Abstract
Deglycosylation of viral glycoproteins has been shown to influence the number of available epitopes and to modulate immune recognition of antigens. We investigated the role played by N-glycans in the immunogenicity of hepatitis C virus (HCV) E1 envelope glycoprotein, a naturally poor immunogen. Eight plasmids were engineered, encoding E1 protein mutants in which the four N-linked glycosylation sites of the protein were mutated separately or in combination. In vitro expression studies showed an influence of N-linked glycosylation on expression efficiency, instability, and/or secretion of the mutated proteins. Immunogenicity of the E1 mutants was studied in BALB/c mice following intramuscular and intraepidermal injection of the plasmids. Whereas some mutations had no or only minor effects on the antibody titers induced, mutation of the fourth glycosylation site (N4) significantly enhanced the anti-E1 humoral response in terms of both seroconversion rates and antibody titers. Moreover, antibody induced by the N4 mutant was able to recognize HCV-like particles with higher titers than those induced by the wild-type construct. Epitope mapping indicated that the E1 mutant antigens induced antibody directed at two major domains: one, located at amino acids (aa) 313 to 332, which is known to be reactive with sera from HCV patients, and a second one, located in the N-terminal domain of E1 (aa 192 to 226). Analysis of the induced immune cellular response confirmed the induction of gamma interferon-producing cells by all mutants, albeit to different levels. These results show that N-linked glycosylation can limit the antibody response to the HCV E1 protein and reveal a potential vaccine candidate with enhanced immunogenicity.
Hepatitis C virus (HCV) is a major cause of chronic liver disease, cirrhosis, and hepatocellular cancer worldwide (1). Vaccine development is therefore essential but has been hampered by the poor understanding of the type of immunity that naturally terminates HCV infection. The identification of viral components involved in the development of neutralizing immunity has been limited in part because the necessary cell culture system to grow the virus and a small-animal model susceptible to HCV infection do not exist. In both humans and chimpanzees, the frequency of persistent infection is high, and virus replication occurs despite the presence of cellular and humoral immune responses (20, 40). Different factors are likely to contribute to viral persistence. These include a weak antiviral immune response of the infected host, hiding of the virus from neutralizing antibodies via its association with lipids, emergence of escape mutants at the level of both B- and T-cell epitopes, and possibly the biased or low level of cytokine production (11).
Recent studies have shown that the development of early, polyclonal, vigorous, and maintained CD4+ and CD8+ T-cell-mediated specific immune responses appears to play a major role in viral clearance for both humans and chimpanzees (13, 19, 23, 28, 29, 48). Nonetheless, it has also been shown in different studies that specific antibodies targeted at hypervariable region 1 (HVR-1) of E2 that are present in the sera of HCV-infected patients or induced following vaccination of animals may be neutralizing (45, 53, 54). In chimpanzees, a recombinant gpE1/gpE2 subunit vaccine has been shown to prevent either acute or chronic infection following challenge with a homologous viral strain and a low infectious dose (25). This protection was linked to both the induction of specific anti-E2 antibody, referred to as neutralizing-of-binding antibodies (43), and of a specific CD4+ T-cell-mediated response (M. Houghton et al., 5th International Meeting on HCV and Related Viruses, abstr. O57, 1998). More recently, therapeutic vaccination of chronically infected chimpanzees using a recombinant E1 protein has resulted in improvement of the liver histology and clearance of viral antigens from the liver of vaccinated animals (G. Maertens et al., 6th International Symposium on Hepatitis C and Related Viruses, p. 74, 1999).
Both HCV envelope proteins are heavily glycosylated. For the prototype H strain (subtype 1a), E1 contains 5 and E2 contains 11 potential glycosylation sites. We have shown, in previous work, that E1 is glycosylated at positions 196, 209, 234, and 305, indicating that the fifth sequon is not used for the addition of N-linked oligosaccharides (21, 33). Among the modifications affecting proteins targeted at the secretory pathway, N-linked glycosylation plays important roles in the folding, stability, biological activity, and antigenicity of proteins (39, 41, 50).
Glycans can influence the immunogenicity of proteins in different ways: through their ability to structurally maintain an appropriate antigenic conformation, through their capacity to shield potential neutralization epitopes (3, 7, 8), and through their ability to alter the proteolytic susceptibility of proteins (47). Oligosaccharides have been shown to limit the neutralizing antibody response to simian immunodeficiency virus and influenza virus by covering portions of B-cell epitopes of the gp120 and the hemagglutinin protein, respectively, while removal of N-linked glycans appears to enhance the production of cytotoxic T lymphocytes (CTL) specific for the human immunodeficiency virus type 1 (HIV-1) envelope protein (16, 42, 52). On the contrary, deletion of some glycans of the HIV-1 gp160 abrogated the in vivo priming of T cells recognizing an epitope close to the deletion sites (46).
In the present study, we investigated whether removal of defined N-linked oligosaccharide chains of the HCV E1 envelope protein could influence the induction of E1-specific humoral and cellular immune responses. The immunogenicity of seven different E1 glycosylation mutants was analyzed in mice and compared with that of the wild-type E1 protein using a DNA-based vaccination approach.
MATERIALS AND METHODS
Plasmid constructs and recombinant viruses.
Plasmids encoding E1 protein mutants were engineered by cloning the cDNA corresponding to the H strain E1 glycoprotein (subtype 1a) into the replicative-form DNA of M13mp18 as previously described (33). Five synthetic oligonucleotide primers were designed so that for each N-glycosylation consensus sequence, Asn-X-Thr/Ser, the Asn-encoding codon was replaced with a Gln-encoding codon. Mutants were named with an N (for the Asn amino acid modification) and a number (related to the position of the Asn amino acid on the polyprotein). Mutants carrying a single mutation are referred to as N1, N2, N3, and N4; those carrying two mutations are referred to as N1-4 (mutation of the N1 and N4 sites) and N2-3 (mutation of the N2 and N3 sites). Finally, a quadruple mutant, carrying mutations at all four recognized glycosylation sites, including a silent mutation at the fifth site, which is known to be unglycosylated (33), was called N1-2-3-4 (Fig. 1). The mutated fragments were amplified by PCR and cloned into the SmaI site of the pCI vector (Promega). All recombinant plasmids were controlled by sequencing.
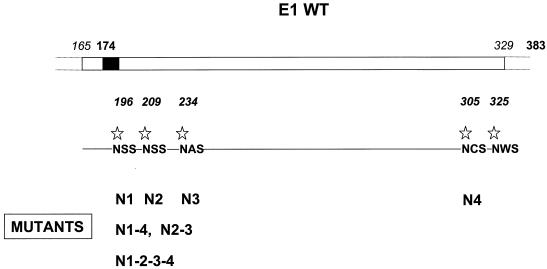
FIG. 1.
