Abstract
DC-SIGN is a C-type lectin expressed on dendritic cells and restricted macrophage populations in vivo that binds gp120 and acts in trans to enable efficient infection of T cells by human immunodeficiency virus type 1 (HIV-1). We report here that DC-SIGN, when expressed in cis with CD4 and coreceptors, allowed more efficient infection by both HIV and simian immunodeficiency virus (SIV) strains, although the extent varied from 2- to 40-fold, depending on the virus strain. Expression of DC-SIGN on target cells did not alleviate the requirement for CD4 or coreceptor for viral entry. Stable expression of DC-SIGN on multiple lymphoid lines enabled more efficient entry and replication of R5X4 and X4 viruses. Thus, 10- and 100-fold less 89.6 (R5/X4) and NL4–3 (X4), respectively, were required to achieve productive replication in DC-SIGN-transduced Jurkat cells when compared to the parental cell line. In addition, DC-SIGN expression on T-cell lines that express very low levels of CCR5 enabled entry and replication of R5 viruses in a CCR5-dependent manner, a property not exhibited by the parental cell lines. Therefore, DC-SIGN expression can boost virus infection in cis and can expand viral tropism without affecting coreceptor preference. In addition, coexpression of DC-SIGN enabled some viruses to use alternate coreceptors like STRL33 to infect cells, whereas in its absence, infection was not observed. Immunohistochemical and confocal microscopy data indicated that DC-SIGN was coexpressed and colocalized with CD4 and CCR5 on alveolar macrophages, underscoring the physiological significance of these cis enhancement effects.
Enveloped viral entry is mediated by interactions between cell surface receptor(s) and the viral envelope (Env) embedded in the virion lipid bilayer. These interactions trigger the requisite conformational changes in the viral Env that eventually lead to fusion between the viral and host cell membranes and delivery of the viral genome into the target cell. Human immunodeficiency virus type 1 (HIV-1) has evolved to use the CD4-coreceptor complex to trigger the conformational events leading to membrane fusion (reviewed in reference 8). In addition to entry receptors, attachment receptors have been described that can modulate the efficiency of entry mediated by the CD4-coreceptor complex (reviewed in reference 37). For example, heparan sulfate proteoglycans (21, 24, 29) and LFA-1 (12) have been reported to interact with viral Env or virion-associated adhesion molecules such as ICAM-1 in a manner that enhances the viral entry process. However, the majority of these interactions are low affinity in nature, having binding constants of 500 nM or greater (36, 37). Unique among these attachment molecules is the calcium-dependent lectin DC-SIGN, which binds to monomeric HIV-1 gp120 with a greater affinity than CD4 (Kd, 1.4 nM versus 4 to 5 nM for CD4) (6). The binding of DC-SIGN to HIV Env is carbohydrate dependent and is most effectively competed off by mannan (6, 13, 27).
DC-SIGN is a type II integral membrane protein originally cloned from a placental cDNA library as a gp120 binding protein (6). It is highly expressed on dendritic cells (DCs) and is largely responsible for HIV-1 attachment to this cell type (13, 14). A homolog of DC-SIGN, termed “DC-SIGNR/L-SIGN,” is expressed on some types of endothelial cells and also serves as a virus attachment factor (3, 28, 33). Virus bound to DC-SIGN-positive cells can be transmitted to cells expressing CD4 and coreceptor, resulting in efficient virus infection in trans (13, 27). DC-SIGN on DCs may serve as a conduit for the transfer of HIV-1 from the submucosa to permissive T cells in secondary lymphoid organs (13, 35). We have shown that DC-SIGN also binds HIV-2 and SIV Envs and thus can be considered a universal attachment factor for primate lentiviruses (27). Despite initial reports that DC-SIGN expression is restricted to DCs, we have found that DC-SIGN is expressed on CD4+ macrophages in the placenta and lung (34; E. Soilleux, L. S. Morris, G. Leslie et al., submitted for publication). The presence of such a high-affinity attachment molecule on permissive cells in vivo prompted us to examine the consequences of DC-SIGN expression on the efficiency of viral entry.
We found that expression of DC-SIGN in cis with CD4 and coreceptor allowed for more efficient entry of HIV and SIV. The ability of DC-SIGN to facilitate infection in cis was most apparent when either CD4 or coreceptor was limiting. In some cases, DC-SIGN expression allowed infection of cells via CD4 and an alternate coreceptor (STRL33) that is otherwise used inefficiently (30). In addition, some T-cell lines engineered to express DC-SIGN required up to 100-fold less of the viral inoculum in order to establish a productive infection. The in vivo significance of this cis-enhancement effect was supported by confocal microscopy data indicating that DC-SIGN was expressed and colocalized with CD4 and CCR5 on primary alveolar macrophages. Thus, DC-SIGN expression or upregulation in vivo can potentially expand viral tropism by allowing viruses to infect cells with limiting amounts of CD4 or coreceptor or by more efficient use of alternative coreceptors.
MATERIALS AND METHODS
Viruses and cells.
DC-SIGN was cloned into the retroviral MIGR1 vector (a kind gift from Warren Pear, University of Pennsylvania) via the HpaI and BamHI site. Expression of DC-SIGN was mediated by the MSCV long terminal repeat promoter and green fluorescent protein (GFP) was expressed in tandem via an internal ribosome entry site (IRES) linker. Retroviruses expressing DC-SIGN were made by cotransfecting HEK 293T producer cells with MIGR1–DC-SIGN, pcGP (expressing gag and pol genes) and pVSV-G (expressing the vesicular stomatitis virus [VSV]-G envelope glycoprotein) by using Geneporter. Forty-eight hours after transfection, the retroviral supernatant was filtered through a 0.22-μm-pore-size filter and stored at −80°C. Retroviral transduction of the indicated cell lines was performed by spinoculation as described previously (23). All cell lines were originally obtained from the American Tissue Type Collection (ATCC). One week after spinoculation, cell lines were sorted for GFP-positive cells to select for stably transduced cells. GFP-positive cell lines were confirmed to be expressing DC-SIGN by costaining with a monoclonal antibody (MAb) against DC-SIGN (MAb 28) (2a). Pseudotyped GFP or luciferase reporter viruses were made as previously described (30).
Infections.
