Abstract
The hypothalamus-pituitary-gonadal (HPG) axis is an important neuroendocrine regulatory center involved in egg-laying process in poultry. However, its mechanism of regulating broodiness behavior and laying performance in geese remains unclear. This study explored the molecular mechanism by which the HPG axis regulates brooding behavior in Wanxi white geese (WWG). The hypothalamus, pituitary, and ovarian tissues of Wanxi white geese were collected at laying and brooding periods for transcriptome sequencing analysis. A total of 240 (BH vs. LH), 319 (BP vs. LP), and 445 (BO vs. LO) differentially expressed genes, and 56 (BH vs. LH), 82 (BP vs. LP), and 48 (BO vs. LO) differentially expressed miRNAs were identified. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis showed that differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) were significantly enriched in hormone level regulation, cell communication, calcium signaling pathway, GnRH signaling pathway, MAPK signaling pathway, Wnt signaling pathway, and other processes. Six DEGs and four DEMs were randomly selected for real-time fluorescence quantitative reverse transcription PCR (RT-qPCR). The results showed that the transcriptome sequencing data were accurate and reliable. In addition, 22 potential hub miRNAs were screened. Dual luciferase reporter assays confirmed the targeting relationship between miR-144-y and DIO3. The results showed that the miRNAs mainly regulated the laying performance and brooding behavior of WWG by mediating the expression of target genes. In this study, we systematically elucidated the mechanisms by which the HPG axis regulates the broodiness behavior and laying performance of WWG at the post-transcriptional level. Several miRNAs and mRNAs associated with the reproductive performance of WWG were identified, providing a crucial reference for the subsequent use of gene editing technologies to breed new varieties and advance the development of WWG breeding industry.
Keywords: Wanxi white goose, Hypothalamus-pituitary-gonadal axis, miRNA, Broodiness behavior, Laying performance
Introduction
Geese are seasonal breeding poultry with short breeding period and strong broodiness (Yao et al., 2019). Broodiness is a unique behavioral trait commonly observed in poultry reproduction and is mainly affected by environmental, endocrinal, and genetic factors. During the brooding process, the ovarian steroid production and cell proliferation and apoptosis processes are reduced in female poultry. This is accompanied by atretic follicles and atrophic ovaries and fallopian tubes (Liu et al., 2018; Yin et al., 2019; Shen et al., 2024), resulting in reduced egg production in geese. Poultry reproductive traits are mainly regulated by the central nervous system (Nakane and Yoshimura, 2019), which is highly dependent on the synthesis and release of sex hormones. As the main branch of the endocrine system, the hypothalamic-pituitary-gonadal (HPG) axis directly controls the secretion of sex hormones and is the basis of reproductive endocrine control in poultry (Guchhait et al., 2018). The hypothalamus is located above the HPG axis and stimulates the anterior pituitary gland to secrete gonadotropin via pulsed secretion of gonadotropin-releasing hormone (GnRH). It also acts on the ovary to secrete estrogen (Sanchez and De Jesus, 2024), facilitating egg production in female poultry. Multiple hormones and neurotransmitters produced by the HPG axis also collectively regulate broodiness in various poultry, including Wanxi white geese (WWG). The hypothalamus releases GnRH and stimulates the secretion of prolactin (PRL), whose levels increase with the degradation of reproductive functions (Pan et al., 2022). If GnRH secretion exceeds a critical threshold, it triggers a negative feedback mechanism that suppresses the activity of GnRH neurons, luteinizing hormone release, and gonadotropin secretion, ultimately impairing follicular development (Grachev et al., 2015). Therefore, an in-depth exploration of the regulatory mechanism of the HPG axis could aid in revealing the factors underlying broodiness and improve the laying performance in WWG.
As an important part of functional genomics studies, transcriptome sequencing technology has been widely used in poultry transcriptome analysis (Gao et al., 2023). Many genes that may affect broodiness have been found in the HPG axis of geese. Through screening the differentially expressed genes (DEGs) in the hypothalamus of Huoyan geese during the laying and brooding periods, it was found that GnRH-related genes Adiponectin Receptor 2 (AdipoR2), Neuregulin 1 (Nrg1), and Neural Cell Adhesion Molecule 1 (NCAM1) may regulate broodiness in geese (Luan et al., 2014). miRNAs are a class of endogenous small non-coding RNA molecules consisting of about 21-25 nucleotides. MiRNAs regulate gene expression by binding to the 3′ or 5′ untranslated regions (UTRs) of the target mRNAs, leading to degradation or translation inhibition of the mRNAs, thereby affecting broodiness and egg-laying performance in poultry. G-miR-146 and G-miR-143* were found to regulate broodiness in poultry (Chen et al., 2014). Moreover, miR-21 (Zhang et al., 2017), miR-205 (Zhang et al., 2019), G-miR-34b-5p, miR-6006, and G-miR-1620 (Liu et al., 2018) are reportedly differentially expressed in the hypothalamus, ovary, and follicles of poultry during the brooding and laying periods. Although several candidate genes and pathways related to broodiness have been screened at the transcriptome level in recent years, studies on the molecular regulatory mechanism of broodiness in WWG are still scarce. Therefore, the interaction between candidate genes, the miRNA-mediated regulation of genes, and the specific molecular mechanism regulating broodiness in WWG are still unclear.
Geese breeding industry plays an important role in China's agricultural and economic development (Akhtar et al., 2021). As one of the high-quality poultry breeding resources in China, WWG exhibits rapid growth, high slaughter rate, and superior meat quality during the early growth stage, with good economic benefits. However, the broodiness behavior causes reduced annual egg production of WWG and obvious reproductive cycle segmentation (broodiness period, laying period) (Wang et al., 2021), which seriously affects the reproductive performance and development of the WWG production industry. By exploring the differentially expressed mRNA and miRNA in WWG at various breeding stages and constructing an mRNA-miRNA interaction network, a combined analysis of mRNA and miRNA can offer a more comprehensive understanding of the mechanisms by which the HPG axis regulates laying performance and brooding behavior in WWG. In this study, the expression profiles of miRNAs and mRNAs in the HPG axis of WWG were explored at laying and brooding periods using RNA-seq technology. The key genes of the HPG axis regulating the broodiness of geese at laying and brooding periods were also screened. Moreover, the miRNA-mRNA interaction network was constructed to understand the molecular regulatory mechanism of WWG broodiness. The results of this study provide a reference for further studies on the miRNA and mRNA related to WWG broodiness and their potential relationship and functional role in reproductive regulation. This could aid in improving the broodiness traits and egg production performance of WWG, thus promoting the development of the geese breeding industry.
Material and methods
Sample collection
The geese used in this study were obtained from Dingyuan Junming Ecological Farm. (Dingyuan, Anhui Province, China) and were reared under standardized feeding management practices in a controlled environment. Based on the laying and brooding behavior of WWG, geese were selected and divided into two groups: laying period and brooding period. Nine laying period geese (n = 9; 3-year-old; 3.2 kg ± 0.2 kg; female) and nine brooding period geese (n = 9; 3-year-old; 3.2 kg ± 0.2 kg; female) were randomly selected and reared separately. Next, three geese from each group were randomly chosen for sample collection. All geese were euthanised via cervical dislocation, and the hypothalamus, pituitary gland, and ovaries were immediately excised and rinsed with phosphate-buffered saline (PBS) buffer. The samples were snap-frozen in liquid nitrogen and stored at -80°C for total RNA extraction. The samples from the brooding period hypothalamus (BH) group were labelled BH1, BH2, and BH3; those from the brooding period pituitary (BP) group were labelled BP1, BP2, and BP3; and those from the brooding period ovary (BO) group were labelled BO1, BO2, and BO3. Similarly, samples from the laying period hypothalamus (LH) group were labelled LH1, LH2, and LH3; those from the laying period pituitary (LP) group were labelled LP1, LP2, and LP3; and those from the laying period ovary (LO) group were labelled LO1, LO2, and LO3. All sequencing libraries were constructed and sequenced by the Gene Denovo Biotechnology Co., Ltd (Guangzhou, China). The specific steps were as follows: Total RNA was extracted using the Trizol kit (Invitrogen, USA) according to the manufacturer's instructions, and its quality was assessed with the Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, USA). The RNA was then purified and repaired using the QiaQuick PCR kit (QIAGEN, China). Transcriptome sequencing libraries were prepared using the Illumina TruSeq kit (Illumina, USA) and sequenced on the Illumina HiSeqTM 4000 (Illumina, USA).
Preparation of ovarian tissue sections
Tissue samples were fixed in 4 % paraformaldehyde for 6-8 h, rinsed with running water for 1 h, and then dehydrated in ethanol solutions with varying concentrations (70 %, 80 %, 85 %, 90 %, 95 %, and 100 %) for 30 mins each. The dehydrated samples were subsequently embedded in paraffin. The embedded samples were sectioned into 4 μm slices and stained with hematoxylin solution (Beyotime, China) for 4 min, followed by eosin staining for 3 min. The stained sections were then dehydrated in ethanol, cleared in xylene for transparency, and finally mounted with resin.