Schematic representation of HCV E1 envelope glycoprotein and its glycosylation sites. Wild-type (WT) E1 maps within aa 174 to 383 on the HCV polyprotein. The black box corresponds to the signal sequence of E1. The five potential glycosylation sites are indicated by stars. The different mutants generated are named with an N for the Asn amino acid mutated and a number referring to the position of the glycosylation site on the map (N1 to N4). Mutants carrying double or quadruple mutations are referred as N1-4, N2-3, and N1-2-3-4.
To generate recombinant vaccinia viruses corresponding to each mutant, fragments containing E1 mutated sequences were PCR amplified from the different pCI constructs and inserted into the EcoRI and SpeI sites of the pTM1 polylinker (37). Recombinant vaccinia viruses were generated by homologous recombination essentially as described elsewhere (27) and plaque purified twice on 143 thymidine kinase-negative cells under bromodeoxyuridine selection (50 μg/ml). Each virus stock, derived from a single plaque isolate, was expanded in CV1L cells at a multiplicity of infection (MOI) of 1 PFU/cell. Virus vTF7-3, a recombinant vaccinia virus expressing the T7 DNA-dependent RNA polymerase (22), was obtained from B. Moss (National Institutes of Health, Bethesda, Md.).
In vitro expression studies following transient transfection.
Expression of E1 protein mutants was examined in cell extracts by Western blotting, capture enzyme-linked immunosorbent assay (ELISA), and immunoprecipitation following transient transfection of NIH 3T3 cells cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% fetal calf serum (FCS), and 4 mM l-glutamine, plus 80 U of penicillin and 80 mg of streptomycin per ml.
For Western blotting and ELISAs, 3 × 106 cells per 100-mm dish were transfected with 4 μg of each plasmid using Lipofectamine Plus (Gibco-BRL). At 48 h, cells were lysed in 50 mM Tris (pH 7.5)–150 mM NaCl–5 mM MgCl2–1% Triton X-100–1.5 μg of aprotinin per ml. For Western blots, samples were heated at 100°C for 5 min in the presence of Laemmli buffer and then analyzed by immunoblotting using a mouse monoclonal anti-E1 antibody (IGH201; Innogenetics) after separation by electrophoresis on sodium dodecyl sulfate (SDS)–13% polyacrylamide. For the capture ELISA, a 1:20 dilution of the lysates was administered to microtiter plates on which antibody IGH201 had been adsorbed. Bound E1 protein was detected by incubation with a biotinylated specific monoclonal antibody (MAb) (IGH200; Innogenetics) followed by an incubation with streptavidin labeled with peroxidase. The final concentration of the detected E1 protein was deduced after comparison with a standard curve established using known concentrations of purified E1 protein.
For immunoprecipitation analysis, 3 × 105 cells per 35-mm well were transfected with 2 μg of each plasmid. At 28 h posttransfection, cells were incubated for 1 h in medium lacking methionine and cysteine and then metabolically labeled with 75 μCi of [35S]Translabel (Amersham) per ml overnight in the same medium. Cells were lysed in 50 mM Tris (pH 7.5)–150 mM NaCl–1 mM EDTA–1% NP-40–10 μg of aprotinin per ml. Clarified lysates were incubated for 1 h with 10 μl of the murine MAb A4 (18) in RIPA buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.05% sodium deoxycholate, 0.1% SDS, 0.5% NP-40, and 10 μg of aprotinin per ml and then with 50 μl of protein G-Sepharose overnight. After the last step, beads were washed three times with RIPA buffer; then samples were boiled for 5 min in the presence of Laemmli buffer and analyzed by electrophoresis on SDS–13% polyacrylamide.
Vaccinia virus expression assays.
CV1 cells were infected with vTF7.3 alone or in combination with recombinant vaccinia viruses, each at an MOI of 5 PFU/cell. After 1 h at 37°C, the inoculum was removed and replaced with DMEM supplemented with 5% FCS. At 1 h postinfection, cells were incubated for 1 h with DMEM lacking methionine and cysteine and then metabolically labeled overnight with 50 μCi of [35S]Translabel. Expression of E1 mutants was then examined in cell extracts by immunoprecipitation as described above.
Immunization protocols.
Six- to 8-week-old female BALB/c mice were purchased from IFFA Credo. All DNA preparations were produced using endotoxin-free purification columns (Qiagen). Immunizations were performed with a gene gun (PowderJect) resulting in the injection of 5 μg of plasmid DNA into the abdominal skin (referred to as intraepidermal [i.e.] injection) and with a syringe in the anterior tibialis muscle (referred to as intramuscular [i.m.] injection) using 100 μg of plasmid DNA per injection. Two immunization schedules were followed. In the first one, DNA was injected five times i.e. and i.m. at weeks 0, 5, 9, 14, and 18, and mice were bled three times at 11, 21, and 29 weeks afer the first injection. In the second one, DNA was injected four times i.e. and i.m. at weeks 0, 2, 4, and 6, and spleens from the vaccinated mice were removed at 9 weeks for analysis.
Detection of anti-E1 antibodies.
Anti-E1 antibodies were first measured using a specific ELISA (Innogenetics). A recombinant E1 protein, expressed and purified from mammalian cells, was adsorbed to microtiter plates, and after blocking, serial dilutions of the mouse sera were added to the wells. The endpoint titer was defined as the serial threefold dilution resulting in an optical density (OD) equal to two times the mean of the background of the assay. Samples were analyzed in duplicate. Sera from mice were examined for the dominant antibody isotype response using a commercial isotyping kit (Pharmingen) according to the manufacturer's instructions.
The reactivity of the anti-E1 antibodies against HCV-like particles (HCV-LPs) derived from a subtype 1a and expressed in insect cells (4) was also analyzed using an ELISA as recently described (5).
Peptide-based epitope mapping.
Two peptides were used in ELISAs to further characterize the immune reactivity of the induced antibodies. One, V1C1V2, is located within the N-terminal part of E1 between amino acids (aa) 192 and 226 (YEVRNVSGMYHVTNDCSNSSIVYEAADMIMHTPGC). The other one, C4, mapped within the C-terminal part of the protein corresponding to aa 313 to 332 (ITGHRMAWDMMMNWSPTTAL). All sera were tested at a 1:100 dilution, and experimental conditions were as previously described (35). Only sera giving absorbance values greater than three times the OD values of negative sera were considered positive.
Elispot assays.
Spleen cells from individual mice were stimulated for 5 days in vitro with 10 μM peptide H16A (aa 312 to 326; HITGHRMAWDMMMNWA [10]), G10A (aa 315 to 323; GHRMAWDMMA [10]), or S9V, an irrelevant HCV peptide (SMVGNWAKV), in alpha minimal essential medium (αMEM) culture medium (Gibco-BRL) supplemented with 10% FCS, 10 mM HEPES buffer, 5 × 10−5 M β-mercaptoethanol, and 4 mM l-glutamine plus 80 U of penicillin, 80 mg of streptomycin, nonessential amino acids (Gibco-BRL), and 10 U of recombinant interleukin-2 (IL-2) (Pedro-Tech EC Ltd.) per ml.
