Abstract
Deletion of a region of the human immunodeficiency virus type 2 (HIV-2) 5′ leader RNA reduces genomic RNA encapsidation to about 5% that of wild-type virus with no defect in viral protein production but severely limits virus spread in Jurkat T cells, indicating that this region contains a major cis-acting encapsidation signal, or psi (Ψ). Being upstream of the major splice donor, it is present on all viral transcripts. We have shown that HIV-2 selects its genomic RNA for encapsidation cotranslationally, rendering wild-type HIV-2 unable to encapsidate vector RNAs in trans . Virus with Ψ deleted, however, encapsidates an HIV-2 vector, demonstrating competition for Gag protein. HIV-2 overcomes the lack of packaging signal location specificity by two novel mechanisms, cotranslational packaging and competition for limiting Gag polyprotein.
Human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively), the etiological agents of AIDS in humans (18), are members of the lentivirus group of retroviruses. Despite a similar genetic organization, homology at the nucleotide level is limited, reflecting different evolutionary origins; HIV-1 is closely related to the chimpanzee group of simian immunodeficiency viruses, and HIV-2 is related to the macaque group (14).
Encapsidation of full-length viral genomic RNA is an essential stage in the life cycle of all retroviruses. In lentiviruses, gene expression is temporally regulated, and full-length RNA is produced only during late stages of infection. This RNA serves as both the viral genome and the mRNA for viral core (Gag) and enzymatic (Pol) gene products. Genomic RNA constitutes approximately 1% of the total RNA in an infected cell but is the major species incorporated into virus particles. Thus, the process of encapsidation is specific. Specificity is mediated by cis-acting signals present within the RNA and by protein factors acting in trans, in particular, the viral Gag polyprotein. The cis-acting signals in all retroviruses are found in the 5′ untranslated leader region of the genome and have been well characterized for a number of viruses, including HIV-1. Deletion and substitution mutagenesis have mapped these signals, known as psi (Ψ) or E, revealing many of them to include stem-loop structures possessing purine-rich sequences at their termini (1, 7, 20, 25, 30, 44). The core Ψ region in HIV-1, stem-loop 3 in the leader RNA, is immediately downstream of the major splice donor, upstream of the gag open reading frame (ORF), and thus present only on the viral genomic RNA. However, other RNA structures situated both upstream and downstream of the splice donor have roles in encapsidation, notably, the dimerization initiation site stem-loop, or stem-loop 1 (5, 6, 8, 25, 26, 32, 44).
The uncleaved HIV-1 Gag polyprotein specifically recognizes and binds to RNAs that contain Ψ (24). It is unlikely that Gag cleavage products are responsible for this recognition, as protease-deficient viruses still encapsidate their genomes efficiently and the majority of proteolytic cleavage occurs after the particle leaves the cell during virion maturation. HIV-1 Gag is able to encapsidate RNA without being translated in cis from the viral genome (34), allowing HIV-1 to be successfully used as a gene vector system (37, 43, 45). The nucleocapsid (NC) domain of Gag, in the context of the uncleaved polyprotein, confers RNA binding specificity via its two zinc finger regions, which are essential for the recognition of Ψ (10, 16, 17, 43, 44, 48). Their disruption abrogates viral replication. The binding of Gag to stem-loop 3 via these domains has been well characterized by both functional and structural studies (4, 8, 19, 33, 40, 49) and appears to cause a change in RNA structure that may enable the subsequent nucleation of ribonucleoprotein complexes via Gag-Gag and nonspecific Gag-RNA interactions.
HIV-2 RNA encapsidation has been less extensively studied (2, 15, 23, 22, 35, 41, 47). Although the components involved are similar to those in HIV-1, the mechanisms appear to be somewhat different. It was previously demonstrated that deletion of regions upstream of the splice donor causes a significant reduction in encapsidation efficiency, whereas those downstream appear to have only a mild effect (23, 22, 35). Thus, in contrast to the situation with HIV-1, all viral messages will contain Ψ. Other authors found downstream regions to be involved (2, 15, 41). It was also demonstrated that there is a nonreciprocal packaging relationship between HIV-1 and HIV-2: wild-type HIV-2 is able to encapsidate only its own RNA, whereas HIV-1 efficiently encapsidates both HIV-1 and HIV-2 vector constructs in addition to its own RNA. When HIV-2 vectors are encapsidated by an HIV-1 helper, both full-length and spliced HIV-2 vector RNAs are encapsidated, confirming that a functional Ψ is upstream of the splice donor in HIV-2 (23).
HIV-2 encapsidates its genomic RNA in a cotranslational manner such that only genomic HIV-2 RNAs which are templates for a full-length Gag polyprotein containing an intact NC region are efficiently incorporated into progeny virions (22). It is thought that this mechanism arises via the newly translated Gag protein binding to its own template RNA at the polysome and has been termed cis-acting encapsidation. This mechanism explains the inability of wild-type HIV-2 to encapsidate either HIV-1 or HIV-2 vectors in trans. However, several published studies have demonstrated that HIV-2 can be used as a vector (2, 41, 47).
In this study, mutagenesis of the leader region of HIV-2 identified a 28-nucleotide region upstream of the major splice donor that, when deleted, reduces encapsidation efficiency to levels comparable to those seen with the most severe Ψ mutations in HIV-1. This deletion severely limits virus replication in Jurkat T cells but causes no apparent defect in transcription or protein expression in COS-1 cells in transient transfection assays, and proteolytic processing of viral polyproteins appears normal. We have observed a competition effect in cotransfection assays between wild-type HIV-2 and Ψ region mutant HIV-2; this effect results in an increase in wild-type encapsidation and a corresponding decrease for the mutant. These findings are consistent with the amount of available Gag polyprotein being the limiting factor for HIV-2 encapsidation. We have further demonstrated that Ψ-containing HIV-2 vectors that are unable either to encapsidate themselves or to be encapsidated by wild-type HIV-2 are nonetheless able to compete for Gag made by Ψ region mutants, leading to efficient incorporation of vector RNA into virus particles. These findings have allowed the design of an HIV-2 vector system in which packaging competition is readily demonstrable in transduction assays. Furthermore, encapsidation of HIV-2 vectors is greatly enhanced by the inclusion of sequences from the gag ORF. Last, we show that it is likely that contamination of vector preparations by helper virus will be eliminated due to the observed competition effect.
MATERIALS AND METHODS
Plasmid construction.
pSVR is an infectious proviral clone of the ROD strain of HIV-2 containing the replication origin of simian virus 40 and has been previously described (35). Restriction sites, where given, are numbered relative to the first nucleotide of the viral RNA. Proviral constructs pSVRΔ1, pSVRΔ2, pSVRΔ3, and pSVRΔ4, containing deletions in the 5′ leader region, have been previously described. The positions of these and newly introduced deletions are shown in Fig. 1A. Deletion mutations in the 5′ leader were introduced by site-directed mutagenesis by the method of Kunkel et al. (29) into a subclone of HIV-2, pGRAXS, which has been previously described (23). The mutagenic oligonucleotide used for construction of the Ψ1 deletion was 5′-GGCAGCGTGGAGCGGGGTGAAGGTAAGTACC-3′, and that used for construction of the DM deletion was 5′-GGCAGTAAGGGCGGCAGGAGCGCGGGCCGAGGTACCAAAGGC-3′. Sequences from the resulting subclones, pGRAXΨ1 and pGRAXDM, containing the deletions were introduced into the provirus by exchanging an AatII (position −1384)-XhoI (position 2032) fragment. The DM-Ψ1 double mutation was constructed by mutating pGRAXDM using the Ψ1 oligonucleotide and introducing this construct into the provirus.
