseminators of AMR [12]. Zhou et al. (2021) found a similar distribution of ARGs in wild fish and water, suggesting an exchange between the two [13]. Ballash et al. (2022) demonstrated that fish from rivers receiving wastewater containing ARGs, antibiotics and AMR bacteria, are effective bioindicators of contaminated waters [10]. However, data on this topic remains scarce.
In Minnesota (MN), United States, lakes and their fish inhabitants have been studied to determine the ecological distribution of contaminants of emerging concern, including antimicrobial compounds [14]. A wide range of concentrations of antimicrobials have been found across lakes with different gradients of anthropogenic impacts. For example, azithromycin, roxithromycin, and miconazole have been found in certain MN lakes, and these compounds have been associated with impacts on aquatic species at the genetic, physiological, and behavioral levels [14,15].
Among fish species living in MN, yellow perch (Perca flavescens) is relevant for subsistence in some Ojibwe communities, and as prey for economically important fish species like walleye (Sander vitreus) [16]. To date, few studies have characterized the microbiome of yellow perch through 16S rRNA gene sequencing [[17], [18], [19]], which cannot accurately discriminate at lower taxonomic levels [20] or detect ARGs across the microbiome (termed the resistome). Few metagenomic studies have characterized the resistome of wild fish living under varying anthropogenic pressures [21,22]. This is a major gap in the scientific literature, especially considering the relevance of wild fish as sentinels of pathogenic bacteria and ARGs, as well as their importance as a subsistence food source for some human populations. To help address this knowledge gap, we sampled intestinal contents from yellow perch across several MN lakes under varying anthropogenic pressure. Our objective was to characterize the resistome and microbiome of yellow perch intestinal contents at both the DNA and RNA levels. We hypothesized that both the DNA and RNA resistome profiles would be more diverse in samples collected from lakes with more anthropogenic impact.
2. Material and methods
2.1. Sample collection and study site
Yellow perch were sampled from five lakes (Shagawa, Trout, Whitewater, Ball Club, and Elbow) in northern MN during two sampling campaigns: September 2018 (N = 96) and July 2019 (N = 68). These lakes were chosen for their accessibility and classification in a previous study [14] as being subjected to different levels of anthropogenic pressure, i.e., receiving wastewater effluent (“effluent-impacted”, Shagawa and Whitewater) or without any shoreline development (“undeveloped”, Trout, Ball Club, and Elbow). Lake location coordinates are available in Supplemental Table 1.
Fish were captured using electrofishing and euthanized by manually applied blunt force trauma (cranial concussion), followed by brain pithing, in accordance with a method described by the American Veterinary Medical Association included in an approved protocol (University of Minnesota IACUC protocol ID: 1803-35736A). Fish selected for sampling were 10–20 cm in length (∼50–150 g in weight), and grossly healthy with no visible lesions or abnormalities. After euthanasia, an incision along the ventral midline was made to remove the intact gastrointestinal tract, and intestinal contents were extracted into frozen 15 ml Falcon tubes. In 2018, samples were preserved in RNAlater (ThermoFisher, USA) (N = 24) or liquid nitrogen (N = 72), while all 2019 samples were preserved in liquid nitrogen. After collection, all samples were stored at −80 °C until nucleic acid extraction.
2.2. Nucleic Acid Extraction and library preparation
Nucleic acids were extracted from intestinal content samples using the DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany) per the manufacturer's instructions. Frozen fecal samples were ground in a prechilled, sterile mortar with liquid nitrogen, using sterile pestles, and then divided for DNA and RNA extractions. All scalpels, spatulas, and forceps were made nuclease-free and prechilled in liquid nitrogen before use. Approximately 100 mg of feces was weighed into prechilled, nuclease-free microfuge tubes to prevent thawing before extraction.
Samples stored in RNAlater were centrifuged, and the supernatant was removed, leaving only the solid feces. For DNA extraction, 100 mg of feces was placed in the PowerBead Pro tube with 800 μl of CD1 solution. RNA was extracted using the RNeasy Power Microbiome Kit (Qiagen), following the manufacturer protocol. Briefly, 100 mg of feces was placed into the PowerBead Tube, and 650 μl PM1 and 6.5 μl β-ME were added to the tube. Both DNA and RNA extraction involved vortexing PowerBead tubes for approximately 10 s to ensure homogenization. Cells were disrupted using a bead beater (BioSpec Products, USA), following an optimized method [23]. DNA samples for extraction were processed with Inhibitor Removal Technology (IRT) to eliminate inhibitors. After vortexing, the PowerBead tubes were centrifuged at 15,000 ×g for 2 min and the supernatant was collected for subsequent steps. DNA extraction steps were automated using the QIAcube Connect instrument (Qiagen), while RNA extractions were performed manually. DNA and RNA concentration were measured with the Qubit 4 Fluorometer (ThermoFisher) and integrity was checked with the TapeStation 4200 (Agilent Technologies, USA).
Due to limited fecal amounts and low quantity and integrity of some nucleic acids, we were able to generate DNA libraries for 38 samples and RNA libraries for 24 samples (Suppl. Table 1). DNA libraries were prepared with 100 ng of input DNA using the QIAseq FX DNA Library Kit (Qiagen). RNA libraries were made with 100 ng of input RNA using the QIAseq Stranded RNA Library Kit (Qiagen) following manufacturer protocols. rRNA was removed using the QIAseq FastSelect –5S/16S/23S Kit (Qiagen) before metatranscriptomic library preparation. To avoid fragmentation due to low RNA integrity, heat fragmentation was omitted. Library quality and quantity were assessed using the TapeStation 4200 and Qubit 4.0. DNA and RNA libraries were pooled separately for paired-end sequencing on S4 flow cells of the NovaSeq 6000 (Illumina) at the University of Minnesota Genomics Center.
Table 1.
Results from the Permutational Analysis of Variance (PERMANOVA) at the phylum, genus, species and OTU levels (microbiome); and at the ARG group level (resistome), using both the DNA and RNA shotgun datasets.
| Microbiome |
Resistome |
||||
|---|---|---|---|---|---|
| Phylum | Genus | Species | OTU | ARG group | |
| DNA shotgun sequencing | |||||
| Anthropogenic pressure | F = 3.26, R2 = 0.08, P = 0.02 | F = 2.41, R2 = 0.06, P = 0.03 | F = 2.50, R2 = 0.06, P = 0.03 | F = 2.50, R2 = 0.06, P = 0.02 | F = 3.25, R2 = 0.08, P = 0.03 |
| Lake | F = 1.41, R2 = 0.11, P = 0.18 | F = 1.33, R2 = 0.10, P = 0.14 | F = 1.33, R2 = 0.10, P = 0.16 | F = 1.33, R2 = 0.10, P = 0.14 | F = 1.21, R2 = 0.09, P = 0.02 |
| RNA shotgun sequencing | |||||
| Anthropogenic pressure | F = 3.74, R2 = 0.12, P = 0.01 | F = 3.00, R2 = 0.10, P = 0.01 | F = 2.94, R2 = 0.11, P = 0.008 | F = 2.94, R2 = 0.11, P = 0.01 | F = 1.73, R2 = 0.08, P = 0.11 |
| Lake | F = 2.08, R2 = 0.21, P = 0.02 | F = 1.84, R2 = 0.20, P = 0.02 | F = 1.67, R2 = 0.19, P = 0.03 | F = 1.67, R2 = 0.19, P = 0.02 | F = 0.41, R2 = 0.05, P = 0.98 |
2.3. Bioinformatic analysis of the resistome and microbiome
An extended description of bioinformatic analysis can be found in supplementary data. DNA and RNA datasets were processed separately. Demultiplexed sequence reads were analyzed with the AMR++/v3.0 pipeline [24], including quality control and removal of host reads (host genome accession No: GCF_004354835.1_PFLA_1.0). Non-host reads were used for taxonomic classification with Kraken2 using default settings [25]. The resulting operational taxonomic unit (OTU) count matrix was used for downstream microbiome analysis.