Equal number of cells from DC-SIGN-transduced cell lines or the parental lines (2 × 105 to 5 × 105 cells/infection) were infected with the indicated amount of viral supernatant. Four hours after infection, cells were washed three times vigorously with 1× phosphate-buffered saline (PBS), and the medium was replaced. For measurement of p24 antigen production, infections were performed in 96 wells in a total volume of 200 μl. On the indicated days after infection, medium was half-exchanged with fresh medium, and the supernatant was stored at −20°C. p24 levels in viral supernatants from the same infection series were analyzed together with a commercial enzyme-linked immunosorbent assay (ELISA) kit. For inhibition experiments, cells were infected as described above or in the presence of an anti-CXCR4 MAb (20 μg of MAb 45701 per ml; R&D Systems, Minneapolis, Minn.) or an anti-CD4 MAb (10 μg of Leu3A per ml), or TAK779 (20 μM; a kind gift from Takeda Pharmaceuticals, Osaka, Japan). Four hours after infection, cells were washed three times vigorously with 1× PBS and the medium was replaced with fresh medium containing the inhibitory reagents as indicated above. On the indicated days after infection, the medium was half-exchanged with fresh medium containing fresh inhibitory reagents as described above. Pseudotyped virus infection was performed as described above, except that analysis was performed 3 days postinfection. The extent of infection was quantified either by fluorescence-activated cell sorter (FACS) analysis of GFP-positive cells or of p24-positive cells (determined by intracellular p24 staining with phycoerythrin-conjugated anti-p24 KC57 clone from Coulter) as previously described (30). For mannan inhibition, infections were performed as described above in the presence of 100 μg of mannan per ml (Sigma).
Measurement of cell-associated viral DNA.
Cell-associated DNA was prepared from 105 infected CEM-SS cells, by lysis in 100 μl of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.2 mM CaCl2, 0.001% Triton X-100, 0.001% sodium dodecyl sulfate (SDS), 1 μg of proteinase K per ml. The lysates were then incubated at 58°C overnight, heat inactivated at 95°C for 15 min, and stored at −80°C. Kinetic (fluorescence monitored) PCR was performed with 1.25 × 104 cell equivalents to quantitate viral gag DNA and cellular β-globin DNA. The sequences of the β-globin forward and reverse primers and the molecular beacon were 5′-CCCTTGGACCCAGAGGTTCT-3′ and 5′-CGAGCACTTTCTTGCCATGA-3′ and GCGAGCATCTGTCCACTCCTGATGCTGTTATGGGCGCTCGC-3′, respectively. The molecular beacon was labeled with JOE (6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein) and DABCYL, at the 5′ and 3′ ends, respectively. The sequences of the gag forward and reverse primers and TaqMan probe were 5′-AAGCAGCAGCTGACACAGGA-3′, 5′-TTTGCCCCTGGATGTTCTG-3′, and 5′-ACAGCAATCAGGTCAGCCAAAATTACCCTATAGT-3′, respectively. The TaqMan probe was labeled at the 5′ end with FAM (fluorochrome 6-carboxyfluorescein) and at the 3′ end with TAMARA (6-carboxy-N′,N′,N′,N′-tetramethylrhodamine). Reactions were individually optimized and carried out in 50-μl volumes containing the following: 50 mM KCl; 10 mM Tris-HCl (pH 8.3); 3 (β-globin) to 5.5 (gag) mM MgCl2; 200 (β-globin) to 300 (gag) μM dATP, dCTP, dGTP, and dTTP; 200 (gag) to 1,000 (β-globin) nM primer; 200 nM probe; 0.025 U of AmpliTaq Gold (PE BioSystems) per μl; and 500 nM carboxy-X-rhodamine (Rox) as a passive reference (Molecular Probes, Eugene, Oreg.). The reaction times and temperatures were 10 min at 95°C and then 40 cycles of 15 s at 95°C and 45 s at 60°C.
A standard curve for HIV-1 DNA copy number was prepared from mixtures of ACH-2 cells (which harbor two HIV-1 proviruses) and CEM-SS cells, by using the lysis procedure described above. To correct for variations in cell numbers and DNA recovery, a standard curve for cellular β-globin was generated by preparing DNA lysate from uninfected CEM-SS cells that had been counted with a hemacytometer. These cell counts were then used to design serial dilutions of DNA lysate in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 ng of poly(rA) per ml. Sequence Detection software, version 3 (PE BioSystems), was used to analyze the kinetic PCR amplification data.
Selecting and obtaining tissue and tissue processing.
All tissues were obtained with Local Research Ethics Committee approval. Fully anonymized histologically normal spleen, lymph node, lung, liver, and placenta of 12 and 40 weeks of gestation was obtained from the Department of Histopathology, Addenbrooke's Hospital, Cambridge, United Kingdom. Tissue was either snap-frozen and kept at −80°C before being processed to cryosections, or was fixed in 10% neutral buffered formalin, followed by paraffin wax embedding and sectioning.
Single immunostaining of paraffin sections.
Sections were immunostained with rabbit anti-DC-SIGN polyclonal serum, with preimmune serum on serial sections as a negative control, exactly as described previously (34). Further serial sections were immunostained with anti-CD14 (Novocastra, Newcastle upon Tyne, United Kingdom) as previously described (34).
Staining of frozen sections for confocal microscopy.
Ten-micrometer cryosections of lung and placenta were fixed for 5 min in 4% paraformaldehyde and then immunostained with mouse monoclonal anti-CD4 (PharMingen, San Diego, Calif.), mouse monoclonal anti-CCR5 (clone CTC5; R&D Systems, Minneapolis, Minn.) or rabbit polyclonal anti-DC-SIGN (34). Each mouse MAb was also used in double immunostaining in combination with rabbit polyclonal anti-DC-SIGN. Primary antibody was added in 1% bovine serum albumin–10% goat serum–10% swine serum in Tris-buffered saline (TBS). Following overnight incubation, sections were rinsed thoroughly in TBS and incubated for 1 h with secondary antibody, which was phycoerythrin-conjugated goat anti-mouse antibody (Sigma, Poole, United Kingdom) and/or fluorescein isothiocyanate (FITC)-conjugated swine anti-rabbit antibody (Dako, Glostrop, Denmark). Sections were rinsed in TBS and mounted in fluorescence mounting medium (Dako, Glostrop, Denmark). As a negative control for the anti-DC-SIGN antiserum, preimmune rabbit serum was used to immunostain serial sections. For the mouse MAbs, the primary antibody was omitted from the negative control slides.
Confocal microscopy.
Images were obtained by using a confocal laser scanning microscope TCS 4D (Leica Lasertechnik, Heidelberg, Germany). All double immunostaining was photographed with sequential scanning techniques.
DC-SIGN–CD4–CCR5 coimmunoprecipitation.
293T cells that stably express CCR5 and CD4 were transfected with pcDNA3–DC-SIGN or a red fluorescent protein (Clontech, Palo Alto, Calif.) (negative control) expression plasmid. Cells were washed three times with cold 1× PBS and divided into two aliquots prior to lysis. One aliquot was lysed in 1 ml of Triton X buffer (300 mM NaCl, 50 mM Tris [pH 7.6], 10% glycerol, 0.5% Triton X, and EDTA-free protease inhibitor from Sigma) at room temperature for 30 min while rocking. The other aliquot was lysed in 1 ml of CHAPSO-based buffer (20 mM Tris [pH 7.5], 100 mM ammonium sulfate, 10% glycerol, 1% CHAPSO, and EDTA-free protease inhibitor from Sigma) at 4°C for 30 min while rotating. The cell debris was settled by centrifuging the samples for 10 min at 14,000 rpm. One hundred microliters of lysate, 2.5 μl of 2 M calcium chloride, 8 μl of antibody, and 50 μl of protein G beads (Pierce) were added to 500 μl of either CHAPSO buffer or Triton X buffer. All samples were rotated overnight at 4°C and run on and SDS-polyacrylamide gel electrophoresis (PAGE) gel. Samples were then blotted for CD4 with rabbit anti-CD4 polyclonal antibodies (previously generated by immunizing New Zealand White rabbits with the recombinant soluble 4 domain, CD4) or a mouse MAb against CCR5 (CTC5; R&D Systems, Minneapolis, Minn.). All lysates were blotted individually for DC-SIGN, CD4, and CCR5 to confirm appropriate expression.