Transcriptome data analysis
To ensure the reliability and accuracy of the data, we first processed raw reads, which entailed removing the low-quality reads, trimming the splice sequences, and eliminating the poor quality bases (Xu et al., 2023). The sequences were then filtered using Seqtk (https://github.com/lh3/seqtk) to get the clean reads (Lin et al., 2022). The filtered clean reads were compared with the reference genome (NCBI_GCF_002166845.1) using Hisat2 (v2.0.4) (Kim et al., 2015) software to identify DEGs and quantify gene expression in the samples. Transcripts were assembled using StringTie (v1.3.5) (Kovaka et al., 2019), and the expression levels of the transcripts were determined using the Fragments per Kb per Million Fragments (FPKM) method. The differential fold change (FC) was calculated based on the threshold parameters of |log2FC| > 1 and P < 0.05 for the DEGs (Hu et al., 2020).
MiRNA data analysis
After quality control of raw sequencing data, miRNA identification and quantification were performed to identify known miRNAs and predict new miRNAs. MiRNA expression of different samples was compared using Transcription per million (TPM) (Zhao et al., 2020) to identify the differentially expressed miRNAs (DEMs). DEMs were identified using the DEseq2 package (Anders and Huber, 2010), with thresholds set at |log2FC| > 0.26 and P < 0.05. Since DEMs may affect physiological processes by regulating specific target genes, target gene prediction was conducted via two methods, namely Miranda (v3.3.a) and TargetScan (v7.0) (Kuijjer et al., 2020). The genes predicted by both methods are considered to be miRNA target genes.
GO and KEGG enrichment analysis
DEGs and the target genes of the DEMs were analysed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using ClusterProfiler (v3.10.1) (Guangchuang et al., 2012) and KOBAS (v2.0) (Chen et al., 2011). The P < 0.05 values indicated significantly enriched, while P < 0.01 indicated highly significantly enriched GO terms and KEGG pathways.
Combined MiRNA-MRNA analysis
To investigate the interactions between DEGs and the miRNA-mediated regulation of genes, we performed a comprehensive analysis of DEMs and their target genes elucidate the miRNA-mRNA interaction. The miRNA-mRNA networks were constructed using Cytoscape (v3.8.0) (Shannon et al., 2003) software.
Validation by real-time fluorescence quantitative reverse transcription PCR (RT- qPCR)
Primers (Supplementary Table S1) were designed using Oligo 7 and miRNA Design software and synthesised by Sangon Biotech Co., Ltd (Shanghai, China). GAPDH and U6 were used as the internal reference genes for mRNAs and miRNAs, respectively, and six DEGs and four DEMs were randomly selected to verify the sequencing results (Li et al., 2014). The mRNAs were quantified using SYBR qPCR Master Mix (EZBioscience, USA), while miRNAs were quantified using miRNA Universal miRNA SYBR qPCR Master Mix (Vazyme, Nanjing, China). The reaction conditions for both mRNAs and miRNA were 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s.
Dual luciferase detection
The DIO3-3′ UTR-WT and DIO3-3′ UTR-MUT plasmids were designed using the vector psiCHECK2 (Promega, USA). 293T cells (Procell, CA) were seeded in a 24-well plate and cultured for 24 h until they reached 80 % confluency. The cells were transfected with Lip3000 (Invitrogen, USA) using 1 ug DNA and 15 pmol of miRNA for co-transfection. After 48 h, the cells were lysed and centrifuged at 12000 × g at 4°C for 10 min. Subsequently, 20 μl of the supernatant was mixed with the firefly luciferase and renal luciferase detection reagents, and luciferase activities were measured at wavelengths of 560 nm for firefly luciferase and 465 nm for renal luciferase activity.
Statistical analyses
The relative expression levels of DEGs and DEMs were normalized to the internal reference genes GAPDH and U6 using the 2-ΔΔCT method (Bustin et al., 2009). All experimental data are expressed as mean ± standard error of the mean (SEM), and statistical significance was analyzed using one-way ANOVA and independent sample t test with SPSS 27.0 software, where P < 0.05 was considered significant. Visual statistical analysis was performed using GraphPad Prism 8 (v8.2.1) and Cytoscape (v3.8.2).
Results
Morphological analysis of ovarian tissue of WWG in different breeding periods
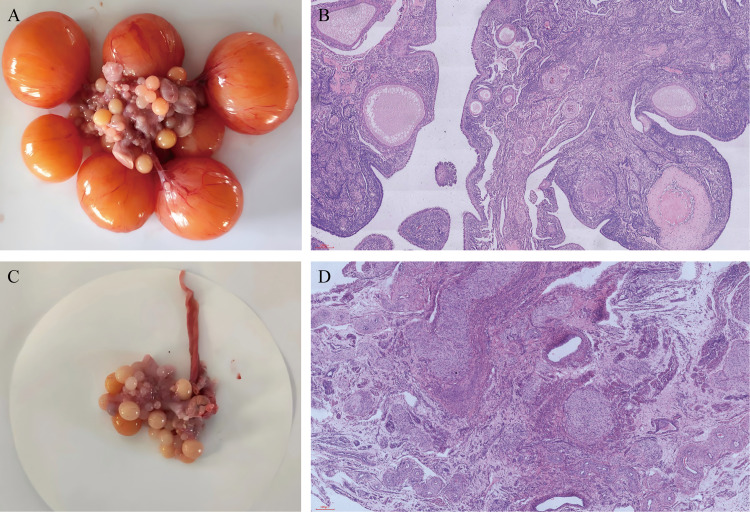
Numerous graded follicles were detected in WWG ovaries during the laying period (Fig. 1 A and B). However, in the WWG ovaries, a significant number of follicles were blocked, indicating a decline in ovarian function (Fig. 1C and D).
Fig. 1.
Morphological changes in the ovaries of WWG during different breeding periods. (A) Ovarian tissue during the laying period. (B)The results of hematoxylin-eosin staining of ovarian tissue during the laying period. (C) Ovarian tissue during the brooding period. (D)The results of hematoxylin-eosin staining of ovarian tissue during the brooding period.
Quality control of sequencing data
We obtained 1,604,901,192 raw sequences from 18 samples. After filtering the data, 1,598,999,134 valid sequences were obtained. The Q20 and Q30 of each sample were above 97 % and 93 %, respectively, and the GC base contents were relatively the same, with stable and balanced base composition. The comparison results showed that more than 83 % of the valid sequences could align with the reference genome of geese (Supplementary Table S2). These results indicated that the quality of the sequencing data was good and could be used for further analysis.
Analysis of DEGs
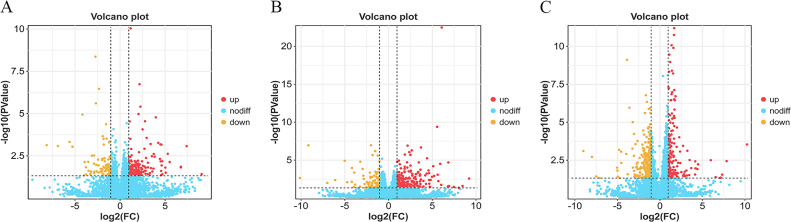
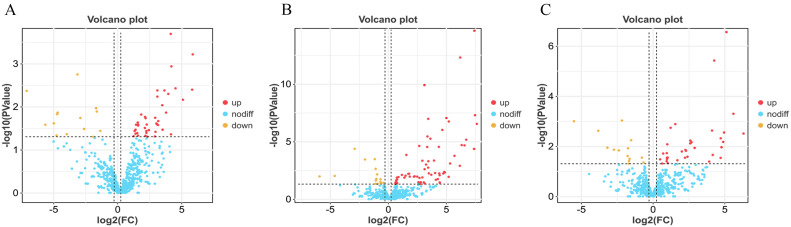
To accurately determine the differences in gene expression between groups, we generated volcano plots to visually observe the DEGs between the two groups. The BH vs. LH comparison group had 152 up-regulated and 88 down-regulated genes (Fig. 2A, Supplementary Table S3), while the BP vs. LP had 239 up-regulated and 80 down-regulated genes (Fig. 2B, Supplementary Table S4). Moreover, there were 161 up-regulated and 284 down-regulated genes in the BO vs. LO comparison group (Fig. 2C, Supplementary Table S5).
Fig. 2.
Distribution map of the number of DEGs between the different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Red indicates up-regulation, while yellow indicates down-regulation.
