Abstract
Fewer than half of individuals with a suspected Mendelian or monogenic condition receive a precise molecular diagnosis after comprehensive clinical genetic testing. Improvements in data quality and costs have heightened interest in using long-read sequencing (LRS) to streamline clinical genomic testing, but the absence of control data sets for variant filtering and prioritization has made tertiary analysis of LRS data challenging. To address this, the 1000 Genomes Project (1KGP) Oxford Nanopore Technologies Sequencing Consortium aims to generate LRS data from at least 800 of the 1KGP samples. Our goal is to use LRS to identify a broader spectrum of variation so we may improve our understanding of normal patterns of human variation. Here, we present data from analysis of the first 100 samples, representing all 5 superpopulations and 19 subpopulations. These samples, sequenced to an average depth of coverage of 37× and sequence read N50 of 54 kbp, have high concordance with previous studies for identifying single nucleotide and indel variants outside of homopolymer regions. Using multiple structural variant (SV) callers, we identify an average of 24,543 high-confidence SVs per genome, including shared and private SVs likely to disrupt gene function as well as pathogenic expansions within disease-associated repeats that were not detected using short reads. Evaluation of methylation signatures revealed expected patterns at known imprinted loci, samples with skewed X-inactivation patterns, and novel differentially methylated regions. All raw sequencing data, processed data, and summary statistics are publicly available, providing a valuable resource for the clinical genetics community to discover pathogenic SVs.
As an initiative to sequence a large set of healthy reference genomes from globally diverse ancestries, the 1000 Genomes Project (1KGP) marked a significant milestone in genomic research, yielding the first sequencing-based map of normal patterns of human genetic variation for filtering and prioritizing candidate disease-causing variants (International HapMap Consortium 2005; The 1000 Genomes Project Consortium 2015; Byrska-Bishop et al. 2022). The impact of 1KGP on our understanding of human genetic diversity has been enormous, and the flagship papers have been cited more than 10,000 times in clinical and basic research studies. The success of the project has been amplified by the use of diverse, high-quality, open-access data sets, and databases such as gnomAD (Koenig et al. 2024) and DECIPHER (Firth et al. 2009) have built on the 1KGP principles for determining the population allele frequency of variants to aid in variant interpretation. Pooling of data from large projects has improved the usefulness of these databases, and analyses of 1KGP data to date have made profound contributions using arrays or short-read sequencing technology. However, these approaches are inherently limited in their ability to identify variants in complex genomic regions or to capture certain types of genetic differences, such as structural variants (SVs), repeat expansions, and epigenetic changes (Chaisson et al. 2019; Ebert et al. 2021; Liao et al. 2023).
SVs—defined as insertions, deletions, duplications, inversions, repeat expansions, and translocations at least 50 bp in size—are major contributors to genetic diversity and disease susceptibility and are more likely to have a larger effect size than single nucleotide variants (SNVs) (Eichler 2019). SV calling using short-read sequencing can be challenging because it detects fewer than half of the ∼25,000 SVs present in an individual, is incapable of fully resolving the complex structure of many SVs, and has low concordance between callers (Cameron et al. 2019; Chaisson et al. 2019; Zhao et al. 2021). These challenges extend into clinical testing where commonly used approaches, such as exome sequencing, have low sensitivity for SV detection, meaning individuals with disease-causing SVs may remain undiagnosed (Hiatt et al. 2021; Miller et al. 2021; Cohen et al. 2022; AlAbdi et al. 2023). Therefore, there is broad interest in using long-read sequencing (LRS) to develop comprehensive catalogs of common human SVs to facilitate improved detection of disease-associated variants (Wojcik et al. 2023).
LRS has increasingly demonstrated its ability to detect and resolve SVs missed by traditional methods. Previous concerns about cost, error rates, sample preparation, and computational tools for both commercially available LRS technologies (Pacific Biosciences, PacBio and Oxford Nanopore Technologies, ONT) have largely been resolved (Logsdon et al. 2020; Wang et al. 2021; Kolmogorov et al. 2023), paving the way for its adoption into clinical settings (Wojcik et al. 2023; Damaraju et al. 2024).
Building on the landmark effort of the 1KGP, the 1000 Genomes Project ONT Sequencing Consortium (1KGP-ONT) is leveraging ONT LRS with the goal of generating high-coverage, high-quality sequencing data from the 1KGP sample set. This international initiative aims to: (1) assess both assembly-based and alignment-based approaches to LRS data analysis; (2) evaluate variants in difficult-to-analyze regions of the genome; and (3) facilitate the identification of SVs not fully characterized by short-read approaches. This effort is complementary to work from other groups performing PacBio LRS of 1KGP samples, such as the Human Pangenome Reference Consortium (HPRC) (Wang et al. 2022) and the Human Genome Structural Variant Consortium (HGSVC) (Ebert et al. 2021), as well as lower coverage and N50 ONT sequencing from Schloissnig et al. (2024). With these collective endeavors, it is increasingly likely that the entire collection will ultimately be sequenced using both LRS platforms. Following 1KGP principles, all data generated through the 1KGP-ONT Consortium are publicly released for immediate incorporation into clinical and basic research projects.
Here, we present our analysis of the first 100 samples sequenced by the 1KGP-ONT Consortium. Because a major goal of the consortium is to develop a catalog of common human SVs for filtering and prioritizing disease-associated SVs, we demonstrate how SV data from a modest number of individuals can be used to filter variants in unsolved cases and identify high-priority regions for follow-up analysis. We also describe variation that would be difficult or impossible to detect or fully resolve using short-read technology, including disease-associated repeat expansions, skewed X-Chromosome inactivation in 46,XX samples, and differentially methylated regions (DMRs) unique to individual samples.
Results
Approximately 3200 cell lines or DNA samples from the 1KGP are available at the National Human Genome Research Institute (NHGRI) Sample Repository for Human Genetic Research housed at the Coriell Institute for Medical Research repositories (Coriell) (International HapMap Consortium 2005; The 1000 Genomes Project Consortium 2015). These anonymized samples, which are not associated with medical or phenotypic data, are from individuals who self-reported ancestry, sex, and good health at the time of sample collection. We selected 100 samples from all five superpopulations based on their absence from other large-scale sequencing efforts (Ebert et al. 2021; Liao et al. 2023; Schloissnig et al. 2024); we did not attempt to balance subpopulations within these samples, and four of the 100 samples represent two parent–child pairs (Fig. 1A; Supplemental Table S1).
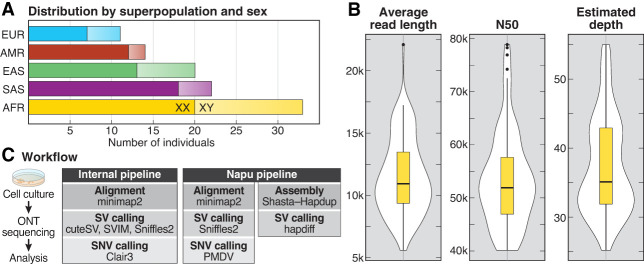
Figure 1.
Summary statistics of samples, sequencing, and small variant detection. (A) Samples selected for sequencing are shown by superpopulation and sex. (B) Violin plots showing average read length, read N50, and average depth of coverage for all 100 samples. (C) DNA was extracted from cells grown from aliquots received from Coriell and sequenced using the R9.4.1 pore. Data were analyzed using both alignment- and assembly-based approaches.
Sequencing pipeline
High molecular weight (HMW) DNA was isolated from lymphoblastoid cell lines (LCLs) cultured in the laboratory, and samples were sequenced using the ONT R9.4.1 pore with an average depth of coverage of 37.4× and read N50 of 53.8 kbp (Fig. 1B; Supplemental Table S2). All samples were processed using two separate pipelines (Fig. 1C). First, an internal alignment pipeline used minimap2 for alignment, Clair3 for small variant calling, and Sniffles2, cuteSV, and SVIM for SV calling (Li 2018; Heller and Vingron 2019; Jiang et al. 2020; Zheng et al. 2022; Smolka et al. 2024). SNV calls from this pipeline were used to ensure sample identity by comparison with previous short-read-based variant calls (Byrska-Bishop et al. 2022). Second, samples were processed using the Nanopore Analysis Pipeline (Napu), which generates assembly-based SV calls using hapdiff after generating a phased de novo assembly using Shasta–Hapdup, minimap2 alignment-based small variant calls using PEPPER-Margin-DeepVariant (PMDV), and minimap2 alignment-based SV calls using Sniffles2 (Shafin et al. 2021; Kolmogorov et al. 2023; Smolka et al. 2024).