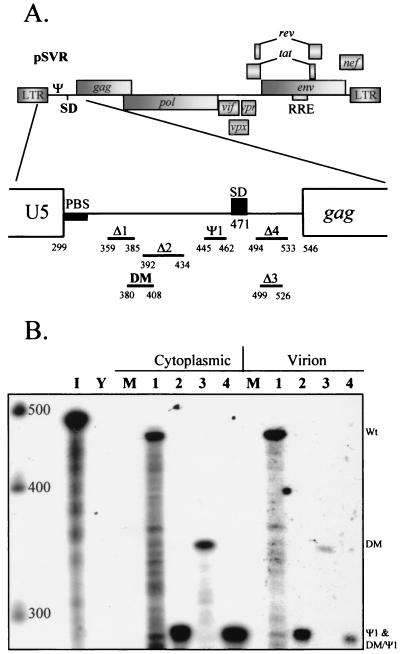
FIG. 1.
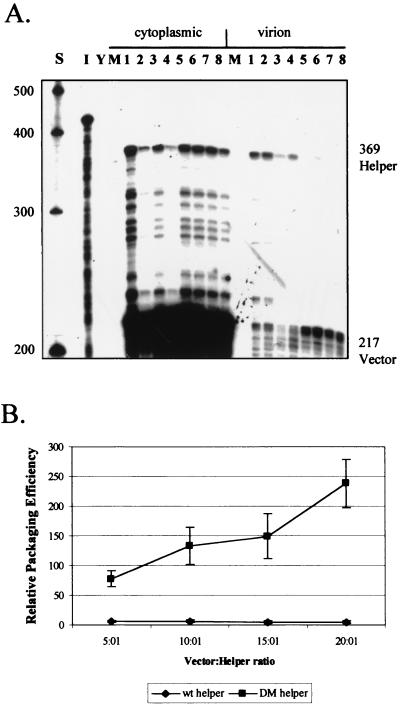
Deletion mutations of the HIV-2 leader region. Deletions were introduced into the infectious proviral clone pSVR by site-directed mutagenesis, and their effects on RNA encapsidation were assessed by RPAs. (A) Schematic representation of the locations of DM and Ψ1 deletions in the HIV-2 leader, as well as those characterized by previous studies. LTR, long terminal repeat; SD, splice donor; RRE, Rev-responsive element; PBS, primer binding site. (B) RPA of RNA from COS-1 cells transiently transfected with new mutants using the KSΨ2KE riboprobe. Wt, wild type. Lanes: 1, pSVR; 2, pSVRΨ1; 3, pSVRDM; 4, pSVRDM/Ψ1; I, input riboprobe diluted 1/100; Y, yeast RNA plus RNase (control); M, RNA from mock-transfected cells. The leftmost lane contains RNA size markers (Century Plus; Ambion).
The HIV-2 vector pSVRΔH is a vector based on pSVR containing a premature stop codon in the capsid (CA) region of the gag ORF. This construct was generated by digestion of a HindIII site (position 1458), subsequent refilling with the Klenow fragment of T4 DNA polymerase, and religation of the DNA. pSVRDMΔH contains the DM deletion in the leader region and the stop codon from pSVRΔH; it was generated by introducing an EcoRV (position 1101)-XhoI (position 2032) fragment from pSVRΔH into pGRAXDM. The AatII (position 11444)-XhoI (position 2032) fragment from this plasmid was then used to replace the same region of pSVR. pSVRΔX was generated by introducing an artificial XbaI site at position 555 by site-directed mutagenesis as described above using the mutagenic oligonucleotide 5′-GGAGATGGGCTCTAGAAACTCCG-3′. Subsequent partial digestion with XbaI allowed removal of almost the entire gag and pol ORFs (positions 555 to 5067). The HIV-2 vectors pSVRΔAX, pSVRΔHX, pSVRΔpol, pSVRΔHΔpol, and pSVRΔpolncm have been previously described (22). pSVRΔNB was generated as follows. An EheI fragment (positions 306 to 5864) was removed from pSVR to generate pSVRΔE. This construct was subsequently digested with NsiI and BstXI, deleting a 550-bp fragment of the env gene (positions 6369 to 6927) but leaving the Rev-responsive element and the rev and tat ORFs intact. A DNA linker containing a SalI site was ligated into this position after blunting with T4 DNA polymerase as described above, generating pSVRΔENBSalI. The EheI fragment was reintroduced into this plasmid, giving pSVRΔNB (see Fig. 7A). pSVRΔNBDM (see Fig. 7A) was generated by replacing the AatII-XhoI (11444 to 2037) region of pSVRΔNB with the same region of pSVRDM. In pSVRΔNBPuroΔE and pSVRΔNBPuroΔH (see Fig. 7A), both based on pSVRΔNB, a SalI fragment from plasmid KSIISVPuro was introduced into the linker site, and an EcoRV fragment (positions 1101 to 2939) was removed or replaced with the same region of pSVRΔH, respectively. pCMV-VSVG contains the vesicular stomatitis virus (VSV) G glycoprotein gene in the context of pCDNA3 (Invitrogen). All plasmids based on HIV-2 proviral sequences were grown in TOPF′10 (Invitrogen) Escherichia coli at 30°C or room temperature to avoid recombination. All other plasmids were grown in DH5α E. coli under standard conditions.
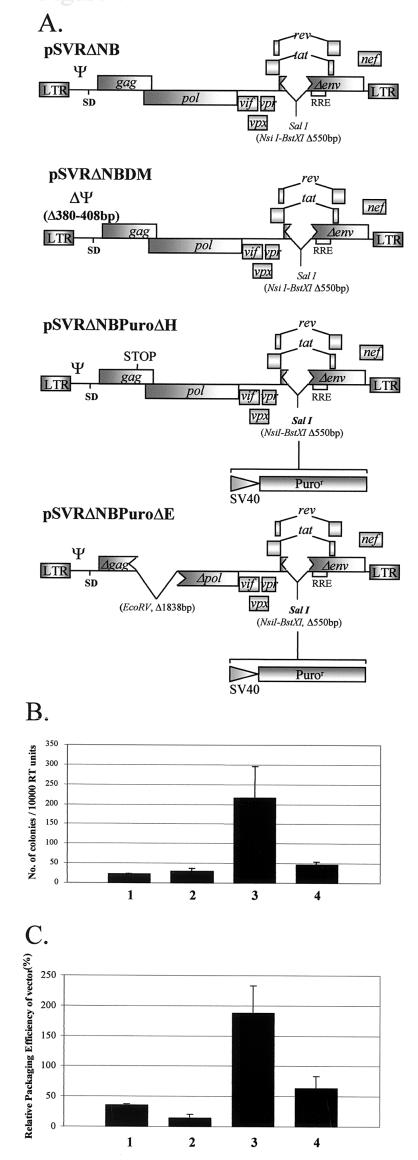
FIG. 7.
First-generation HIV-2 vector system. (A) Structures of two HIV-2 helper constructs as well as two HIV-2 vectors containing a puromycin resistance gene cassette under the control of the simian virus 40 promoter. See the legend to Fig. 1 for definitions. (B) Results of transduction experiments with HeLa CD4+ LTR-βgal cells and different combinations of the helper and the vector described above, pseudotyped with the VSV G glycoprotein envelope. Results are the means of three experiments and are expressed as CFU/10,000RTU; error bars represent the standard error of the mean between experiments. Bars: 1, pSVRΔNB-pSVRΔNBPuroΔH; 2, pSVRΔNB-pSVRΔNBPuroΔE; 3, pSVRDM-pSVRΔNBPuroΔH; 4, pSVRDM-pSVRΔNBPuroΔE. (C) Quantification of vector RNA encapsidation by an RPA using the SKH2CA riboprobe and different helper-vector combinations. Results are the means of three separate experiments; error bars represent the standard error of the mean between experiments. Bars are as described for panel B.