For resistome analysis, non-host reads were aligned to ARG sequences in the MEGARES/v3.0 database using default settings, including a gene fraction coverage cutoff of 0.80 [24]. Single Nucleotide Polymorphism (SNP) confirmation was included with the ‘--snp Y' flag. The resulting count matrix was used for downstream analysis. For microbiome and resistome analyses, the respective count matrices, taxonomy tables, and sample metadata were used to create a phyloseq object with the Phyloseq v1.36 package. All analyses were performed in R/v4.1.0. To explore ARGs of public health concern, relevant detected ARG sequences within MEGARES were compared with NCBI sequences using the nucleotide basic local alignment tool (BLASTn) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.4. Bioinformatic procedures to assemble genomes from metagenomes
DNA shotgun sequences from undeveloped and effluent-impacted lakes underwent de novo assembly with MEGAHIT/v1.2.9 to generate contiguous sequences (contigs) with a minimum size of 1000 base pairs. The resulting assemblies were processed through the GeNomad/v1.8.0 pipeline using the end-to-end command for plasmidome and virome identification [26]. Subsequently, individual fasta files were used to construct a contig fasta database using Anvi'o/v8.0 [27], where k-mer frequencies and open reading frames were computed and identified via Prodigal/v2.6.3. Next, Hidden Markov Models were generated to annotate bacterial single-copy core genes from the Genome Taxonomy Database (GTDB) (Bacteria_71) through HMMER/v3.4. Contigs were indexed using Bowtie2/v2.3.4.1, and resulting index files were aligned to contigs, yielding a BAM file for each sample. Binning of contigs was conducted with anvi-cluster-contigs using Metabat2 [28]. Thresholds for construction of metagenome assembled genomes (MAGs) were set at >70 % completion and < 10 % redundancy. Finally, GeNomad was run on each individual MAG to identify plasmids and viruses within each set of lakes. Additionally, we transformed the GeNomad output of protein family (Pfam) annotations to gene ontology (GO) terms to elucidate the functional potential of identified plasmids and viruses. The redundancy of plasmidome and virome GO profiles was simplified based on semantic similarity groupings as specified with rrvgo/v1.4.4. The simplified profiles were used to perform principal coordinates analysis (PCoA), and the first two coordinates of the PCoA were visualized with the rrvgo package.
2.5. Statistical analysis
Primary independent variables of interest were lake ID (i.e., Shagawa, Trout, Whitewater, Elbow, Ball Club), and anthropogenic pressure (effluent-impacted versus undeveloped). To compare diversity within samples, alpha diversity measurements (i.e., richness, Shannon diversity, Pielou's evenness index) were calculated using the phyloseq and microbiome/v1.14 packages, following aggregation of counts at the phylum, genus and species levels. Differences in alpha diversity between anthropogenic pressures were assessed with non-parametric Wilcoxon rank-sum tests (W), with a significance level of P < 0.05.
To compare diversity in the microbial community structure between groups (i.e., beta diversity), we calculated Bray-Curtis dissimilarities with the phyloseq package, and used these distance values to perform ordination using the non-metric multidimensional scaling (NMDS) method. Ordinations were visualized with the ggplot2/v3.5.1 package. To test whether variance in beta diversity was significantly associated with lake ID or anthropogenic pressure, we performed Permutational Analysis Of Variance (PERMANOVA, adonis2 function in vegan R package). A value of P < 0.05 was considered statistically significant.
The same statistical tests were conducted on the resistome count matrix, including alpha and beta diversity outcomes. Alpha diversity indices were computed at the ARG group level, while beta diversity was performed at the ARG accession level, using the MEGARES ontology [24]. The differential relative abundance of ARG groups between lakes with varying anthropogenic pressure was calculated using the MaAsLin2/v1.4 package. To ensure robust results, taxa with a relative abundance lower than 0.001 % or sample prevalence lower than 10 % were excluded. Feature counts were normalized with the cumulative sum scaling (CSS) method implemented in metagenomeSeq/1.36 package and transformed to the logarithmic scale. The Benjamini-Hochberg method was used to calculate adjusted P-values, which were considered significant at <0.05.
3. Results
3.1. Sequencing results
Out of 164 total samples collected in 2018 and 2019, complete sequencing was achieved for 24 and 38 samples for RNA and DNA shotgun samples, respectively. RNA shotgun data generated 2.76B raw reads, averaging 114.98 M reads per sample (median: 116.27 M, range: 56.88 M - 163.89 M reads per sample). Read quality filtering removed an average of 1.9 % of raw reads from each sample (range: 0.7–5.9 %), and subsequent filtering of host-aligned reads resulted in an average of 42.2 % of reads per sample for further analysis (range: 24.7–48.3 %). For the resistome analysis, an average of 5.6 % of reads aligned to ARGs (median: 1.9 %, range: 0.3–22.0 % per sample). Ninety-eight unique ARGs were identified from 19 classes, 24 mechanisms, and 31 groups. For microbiome analysis, an average of 18.4 % of reads were classified per sample (median: 36.9 %, range: 14.9–49.9 %) resulting in 56,540 OTUs: 55 % Eukaryota, 35 % Bacteria, and 10 % “Others” (i.e., Archaea, viroids, viruses).
DNA shotgun data produced 2.65B raw reads, averaging 69.99 M reads per sample (median: 69.13 M, range: 47.17 M - 96.26 M). Quality filtering removed 0.33 % of reads per sample (range: 0.21–0.49 %), and subsequent filtering of host-aligned reads resulted in an average of 45.67 % of reads per sample for further analysis (range: 43.42–48.39 %). For resistome analysis, an average of 0.1 % of reads aligned to ARGs (median: 0.06 % range: 0.0004–0.3 % per sample). A total of 1830 ARGs were identified from 50 classes, 155 mechanisms, and 659 groups. For microbiome analysis, 15.0 % of reads were classified per sample (median: 14.3, range: 5.9–22.9 %), resulting in 259,431 OTUs: 74.5 % Eukaryota, 17 % Bacteria, and 7.9 % ‘Others’.