Small variant accuracy
We evaluated the performance of our variant-calling pipelines by comparing small variant calls (SNVs and indels <50 bp) to those generated by prior studies and using orthogonal short- and long-read sequencing technologies. We first compared the ONT sequencing of five samples (outside of our 1KGP cohort) to Genome in a Bottle (GIAB) benchmarking data and the HiFi PacBio data from the Human Pangenome Research Project. Restricting analysis to the GIAB high-confidence regions for HG002 resulted in F1 scores >0.984 for SNVs and >0.699 for indels for both data sets (Supplemental Table S3). However, these values were highly influenced by the presence of homopolymers in the ONT data (Harvey et al. 2023; Kolmogorov et al. 2023). When homopolymers were removed from the analysis, F1 scores increased to >0.984 for SNVs and >0.874 for indels (Supplemental Fig. S1; Supplemental Table S4). Next, we compared ONT data from our 1KGP cohort to complementary Illumina data. We observed an average F1 score of 0.982 for SNVs and 0.878 for indels outside of homopolymers. (Supplemental Fig. S2; Supplemental Table S5). These results validated that both variant-calling approaches (Clair3 and PMDV) produced high-quality small variant calls concordant with prior studies (Kolmogorov et al. 2023).
Genome assembly
We performed de novo genome assemblies for each of the 100 samples using both the Napu pipeline (which runs Shasta–Hapdup) and Flye (Shafin et al. 2020; Kolmogorov et al. 2023). In general, we found that Flye assemblies had a higher contig NG50 than Shasta–Hapdup assemblies (Fig. 2A), and results were robust to read N50 differences (Fig. 2B). We saw similar contig NG50 patterns when our analysis included the five benchmarking genomes with similar average depth of coverage and read N50. The assembled genomes were highly complete, with each assembly covering ∼93.5% (Flye) or 93.6% (Shasta–Hapdup) of the GRCh38 reference genome (Supplemental Fig. S3) with a consensus accuracy similar to previously published studies using the R9 pore (Fig. 2C; Kolmogorov et al. 2023).
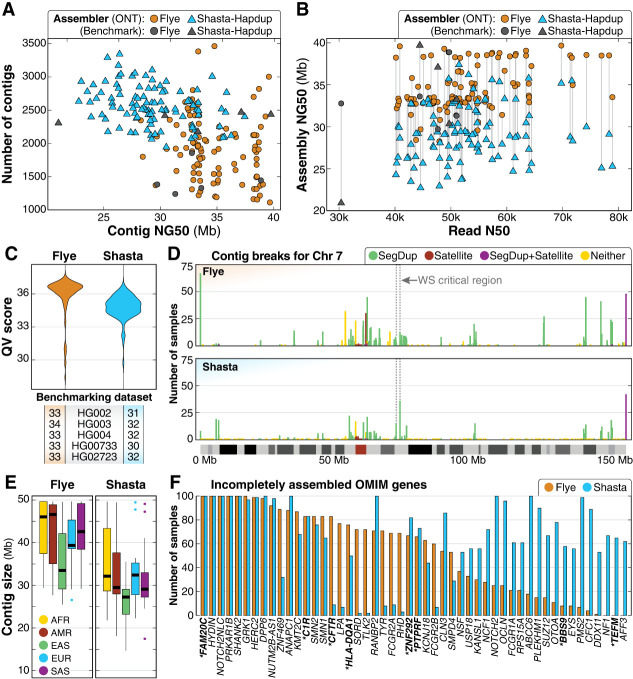
Figure 2.
Summary of de novo assembly results. (A) Contig NG50 compared to the total number of contigs shows that haploid assemblies generated by Flye are longer and have fewer contigs than Shasta–Hapdup. No contig NG50 generated by Flye exceeds 40 Mbp. Assemblies for each benchmarking sample show similar statistics. (B) Assembly NG50 does not significantly improve with higher read N50. (C) QV scores for both Flye and Shasta–Hapdup assemblies, and the five benchmarking genomes. (D) Count of contig breaks for all 100 samples on Chromosome 7 shows that while assembly breaks cluster there are a large number of single breaks spread across the chromosome. The 1.5–1.8 Mbp Williams–Beuren syndrome critical region is indicated with a dashed box and is flanked by clusters of assembly breaks within segdups (Morris 1993). (E) Contig sizes filtered for contigs longer than 1 Mbp for each superpopulation. (F) OMIM genes incompletely assembled in 50 or more samples using Flye or Shasta–Hapdup. For Shasta–Hapdup, if one haplotype was completely assembled in a sample but the other was incomplete, the gene is counted as incompletely assembled. Assembly of five genes (FAM20C, HYDIN, NOTCH2NLC, PRKAR1B, and SHANK2) was incomplete for all 100 samples using both assemblers. Genes that are not in or do not contain a segdup are in bold with an asterisk.
We investigated why many of the Flye assemblies had similar contig NG50 values by plotting the contig breakpoints for both the Shasta–Hapdup and Flye assemblies. Among the 100 Flye assemblies, 97.1% of assembly breaks occurred within regions annotated as segmental duplications (segdups), satellite sequences, or both, while 2.9% occurred within nonrepetitive sequence (Supplemental Table S6). Among the 2.9% of assembly breaks in nonrepetitive sequence, 90% were seen in only one sample, suggesting stochastic artifacts of the assembly process. A focused analysis of Chromosome 7 revealed an increased number of contig breaks in the telomeric and pericentromeric regions for both Flye and Shasta–Hapdup assemblies (Fig. 2D) and at positions flanking well-described recurrent copy number changes associated with disease (Morris 1993). Visual analysis of breaks in nonrepetitive sequence did not reveal sample-specific differences that would easily explain the break in assembly, such as a duplication, inversion, or increased number of SNVs, suggesting that local sequence variation did not influence the position of assembly breaks in nonrepetitive regions (Supplemental Fig. S4). A list of assembly breaks in 20 or more samples from either the Flye or Shasta–Hapdup assemblies genome-wide is available (Supplemental File S1).
We then evaluated contig size across superpopulation groups and the assembly of disease-associated OMIM genes. The median contig size per sample excluding contigs <1 Mbp (Fig. 2E) was higher for African ancestry samples. This was expected given the higher genetic diversity in individuals of African ancestry, which results in a higher number of distinct sequences leading to longer and more contiguous sequences in the assembly. Next, we examined how well disease-associated genes were assembled in these samples. Among 4615 disease-associated OMIM genes (excluding genes on the X and Y Chromosomes), we found that 97% (4492/4615) and 97% (4475/4615) of genes in the Flye or Shasta–Hapdup assemblies, respectively, were completely and correctly assembled (i.e., they were spanned by a single, complete contig) in at least 95 out of 100 samples (Supplemental File S2). Among the 200 assemblies (100 Flye and 100 Shasta–Hapdup), we found that five OMIM genes were incompletely assembled in all 200 assemblies and another 45 OMIM genes were incompletely assembled in at least 50 or more of the 200 assemblies (Fig. 2F). We observed more incompletely assembled genes in the Shasta–Hapdup assemblies, partly due to the requirement for a single gene to be entirely spanned by a single contig in both haplotypes for it to be considered fully assembled.
We subsequently applied PanGenome Graph Building (PGGB) to construct chromosome-level pangenome graphs from the 100 Shasta–Hapdup assemblies and generate multisample variant calls including all types of variants (Garrison et al. 2023). To investigate the differences between assembly approaches, we performed principal component analysis (PCA) on a Chromosome 20 pangenome graph created by combining the 100 Shasta–Hapdup assemblies with 44 assemblies from the HPRC (Liao et al. 2023). The PCA showed a clear separation between the two pangenomes (Supplemental Fig. S5A). However, a PCA based on the euchromatic, noncentromeric fraction of the Chromosome 20 graph demonstrates that this difference is primarily due to the improved resolution of highly repetitive sequences by the HiFi-based HPRC assemblies (Supplemental Fig. S5B), supporting the high-quality nature of our assemblies.
Variation within active transposable elements
The largely repetitive and polymorphic nature of active transposable elements, especially full-length long interspersed element 1 (LINE-1) and endogenous retroviruses (ERVs), makes them challenging to fully resolve and characterize using short-read assemblies (Yang et al. 2024). We anticipated that long-read assemblies would allow us to overcome these challenges. Using RepeatMasker (https://www.repeatmasker.org), we identified interspersed repeats in the 100 Shasta–Hapdup assemblies and found that the fraction of major interspersed repeats differs by no more than 3% compared to that of the T2T-CHM13 assembly (Supplemental Table S7; Nurk et al. 2022). Furthermore, there was minimal variation among the 100 assemblies in interspersed repeat content.