Plasmids used for the generation of antisense riboprobes for use in RNase protection assays (RPAs) were generated as follows. Plasmids KS2ΨKE and KS2ES have been previously described (23, 22). They generate antisense transcripts of regions of the HIV-2 genome corresponding to positions 306 to 751 and 4915 to 5284, respectively, and are in the context of the pBluescript KSII(+) transcription vector (Stratagene). Plasmid KS2ΨΕP generates an antisense probe for the viral sequence between EheI (position 306) and PstI (position −286) and is also in the context of pBluescript. Plasmid SKH2CA generates an antisense probe for the CA region of the gag ORF. In vitro transcription of linearized template DNA was carried out using T3 or, in the case of SKH2CA, T7 RNA polymerase and a riboprobe transcription system (Promega).
Cell culturing and transfection.
COS-1 simian epithelioid cells were maintained in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal calf serum, penicillin, and streptomycin. Cells were transfected in 10-cm-diameter dishes by the DEAE-dextran method (36) with a total of 10 μg of DNA. Cells and supernatants were harvested 44 to 48 h later, and virus production was assessed with a reverse transcriptase (RT) assay (42). Jurkat T cells were maintained in RPMI-10 medium (Gibco BRL) supplemented with 10% fetal calf serum, penicillin, and streptomycin. HeLa CD4+ LTR-βgal cells were maintained in Dulbecco's modified Eagle's medium as previously described (39).
Protein analysis.
COS-1 cells were metabolically labeled with [35S]methionine (>1,000 Ci/mmol) (Amersham) from 44 to 48 h posttransfection. Viral proteins were harvested from cellular and virion fractions and visualized as previously described (22).
T-cell replication assay.
Ten milliliters of supernatants from transfected COS-1 cells was removed 48 h posttransfection and passed through a 0.45-μm-pore-size filter into a tube containing 5 ml of 30% polyethylene glycol 8000 in 0.4 M NaCl. The contents were mixed by inversion and allowed to stand overnight at 4°C. On the next day, virions were pelleted by centrifugation at 2,000 rpm in a bench-top centrifuge rotor (MSE 43124-129) at 4°C for 40 min. The pellets were resuspended in 0.5 ml of TNE (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1 mM EDTA [pH 7.5]), and a 10-μl sample was removed to measure particle production in an RT assay. The remainder was layered over 0.5 ml of 20% sucrose in TNE. Virions were purified by centrifugation at 40,000 rpm in a Beckman TLA-45 rotor at 4°C for 2 h. Pelleted virus was resuspended in 100 μl of RPMI-10 medium, and an amount equivalent to 500,000 U of RT activity was added to 50,000 Jurkat T cells in one well of a U-bottom 96-well culture plate; the final volume was 200 μl. Any given well received virus from only one transfection supernatant; virus was not pooled at any stage. Replication was followed every 3 to 4 days by an RT assay. A 10-μl sample was removed from each well for the assay. Fresh medium was added to the original volume; fresh Jurkat cells were not added during the assay.
RNA isolation.
Cytoplasmic and virion RNAs were harvested, purified, DNase treated, and stored as previously described (23, 22).
RPAs.
32P-labeled antisense riboprobes were transcribed in vitro from linearized DNA templates using the riboprobe system and T3 or T7 RNA polymerase. Riboprobes were purified from 5% polyacrylamide–8 M urea gels prior to use.
Reagents for RPAs were obtained from a commercially available kit (Ambion). RNA inputs were normalized for all reactions. For cytoplasmic RNA, the sample concentration was determined by spectophotometry, and the same amount was included in each tube, typically 1 μg. Virion RNA input was normalized to RT activity, with an equivalent of 50,000 U being the standard amount used per reaction. RNA was coprecipitated with 2 × 105cpm of riboprobe and 3 μg of carrier RNA from Torrula yeast (Ambion). Hybridization and subsequent nuclease protection were carried out according to the manufacturer's instructions. Pelleted RNA was resuspended in RNA loading buffer (Ambion), separated on a 5% polyacrylamide–8 M urea gel, visualized by autoradiography, and quantified using a real-time Instant Imager (Packard). Size determination of fragments was achieved by running 32P-labeled RNA markers made using a Century Marker template set (Ambion) in parallel.
For each experiment, a separate RPA was performed using the same RNA inputs but probing for viral plasmid DNA using a probe generated from plasmid KS2ΨΕP. In addition, a probe for human β-actin RNA (Ambion) was included in the reaction to control for variations in cytoplasmic RNA input. Any DNA contamination or variations in the β-actin signal were accounted for when calculating encapsidation efficiencies, taken as the ratio of virion to cytoplasmic RNAs of a mutant relative to the wild type.
Transduction and selection of HeLa CD4+ LTR-βgal cells.
Supernatants from COS-1 cells transfected with helper, vector, env-expressor, or empty env-expressor backbone as well as mock-transfected cells were harvested as described above, except that the resulting pellet was resuspended in 100 μl of Dulbecco's modified Eagle's medium. The RT activity of the resulting vector preparations was determined as described above, and the amount added to a 12-well dish of cells was normalized in this way. Each well was at 20% confluence when the vector was added. After 3 days, the medium was replaced with selection medium containing the appropriate antibiotics for maintaining the cell line, as well as 1 μg of puromycin/ml. Cells were maintained under selection until all in the mock-transduced wells were dead. The contents of the wells were fixed and stained for β-galactosidase as previously described (39), and the number of colonies in each well was counted. Transduction efficiencies were expressed as CFU per 10,000 U of RT activity (CFU/10,000RTU).
Computer analysis of RNA structure.
Free-energy-based RNA folding predictions were performed using the Mfold program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html), written by M. Zuker.
RESULTS
Identification of a 28-nucleotide sequence that is located upstream of the splice donor and that contains the core Ψ region of HIV-2.
Deletions in the 5′ leader of HIV-2 were designed based on available structural information generated by computer modeling and biochemical analysis of the HIV-2 leader RNA (3, 9). Previously described deletion mutants Δ1, Δ2, Δ3, and Δ4 were also used (35). The first new deletion, Ψ1, was designed to remove a predicted stem-loop from positions 445 to 462. The second, DM, encompassing positions 380 to 408, overlaps deletions that we have previously shown to affect encapsidation (Fig. 1A). Both are located upstream of the major splice donor (position 472). A double-deletion mutant of both regions, DM-Ψ1, was also constructed. Proviral clones containing these mutations were used to transiently transfect COS-1 cells. RNAs from cytoplasmic and virion fractions were then analyzed by RPAs to assess any effects of the deletions on encapsidation. The results are shown in Fig. 1B. The Ψ1 mutation had only a very minor effect on encapsidation efficiency, whereas the DM deletion had a profound effect on the level of RNA incorporated into progeny virions; the effect of the latter was considerably greater than the effect of the previously described Δ2 deletion, which reduced encapsidation to about 20% the level of wild-type HIV-2. Relative packaging efficiencies (ratio of virion RNA to cytoplasmic RNA, relative to that of the wild type) were 73% ± 7.8% (mean and standard error) for Ψ1 and 5.7% ± 1.6% for DM. These results are consistent with the region deleted by the DM mutation containing the core Ψ element of the virus. In addition, the double mutation had a similar phenotype, confirming that the DM deletion causes a profound defect and that the Ψ1 deletion causes no additional defect in encapsidation. There is also no apparent lack of RNA available for encapsidation in the mutants relative to the wild type.
Deletions in the 5′ leader cause no defect in viral protein production or subsequent processing.