3.2. Alpha diversity
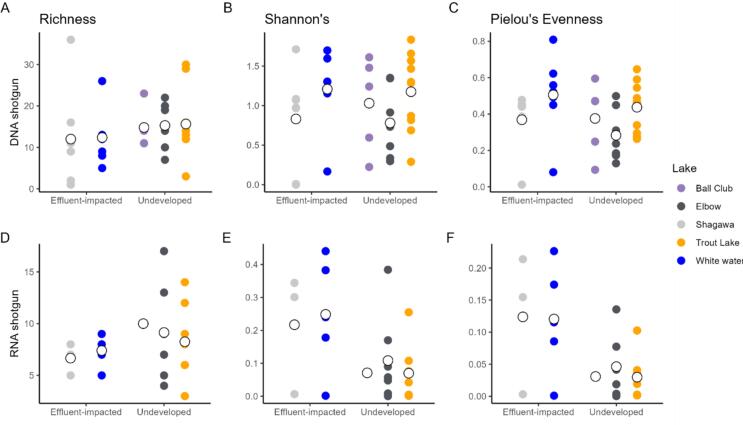
In the RNA shotgun dataset, samples from undeveloped lakes had higher resistome richness but lower Shannon's and Pielou evenness, as compared to impacted lakes (Shannon: W = 95, P = 0.06, Pielou's evenness: W = 98, P < 0.05). For the DNA shotgun dataset, resistome richness was significantly higher in undeveloped lakes (W = 103, P < 0.05, Fig. 1A, D). Microbiome diversity in RNA shotgun data was higher in undeveloped lakes at the phylum (Shannon: W = 12, P < 0.001, Pielou evenness: W = 10.1 P < 0.01) and genus levels (Shannon: W = 20, P < 0.01) (Suppl. Fig. 1B, C, E). In DNA shotgun data, microbiome alpha diversity was higher in samples from undeveloped lakes, however this was not statistically significant (richness, Shannon, Pielou evenness: P > 0.05, Suppl. Fig. 2).
Fig. 1.
Resistome alpha diversity in the RNA shotgun data (A, B, C) and the DNA shotgun data (D, E, F). Indices shown: richness (A, D), Shannon diversity index (B, E), and Pielou's evenness index (C, F). Each dot represents a single sample, and observations are grouped by anthropogenic pressure (x-axes), and colored by lake. The larger empty circles represent the mean value for each lake.
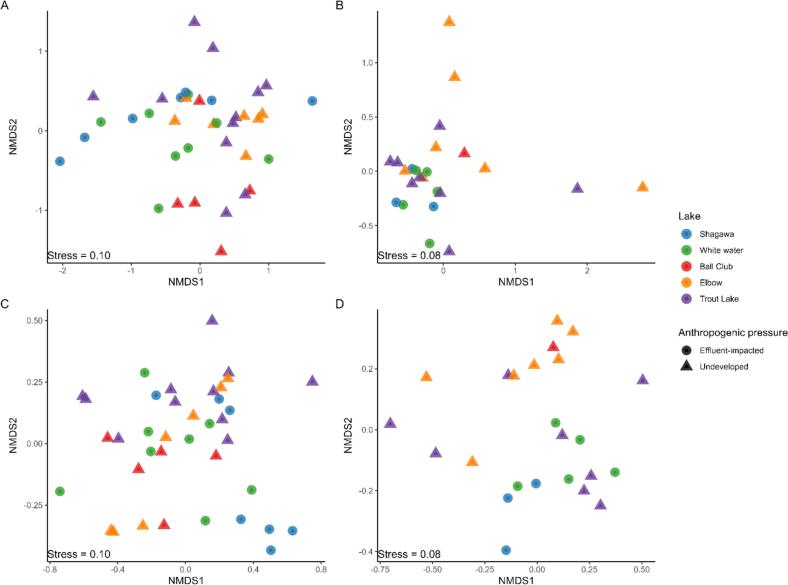
Fig. 2.
Non-metric Multidimensional Scaling (NMDS) analysis of the resistome and microbiome. A) NMDS of DNA shotgun dataset at the ARG level. B) NMDS of RNA shotgun dataset at the ARG level. C) NMDS of DNA shotgun dataset at the OTU level. D) NMDS of RNA shotgun dataset at the OTU level.
3.3. Beta diversity
PERMANOVA analysis indicated significant differences in resistome beta diversity from DNA shotgun data by lake and by anthropogenic pressure (PERMANOVA results for lake ID: R2 = 0.09, P < 0.05, anthropogenic pressure: R2 = 0.08, P < 0.05) (Table 1, Fig. 2A), while these differences were not statistically significant for the RNA shotgun dataset (P > 0.05) (Fig. 2B). With regards to microbiome beta diversity, we found significant differences in the DNA shotgun data by anthropogenic pressure at all taxonomic levels (P < 0.05) (Fig. 2C); and in the RNA shotgun data by both lake ID and anthropogenic pressure (P < 0.05) (Fig. 2D) (Table 1).
3.4. Taxonomic composition and differential abundance
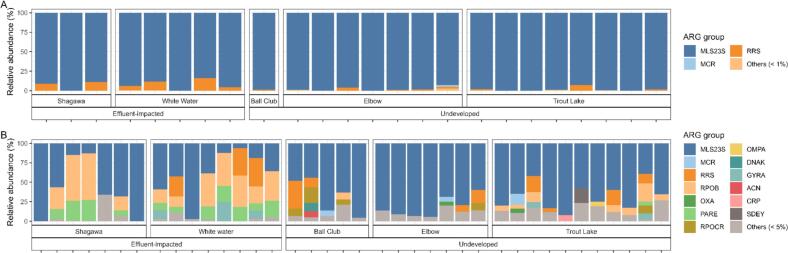
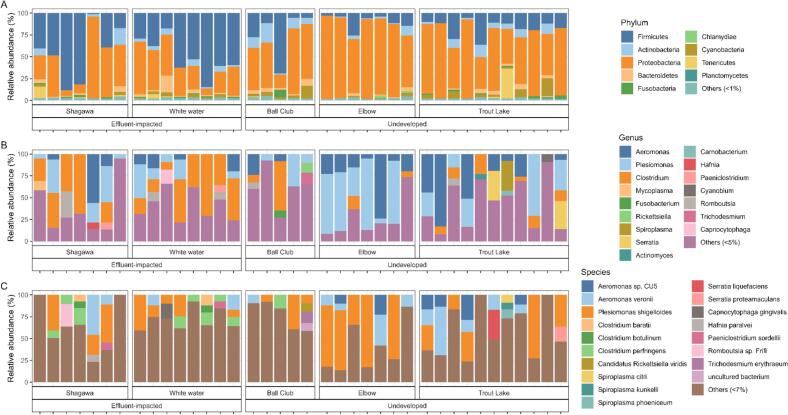
In both the RNA and DNA shotgun datasets, the most abundant ARG group was MLS23S (Fig. 3A–B). MaAslin2 differential abundance testing showed higher abundances of parE and rpoB ARG groups in effluent-impacted lakes compared to undeveloped lakes (log10 = 3.47, SE = 0,93, adjusted P < 0.05, and log10 = 3.55, SE = 0.93, adjusted P < 0.05, respectively). The dominant phyla in both ‘omics datasets were Proteobacteria, Firmicutes and Actinobacteria (Fig. 4A). In effluent-impacted lakes, Firmicutes comprised 51 % of all taxa, followed by Proteobacteria (39 %), and Actinobacteria (1.5 %). In undeveloped lakes, the dominant phyla were Proteobacteria (77 %) and Firmicutes (13 %). At the genus level, the predominant taxa across all samples were Plesiomonas (27.6 %), Clostridium (20.6 %), and Aeromonas (18.9 %) (Fig. 4B). The three most abundant species across all samples included P. shigelloides (27.6 %), A. veronii (10.7 %), and C. perfringens (4.3 %) (Fig. 4C).