Gamma interferon (IFN-γ)- and IL-4-producing cells were quantified by cytokine-specific enzyme-linked immunospot assay (Elispot). Ninety-six-well nitrocellulose-backed plates (Multiscreen; Millipore) were coated with 100 μl of anti-mouse IFN-γ MAb (10 μg/ml; Pharmingen) overnight at 4°C, then washed with phosphate-buffered saline (PBS) and blocked for 1 h with αMEM. Splenocytes (5 × 105 to 10 × 105/well) were cultured for 18 h in αMEM alone (negative control) or with 10 μg of selected peptide or 5 μg of concanavalin A (positive control) per ml in the presence of IL-2 (5 U/ml) in triplicate wells. After washing with PBS followed by PBS–0.05% Tween, biotinylated anti-IFN-γ MAb at 10 μg/ml was added and incubated for 2 h at room temperature. Plates were washed in PBS–0.05% Tween, and streptavidin-conjugated horseradish peroxidase (Southern Biotechnology; 1:1,000 dilution) was added for 1 h at room temperature. After rewashing, spots representing individual cytokine-producing cells were visualized by developing with the substrate 3-amino-9-ethylcarbazole and then counted using a Microvision Instruments KL1500 electron microscope.
Depletion of CD4+ or CD8+ cells was performed after 5 days of stimulation with anti-mouse CD4 (clone GK1.5; Southern Biotechnology Associates) or anti-mouse CD8 (clone 53-6.7; Southern Biotechnology Associates) antibodies using magnetic cell sorting (MACS; Miltenyi Biotec). The efficiency of depletions was assessed by flow cytometry.
RESULTS
In vitro characterization of HCV E1 glycosylation mutants.
N-linked glycosylation can play an important role in the stability, solubility, and secretion of glycoproteins and in their capacity to interact with the lectin-based chaperone system in the endoplasmic reticulum (ER), all properties influencing protein folding (41). To determine if the mutation of glycosylation sites of HCV E1 affects some of these properties, expression of E1 mutants lacking one or several glycans was analyzed (Fig. 1).
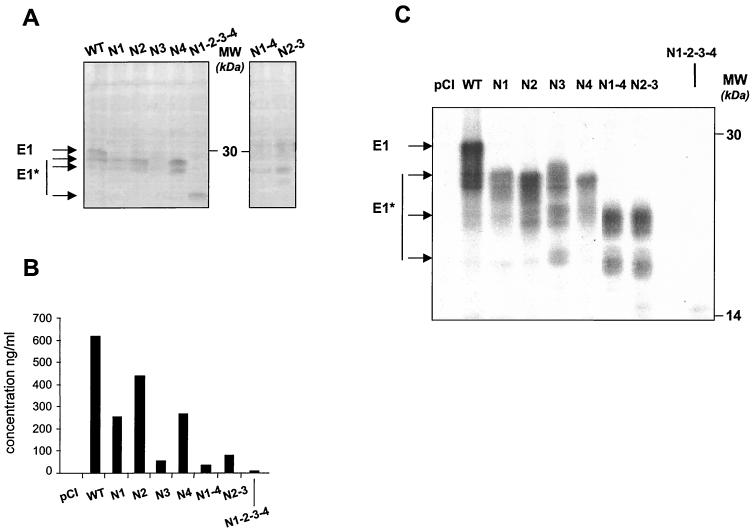
First, E1 mutants were expressed in NIH 3T3 cells following transient transfection with the corresponding plasmids and analyzed by Western blotting. As shown in Fig. 2A, all mutated proteins were expressed and recognized by a MAb (IGH201) targeting an epitope located between aa 212 and 224 on the polyprotein, which corresponds to a domain outside of the N-linked glycosylation sites. Their electrophoretic mobilities were compared with that of a wild-type E1 protein truncated at position 329 (wild type) and found to be compatible with E1 proteins lacking one (N1, N2, N3, and N4), two (N1-4 and N2-3), or four (N1-2-3-4) glycans.
FIG. 2.
In vitro expression studies following transient transfection of NIH 3T3 cells with the DNA-encoding E1 mutants. (A) Western blot. (B) Quantitative determination of intracellular expression of E1 mutants by capture ELISA. (C/) Immunoprecipitation (see Materials and Methods). The figure shows representative experiments. HCV proteins are indicated (E1* refers to the deglycosylated forms of E1), and positions of protein size markers are shown.
As previously observed, the wild-type E1 and the different mutants were resolved as a multiband protein, representing proteins that differ in their degree of glycosylation (17). The intensity of the bands detected for the N3 and N1-4 mutants appeared to be lower than the others, suggesting either that their expression is less efficient or that these proteins are more prone to degradation or, alternatively, more poorly recognized by MAb IGH201. As samples were denatured in Laemmli buffer before separation by SDS-polyacrylamide gel electrophoresis, the lack of detection of the N3 and N1-4 proteins is unlikely to be related to a distinct folding of the proteins or to a lower solubility.
Quantitative determination of the intracellular expression of the wild-type and mutated E1 proteins (performed on the same samples as those used for Western blot analysis) by a capture ELISA using MAb IGH201 showed differences between the amounts of antigen detected for the different transfected plasmids (Fig. 2B). For the wild-type, N1, N2, and N4 antigens, the concentration obtained ranged between 251 and 616 ng/ml of cellular extract tested, whereas detection of mutants N3, N1-4, and N2-3 led to concentrations lower than 80 ng/ml. A complete lack of detection of N1-2-3-4 was noted. The poor detection or lack of detection of some mutants by this technique may simply be due to the fact that these antigens adopted a specific folding which could have masked the epitope recognized by the capture antibody IGH201 or the detecting antibody IGH200. Another explanation would be that the lack of detection of these mutants could be related to the ELISA system itself, in particular to the insolubility of the different antigens in the buffer used.
Immunoprecipitation studies were then carried out with another MAb (A4) targeted at an epitope located between aa 197 and 207 (Fig. 2C). This assay, although not a quantitative one, revealed fewer differences in the detection of the different mutants except for N1-2-3-4, which could still not be detected. This method, which was less denaturing for the tested antigens, provided data indicating either that the accessibility of the A4 epitope is indeed suppressed in the mutant N1-2-3-4 or that this antigen is insoluble under the conditions used.
Altogether, these data indicate that (i) all E1 mutants were expressed; (ii) mutation of all glycosylation sites (N1-2-3-4) considerably affected the conformation or the nature (solubility) of the protein; and (iii) transfection of plasmids encoding mutants N3, N1-4, and N2-3 led to the detection of smaller amounts of proteins compared with the wild-type E1 or other mutants, suggesting either a lower expression efficiency or an instability of these proteins.
Mutation of glycosylation sites affects the secretion of E1.
To determine whether the absence of oligosaccharide chains influences the secretion of the resulting E1 protein, the different E1 mutants were expressed using recombinant vaccinia viruses. This expression system, which allows high antigen production, was used because we were unable to detect E1 proteins in supernatants after transient transfection with the different engineered plasmids.