Among the youngest polymorphic interspersed repeats that are too long to resolve with short reads (Chaisson et al. 2019), LINE-1s (∼6000 bp) are the only types that are actively expanding in the human genome. We found that the total base pairs of LINE-1 sequence (including young and old LINE-1s) in the 100 assemblies (496 Mbp average) is lower than observed in the CHM13 T2T assembly (512 Mbp), likely due to LINE-1s within unassembled regions. To measure the ability of these ONT-based assemblies to resolve young LINE-1s, we calculated the number of the youngest LINE-1 elements (L1HS) and the number of full-length (≥6 kbp) L1HS elements. Overall, we found similar numbers of L1HS and full-length L1HS sequences compared to HG002 and HG005 from GIAB and the CHM13 T2T assembly (Supplemental Fig. S6). Although HERV-Ks (∼9000 bp) are unlikely to be actively replicating in modern humans, like LINE-1s, they are known to be polymorphic in the human population (Subramanian et al. 2011; Li et al. 2019). Therefore, we also counted the number of full-length HERV-Ks (HERVK-int) and found that the number per genome is similar among the 100 assemblies and CHM13 T2T, HG002, and HG005. This demonstrates that these assemblies are of sufficient quality to resolve the youngest long interspersed repeats and that there is variation in the number of these insertions among different human populations.
Structural variant analysis
We called SVs using four alignment-based and one assembly-based method (see Methods) and compared them to a known set of SV calls generated by the HPRC (Liao et al. 2023). From three of the five genomes used for small variant benchmarking (HG002/NA24385, HG00733, and HG02723) we identified an average of 23,732 SVs across all five callers. This is similar to the average of 22,755 SVs among 15 human genomes assembled by Audano et al. (2019) but less than those predicted by the HPRC and HGSVC (Ebert et al. 2021; Liao et al. 2023). The greater number than Audano et al. (2019) is expected given that those were called with older PacBio chemistries (RSII CLR) and an approach, SMRT-SV, that excluded SV calls in some pericentromeric regions or regions where variant calls were considered less reliable. Benchmarking against the HPRC Sniffles2 SV calls (Liao et al. 2023) and restricting calls to regions within the GIAB HG002 SV Tier1 v0.6 benchmarking regions (GIAB Tier1 Regions) (Zook et al. 2020) revealed F1 scores >90% for both methods among all three samples (Fig. 3A). When comparing genome-wide SV calls (not restricted to the GIAB Tier1 regions), our F1 score decreased to ∼70% for all three samples, suggesting difficulty in generating concordant SV calls in low complexity or repetitive regions of the genome (Supplemental Table S8).
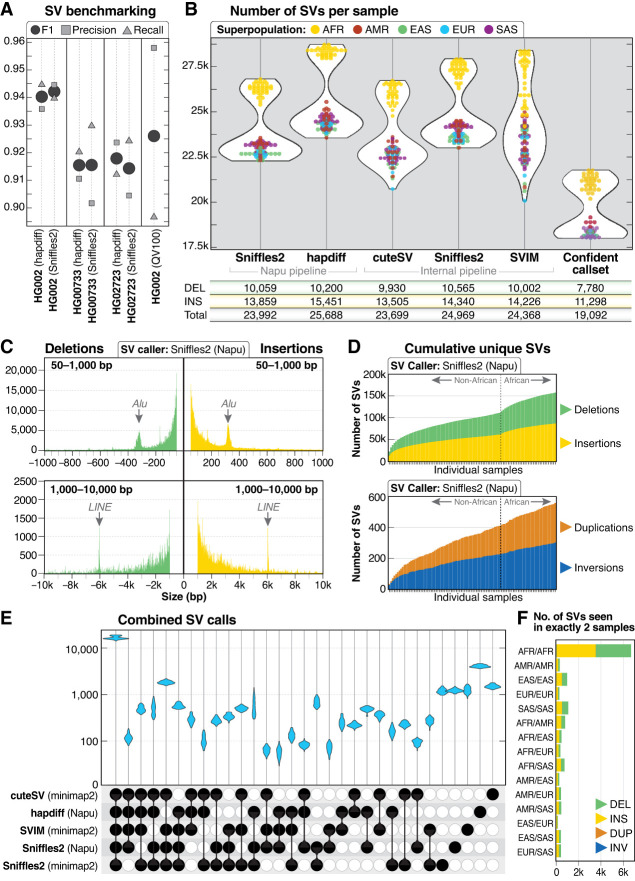
Figure 3.
SV call set. (A) SV calls were benchmarked against HPRC Sniffles2 SV calls within the GIAB HG002 SV Tier1 benchmarking regions. (B) A similar number of genome-wide SVs were identified by all five callers used in this study. The confident call set is defined as variants called by hapdiff and at least two unique alignment-based callers. For each call set, the average number of deletions (DEL), insertions (INS), and total SVs (including INV, DUP, and BND events) per sample is shown. (C) Histogram of insertion and deletion counts stratified by size. The peak ∼300 bp represents Alu insertions or deletions, and the peak ∼6 kbp represents LINE insertions or deletions. (D) Cumulative novel SVs per sample. The frequency of new SVs observed increases when samples from individuals of African ancestry are included. (E) Upset plot of overlap among SV callers after merging with Jasmine. For each sample, five VCF files were merged, demonstrating that the majority of calls in each sample were called by all five callers. (F) Among 113,696 SVs from the Jasmine-merged confident call set, 12,432 were found in exactly two samples, with 6181 (50%) of those calls in pairs in which both samples are from the African superpopulation.
We observed high per-caller concordance between the number of SV calls from the three benchmarking genomes and the 100 genomes presented here (Fig. 3B). Across the five callers, we identified an average of 24,543 SVs per sample (min: 20,068, max: 28,734), similar to the 23,000–28,000 SVs per sample reported by the HGSVC (Ebert et al. 2021). Consistent with prior work, we observed more total SV calls in samples from the African superpopulation (The 1000 Genomes Project Consortium 2015; Audano et al. 2019; Ebert et al. 2021). The distribution of insertions and deletions called in this data set was also as expected, with an Alu peak ∼300 bp and LINE peak ∼6 kbp (Fig. 3C). A generally proportional number of SVs per chromosome was observed and, on average, more insertion than deletion events were identified per chromosome for all SV callers (Supplemental Fig. S7). The genome-wide distribution of total SV events was as expected, with more insertions and deletions near the telomeres and centromeres (Supplemental Fig. S8). We identified an increasing number of novel SVs, excluding breakends (BNDs), for each additional sample sequenced among all SV callers (Fig. 3D).
Because the primary goal of our study is to identify and catalog high-quality SVs among the 1KGP samples, we merged the SVs from each of the five SV callers per sample using Jasmine (Kirsche et al. 2023). We observed high concordance between SV callers across all samples (Fig. 3E), with an average of 16,722 SVs per sample called by all callers and no individual sample having an SV type that was noticeably higher or lower than other samples within the same superpopulation (Supplemental Fig. S9A). An average of 20,242, 22,685, 25,540, and 34,796 SVs were called by at least four, three, two, or one callers, respectively (Supplemental Fig. S9B).
The SVs called exclusively by hapdiff represent the majority of SVs called by a single caller. Because hapdiff was the only assembly-based caller in our data set, we examined whether these calls represented false positives or SVs in regions where alignment may be challenging. Our analysis found that of the 407,779 SVs (excluding BNDs) called only by hapdiff across all 100 samples, 151,575 (37.1%) were fully or partially within a segdup or within 1000 bp of a segdup, suggesting that they may be in complex copy-number polymorphic regions of the genome, and thus potential artifacts because of their proximity to a segdup. Of the SVs that were not fully within, partially within, or within 1000 bp of a segdup, 119,255 (46.5% of the remaining SVs) overlap a variable number tandem repeat (VNTR) region. Analysis of SVs called only by hapdiff did not reveal any individual sample or population outliers (Supplemental Fig. S9C), and visual analysis of 30 randomly selected SVs from this set found that 28/30 were likely false-positive calls (Supplemental Fig. S10). This suggests that difficult-to-assemble regions are a major source of false-positive assembly-based SV calls and that annotating SV calls with information about genomic context might provide insight into the confidence of these calls.
An SV frequency call set was generated that represented SVs called by all five callers (100,915 total SVs), four or more (119,805 total SVs), three or more (133,766 total SVs), two or more (155,407 total SVs), or at least one caller (252,954 total SVs). Among the 100 samples described here, there were a total of 113,696 shared or unique high-confidence SVs (SVs identified by hapdiff and two or more unique callers, excluding BNDs), with 32% found in only one sample (36,096 of 113,696). We found that 12,432 (11%) of these shared SVs were seen in exactly two samples, and that approximately half of these shared SVs were in samples only from the African superpopulation (Fig. 3F), similar to previous analysis (The 1000 Genomes Project Consortium 2015). Among 50,458 high-confidence SVs that intersect protein-coding genes, 97% (49,142/50,458) are within or include intronic sequence, 3.3% (1654/50,458) are within or include coding sequence, and 2.0% (992/50,458) are within or include a 5′ or 3′ untranslated region (UTR).