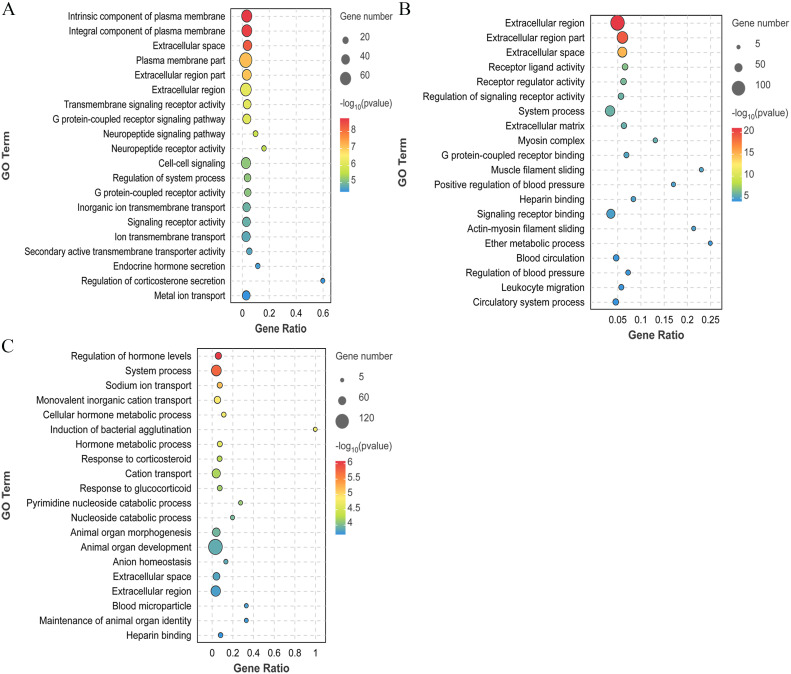
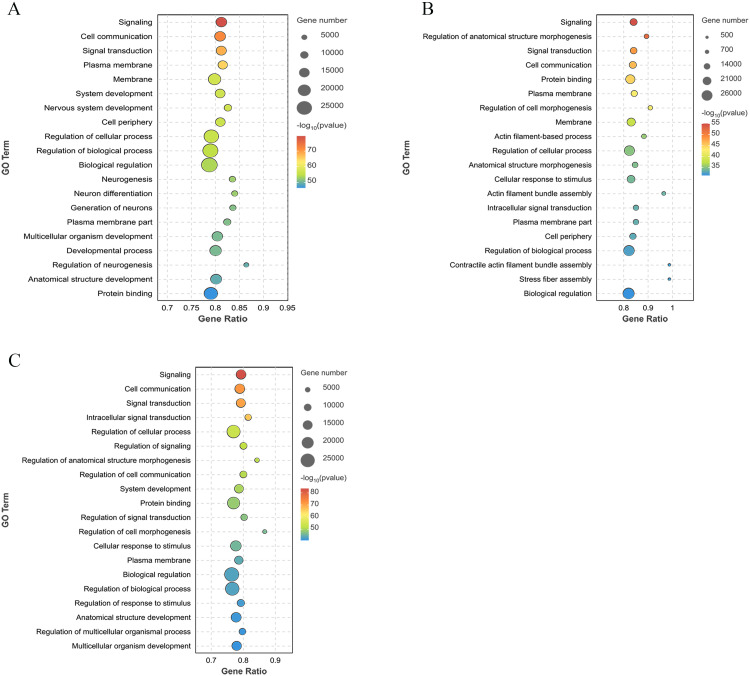
GO functional enrichment analysis of the DEGs
GO enrichment analysis was performed on the selected DEGs. In the BH vs. LH group, the DEGs were significantly enriched in 142 GO entries (P < 0.05), mainly the G protein-coupled receptor signaling pathway, cell-cell signaling, endocrine hormone secretion, and regulation of corticosterone secretion (Fig. 3A). For the BP vs. LP group, the DEGs were significantly enriched in 54 GO entries (P < 0.05), mainly receptor ligand activity, receptor activity regulation, myosin complex, and G protein-coupled receptor binding (Fig. 3B). The BO vs. LO group exhibited significant enrichment of the DEGs in 20 GO entries (P < 0.05), including hormone level regulation, ion transport, nucleoside catabolism, and animal organ development (Fig. 3C).
Fig. 3.
GO enrichment analysis plot of DEGs between different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Bubble represents the number of genes, while the color indicates the P value.
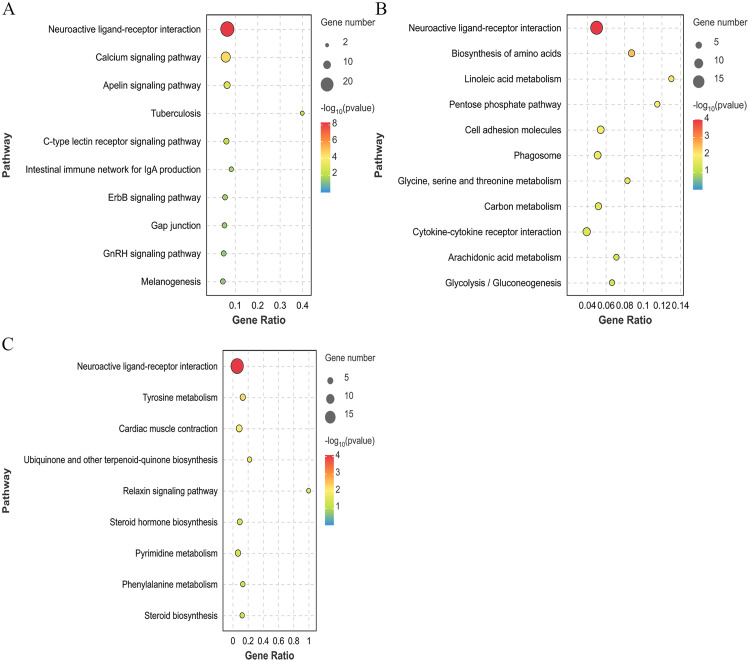
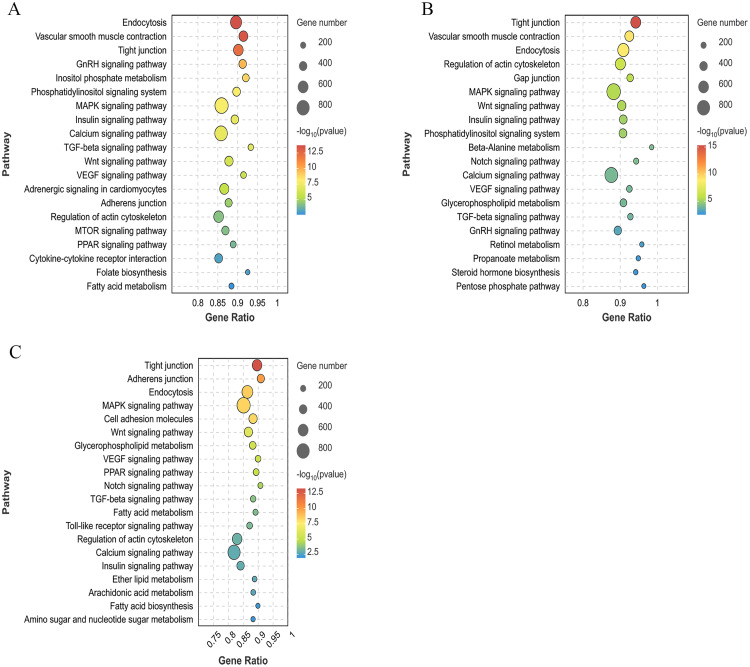
KEGG enrichment analysis of the DEGs
Through KEGG enrichment analysis, we identified 70 DEGs in the BH vs. LH group, which significantly were enriched in 10 signaling pathways (P < 0.05), including calcium signaling, Apelin signaling, and GnRH signaling pathways (Fig. 4A). There were 80 DEGs in the BP vs. LP group, which were significantly enriched in 11 signal pathways (P < 0.05), including amino acid biosynthesis, linoleic acid metabolism, and carbon metabolism pathways (Fig. 4B). Moreover, 104 DEGs in the BO vs. CL group were significantly enriched in 9 signal pathways (P < 0.05), including tyrosine metabolism, steroid hormone biosynthesis, and pyrimidine metabolism pathways, among others (Fig. 4C).
Fig. 4.
KEGG enrichment analysis plot of DEGs among different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Bubble represents the number of genes, while the color represents the P value.
Analysis of the DEMs
To accurately determine the differences in miRNA expression between groups, we generated a volcano plot for visual evaluation. There were 56 DEMs (42 up-regulated and 14 down-regulated) in the BH vs. LH group (Fig. 5A, Supplementary Table S6) and 82 DEMs (67 up-regulated and 15 down-regulated) BP vs. LP group (Fig. 5B, Supplementary Table S7). In the BO vs. LO group, 34 DEMS were up-regulated, and 14 were down-regulated (Fig. 5C, Supplementary Table S8).
Fig. 5.
Distribution map of the number of DEMs between different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Red indicates up-regulation, while yellow indicates down-regulation.
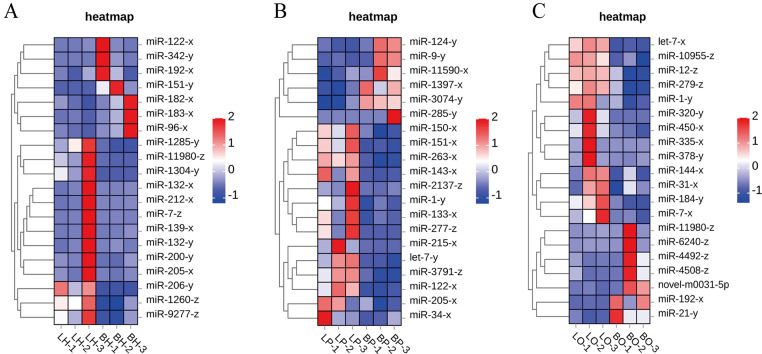
Cluster plot of the DEMs
Cluster analysis of DEMs was conducted across different groups (Fig. 6), and the heatmap further revealed the differential expression patterns of miRNAs associated with the HPG axis in WWG during the laying and brooding periods.
Fig. 6.
Cluster plot of DEMs among different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Red indicates up-regulation, while blue indicates down-regulation.
GO functional enrichment analysis of the DEMs
In the BH vs. LH group, the target genes of the DEMs were significantly enriched in the signaling, nervous system development, and biological regulation processes (P < 0.01, Fig. 7A). However, in the BP vs. LP group, the target genes of the DEMs were significantly enriched in protein binding, cellular process regulation, and cellular response to stimuli processes, among others (P < 0.01, Fig. 7B). The target genes of the DEMs were also significantly enriched in the protein binding, biological regulation, and multicellular biosynthesis processes in the BO vs. CL groups (P < 0.01, Fig. 7C). The results showed that most of the DEMs were related to cellular components and biological regulatory processes.