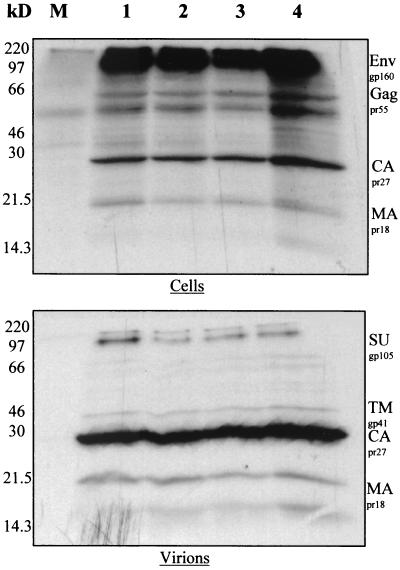
To ensure that the effects observed for deletions on encapsidation were not due to aberrant protein production, COS-1 cells transfected with wild-type or mutant proviruses were metabolically labeled with [35S]methionine, and viral proteins were immunoprecipitated from cellular and virion fractions using pooled immune sera from HIV-2-infected individuals (Medical Research Council AIDS Reagent Project). The results are shown in Fig. 2. From a comparison of mutant and wild-type proviruses, it is clear that protein production was not affected by these deletion mutations. In the cellular fraction, significant amounts of viral Gag and Env polyprotein precursors were apparent; these had been predominantly cleaved into mature proteins in the virions present in the supernatant. This result indicates that no apparent defect in the posttranslational processing of viral proteins is caused by these deletions. Encapsidation defects are therefore unlikely to be caused by a reduced availability of Gag polyprotein for encapsidation or any defect in particle release from the cell surface. These observations were confirmed by Western blotting using a monoclonal antibody to HIV-2 CA protein (Chemicon) and by measuring RT activities in culture supernatants (data not shown).
FIG. 2.
Protein production by new HIV-2 deletion mutants. COS-1 cells transfected with proviral clones were metabolically labeled with [35S]methionine, and viral proteins were immunoprecipitated with pooled HIV-2-positive patient sera. Proteins from cellular and virion fractions were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. Lanes: M, protein from mock-transfected cells; 1, pSVR; 2, pSVRΨ1; 3, pSVRDM; 4, pSVRDM/Ψ1. gp, glycoprotein; pr, protein; MA, matrix; Env, envelope; Gag, group antigen; SU, surface; TM, transmembrane.
Deletion of the core Ψ region of HIV-2 severely limits viral replication in Jurkat T cells.
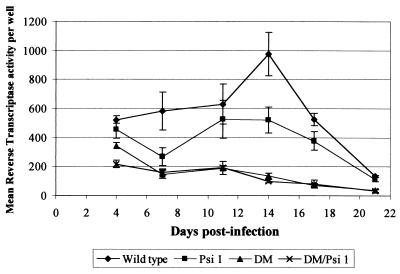
We examined the effects of our new deletions on viral replication in a physiologically relevant cell type. Studying infection rather than transfection can identify preintegration defects that may be caused by deletions, as has been observed previously (35). Supernatants from transfected COS-1 cells were prepared as described above and used to infect Jurkat T cells in replicate assays. Virion particle production was measured over time as RT activity present in supernatants. The results are shown in Fig. 3. Cultures infected with wild-type virus showed a gradual increase in particle production that peaked at 14 days postinfection (dpi). After this point, particle production decreased, probably due to a decline in surviving susceptible cell populations, there being no fresh cells added to the assay. Cells infected with Ψ1 mutant virus displayed an intermediate replication phenotype, with no discernible peak at 14 dpi. This result is consistent with the same mild encapsidation defect observed in COS-1 cells retarding virus spread due to the release of fewer infectious particles into the culture. Virus production from cultures infected with virus containing the DM deletion, both alone and in the context of the double mutant, was severely reduced. There was a gradual decline from an already low initial level of particle production at 4 dpi to levels of RT activity at 21 dpi barely measurable above the background. This result indicates that the virus was unable to spread efficiently beyond cells that were infected by the original inoculum. In addition, early RT readings indicated that infection was initiated successfully, so it is unlikely that the DM deletion interferes with early events in the virus life cycle. The DM deletion therefore causes a replication phenotype in permissive cells that is predominantly attributable to a defect in RNA encapsidation.
FIG. 3.
Replication of HIV-2 deletion mutants in Jurkat T cells. Replicate wells of 50,000 Jurkat T cells were infected with normalized amounts of concentrated supernatants from provirus-transfected COS-1 cells. Every 3 to 4 days, a 10-μl sample of medium was removed prior to feeding of the cells, and viral replication was assessed by measuring the RT activity of the sample.
Competition with wild-type virus heightens the encapsidation defects of Ψ region mutants.
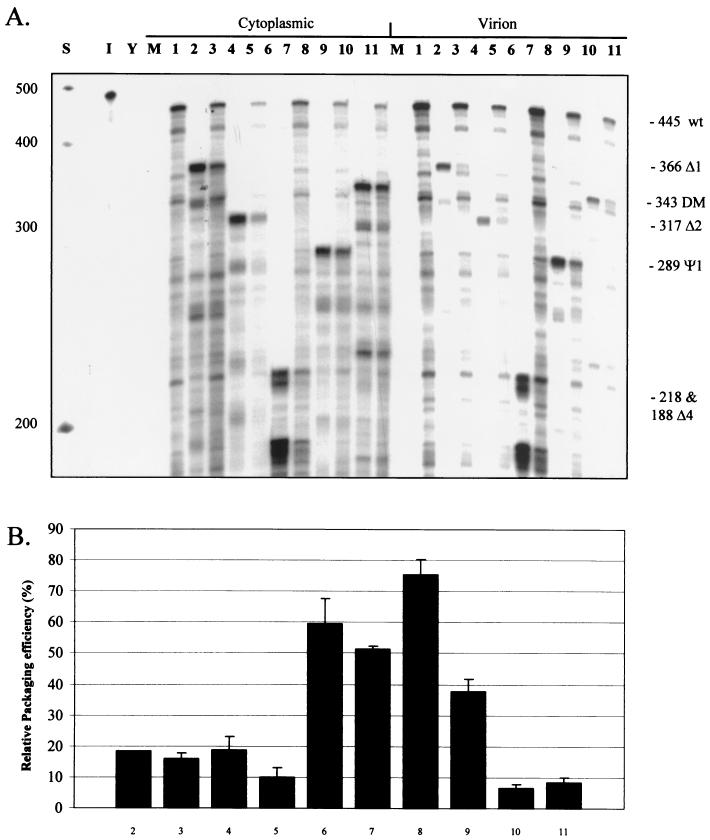
We used cotransfection with wild-type virus as an internal control for levels of RNA during encapsidation studies, as it enables mutant viruses to be normalized to the wild-type virus when calculating encapsidation efficiencies (22, 33). We found, however, that the encapsidation efficiencies of cotransfected mutants possessing deletions located upstream of the splice donor were consistently reduced compared to when they were transfected alone. A representative RPA of such experiments is shown in Fig. 4A. The largest reduction in encapsidation caused by competition was observed for the Ψ1 mutant; efficiency relative to that of the wild type was reduced from about 70% to 40% (Fig. 4B). Both deletion mutants Δ1 and Δ2 showed reductions, albeit less marked, although these viruses are already quite severely deficient in encapsidation. A deletion located downstream of the splice donor, Δ4, also showed a slight decrease in encapsidation efficiency in competition, despite having only a mild effect on encapsidation itself. Encapsidation efficiency in the DM mutant is already so profoundly impaired even in the absence of competition that detection of any change is beyond the level of sensitivity of the assay. It appears, based on these observations, that Ψ region mutants are less efficient at targeting de novo synthesized Gag back to their own RNA in cis than is wild-type HIV-2. Furthermore, wild-type HIV-2 RNA with an intact Ψ region is able to compete for this Gag, causing a reduction in mutant encapsidation efficiency.
FIG. 4.