Fig. 3.
Relative abundance of A) ARG groups within the RNA shotgun dataset and B) DNA shotgun dataset. Each column represents one sample, and samples are grouped by anthropogenic pressure and lake.
Fig. 4.
Relative Abundance of the DNA shotgun sequence dataset. A) Relative abundance at phylum level. B) Relative abundance at genus level. C) Relative abundance at species level. Each column represents one sample, and samples are grouped by anthropogenic pressure and lake.
3.5. Identification of mcr sequences with BLASTn and MAG reconstruction
To identify the specific mcr genes detected in the DNA dataset (Fig. 3A–B), we compared the sequences of detected mcr genes to those in the NCBI database using BLASTn. Results showed alignments to mcr-7, mcr-3 and mcr-9 genes from Aeromonas, and Enterobacteriaceae family members including Salmonella, Escherichia, and Klebsiella.
In effluent-impacted lakes, 1.4 million contigs were assembled from 15 samples, generating 178 bins with Metabat2. From these, 10 MAGs were selected based on predefined thresholds of completion and redundancy [29], with 70 % (7/10) identified at the species level, including Plesiomonas shigelloides, Brevinema andersonii, and Paenibacillus alvei (Suppl. Table 2, Suppl. File 3). Additionally, GeNomad identified 23,865 plasmids and 5532 viruses across the samples from effluent-impacted lakes. In undeveloped lakes, 1.7 million contigs from 21 samples led to the assembly of 287 bins, with 31 MAGs meeting the criteria. Of these, 48 % (15/31) were identified at the species level, including Limnosomonas limnophila, Enteroccocus rivorum, and Neorickettsia helminthoeca (Suppl. Table 2, Suppl. File 3). A total of 15,657 plasmids and 6922 viruses were identified in undeveloped lake samples. A large proportion of MAGs had poor taxonomic representation, with 53 % (22/41) classified at the species level and 17 % (7/41) classified at the domain level.
Functional enrichment analysis with GeNomad revealed diverse viromes in the DNA shotgun data. The viromic functional profiles in both effluent-impacted and undeveloped lakes were dominated by elements involved in viral replication, recombination, transcription, and transmembrane transport (Suppl. Fig. 3A–B). The plasmidome profile in samples collected from effluent-impacted lakes had a higher proportion of sequences associated with response to antibiotics and protein secretion. The plasmidome in samples from undeveloped lakes had a relatively high representation of sequences associated with protein transport/secretion, and cellular response to DNA damage (Suppl. Fig. 3C–D).
4. Discussion
We characterized the gut microbiome and resistome of yellow perch sampled in effluent-impacted and undeveloped lakes in MN. Previous research suggests that wildlife plays an important role in the acquisition and dissemination of AMR [8,12], and we focused specifically on a wild fish population with importance for both the lake ecosystem and for indigenous lifeways in MN.
4.1. Metagenomic sequencing revealed diverse resistome patterns across lakes under varying anthropogenic pressures, while metatranscriptomic sequencing revealed a narrower range of actively transcribed ARG groups
Our results suggest that the resistome composition of fish GI contents differs between lakes, and may be associated with differences in anthropogenic pressure. Xue et al. (2021) used 16S rRNA gene sequencing and ARG quantification in fish from rivers, finding an enrichment of ARGs in fish gut microbiomes living downstream from effluent-impacted water versus fish living in waters upstream [8]. Similarly, Guan et al. (2022) found comparable results in using DNA shotgun sequencing [22]. However, we could not identify any other publications comparing fish microbiomes and resistomes across anthropogenic pressure gradients using metagenomics and metatranscriptomics.
DNA shotgun sequencing identifies ARGs present in a sample but cannot differentiate actively transcribed ARGs at the time of sampling [30,31]. To address this, we performed metatranscriptomics. Based on this paired DNA-RNA analysis, we observed a relatively diverse resistome profile at the DNA level, but this diversity was not fully reflected within the RNA dataset (Fig. 1a–f). This suggests that not all ARGs within the microbiome were actively transcribed during sampling, indicating conditional transcription of ARGs and/or differences in the overall metabolic activity of different subpopulations within the microbiome. The initiation of transcription is likely multi-factorial, and could include activation of generic stress responses and/or response to antimicrobial compounds within the lake. The SOS response in particular has been associated with increased mutation rates within genes that can confer antimicrobial resistance, which includes the MLS 23S gene that was predominant within the metatranscriptome profile [32]. Despite precautions to preserve RNA, its inherent instability may have led to degradation during the workflow, potentially biasing our metatranscriptomic ARG profile [33].
Regarding the resistome, our results showed a high prevalence of MLS23S in both DNA and RNA shotgun sequencing datasets (Fig. 3). This dominance, previously described in other fish species, suggests MLS groups may be present in plasmids [22]. The MLS23S gene confers macrolide resistance through a 23S rRNA gene mutation, reported in many pathogens, including Escherichia coli, Helicobacter pylori, Mycobacterium sp., and Mycoplasma sp. [34]. Detecting mutation-based ARGs within ‘omics datasets is challenging due to the need for SNP verification, which we addressed with a bioinformatic SNP verification workflow [24]. Our study was not designed to identify the origin of detected ARGs such as the predominant MLS23S ARG, but previous studies have also reported a relatively high prevalence of MLS23S in samples from pristine environments [35].
Additionally, we observed the mcr gene group in both lake categories, with higher relative abundance in samples from undeveloped lakes (Fig. 3). This ARG group can be concerning due to its potential to confer resistance to colistin, an antimicrobial of last resort [36]; however, some variants are less clinically relevant than others, most notably mcr-9 [37]. First described as mcr-1 in E. coli isolates from human, chicken and swine in China in 2015 [38], variants mcr-2 through mcr-9 have since been found in isolates from humans, animals and the environment [39]. The mcr variants are also widely distributed within metagenomic datasets [40]. In our study, sequences matching variants mcr-3, mcr-7 and mcr-9 appeared in both RNA and DNA datasets, with mcr-3 being the most common. mcr-3, first reported in a human E. coli isolate [41], has been reported to be prevalent in water samples near human communities in South Africa [42] and is often reported within isolates of Aeromonas spp. [42], a relatively abundant taxon in our samples (Fig. 4B–C). mcr-9, more commonly reported than mcr-3, was the most frequent mcr variant reported in a recent analysis of >214,000 metagenomic samples [40]. Therefore, like mcr-3, its presence within our sample set is not unexpected. We were not able to find existing data regarding mcr prevalence within environmental or animal samples collected in Minnesota, but Martiny et al. reported that mcr-4 had a higher prevalence than other mcr variants in samples collected from Lake Huron [40].