Fig. 7.
GO enrichment analysis of DEMs target genes between different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Bubble represents the number of genes, while the color represents the P value.
KEGG enrichment analysis of the DEMs
The KEGG enrichment analysis results showed that the target genes of the DEMs in the BH vs. LH group were significantly enriched in the GnRH signaling pathway, inositol phosphate metabolism, MAPK signaling pathway, cytokine-cytokine receptor interaction, and other pathways (P < 0.01, Fig. 8A). In the BP vs. LP group, the target genes of the DEMs were significantly enriched in the pathways such as gap junction, Wnt signaling, Notch signaling, and steroid hormone biosynthesis pathways (P < 0.01, Fig. 8B). Additionally, the target genes of the DEMs in the BO vs. CL group were significantly enriched in the glycerophospholipid metabolism, VEGF signaling, PPAR signaling, TGF-β signaling, and other pathways (P < 0.01, Fig. 8C).
Fig. 8.
KEGG enrichment analysis of DEMs target genes between different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Bubble represents the number of genes, while the color represents the P value.
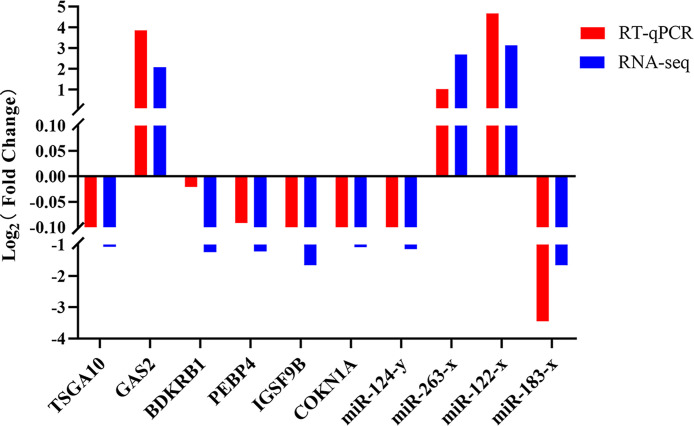
Sequencing validation by RT-qPCR
To evaluate the accuracy of sequencing data, we randomly selected 10 DEGs and DEMs (IGSF9B, CDKN1A, TSGA10, PEBP4, BDKRB1, GAS2, miR-124-y, miR-263-x, miR-122-x, and miR-183-x) for RT-qPCR analysis, with each assay repeated three times. The internal reference gene demonstrated stable expression across multiple samples. The results showed that the expression trends of DEGs and DEMs in different groups of sample tissues were consistent with those of transcriptome sequencing data, indicating that the RNA-seq data were reliable (Fig. 9).
Fig. 9.
RT-qPCR validation of DEGs and DEMs. Red represents RT-qPCR results, while blue represents RNA-seq results.
Construction of the MiRNA-MRNA interaction networks
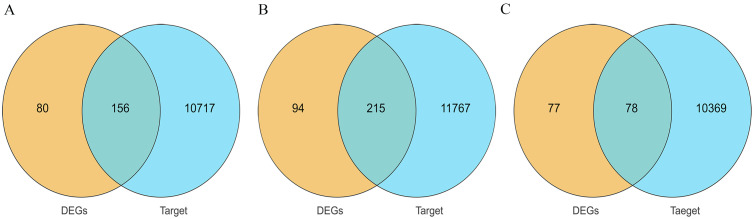
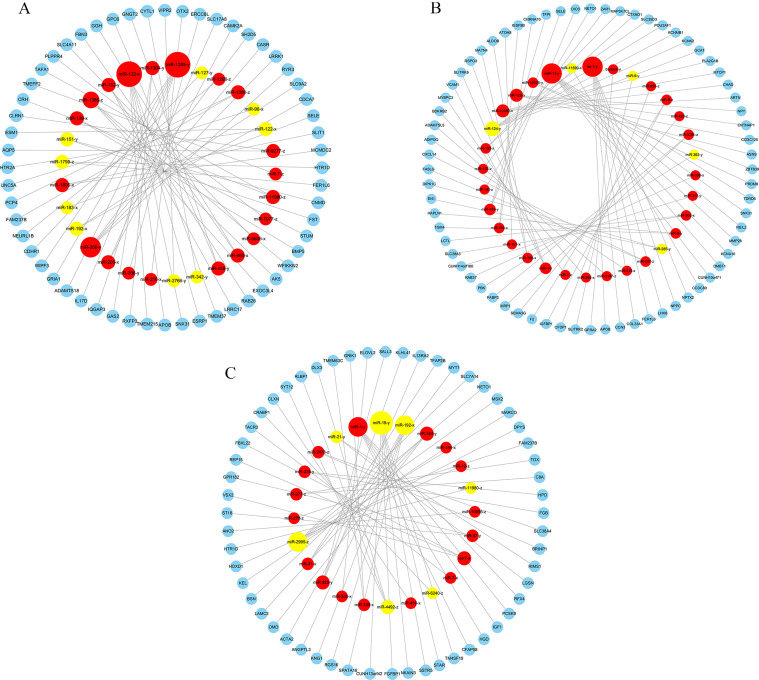
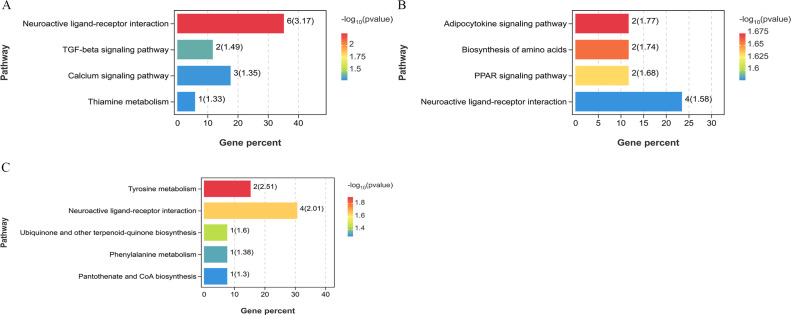
To further analyze the relationship between miRNA and mRNA, we used the screened DEGs and DEMs to construct an interactive co-expression network. In the BH vs. LH group, 10,873 target genes were predicted to intersect with 236 DEGs, and 156 genes were obtained for subsequent analysis (Fig. 10A). For the BP vs. LP group, 11982 target genes were predicted to intersect with 309 DEGs, and 215 genes were obtained for subsequent analysis (Fig. 10B). Similarly, out of 10,477 target genes predicted to intersect with 155 DEGs in the BO vs. LO group, 78 genes were obtained for subsequent analysis (Fig. 10C). By increasing the screening threshold (|log2FC| >1.58, P < 0.05) and excluding genes that are not officially named and cannot be fully verified experimentally, we constructed the mRNA-miRNA interaction network. The miRNA-mRNA interaction network in the BH vs. LH group consisted of 85 nodes and 56 edges, and the nodes contained 29 miRNAs and 56 mRNAs (Fig. 11A). For the BP vs. LP group, the miRNA-mRNA interaction network consisted of 100 nodes and 68 edges, and the nodes had 32 miRNAs and 68 mRNAs (Fig. 11B). Moreover, the miRNA-mRNA interaction network in the BO vs. LO group consisted of 80 nodes and 56 edges, and the nodes comprised 24 miRNAs and 56 mRNAs (Fig. 11C). In the interaction network, the nodes represent mRNA and miRNA, while the edges between the nodes indicate the functional interaction between them. KEGG functional enrichment analysis of the target genes of DEMs in the BH vs. LH group showed that the target genes were significantly enriched in neuroactive ligand-receptor interaction, TGF-β signaling pathway, calcium signaling pathway, and other pathways (P < 0.05, Fig. 12A). In the BP vs. LP group, the target genes of DEMs were significantly enriched in amino acid biosynthesis, PPAR signaling pathway, neuroactive ligand-receptor interaction pathways (P < 0.05, Fig. 12B). Additionally, the target genes of DEMs in the BO vs. LO group were significantly enriched in pathways such as tyrosine metabolism, neuroactive ligand-receptor interaction, and phenylalanine metabolism pathways (P < 0.05, Fig. 12C).
Fig. 10.
Intersecting genes between DEGs and predicted target genes of DEMs. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Yellow represents DEGs, blue represents the target gene of DEMs, and the intersection represents the common genes between the two.
Fig. 11.
The miRNA-mRNA interaction network between different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. Blue represents DEGs, red represents up-regulated DEMs, and yellow represents down-regulated DEMs.
Fig. 12.
The KEGG enrichment analysis diagram of the interaction network between different groups. (A) BH vs. LH group. (B) BP vs. LP group. (C) BO vs. LO group. The color represents the P value.