To investigate the functional significance of SVs on gene expression, we performed an SV-eQTL analysis using the merged SV call set and the recently published MAGE data set, which includes RNA-seq data from 731 samples from the 1KGP cohort (Supplemental Fig. S11A; Taylor et al. 2023). Among 65 samples shared between MAGE and this study, we found 153 significant SV-eQTLs (Q-value < 0.05), of which 37 were previously found using a collection of 31 diverse LRS-based genomes (Supplemental Fig. S11B). This includes a 484 bp insertion associated with ZNF79, a gene implicated in neurological diseases (Supplemental Fig. S11C,D; Bu et al. 2021). This analysis also revealed several new significant associations, including an 81 bp deletion not previously detected (Kirsche et al. 2023) that is associated with the NAPRT gene, an important factor in cancer susceptibility (Supplemental Fig. S11E,F; Duarte-Pereira et al. 2021). To further explore the application of the variant call set for SV-eQTL discovery, we genotyped the SVs in all 731 MAGE individuals using their matched short-read genomic data from Byrska-Bishop et al. (2022). Using the 65 samples common to both the 1KGP-ONT and MAGE data sets, we found the genotype consistency was >98% between the short- and long-read data sets after filtering for tandem repeats and Hardy–Weinberg consistency (Supplemental Fig. S11G). Across all 731 samples, we identified 1324 significant SV-eQTLs, of which 1258 were uniquely in the short-read data, including a 2716 bp deletion associated with GBP3, a gene implicated in infectious diseases and immune responses (Supplemental Fig. S11H; Tretina et al. 2019).
Structural variation within medically relevant genes
Sequencing of samples from all five superpopulations allowed us to evaluate population-specific SVs intersecting genes associated with an OMIM phenotype (n = 4866) and revealed 349 high-confidence SVs in or including at least one defined exon (Supplemental Fig. S12A; Supplemental Table S9). These events ranged in size from 50 bp (deletions in TNFRSF13C and TF and insertion in IMPG2) to 87,776 bp (a deletion that fully includes IGHM). Visual analysis of 30 randomly selected events confirmed that all were likely true positives. These 349 SVs are distributed across all chromosomes and impact 335 exons in 236 unique OMIM genes, with 123 of those 335 exons containing ClinVar variants that are annotated as pathogenic or likely pathogenic (Supplemental Fig. S12B). We found that 150/349 (43%) of these SVs were found in only one sample, and no single sample had more than six unique SVs (HG01369). Three SVs (a 458 bp insertion in ABCC11, a 243 bp insertion in XYLT1, and a 118 bp insertion in MED13L) were seen in all 100 samples, suggesting the reference genome represents a minor allele at these positions. Indeed, GRCh38 has been patched to include a similar insertion in XYLT1. Of the 38 SVs observed in only two samples, 76% (29/38) were superpopulation-specific with 55% of those (16/29) seen in samples from the African superpopulation. We observed four SVs spanning multiple genes, some of which are known population variants. This includes a 22.8 kbp deletion spanning HBB, HBD, and HBG1 associated with beta thalassemia (Huisman et al. 1972) (MIM: 613985) and two samples with a 19,304 bp deletion including HBA1 and HBA2 commonly referred to as the Southeast Asian deletion (Farashi and Harteveld 2018) (MIM: 604131) (Supplemental Fig. S13).
We did not expect to find rare SVs in X-linked OMIM genes in 46,XY samples, since those events would be more likely to be associated with a disease. However, we did find five such events in at least one 46,XY sample. Of these, four were in a 3′ UTR and were observed in at least two 46,XX samples. One of the four events, found in only one sample, was an ∼141 bp insertion in exon 15 of RPGR (OMIM: 312610), a gene associated with several X-linked conditions including retinitis pigmentosa, cone-rod dystrophy, and macular degeneration (Fahim et al. 1993). A similar insertion at this position has been reported twice in ClinVar as a variant of uncertain significance (VUS) associated with primary ciliary dyskinesia, once as a 141 bp insertion (ClinVar entry 2121719) and once as a 69 bp insertion (ClinVar entry 1975740). Evaluation of the short-read sequencing data for this sample at this position did not clearly demonstrate the insertion, but the insertion consists of only C- and T-nucleotides, which would make it difficult to align and evaluate using short-read technology (Supplemental Fig. S14). The presence of this insertion in a 46,XY 1KGP sample suggests that this variant may be present at a higher allele frequency than expected, is difficult to reliably call using short-read technology, or could be associated with a later onset of the associated phenotype.
A substantial number of high-confidence SVs were observed in regions of the genome difficult to evaluate using short reads, meaning they may be filtered by variant annotation pipelines. For example, 42% (47,315/113,696) of the high-confidence SVs occur fully outside of the GIAB Tier 1 regions, and visual inspection of 30 events confirmed the presence of an SV. We also identified 407 high-confidence SVs within coding regions defined as unreliable for variant identification using short-read sequencing based on analysis of gnomAD data (Hijikata et al. 2024). Finally, 9788 of the high-confidence insertions were ≥500 bp, which may preclude accurate resolution of these events and limit our understanding of their impact on gene expression or splicing when evaluated using short-read technology.
Cytochrome P450 (CYP) genes impact drug response and are among the gene sets that are challenging to interrogate using short-read technologies and may require separate variant calling approaches to fully evaluate (Zanger and Schwab 2013, Lee et al. 2019). Within this data set, LRS enabled better resolution of full gene deletion and duplication SV events in highly polymorphic CYP pharmacogenes such as CYP2D6, a pharmacogene involved in the metabolism of over 20% of clinically prescribed medications (Zanger and Schwab 2013). For example, we identified one individual (HG02396) with a CYP2D6 gene deletion (*5) on one haplotype and a hybrid tandem arrangement (*36 + *10)—shown via an insertion—on the second haplotype (Supplemental Fig. S15A). In the equivalent short-read WGS data, it can be difficult to identify both the gene deletion and the hybrid tandem star allele in the same individual using specialized short-read genotyping tools (Twesigomwe et al. 2023). Analysis of a known complex CYP2B6 star allele (CYP2B6*29) showed that it was called by hapdiff but not the alignment-based callers, demonstrating that some of these complex alleles may not be represented in our initial high-confidence SV set (Supplemental Fig. S15B; Twesigomwe et al. 2024).
We used Jasmine to test whether the SVs identified in these 100 samples could be used to accurately filter SVs in 16 cases with known disease-associated SVs identified by whole-genome (eight cases) or targeted (eight cases) ONT sequencing (Supplemental Table S10; Miller et al. 2021; Wilderman et al. 2024). Among the eight cases that had undergone whole-genome LRS, filtering reduced the average number of SVs called by Sniffles2 by 93% (from 22,743 to 1664), and in all 16 cases the pathogenic SV was retained after filtering. Subsequent annotation of the filtered SVs (i.e., if the SV intersects with a gene, if that gene is associated with an OMIM phenotype, if the SV is exonic, if the SV is within a segmental duplication or low complexity region, etc.) allowed us to substantially further narrow the output candidate SVs. This demonstrates that the high-confidence SV calls can be used to filter SVs in cases with high suspicion of a monogenic condition.
Analysis of disease-associated repeat expansions
Tandem repeat expansions (e.g., short tandem repeats [STRs] and VNTRs) at more than 60 loci have been implicated in human diseases such as the GGC expansion in the 5′ UTR of XYLT1 (MIM: 608124) associated with Baratella–Scott syndrome (MIM: 300881) (Hannan 2018; Depienne and Mandel 2021). Pathogenic repeat expansions associated with monogenic disease can be difficult to precisely size or fully sequence-resolve using short-read sequencing, meaning clinically relevant interruptions in the repeat may not be easily identified (Chaisson et al. 2023; Tanudisastro et al. 2024). Thus, there is interest in using LRS to evaluate repeat expansions genome-wide and at clinically relevant loci (Sulovari et al. 2019; Reis et al. 2023; Dolzhenko et al. 2024).
We used vamos (Ren et al. 2023) to perform genome-wide haplotype-resolved analysis of 562,005 loci—including 66 disease-associated loci—consisting of both simple and complex repeat units, and identified pathogenic-sized expansions in RFC1, ATXN10, FGF14, and ATXN80S (Fig. 4A; Supplemental Figs. S16–S19; Supplemental File S3; Hiatt et al. 2024). We also identified alleles over the pathogenic threshold but with a benign motif in SAMD12, BEAN1, and DAB1, as well as several alleles at AR where the total repeat count was over the threshold but the CAG motif was only a portion of the region.