QFACS analysis.
For quantitative FACS (QFACS) analysis, DC-SIGN expression on stably transduced cell lines was quantified as previously described (18, 27), with the exception that a MAb against DC-SIGN (DC028) (2a) was used instead of an anti-AU1 antibody against AU1-tagged DC-SIGN. Since the same secondary reagent was used in this study (phycoerythrin-conjugated Fab goat anti-mouse from Caltag, Burlingame, Calif.), the levels of DC-SIGN quantified can be reasonably compared. However, the confidence in the absolute number of antibody binding sites obtained would not be as great as that obtained with a directly conjugated primary antibody.
RESULTS
cis expression of DC-SIGN enhances viral infection.
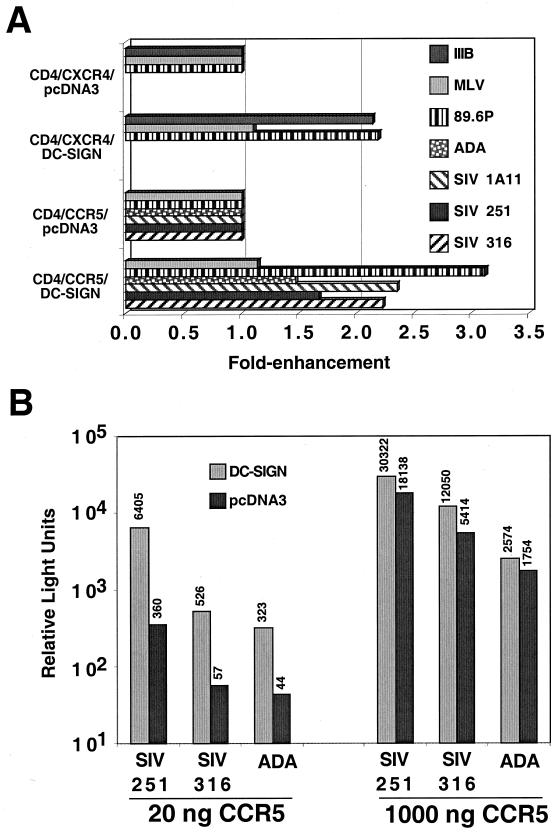
The presence of viral attachment molecules on permissive cells may affect the efficiency of viral entry (37). Since we recently found that DC-SIGN is expressed not only on DCs, but also on CD4+ macrophages in the placenta and lung (34; E. Soilleux, L. S. Morris, G. Leslie et al., submitted for publication), we sought to examine the consequences of DC-SIGN expression on infection efficiency. Using transfected 293T cells expressing high levels of CD4 and coreceptor, we found that cis expression of DC-SIGN reproducibly enhanced HIV-1 and SIV infection by two- to threefold (Fig. 1A). This cis enhancement was specific to HIV-1 and SIV Envs, because infection by murine leukemia virus (MLV) pseudotypes was not affected by the presence of DC-SIGN. Since cell surface densities of CD4 and CCR5 can affect significantly the efficiency of viral entry (26, 30), we sought to determine if the DC-SIGN enhancement effect was more prominent when CCR5 levels were reduced. Indeed, when target cells were transfected with 50-fold less CCR5 expression plasmid than the experiment indicated above, the presence of DC-SIGN was able to enhance R5 HIV-1 and SIV entry by more than 10-fold (Fig. 1B), even though the absolute amount of virus entering these cells was considerably less (compare Fig. 1A and B). Thus, coexpression of DC-SIGN along with CD4 and an appropriate coreceptor enhances virus infection, particularly when CCR5 levels are limiting for virus entry.
FIG. 1.
cis expression of DC-SIGN enhances viral infection. (A) One microgram each of plasmids expressing CD4, coreceptor (CCR5 or CXCR4), and DC-SIGN or vector alone (pcDNA3) was transfected into 293T cells in each well of a 24-well plate. Twenty hours postinfection, the transfected cells were infected with the indicated pseudotyped luciferase reporter viruses. Cells were lysed 4 days postinfection, and luciferase activity was detected as described in Materials and Methods. Results are not shown for ADA and SIV pseudotypes on CXCR4-transfected cells and IIIB pseudotypes on CCR5-transfected cells because they did not result in reproducible infections above the background in the presence or absence of DC-SIGN. Results are representative of three experiments performed in duplicates or triplicates. (B) Infections were performed as in panel A, except that target cells were transfected with the equivalent of 20 ng of CCR5 expression plasmid where indicated. Shown here are averages from infections performed in duplicates. Average raw relative light units are indicated above each bar, so that results from panels A and B can be compared directly.
DC-SIGN expression in T-cell lines enhances virus replication.