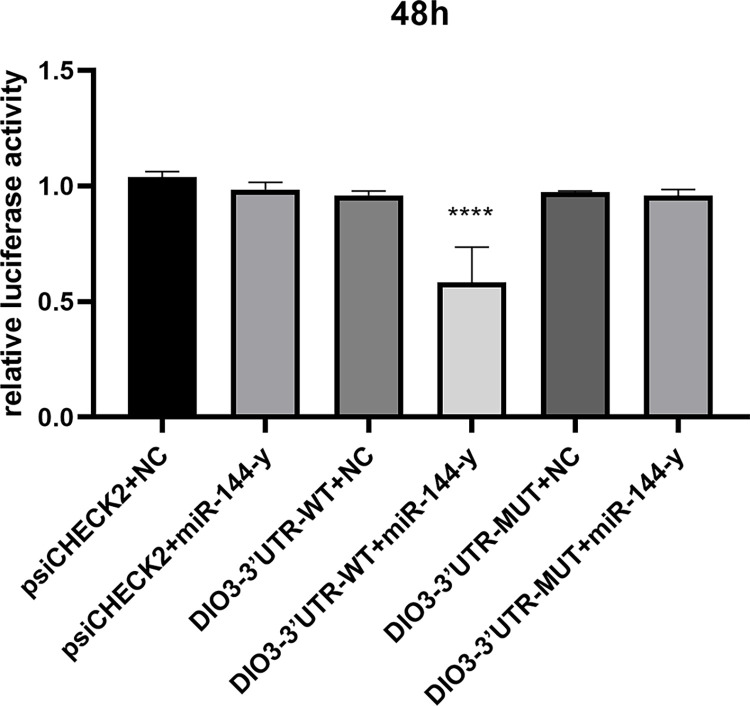
Dual luciferase reporter gene detection
The miR-144-y and DIO3-3′ UTR wild-type reporter vectors, DIO3-3′ UTR-WT and miR-144-y, were co-transfected into 293T cells. The dual luciferase activity ratio of the co-transfection of miR-144-y and DIO3-3′ UTR-WT was significantly down-regulated compared to the control group (P < 0.05). The results indicate a targeting relationship between miR-144-y and DIO3-3′ UTR-WT (Fig. 13).
Fig. 13.
The dual luciferase reporter assay used to detect the predicted target gene DIO3 of miR-144-y. * * * * P < 0.0001, extremely significant difference.
Discussion
Broodiness seriously affects the laying performance of WWG. During the brooding period, the ovaries of the female geese are degenerated, the follicles are shrunken and blocked, and egg production is halted. Environmental factors such as light and temperature can also affect the brooding behavior of poultry by altering the hormonal levels in the HPG axis. PRL secreted by the anterior pituitary is a key hormone that regulates broodiness. The increased secretion levels of GnRH and vasoactive intestinal peptide (VIP) by the hypothalamus increase PRL levels, inhibit the secretion of follicle stimulating hormone (FSH) and luteinizing hormone, degrade the ovaries and fallopian tubes, and trigger broodiness (Pan et al., 2022). However, the transcriptional regulatory mechanism of miRNAs and mRNAs in the HPG axis concerning the broodiness behavior of Wanxi white geese is still unclear.
In this study on the histomorphological analysis of ovarian tissues during the brooding and laying periods found that there were many atresia follicles in the ovaries of WWG during the brooding period, during which the follicular development had stopped (Fig. 1). Additionally, there were several primary, secondary, and mature in the ovarian cortex during the laying period, during which the follicle development was obvious, and the follicles were many. Therefore, it is speculated that the changes in the ovarian tissue structure at different breeding periods can affect the laying performance of WWG. In this study, the hypothalamus, pituitary, and ovarian tissues of WWG in the brooding and laying periods were selected for the differential expression analysis of mRNA and miRNA. The study found 240, 319, and 445 DEGs in the BH vs. LH, BP vs. LP, and BO vs. LO comparison groups, respectively. The GO database was used to analyze the molecular biological process of DEGs. It was found that the DEGs in the three comparison groups were mainly significantly enriched in biological regulation, catalytic activity, and cell composition processes, among others. In addition, KEGG results showed that DEGs in the three control groups were significantly enriched in multiple signaling pathways associated with reproductive performance. These pathways included neuroactive ligand-receptor interaction, calcium signaling, ECM-receptor interaction, and cell adhesion molecules signaling pathway. All four signaling pathways are associated with signal transduction, with the neuroactive ligand-receptor interaction pathway being present in all three groups. Notably, neuroactive ligand-receptor interaction is the basis of the nervous system function, which involves signal transmission and cell communication. It regulates the brooding behavior of poultry by controlling the binding affinity of PRL, VIP, GnRH, and the corresponding receptors. The calcium signaling pathway is a very important signal transduction channel in cells, which is closely related to cell growth, differentiation, metabolism, and reproduction. The pathway regulates the PRL secretion based on the calcium ion concentration. The fluctuation of calcium ions can trigger dopamine and 5-hydroxytryptamine release and affect the brooding behavior by binding the corresponding receptors. Moreover, the calcium signaling pathway regulates the cell cycle, affecting cell proliferation and differentiation and inhibiting the apoptosis of gonadal cells, thus indicating its close association with follicular development and oocyte maturation (Li et al., 2022). In the animal reproductive system, the ECM-receptor interaction plays an important role in the brooding process. The ECM-receptor interaction affects the formation, development, and ovulation of follicles by regulating the extracellular matrix (Hu et al., 2019). The adhesion signaling pathway can also affect the selection and formation of follicles. Specific components of the extracellular matrix affect follicular development by interacting with adhesion receptors on the surface of cells around follicles (Kulus et al., 2023). Therefore, the hypothalamus and pituitary gland, as crucial components of the nervous and endocrine systems, play a vital role in maintaining normal physiological activities (Adamantidis and de Lecea, 2023). Interestingly, CGA, VIPR2, CRH, CRHR2, DRD1, and DRD2 were significantly differentially expressed in the hypothalamus and pituitary within the HPG axis. CGA is closely related to the secretion of FSH and luteinizing hormone that promote follicular maturation. (Thompson and Kaiser, 2014). As a VIP receptor, VIPR2 is mainly involved in the signal transduction pathway of hens reproductive regulation. (Sun et al., 2021). CRH and CRHR2 are members of the corticotropin-releasing factor family, which promotes the production of sex hormones by synthesizing the precursors. CRH is also involved in follicular maturation and ovulation in the ovaries (Wypior et al., 2011). DRD1 and DRD2 are dopamine receptors, which can regulate the secretion of PRL in poultry through dopamine, and low levels of DRD1 are associated with the occurrence of broodiness (Xu et al., 2010). In this study, the expression of CGA, VIPR2, CRH, CRHR2, and DRD2 were significantly up-regulated in the hypothalamus of WWG during the laying period. The expression levels of DRD1 in pituitary tissues of WWG were significantly lower in the brooding period than in the laying period. These DEGs may be involved in signal transduction within the HPG axis, thereby regulating the broodiness behavior of WWG.
A total of 56 (BH vs. LH), 82 (BP vs. LP), and 48 (BO vs. LO) DEMs were identified via the differential expression analysis of miRNA. The GO enrichment analysis of the DEMs showed that the target genes of the three comparison groups of DEMs were mainly significantly enriched in biological processes such as biological regulation, transcription factor activity, and cell junction (P < 0.05). KEGG enrichment results showed that GnRH, MAPK, Wnt, calcium, and TGF-β signaling pathways were significantly enriched among the signaling pathways shared by the three groups of DEMs target genes (P < 0.05). Among these, the GnRH signaling pathway mainly regulates the secretion of gonadal hormones, LH and FSH, in the reproductive system (Stamatiades and Kaiser, 2018). The MAPK signaling pathway is an important signal transduction pathway in cells, which regulates cell proliferation, differentiation, apoptosis, and cell stress response (Sun et al., 2015). Wnt signaling activates multiple signaling cascades by binding the Frizzled receptors and LRP co-receptors on the surfaces of cells found in the β-catenin-dependent and β-catenin-independent pathways, thus affecting intercellular communication, embryonic and tissue development (Rim et al., 2022). The calcium signaling pathway regulates various cellular functions, including nerve conduction, cell proliferation, and apoptosis (Li et al., 2022). The inactivation of the TGF-β signaling pathway leads to granular cell apoptosis and follicular atresia, affecting follicular growth and development (Zhou et al., 2015). Therefore, the hypothalamic-pituitary-ovarian axis is a complete neuroendocrine regulation system, which plays an important role in regulating the ovarian development and laying behavior in female geese. Interestingly, based on these signaling pathways, we identified 10 DEMs (miR-21-y, miR-139-x, miR-144-x, let-7-y, miR-1805-y, miR-335-x, miR-2766-y, miR-132-y, miR-361-x, and miR-460-x) that were significantly differentially expressed in the hypothalamus, pituitary, and ovarian tissues of WWG (Supplementary Table S10). In addition to regulating follicular development by targeting AMH, miR-21-y inhibits FSH-stimulated follicular growth and maintains follicular reserve (Li et al., 2020). The common target gene of miR-132-y, miR-335-x, and miR-361-x, GDF9, regulates follicular development and promotes the transition from primary to secondary follicles (Emori and Sugiura, 2014). Additionally, the common target gene of miR-1805-y, miR-335-x, and miR-2766-y, FSHB, encodes the β subunit of follicle-stimulating hormone, regulates FSH level and interacts with luteinizing hormone to induce rat oocyte production (Stamatiades et al., 2019). The target gene of miR-460-x, GnRH1, encodes a proprotein, which is hydrolyzed to produce GnRH-related peptides to promote poultry luteal formation and FSH release (Patil et al., 2022). The common target gene of miR-139-x, miR-144-x, and let-7-y, ESR1, encodes the estrogen receptor, a ligand-activated transcription factor that regulates the transcription of estrogen-induced genes. The function of estrogen (E2) is mainly mediated through the estrogen receptor (ESR), which promotes ovarian development in female individuals and facilitates the redevelopment of ovaries after spawning. In addition, E2 is transported to the liver via the bloodstream, where it stimulates the synthesis and secretion of vitellogenin (VTG). VTG is subsequently decomposed into vitellin in oocytes providing essential nutrients for embryonic development (Henriques et al., 2022; Knoedler et al., 2022; Ye et al., 2022). In this study, the expression level of miR-21-y was significantly up-regulated in the ovaries of WWG during the brooding period. During the laying period, the expression levels of miR-139-x, miR-1805-y, miR-132-y, miR-460-x, let-7-y, miR-2766-y, miR-361-x, miR-21-y, miR-144-x, and miR-335-x were up-regulated in the hypothalamus, pituitary, and ovaries of WWG. These miRNAs may promote follicular maturation and ovulation in WWG, suggesting their roles as candidate miRNAs associated with reproduction. Moreover, there has been no in-depth research on the relationship between miR-460-x, miR-139-x, let-7-y, and miR-144-x, and poultry reproduction. We speculate that these miRNAs may affect the brooding behavior and laying performance of WWG by regulating the expression level of their corresponding target genes.