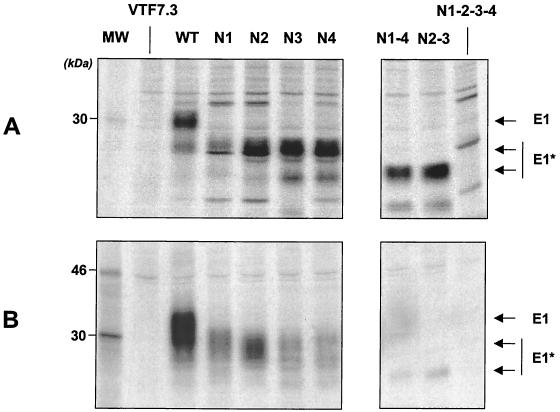
Intracellular and secreted E1 mutant proteins, truncated at aa 329, were identified by immunoprecipitation with MAb A4 (Fig. 3). The intracellular form of the E1 mutants was detected in all cases, although the level of expression of E1-N1 appeared lower than that of other mutants. Secretion was observed for the wild-type protein and the single mutants N1, N2, N3, and N4, although secretion of N3 and N4 proteins appeared to be reduced. Mutants N1-4, N2-3, and N1-2-3-4 were nearly undetectable, showing that mutation of at least two glycosylation sites abrogates the secretion of the E1 protein. Molecular weights of the mutated E1 proteins were higher for the secreted forms than for the intracellular forms, suggesting, as has been reported previously for the wild-type protein, that the secreted proteins underwent additional modification during secretion. This demonstrates that the proteins were indeed secreted and not released after cell death.
FIG. 3.
Immunoprecipitation of E1 mutants expressed by recombinant vaccinia viruses. CV1 cell monolayers infected with vTF7.3 alone or coinfected with vTF7.3 and recombinant vaccinia viruses expressing the different E1 mutants were labeled with [35S]methionine. E1 proteins present in the cell lysates (A) or the corresponding supernatants (B) of infected cells were immunoprecipitated with the anti-E1 specific MAb A4 as described in Materials and Methods. HCV proteins are indicated on the right (E1* refers to the deglycosylated forms of E1), and positions of the 14C-labeled protein markers are shown (lane MW).
Degree of glycosylation of E1 influences antibody response induced in immunized mice.
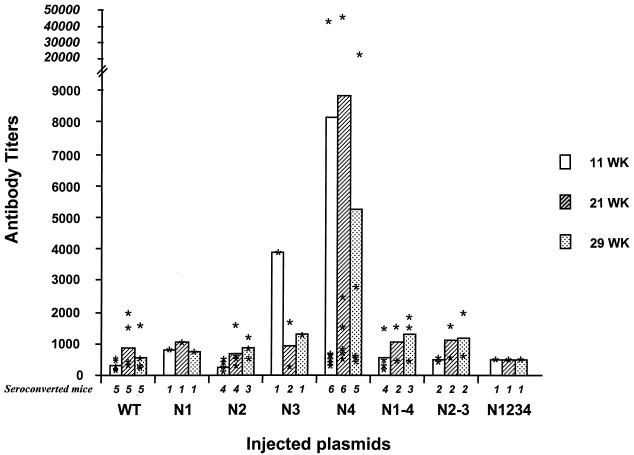
The ability of the different E1 mutants to induce an antibody response was evaluated in BALB/c mice following concomitant i.e. and i.m. immunizations. Sera obtained from mice at 11, 21, and 29 weeks after the first injection were tested for reactivity against a recombinant wild-type E1 protein (Fig. 4). Sera from mice immunized by the N2 and N1-4 plasmids showed a reactivity similar to that obtained from mice that received the wild-type E1-expressing plasmid in terms of either seroconversion rates or antibody titers induced. Sera from animals injected with the N1, N3, N2-3, and N1-2-3-4 plasmids showed that only a very small number of mice seroconverted. In contrast, the group of N4-immunized mice was the only group for which 100% of the animals seroconverted as early as the first time point studied (11 weeks after the first injection). Moreover, antibody titers tended to be higher in the N4-immunized mouse group (mean and median antibody titers, 1:8,837 and 1:1,105 at 21 weeks post-first injection) compared with the wild-type E1-immunized mice (1:872 and 1:380), although the differences were not statistically significant (P > 0.5).
FIG. 4.
Anti-E1 antibody titers and seroconversion rates. Each plasmid encoding E1 wild type (WT) or an E1 mutant was injected into groups of six mice. Titers, given for seroconverted mice only, are represented for each individual mouse with stars, while mean titers are indicated by bars. Titers are shown for sera obtained 11, 21, and 29 weeks postimmunization. The number of seroconverted mice is indicated at the bottom of each bar. Results are shown as the reciprocal of the serum dilution resulting in an OD equal to two times the mean of the background of the assay.
To confirm that the N4 mutant was able to enhance the antibody response induced in mice, a second immunization protocol was performed following the same schedule but including only two groups of vaccinated mice, one with the wild-type E1 and one with the N4-expressing DNA. A larger number of animals (14 for wild-type E1 and 12 for N4) (Table 1) was included in this protocol. Comparison of the antibody response induced in these two groups showed a significantly higher seroconversion rate in the N4-immunized mice (11 of 12 versus 7 of 14, P = 0.011, test of proportion) as well as antibody titers that tended to be higher (mean and median antibody titers, 1:6,616 and 1:485) than those induced by wild-type E1 plasmid (1:983 and 1:196). Isotype of the induced immunoglobulin G (IgG) was analyzed and found to be comparable, including a mix of IgG1 and IgG2a in both groups (Table 1).
TABLE 1.
Antibody response against E1 protein and HCV-LPs at 21 weeks after first immunization
| Mouse group | E1 protein
|
HCV-LP titera | ||
|---|---|---|---|---|
| No. seroconverted | Titera (mean/median) | Isotype | ||
| 836 | IgG1 | — | ||
| WT | 7/14 | — | — | |
| 4,766 | IgG2a, IgG1 | 1,000 | ||
| 181 | — | |||
| — | — | |||
| — | 500 | |||
| — | — | |||
| 193 983/196 | — | |||
| 521 | IgG2a, IgG1 | — | ||
| — | 1,000 | |||
| 186 | — | |||
| 196 | — | |||
| — | — | |||
| — | — | |||
| 485 | IgG1 | — | ||
| N4 | 11/12 | 196 | 1,000 | |
| 521 | IgG1 | — | ||
| >60,000 | IgG1, IgG2a | 8,000 | ||
| 185 | — | |||
| 395 6,616/485 | 1,000 | |||
| — | 1,000 | |||
| 4,000 | IgG2a, IgG1 | 1,000 | ||
| 572 | IgG2a, IgG1 | 1,000 | ||
| 5,849 | IgG1, IgG2a | — | ||
| 412 | IgG1, IgG2a | — | ||
| 161 | — | |||
Titers are shown as the reciprocal of the serum dilution resulting in an OD equal to two times the mean of the background of the assay. —, nonseroconverted mouse.