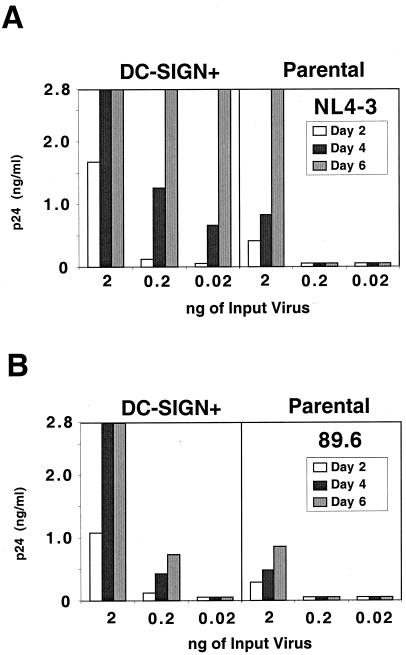
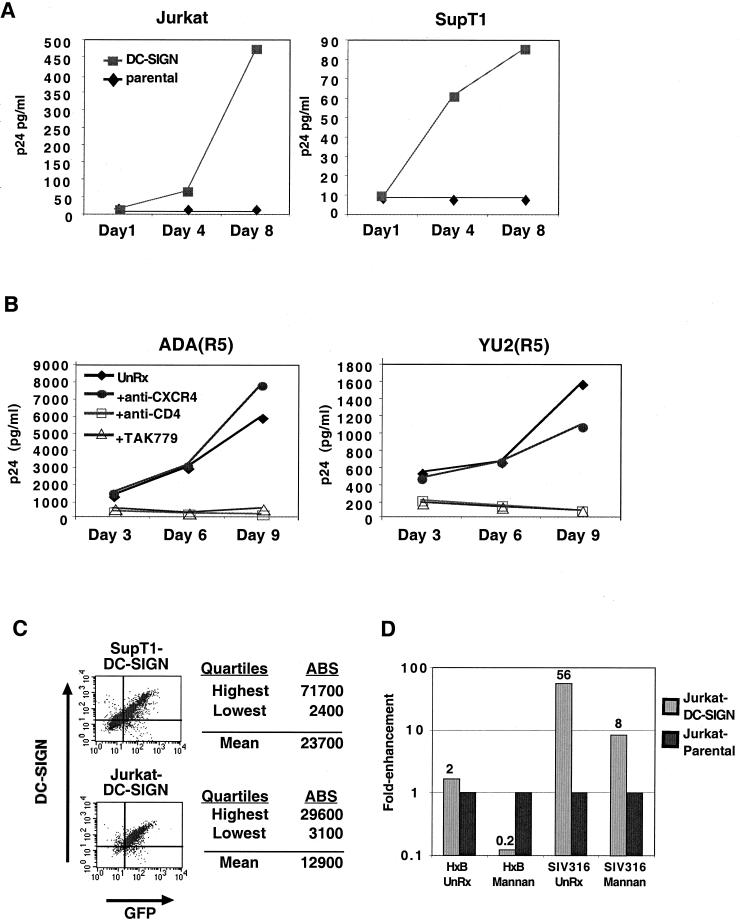
In order to examine the effect of DC-SIGN cis enhancement on more relevant cells types, a variety of T-cell lines were transduced with a murine stem cell retrovirus vector (MIGR1–DC-SIGN–GFP) expressing DC-SIGN in tandem with GFP via an IRES linker. We have previously shown that most T-cell lines commonly used to propagate HIV express tens of thousands of CD4 and CXCR4 molecules per cell, with the exception of Jurkat cells, which express less than a thousand CD4 molecules per cell (18). Jurkat cells also express less than 1,000 copies of CCR5 per cell, but do express high levels of CXCR4 (18). Therefore, we hypothesized that the effect of DC-SIGN enhancement might be most pronounced in this cell line. Indeed, we found that for a given viral inoculum, DC-SIGN-transduced Jurkat cells supported faster viral replication kinetics for both an X4 (NL4–3) and an R5X4 (89.6) viral strain (Fig. 2A and B, respectively). In addition, up to 100-fold less virus (NL4–3) was required for productive replication in DC-SIGN-transduced Jurkat cells (Fig. 2A). Replication of HIV-1 NL4–3 was also modestly enhanced (two- to fivefold increase in peak p24 antigen values) when DC-SIGN was expressed in SupT1, Molt4 Clone8, and PM1 cells (data not shown), all of which express high levels of CD4 and CXCR4 (18). Therefore, DC-SIGN expression enhances CXCR4-dependent virus replication in several T-cell lines, with its effects being more pronounced when CD4 expression levels are limiting.
FIG. 2.
DC-SIGN-expressing cell lines can support more efficient viral replication. Parental Jurkat cells or DC-SIGN-transduced Jurkat cells were spinoculated with 2, 0.2, or 0.02 ng of NL4-3 (X4 [A]) or 89.6 (R5X4 [B]) in 200 μl of medium. Target cells were vigorously washed three times with medium 4 h postinfection. Culture supernatants were half-exchanged with fresh medium on days 2, 4, and 6, and p24 levels were determined with a commercial ELISA kit. Experiments were repeated three times with replication curves taken out to 10 days postinfection. In every case, peak p24 antigen levels in DC-SIGN-transduced Jurkat cells were at least 5- to 10-fold more than that found in Jurkat parental cells. Shown here is one representative experiment. Values greater than 2.8 ng/ml are shown as 2.8 ng/ml on the graph.
We have previously shown that Jurkat, SupT1, and Molt4 Clone 8 cells have very low levels of CCR5 (<1,000 molecules), below the threshold required to support replication by most R5 viral strains (18). We therefore sought to determine if DC-SIGN-transduced Jurkat and SuptT1 cells can support replication by commonly used CCR5-tropic strains. Figure 3A shows that DC-SIGN-transduced SupT1 and Jurkat cells acquired the ability to support productive replication by ADA, an R5-tropic strain. To ensure that this DC-SIGN-enhanced viral entry was CD4 and CCR5 dependent, two R5 virus strains were used to infect Jurkat–DC-SIGN cells in the presence or absence of CD4 and coreceptor antagonists. Figure 3B shows that both ADA and YU2 were able to replicate in Jurkat–DC-SIGN-positive cells and that this replication was completely inhibited by a neutralizing CD4 antibody (Leu3a) or by the CCR5 antagonist TAK779 (1). Addition of an anti-CXCR4 antibody had no appreciable affect on viral replication. Therefore, the presence of DC-SIGN allowed for more efficient usage of low levels of CCR5 for viral entry on SupT1 cells. In the case of Jurkat cells, cis expression of DC-SIGN enabled virus to enter despite low levels of both CD4 and CCR5. Since DC-SIGN did not allow these viruses to enter Jurkat or SupT1 cells by using CXCR4, DC-SIGN had, in effect, expanded the cellular tropism of these viral strains without altering their coreceptor preference.
FIG. 3.
DC-SIGN can enhance viral entry via limiting levels of CCR5. (A) A total of 105 DC-SIGN-transduced Jurkat or SupT1 cells were infected with 0.2 ng of ADA as in Fig. 2. p24 antigen levels in the culture supernatant were determined at days 1, 4, and 8 postinfection. (B) DC-SIGN-transduced Jurkat cells were infected with two different strains of R5 viruses (2 ng each of ADA and YU2) in the absence (UnRx) or presence of anti-CXCR4 (20 μg of MAb 45701 per ml), anti-CD4 (10 μg of Leu3A per ml), or TAK779 (20 μM). Culture supernatants were half-exchanged on days 3, 6, and 9 with fresh growth media and the appropriate blocking agents as indicated. p24 antigen levels were determined with a commercial ELISA kit. The results shown are averages of experiments done in triplicate. (C) Retrovirally (MIGR1–DC-SIGN–GFP) transduced SupT1 and Jurkat cells were stained for DC-SIGN with DC028, and the number of antibody binding sites was determined by QFACS analysis with the Quantum Simply Cellular kit (Sigma). Note that the MIGR1 vector expresses DC-SIGN in tandem with GFP via an IRES linker. (D) cis enhancement effect on DC-SIGN-transduced Jurkat cells can be inhibited by mannan. HxB or SIV316 psuedotyped GFP reporter viruses were used to infect parental Jurkat or DC-SIGN-transduced Jurkat cells in the presence of absence of 100 μg of mannan per ml as described in Materials and Methods. Productive infection was determined by staining for intracellular p24 antigen 3 days postinfection. Results are presented as fold enhancement: that is, the percentage of p24+ cells obtained with Jurkat-DC-SIGN cells divided by the percentage of p24+ cells obtained with Jurkat parental cells. For comparison, the fold enhancement obtained with parental Jurkat cells is shown and is, by definition, normalized to a value of 1.