The presence of antimicrobial compounds in MN lakes and ARGs in aquatic environments has been previously reported [4,14,15], and environmental antibiotics can contribute to the emergence and persistence of ARGs [8,11]. Previous research in MN lakes reported the macrolides azithromycin and roxithromycin in effluent-impacted and undeveloped lakes, respectively [14,15]. We found higher relative abundance of rpoB and parE, with known resistance-conferring mutations, in effluent-impacted lakes. These ARGs confer resistance to rifampicin and fluoroquinolones, respectively. Previous studies in MN lakes have detected fluoroquinolones, particularly ciprofloxacin, in undeveloped lakes and sarafloxacin in effluent-impacted lakes [14]. Further research is needed to integrate antimicrobial contaminant detection with the study of antimicrobial metabolites, AMR and microorganisms through integrated ‘omics analysis.
Future ‘omics based studies should carefully address challenges with samples of wild-caught fish, particularly preserving RNA for metatranscriptomic analysis [33]. We excluded numerous samples due to preservation issues (in particular, the use of liquid nitrogen within field conditions), and insufficient DNA and RNA yields. Many fish had little or no fecal matter in their GI tracts, worsening the low yield problem and resulting in high amounts of host DNA/RNA in our datasets. Future studies should consider use of protocols for low biomass samples and include negative and positive controls to assess contamination and sequencing performance [43,44]. For resistome studies, target-enrichment protocols can decrease host-associated nucleic acids while increasing the relative abundance of ARG-associated nucleic acids. However, these protocols bias the non-ARG content of the sample, thus precluding microbiome analyses [45].
4.2. Lake-level factors beyond anthropogenic pressure influence the intestinal microbiome and resistome of yellow perch
Few studies have characterized the metagenome of wild fish, with most using targeted 16S rRNA gene sequencing for microbiome analysis [46]. Even fewer studies focus on the microbiome of yellow perch [[17], [18], [19]]. Recent studies reported that cadmium exposure, a metal contaminant in wastewater, impacted the gut microbiome diversity of juvenile yellow perch [17,18]. Another study showed that dietary exposure to microplastics increased the phyla Proteobacteria and Bacteroidetes in the gut microbiome [19]. In our study, we found that richness and Shannon's diversity were higher in samples from undeveloped lakes, but this was statistically significant only at the phylum and genus levels from the RNA dataset. Beta diversity analysis revealed significant differences due to anthropogenic pressure and lake ID, with lake ID explaining most variation. This suggests that factors beyond anthropogenic pressures contribute to lake-to-lake variation in the yellow perch GI microbiome, including lake-specific microclimate, water composition, and dietary sources [47]. Environmental pollution, such as wastewater drainage into pristine environments, also shapes the gut microbiome of wild fish [48]. More research is needed to confirm the causal role of anthropogenic inputs in shaping the microbiome of yellow perch.
4.3. Fish from effluent-impacted lakes contained higher levels of bacteria of public health concern
In our study, the intestinal microbiome was dominated by Proteobacteria, Firmicutes and Actinobacteria, consistent with previous reports of the gut microbiome in teleost fish [49]. At the genus level, Plesiomonas, Aeromonas, and Clostridium were dominant in effluent-impacted lakes. Notably, P. shigelloides and A. veronii, associated with fish diseases [[50], [51], [52]] and potential zoonotic threats [53,54] were found in higher relative abundance in these samples as compared to samples from non-impacted lakes. Aeromonas, a proposed indicator of AMR in aquatic environments [55], was relatively highly abundant within the yellow perch gut microbiome in our study [12,56].
4.4. Opportunistic, poorly characterized, and unknown bacteria were recovered from intestinal samples of yellow perch
To date, no studies have reported MAGs recovered from yellow perch gut samples. While the genus Aeromonas was relatively abundant in our samples, we could not retrieve MAGs with sufficient completeness and redundancy. However, we retrieved MAGs from P. shigelloides in effluent-impacted lakes. This species was reported to be resistant to lincomycin and susceptible to sulfamethoxazole-trimethoprim and ciprofloxacin [57], and has been described as a carrier of ARGs for tetracycline and florfenicol in tilapia aquaculture in Brazil [58]. Our P. shigelloides MAG contained five plasmids related to secretion systems and one virus from the class Caudoviricetes.
Several of our retrieved MAGs have been previously described and isolated from natural water environments (Suppl. Table 2). Notably, the Paenibacillus alvei MAG contained the plasmid conjugation gene TcpC. The transfer clostridial plasmid (Tcp) conjugation system has been deeply characterized in the gram-positive pathogen Clostridium perfringens and is important for the spreading of virulence and resistance determinants [59]. Several MAGs in our samples harbored plasmids and viruses known to be involved in AMR. The ability to utilize non-reference-based methods such as MAGs is critical for robust metagenomic analysis, especially for underrepresented samples that tend to suffer from low classification rates based on current genomic databases. Future fish microbiome studies should consider a MAG approach to enhance the knowledge of underexplored fish-associated bacteria.
4.5. Limitations and future steps
A major limitation of our study was the small number of lakes, compounded by the significant variation in microbiome and resistome profiles linked to lake ID. Future studies should increase the number of lakes, particularly if examining lake-level factors such as anthropogenic pressures. Given the importance of lake ID in beta-diversity of our samples, we recommend that future studies also include more extensive metadata collection and analysis for a larger number of lakes; important metadata factors include weather patterns, water quality parameters, and levels of anthropogenic-origin compounds within the water. Another limitation of our study was the low relative abundance of ARGs and microbes within the RNA and DNA shotgun datasets. This aligns with reports that ARGs often represent less than <1 % of total sequence data, usually below <0.1 % [45]. Additionally, our microbiome analysis classification rates were very low, even compared to other understudied host species [60]. Future studies could improve these rates through molecular-based enrichment methods for resistome analysis [45] and agnostic bioinformatic methods for identifying novel microbial genomes, such as MAGs [22]. Finally, linking ARGs to bacterial hosts remains challenging with current metagenomic and metatranscriptomic data, as no robust bioinformatic methods exist for this analysis [61]. Long-read sequencing could help identify bacterial hosts of transcribed and non-transcribed ARGs, offering a more complete understanding of why certain ARGs are maintained and/or transcribed within the wild perch population, under varying environmental conditions.
5. Conclusions
This study characterizes the DNA- and RNA-level resistome and microbiome of yellow perch across northeastern Minnesota lakes under different anthropogenic pressures. We observed that fish from lakes impacted by wastewater effluent had distinct resistome and microbiome profiles compared to those living in undeveloped lakes, characterized by lower richness in both the microbiome and resistome at both the DNA and RNA levels. MLS23S genes dominated the resistome at both the DNA and RNA levels, and colistin resistance mcr genes were detected both in the DNA and RNA datasets, with higher detection in samples from undeveloped lakes. Metagenomic reconstruction revealed a relatively large number of un- or under-characterized bacterial genomes, highlighting the lack of representative genomes for many microbes found in wild fish samples. These findings emphasize the need for continued research to better understand the effects of human activity on antimicrobial resistance in aquatic environments, with broader implications for environmental and public health monitoring.
The following are the supplementary data related to this article.
A) Metadata from the samples used in the DNA shotgun dataset and B) metadata from the samples used in the RNA shotgun dataset.
MAGs with >75 % completion and < 10 % redundancy from different anthropogenic pressures.