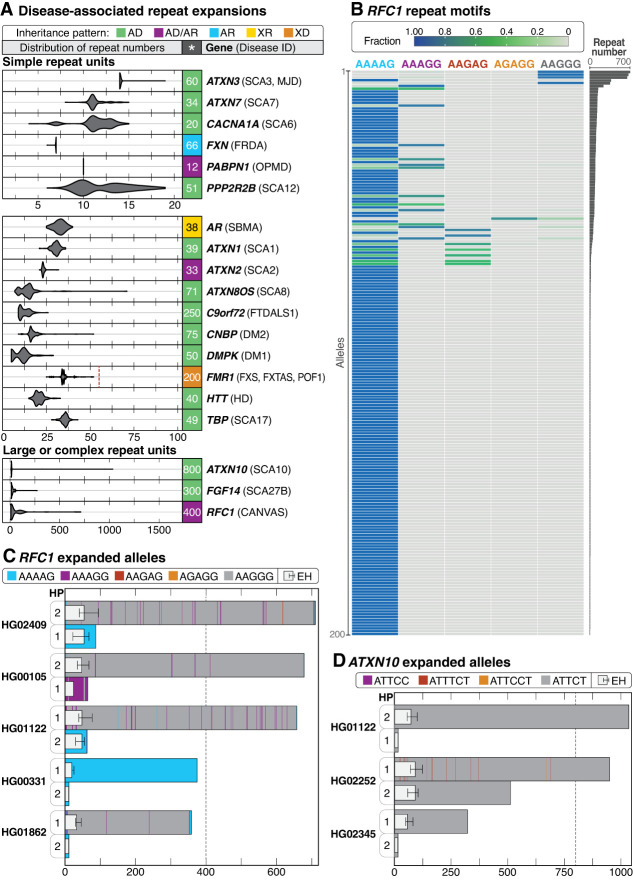
Figure 4.
Evaluation of repeat expansions known to be associated with Mendelian conditions. (A) Haplotype-resolved repeat expansions of selected repeat loci for simple and complex repeat units. Pathogenic repeat size is shown to the right of each plot (*), the associated condition is in parentheses, and the full name of each condition can be found in Supplemental Table S11. The pathogenic repeat size for FMR1 is listed as 200 repeats, but a dashed vertical line represents the 55-repeat threshold that puts 46,XX and 46,XY individuals at risk for fragile X-associated tremor/ataxia syndrome (FXTAS, MIM #300623) and 46,XX individuals at risk of fragile X-associated primary ovarian insufficiency (POF1/FXPOI, MIM #311360). (AD) autosomal dominant, (AD/AR) autosomal dominant/recessive, (AR) autosomal recessive, (XR) X-linked recessive, (XD) X-linked dominant. (B) Among 200 haplotypes (y-axis), an expansion in RFC1 near or over 400 repeat units was seen in five haplotypes. AAGGG is the most common pathogenic repeat expansion; additional pathogenic expansions include ACAGG (not shown), and a mixed AAAGG/AAGGG expansion (Cortese et al. 1993). (C) Haplotype (HP)-resolved detail of RFC1 repeat expansions in five samples with an expansion of one allele. Haplotypes are assigned arbitrarily. The dotted line represents the position of full penetrance alleles typically seen at 400 repeat units. (D) Three samples with expansions in ATXN10 larger than 280 ATTCT repeats were observed. The dotted line at 800 repeat units represents the position of the lower end of the full penetrance range. ExpansionHunter (EH) estimates are overlayed atop the bar plots in (C) and (D), placed on HP1 or HP2 based on their length.
Expansions in RFC1, which are associated with autosomal recessive cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS, MIM #614575), were observed in five samples ranging from 359 to 712 repeat units in size (Fig. 4B). Pathogenic expansions in this gene are typically 400 repeat units or larger and are motif-dependent, with AAGGG being the most common pathogenic expansion (Cortese et al. 2019; Beecroft et al. 2020; Scriba et al. 2020). Our observation that some of these samples carried the AAGGG repeat unit while others carried a nonpathogenic repeat unit, such as AAAAG, was similar to recent work that identified expansions in RFC1 of varying repeat motifs in 5/100 HPRC samples (Fig. 4C; Dolzhenko et al. 2024). That we observed an expansion in 5% of samples was not unexpected, as the carrier frequency of RFC1 expansions has been reported at 1%–5% across at least two populations (Akçimen et al. 2019; Fan et al. 2020).
Expansions in ATXN10 are associated with autosomal dominant spinocerebellar ataxia type 10 (SCA10, MIM #603516), a slowly progressive ataxia with typical age of onset between 12 and 48 years and full penetrance alleles varying from 800 to 4500 ATTCT repeats (Matsuura and Ashizawa 1993; Alonso et al. 2006; Raskin et al. 2007). Two of the 100 samples were heterozygous for ATXN10 alleles larger than 800 motifs, one of which had a second allele with 511 repeat units (Fig. 4D). In addition, two other samples harbored expansions close to or larger than 280 repeat units, which has been reported as causative in one individual with ataxia (Matsuura et al. 2006). However, three of the four large alleles are purely ATTCT, and evidence suggests that interruptions of ATTCC are necessary for the allele to be pathogenic (Morato Torres et al. 2022).
To determine whether any of the expanded RFC1 and ATXN10 alleles would be identified using short-read data, we ran ExpansionHunter on short-read data from all affected samples (Dolzhenko et al. 2019). In all cases, when an expanded allele was present, the corresponding ExpansionHunter estimate was larger than the normal allele but, in most cases, still significantly underestimated the size of the expansion (Fig. 4C,D; Supplemental Table S11). For example, in ATXN10, LRS identified a normal allele (15 repeat units) and an expansion of more than 1000 repeat units in HG01122. The ExpansionHunter estimates for this sample are 15 (range 15–15) and 73 (range 56–101) repeat units, thus the normal allele was correctly estimated but the expanded allele was markedly underestimated.
Evaluation of genome-wide methylation patterns and identification of novel DMRs
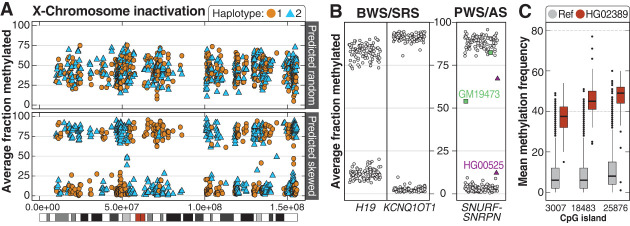
An advantage of LRS is the ability to simultaneously capture both DNA sequence and modification information, allowing for simultaneous evaluation of how changes in sequence, such as a repeat expansion, may alter the local epigenetic landscape. We evaluated methylation both genome-wide and at loci associated with imprinting disorders. Among 69 of the 70 46,XX samples sequenced, we found that 39% (27/69) had X-Chromosome methylation patterns suggestive of skewed X-inactivation (Fig. 5A; Supplemental Table S12).
Figure 5.
Patterns of methylation among the 1000 Genomes samples. (A) Among 69 46,XX samples, 42 had mixed X-Chromosome inactivation (top, example from HG01414), while 27 were skewed (bottom, example from HG01801). The color differences are related to breaks in phasing and do not suggest methylation is mixed along a single haplotype. (B) Haplotype-resolved methylation fraction is shown for three imprinted loci associated with four imprinting disorders. Methylated (>75%) or unmethylated (<25%) fraction at IC1 in H19 and IC2 in KCNQ1OT1. Haplotype-resolved methylation fraction is also shown for the CpG island within SNURF-SNRPN that is evaluated when testing for PWS or AS. Two samples have either gain (GM19473) or loss (HG00525) of methylation at this locus. (C) Unique methylation differences within defined CpG islands were identified in individual samples. An example from HG02389 shows three CpG sites with increased methylation (red boxes) compared to controls (gray).
We then performed genome-wide PCA of methylation to evaluate whether samples would correlate with ancestry or if patterns of X-inactivation would be apparent (Supplemental Fig. S20). This analysis revealed that GM18864 clustered with 46,XY samples despite being reported as 46,XX. Because we validated each sample using SNVs from short-read sequencing, we wondered whether this sample had lost an X Chromosome. We found that the average X Chromosome depth of coverage was ∼55% of the full-length autosomes in the LRS data and ∼75% in the short-read data, confirming the loss of an X Chromosome in this sample (Pedersen et al. 2020).