Reduction of mutant encapsidation efficiencies in competition with wild-type HIV-2. COS-1 cells were transfected with leader region mutant HIV-2 proviruses either alone or with an equal amount of wild-type HIV-2 provirus. RNA was collected, and encapsidation efficiencies were assessed by RPAs. (A) Representative RPA using the KSΨ2KE probe with cytoplasmic and virion RNAs from transfected COS-1 cells. Lanes: S, markers; I, input probe diluted 1/100; Y, yeast RNA plus RNase; M, RNA from mock-transfected cells; 1, pSVR; 2, pSVRΔ1; 3, pSVR plus pSVRΔ1; 4, pSVRΔ2; 5, pSVR plus pSVRΔ2; 6, pSVRΔ4; 7, pSVR plus pSVRΔ4; 8, pSVRΨ1; 9, pSVR plus pSVRΨ1; 10, pSVRDM; 11, pSVR plus pSVRDM. wt, wild type (pSVR); mutant proviruses are abbreviated by their deletion names, e.g., Δ1 represents pSVRΔ1. (B) Bar chart showing quantification of encapsidation efficiencies of mutants, with and without competition, relative to wild-type virus. Results are averages of at least three separate experiments (numbers vary for each mutant); error bars represent the standard error of the mean between experiments. Numbering is as described for panel A.
Levels of Gag polyprotein are limiting for encapsidation in HIV-2.
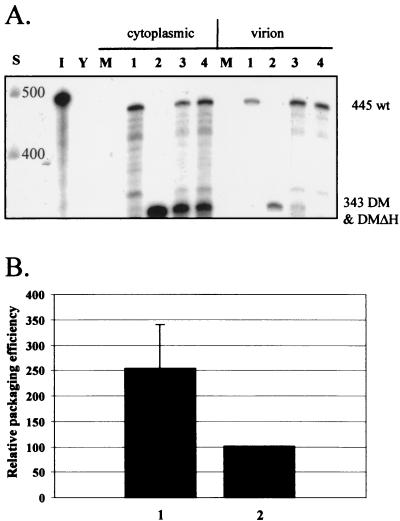
If competition effects decrease the encapsidation of Ψ region mutants, then a logical question to ask is whether there is a corresponding increase in the encapsidation of wild-type RNA. To address this question, an HIV-2 provirus that contained the DM deletion and a premature stop codon in the Gag ORF was constructed: pSVRDMΔH. It was previously shown that HIV-2 vectors containing this stop mutation synthesize a truncated Gag polyprotein that is unable to incorporate RNA into virions (22). Such vectors are also unable to be efficiently encapsidated by wild-type HIV-2 in trans. Cotransfection experiments were performed to compare the encapsidation of wild-type HIV-2 RNA in competition with this virus or with pSVRDM, which is able to make its own full-length Gag. Twice the amount of Gag should be available in the cell in the latter. The results obtained in RPAs are shown in Fig. 5. As expected, wild-type HIV-2 in competition with a DM mutant that is able to make full-length Gag was encapsidated about twice as efficiently as wild-type HIV-2 competing with a DM virus that cannot do so. As the levels of wild-type RNA available for encapsidation in the cytoplasm will be the same in the two situations, it follows that the increase in efficiency observed is due to their being twice the amount of Gag present. The availability of Gag is therefore limiting for HIV-2 RNA encapsidation.
FIG. 5.
Gag availability is limiting for HIV-2 RNA encapsidation. The encapsidation of wild-type HIV-2 was assessed by RPAs in cotransfections with a DM mutant virus that could produce Gag protein compared to one that could not. (A) Representative RPA using the KSΨ2KE riboprobe and showing an increase in wild-type encapsidation in competition with a DM virus that makes its own Gag protein. Lanes: S, RNA size markers (Century Plus); I, input probe diluted 1/100; Y, yeast RNA plus RNase; M, RNA from mock-transfected cells; 1, pSVR; 2, pSVRDM; 3, pSVR plus pSVRDM; 4, pSVR plus pSVRDMΔH. wt, wild type (pSVR); DMΔH, pSVRDMΔH; DM, pSVRDM. (B) Bar chart showing quantification of experiments. The encapsidation efficiency of pSVR in competition with a non-Gag-producing virus, pSVRDMΔH, is taken as 100%. Results are averages of four separate experiments; error bars represent the standard error of the mean between experiments. Bars: 1, pSVR-pSVRDM; 2, pSVR-pSVRDMΔH.
Competition for limiting amounts of Gag allows trans-acting encapsidation of HIV-2 vectors.
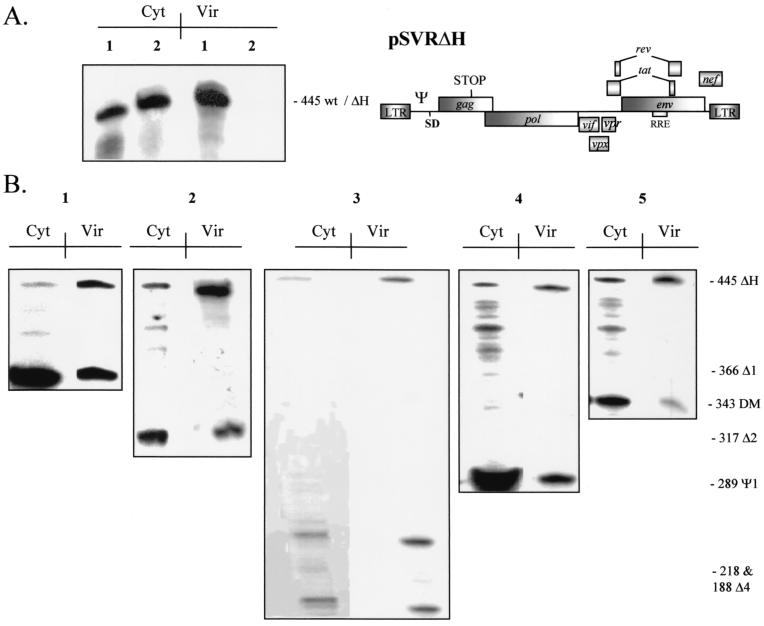
It was previously shown that wild-type HIV-2 is unable to efficiently encapsidate vectors in trans due to the use of a cotranslational method of selecting its genomic RNA for encapsidation, termed cis-acting encapsidation. HIV-1, however, is able to do this efficiently and predominantly uses a trans-acting mechanism to select its genome for encapsidation, a strategy made possible due to the location in HIV-1 of the core Ψ region downstream of the major splice donor. The results of our competition experiments indicated that the Gag being competed for was that made by the HIV-2 Ψ region mutants. This meant that such Gag was not being efficiently targeted in cis to its template RNA and was therefore available to trans pathways. We reasoned, then, that an HIV-2 vector possessing intact Ψ, pSVRΔH (see Materials and Methods), may be able to compete for this Gag in a fashion similar to that of the wild type, although being unable to encapsidate itself (Fig. 6A). To test this notion, we cotransfected HIV-2 Ψ region mutants with a vector containing the stop mutation in Gag described above. Representative results are shown in Fig. 6B. All of the HIV-2 Ψ region mutants tested were able to efficiently incorporate vector RNA into virions, in contrast to the analogous experiments, in which wild-type HIV-2 was used as a helper virus. In addition, mutants with deletions downstream of the splice donor were also able to efficiently encapsidate vector RNA in trans.
FIG. 6.
Packaging region HIV-2 mutants are able to encapsidate HIV-2 vectors efficiently in trans. (A) Representative RPA with the KSΨ2KE riboprobe showing encapsidation of wild-type HIV-2 pSVR and HIV-2 vector RNA pSVRΔH. Lanes: 1, pSVR; 2, pSVRΔH. Cyt, cytoplasmic RNA; Vir, virion RNA. The structure of pSVRΔH is shown at the right. wt, wild type. See the legend to Fig. 1 for other definitions. (B) Representative RPAs showing trans-acting encapsidation of pSVRΔH by packaging region mutants. Lanes: 1, pSVRΔ1 plus pSVRΔH; 2, pSVRΔ2 plus pSVRΔH; 3, pSVRΔ4 plus pSVRΔH; 4, pSVRΨ1 plus pSVRΔH; 5, pSVRDM plus pSVRΔH. Cyt, cytoplasmic RNA; Vir, virion RNA.
