Abstract
Human immunodeficiency virus type 1 (HIV-1) Nef protein exerts several effects, both on infected cells and as a virion protein, which work together to enhance viral replication. One of these activities is the ability to enhance infectivity and the formation of proviral DNA. The mechanism of this enhancement remains incompletely understood. We show that virions with nef deleted can be restored to wild-type infectivity by stimulating intravirion reverse transcription. Particle composition and measures of reverse transcriptase activity remain the same for Nef+ and Nef− virions both before and after natural endogenous reverse transcription (NERT) treatment. The effect of NERT treatment on virions pseudotyped with murine leukemia virus envelope protein was similar to that on particles pseudotyped with HIV-1 envelope protein. However, virions pseudotyped with vesicular stomatitis virus G envelope protein showed no influence of Nef on NERT enhancement of infectivity. These observations suggest that Nef may function at a level prior to reverse transcription. Since NERT treatment results in partial disassembly of the viral core, we speculate that Nef may function at the level of core particle disassembly.
The nef gene of human immunodeficiency virus type 1 (HIV-1) modulates the viral life cycle in several distinct ways (reviewed in references 10, 17, 22, 38, and 40). The absence of a functional nef gene in virus infecting both humans and rhesus monkeys diminishes viral loads and significantly increases the time of clinical progression of disease (11, 28). Expression of nef as a transgene in mice produces many of the pathological effects seen in AIDS (12, 21). In vitro studies demonstrate that Nef can affect multiple cellular functions that help explain how it modulates pathogenicity. Nef promotes the removal of CD4 molecules from the surface of infected cells (4, 16, 35) and downregulates the surface expression of major histocompatibility complex class I molecules (33, 34). These properties are structurally separate features of the Nef molecule (3, 19). Nef also exerts a profound effect on infected cells by altering cell signaling pathways (9, 23, 49) and inducing chemokine release from infected macrophages (47).
One key feature of Nef's influence on pathogenesis is that it can enhance the intrinsic infectivity of viral particles (7, 18, 26, 36, 37, 45). Viruses which have nef deleted are from 4- to 40-fold less infectious than wild-type (WT) HIV-1 in tissue culture systems (7, 37). Enhancement of viral infectivity appears to be linked to an increase in the initiation of proviral DNA (2, 43). However, the exact nature of how Nef enhances infectivity and proviral DNA synthesis remains elusive.
Several pieces of evidence from prior studies may be relevant in explaining Nef's ability to enhance proviral DNA synthesis and infectivity. Nef is a virion protein (39) and is present in the viral core (30). Although Nef can be cleaved by viral protease to liberate the C-terminal core domain, this cleavage does not appear to correlate with the ability to stimulate virion infectivity (6). Viruses with nef deleted can be restored to WT infectivity by coexpression of Nef in the virus producer cells but not target cells (2, 7). Pseudotyping of HIV-1 virions by vesicular stomatitis virus G (VSV-G) envelope glycoprotein targets viral entry to the endocytic pathway and suppresses the requirement for Nef as well as sensitivity to cyclosporine (2). Recently, it has been shown that Nef may enhance proviral DNA formation by increasing delivery of virions to the cytoplasm of infected cells (42). This study suggests that Nef may function as an entry factor, which may involve interactions with viral envelope protein. In another recent study Nef+ and Nef− virions were allowed to undergo intravirion fusion by pseudotyping a donor particle with gp160 and a target particle with CD4 (56). Nef+ donor virus could enhance the infectivity of a Nef− target particle after fusion. An interesting observation from this study was that the ability to complement nef was dependent on envelope glycoprotein but did not appear to be acting solely at the level of enhancing membrane fusion. One possible explanation for this observation is that Nef may act by altering the composition of the lipid rafts from which HIV-1 particles bud. Other studies have also implicated Nef in altering the composition of membrane rafts as a possible mechanism for enhancement of infectivity (8, 31). It has been shown that cell surface expression of CD4 during virus production can lead to a reduction in infectivity by blocking Env incorporation (31). Since one of Nef's functions is to downregulate CD4 expression, it can also alter infectivity through such a mechanism. It has also been noted that there is significant overlap in mutations known to affect cell sorting of CD4 and enhancement of infectivity (8, 42). Both functions appear to be highly dependent on a functional dileucine motif of Nef. All of these studies suggest that Nef may function at some level prior to reverse transcription to allow an increase in proviral DNA formation.
Disassembly of HIV-1 normally occurs as a part of the entry process and, in contrast to some other enveloped viruses, does not appear to require a pH-dependent step (25). It has been shown that HIV-1 particles pseudotyped with envelope proteins that fuse at low pH do not require Nef for enhanced infectivity (5). One method that can be used to uncouple disassembly from the entry process is treating virions by natural endogenous reverse transcription (NERT) (14, 52–55). In this procedure HIV-1 virions are exposed to buffer containing high concentrations of deoxynucleoside triphosphates (dNTPs) and spermidine. The dNTPs enter the virion via the amphipathic domains of the gp41 on the virion (51). Detailed electron microscopy studies have shown that treatment of HIV-1 virions by NERT results in partial disassembly of the viral core (52). This disassembly also results in disruption of a structure known as the core-envelope linkage (CEL), which is an attachment between the smaller end of the core and the envelope (24). NERT treatment can restore vif deletion viruses to WT infectivity (13), which is important since the Vif protein is also thought to be involved in the formation and stabilization of the early reverse transcription complex. Since NERT treatment can rescue the activity of vif deletion virions and it appears that NERT can induce partial disassembly and initiation of reverse transcription, we were interested in how NERT might affect nef deletion virions. In this study, we have treated Nef+ and Nef− virions to induce NERT. We show that pretreatment by NERT can restore infectivity of nef deletion virions to WT levels.