The cis enhancement effect of DC-SIGN on these cell lines was observed at levels of DC-SIGN expression that are at or near the threshold levels of DC-SIGN (Fig. 3C) previously shown to be required for efficient infection in trans (27). Figure 3C shows the FACS analysis of DC-SIGN expression on SupT1–DC-SIGN and Jurkat–DC-SIGN cells, and quantitative FACS analysis was used to determine the corresponding number of DC-SIGN antibody binding sites on these cells. The ability of mannan to markedly decrease the infection efficiency in DC-SIGN-transduced cell lines, but not in the parental lines, further confirms that the cis enhancement effect is due to DC-SIGN (Fig. 3D). The DC-SIGN-mediated cis enhancement effect on SIV316 entry into Jurkat cells may be more pronounced because SIV316 entry into Jurkat cells would occur via the low level of CCR5 (18) or STRL33/CXCR6 (11) present on Jurkat cells (see Fig. 5 below).
FIG. 5.
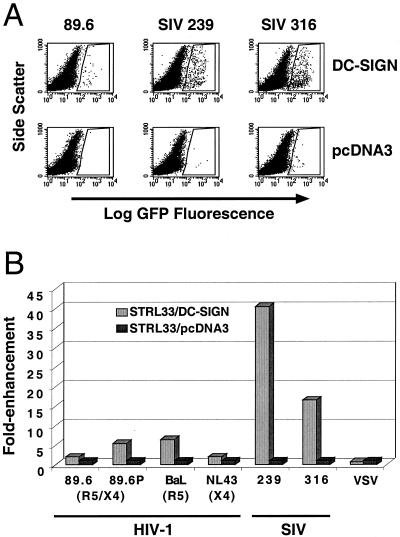
cis enhancement of viral infection via alternate coreceptors. CD4 and STRL33 expression plasmids were transfected into 293T cells either with DC-SIGN or pCDNA3 as a DNA control. The equivalent of only 20 ng of STRL33 expression vector was transfected into each 12-well plate. GFP-reporter viruses pseudotyped with HIV-1 (89.6, 89.6P, BaL, and NL4-3), SIV (239 and 316), and VSV Envs were used to infect these cells 24 h posttransfection. (A) FACs analysis of infected cells. The GFP-positive cells, which are boxed in the figure, indicate productively infected cells. Shown here is one representative experiment out of two. Raw data plots for 89.6P and SIV infections are shown for illustrative purposes. (B) Quantitative analysis of results presented in panel A. Fold enhancement is the percentage of GFP-positive cells obtained with CD4–STRL33–DC-SIGN-transfected cells divided by the percentage of GFP-positive cells obtained with CD4-STRL33-pcDNA3-transfected cells. For comparison, the fold enhancement obtained with CD4-STRL33-pcDNA3-transfected cells is shown and is, by definition, normalized to a value of 1.
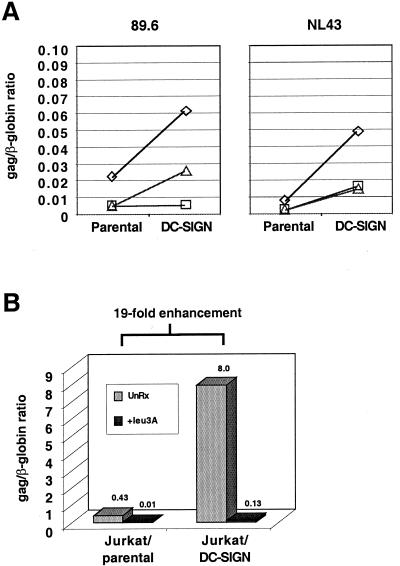
Increased viral replication in DC-SIGN-transduced cell lines is due to enhanced viral entry and is CD4 dependent.
We have previously shown that virus adsorption onto cells is markedly enhanced in the presence of DC-SIGN (27, 28). However, in order to determine if DC-SIGN-enhanced viral replication is due to more efficient viral entry, a quantitative real-time PCR assay for reverse-transcribed products was performed at 20 h postinfection. With the same amount of viral inoculum, we found that there was a significant increase in the number of viral gag DNA copies per cell (represented by the gag/β-globin ratio) in various DC-SIGN-transduced cell lines when compared to their respective parental lines during the first round of infection (Fig. 4A). DC-SIGN-enhanced viral entry was CD4 dependent, because the gag/β-globin ratio was reduced from 8.0 to 0.13 in the presence of Leu3a, a neutralizing anti-CD4 antibody (Fig. 4B). Thus, DC-SIGN itself cannot mediate entry in the absence of CD4, consistent with previous reports (13), and its enhancement effects are at the level of virus entry.
FIG. 4.
DC-SIGN-enhanced viral entry is CD4 dependent. Five nanograms of NL4-3 or 89.6 was spinoculated onto Jurkat (open diamonds), THP-1 (open square), and Molt4 Clone 8 (open triangle) cells and their DC-SIGN-transduced counterparts. Quantitative real-time PCR for gag DNA and cellular β-globin DNA copies was performed 20 h postinfection. (A) Results are presented as the ratio of Gag to β-globin DNA copies, which is indicative of the number of gag DNA copies per cell. (B) The experiment was repeated as in panel A for NL4-3 on Jurkat cells (parental versus DC-SIGN transduced) with 50 ng of viral inoculum. For Leu3A inhibition, target cells were preincubated for 30 min with 10 μg of purified antibody per ml before viral infection. Note that the Gag/β-globin ratio increased from 0.43 to 8 (19-fold) when the same amount of viral inoculum was used to infect the same number of Jurkat parental versus Jurkat DC-SIGN-transduced cells. In both cases, Leu3A inhibited viral entry by more than 95%.
cis enhancement of viral infection via alternate coreceptors.
Alternate coreceptors such as STRL33 tend to support virus infection less efficiently than CCR5 and CXCR4. We have shown that STRL33 is expressed on some CD4+ T cells at levels (<10,000 molecules per cell) below that needed to support efficient entry for most virus strains (30). Therefore, we determined whether DC-SIGN could enhance infection when coexpressed with this alternate coreceptor. By using cells expressing STRL33 at levels previously determined to result in little or no infection by either SIV or HIV Env pseudotypes (30), we found that expression of DC-SIGN-enhanced virus infection between 2- and 40-fold, depending on the viral Env used (Fig. 5A and B). Infection by VSV pseudotypes was not affected by the presence of DC-SIGN (Fig. 5B). A similar level of enhancement was obtained when luciferase rather than GFP reporter viruses were used (data not shown). Thus, the ability of DC-SIGN to enhance viral infection when expressed in cis extends to coreceptors other than CCR5 and CXCR4.