HIV-2 vector transduction studies demonstrate encapsidation competition.
Having demonstrated efficient incorporation of vector RNA into HIV-2 particles by RPAs, we decided to investigate whether the observed competition would translate into a system involving the transduction of target cells. We designed novel HIV-2 vectors containing the puromycin resistance selectable marker based on an env deletion HIV-2 provirus, pSVRΔNB (see Materials and Methods). Both vectors had the same deletion in env as the parental plasmid and had the puromycin resistance cassette at this locus. The first vector, pSVRΔNBPuroΔH, contained the same premature stop codon as pSVRΔH, which was used in the RPA studies. The second vector, pSVRΔNBPuroΔE, contained a large deletion that removed the majority of the gag and pol ORFs. In order to assess competition effects, help was provided either by parental pSVRΔNB or by pSVRΔNBDM, which contains the DM deletion. The structures of the constructs used are shown in Fig. 7A. Vectors were pseudotyped with the VSV G glycoprotein, and their ability to transduce HeLa CD4+ LTR-βgal cells was assessed.
Concentrated supernatants were prepared from COS-1 cells transiently transfected (see Materials and Methods) with 5 μg each of vector and helper plasmids along with 2 μg of VSV G glycoprotein expression construct, pCMV-VSVG (see Materials and Methods), or empty vector. Twelve-well dishes containing HeLa CD4+ LTR-βgal cells at 20% confluence were transduced with COS-1 cell supernatants containing equivalent amounts of RT activity. Three days posttransduction, selection media containing puromycin were applied to the cells. Selection was maintained until all mock-transduced cells were dead. Puromycin resistance was not seen in envelope-negative transduced control cells after selection (data not shown), indicating that the ΔNB deletion is sufficient to abrogate the function of the HIV-2 Env glycoprotein. Cells were fixed and stained as described above, the number of colonies was counted, and the results were expressed as CFU/10,000RTU (Fig. 7B).
The DM deletion construct was a far more efficient helper than the wild type. This result is in accordance with the data for encapsidation described above. Furthermore, there was a difference between the vector that contained a stop mutation in gag and the vector with the deletion, the former giving far higher titers. In order to confirm that the differences in titers corresponded to the differences in the encapsidation efficiencies of the vectors, analogous COS-1 cell transfections were assessed by RPAs (Fig. 7C). The relative encapsidation efficiencies of the vectors did indeed correspond to the vector titers, with the most efficient combination being a DM deletion helper vector encapsidating a vector without a large deletion in the gag ORF.
The results confirmed that an HIV-2 helper containing intact Ψ is unable to perform efficiently in vector systems, due to the cotranslational encapsidation mechanism used by the virus. Competition for limiting Gag polyprotein, however, allows the production of comparatively high-titer vector preparations using a Ψ deletion helper.
Inclusion of sequences from gag may enhance vector encapsidation.
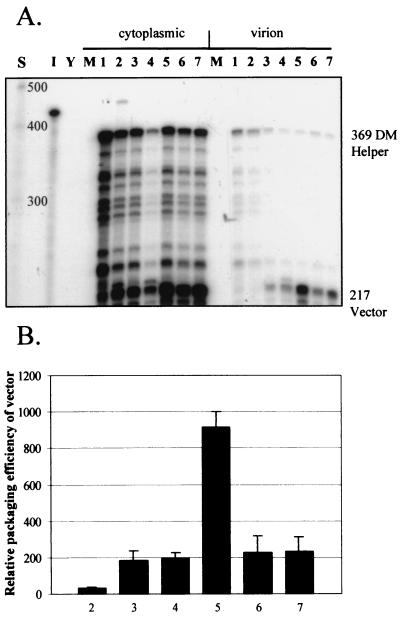
The differences in both titers and encapsidation efficiencies between different puromycin-resistant vectors might implicate cis-acting signals present in the gag ORF. It was previously shown that there is no effect of including such regions when wild-type HIV-2 encapsidates vector RNAs (22). We tested the ability of a DM mutant virus to encapsidate a panel of HIV-2 vectors that contained different lengths of the gag ORF. All had the pol ORF deleted. Equal amounts (5 μg) of vector and pSVRDM were transfected into COS-1 cells, and RNA encapsidation was assessed by RPAs (Fig. 8).
FIG. 8.
trans-Acting encapsidation by pSVRDM of HIV-2 vectors containing various amounts of the gag ORF. (A) Representative RPA using the KS2ES riboprobe and cytoplasmic and virion RNAs from COS-1 cells transfected with equal amounts of pSVRDM and vector. Lanes: S, RNA size markers (Century Plus); I, input probe diluted 1/100; Y, yeast RNA plus RNase; M, RNA from mock-transfected cells; 1, pSVRDM; 2, pSVRDM plus pSVRΔX; 3, pSVRDM plus pSVRΔAX; 4, pSVRDM plus pSVRΔHX; 5, pSVRDM plus pSVRΔpol; 6, pSVRDM plus pSVRΔHΔpol; 7, pSVRDM plus pSVRΔpolncm. (B) Quantification of vector encapsidation efficiencies, relative to that of the DM helper, in the experiments detailed above. Results are averages of at least two separate experiments; error bars represent the standard error of the mean between experiments. Numbering is as described for panel A.
As shown previously, the HIV-2 vector containing an intact gag ORF, pSVRΔpol, is capable of efficiently encapsidating its own RNA and serves as a positive control. In contrast, pSVRΔHΔpol, which contains the premature stop codon, is efficiently encapsidated by Gag provided by the DM mutant helper, albeit to a lesser extent. This construct contains the entire gag ORF and so possesses any cis-acting signals contained therein. Removal of sequences up to and including the 3′ region of the matrix also has no detrimental effect on vector encapsidation, as constructs pSVRΔHX and pSVRΔAX are both packaged to the same level as the above constructs (22, 23). In contrast to the other vectors, pSVRΔX is encapsidated very poorly by the DM mutant helper. This vector has a deletion of almost the entire gag ORF, starting from near the ATG (position 555). This result indicates that there may be a signal in the 5′ part of gag, specifically in the matrix, that enhances encapsidation or, alternatively, that ribosomal scanning of this region may be important in promoting the correct folding of RNA structures present in the leader or in gag itself. In this way, translation and encapsidation may be linked in the HIV-2-infected cell.
Limiting Gag availability could allow complete removal of helper virus from HIV-2 vector preparations.
No single deletion in any lentiviral system completely abrogates the encapsidation of viral RNA. This fact is probably due to functional redundancy in packaging signals. Contamination of prospective therapeutic vector preparations with helper virus sequences is therefore a major biosafety issue. We reasoned that even though the DM deletion did not completely abrogate HIV-2 encapsidation, the fact that Gag levels appeared to be limiting might allow the complete removal of helper RNAs by competition. To investigate this notion, we cotransfected COS-1 cells with increasing amounts of stop codon-containing vector, pSVRΔHΔpol, along with a fixed amount of either pSVR or pSVRDM and analyzed the effects on encapsidation by RPAs (Fig. 9). A compensatory amount of non-HIV-2 stuffer DNA, pBluescript KSII(+), was transfected where necessary in order to bring the amount of total DNA used to 21 μg in each instance.
FIG. 9.