MATERIALS AND METHODS
Plasmid and viral constructs.
A single-round infectivity assay was developed from the viral clone pNL4-3. An env deletion variant of this clone designated pNL4-3KFS was produced by insertion of KpnI linkers into the env reading frame, introducing a frameshift mutation (gift of Eric Freed, National Institutes of Health [NIH]) (15). A further mutation of pNL4-3KFS to delete nef was produced by insertion of tandem stop codons at the beginning of the nef reading frame, and this construct was designated pNL4-3KFSΔNef (gift of Judith Levin, NIH). To produce infectious NL4-3KFS or NL4-3KFSΔNef virus stocks, HeLa cells were cotransfected with either pNL4-3KFS or pNL4-3KFSΔNef plasmid and the envelope plasmid pIIIenv3-1 (gift of Eric Freed) (44). In some cases the HIV-1 Env plasmid was replaced with pHCMV-G (50) (kindly provided by J. Burns, University of California, San Diego) to pseudotype particles with the VSV-G envelope glycoprotein or with pSVAMLVenv (32) to pseudotype particles with the amphotrophic murine leukemia virus (MLV) envelope glycoprotein. The transfections were done using Effectene (Qiagen) according to the manufacturer's protocol. Transfections were carried out in six-well plates using 1 μg of viral plasmid and 83 ng of envelope plasmid per well. Cells were incubated for 16 h at 37°C and then refed to remove the Effectene and any residual plasmid. Inoculated cells were then incubated for an additional 36 h before supernatants containing the pseudotyped viral particles were collected. Typically transfections produced from 5 to 20 ng of viral p24 antigen per ml as determined by enzyme-linked immunosorbent assay (National Cancer Institute AIDS Vaccine Program). Viral particles were treated with 20 μg of DNase I (Roche) (2,000 U/mg) per ml for 30 min at 37°C to remove any residual plasmid DNA prior to storage. Using this system, the virus produced will infect HeLa CD4+ cells and undergo a single round of replication, producing progeny that lack envelope proteins.
Single-round infection assay.
The relative infectivity of viral particles was determined by the multinuclear activation of a galactosidase indicator (MAGI) assay (29). HeLa-CD4+-LTR-βgal cells (NIH AIDS Reagent Program, catalog no. 1470) were maintained in Dulbecco modified Eagle medium supplemented with 5% fetal bovine serum, 0.1 mg of G418 per ml, 0.05 mg of hygromycin B per ml, l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Assays were done in six-well plates seeded with 2 × 105 cells per well the day before the assay. Each well was infected with pseudotyped virus at a concentration of 1 ng of p24 of either NL4-3KFS or NL4-3KFSΔNef per ml and incubated for 48 h at 37°C. Cells were then fixed with 0.2% glutaraldehyde and 1% formaldehyde for 5 min at room temperature. The cells were then washed twice with phosphate-buffered saline (PBS) and stained in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution (0.4 mg of X-Gal per ml dissolved in dimethylformamide–4 mM potassium ferricyanide–4 mM potassium ferrocyanide–2 mM MgCl2 in PBS). The staining was allowed to continue for 50 min at 37°C, and then cells were washed twice with PBS. Results were scored as the total number of blue cells per nanogram of p24 equivalent of virus added.
Immunoblot analysis of viral particles.
Particle composition was determined by Western blotting of disrupted whole virions. Ten milliliters of supernatant from cells transfected as described above was pelleted by centrifugation at 26,000 × g for 1 h and contained 200 ng of p24. Fifty nanograms of p24 equivalent of virus was added to sodium dodecyl sulfate (SDS) loading buffer and heated to 100°C before being loaded onto an SDS–7.5% polyacrylamide gel. The separated proteins were detected by ECL (Amersham) using human HIV immunoglobulin (AIDS Reagent Program, catalog no. 3957) and protein A conjugated to horseradish peroxidase.
Detection of proviral DNA by PCR.
MAGI cells were infected as described above. After 48 h at 37°C cells, were harvested by trypsinization. The cells were pelleted and then resuspended in PBS supplemented with 5 mM magnesium. The resuspended pellet was then treated with DNase I (Roche; 20 μg/ml) for 30 min at 37°C. The treated cells were then washed twice in PBS and left as a cell pellet. The cell pellets were lysed and DNA was isolated using DNAeasy reagents from Qiagen according to the manufacturer's protocol. The final DNA extracts were adjusted to 150 μl in volume. Uninfected control cells were also processed as described above.