In this study, DEMs and DEGs data were combined to construct a miRNA-mRNA interaction network to further screen candidate miRNAs and mRNAs that regulate the brooding behavior and egg production performance of WWG. In the miRNA-mRNA interaction network comparing the BH and LH groups, we identified five miRNAs (miR-200-y, miR-205-x, miR-206-y, miR-96-x, and miR-122-x) that may be associated with broodiness behavior and egg production performance in WWG (Supplementary Table S11). miR-200 could promote oocyte maturation and oocyte excretion by regulating luteinizing hormone levels (Xiong et al., 2020). The target genes of miR-200-y, RYR3, and CASR, are closely related to animal reproduction. RYR3 regulates the release of intracellular calcium ions, which in turn affects the maturation, fertilization, and early embryonic development of mouse germ cells (Dabertrand et al., 2007). CASR regulates gonadotropin levels, stimulating the pituitary gland to secrete FSH and luteinizing hormone, thereby promoting porcine oocyte maturation (Liu et al., 2015). The target gene of miR-205-x, CAMK2A, is mainly involved in the calcium signaling pathway, affecting the development and function of germ cells (Tahir et al., 2021). Calcium concentration is critical for regulating the release of neurotransmitters and endocrine hormones, and ovarian degeneration is closely associated with calcium levels (Bae and Channing, 1985). MiR-96 is a key mediator of luteinizing hormone release (Mohammed et al., 2017). The protein encoded by UNC5A, the target gene of miR-96-x, belongs to the UNC5 receptor family and plays a role in axon guidance and apoptosis. UNC5A mainly regulates the migration or apoptosis of germ cells (Akkermans et al., 2022). Furthermore, miR-122 can regulate the expression of luteinizing hormone receptor (LHR) in poultry ovaries, and its overexpression can inhibit the expression of LHR and affect follicular development (Menon et al., 2018). The target gene of miR-122-x, HTR2A, encodes serotonin receptor 2A, and the serotonin system plays a role in poultry reproductive behavior and sexual function (Ding et al., 2021). In the miRNA-mRNA interaction network comparing the BP and LP groups, seven miRNAs (let-7-y, miR-144-y, miR-150-x, miR-205-x, miR-215-x, miR-9-y, and miR-124-y) were identified as potentially associated with broodiness and egg-laying performance (Supplementary Table S12). Notably, let-7 negatively regulates the expression of steroidogenic acute regulatory protein (StAR), which is a rate-limiting enzyme in progesterone synthesis. The biosynthesis of all steroid hormones is closely linked to StAR, which facilitates the transfer of cholesterol to the inner mitochondrial membrane. This process is catalyzed by cytochrome P450 family enzymes (CYPs) and hydroxylated steroid dehydrogenases (HSDs), leading to the production of various sex hormones, mainly E2 and progesterone. These hormones are then transported back to the hypothalamus and pituitary gland via the bloodstream to regulate the synthesis and secretion of GnRH, FSH, and luteinizing hormone (Tremblay et al., 2001; Munro et al., 2018; Chen et al., 2023). The target gene of miR-144-y, DIO3, encodes a thyroid hormone inactivation enzyme, which catalyzes the metabolism of thyroid hormone. Thyroid hormone is one of the key hormones regulating seasonal reproduction and thus directly affects follicular development in seasonal breeding animals (Yoshimura, 2013). MiR-150 and miR-205 are closely related to follicular development (Song et al., 2021), while miR-215 is associated with early embryonic development. Inhibiting the expression of miR-215 might adversely affect the embryonic development (Ibrahim et al., 2015). MiR-9 can inhibit the expression of the D2r gene and its splicing variant D2, resulting in increased synthesis and secretion of PRL, eventually affecting the brooding behavior (Gangisetty et al., 2017). Additionally, miR-124 inhibits the expression of Sox9 in the developing mice ovarian cells. Sox9 promotes testicular development and inhibits ovarian development (Real et al., 2013). The target gene of miR-124-y, KCNK2, regulates cell membrane potential and the maturation and development of germ cells (Hur et al., 2009). In the BO vs. LO group, five miRNAs (miR-320-y, miR-339-x, miR-7-x, and miR-21-y) related to brooding behavior and laying performance were identified in the miRNA-mRNA interaction network (Supplementary Table S13). The transcription factor encoded by the target gene of miR-320-y, SALL3, plays a central role in the development of dagu hens ovaries (Zhu et al., 2018). TACR3, the target gene of miR-7-x, is a neurokinin receptor that is regulated by ovarian steroids and involved in the regulation of PRL secretion (Czelejewska et al., 2020). MiR-21 regulates the apoptosis of norepinephrine-mediated granulosa cells and inhibits rat ovarian granulosa cell apoptosis by targeting Smad7 (Zhang et al., 2017). The target gene of miR-21-y, RIMS1, is related to placental methylation and the release of neurotransmitters, which may affect the function of the nervous system (Xu et al., 2017). During the laying period, the expression of miR-200-y, miR-205-x, and miR-206-y was up-regulated, while the expression of miR-96-x and miR-122-x was down-regulated in the hypothalamus of WWG. In the pituitary of WWG during laying period, the expression levels of let-7-y, miR-144-y, miR-150-x, miR-205-x, and miR-215-x were up-regulated, whereas the expression of miR-9-y and miR-124-y was down-regulated. The expression levels of miR-320-y, miR-339-x, and miR-7-x were up-regulated, while miR-21-y was down-regulated in the ovaries of WWG during the laying period. In summary, we hypothesize that these miRNAs influence the broodiness and egg laying behavior of Wanxi white geese by regulating the expression level of target genes.
To further validate the targeting relationship between the key differential miRNAs identified by the miRNA-mRNA interaction network and their corresponding target genes, we focused on the differential expression of miR-144-y in the pituitary gland of broody WWG. Given its potential involvement in the synthesis and secretion of sex hormones, we noted that its candidate target gene, DIO3, may also play a role in regulating follicular development. In this study, 293T cells were used to predict the binding site of miR-144-y to the 3′ UTR of DIO3. We constructed dual luciferase wild-type and mutant vectors containing the binding sites, and transfected these vectors into 293T cells for dual luciferase activity detection to verify the reliability of the binding site. The results demonstrated a targeting relationship between miR-144-y and DIO3-3′ UTR-WT.
Conclusions
In this study, we conducted a comprehensive analysis of DEGs and DEMs in the hypothalamus, pituitary, and ovaries of Wanxi white geese during the laying period and broodiness period utilizing transcriptome sequencing technology. We identified several signaling pathways related to laying performance and broodiness behavior. The interaction between DEGs and DEMs were determined by constructing a mRNA-miRNA interaction network, which led to the identification of several candidate miRNAs that influence the laying performance and broodiness behavior of WWG. At the same time, the targeting relationship between miR-144-y and DIO3 was verified using a dual luciferase reporter gene. This study revealed the molecular mechanism underlying the HPG axis-mediated post-transcriptional regulation of the brooding behavior and laying performance of WWG. The results provide a theoretical basis for improving the laying performance of geese and accelerating the breeding of new breeds.
Author contributions
X.W. Tong: Writing - original draft, Conceptualization, Data curation, Formal analysis, Methodology, Investigation, Visualization. X.J. Li: Conceptualization, Methodology, Project administration, Funding acquisition, Software, Writing - review & editing. Y.H. Wang: Writing - original draft, Validation, Visualization. F. Xie: Formal analysis, Validation, Visualization. R.D. Li: Writing - original draft, Validation, Visualization. M. Ren: Project administration, Resources, Supervision. Q.Q. Hu: Writing - review & editing, Supervision. S.H. Li: Conceptualization, Methodology, Project administration, Funding acquisition, Resources, Supervision.
Ethics approval
Experiments were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals and approved by the Animal Research Committee of Anhui Science and Technology University (license number 2023-014).
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work the author(s) did not use any Al and Al-assisted technologies.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
Data and model availability statement
The original sequencing data for the hypothalamus, pituitary, and ovary of Wanxi White Goose have been uploaded to the NCBI database under BioProject ID: PRJNA1169106.
Acknowledgements
This work was supported by grants from the talent introduction project of Anhui Science and Technology University (DKYJ202105), the Anhui Province Science and Technology Major Project (17030701004), the Local Goose Gene Bank in Anhui Province, the Science and Technology Project of Chuzhou City, Anhui Province (2022ZN002), the Veterinary Science Peak Discipline Project of Anhui Science and Technology University (XK-XJGF002), and the Establishment and Application of a Reverse Genetic Operating Platform for Goose Paramyxovirus (KJ2019A0801).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2024.104510.