DC-SIGN is expressed on permissive CD4+ CCR5+ cells in situ
Since DC-SIGN can enhance viral entry on permissive cells expressing low levels of CD4 or coreceptor, we sought to obtain evidence of DC-SIGN expression on such candidate permissive cells in vivo. We have previously shown that DC-SIGN is expressed on both maternal decidual macrophages and fetal Hofbauer cells in human placenta (34) and on some human alveolar macrophages (E. Soilleux, L. S. Morris, G. Leslie et al., submitted for publication). Both alveolar and placental tissue macrophages have been reported to express very low levels of CD4 and to be targets for HIV infection in vivo (16, 17, 20, 22, 25, 31). In the case of SIV, macrophage tropism is often associated with changes in the viral Env protein that either make it CD4 independent or better able to infect cells that express low levels of CD4 (2, 22; B. Puffer, S. Pohlmann, A. L. Edinger et al., submitted for publication). Therefore, DC-SIGN expression may contribute to the establishment of a viral reservoir in these cells by allowing more efficient entry.
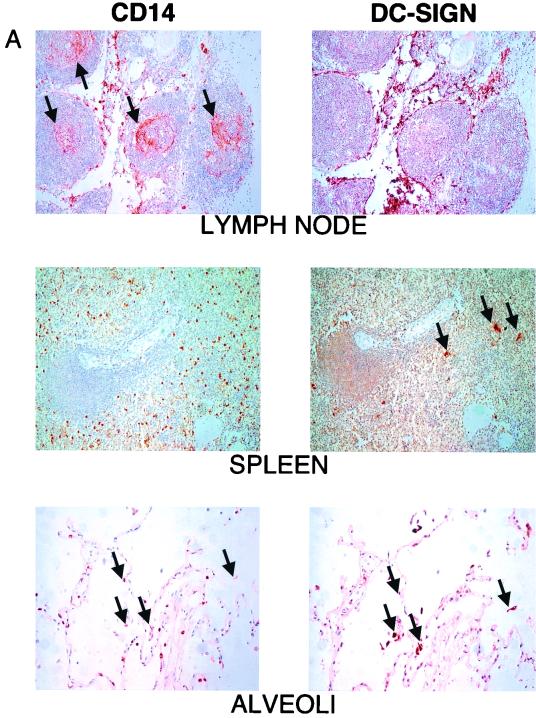
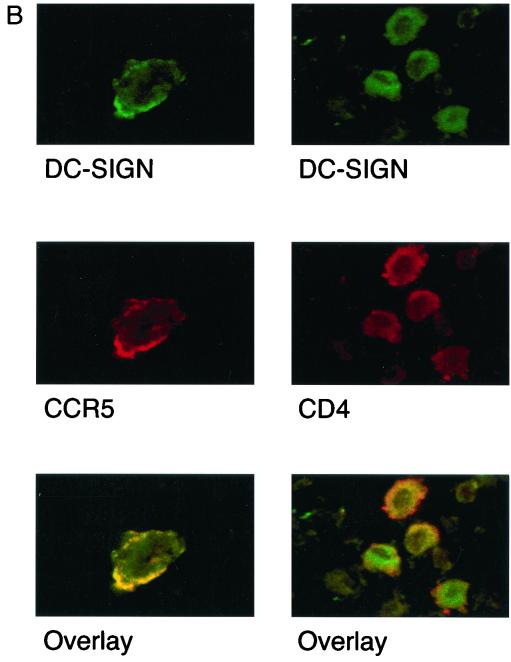
Accordingly, we performed immunohistochemistry on various tissues to determine if DC-SIGN was expressed on other macrophage populations in situ. We found that while CD14+ macrophages from lymph node, spleen, and liver were DC-SIGN negative (data not shown) (Fig. 6A), DC-SIGN expression could clearly be detected on placental, decidual, and alveolar macrophages (34) (Fig. 6A). It is known that alveolar macrophages are heterogeneous with regard to their morphology, immunophenotype (CD14 expression), and function (15, 39), and we note that DC-SIGN was only coexpressed with a subset of CD14low alveolar macrophages (Fig. 6A). Strikingly, when confocal microscopy was performed on alveolar macrophages to determine coexpression of CCR5 and CD4 with DC-SIGN, we found physical colocalization of DC-SIGN and CCR5, and, to a lesser extent, CD4 (Fig. 6B). In Fig. 6B, essentially 100% of CCR5+ membrane areas (red) were also positive for DC-SIGN (yellow overlap). The physical colocalization of DC-SIGN with CCR5 and CD4 may in part explain the cis enhancement effect of DC-SIGN. However, we were unable to determine that DC-SIGN is physically associated with CCR5 or CD4 in coimmunoprecipitation experiments in transiently transfected cells, at least under the conditions examined (see Materials and Methods) (data not shown).
FIG. 6.
DC-SIGN expression on permissive tissue macrophages. Immunohistochemistry was performed on tissue sections as indicated. Staining for CD14 (macrophage marker) and DC-SIGN was performed on 5-μm serial sections in order to show costaining of CD14 and DC-SIGN on the same cell. (A) (Lymph node) CD14+ macrophages (arrows in left panel) in the germinal centers appear negative for DC-SIGN. (DC-SIGN expression is indicated by brown cells in the right panel.) (Spleen) Numerous CD14+ macrophages are scattered throughout the red and white pulp (left panel); the few DC-SIGN-positive cells (right panel, arrows) appear CD14−. (Alveoli) DC-SIGN-positive cells in the alveoli also express low levels of CD14 (arrows in right and left panels point to cells staining for both markers). Alveolar macrophages are a heterogeneous population and can vary in their amount of CD14 expression. DC-SIGN-positive cells in the alveoli appear to be restricted to CD14low cells (arrows). (B) Confocal microscopy performed on alveolar macrophages showing expression of DC-SIGN (red) with CCR5 (green) and CD4 (green). Note the almost complete colocalization of DC-SIGN with CCR5 (yellow overlap).
DISCUSSION
Binding of the HIV-1 Env protein to CD4 and a coreceptor is required for virus entry. However, attachment of virus to the cell surface can occur via low-affinity interactions with a relatively large number of cellular molecules (reviewed in reference 37). By concentrating virus on the cell surface, attachment factors may make subsequent receptor engagement more efficient. Among attachment factors, the C-type lectin DC-SIGN is unusual in that it binds to gp120 with very high affinity (6) and is particularly efficient at retaining bound virus in an infectious state for prolonged periods of time and in presenting bound virus to receptor-positive cells in trans (13). Because DC-SIGN is expressed at high levels on some types of DCs, it has been suggested that HIV-1 may bind to DCs via DC-SIGN and, through the normal trafficking patterns of this antigen-presenting cell, be delivered to lymphoid organs that serve as the major site for virus replication in vivo (13).