Proviral DNA was detected by traditional PCR using the following primer set to detect strong-stop DNA: forward primer 5′-GGC TAA CTA GGG AAC CCA CTG CTT and reverse primer 5′-CTG CTA GAG ATT TTC CAC ACT GAC, which amplify region 496 to 635 of NL4-3 (GenBank accession no. AF070521). The amount of DNA from each cell was normalized by detection of β-globin (forward primer 5′-TCT ACC CTT GGA CCC AGA GG and reverse primer 5′-CTG AAG TTC TCA GGA TCC ACG). Quantitative results were obtained using I-cycler real-time PCR (Bio-Rad) and SYBR green (Perkin-Elmer) core reagents. In this case, the amount of cellular DNA present in the preparations adversely affected measurement using the β-globin primer set, so the amount of DNA in each preparation was confirmed using the β-actin Taqman control kit from Perkin-Elmer instead. Plasmid DNAs from molecular clones of NL4-3 were used as standards for quantitation of proviral DNA, and the human genomic DNA from the Perkin-Elmer kit was used for standards for β-actin.
To detect the progress of proviral DNA during endogenous reverse transcription (ERT) and NERT reactions, we also included the following primer set which would amplify late viral products: forward primer 5′-GGC TAA CTA GGG AAC CCA CTG CTT and reverse primer 5′-ATA CCG ACG CTC TCG CAC CCA T, which amplify region 496 to 811 of NL4-3 (GenBank accession no. AF070521). In these cases, the ERT and NERT reactions were allowed to proceed for 4 h. At this time point the amount of early NERT product had already reached saturation, so additional time points at 45, 120, and 240 min were included for the early primer set.
RT assays.
Three types of reverse transcriptase (RT) assays were employed in this study to test different aspects of reverse transcription. The first assay is an exogenous RT assay using detergent-treated virions and a poly(rA)-oligo(dT) template. This assay measures the intrinsic enzymatic activity of the RT in particles. The assay was done essentially as previously described (27). The RT assay mixture contained 50 mM Tris-HCl (pH 7.8), 75 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 5 μg of poly(rA)-oligo(dT) (Calbiochem) per ml, 0.05% NP-40, 1 mM EDTA, and 10 μCi of [α-32P]dTTP (Amersham) per ml. For each assay, 5 μl of supernatant from each transfection was removed and mixed with 25 μl of RT assay mixture. Each sample was then incubated at 37°C, and 6-μl aliquots were removed at 1, 10, 30, and 60 min and spotted on DE81 paper (Whatman). The filters were dried and washed four times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 to 10 min each. The filters were then washed twice in 95% ethanol for 1 min each. Filters were then dried, and radioactivity was counted by liquid scintillation. Counts were normalized by p24 content prior to plotting.
The second assay is a classical ERT assay (48) using detergent-treated virions and the endogenous tRNA primer and viral template. This assay measures the ability of each virus to initiate reverse transcription on a viral template. Virus-containing supernatants from transfection of KFS or KFSΔNef plasmids were normalized by p24 content. Particles (5 ng) were pretreated with 30 U of micrococcal nuclease (Roche) for 1 h at 37°C in 50 μl of MN buffer (50 mM Tris HCl [pH 7.8], 5 mM NaCl, 2.5 mM CaCl2). To inactivate the micrococcal nuclease but not viral RT, EGTA was added to a final concentration of 2 mM and then 50 μl of endogenous buffer (50 mM Tris HCl [pH 7.5], 60 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 10 μCi of [α-32P] dATP, and 0.05% NP-40) was added to each reaction mixture. The mixture was incubated at 37°C overnight. The products were analyzed by PCR using early and late primers as described above.
The third assay is the NERT reaction. Viral particles were normalized by p24 content, and 4 ng of each stock was treated with NERT cocktail (1 mM dNTPs [Roche], 30 μM spermidine [pH 7.2] [Sigma], 2.5 mM MgCl2) for 4 h at 37°C as previously described (13). One nanogram of treated virions was used to infect MAGI cells for infectivity measurement or was directly tested by PCR using early and late PCR primers to determine the progress of the reaction. In some cases the products of the NERT reaction were used to test intrinsic RT activity after NERT treatment. In these cases, samples were removed after 4 h of NERT treatment and diluted 1:50 to reduce the relatively high concentration of cold dNTPs. The samples were then treated as described above for the exogenous RT assay.
RESULTS
Deletion of Nef does not grossly affect particle composition.