Extended material and methods and supplementary figures. Detailed steps used during wet and dry lab procedures. Supplementary Fig. 1. Microbiome alpha diversity in the RNA shotgun data. Supplementary Fig. 2. Microbiome alpha diversity in the DNA shotgun data. Supplementary Fig. 3. Multidimensional scaling of semantic similarities of the GO terms enriched in A) Effluent-impacted lakes virome, B) undeveloped lakes virome, C) Effluent-impacted lakes plasmidome, D) undeveloped lakes plasmidome.
Supplementary File 2. BLASTn.MCRsearch.zip. Search strategies used with BLASTn to determine the specific ARG mcr.
Supplementary File 3. Anvio_reportsMAGs.zip Reports from assemblies generated with Anvi'o software for effluent-impacted and undeveloped lakes.
Funding
This work was supported by the Norwegian Centennial Chair Transatlantic Research Program and College of Veterinary Medicine, Signature Programs Funding Grant Number MN-62-120.
CRediT authorship contribution statement
Omar Jimenez-Lopez: Writing – original draft, Visualization, Validation, Software, Formal analysis, Data curation. Tui Ray: Writing – review & editing, Validation, Methodology, Investigation. Christopher Dean: Writing – review & editing, Investigation. Ilya Slizovskiy: Investigation. Jessica Deere: Writing – review & editing, Investigation. Tiffany Wolf: Writing – review & editing, Supervision, Methodology, Funding acquisition. Seth Moore: Writing – review & editing, Supervision, Resources, Investigation, Funding acquisition. Alexander Primus: Writing – review & editing, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization. Jennifer Høy-Petersen: Writing – review & editing, Investigation. Silje Finstad: Writing – review & editing, Investigation. Jakob Mo: Writing – review & editing, Investigation. Henning Sørum: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization. Noelle Noyes: Writing – review & editing, Validation, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
I have nothing to declare. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was completed in part with resources provided by the University of Minnesota Genomics Center and the Minnesota Supercomputing Institute (MSI) at the University of Minnesota. The research was funded by the Norwegian Centennial Chair at the University of Minnesota, and authors were funded by the College of Veterinary Medicine Signature Programs and the MnDrive Global Food Ventures Professional Development Program, both at the University of Minnesota.
Footnotes
This work was conducted in collaboration with the Grand Portage Band of Lake Superior Chippewa, an indigenous nation that proudly exercises rights to subsistence foods through inherent and treaty-recognized sovereignty. The Grand Portage Band recognizes that pharmaceuticals and legacy contaminants in the environment pose risks to ecosystem health and indigenous lifeways, thereby impacting food sovereignty, security and challenging resilience.
Data sharing
The raw sequence data for this study was uploaded to the Sequence Read Archive (SRA) under BioProject accession number PRJNA1141435.
References
- 1.D’Costa V.M., King C.E., Kalan L., Morar M., Sung W.W.L., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R., Golding G.B., Poinar H.N., Wright G.D. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.Abushaheen M.A., Muzaheed A.J., Fatani M., Alosaimi W., Mansy M., George S., Acharya S., Rathod D.D., Divakar C., Jhugroo S., Vellappally A.A., Khan J., Shaik P. Jhugroo. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month. 2020;66 doi: 10.1016/j.disamonth.2020.100971. [DOI] [PubMed] [Google Scholar]
- 3.Munk P., Brinch C., Møller F.D., Petersen T.N., Hendriksen R.S., Seyfarth A.M., Kjeldgaard J.S., Svendsen C.A., van Bunnik B., Berglund F., Larsson D.G.J., Koopmans M., Woolhouse M., Aarestrup F.M. Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat. Commun. 2022;13:7251. doi: 10.1038/s41467-022-34312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bueno I., Beaudoin A., Arnold W.A., Kim T., Frankson L.E., LaPara T.M., Kanankege K., Wammer K.H., Singer R.S. Quantifying and predicting antimicrobials and antimicrobial resistance genes in waterbodies through a holistic approach: a study in Minnesota, United States. Sci. Rep. 2021;11:18747. doi: 10.1038/s41598-021-98300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seiler C., Berendonk T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurenbach B., Marjoshi D., Amábile-Cuevas C.F., Ferguson G.C., Godsoe W., Gibson P., Heinemann J.A. Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. mBio. 2015;6 doi: 10.1128/mbio.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.I. Alav, M.M.C. Buckner, Non-antibiotic compounds associated with humans and the environment can promote horizontal transfer of antimicrobial resistance genes, Crit. Rev. Microbiol. (n.d.) 1–18. doi: 10.1080/1040841X.2023.2233603. [DOI] [PMC free article] [PubMed]
- 8.Xue X., Jia J., Yue X., Guan Y., Zhu L., Wang Z. River contamination shapes the microbiome and antibiotic resistance in sharpbelly (Hemiculter leucisculus) Environ. Pollut. 2021;268 doi: 10.1016/j.envpol.2020.115796. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Du S., Liu D., Dong D., Zhang W., Guo Z. Antibiotics in fish caught from ice-sealed waters: spatial and species variations, tissue distribution, bioaccumulation, and human health risk. Sci. Total Environ. 2022;821 doi: 10.1016/j.scitotenv.2022.153354. [DOI] [PubMed] [Google Scholar]
- 10.Ballash G.A., Baesu A., Lee S., Mills M.C., Mollenkopf D.F., Sullivan S.M.P., Lee J., Bayen S., Wittum T.E. Fish as sentinels of antimicrobial resistant bacteria, epidemic carbapenemase genes, and antibiotics in surface water. PLoS One. 2022;17 doi: 10.1371/journal.pone.0272806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi A., Singh B., Billekallu Thammegowda N.K., Singh N.K. Shotgun metagenomics offers novel insights into taxonomic compositions, metabolic pathways and antibiotic resistance genes in fish gut microbiome. Arch. Microbiol. 2019;201:295–303. doi: 10.1007/s00203-018-1615-y. [DOI] [PubMed] [Google Scholar]
- 12.Arnold K.E., Williams N.J., Bennett M. ‘Disperse abroad in the land’: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 2016;12:20160137. doi: 10.1098/rsbl.2016.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z.-C., Lin Z.-J., Shuai X.-Y., Zheng J., Meng L.-X., Zhu L., Sun Y.-J., Shang W.-C., Chen H. Temporal variation and sharing of antibiotic resistance genes between water and wild fish gut in a peri-urban river. J. Environ. Sci. 2021;103:12–19. doi: 10.1016/j.jes.2020.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Deere J.R., Streets S., Jankowski M.D., Ferrey M., Chenaux-Ibrahim Y., Convertino M., Isaac E.J., Phelps N.B.D., Primus A., Servadio J.L., Singer R.S., Travis D.A., Moore S., Wolf T.M. A chemical prioritization process: applications to contaminants of emerging concern in freshwater ecosystems (Phase I) Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.146030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deere J.R., Jankowski M.D., Primus A., Phelps N.B.D., Ferrey M., Borucinska J., Chenaux-Ibrahim Y., Isaac E.J., Singer R.S., Travis D.A., Moore S., Wolf T.M. Health of wild fish exposed to contaminants of emerging concern in freshwater ecosystems utilized by a Minnesota Tribal community. Integr. Environ. Assess. Manag. 2024;20:846–863. doi: 10.1002/ieam.4822. [DOI] [PubMed] [Google Scholar]