Next, we evaluated methylation patterns at two disease-associated loci: 11p15.5, which is associated with both Beckwith–Wiedemann syndrome (BWS, MIM #130650) and Silver–Russell syndrome (SRS, MIM #180860) (Saal et al. 1993; Shuman et al. 1993); and 15q11.2-q13, associated with Prader–Willi syndrome (PWS, MIM #176270) and Angelman syndrome (AS, MIM #105830) (Dagli et al. 1993; Driscoll et al. 1993). For the 11p15.5 region, we found that in all samples, one haplotype was completely methylated while the other was completely unmethylated at imprinting centers IC1 and IC2 (Fig. 5B). Evaluation of haplotype-resolved methylation at the SNURF-SNRPN locus on 15q11.2 revealed two samples, GM19473 and HG00525, where one haplotype was 25%–75% methylated. Visual evaluation of these samples showed that one haplotype of GM19473 had increased methylation while one haplotype of HG00525 had reduced methylation, which was unexpected and further demonstrates that changes in methylation can occur throughout the genome in these cell lines, even at well-established DMRs (Supplemental Fig. S21).
We used Methylation Operation Wizard (MeOW) (Zalusky and Miller 2024) to analyze differences in methylation at CpG sites genome-wide and identified 134 CpGs with methylation differences across 37 samples, with a median of two DMRs per sample (Supplemental Table S13). As an example, three DMRs were found in HG02389 (Fig. 5C), including a hypermethylated CpG in SLC29A3 not present in controls (Supplemental Fig. S22). We observed both hypermethylation (86 CpGs) and hypomethylation (48 CpGs) among the 134 CpGs and identified four samples with more than 10 DMRs (Supplemental Fig. S23). Among the 15 samples from the African superpopulation with a DMR, there was an enrichment of expression outliers near the DMR with increasingly stringent Z-score thresholds, suggesting associated changes in gene expression (Supplemental Fig. S24).
Discussion
Current approaches to clinical genetic testing are incomplete as they are unable to capture the full spectrum of disease-causing variation (Wojcik et al. 2023). This is because: (1) new technologies, such as LRS, are not yet widely implemented in clinical labs; (2) computational tools are not yet able to efficiently capitalize on the data provided by these new technologies, and those that can have substantial computational requirements; and (3) databases are not yet available for filtering and prioritizing variants identified using new technologies. The 1KGP-ONT Consortium plans to sequence at least 800 1KGP samples to generate a more complete catalog of variation, especially rare yet presumably benign variants across the 1KGP populations. While the expanded collection will enable a more accurate estimate of allele frequency for challenging variants and add information about haplotype-resolved epigenetic variation, we acknowledge that this cohort represents a limited representation of human diversity, notably excluding individuals of indigenous Australian and Middle Eastern ancestries.
Here, we describe the initial analysis of the first 100 samples sequenced to an average of 30× depth of coverage and average read N50 >50 kbp, which was possible because of the use of HMW DNA isolated directly from cell culture (Fig. 1). This resulted in high sensitivity for SV detection—especially larger duplications and repeat expansions—using both assembly- and alignment-based approaches. We identified an average of 24,543 SVs per sample, similar to the prior analysis of other 1KGP samples by the HGSVC and HPRC (Ebert et al. 2021; Liao et al. 2023). Our efforts complement recent work that identified ∼16,000 SVs from ∼1000 1KGP samples sequenced to the lower average depth of coverage (15×) and median read length (6.2 kbp) (Schloissnig et al. 2024). While the difference in total SVs underscores the advantage of sequencing HMW DNA, further analysis will be required to fully assess the significance of the differences between these data sets.
We performed one of the most comprehensive benchmarking analyses to date of SNVs, indels, and SVs using data from the ONT platform. Consistent with prior studies, data generated on the ONT platform has a higher recall and precision than Illumina-based approaches for SNVs in well-characterized genomic regions and performs well for indels, specifically outside of homopolymers (Kolmogorov et al. 2023). Because all data from these first 100 samples were generated on the R9.4.1 pore, we anticipate that improvements in chemistry, such as the use of the R10.4.1 pore, will reduce context-specific errors and result in improved concordance with truth sets. Because of this, we have transitioned ongoing sequencing to the R10.4.1 pore. SV benchmarking also revealed high F1 scores for three samples for which orthogonal calls were available, highlighting how the R9.4.1 pore is sufficient for this application. Over time, we anticipate additional updates to ONT chemistry or software, and plan to evaluate each change carefully before data reanalysis or changing the chemistry used for this effort.
SVs were called using four alignment-based and one assembly-based caller. After merging, a high-confidence SV call set comprising 124,927 SVs was generated that we show can be used for filtering and variant prioritization. Genome-wide evaluation of these high-confidence SVs revealed 349 that were within or encompassed an exon of a medically relevant gene. The low number of SVs intersecting medically relevant genes was reassuring, as we expect there to be selection against these events within coding regions of the genome. Nevertheless, we did identify one SV—an ∼141 bp insertion in exon 15 of RPGR, a gene with an X-linked phenotype—in a 46,XY sample near two similar insertions that have been reported as VUSs in ClinVar. Because the 1KGP samples came from presumably healthy individuals, it could be that this event is associated with a later onset of an associated phenotype or that the insertion is benign. Identification of this insertion in a 1KGP sample is valuable as it may lead to functional studies that clarify the nature of the variant. Analogous to what has been reported for the relatively common occurrence of single nucleotide loss-of-function mutations in otherwise healthy individuals, the presence of an SV in a gene does not necessarily imply the variant is pathogenic (MacArthur et al. 2012). Indeed, early studies of human population samples using SNP microarrays identified extremely rare copy number variants >500 kbp in length among individuals without overt disease (Cooper et al. 2011).
Genome-wide evaluation of select repeat expansions revealed expansions in complex alleles not previously reported and difficult to identify using short-read technology (Fig. 4). We identified repeat expansions associated with diseases that are difficult to fully interpret because the individuals recruited to the 1KGP were presumably healthy. These individuals may be at risk of developing symptoms later in life, or they may be carrying alleles that are benign because of nonpathogenic motif composition or sequence interruptions that we did not detect. Alternatively, these expansions may simply be an artifact of the cell culture process and should be considered when these samples are used in other experiments or when these data are used for variant filtering and prioritization. We anticipate that comparison of this data set to larger efforts, such as All of Us, will allow us to better understand whether these variants represent artifact from the cell culture process or true human genetic variation.
Finally, we evaluated patterns of methylation genome-wide and at loci associated with disease. We observed large-scale changes, such as skewed X-inactivation, in over one-third of 46,XX samples as well as unique changes, such as novel differential methylation that correlates with changes in local gene expression. These changes provide a mechanism by which distinct signals from samples maintained in cell culture can be explained and demonstrate the potential limitations of using immortalized cell lines to infer epigenetic signatures.
Sequencing of 1KGP samples is ongoing and we expect the analysis of a larger number of samples to further refine many of the findings in this study. Most analysis presented here was performed using GRCh38 as a reference due to its widespread use in clinical and research laboratories; work is ongoing to evaluate the impact of the more complete CHM13 T2T genome on variant calling (Nurk et al. 2022). Overall, we anticipate that the data set provided here will hasten the use of LRS to evaluate individuals with suspected Mendelian conditions for whom a precise molecular diagnosis remains elusive. This work not only provides valuable resources for candidate variant filtering and analysis but also emphasizes the critical need for ongoing investment in technology, software, and database development to fully realize the benefits of LRS. The more comprehensive analysis that can be performed using LRS—such as the identification and resolution of complex SVs, improved phasing, and incorporation of associated methylation information—will allow clinical and research teams to stop focusing on “what's the next best test” when evaluating an individual with a suspected genetic condition and instead focus on interpreting those variants that were previously difficult to detect or that may involve a novel gene. Together, these efforts will lead to improved clinical outcomes, new gene–phenotype associations, the use of novel therapies, and an end to the diagnostic odyssey for many of the individuals and their families who are living with an unsolved or incompletely understood genetic condition.
Methods
DNA extraction, sequencing, alignment, validation, and variant calling
DNA for sequencing was isolated from B lymphocytes obtained from the NHGRI Sample Repository at the Coriell Institute for Medical Research. After sequencing and quality checks (Supplemental Table S2), an internal alignment pipeline and the Napu pipeline were run before variant calling and annotation (Kolmogorov et al. 2023). Additional details can be found in Supplemental Methods.
SNV and indel benchmarking and comparison with Illumina data
Original sequencing data for five benchmarking samples was base called with Dorado 0.5.0 (ONT) and downsampled to match the depth of coverage of the 100 study samples, then processed with both the internal alignment pipeline and the Napu pipeline. Long-read SNV and indel calls from the HPRC and GIAB (Shafin et al. 2020; Liao et al. 2023) and short-read SNV and indel calls from GIAB were obtained (Wagner et al. 2022) and preprocessed. Benchmarking comparisons and comparisons with Illumina data were conducted using hap.py (https://github.com/Illumina/hap.py), with analysis limited to high-confidence regions.