Impact of oligosaccharide chains on qualitative aspects of the anti-E1-specific humoral response. (i) Two major epitopes are recognized.
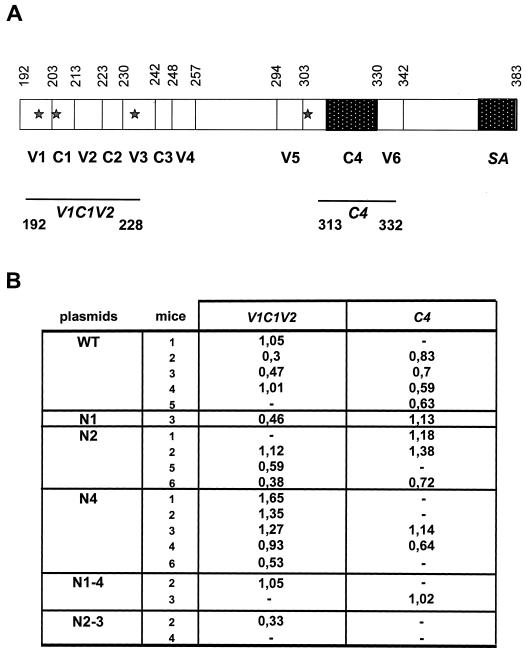
As indicated in Fig. 5, the HCV E1 protein is composed of four conserved amino acid regions (C1 to C4) interspersed with six variable domains (V1 to V6). To perform fine epitope mapping, reactivity of the mice sera was analyzed with different peptides spanning over 75% of the E1 protein. As shown in Fig. 5, whatever DNA-encoded E1 mutant was injected, antibodies induced displayed a reactivity against two different peptides, V1C1V2 and C4. There was a correlation between the overall antibody titers detected in ELISA (Fig. 4) and the reactivity against the V1C1V2 peptide. Surprisingly, this correlation did not exist for the C4 peptide and N4-immunized mice; even some of those which displayed the highest anti-E1 antibody titers in ELISA (Fig. 4) did not recognize the C4 epitope. These results suggest that of the E1 domains screened by our peptide analysis, the major determinant recognized by N4-derived sera is located at the N-terminal part of E1, while that recognized by wild-type E1 or the other mutant-induced sera showed a good overall reactivity to both the N- and C-terminal epitopes.
FIG. 5.
Epitope mapping. (A) Schematic representation of E1 protein with the delineation of its four conserved amino acid sequences (C1 to C4) and its six variable hydrophilic regions (V1 to V6). The four glycosylated sites are indicated by stars. SA, signal anchor domain. Peptides V1C1V2 and C4 used for ELISA are indicated. (B) Reactivity of mouse sera against V1C1V2 and C4 peptides. The binding results obtained are given as OD values. Results are given for seroconverted mice only.
(ii) Mutation of the fourth glycosylation site (N4) enhances the ability of induced anti-E1 antibodies to recognize HCV-LPs.
The lack of a convenient and reproducible tissue culture system for HCV infection precludes a reliable analysis of the neutralizing capacity of the induced antibodies. Structural and antigenic composition of HCV-LPs assembled in insect cells has been proposed to be similar to those of putative virions (4). The HCV envelope proteins present in the HCV-LPs are presumably presented in a native, virion-like conformation and may therefore interact with anti-envelope antibody directed at conformational epitopes (6). The reactivity of anti-E1 antibody induced by the wild-type DNA-encoded E1 protein or the N4 DNA mutant against HCV-LPs was investigated using a recently developed ELISA (6). As shown in Table 1, the N4 plasmid induced antibody responses significantly enhanced over the E1 wild-type-encoding plasmid in terms of seroconversion rates (6 of 12 [N4] versus 3 of 14 [wild type]) and antibody titers (N4 ranging from 1:500 to 1:1,000, wild-type E1 ranging from 1:1,000 to 1:8,000; P = 0.048, permutation test based on ratio of variances).
E1 mutants are able to induce cell-mediated immune responses similar to that observed with wild-type E1.
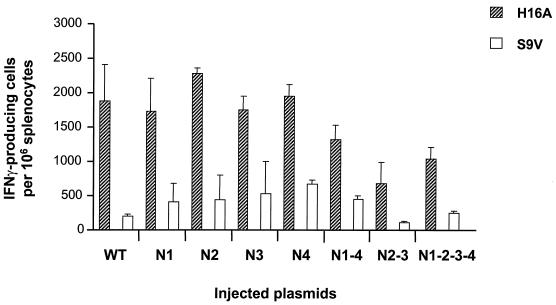
The cellular immune response induced by the different E1 mutants was analyzed by quantification of cytokine-producing T cells in vitro. Spleens of mice were removed 2 weeks after the final injection. Splenocytes were incubated in the presence of two different peptides: H16A, which has been shown to contain a CD8+ and a CD4+ epitope in the context of a genotype 1b sequence, and G10A, which corresponds to the CD8+ epitope present in the H16A peptide (30). The H16A peptide used here derives from a subtype 1a E1 sequence and contains two mutations located in the CD4+ determinant compared with the 1b peptide originally described in the literature (Fig. 6 legend).
FIG. 6.
Elispot assay. Each plasmid encoding wild-type (WT) or mutant E1 was injected into groups of five mice. The IFN-γ-producing T-cell frequency was determined for all mice in an Elispot assay using splenocytes following a 5-day in vitro stimulation with the H16A peptide (HITGHRMAWDMMMNWA; the underlined I and T correspond to nonconserved amino acids between genotypes 1a and 1b, and the A in italic was added for synthesis convenience) or the irrelevant peptide S9V and IL-2. Results are given as number of spots (corresponding to IFN-γ-producing T cells) per 106 splenocytes.
IFN-γ- and IL-4-producing cells were quantified by Elispot after 5 days of in vitro stimulation in the presence of each peptide at 10 μM (neither IFN-γ nor IL-4 was ever detected in ex vivo experiments). As shown in Fig. 6, IFN-γ-producing T cells were detected for all E1 mutants, albeit to different levels, whereas IL-4-producing cells were never detectable (data not shown). It is interesting that the observed IFN-γ-producing peptide-specific recall response observed was IL-2 dependent, as this cytokine was indeed included in the culture medium used. Such detection was highly specific for the H16A peptide, as basically no or a very few IFN-γ-producing T cells could be detected in the presence of G10A (data not shown) or the irrelevant peptide S9V. Characterization of the phenotype of the IFN-γ-producing T cells was performed after in vitro depletion of either CD4+ or CD8+ cells. Depletion of CD8+ cells significantly reduced the number of IFN-γ-producing T cells, whereas it was maintained after CD4+ cell depletion, indicating that IFN-γ-producing T cells presented mainly a CD8 phenotype (data not shown).