- 16.Bell S.P.M. Stults J., Baule W., Nasser E., Gibbons E., Fougerat M. Climate Change Vulnerability Assessment and Adaptation Plan: 1854 Ceded Territory Including the Bois Forte, Fond du Lac, and Grand Portage Reservations. 2016. https://scholar.google.com/scholar_lookup?title=Climate%20Change%20Vulnerability%20Assessment%20and%20Adaptation%20Plan%3A%201854%20Ceded%20Territory%20Including%20the%20Bois%20Forte%2C%20Fond%20du%20Lac%2C%20and%20Grand%20Portage%20Reservations&publication_year=2016&author=M.%20Stults&author=S.%20Petersen&author=J.%20Bell&author=W.%20Baule&author=E.%20Nasser&author=E.%20Gibbons&author=M.%20Fougerat
- 17.Cheaib B., Seghouani H., Ijaz U.Z., Derome N. Community recovery dynamics in yellow perch microbiome after gradual and constant metallic perturbations. Microbiome. 2020;8:1–19. doi: 10.1186/s40168-020-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheaib B., Seghouani H., Llewellyn M., Vandal-Lenghan K., Mercier P.-L., Derome N. The yellow perch (Perca flavescens) microbiome revealed resistance to colonisation mostly associated with neutralism driven by rare taxa under cadmium disturbance. Anim. Microbiome. 2021;3:1–19. doi: 10.1186/s42523-020-00063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X., Deng D.-F., Huang F., Casu F., Kraco E., Newton R.J., Zohn M., Teh S.J., Watson A.M., Shepherd B., Ma Y., Dawood M.A.O., Rios Mendoza L.M. Chronic exposure to high-density polyethylene microplastic through feeding alters the nutrient metabolism of juvenile yellow perch (Perca flavescens) Anim. Nutr. 2022;9:143–158. doi: 10.1016/j.aninu.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brumfield K.D., Huq A., Colwell R.R., Olds J.L., Leddy M.B. Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One. 2020;15 doi: 10.1371/journal.pone.0228899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.-M., Holmes E.C., Chen X., Tian J.-H., Lin X.-D., Qin X.-C., Gao W.-H., Liu J., Wu Z.-D., Zhang Y.-Z. Diverse and abundant resistome in terrestrial and aquatic vertebrates revealed by transcriptional analysis. Sci. Rep. 2020;10:18870. doi: 10.1038/s41598-020-75904-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan Y., Xue X., Jia J., Li X., Xing H., Wang Z. Metagenomic assembly and binning analyses the prevalence and spread of antibiotic resistome in water and fish gut microbiomes along an environmental gradient. J. Environ. Manag. 2022;318 doi: 10.1016/j.jenvman.2022.115521. [DOI] [PubMed] [Google Scholar]
- 23.Ray T., Gaire T.N., Dean C.J., Rowe S., Godden S.M., Noyes N.R. The microbiome of common bedding materials before and after use on commercial dairy farms. Anim. Microbiome. 2022;4:18. doi: 10.1186/s42523-022-00171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonin N., Doster E., Worley H., Pinnell L.J., Bravo J.E., Ferm P., Marini S., Prosperi M., Noyes N., Morley P.S., Boucher C. MEGARes and AMR++, v3.0: an updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 2023;51:D744–D752. doi: 10.1093/nar/gkac1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camargo A.P., Roux S., Schulz F., Babinski M., Xu Y., Hu B., Chain P.S.G., Nayfach S., Kyrpides N.C. Identification of mobile genetic elements with geNomad. Nat. Biotechnol. 2023:1–10. doi: 10.1038/s41587-023-01953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eren A.M., Sogin M.L., Morrison H.G., Vineis J.H., Fisher J.C., Newton R.J., McLellan S.L. A single genus in the gut microbiome reflects host preference and specificity. ISME J. 2015;9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang D.D., Li F., Kirton E., Thomas A., Egan R., An H., Wang Z. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7 doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Genome Standards Consortium, Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K., Schulz F., Jarett J., Rivers A.R., Eloe-Fadrosh E.A., Tringe S.G., Ivanova N.N., Copeland A., Clum A., Becraft E.D., Malmstrom R.R., Birren B., Podar M., Bork P., Weinstock G.M., Garrity G.M., Dodsworth J.A., Yooseph S., Sutton G., Glöckner F.O., Gilbert J.A., Nelson W.C., Hallam S.J., Jungbluth S.P., Ettema T.J.G., Tighe S., Konstantinidis K.T., Liu W.-T., Baker B.J., Rattei T., Eisen J.A., Hedlund B., McMahon K.D., Fierer N., Knight R., Finn R., Cochrane G., Karsch-Mizrachi I., Tyson G.W., Rinke C., Lapidus A., Meyer F., Yilmaz P., Parks D.H., Murat Eren A., Schriml L., Banfield J.F., Hugenholtz P., Woyke T. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017;35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashiardes S., Zilberman-Schapira G., Elinav E. Use of metatranscriptomics in microbiome research. Bioinform. Biol. Insights. 2016;10:BBI.S34610. doi: 10.4137/BBI.S34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosalbes M.J., Durbán A., Pignatelli M., Abellan J.J., Jiménez-Hernández N., Pérez-Cobas A.E., Latorre A., Moya A. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J.K. Crane, C.L. Alvarado, M.D. Sutton, Role of the SOS response in the generation of antibiotic resistance in vivo, Antimicrob. Agents Chemother. 65 (n.d.) e00013–21. doi: 10.1128/AAC.00013-21. [DOI] [PMC free article] [PubMed]
- 33.Reck M., Tomasch J., Deng Z., Jarek M., Husemann P., Wagner-Döbler I., On behalf of COMBACTE consortium Stool metatranscriptomics: a technical guideline for mRNA stabilisation and isolation. BMC Genomics. 2015;16:494. doi: 10.1186/s12864-015-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vester B., Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 2001;45:1–12. doi: 10.1128/aac.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provencher J., George P.B.L., Thaler M., Vincent W.F., Duchaine C., Culley A.I., Girard C. Microbial antibiotic resistance genes across an anthropogenic gradient in a Canadian High Arctic watershed. Sustain. Microbiol. 2024;1 doi: 10.1093/sumbio/qvae021. qvae021. [DOI] [Google Scholar]
- 36.Paterson D.L., Harris P.N.A. Colistin resistance: a major breach in our last line of defence. Lancet Infect. Dis. 2016;16:132–133. doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 37.Tyson G.H., Li C., Hsu C.-H., Ayers S., Borenstein S., Mukherjee S., Tran T.-T., McDermott P.F., Zhao S. The mcr-9 gene of Salmonella and Escherichia coli is not associated with Colistin resistance in the United States. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/aac.00573-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., Yu L.-F., Gu D., Ren H., Chen X., Lv L., He D., Zhou H., Liang Z., Liu J.-H., Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 39.Hussein N.H., AL-Kadmy I.M.S., Taha B.M., Hussein J.D. Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol. Biol. Rep. 2021;48:2897–2907. doi: 10.1007/s11033-021-06307-y. [DOI] [PubMed] [Google Scholar]