Titration of helper encapsidation by competition for limiting amounts of Gag polyprotein. COS-1 cells were transfected with 1 μg of helper virus, either pSVR or pSVRDM, and increasing amounts of HIV-2 vector pSVRΔHΔpol along with stuffer DNA. RNA from cytoplasmic and virion fractions was then analyzed by RPAs. (A) Representative RPA with the KS2ES riboprobe and RNA from titration experiments. Lanes: S, RNA size markers (Century Plus); I, input probe diluted 1/100; Y, yeast RNA plus RNase; M, RNA from mock-transfected cells; 1, 1 μg of pSVR plus 5 μg of pSVRΔHΔpol; 2, 1 μg of pSVR plus 10 μg of pSVRΔHΔpol; 3, 1 μg of pSVR plus 15 μg of pSVRΔHΔpol; 4, 1 μg of pSVR plus 20 μg of pSVRΔHΔpol; 5, 1 μg of pSVRDM plus 5 μg of pSVRΔHΔpol; 6, 1 μg of pSVRDM plus 10 μg of pSVRΔHΔpol; 7, 1 μg of pSVRDM plus 15 μg of pSVRΔHΔpol; 8, 1 μg of pSVRDM plus 20 μg of pSVRΔHΔpol. (B) Quantification of vector encapsidation efficiencies in the experiments detailed above. wt, wild type. Results are the averages of three separate experiments; error bars represent the standard error of the mean between experiments.
As expected, the vector was efficiently encapsidated in trans only by the DM deletion virus. Even at vector/helper ratios of 20:1, wild-type HIV-2 does not efficiently encapsidate vector RNAs, indicating that the coupling of translation and encapsidation in HIV-2 is very strong indeed. In contrast, the amount of vector encapsidated by pSVRDM increases slowly as the vector/helper ratio increases. In addition, the efficiency of vector encapsidation is reduced, even at 5:1, compared to when the latter two are transfected in equal amounts. This result is due to there being an enormous amount of vector RNA present in the cytoplasm that is unable to be encapsidated by the limiting amounts of Gag present. Instead, the cause of the apparent increase in vector encapsidation is a reduction in the amount of pSVRDM RNA being encapsidated. The shift in virion RNA/cytoplasmic RNA ratios between the helper and the vector therefore leads to an apparent increase in the encapsidation efficiency of the vector. Although the levels of pSVRDM RNA in the virion fraction of these experiments is not reduced to zero, the levels are only just measurable above the background, whereas wild-type HIV-2 maintains a high level of encapsidation. These experiments therefore indicate that it would be theoretically possible to completely titrate out the encapsidation of a DM deletion helper from HIV-2 vector preparations.
DISCUSSION
We have identified a small deletion mutation in the 5′ leader region of HIV-2 that dramatically reduces the encapsidation of genomic RNA. The deletion removes sequences upstream of the major splice donor of the virus and as such are present on genomic as well as spliced virion mRNAs. It was previously reported that the virus discriminates between the various viral and cellular RNA species using a predominantly cotranslational mechanism, in which only RNAs coding for Gag polyprotein are efficiently selected for encapsidation. Although this scenario is different from that of HIV-1, there are precedents from other viral systems. HIV-2 is closely related to macaque simian immunodeficiency virus (SIVmac). A study in which SIVmac RNA was encapsidated by HIV-1 Gag protein expressed in trans revealed that deletion of sequences upstream of the splice donor in SIVmac significantly reduced encapsidation, whereas those downstream of the splice donor did not (46). Although this was not a system representative of the in vivo situation, we have also generated similar results with systems in which SIVmac encapsidates its own RNA, and these results have been confirmed by other investigators. It is therefore likely that SIVmac uses a mechanism analogous to that of HIV-2 to select its genome for encapsidation.
In simple avian retroviruses, such as Rous sarcoma virus, Ψ has also been mapped to a region of the 5′ leader located upstream of the splice donor that contains three viral ORFs (21, 27, 28). A close link between RNA encapsidation and translation has been postulated for Rous sarcoma virus (12, 11). Furthermore, it has been shown that in both HIV-1 and HIV-2, genomic RNA appears not to be sorted into separate pools that are fated to be either translated or encapsidated (13), contrary to the situation in murine retroviruses (31). Conceivably, Gag binds to an HIV-2 genomic RNA as it is being translated and eventually reaches a sufficient concentration to prevent translation by inhibition of ribosomal scanning. The RNA would then be targeted sequentially for incorporation into progeny virions. Interestingly, it has been reported that an internal ribosome entry site is present in the 5′ leader of SIVmac downstream of the splice donor (38). This would presumably allow translation to continue for a time as Gag bound to Ψ and other upstream regions. There is, however, no current evidence for an internal ribosome entry site in HIV-2 and, in fact, regions downstream of the splice donor have been reported to contain a negative regulator of gene expression (15).
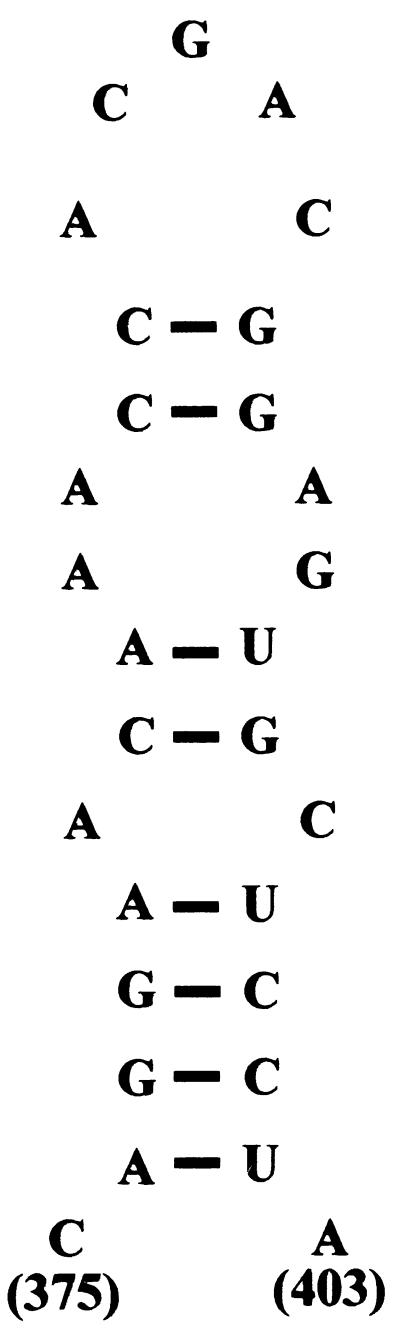
Limited information is available on the RNA structures present within the 5′ leader of HIV-2. The region between the transcription start site and the primer binding site stem-loop has been subjected to secondary structure modeling, yet the only published structure for the remainder of the leader is a computer prediction (3). One study mapped the binding of HIV-1 Gag and NC proteins fused with glutathione S-transferase to in vitro transcribed HIV-2 leader RNAs (9). Specific in vitro binding of Gag to regions of the RNA both upstream and downstream of the splice donor was shown, in agreement with the results of encapsidation studies performed by other authors. The region upstream of the splice donor that was bound by HIV-1 Gag corresponded to the region deleted by the Ψ1 mutation, which was shown in this study to have relatively little effect on encapsidation. From the published structure, the DM deletion would remove a nonstructured region of the leader between the primer binding site stem-loop and a stem-loop that contains a palindromic sequence at its terminus, postulated as being the dimerization initiation site of HIV-2, although no direct evidence exists for this. The DM deletion removes purine-rich motifs that could be present on a stem-loop structure not predicted by the computer model. We are currently undertaking our own secondary structure analysis of the region to address this possibility. Our own preliminary computer analysis of the region spanned by the DM deletion identifies a stable stem-loop structure (Fig. 10) which includes sequences between positions 375 and 403, a region nearly completely removed by the DM mutation. We cannot rule out, however, the possibility that the DM deletion disrupts proper folding of adjacent RNA structures or that it disrupts the binding of a cellular factor to the RNA, as these are concerns raised by all studies involving deletion mutagenesis.
FIG. 10.
Alternative secondary structure for the region of the DM deletion in the HIV-2 leader generated with the Mfold program. The free energy of the structure was calculated to be −7.6 kcal/mol.