DISCUSSION
It has often been thought that oligosaccharides on envelope proteins may form a barrier to help shield viruses from immune recognition, thus limiting effective antibody responses or affecting the proteolytic degradation of these proteins and reducing T-cell recognition by the host (16, 42). However, in some cases, elimination of glycans can alter the antigenic conformation without uncovering hidden neutralization epitopes and broadening the immune response (9, 46). We have now shown that mutation of one specific N-linked glycosylation site of the HCV envelope glycoprotein E1, located at position 305 on the HCV polyprotein (N4), importantly increases the antibody response to this protein in terms of seroconversion rate and antibody titers, particularly those measured against HCV-LPs. In addition, this mutant retains the capacity to induce IFN-γ-secreting T cells.
We performed extensive in vitro characterization of the different mutants generated as far as the expression level, folding, solubility, and secretion of the proteins were concerned. Such characterization was important to better appreciate the in vivo results that were generated after vaccination of mice. We showed that different mutations could affect, albeit to various levels, the recognition of the mutated E1 antigens by specific anti-E1 antibodies. One mutant, the quadruple mutant N1-2-3-4, remained difficult to detect except in cell lysates with the antibody IGH201. This may indicate that its conformation or solubility was considerably altered compared to the wild-type E1 or the other mutants.
For all constructs generated, E1 was expressed with its signal peptide, which guides it through the ER. It has been shown that the C-terminal transmembrane domain of the protein acts as a signal for retention in this compartment, and poor secretion has only been observed for C-terminally truncated forms ending at aa 311 (1a strain) (34) or, alternatively, for a 1b strain, at aa 340 when a deletion is introduced between aa 262 and 290 (32). We found here that a longer form of E1, truncated at aa 329, was secreted but that its oligosaccharide chains play an important role in this secretion. Mutation at one position could decrease the level of E1 present in the cell supernatant, particularly for positions N3 and N4. The amount of secreted E1 was apparently even more reduced when two or more glycosylation sites were mutated. Although mutations did not seem to affect the stability of the mutants in the intracellular compartment, we could not exclude the possibility that proteins were unstable in the supernatants of cell cultures. Overall, our results indicated that all mutants were well expressed in the cells and that their secretion (or stability after secretion) was in all cases affected compared with the wild-type E1. The level of this alteration appears to be quite similar for all single mutants.
The mechanisms involved in the induction of immune responses following DNA vaccination are as yet unclear. One point of extensive investigation has been to determine the impact of the level of antigen secretion on the induction of specific antibodies. Some reports have shown that secreted DNA-expressed antigens induce optimum immune responses, whereas in other reports, such as for the influenza virus H1 protein, secretion of the antigen reduced the induction of antibodies (26, 49). Our results suggest that the enhanced immunogenicity of the E1 N4 mutant was not related to an increased secretion of the antigen, at least if it is assumed that the observations made from our in vitro analysis reflect those that may take place following in vivo vaccination with DNA. Such observations would rather argue in favor of the introduction, following mutation of the N4 site, of a conformational change that revealed or enhanced the exposure of an important determinant.
In comparison with the wild-type E1 protein, no particular alteration of the folding of the N4 antigen was revealed by the different in vitro analyses that we performed. However, it was reported previously that mutation of the fourth glycosylation site dramatically reduced the efficiency of the formation of noncovalent E1-E2 complexes, suggesting that the presence of glycans at site 4 is important for stabilizing the E1 structure or favoring its proper folding, at least in the presence of E2 (33). Thus, when E1 folds without glycans on site 4, sufficient conformational changes are likely to occur, revealing some B-cell epitopes. We tested mouse sera (Table 1) for reactivity to a 23-mer peptide that spans glycosylation site 4, but antibodies induced in wild-type E1- as well as N4-immunized mice showed little or no reactivity to this peptide (data not shown). These data suggested that rather than revealing a B-cell epitope shielded by glycans on the N4 site, elimination of this glycosylation site resulted in an altered local antigenic conformation uncovering an epitope(s) broadening the immune response.
We tried to investigate more extensively the qualitative specificity of the antibodies induced by the N4 mutant versus that obtained with the wild-type E1 protein. First, we performed epitope mapping with different peptides spanning the E1 sequence. For the N4-induced antibody, we found essentially a reactivity at one N-terminal epitope, V1C1V2, while that against the C-terminal epitope, C4, was reduced compared with that of wild-type E1-induced sera. It has been shown that the C4 region contains a genotype-cross-reactive and highly immunogenic epitope and that the prevalence of antibodies reacting to this epitope is high in sera from chronically infected hepatitis patients (14). In contrast, chronically infected chimpanzees vaccinated with the E1 protein develop a humoral response targeted principally against the N-terminal region of the protein (V1C1V2 domain) (E. Depla, personal communication). The capacity of the N4-E1 mutant to preferentially induce antibody directed at this determinant should make this antigen a good component in a therapeutic vaccine formulation.
It was previously reported that the IgG antibody response to E1 (and other HCV antigens except the core [12]) is highly restricted to the IgG1 isotype during HCV infection in patients, whereas E1-vaccinated chimpanzees developed a mixture of IgG1, IgG2, and IgG4 antibodies (31). Here, a mix of IgG1 and IgG2a antibody was detected following immunization with N4 as well as with the wild-type E1 antigen. In mice, antibodies of the IgG1 subclass are linked to a TH2-mediated type of response. Such response could be induced by the secreted part of the antigen, expected to be taken up by antigen-presenting cells and processed as an exogenous antigen for antibody production. The additional IgG2a component could be due to the transport of the antigen from the ER to the cytosol (2), which has been described previously for E1 (44).
As there is no productive cell culture system available to test the neutralization capacity of antibodies induced against HCV envelope proteins, we tested the ability of N4-induced antibodies to react with HCV-LPs. In contrast to previously described serologic assays, the HCV envelope proteins of HCV-LPs are presumably presented in a native, virion-like conformation and represent today an alternative system with which to study the capacity of antibodies to be directed at either linear or conformational epitopes that may represent neutralizing epitopes. Interestingly, wild-type E1-induced antibodies were able to recognize HCV-LPs, but the N4 mutant significantly enhanced this response in terms of both seroconversion rates and antibody titers. One possible explanation for this observation is that the oligosaccharide chain at the N4 position limits but does not prevent antibody responses to a particular epitope(s) present in the HCV-LPs. When glycans are absent, antibody to this epitope(s) may be induced at higher titers. Induction of anti-viruslike particle antibodies by a vaccine is an interesting property, as it has been suggested that viral clearance during IFN-α therapy is associated with existing high-titer anti-HCV-LP antibodies (6).
Accumulating data obtained from the study of chronically HCV-infected patients and self-limited carriers show quite clearly that viral clearance is associated with a strong CD4+ helper T-cell response (15, 23, 36), although CD8+ CTL-mediated responses might play a role in HCV eradication (24, 29, 38, 48). Analysis of the cytokine profile from bulk cultures as well as CD4+ T-cell clones from patients with acute hepatitis reveals that viral clearance is correlated with a T-helper type I profile (IL-2 and IFN-γ production) (51). In the present study, we analyzed the cellular immune responses induced by the different E1 glycosylation mutants using an Elispot assay. We detected for all plasmids specific IFN-γ-producing T cells which presented mainly a CD8+ phenotype and were induced via a CD4+-dependent pathway. We did not observe any enhancement of the capacity of the different E1 mutants to specifically induce the production of IFN-γ. Thus, while E1 mutants displayed various capacities to induce specific antibodies, they appear to overall conserve the ability to induce IFN-γ-producing cells. Determination of whether the mutation of N-glycosylation affects the proteolytic degradation of E1 will need further studies.