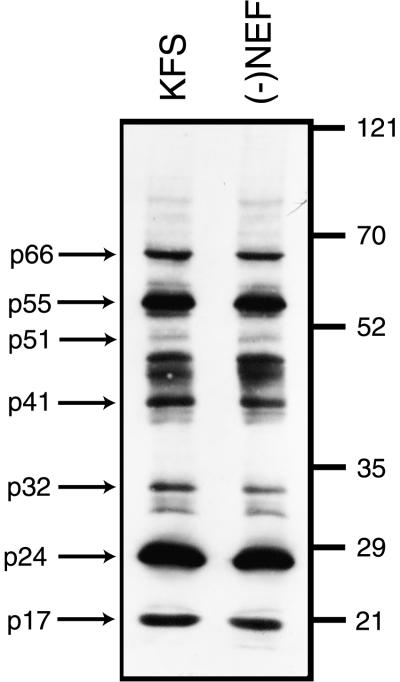
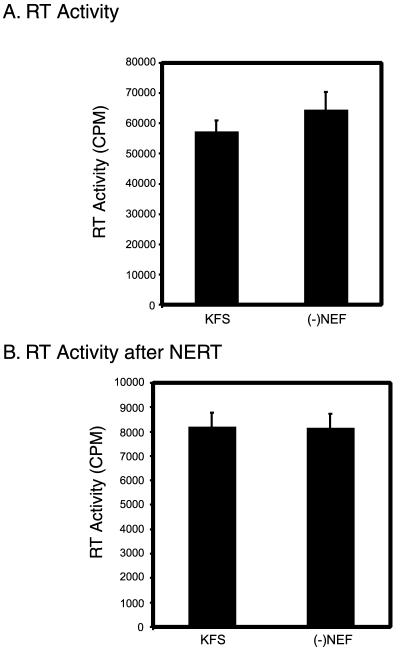
Western blot analysis of whole viral particles did not reveal any change in protein composition from Nef+ and Nef− virions (Fig. 1). One way to explain the increase in proviral DNA synthesis from virions that contain Nef could be that there is a change in p24 relative to RT. Since particle numbers are generally normalized by p24 content, the increase in proviral DNA could simply reflect a larger number of particles containing less total p24. However, a comparison of overall content shows almost identical levels of detectable proteins in each type of virion. Significantly, the amounts of p24 were essentially the same in each virus preparation, and the amounts of p66 and p51 of RT were the same (Fig. 1). This is also confirmed by the observation that the RT activity of the same amount of virions as judged by p24 content is essentially the same (Fig. 2A). This suggests that Nef is not influencing proviral DNA synthesis through an artifactual change in viral composition.
FIG. 1.
Deletion of nef does not affect virion composition. Equal amounts of virions as determined by p24 content were loaded onto an SDS–7.5% polyacrylamide gel. The gel was blotted, and HIV-1 proteins were detected using human antiglobulin to HIV-1. The relative amounts of each protein detected in virions containing Nef (KFS) and in virions in which Nef was absent [(−)Nef] appear to be identical. Numbers on the right are molecular weights in thousands.
FIG. 2.
Deletion of nef does not affect intrinsic RT activity or residual RT activity after NERT treatment. (A) The intrinsic RT activity of Nef-containing virions (KFS) and virions lacking Nef [(−)NEF] were tested using detergent-treated virions and an exogenously added poly(rA)-oligo(dT) template. In both cases the RT activity was essentially the same. (B) Intrinsic RT activity after NERT treatment was also determined for Nef-containing virions and virions lacking Nef. The samples were diluted 1:50 to lower the relatively high concentration of dNTPs carried over from the NERT reaction, and this may be responsible for the lower total counts. The error bars represent the standard deviations of at least triplicate measurements.
Intrinsic RT activities of Nef+ and Nef− virions are the same.
If Nef were somehow acting in the particle to directly augment the catalytic activity of RT, one would expect that the intrinsic activities of RT from the two types of particles could be different. Therefore, we tested the intrinsic RT activities from Nef+ and Nef− virions using a poly(rA)-oligo(dT) template (Fig. 2A). In this assay the particles are disrupted by treatment with NP-40 and tested using an exogenously added template. As can be seen in Fig. 2A, the activities of identical amounts of virus were essentially the same. This suggests that Nef does not change the intrinsic catalytic activity of the RT in the virions.
RT activities after NERT treatment of Nef+ and Nef− virions are the same.
To ensure that NERT treatment was not somehow altering the activities of the RT present in Nef+ and Nef− virions in a differential way, we tested the RT activities of NERT-treated virions. Both Nef+ and Nef− virions were allowed to undergo the NERT reaction. After NERT treatment, the virions were diluted 1:50 to lower the high concentration of cold dNTPs in the NERT cocktail. The virions were then tested for exogenous RT activity using the standard poly(rA)-oligo(dT) substrate. The results (Fig. 2B) show that the RT activities after NERT were essentially the same for both types of virions. The kinetics of this reaction over 12 h were also essentially identical (data not shown).