Appendix. Supplementary materials
References
- Adamantidis A.R., de Lecea L. Sleep and the hypothalamus. Science. 2023;382:405–412. doi: 10.1126/science.adh8285. [DOI] [PubMed] [Google Scholar]
- Akhtar M.F., Shafiq M., Ali I. Improving gander reproductive efficacy in the context of globally sustainable goose production. Animals (Basel) 2021;12 doi: 10.3390/ani12010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkermans O., Delloye-Bourgeois C., Peregrina C., Carrasquero-Ordaz M., Kokolaki M., Berbeira-Santana M., Chavent M., Reynaud F., Raj R., Agirre J., Aksu M., White E.S., Lowe E., Ben A.D., Zaballa S., Huo J., Pakos I., McCubbin P., Comoletti D., Owens R.J., Robinson C.V., Castellani V., Del T.D., Seiradake E. GPC3-Unc5 receptor complex structure and role in cell migration. Cell. 2022;185:3931–3949. doi: 10.1016/j.cell.2022.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae I.H., Channing C.P. Effect of calcium ion on the maturation of cumulus-enclosed pig follicular oocytes isolated from medium-sized graafian follicles. Biol. Reprod. 1985;33:79–87. doi: 10.1095/biolreprod33.1.79. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen F., Li J., Zhang H., Xu J., Tao Z., Shen J., Shen J., Lu L., Li C. Identification of differentially expressed known and novel miRNAs in broodiness of goose. Mol. Biol. Rep. 2014;41:2767–2777. doi: 10.1007/s11033-014-3131-8. [DOI] [PubMed] [Google Scholar]
- Chen J., Liu K., Liu W., Yeung W.S. The involvement of let-7 in hCG-induced progesterone synthesis via regulating p27(Kip1) and p21(Cip1) expression. Mol. Cell. Endocrinol. 2023;573 doi: 10.1016/j.mce.2023.111970. [DOI] [PubMed] [Google Scholar]
- Chen X., Xizeng M., Jiaju H., Yang D., Jianmin W., Shan D., Lei K., Ge G., Chuan-Yun L., Liping W. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic. Acids. Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czelejewska W., Zmijewska A., Dziekonski M., Okrasa S. The potential implication of neurokinin B in the modulation of prolactin secretion at the pituitary level in cyclic gilts. J. Physiol. Pharmacol. 2020;71 doi: 10.26402/jpp.2020.2.10. [DOI] [PubMed] [Google Scholar]
- Dabertrand F., Fritz N., Mironneau J., Macrez N., Morel J.L. Role of RYR3 splice variants in calcium signaling in mouse nonpregnant and pregnant myometrium. Am. J. Physiol. Cell Physiol. 2007;293:C848–C854. doi: 10.1152/ajpcell.00069.2007. [DOI] [PubMed] [Google Scholar]
- Ding L., Maloney S.K., Wang M., Rodger J., Chen L., Blache D. Association between temperament related traits and single nucleotide polymorphisms in the serotonin and oxytocin systems in Merino sheep. Genes Brain Behav. 2021;20:e12714. doi: 10.1111/gbb.12714. [DOI] [PubMed] [Google Scholar]
- Emori C., Sugiura K. Role of oocyte-derived paracrine factors in follicular development. Anim Sci J. 2014;85:627–633. doi: 10.1111/asj.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangisetty O., Jabbar S., Wynne O., Sarkar D.K. MicroRNA-9 regulates fetal alcohol-induced changes in D2 receptor to promote prolactin production. J. Endocrinol. 2017;235:1–14. doi: 10.1530/JOE-17-0135. [DOI] [PubMed] [Google Scholar]
- Gao G., Zhang H., Ni J., Zhao X., Zhang K., Wang J., Kong X., Wang Q. Insights into genetic diversity and phenotypic variations in domestic geese through comprehensive population and pan-genome analysis. J Anim Sci Biotechnol. 2023;14:150. doi: 10.1186/s40104-023-00944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachev P., Li X.F., Goffin V., O'Byrne K.T. Hypothalamic prolactin regulation of luteinizing hormone secretion in the female rat. Endocrinology. 2015;156:2880–2892. doi: 10.1210/en.2015-1040. [DOI] [PubMed] [Google Scholar]
- Guchhait R., Chatterjee A., Mukherjee D., Pramanick K. Seasonal ovarian development in relation to the gonadotropins, steroids, aromatase and steroidogenic factor 1 (SF-1) in the banded gourami, Trichogaster fasciata. Gen. Comp. Endocrinol. 2018;268:40–49. doi: 10.1016/j.ygcen.2018.07.014. [DOI] [PubMed] [Google Scholar]
- Henriques P.C., Aquino N., Campideli-Santana A.C., Silva J.F., Araujo-Lopes R., Franci C.R., Coimbra C.C., Szawka R.E. Hypothalamic expression of estrogen receptor isoforms underlies estradiol control of luteinizing hormone in female rats. Endocrinology. 2022;163 doi: 10.1210/endocr/bqac101. [DOI] [PubMed] [Google Scholar]
- Hu S., Yang S., Lu Y., Deng Y., Li L., Zhu J., Zhang Y., Hu B., Hu J., Xia L., He H., Han C., Liu H., Kang B., Li L., Wang J. Dynamics of the transcriptome and accessible chromatin landscapes during early goose ovarian development. Front. Cell Dev. Biol. 2020;8:196. doi: 10.3389/fcell.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Liang X., Ren X., Shi Y., Su H., Li Y., Du K., Wang J., Jia X., Chen S., Lai S. Integrated analysis of mRNA and miRNA expression profiles in the ovary of oryctolagus cuniculus in response to gonadotrophic stimulation. Front Endocrinol (Lausanne) 2019;10:744. doi: 10.3389/fendo.2019.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur C.G., Choe C., Kim G.T., Cho S.K., Park J.Y., Hong S.G., Han J., Kang D. Expression and localization of two-pore domain K(+) channels in bovine germ cells. Reproduction. 2009;137:237–244. doi: 10.1530/REP-08-0035. [DOI] [PubMed] [Google Scholar]
- Ibrahim S., Salilew-Wondim D., Rings F., Hoelker M., Neuhoff C., Tholen E., Looft C., Schellander K., Tesfaye D. Expression pattern of inflammatory response genes and their regulatory micrornas in bovine oviductal cells in response to lipopolysaccharide: implication for early embryonic development. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoedler J.R., Inoue S., Bayless D.W., Yang T., Tantry A., Davis C.H., Leung N.Y., Parthasarathy S., Wang G., Alvarado M., Rizvi A.H., Fenno L.E., Ramakrishnan C., Deisseroth K., Shah N.M. A functional cellular framework for sex and estrous cycle-dependent gene expression and behavior. Cell. 2022;185:654–671. doi: 10.1016/j.cell.2021.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019;20:278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijjer M.L., Fagny M., Marin A., Quackenbush J., Glass K. PUMA: PANDA using MicroRNA associations. Bioinformatics. 2020;36:4765–4773. doi: 10.1093/bioinformatics/btaa571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulus J., Kranc W., Kulus M., Bukowska D., Piotrowska-Kempisty H., Mozdziak P., Kempisty B., Antosik P. New gene markers of exosomal regulation are involved in porcine granulosa cell adhesion, migration, and proliferation. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241411873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu H., Li Y., Yang M., Qu C., Zhang Y., Liu Y., Zhang X. Identification of suitable endogenous control genes for quantitative RT-PCR analysis of miRNA in bovine solid tissues. Mol. Biol. Rep. 2014;41:6475–6480. doi: 10.1007/s11033-014-3530-x. [DOI] [PubMed] [Google Scholar]
- Li X., Lin B., Zhang X., Shen X., Ouyang H., Wu Z., Tian Y., Fang L., Huang Y. Comparative transcriptomics in the hypothalamic-pituitary-gonad axis of mammals and poultry. Genomics. 2022;114 doi: 10.1016/j.ygeno.2022.110396. [DOI] [PubMed] [Google Scholar]
- Li X., Xie J., Wang Q., Cai H., Xie C., Fu X. miR-21 and Pellino-1 expression profiling in autoimmune premature ovarian insufficiency. J. Immunol. Res. 2020 doi: 10.1155/2020/3582648. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Yan X., Wang H., Su Y., Zhu W. Effects of lactic acid bacteria-fermented formula milk supplementation on ileal microbiota, transcriptomic profile, and mucosal immunity in weaned piglets. J Anim Sci Biotechnol. 2022;13:113. doi: 10.1186/s40104-022-00762-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu G.Q., Fu X.W., Mo X.H., Zhao L.H., Hu H.M., Zhu S.E., Hou Y.P. The extracellular calcium-sensing receptor (CASR) regulates gonadotropins-induced meiotic maturation of porcine oocytes. Biol. Reprod. 2015;93:131. doi: 10.1095/biolreprod.115.128579. [DOI] [PubMed] [Google Scholar]
- Liu L., Xiao Q., Gilbert E.R., Cui Z., Zhao X., Wang Y., Yin H., Li D., Zhang H., Zhu Q. Whole-transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis. Sci. Rep. 2018;8:7231. doi: 10.1038/s41598-018-25103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan X., Cao Z., Li R., Liu M., Hu J. Differential expression profiling of hypothalamus genes in laying period and ceased period Huoyan geese. Mol. Biol. Rep. 2014;41:3401–3411. doi: 10.1007/s11033-014-3202-x. [DOI] [PubMed] [Google Scholar]