Results described here should allow optimal design of E1-based vaccines. Our data suggest that N-oligosaccharides on the HCV E1 envelope protein, in a natural situation, could limit the humoral immune response developed by the host and help shield the virus from immune recognition. Although the E1 glycosylation mutants that we studied here were out of the context of a full-length HCV polyprotein (34), it is tempting to speculate that such shielding could contribute to the ineffectiveness of the natural immune response to HCV.
ACKNOWLEDGMENTS
We are grateful to the Aventis Pasteur corporation for the use of the spot counter.
This work was supported by grants from bioMérieux, the ARC (Association pour la Recherche sur le Cancer), and the European Community (QLK2-CT-1999-00356).
REFERENCES
- 1.Alter M J. Epidemiology of hepatitis C. Eur J Gastroenterol Hepatol. 1996;8:319–323. doi: 10.1097/00042737-199604000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Bacik I, Snyder H L, Anton L C, Russ G, Chen W, Bennink J R, Urge L, Otvos L, Dudkowska B, Eisenlohr L, Yewdell J W. Introduction of a glycosylation site into a secreted protein provides evidence for an alternative antigen processing pathway: transport of precursors of major histocompatibility complex class I-restricted peptides from the endoplasmic reticulum to the cytosol. J Exp Med. 1997;186:479–487. doi: 10.1084/jem.186.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Back N K, Smit L, De Jong J J, Keulen W, Schutten M, Goudsmit J, Tersmette M. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199:431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- 4.Baumert T F, Ito S, Wong D T, Liang T J. Hepatitis C virus structural proteins assemble into virus-like particles in insect cells. J Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumert T F, Vergalla J, Satoi J, Thomson M, Lechmann M, Herion D, Greenberg H B, Ito S, Liang T J. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology. 1999;117:1397–1407. doi: 10.1016/s0016-5085(99)70290-8. [DOI] [PubMed] [Google Scholar]
- 6.Baumert T F, Wellnitz S, Aono S, Satoi J, Herion D, Tilman Gerlach J, Pape G R, Lau J Y, Hoofnagle J H, Blum H E, Liang T J. Antibodies against hepatitis C virus-like particles and viral clearance in acute and chronic hepatitis C. Hepatology. 2000;32:610–617. doi: 10.1053/jhep.2000.9876. [DOI] [PubMed] [Google Scholar]
- 7.Benjouad A, Gluckman J C, Montagnier L, Bahraoui E. Specificity of antibodies produced against native or desialylated human immunodeficiency virus type 1 recombinant gp160. J Virol. 1993;67:1693–1697. doi: 10.1128/jvi.67.3.1693-1697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolmstedt A, Hemming A, Flodby P, Berntsson P, Travis B, Lin J P, Ledbetter J, Tsu T, Wigzell H, Hu S L, et al. Effects of mutations in glycosylation sites and disulfide bonds on processing, CD4-binding and fusion activity of human immunodeficiency virus envelope glycoproteins. J Gen Virol. 1991;72:1269–1277. doi: 10.1099/0022-1317-72-6-1269. [DOI] [PubMed] [Google Scholar]
- 9.Bolmstedt A, Sjolander S, Hansen J E, Akerblom L, Hemming A, Hu S L, Morein B, Olofsson S. Influence of N-linked glycans in V4–V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12:213–220. doi: 10.1097/00042560-199607000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Bruna-Romero O, Lasarte J J, Wilkinson G, Grace K, Clarke B, Borras-Cuesta F, Prieto J. Induction of cytotoxic T-cell response against hepatitis C virus structural antigens using a defective recombinant adenovirus. Hepatology. 1997;25:470–477. doi: 10.1002/hep.510250236. [DOI] [PubMed] [Google Scholar]
- 11.Cerny A, Chisari F V. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology. 1999;30:595–601. doi: 10.1002/hep.510300312. [DOI] [PubMed] [Google Scholar]
- 12.Chen M, Sallberg M, Sonnerborg A, Weiland O, Mattsson L, Jin L, Birkett A, Peterson D, Milich D R. Limited humoral immunity in hepatitis C virus infection. Gastroenterology. 1999;116:135–143. doi: 10.1016/s0016-5085(99)70237-4. [DOI] [PubMed] [Google Scholar]
- 13.Cooper S, Erickson A L, Adams E J, Kansopon J, Weiner A J, Chien D Y, Houghton M, Parham P, Walker C M. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 14.Depraetere S, Leroux-Roels G. Hepatitis C virus envelope proteins: immunogenicity in humans and their role in diagnosis and vaccine development. Viral Hepatitis. 1999;5:113–146. [Google Scholar]
- 15.Diepolder H M, Gerlach J-T, Zachoval R, Hoffmann R M, Jung M-C, Wierengaz E A, Scholz S, Santantonio T, Houghton M, Southwood S, Sette A, Pape G R. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doe B, Steimer K S, Walker C M. Induction of HIV-1 envelope (gp120)-specific cytotoxic T lymphocyte responses in mice by recombinant CHO cell-derived gp120 is enhanced by enzymatic removal of N-linked glycans. Eur J Immunol. 1994;24:2369–2376. doi: 10.1002/eji.1830241017. [DOI] [PubMed] [Google Scholar]
- 17.Dubuisson J, Duvet S, Meunier J C, Op De Beeck A, Cacan R, Wychowski C, Cocquerel L. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J Biol Chem. 2000;275:30605–30609. doi: 10.1074/jbc.M004326200. [DOI] [PubMed] [Google Scholar]
- 18.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckels D D, Wang H, Bian T H, Tabatabai N, Gill J C. Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol Rev. 2000;174:90–97. doi: 10.1034/j.1600-0528.2002.017403.x. [DOI] [PubMed] [Google Scholar]
- 20.Erickson A L, Houghton M, Choo Q L, Weiner A J, Ralston R, Muchmore E, Walker C M. Hepatitis C virus-specific CTL responses in the liver of chimpanzees with acute and chronic hepatitis C. J Immunol. 1993;151:4189–4199. [PubMed] [Google Scholar]
- 21.Fournillier-Jacob A, Cahour A, Escriou N, Girard M, Wychowski C. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J Gen Virol. 1996;77:1055–1064. doi: 10.1099/0022-1317-77-5-1055. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlach J T, Diepolder H M, Jung M C, Gruener N H, Schraut W W, Zachoval R, Hoffmann R, Schirren C A, Santantonio T, Pape G R. Recurrence of hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology. 1999;117:933–941. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 24.Hiroishi K, Kita H, Kojima M, Okamoto H, Moriyama T, Kaneko T, Ishikawa T, Ohnishi S, Aikawa T, Tanaka N, Yazaki Y, Mitamura K, Imawari M. Cytotoxic T lymphocyte response and viral load in hepatitis C virus infection. Hepatology. 1997;25:705–712. doi: 10.1002/hep.510250336. [DOI] [PubMed] [Google Scholar]