We have found that DC-SIGN is expressed on certain types of macrophages in vivo, including macrophages in human placenta (34) and alveolar macrophages (Fig. 6A) (E. Soilleux, L. S. Morris, G. Leslie et al., submitted for publication), In addition, DC-SIGN expression can be induced on monocyte-derived macrophages by treatment with interleukin 13, raising the possibility that DC-SIGN may sometimes be expressed on additional cell types in vivo that can support virus replication (Soilleux et al., submitted). Therefore, it becomes important to determine if the presence of this efficient virus attachment factor on macrophages impacts virus entry not only in trans, but in cis as well.
We found that DC-SIGN expression in cis enhanced the efficiency of virus infection, especially when CD4 and coreceptor levels were limiting. The ability of DC-SIGN to allow R5 viral infection of T-cell lines such as Jurkat and SupT1 cells was particularly notable, since these cells express vanishingly small amounts of surface CCR5 (18) and are otherwise refractory to R5 virus entry (5, 7). This finding also shows that expression of DC-SIGN in cis can, in effect, change viral tropism without affecting coreceptor usage. The ability of DC-SIGN, and perhaps other attachment factors as well, to enhance virus infection in cis when receptor levels are limiting may not be appreciated, since cell lines commonly used to assess which coreceptors are used by a given virus strain typically express tens of thousands of CD4 and coreceptor molecules (18). In contrast, receptor levels on primary cell types are often much lower. For example, freshly isolated primary CD4+ lymphocytes generally have less than 10,000 CCR5 and CXCR4 antibody binding sites per cell (18). Therefore, the ability of DC-SIGN to enhance virus infection in cis demonstrates that attachment factors can help virus infect cells under coreceptor-limiting conditions.
We also found that DC-SIGN can enhance virus infection in cis when CD4 expression levels are low. In vivo, CD4 levels can be limiting for virus infection under some conditions, especially on some types of tissue macrophages. Alveolar macrophages, for example, express very low levels of CD4 (2, 19, 22). Interestingly, we found that a significant fraction of these cells also coexpress DC-SIGN and CCR5, thus providing a cellular environment whereby the cis enhancement effect of DC-SIGN can come into play. It is interesting to note that alveolar macrophages have been proposed to be a reservoir in late stages of disease, because there is a significant increase of HIV-1 RNA in alveolar macrophages, but not monocytes from subjects with AIDS (32). Among HIV-positive asymptomatic subjects, HIV-1 was undetectable or at low levels in both blood monocytes and alveolar macrophages (32). Thus, whether progressive HIV disease (as a chronic inflammatory state) can lead to tissue microenvironments that favor DC-SIGN upregulation is a matter for future studies.
In addition to enhancing virus infection via the major coreceptors when expressed in cis, DC-SIGN also made infection via an alternate coreceptor more efficient. Although more than 10 chemokine receptors or 7TM GPCRs (other than CCR5 and CXCR4) have been reported to have coreceptor activity in vitro, some of these receptors are not expressed on CD4+ cell types in vivo, or are expressed at levels that do not support efficient infection, at least for most virus strains (reviewed in references 4, 9, and 10). The alternate coreceptor, STRL33, is an example of this. When overexpressed on cell lines, STRL33 can mediate efficient infection by a number of HIV-1, HIV-2, and SIV strains (11, 30). However, STRL33 expression in vivo is limited to only subsets of CD4+ T cells and NK cells (30, 38, 40), where on average less than 10,000 molecules are expressed at the cell surface (30). While comparable to CCR5 and CXCR4 expression, this level of STRL33 expression is below the threshold needed for most viruses to utilize this coreceptor (30). Our finding that DC-SIGN can allow usage of alternate coreceptors like STRL33 at expression levels that normally do not support viral entry raises the possibility that the panoply of coreceptors present on permissive cell populations in vivo can actually be used for entry if DC-SIGN was upregulated on the same cells. These findings may have implications for viral pathogenesis in vivo if conditions exist that can upregulate DC-SIGN, or perhaps other attachment factors, in certain tissue microenvironments. To this end, we found that only macrophages in certain tissues (e.g., lung and placenta) express DC-SIGN (Fig. 6A) (34), indicating that microenvironmental cues play a role in regulating the expression of DC-SIGN.
The precise mechanism by which DC-SIGN expression enhances infection in cis is unknown at present. However, the high affinity of DC-SIGN for HIV-1 Env suggests that DC-SIGN may serve to anchor the virion to the cell surface and thus increase the local concentration of viral particles at the plasma membrane. In situations where CD4 or coreceptor is limiting, this local concentration effect will likely increase the probability of the virion encountering its requisite entry factors. The colocalization of DC-SIGN with CD4 and CCR5 on the surface of alveolar macrophages suggests that the cis enhancement effect may be partially due to the ability of DC-SIGN to concentrate virions in physical proximity to its cognate receptors. Whether there are direct interactions between DC-SIGN and CD4 and CCR5, both of which are glycosylated, or whether these proteins partition into membrane microdomains remains to be determined.
In summary, our studies provide evidence that DC-SIGN can enhance infection in cis in addition to its reported trans infection properties. Our findings indicate that under some conditions, viral tropism can be modulated by a molecule other than a coreceptor. Since DC-SIGN is not commonly expressed on in vitro-cultured PBMCs and macrophages, studies of viral tropism with in vitro-cultured primary cells, which are at best a poor mimic of the complex microenvironmental milieu should be interpreted with some degree of caution. Whether the phenomenon of DC-SIGN cis enhancement plays a major role in the pathogenesis of HIV disease awaits confirmation by studying DC-SIGN expression in animal models and by the use of antibodies or other inhibitors of virus–DC-SIGN interactions.
ACKNOWLEDGMENTS
We acknowledge the support of the UCLA AIDS Institute and the flow cytometry core (UCLA CFAR grant, NIH AI-28697). B.L. is a recipient of the Burroughs Wellcome Fund Career Development Award and is supported by NIH grant HL 03923 and a Frontiers of Science Award from UCLA. E.S. is supported by a Medical Research Council Clinical Training Fellowship and by the Sackler Foundation. N.C. is supported by the Medical Research Council and the Cancer Research Campaign. M.H.M. is supported by NIH grant AI46942. U.O. is supported by NIH grant HL03984-L. R.W.D. is an Elizabeth Glaser Scientist supported by the Pediatrics AIDS Foundation and a recipient of the Burroughs Wellcome Fund Translational Research Award and is supported by NIH grant AI40880.