De novo genome assembly and evaluation
Flye (v2.9.2) (Kolmogorov et al. 2019) and Napu (Shasta–Hapdup) (Kolmogorov et al. 2023) were used for haploid and diploid genome assembly then aligned to the GRCh38 reference genome using minimap2 (v2.24) (Li 2018), with starts and ends of aligned contigs determined using BEDTools (v2.3.0) (Quinlan and Hall 2010). Assembly breakpoints were characterized using precomputed segdup and RepeatMasker positions downloaded from UCSC (Bailey et al. 2002; Kent et al. 2002) then categorized as Satellite, SegDup, SegDup + Satellite, or Neither.
SV analysis, merging, and benchmarking
SV calls were parsed using BCFtools (Danecek et al. 2021) for variants that passed filtering criteria, were ≥50 bp, and were assigned to a full-length chromosome. SVs were counted by type and length per sample and caller. Novel SVs per sample were calculated through iterative merging by Jasmine. To benchmark SV calling methods, ONT data from HG002/NA24385, HG00733, and HG02723 were processed using the Napu pipeline. SV calls for Sniffles2 and hapdiff were benchmarked to the HPRC (truth) calls using Truvari (v4.1.0) (English et al. 2022). The GIAB HG002 SV Tier1 benchmarking BED was used to define regions for inclusion. Additionally, we benchmarked HG002 SV calls against the draft GIAB T2TQ100 HG002 GRCh38 SV benchmark. SVs per individual were analyzed for multicaller concordance based on Jasmine merging. SVs meeting a threshold of support (described in Supplemental Methods) were reported as high confidence. SVs from this high-confidence call set were further annotated with functionally relevant genomic information (i.e., intersection with exonic regions, genes associated with OMIM phenotypes, centromeric/telomeric regions, etc.) as defined by GENCODE release 45.
Filtering and prioritization of SVs
Sniffles2 SV calls from cases known to have a disease-causing SV were preprocessed as above and merged using Jasmine with Sniffles2 SV calls from the Napu pipeline from the 100 samples.
Pangenome construction
Contigs from the Shasta–Hapdup assemblies were partitioned by chromosome by mapping them against the human reference genomes using WFMASH (v0.12.6, commit 0b191bb) pangenome aligner (Marco-Sola et al. 2021).
eQTL analysis
We applied the SV-eQTL analysis from Kirsche et al. (2023) to the 65 samples with both long-read DNA and short-read RNA data from MAGE and analyzed them as described in Supplemental Methods.
Tandem repeat genotyping
Repeats were genotyped using vamos v1.2.6 (Ren et al. 2023). A BED file with the coordinates and metadata for each STRchive locus is provided.
Methylation analysis
Haplotype-resolved, whole-genome methylation pileup files were generated using Modkit v0.1.11 (ONT) from the PMDV haplotagged BAM file. For X-Chromosome analysis, the average fraction of methylated reads was calculated for each CpG island. CpG islands at disease-associated loci were subsetted and the average fraction of reads methylated was calculated per sample and per haplotype. Unique DMRs were identified using MeOW by a leave-one-out analysis (Zalusky and Miller 2024).
Data access
Data for all samples sequenced as part of the 1000 Genomes Project ONT Sequencing Consortium are publicly available at https://s3.amazonaws.com/1000g-ont/index.html and scripts used in the analysis can be found at GitHub (https://github.com/millerlaboratory/1000g_ONT) and as Supplemental Scripts. Data from the 100 samples reported here, as well as summary analysis data, are available at https://s3.amazonaws.com/1000g-ont/index.html?prefix=ALIGNMENT_AND_ASSEMBLY_DATA/FIRST_100/. Data and code related to pangenome analyses are available at GitHub (https://github.com/AndreaGuarracino/1000G-ONT-F100-PGGB).
Supplemental Material
Acknowledgments
S.B.G. is supported by NIH grant 5T32HG000035-29; W.D.C. is a recipient of a postdoctoral fellowship from FWO (12ASR24N); E.G. and A.G. are supported by NIH grants R01HG013017 and U01DA057530 and NSF grant 2118744; S.G. is supported by NIH grant 5R50CA243890; T.D.J. is supported by NIH grant T32HG000044; M.K. is supported by Intramural NIH funding; S.B.M., T.D.J., and E.R. are supported by NIH grant U01HG011762; M.C.S. is supported by NIH grants U24HG010263, R03CA272952, and U01CA253481 and the Lustgarten Foundation grant 90101412; F.J.S. is supported by NIH grants 1U01HG011758-01, 1UG3NS132105-01, and U01AG058589; A.A.S. is supported by an NSF Postdoctoral Research Fellowship in biology (NSF 22-623); R.N.M. and L.Y. are supported by NIH grants 5R35GM142733-03 and 5R21AI174130-02; E.E.E. is supported by NIH grant HG010169 and is an investigator of the Howard Hughes Medical Institute; D.E.M. is supported by the NIH Director's Early Independence Award DP5OD033357. The GREGoR Consortium is funded by the National Human Genome Research Institute of the National Institutes of Health, through the following grants: U01HG011758, U01HG011755, U01HG011745, U01HG011762, U01HG011744, and U24HG011746. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. No human subjects, live vertebrates, or higher invertebrate research was undertaken as part of this manuscript. Certain commercial equipment, instruments, or materials are identified to adequately specify experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose.
Author contributions: Conceptualization: E.E.E. and D.E.M. Data curation: J.A.G., S.B.G., N.D., M.P.G.Z., K.H., and H.D. Formal analysis: J.A.G., S.B.G., N.D., D.T., L.Y., P.A.R., W.D.C., N.D.O., A.G., Q.L., W.E.C., A.O.B., A.C., A.H., R.L.M., M.R., T.D.J., E.R., J.M.Z., E.G., M.C.S., R.N.M., E.E.E., and D.E.M. Funding acquisition: E.E.E. and D.E.M. Investigation: M.P.G.Z., K.H., J.G., Z.B.A., S.H.R.S., S.A.W., and M.S. Methodology: J.A.G., S.B.G., N.D., M.P.G.Z., K.H., W.D.C., C.E.R., A.H., S.G., W.R.M., M.K., H.D., M.L., M.J., H.D., E.E.E., and D.E.M. Resources: M.C.Z., M.L., M.J., E.E.E., and DE.M. Supervision: E.E.E. and D.E.M. Visualization: J.A.G., S.B.G., N.D., D.T., L.Y., W.D.C., A.G., A.L.M., E.G., and R.N.M. Writing—original draft: J.A.G., S.B.G., N.D., M.P.G.Z., D.T., L.Y., W.D.C., A.L.M., J.G., S.H.R.S., W.E.C., R.N.M., E.E.E., and D.E.M. Writing—review and editing: J.A.G., S.B.G., N.D., K.H., A.A.S., N.D.O., A.G., Q.L., A.L.M., C.G.J., W.E.C., A.O.B., A.C., C.E.R., M.R., K.E.P., C.R.P., C.Z., S.G., T.D.J., W.R.M., F.J.S., S.B.M., E.G., M.K., M.C.S., H.D., M.C.Z., M.L., M.J., E.E.E., and D.E.M.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279273.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
W.D.C., M.L., F.J.S., and D.E.M. have received research support and/or consumables from ONT. W.D.C., J.G., S.B.G., F.J.S., and D.E.M. have received travel funding to speak on behalf of ONT. D.E.M. is on a scientific advisory board at ONT and holds stock options in MyOme. F.J.S. has received research support from Illumina, Genentech, and PacBio. S.B.M. is an advisor to BioMarin, MyOme, and Tenaya Therapeutics. E.E.E. is a scientific advisory board member of Variant Bio, Inc.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akçimen F, Ross JP, Bourassa CV, Liao C, Rochefort D, Gama MTD, Dicaire M-J, Barsottini OG, Brais B, Pedroso JL, et al. 2019. Investigation of the RFC1 repeat expansion in a Canadian and a Brazilian ataxia cohort: identification of novel conformations. Front Genet 10: 1219. 10.3389/fgene.2019.01219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlAbdi L, Shamseldin HE, Khouj E, Helaby R, Aljamal B, Alqahtani M, Almulhim A, Hamid H, Hashem MO, Abdulwahab F, et al. 2023. Beyond the exome: utility of long-read whole genome sequencing in exome-negative autosomal recessive diseases. Genome Med 15: 114. 10.1186/s13073-023-01270-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso I, Jardim LB, Artigalas O, Saraiva-Pereira ML, Matsuura T, Ashizawa T, Sequeiros J, Silveira I. 2006. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology 66: 1602–1604. 10.1212/01.wnl.0000216266.30177.bb [DOI] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- Beecroft SJ, Cortese A, Sullivan R, Yau WY, Dyer Z, Wu TY, Mulroy E, Pelosi L, Rodrigues M, Taylor R, et al. 2020. A Māori specific RFC1 pathogenic repeat configuration in CANVAS, likely due to a founder allele. Brain 143: 2673–2680. 10.1093/brain/awaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu S, Lv Y, Liu Y, Qiao S, Wang H. 2021. Zinc finger proteins in neuro-related diseases progression. Front Neurosci 15: 760567. 