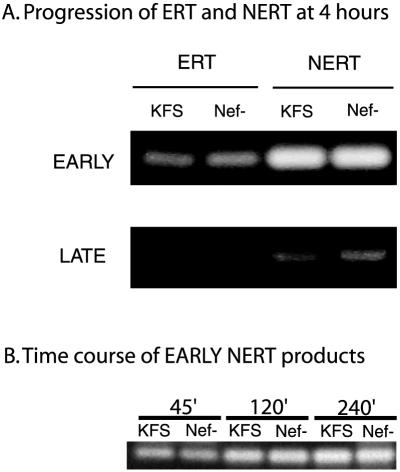
To see whether the progress of ERT or NERT reactions was influenced by the presence of Nef, we performed PCR analysis of both ERT and NERT reactions using primers that would detect early and late proviral DNA products (Fig. 3A). The progress of replication appears to be the same in Nef+ and Nef− virions in both the ERT and NERT reactions. The earliest time that late products could be detected was 4 h. At this time there were no late products visible in the ERT reaction mixture and only minimal products in the NERT reaction mixture. Unfortunately, at this time point the early NERT products had already reached saturation (Fig. 3A). To confirm that the amounts of proviral DNA formed by KFS and Nef− virions were the same at earlier times points, we performed PCR on samples removed from the NERT reaction mixture at 45, 120, and 240 min (Fig. 3B). Results from the time course confirm that the amounts of early products at earlier time points were also the same. Together, these data confirm that the presence of Nef does not appear to influence directly the course of reverse transcription on the actual viral template. These data also confirm that the NERT reaction proceeds to similar extents in both Nef+ and Nef− virions and does not introduce a bias in the later MAGI assay for infectivity.
FIG. 3.
The progression of reverse transcription is not affected by nef deletion. (A) The progress of reverse transcription in ERT and NERT reactions was monitored by PCR with primers to detect both early and late proviral DNA products. In both cases the relative amount of proviral DNA appears to be the same. Both sets of samples were run for 40 cycles. (B) To confirm that the early NERT products were the same at earlier time points, samples were also included at 45, 120, and 240 min, using the same PCR protocol for 40 cycles. Samples were run in triplicate, and a representative experiment is shown.
NERT treatment restores WT infectivity to Nef− virions.
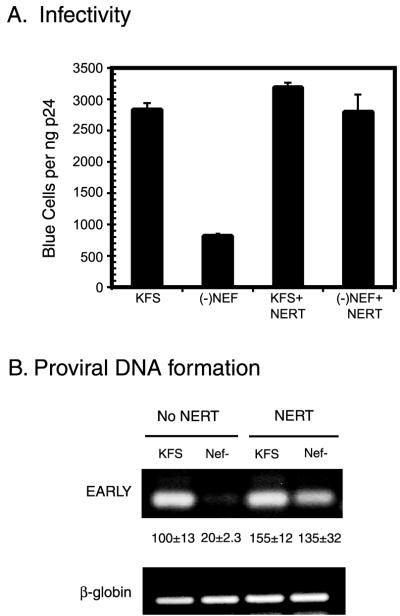
Using the MAGI cell assay, we determined the infectivities of Nef+ and Nef− virions. As has been noted by others, we saw about a fivefold decrease in infectivity of Nef− virions compared to Nef+ virions (2) (Fig. 4A). After treatment by NERT the infectivity of Nef− virions was restored to Nef+ levels (Fig. 4A). As previously reported, we saw a modest increase in infectivity of NERT-treated Nef+ virions over nontreated virions (13). The increase in infectivity was mirrored by an increase in the amount of proviral DNA present in the infected cells (Fig. 4B). This was evident both qualitatively by traditional PCR and upon quantitation by real-time PCR (Fig. 4B). The quantitation showed that Nef+ virions increased approximately 50% in infectivity, while Nef− virions increased approximately 500%.
FIG. 4.
The infectivity of virions with nef deleted can be restored after NERT treatment. (A) Nef-containing virions (KFS) and virions lacking Nef [(−)Nef] were tested for infectivity by the MAGI cell assay both before and after NERT treatment. After NERT treatment the virions lacking Nef [(−)Nef+NERT] were restored to levels similar to those of Nef-containing virions (KFS+NERT). Note that some enhancement of infectivity occurs even in Nef-containing virions. The results represent three independent experiments, and the error bars are the standard deviations from those experiments. (B) Proviral DNA was quantitated by PCR using the early primer set. β-Globin was used as a control for total cellular DNA added to each reaction mixture. The numbers between the lanes are values obtained from real-time PCR using the early primer set in the presence of SYBR green. In this case a β-actin Taqman probe was used to control cell number (see Materials and Methods). Samples were run in triplicate, and the data shown are the means ± one standard deviation. The traditional PCR was run for 40 cycles, and real-time data were collected for 50 cycles.
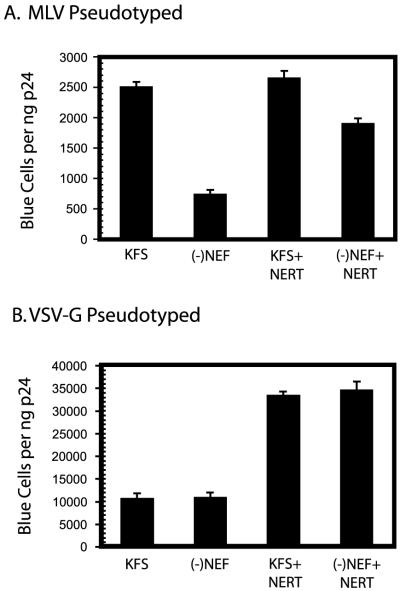
NERT enhancement of virions pseudotyped with MLV or VSV-G.