Different regions of the 5′ leader have been shown by other investigators to affect encapsidation. One report focused on deletions downstream of the splice donor, although the effects of the deletions were possibly affected by abnormal protein production by the mutants (15). Another report showed that a very large deletion that removed the majority of the sequence between the splice donor and the gag ATG affected encapsidation (41). It is unlikely that this deletion removed only a specific signal present in the leader, as it would have had a profound effect on the overall structure of the region due to its size. In addition, the studies were performed using a nonprototypic strain of HIV-2 which is nonpathogenic in macaques. It seems likely that, in common with HIV-1, elements located both upstream and downstream of the splice donor may play a role in RNA encapsidation, although we propose that the core Ψ element is upstream of the splice donor in HIV-2. This proposal is further supported by the strong defect that we observed in virus spread in permissive cells for the DM mutant virus.
The use of a cotranslational packaging mechanism by wild-type HIV-2 would seemingly preclude it from use as a gene vector system. Others have, however, reported trans-acting encapsidation of HIV-2 vector RNA. We have shown in this study that this occurs due to a failure of Ψ region mutant RNA to efficiently capture newly made cognate Gag polyprotein. The Gag protein thus made is therefore available in trans to other RNAs that contain an intact packaging signal. Thus, competition with wild-type HIV-2 was seen to reduce the encapsidation efficiency of Ψ region mutants, and a corresponding increase was shown for encapsidation of the wild type RNA. These results are consistent with there being a limiting amount of Gag available at the correct subcellular location for encapsidation in an HIV-2-infected cell. HIV-1 appears able to produce Gag in large quantities that will bind to RNAs anywhere in the cytosol that contain Ψ through a trans-acting pathway. This model implicates signals that direct HIV-2 Gag to specific subcellular compartments and, similarly those that enable HIV-1 Gag to access a greater range of locations. This conclusion is supported by the fact that chimeric HIV-2 containing HIV-1 NC in place of the native domain is able to encapsidate vector RNAs in a manner similar to that of HIV-1.
Having Gag as a limiting factor for encapsidation might be beneficial in the development of gene vector systems based on HIV-2. Such systems are essentially dependent on a Ψ deletion helper virus being able to encapsidate a Ψ-positive vector RNA in trans. In lentiviral systems, no single deletion in wild-type virus completely abolishes encapsidation of the helper virus, implying functional redundancy in the process. This fact raises biosafety concerns, there being an increased chance of generating replication-competent virus in the vector fraction. We have shown that uncoupling of the cotranslational encapsidation mechanism of HIV-2 is possible by deleting Ψ, and viruses and vectors that contain intact Ψ can compete for Gag made by such a virus. Given that vector RNA will be far more efficient at competing for this limiting amount of Gag and that RNA is present in excess, the vector is encapsidated at the expense of the helper; this indeed appears to be the case in this investigation. We propose that, by further overexpressing HIV-2 vectors relative to an HIV-2 Ψdeletion helper, incorporation of helper RNA into virions could be completely abolished by a titration effect. HIV-2 vectors have already been shown to be capable of transducing nondividing cells, indicating that the development of a system such as that described above would be an important step toward the development of effective and safe lentiviral vectors for gene therapy.
ACKNOWLEDGMENTS
This work was supported by AVERT, the Medical Research Council, and the Sykes' Trust.
We thank Nijsje Dorman for helpful discussions. Jane F. Allen is supported by a Wellcome Trust research career development fellowship.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arya S K, Zamani M, Kundra P. Human immunodeficiency virus type 2 lentivirus vectors for gene transfer: expression and potential for helper virus-free packaging. Hum Gene Ther. 1998;9:1371–1380. doi: 10.1089/hum.1998.9.9-1371. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout B, Schoneveld I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993;21:1171–1178. doi: 10.1093/nar/21.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel J L. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–295. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R D, Hammarskjold M L, Helga-Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damgaard C K, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donze O, Spahr P F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman N, Lever A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J Virol. 2000;74:11413–11417. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franchini G, Collalti E, Arya S K, Fenyo E M, Biberfeld G, Zagury J F, Kanki P J, Wong-Staal F, Gallo R C. Genetic analysis of a new subgroup of human and simian T-lymphotropic retroviruses: HTLV-IV, LAV-2: SBL-6669, and STLV-IIIAGM. AIDS Res Hum Retrovir. 1987;3:11–17. doi: 10.1089/aid.1987.3.11. [DOI] [PubMed] [Google Scholar]
- 15.Garzino-Demo A, Gallo R C, Arya S K. Human immunodeficiency virus type 2 (HIV-2): packaging signal and associated negative regulatory element. Hum Gene Ther. 1995;6:177–184. doi: 10.1089/hum.1995.6.2-177. [DOI] [PubMed] [Google Scholar]
- 16.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong-Staal F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. doi: 10.1038/312166a0. [DOI] [PubMed] [Google Scholar]
- 19.Harrison G P, Lever A M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi T, Ueno Y, Okamoto T. Elucidation of a conserved RNA stem-loop structure in the packaging signal of human immunodeficiency virus type 1. FEBS Lett. 1993;327:213–218. doi: 10.1016/0014-5793(93)80172-q. [DOI] [PubMed] [Google Scholar]
- 21.Katz R A, Terry R W, Skalka A M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye J F, Lever A M. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J Virol. 1999;73:3023–3031. doi: 10.1128/jvi.73.4.3023-3031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaye J F, Lever A M. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J Virol. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaye J F, Lever A M. trans-Acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaye J F, Richardson J H, Lever A M. cis-Acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H J, Lee K, O'Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 27.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyama T, Harada F, Kawai S. Characterization of a Rous sarcoma virus mutant defective in packaging its own genomic RNA: biochemical properties of mutant TK15 and mutant-induced transformants. J Virol. 1984;51:154–162. doi: 10.1128/jvi.51.1.154-162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 30.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCann E M, Lever A M. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J Virol. 1997;71:4133–4137. doi: 10.1128/jvi.71.5.4133-4137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortlock D, Keller E B, Ziegra C J, Suter M M. High efficiency transfection of monkey COS-1 cells. J Tissue Culture Methods. 1993;15:176–180. [Google Scholar]
- 37.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 38.Ohlmann T, Lopez-Lastra M, Darlix J L. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J Biol Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 39.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pappalardo L, Kerwood D J, Pelczer I, Borer P N. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J Mol Biol. 1998;282:801–818. doi: 10.1006/jmbi.1998.2046. [DOI] [PubMed] [Google Scholar]
- 41.Poeschla E, Gilbert J, Li X, Huang S, Ho A, Wong-Staal F. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J Virol. 1998;72:6527–6536. doi: 10.1128/jvi.72.8.6527-6536.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potts B J. “Mini” reverse transcriptase (RT) assay. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. pp. 103–106. [Google Scholar]
- 43.Poznansky M, Lever A, Bergeron L, Haseltine W, Sodroski J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. J Virol. 1991;65:532–536. doi: 10.1128/jvi.65.1.532-536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson J H, Child L A, Lever A M. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson J H, Kaye J F, Child L A, Lever A M. Helper virus-free transfer of human immunodeficiency virus type 1 vectors. J Gen Virol. 1995;76:691–696. doi: 10.1099/0022-1317-76-3-691. [DOI] [PubMed] [Google Scholar]
- 46.Rizvi T A, Panganiban A T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadaie M R, Zamani M, Whang S, Sistron N, Arya S K. Towards developing HIV-2 lentivirus-based retroviral vectors for gene therapy: dual gene expression in the context of HIV-2 LTR and Tat. J Med Virol. 1998;54:118–128. doi: 10.1002/(sici)1096-9071(199802)54:2<118::aid-jmv9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeffman A, Hassard S, Varani G, Lever A. The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the gag polyprotein. J Mol Biol. 2000;297:877–893. doi: 10.1006/jmbi.2000.3611. [DOI] [PubMed] [Google Scholar]