- Menon K., Menon B., Gulappa T. Regulation of luteinizing hormone receptor mRNA expression in the ovary: the role of miR-122. Vitam. Horm. 2018;107:67–87. doi: 10.1016/bs.vh.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed B.T., Sontakke S.D., Ioannidis J., Duncan W.C., Donadeu F.X. The adequate Corpus luteum: miR-96 promotes luteal cell survival and progesterone production. J. Clin. Endocrinol. Metab. 2017;102:2188–2198. doi: 10.1210/jc.2017-00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro A.W., McLean K.J., Grant J.L., Makris T.M. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem. Soc. Trans. 2018;46:183–196. doi: 10.1042/BST20170218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane Y., Yoshimura T. Photoperiodic regulation of reproduction in vertebrates. Annu Rev Anim Biosci. 2019;7:173–194. doi: 10.1146/annurev-animal-020518-115216. [DOI] [PubMed] [Google Scholar]
- Pan J.Q., Liufu S., Sun J.F., Chen W.J., Ouyang H.J., Shen X., Jiang D.L., Xu D.N., Tian Y.B., He J.H., Huang Y.M. Long-day photoperiods affect expression of OPN5 and the TSH-DIO2/DIO3 pathway in Magang goose ganders. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.102024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.A., Lila A.R., Shah N., Ekbote A.V., Shah R., Bhandare V.V., Sarathi V., Arya S., Memon S.S., Kunwar A., Bandgar T. GNRH1 Variants in congenital hypogonadotropic hypogonadism: single-center experience and systematic literature review. Neuroendocrinology. 2022;112:723–732. doi: 10.1159/000521558. [DOI] [PubMed] [Google Scholar]
- Real F.M., Sekido R., Lupianez D.G., Lovell-Badge R., Jimenez R., Burgos M. A microRNA (mmu-miR-124) prevents Sox9 expression in developing mouse ovarian cells. Biol. Reprod. 2013;89:78. doi: 10.1095/biolreprod.113.110957. [DOI] [PubMed] [Google Scholar]
- Rim E.Y., Clevers H., Nusse R. The Wnt pathway: from signaling mechanisms to synthetic modulators. Annu. Rev. Biochem. 2022;91:571–598. doi: 10.1146/annurev-biochem-040320-103615. [DOI] [PubMed] [Google Scholar]
- Sanchez Jimenez J.G., De Jesus O. StatPearls. StatPearls Publishing; 2023. Hypothalamic dysfunction. [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Zhao X., He H., Zhang Y., Zhu Q., Yin H. Transcriptome profiling reveals SLC5A5 regulates chicken ovarian follicle granulosa cell proliferation, apoptosis, and steroid hormone synthesis. Poult. Sci. 2024;103 doi: 10.1016/j.psj.2023.103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P., Yue Q., Fu Q., Li X., Li X., Zhou R., Chen X., Tao C. Integrated analysis of miRNA-mRNA interaction in ovaries of Turpan Black Sheep during follicular and luteal phases. Reprod. Domest. Anim. 2021;56:46–57. doi: 10.1111/rda.13848. [DOI] [PubMed] [Google Scholar]
- Stamatiades G.A., Carroll R.S., Kaiser U.B. GnRH-A key regulator of FSH. Endocrinology. 2019;160:57–67. doi: 10.1210/en.2018-00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatiades G.A., Kaiser U.B. Gonadotropin regulation by pulsatile GnRH: signaling and gene expression. Mol. Cell. Endocrinol. 2018;463:131–141. doi: 10.1016/j.mce.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Chen X., Zhao J., Ma C., Yan C., Liswaniso S., Xu R., Qin N. Transcriptome comparative analysis of ovarian follicles reveals the key genes and signaling pathways implicated in hen egg production. Bmc Genomics [Electronic Resource] 2021;22:899. doi: 10.1186/s12864-021-08213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Liu W.Z., Liu T., Feng X., Yang N., Zhou H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- Tahir M.S., Porto-Neto L.R., Gondro C., Shittu O.B., Wockner K., Tan A., Smith H.R., Gouveia G.C., Kour J., Fortes M. Meta-analysis of heifer traits identified reproductive pathways in Bos indicus cattle. Genes (Basel) 2021;12 doi: 10.3390/genes12050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson I.R., Kaiser U.B. GnRH pulse frequency-dependent differential regulation of LH and FSH gene expression. Mol. Cell. Endocrinol. 2014;385:28–35. doi: 10.1016/j.mce.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M.R., Lin S.X., Poirier D. Chemical synthesis of 16beta-propylaminoacyl derivatives of estradiol and their inhibitory potency on type 1 17beta-hydroxysteroid dehydrogenase and binding affinity on steroid receptors. Steroids. 2001;66:821–831. doi: 10.1016/s0039-128x(01)00116-7. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang L., Zhang Y., Yao Y., Zhao W., Xu Q., Chen G. Characterization of ovarian morphology and reproductive hormones in Zhedong white geese (Anser cygnoides domesticus) during the reproductive cycle. J. Anim. Physiol. Anim. Nutr. (Berl.) 2021;105:938–945. doi: 10.1111/jpn.13494. [DOI] [PubMed] [Google Scholar]
- Wypior G., Jeschke U., Kurpisz M., Szekeres-Bartho J. Expression of CRH, CRH-related peptide and CRH receptor in the ovary and potential CRH signalling pathways. J. Reprod. Immunol. 2011;90:67–73. doi: 10.1016/j.jri.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Xiong S., Tian J., Ge S., Li Z., Long Z., Guo W., Huang P., He Y., Xiao T., Gui J.F., Mei J. The microRNA-200 cluster on chromosome 23 is required for oocyte maturation and ovulation in zebrafishdagger. Biol. Reprod. 2020;103:769–778. doi: 10.1093/biolre/ioaa125. [DOI] [PubMed] [Google Scholar]
- Xu H., Shen X., Zhou M., Fang M., Zeng H., Nie Q., Zhang X. The genetic effects of the dopamine D1 receptor gene on chicken egg production and broodiness traits. BMC Genet. 2010;11:17. doi: 10.1186/1471-2156-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Barlow G.M., Cui J., Wang E.T., Lee B., Akhlaghpour M., Kroener L., Williams J.R., Rotter J.I., Chen Y.I., Goodarzi M.O., Pisarska M.D. Comparison of genome-wide and gene-specific DNA methylation profiling in first-trimester chorionic villi from pregnancies conceived with infertility treatments. Reprod Sci. 2017;24:996–1004. doi: 10.1177/1933719116675056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Mu R., Gegen T., Ran T., Wu Q., Wen D., Wang F., Chen Z. Transcriptome analysis of hypothalamus and pituitary tissues reveals genetic mechanisms associated with high egg production rates in Changshun green-shell laying hens. Bmc Genomics [Electronic Resource] 2023;24:792. doi: 10.1186/s12864-023-09895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Yang Y.Z., Gu T.T., Cao Z.F., Zhao W.M., Qin H.R., Xu Q., Chen G.H. Comparison of the broody behavior characteristics of different breeds of geese. Poult. Sci. 2019;98:5226–5233. doi: 10.3382/ps/pez366. [DOI] [PubMed] [Google Scholar]
- Ye Z., Zhao T., Wei Q., Lin H., Zhang Y., Li S. Distinct roles of estrogen receptors in the regulation of vitellogenin expression in orange-spotted grouper (Epinephelus coioides) Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23158632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L.Q., Ran J.S., Li J.J., Ren P., Zhang X.X., Liu Y.P. [Genetic regulation of broodiness in poultry] Yi Chuan. 2019;41:391–403. doi: 10.16288/j.yczz.18-316. [DOI] [PubMed] [Google Scholar]
- Yoshimura T. Thyroid hormone and seasonal regulation of reproduction. Front. Neuroendocrinol. 2013;34:157–166. doi: 10.1016/j.yfrne.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang L., Gao J., Cui S. miR-21 is involved in norepinephrine-mediated rat granulosa cell apoptosis by targeting SMAD7. J. Mol. Endocrinol. 2017;58:199–210. doi: 10.1530/JME-16-0248. [DOI] [PubMed] [Google Scholar]
- Zhang P., Wang J., Lang H., Wang W., Liu X., Liu H., Tan C., Li X., Zhao Y., Wu X. MicroRNA-205 affects mouse granulosa cell apoptosis and estradiol synthesis by targeting CREB1. J. Cell. Biochem. 2019;120:8466–8474. doi: 10.1002/jcb.28133. [DOI] [PubMed] [Google Scholar]
- Zhao S., Ye Z., Stanton R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA. 2020;26:903–909. doi: 10.1261/rna.074922.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu J., Pan Z., Du X., Li X., Ma B., Yao W., Li Q., Liu H. The let-7g microRNA promotes follicular granulosa cell apoptosis by targeting transforming growth factor-beta type 1 receptor. Mol. Cell. Endocrinol. 2015;409:103–112. doi: 10.1016/j.mce.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Zhu H., Qin N., Tyasi T.L., Jing Y., Liu D., Yuan S., Xu R. Genetic effects of the transcription factors-sal-like 1 and spalt-like transcription factor 3 on egg production-related traits in Chinese Dagu hens. J Exp Zool A Ecol Integr Physiol. 2018;329:23–28. doi: 10.1002/jez.2156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.