To determine the effect of different types of envelope glycoproteins on the enhancement of infectivity, Nef+ and Nef− virions were pseudotyped with either MLV or VSV-G envelope glycoprotein and treated by NERT. The virions pseudotyped with MLV envelope glycoprotein showed a pattern of infectivity similar to that of virions with HIV-1 envelope (Fig. 5A). The Nef− virus was about fourfold less infectious than Nef+ virus before NERT treatment (Fig. 5A). After NERT treatment, the Nef− virus showed a substantial increase in infectivity, although it did not restore infectivity to the levels seen with the Nef+ virus (Fig. 5A) in repeated experiments. As might be expected, virions pseudotyped with VSV-G envelope glycoprotein showed no difference in infectivity between Nef+ and Nef− virions (Fig. 5B). NERT treatment resulted in about a 3.5-fold increase in infectivity for both Nef+ and Nef− virions, and there was no apparent effect of the presence of Nef in these experiments (Fig. 5B).
FIG. 5.
Effect of pseudotyping with other envelope glycoproteins. KFS and Nef− virions were pseudotyped with either amphotrophic MLV envelope glycoprotein (A) or VSV-G envelope glycoprotein (B) and tested for infectivity with or without NERT treatment using the MAGI assay. Results represent triplicate measurements, and the bars represent the standard deviations of those measurements.
DISCUSSION
The goal of this study was to better understand the role that Nef plays in the enhancement of infectivity and proviral DNA formation. Many possible mechanisms could account for such effects, and it is entirely possible that more than one mechanism may be involved. One of the most direct ways that Nef could influence proviral DNA formation would be to enhance the catalytic activity or amount of RT present in the virion. Alternatively, Nef could enhance the progression of the process of reverse transcription of viral RNA. However, from evidence presented in this study, Nef does not appear to directly modulate the process of reverse transcription, at least as far as can be measured in these assays. This suggests that Nef may act on a step in replication independent of reverse transcription.
It is known that Nef can affect the infectivity of virions in a CD4-dependent manner by both enhancing virion release (41) and relieving a potential block of Env incorporation by CD4 (31). However, in the present study, Nef+ and Nef− viral particles were produced in cells which lack CD4, which should eliminate these CD4-dependent effects. Therefore, NERT treatment appears to act on CD4-independent effects of Nef.
Another possibility is that Nef could be acting to enhance the delivery of replicating complexes to the cytoplasm. A recent study (42) demonstrated an increase in cytoplasmic delivery of Nef+ virions over Nef− virions as measured by cytoplasmic p24 antigen content. One explanation offered for this effect is that the expression of Nef in producer cells somehow modifies the viral envelope protein to allow it to become more efficient during attachment and entry. A possible mechanism is given by the observation that Nef can enhance the phosphorylation of matrix protein (MA) (46) and phosphorylated forms of MA could alter the function of envelope protein. An interesting observation from this study is that mutations which are known to affect CD4 downregulation (LL164, WL57, and DD174) or diminished binding to SH3 domains (P69, P72, and P75) also affect the enhancement of infectivity even if the viral particles are produced in cells that lack CD4.
More evidence of Nef's action is given in another recent study in which Nef+ and Nef− virions were allowed to undergo intravirion fusion prior to infection of cells (56). The fusion of virions was accomplished by producing donor virions pseudotyped with gp160 and target virions that were pseudotyped with CD4. In this study, the infectivity of Nef− virions could be restored by fusion to Nef+ virions. A surprising observation from this study was that expression of Nef during the production of target virions had no effect on infectivity of virions, while expression of Nef during production of donor virions increased infectivity. The effect of Nef appeared to be dependent on the presence of envelope protein. In addition, when donor particles were pseudotyped with both HIV-1 and VSV-G envelope proteins and allowed to infect cells that lack CD4, there was still some residual effect of Nef on infectivity. These observations suggest that the effect of Nef is somehow envelope protein dependent, and yet they were not completely explained by an enhancement of virus-cell fusion alone.
In the present study we show that NERT treatment stimulates the infectivity of Nef− virions. Two directly observable things happen to NERT-treated virions: proviral DNA is elongated and the core particle partially disassembles. Although elongation of proviral DNA gives virions a “head start” in the process of reverse transcription, it appears that the progress of reverse transcription is the same for Nef+ and Nef− virions. Therefore, it seems more likely that the partial disassembly of the core is involved in the enhancement of infectivity. We know very little about what cues are used to trigger the disassembly process. It is possible that core disassembly and the envelope protein could be linked such that Nef must interact with the envelope protein to help trigger the process of disassembly. The observation that HIV-1 virions can be pseudotyped with other envelope proteins and remain infectious (1, 2) suggests that disassembly can occur independently of the type of envelope present. Indeed, the observation that during the NERT reaction cores can disassemble and yet virions remain infectious is evidence that entry and disassembly can be uncoupled. Yet, it is evident from electron microscopy studies that it is difficult to find intact cores even early after attachment and entry (20). This suggests that core disassembly must occur very early after or concurrently with attachment and fusion of envelope and cell membrane. Electron microscopy studies have revealed the presence of a structure know as the CEL (24, 52). This structure is a physical attachment of the smaller end of the core and the inner surface of the viral envelope. Although the function of the CEL is unknown, it does provide for a physical link between the core and envelope with which Nef could interact. If the CEL represented a type of switch to sense attachment and entry, it could provide a signal for disassembly to occur. Nef could play a role in mediating this reaction and thus aid the process of core disassembly. In this fashion Nef could be indirectly aiding in core disassembly in a manner that is dependent on the presence of envelope protein. In the case of the NERT-treated virions, disassembly is started while the virions are still intact. This premature disassembly could overcome the need for Nef during the entry process. A similar situation would exist when HIV-1 particles are pseudotyped with VSV-G envelope protein. In this case, targeting to the endosomal pathway could trigger disassembly through an alternate mechanism such as a change in pH, again bypassing the need for Nef to enhance the trigger process.