REFERENCES
- 1.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Baribaud F, Pohlmann S, Sparwasser T, Kimata M T, Choi Y K, Haggarty B S, Ahmad N, Macfarlan T, Edwards T G, Leslie G J, Arnason J, Reinhart T A, Kimata J T, Littman D R, Hoxie J A, Doms R W. Functional and antigenic characterization of human, rhesus macaque, pigtailed macaque, and murine DC-SIGN. J Virol. 2001;75:10281–10289. doi: 10.1128/JVI.75.21.10281-10289.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashirova A A, Geijtenbeek T B, van Duijnhoven G C, van Vliet S J, Eilering J B, Martin M P, Wu L, Martin T D, Viebig N, Knolle P A, KewalRamani V N, van Kooyk Y, Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 5.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R W. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis B M, Scharnowske S, Watson A J. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1992;89:8356–8360. doi: 10.1073/pnas.89.17.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dejucq N, Simmons G, Clapham P R. Expanded tropism of primary human immunodeficiency virus type 1 R5 strains to CD4+ T-cell lines determined by the capacity to exploit low concentrations of CCR5. J Virol. 1999;73:7842–7847. doi: 10.1128/jvi.73.9.7842-7847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doms R W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 9.Doms R W, Trono D. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 2000;14:2677–2688. doi: 10.1101/gad.833300. [DOI] [PubMed] [Google Scholar]
- 10.Edinger A L, Clements J E, Doms R W. Chemokine and orphan receptors in HIV-2 and SIV tropism and pathogenesis. Virology. 1999;260:211–221. doi: 10.1006/viro.1999.9819. [DOI] [PubMed] [Google Scholar]
- 11.Edinger A L, Hoffman T L, Sharron M, Lee B, O'Dowd B, Doms R W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 12.Fortin J-F, Cantin R, Lamontagne G, Tremblay M. Host-derived ICAM-1 glycoproteins is incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol. 1997;5:3588–3596. doi: 10.1128/jvi.71.5.3588-3596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geijtenbeek T B, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, KewalRamani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 14.Geijtenbeek T B, Torensma R, van Vliet S J, van Duijnhoven G C, Adema G J, van Kooyk Y, Figdor C G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 15.Haugen T S, Nakstad B, Skjonsberg O H, Lyberg T. CD14 expression and binding of lipopolysaccharide to alveolar macrophages and monocytes. Inflammation. 1998;22:521–532. doi: 10.1023/a:1022302228051. [DOI] [PubMed] [Google Scholar]
- 16.Kesson A M, Fear W R, Williams L, Chang J, King N J, Cunningham A L. HIV infection of placental macrophages: their potential role in vertical transmission. J Leukoc Biol. 1994;56:241–246. doi: 10.1002/jlb.56.3.241. [DOI] [PubMed] [Google Scholar]
- 17.Lebargy F, Branellec A, Deforges L, Bignon J, Bernaudin J F. HIV-1 in human alveolar macrophages from infected patients is latent in vivo but replicates after in vitro stimulation. Am J Respir Cell Mol Biol. 1994;10:72–78. doi: 10.1165/ajrcmb.10.1.8292383. [DOI] [PubMed] [Google Scholar]
- 18.Lee B, Sharron M, Montaner L J, Weissman D, Doms R W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewin S R, Sonza S, Irving L B, McDonald C F, Mills J, Crowe S M. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res Hum Retroviruses. 1996;12:877–883. doi: 10.1089/aid.1996.12.877. [DOI] [PubMed] [Google Scholar]
- 20.Martin A W, Brady K, Smith S I, DeCoste D, Page D V, Malpica A, Wolf B, Neiman R S. Immunohistochemical localization of human immunodeficiency virus p24 antigen in placental tissue. Hum Pathol. 1992;23:411–414. doi: 10.1016/0046-8177(92)90088-k. [DOI] [PubMed] [Google Scholar]
- 21.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type I attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori K, Rosenzweig M, Desrosiers R C. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Doherty U, Swiggard W J, Malim M H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel M, Yanagishita M, Roderiquez G, Bou-Habib D C, Oravecz T, Hascall V C, Norcross M A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res Hum Retroviruses. 1993;9:167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- 25.Plata F, Garcia-Pons F, Ryter A, Lebargy F, Goodenow M M, Dat M H, Autran B, Mayaud C. HIV-1 infection of lung alveolar fibroblasts and macrophages in humans. AIDS Res Hum Retroviruses. 1990;6:979–986. doi: 10.1089/aid.1990.6.979. [DOI] [PubMed] [Google Scholar]
- 26.Platt E J, Wehrly K, Kuhnman S E, Chesbro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohlmann S, Baribaud F, Lee B, Leslie G J, Sanchez M D, Hiebenthal-Millow K, Munch J, Kirchhoff F, Doms R W. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohlmann S, Soilleux E J, Baribaud F, Leslie G J, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib D C, Mostowski H, Norcross M A. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol. 1995;69:2233–2239. doi: 10.1128/jvi.69.4.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharron M, Pohlmann S, Price K, Lolis E, Tsang M, Kirchhoff F, Doms R W, Lee B. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood. 2000;96:41–49. [PubMed] [Google Scholar]
- 31.Sheikh A U, Polliotti B M, Miller R K. Human immunodeficiency virus infection: in situ polymerase chain reaction localization in human placentas after in utero and in vitro infection. Am J Obstet Gynecol. 2000;182:207–213. doi: 10.1016/s0002-9378(00)70514-x. [DOI] [PubMed] [Google Scholar]
- 32.Sierra-Madero J G, Toossi Z, Hom D L, Finegan C K, Hoenig E, Rich E A. Relationship between load of virus in alveolar macrophages from human immunodeficiency virus type 1-infected persons, production of cytokines, and clinical status. J Infect Dis. 1994;169:18–27. doi: 10.1093/infdis/169.1.18. [DOI] [PubMed] [Google Scholar]
- 33.Soilleux E J, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 34.Soilleux, E. J., L. S. Morris, S. Poehlmann, B. Lee, R. W. Doms, J. Trowsdale, and N. Coleman. DC-SIGN expression analysis in the adult fetus and placenta: implications for the vertical transmission of HIV. J. Pathol., in press. [DOI] [PubMed]
- 35.Steinman R M. DC-SIGN: a guide to some mysteries of dendritic cells. Cell. 2000;100:491–494. doi: 10.1016/s0092-8674(00)80684-4. [DOI] [PubMed] [Google Scholar]
- 36.Tominaga Y, Kita Y, Satoh A, Asai S, Kato K, Ishikawa K, Horiuchi T, Takashi T. Affinity and kinetic analysis of the molecular interaction of ICAM-1 and leukocyte function-associated antigen-1. J Immunol. 1998;161:4016–4022. [PubMed] [Google Scholar]
- 37.Ugolini S, Mondor I, Sattentau Q J. HIV-1 attachment: another look. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 38.Unutmaz D, Xiang W, Sunshine M J, Campbell J, Butcher E, Littman D R. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J Immunol. 2000;165:3284–3292. doi: 10.4049/jimmunol.165.6.3284. [DOI] [PubMed] [Google Scholar]
- 39.van Hal P T, Wijkhuijs J M, Mulder P G, Hoogsteden H C. Proliferation of mature and immature subpopulations of bronchoalveolar monocytes/macrophages and peripheral blood monocytes. Cell Prolif. 1995;28:533–543. doi: 10.1111/j.1365-2184.1995.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 40.Wilbanks A, Zondlo S C, Murphy K, Mak S, Soler D, Langdon P, Andrew D P, Wu L, Briskin M. Expression cloning of the STRL33/BONZO/TYMSTR ligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol. 2001;166:5145–5154. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]