- 25.Houghton M. Strategies and prospects for vaccination against the hepatitis C viruses. Curr Top Microbiol Immunol. 2000;242:327–339. doi: 10.1007/978-3-642-59605-6_15. [DOI] [PubMed] [Google Scholar]
- 26.Inchauspé G, Vitvitski L, Major M E, Jung G, Spengler U, Maisonnas M, Trépo C. Plasmid DNA expressing a secreted or a nonsecreted form of hepatitis C virus nucleocapsid: comparative studies of antibody and T-helper responses following genetic immunization. DNA Cell Biol. 1997;16:185–195. doi: 10.1089/dna.1997.16.185. [DOI] [PubMed] [Google Scholar]
- 27.Kieny M P, Lathe R, Drillien R, Spehner D, Skory S, Schmitt D, Wiktor T, Koprowski H, Lecocq J P. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature. 1984;312:163–166. doi: 10.1038/312163a0. [DOI] [PubMed] [Google Scholar]
- 28.Lechner F, Gruener N H, Urbani S, Uggeri J, Santantonio T, Kammer A R, Cerny A, Phillips R, Ferrari C, Pape G R, Klenerman P. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol. 2000;30:2479–2487. doi: 10.1002/1521-4141(200009)30:9<2479::AID-IMMU2479>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Lechner F, Wong D K, Dunbar P R, Chapman R, Chung R T, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker B D. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Diaz de Cerio A L, Casares N, Lasarte J J, Sarobe P, Perez-Mediavilla L A, Ruiz M, Prieto J, Borras-Cuesta F. Th1 but not Th0 cell help is efficient to induce cytotoxic T lymphocytes by immunization with short synthetic peptides. Int Immunol. 1999;11:2025–2033. doi: 10.1093/intimm/11.12.2025. [DOI] [PubMed] [Google Scholar]
- 31.Maertens G, Ducatteeuw A, Priem S, van Eerd P, Roskams T, Van Hoek K, Rockel I, Vandeponseele P, Bosman F, Venneman A, Buyse M-A, Stoops E, Fuller S, van Doorn L-J, Kos A, Depla E. Therapeutic vaccination of chronically infected chimpanzees with the HCV E1 protein, p. 74. 6th International Symposium on Hepatitis C and Related Viruses. Bethesda, Md: National Institutes of Health; 1999. [Google Scholar]
- 32.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 33.Meunier J C, Fournillier A, Choukhi A, Cahour A, Cocquerel L, Dubuisson J, Wychowski C. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. J Gen Virol. 1999;80:887–896. doi: 10.1099/0022-1317-80-4-887. [DOI] [PubMed] [Google Scholar]
- 34.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 35.Mink M A, Benichou S, Madaule P, Tiollais P, Prince A M, Inchauspe G. Characterisation and mapping of a B-cell immunogenic domain in hepatitis C virus E2 glycoprotein using a yeast library. Virology. 1994;200:246–255. doi: 10.1006/viro.1994.1182. [DOI] [PubMed] [Google Scholar]
- 36.Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, Rumi M G, Houghton M, Fiaccadori F, Ferrari C. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the antiviral cell mediated immune responses. J Clin Investig. 1996;98:706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 38.Nelson D R, Marousis C G, Ohno T, Davis G L, Lau J Y N. Intrahepatic hepatitis C virus-specific cytotoxic T lymphocyte activity and response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1998;28:225–230. doi: 10.1002/hep.510280129. [DOI] [PubMed] [Google Scholar]
- 39.Opdenakker G, Rudd P-M, Ponting C P, Dwek R A. Concepts and principles of glycobiology. FASEB J. 1993;7:1330–1337. doi: 10.1096/fasebj.7.14.8224606. [DOI] [PubMed] [Google Scholar]
- 40.Prince A M, Brotman B, Huima T, Pascual D, Jaffery M, Inchauspe G. Immunity in hepatitis-C infection. J Infect Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- 41.Rademacher T G, Parekh R B, Dwek R A. Glycobiology. Annu Rev Biochem. 1988;57:786–830. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- 42.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 43.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L C, Dong M C, Weiner A J, Lau J Y N, Choo Q-L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selby M J, Glazer E, Masiartz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu Y K, Igarashi H, Kiyohara T, Cbezon T, Farci P, Purcell R H, Yoshikura H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 46.Sjölander S, Bolmstedt A, Akerblom L, Horal P, Olofsson S, Morein B, Sjölander A. N-linked glycans in the CD4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of T cells recognizing an epitope located in their vicinity. Virology. 1996;215:124–133. doi: 10.1006/viro.1996.0015. [DOI] [PubMed] [Google Scholar]
- 47.Sodora D L, Cohen G H, Muggeridge M I, Eisenberg R J. Absence of asparagine-linked oligosaccharides from glycoprotein D of herpes simplex virus type 1 results in a structurally altered but biologically active protein. J Virol. 1991;65:4424–4431. doi: 10.1128/jvi.65.8.4424-4431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takaki A, Wiese M, Maertens G, Depla E, Seifert U, Liebetrau A, Miller J L, Manns M P, Rehermann B. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med. 2000;6:578–582. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 49.Torres C A T, Yang K, Mustafa F, Robinson H L. DNA immunization: effect of secretion of DNA-expressed hemagglutinins on antibody responses. Vaccine. 2000;18:805–814. doi: 10.1016/s0264-410x(99)00345-x. [DOI] [PubMed] [Google Scholar]
- 50.Trombetta E S, Helenius A. Lectins as chaperones in glycoprotein folding. Curr Opin Struct Biol. 1998;8:587–592. doi: 10.1016/s0959-440x(98)80148-6. [DOI] [PubMed] [Google Scholar]
- 51.Tsai S L, Liaw Y F, Chen M H, Huang C Y, Kuo G C. Detection of type 2-like T-helper cells in hepatitis C virus infection: implications for hepatitis C virus chronicity. Hepatology. 1997;25:449–458. doi: 10.1002/hep.510250233. [DOI] [PubMed] [Google Scholar]
- 52.Wiley D C, Wilson I A, Skehel J J. In: Biological macromolecules and assemblies. McPherson A, editor. Vol. 1. New York, N.Y: John Wiley and Sons; 1984. pp. 299–336. [Google Scholar]
- 53.Zibert A, Kraas W, Meisel H, Jung G, Roggendorf M. Epitope mapping of antibodies directed against hypervariable region 1 in acute self-limiting and chronic infections due to hepatitis C virus. J Virol. 1997;71:4123–4127. doi: 10.1128/jvi.71.5.4123-4127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]