We have shown that the influence of Nef on infectivity and proviral DNA formation can be negated if reverse transcription is allowed to proceed inside the intact virion. We feel that this suggests a role for Nef in the processing of the core particle to allow the more efficient formation of the active reverse transcription complex. One way to explain this would be if Nef had some influence on the ability of core particles to disassemble. In light of recent advances in our understanding of this key process in viral replication, it should be possible to formulate new tests of the present theories and, in doing so, increase our overall understanding of HIV-1 replication.
ACKNOWLEDGMENTS
We thank Eric Freed for pNL4-3KFS, Judith Levin for pNL4-3Δnef, and Jane Burns for pHCMV-G. We also thank Craig Bond for helpful comments and suggestions.
This work was supported by Public Health Service grants G-12-RR03034 (NCRR), K22-HD-1228 (NICHD), and S-06-GM08248 (NIGMS).
REFERENCES
- 1.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akari H, Arold S, Fukumori T, Okazaki T, Strebel K, Adachi A. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol. 2000;74:2907–2912. doi: 10.1128/jvi.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson S, Shugars D C, Swanstrom R, Garcia J V. Nef from primary isolates of human immunodeficiency virus type 1 suppresses surface CD4 expression in human and mouse T cells. J Virol. 1993;67:4923–4931. doi: 10.1128/jvi.67.8.4923-4931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chazal N, Singer G, Aiken C, Hammarskjold M L, Rekosh D. Human immunodeficiency virus type 1 particles pseudotyped with envelope proteins that fuse at low pH no longer require Nef for optimal infectivity. J Virol. 2001;75:4014–4018. doi: 10.1128/JVI.75.8.4014-4018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y L, Trono D, Camaur D. The proteolytic cleavage of human immunodeficiency virus type 1 Nef does not correlate with its ability to stimulate virion infectivity. J Virol. 1998;72:3178–3184. doi: 10.1128/jvi.72.4.3178-3184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowers M Y, Spina C A, Kwoh T J, Fitch N J, Richman D D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig H M, Pandori M W, Guatelli J C. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci USA. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R, Greene W C. Functions of the auxiliary gene products of the human immunodeficiency virus type 1. Virology. 1990;178:1–5. doi: 10.1016/0042-6822(90)90373-y. [DOI] [PubMed] [Google Scholar]
- 11.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 12.Dickie P. Nef modulation of HIV type 1 gene expression and cytopathicity in tissues of HIV transgenic mice. AIDS Res Hum Retroviruses. 2000;16:777–790. doi: 10.1089/088922200308774. [DOI] [PubMed] [Google Scholar]
- 13.Dornadula G, Yang S, Pomerantz R J, Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dornadula G, Zhang H, Bagasra O, Pomerantz R J. Natural endogenous reverse transcription of simian immunodeficiency virus. Virology. 1997;227:260–267. doi: 10.1006/viro.1996.8317. [DOI] [PubMed] [Google Scholar]
- 15.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 17.Geleziunas R, Miller M D, Greene W C. Unraveling the function of HIV type 1 Nef. AIDS Res Hum Retroviruses. 1996;12:1579–1582. doi: 10.1089/aid.1996.12.1579. [DOI] [PubMed] [Google Scholar]
- 18.Glushakova S, Grivel J C, Suryanarayana K, Meylan P, Lifson J D, Desrosiers R, Margolis L. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J Virol. 1999;73:3968–3974. doi: 10.1128/jvi.73.5.3968-3974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldsmith M A, Warmerdam M T, Atchison R E, Miller M D, Greene W C. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grewe C, Beck A, Gelderblom H R. HIV: early virus-cell interactions. J Acquir Immune Defic Syndr. 1990;3:965–974. [PubMed] [Google Scholar]
- 21.Hanna Z, Kay D G, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 22.Harris M. From negative factor to a critical role in virus pathogenesis: the changing fortunes of Nef. J Gen Virol. 1996;77:2379–2392. doi: 10.1099/0022-1317-77-10-2379. [DOI] [PubMed] [Google Scholar]
- 23.Herna R G, Saksela K. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front Biosci. 2000;5:D268–D283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- 24.Hoglund S, Ofverstedt L G, Nilsson A, Lundquist P, Gelderblom H, Ozel M, Skoglund U. Spatial visualization of the maturing HIV-1 core and its linkage to the envelope. AIDS Res Hum Retroviruses. 1992;8:1–7. doi: 10.1089/aid.1992.8.1. [DOI] [PubMed] [Google Scholar]
- 25.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 71–120. [PubMed] [Google Scholar]
- 26.Jamieson B D, Aldrovandi G M, Planelles V, Jowett J B, Gao L, Bloch L M, Chen I S, Zack J A. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J Virol. 1994;68:3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keith W, Peden C, Martin M. Virological and molecular genetic techniques. In: Karn J, editor. HIV. 1. Virology and immunology. Oxford, United Kingdom: Oxford University Press; 1995. pp. 23–25. [Google Scholar]