- 40.Martiny H.-M., Munk P., Brinch C., Szarvas J., Aarestrup F.M., Petersen T.N. Global distribution of mcr gene variants in 214K metagenomic samples. mSystems. 2022;7:e00105–e00122. doi: 10.1128/msystems.00105-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roer L., Hansen F., Stegger M., Sönksen U.W., Hasman H., Hammerum A.M. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 2017;22:30584. doi: 10.2807/1560-7917.ES.2017.22.31.30584. [DOI] [PubMed] [Google Scholar]
- 42.Snyman Y., Whitelaw A.C., Barnes J.M., Maloba M.R.B., Newton-Foot M. Characterisation of mobile colistin resistance genes (mcr-3 and mcr-5) in river and storm water in regions of the Western cape of South Africa. Antimicrob. Resist. Infect. Control. 2021;10:96. doi: 10.1186/s13756-021-00963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean C.J., Deng Y., Wehri T.C., Pena-Mosca F., Ray T., Crooker B.A., Godden S.M., Caixeta L.S., Noyes N.R. The impact of kit, environment, and sampling contamination on the observed microbiome of bovine milk. mSystems. 2024 doi: 10.1128/msystems.01158-23. e01158–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clokie B.G.J., Elsheshtawy A., Albalat A., Nylund A., Beveridge A., Payne C.J., MacKenzie S. Optimization of low-biomass sample collection and quantitative PCR-based titration impact 16S rRNA microbiome resolution. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02255-22. e02255–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noyes N.R., Weinroth M.E., Parker J.K., Dean C.J., Lakin S.M., Raymond R.A., Rovira P., Doster E., Abdo Z., Martin J.N., Jones K.L., Ruiz J., Boucher C.A., Belk K.E., Morley P.S. Enrichment allows identification of diverse, rare elements in metagenomic resistome-virulome sequencing. Microbiome. 2017;5:142. doi: 10.1186/s40168-017-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legrand T.P.R.A., Wynne J.W., Weyrich L.S., Oxley A.P.A. A microbial sea of possibilities: current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquac. 2020;12:1101–1134. doi: 10.1111/raq.12375. [DOI] [Google Scholar]
- 47.Kashinskaya E.N., Simonov E.P., Kabilov M.R., Izvekova G.I., Andree K.B., Solovyev M.M. Diet and other environmental factors shape the bacterial communities of fish gut in an eutrophic lake. J. Appl. Microbiol. 2018;125:1626–1641. doi: 10.1111/jam.14064. [DOI] [PubMed] [Google Scholar]
- 48.Evariste L., Barret M., Mottier A., Mouchet F., Gauthier L., Pinelli E. Gut microbiota of aquatic organisms: a key endpoint for ecotoxicological studies. Environ. Pollut. 2019;248:989–999. doi: 10.1016/j.envpol.2019.02.101. [DOI] [PubMed] [Google Scholar]
- 49.Llewellyn M.S., Boutin S., Hoseinifar S.H., Derome N. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00207. https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2014.00207 (accessed February 22, 2024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khurana H., Singh D.N., Singh A., Singh Y., Lal R., Negi R.K. Gut microbiome of endangered Tor putitora (Ham.) as a reservoir of antibiotic resistance genes and pathogens associated with fish health. BMC Microbiol. 2020;20:249. doi: 10.1186/s12866-020-01911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sreedharan K., Philip R., Singh I.S.B. Isolation and characterization of virulent Aeromonas veronii from ascitic fluid of oscar Astronotus ocellatus showing signs of infectious dropsy. Dis. Aquat. Org. 2011;94:29–39. doi: 10.3354/dao02304. [DOI] [PubMed] [Google Scholar]
- 52.Dong H.T., Techatanakitarnan C., Jindakittikul P., Thaiprayoon A., Taengphu S., Charoensapsri W., Khunrae P., Rattanarojpong T., Senapin S. Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.) J. Fish Dis. 2017;40:1395–1403. doi: 10.1111/jfd.12617. [DOI] [PubMed] [Google Scholar]
- 53.Li T., Raza S.H.A., Yang B., Sun Y., Wang G., Sun W., Qian A., Wang C., Kang Y., Shan X. Aeromonas veronii infection in commercial freshwater fish: a potential threat to public health. Animals. 2020;10:608. doi: 10.3390/ani10040608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janda J.M., Abbott S.L., McIver C.J. Plesiomonas shigelloides revisited. Clin. Microbiol. Rev. 2020;29:349–374. doi: 10.1128/cmr.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grilo M.L., Sousa-Santos C., Robalo J., Oliveira M. The potential of Aeromonas spp. from wildlife as antimicrobial resistance indicators in aquatic environments. Ecol. Indic. 2020;115 doi: 10.1016/j.ecolind.2020.106396. [DOI] [Google Scholar]
- 56.Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duman M., García Valdés E., Ay H., Altun S., Saticioglu I.B. Description of a novel fish pathogen, Plesiomonas shigelloides subsp. oncorhynchi, Isolated from Rainbow Trout (Oncorhynchus mykiss): first genome analysis and comparative genomics. Fishes. 2023;8:179. doi: 10.3390/fishes8040179. [DOI] [Google Scholar]
- 58.Martins A.F.M., Pinheiro T.L., Imperatori A., Freire S.M., Sá-Freire L., Moreira B.M., Bonelli R.R. Plesiomonas shigelloides: a notable carrier of acquired antimicrobial resistance in small aquaculture farms. Aquaculture. 2019;500:514–520. doi: 10.1016/j.aquaculture.2018.10.040. [DOI] [Google Scholar]
- 59.Wisniewski J.A., Rood J.I. The Tcp conjugation system of Clostridium perfringens. Plasmid. 2017;91:28–36. doi: 10.1016/j.plasmid.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Riiser E.S., Haverkamp T.H.A., Varadharajan S., Borgan Ø., Jakobsen K.S., Jentoft S., Star B. Metagenomic shotgun analyses reveal complex patterns of intra- and interspecific variation in the intestinal microbiomes of codfishes. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.02788-19. e02788–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slizovskiy I.B., Mukherjee K., Dean C.J., Boucher C., Noyes N.R. Mobilization of antibiotic resistance: are current approaches for colocalizing resistomes and mobilomes useful? Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01376. https://www.frontiersin.org/articles/10.3389/fmicb.2020.01376 (accessed December 3, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Metadata from the samples used in the DNA shotgun dataset and B) metadata from the samples used in the RNA shotgun dataset.
MAGs with >75 % completion and < 10 % redundancy from different anthropogenic pressures.
Extended material and methods and supplementary figures. Detailed steps used during wet and dry lab procedures. Supplementary Fig. 1. Microbiome alpha diversity in the RNA shotgun data. Supplementary Fig. 2. Microbiome alpha diversity in the DNA shotgun data. Supplementary Fig. 3. Multidimensional scaling of semantic similarities of the GO terms enriched in A) Effluent-impacted lakes virome, B) undeveloped lakes virome, C) Effluent-impacted lakes plasmidome, D) undeveloped lakes plasmidome.
Supplementary File 2. BLASTn.MCRsearch.zip. Search strategies used with BLASTn to determine the specific ARG mcr.
Supplementary File 3. Anvio_reportsMAGs.zip Reports from assemblies generated with Anvi'o software for effluent-impacted and undeveloped lakes.
Data Availability Statement
The raw sequence data for this study was uploaded to the Sequence Read Archive (SRA) under BioProject accession number PRJNA1141435.