- 28.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 29.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotov A, Zhou J, Flicker P, Aiken C. Association of Nef with the human immunodeficiency virus type 1 core. J Virol. 1999;73:8824–8830. doi: 10.1128/jvi.73.10.8824-8830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 32.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Gall S, Erdtmann L, Benichou S, Berlioz-Torrent C, Liu L, Benarous R, Heard J M, Schwartz O. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity. 1998;8:483–495. doi: 10.1016/s1074-7613(00)80553-1. [DOI] [PubMed] [Google Scholar]
- 34.Le Gall S, Prevost M C, Heard J M, Schwartz O. Human immunodeficiency virus type I Nef independently affects virion incorporation of major histocompatibility complex class I molecules and virus infectivity. Virology. 1997;229:295–301. doi: 10.1006/viro.1996.8417. [DOI] [PubMed] [Google Scholar]
- 35.Mariani R, Skowronski J. CD4 down-regulation by nef alleles isolated from human immunodeficiency virus type 1-infected individuals. Proc Natl Acad Sci USA. 1993;90:5549–5553. doi: 10.1073/pnas.90.12.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller M D, Feinberg M B, Greene W C. The HIV-1 nef gene acts as a positive viral infectivity factor. Trends Microbiol. 1994;2:294–298. doi: 10.1016/0966-842x(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 37.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oldridge J, Marsh M. Nef—an adaptor adaptor? Trends Cell Biol. 1998;8:302–305. doi: 10.1016/s0962-8924(98)01318-x. [DOI] [PubMed] [Google Scholar]
- 39.Pandori M W, Fitch N J, Craig H M, Richman D D, Spina C A, Guatelli J C. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piguet V, Trono D. The Nef protein of primate lentiviruses. Rev Med Virol. 1999;9:111–120. doi: 10.1002/(sici)1099-1654(199904/06)9:2<111::aid-rmv245>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 41.Ross T M, Oran A E, Cullen B R. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol. 1999;9:613–621. doi: 10.1016/s0960-9822(99)80283-8. [DOI] [PubMed] [Google Scholar]
- 42.Schaeffer E, Geleziunas R, Greene W C. Human immunodeficiency virus type 1 Nef functions at the level of virus entry by enhancing cytoplasmic delivery of virions. J Virol. 2001;75:2993–3000. doi: 10.1128/JVI.75.6.2993-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz O, Marechal V, Danos O, Heard J M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sodroski J, Goh W C, Rosen C, Campbell K, Haseltine W A. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature. 1986;322:470–474. doi: 10.1038/322470a0. [DOI] [PubMed] [Google Scholar]
- 45.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swingler S, Gallay P, Camaur D, Song J, Abo A, Trono D. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J Virol. 1997;71:4372–4377. doi: 10.1128/jvi.71.6.4372-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swingler S, Mann A, Jacque J, Brichacek B, Sasseville V G, Williams K, Lackner A A, Janoff E N, Wang R, Fisher D, Stevenson M. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat Med. 1999;5:997–1003. doi: 10.1038/12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Temin H M, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 49.Wang J K, Kiyokawa E, Verdin E, Trono D. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc Natl Acad Sci USA. 2000;97:394–399. doi: 10.1073/pnas.97.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yee J K, Friedmann T, Burns J C. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43A:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, Dornadula G, Alur P, Laughlin M A, Pomerantz R J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc Natl Acad Sci USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Dornadula G, Orenstein J, Pomerantz R J. Morphologic changes in human immunodeficiency virus type 1 virions secondary to intravirion reverse transcription: evidence indicating that reverse transcription may not take place within the intact viral core. J Hum Virol. 2000;3:165–172. [PubMed] [Google Scholar]
- 53.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, Dornadula G, Pomerantz R J. Natural endogenous reverse transcription of HIV-1. J Reprod Immunol. 1998;41:255–260. doi: 10.1016/s0165-0378(98)00062-x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Dornadula G, Pomerantz R J. Natural endogenous reverse transcription of HIV type 1. AIDS Res Hum Retroviruses. 1998;14(Suppl. 1):S93–S95. [PubMed] [Google Scholar]
- 56.Zhou J, Aiken C. Nef enhances human immunodeficiency virus type 1 infectivity resulting from intervirion fusion: evidence supporting a role for nef at the virion envelope. J Virol. 2001;75:5851–5859. doi: 10.1128/JVI.75.13.5851-5859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]