10.3389/fnins.2021.760567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrska-Bishop M, Evani US, Zhao X, Basile AO, Abel HJ, Regier AA, Corvelo A, Clarke WE, Musunuri R, Nagulapalli K, et al. 2022. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell 185: 3426–3440.e19. 10.1016/j.cell.2022.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DL, Di Stefano L, Papenfuss AT. 2019. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun 10: 3240. 10.1038/s41467-019-11146-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, Gardner EJ, Rodriguez OL, Guo L, Collins RL, et al. 2019. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10: 1784. 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sulovari A, Valdmanis PN, Miller DE, Eichler EE. 2023. Advances in the discovery and analyses of human tandem repeats. Emerg Top Life Sci 7: 361–381. 10.1042/ETLS20230074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ASA, Farrow EG, Abdelmoity AT, Alaimo JT, Amudhavalli SM, Anderson JT, Bansal L, Bartik L, Baybayan P, Belden B, et al. 2022. Genomic answers for children: dynamic analyses of >1000 pediatric rare disease genomes. Genet Med 24: 1336–1348. 10.1016/j.gim.2022.02.007 [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. 2011. A copy number variation morbidity map of developmental delay. Nat Genet 43: 838–846. 10.1038/ng.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese A, Reilly MM, Houlden H. 1993. RFC1 CANVAS/spectrum disorder. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [PubMed] [Google Scholar]
- Cortese A, Simone R, Sullivan R, Vandrovcova J, Tariq H, Yau WY, Humphrey J, Jaunmuktane Z, Sivakumar P, Polke J, et al. 2019. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat Genet 51: 649–658. 10.1038/s41588-019-0372-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli AI, Mathews J, Williams CA. 1993. Angelman syndrome. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [Google Scholar]
- Damaraju N, Miller AL, Miller DE. 2024. Long-read DNA and RNA sequencing to streamline clinical genetic testing and reduce barriers to comprehensive genetic testing. J Appl Lab Med 9: 138–150. 10.1093/jalm/jfad107 [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10: giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Mandel J-L. 2021. 30 years of repeat expansion disorders: what have we learned and what are the remaining challenges? Am J Hum Genet 108: 764–785. 10.1016/j.ajhg.2021.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E, Deshpande V, Schlesinger F, Krusche P, Petrovski R, Chen S, Emig-Agius D, Gross A, Narzisi G, Bowman B, et al. 2019. Expansionhunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics 35: 4754–4756. 10.1093/bioinformatics/btz431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E, English A, Dashnow H, De Sena Brandine G, Mokveld T, Rowell WJ, Karniski C, Kronenberg Z, Danzi MC, Cheung WA, et al. 2024. Characterization and visualization of tandem repeats at genome scale. Nat Biotechnol. 42: 1606–1614. 10.1038/s41587-023-02057-3 [DOI] [PubMed] [Google Scholar]
- Driscoll DJ, Miller JL, Cassidy SB. 1993. Prader-Willi syndrome. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [Google Scholar]
- Duarte-Pereira S, Fajarda O, Matos S, Oliveira JL, Monteiro Silva R. 2021. NAPRT expression regulation mechanisms: novel functions predicted by a bioinformatics approach. Genes (Basel) 12: 2022. 10.3390/genes12122022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P, Audano PA, Zhu Q, Rodriguez-Martin B, Porubsky D, Bonder MJ, Sulovari A, Ebler J, Zhou W, Serra Mari R, et al. 2021. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372: eabf7117. 10.1126/science.abf7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE. 2019. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med 381: 64–74. 10.1056/NEJMra1809315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Menon VK, Gibbs RA, Metcalf GA, Sedlazeck FJ. 2022. Truvari: refined structural variant comparison preserves allelic diversity. Genome Biol 23: 271. 10.1186/s13059-022-02840-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahim AT, Daiger SP, Weleber RG. 1993. Nonsyndromic retinitis pigmentosa overview. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [Google Scholar]
- Fan Y, Zhang S, Yang J, Mao C-Y, Yang Z-H, Hu Z-W, Wang Y-L, Liu Y-T, Liu H, Yuan Y-P, et al. 2020. No biallelic intronic AAGGG repeat expansion in RFC1 was found in patients with late-onset ataxia and MSA. Parkinsonism Relat Disord 73: 1–2. 10.1016/j.parkreldis.2020.02.017 [DOI] [PubMed] [Google Scholar]
- Farashi S, Harteveld CL. 2018. Molecular basis of α-thalassemia. Blood Cells Mol Dis 70: 43–53. 10.1016/j.bcmd.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Vooren SV, Moreau Y, Pettett RM, Carter NP. 2009. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet 84: 524–533. 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Guarracino A, Heumos S, Villani F, Bao Z, Tattini L, Hagmann J, Vorbrugg S, Marco-Sola S, Kubica C, et al. 2023. Building pangenome graphs. bioRxiv 10.1101/2023.04.05.535718 [DOI] [PubMed] [Google Scholar]
- Hannan AJ. 2018. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 19: 286–298. 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]
- Harvey WT, Ebert P, Ebler J, Audano PA, Munson KM, Hoekzema K, Porubsky D, Beck CR, Marschall T, Garimella K, et al. 2023. Whole-genome long-read sequencing downsampling and its effect on variant-calling precision and recall. Genome Res 33: 2029–2040. 10.1101/gr.278070.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller D, Vingron M. 2019. SVIM: structural variant identification using mapped long reads. Bioinformatics 35: 2907–2915. 10.1093/bioinformatics/btz041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt SM, Lawlor JMJ, Handley LH, Ramaker RC, Rogers BB, Partridge EC, Boston LB, Williams M, Plott CB, Jenkins J, et al. 2021. Long-read genome sequencing for the molecular diagnosis of neurodevelopmental disorders. HGG Adv 2: 100023. 10.1016/j.xhgg.2021.100023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt L, Weisburd B, Dolzhenko E, VanNoy GE, Kurts EN, Rehm HL, Quinlan A, Dashnow H. 2024. STRchive: a dynamic resource detailing population-level and locus-specific insights at tandem repeat disease loci. medRxiv 10.1101/2024.05.21.24307682 [DOI] [Google Scholar]
- Hijikata A, Suyama M, Kikugawa S, Matoba R, Naruto T, Enomoto Y, Kurosawa K, Harada N, Yanagi K, Kaname T, et al. 2024. Exome-wide benchmark of difficult-to-sequence regions using short-read next-generation DNA sequencing. Nucleic Acids Res 52: 114–124. 10.1093/nar/gkad1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman TH, Wrightstone RN, Wilson JB, Schroeder WA, Kendall AG. 1972. Hemoglobin Kenya, the product of fusion of amd polypeptide chains. Arch Biochem Biophys 153: 850–853. 10.1016/0003-9861(72)90408-0 [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. 2005. A haplotype map of the human genome. Nature 437: 1299–1320. 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Liu Y, Jiang Y, Li J, Gao Y, Cui Z, Liu Y, Liu B, Wang Y. 2020. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol 21: 189. 10.1186/s13059-020-02107-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsche M, Prabhu G, Sherman R, Ni B, Battle A, Aganezov S, Schatz MC. 2023. Jasmine and iris: population-scale structural variant comparison and analysis. Nat Methods 20: 408–417. 10.1038/s41592-022-01753-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig Z, Yohannes MT, Nkambule LL, Zhao X, Goodrich JK, Kim HA, Wilson MW, Tiao G, Hao SP, Sahakian N, et al. 2024. A harmonized public resource of deeply sequenced diverse human genomes. Genome Res 34: 796–809. 10.1101/gr.278378.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Billingsley KJ, Mastoras M, Meredith M, Monlong J, Lorig-Roach R, Asri M, Alvarez Jerez P, Malik L, Dewan R, et al. 2023. Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. Nat Methods 20: 1483–1492. 10.1038/s41592-023-01993-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wheeler MM, Patterson K, McGee S, Dalton R, Woodahl EL, Gaedigk A, Thummel KE, Nickerson DA. 2019. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med 21: 361–372. 10.1038/s41436-018-0054-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lin L, Malhotra R, Yang L, Acharya R, Poss M. 2019. A computational framework to assess genome-wide distribution of polymorphic human endogenous retrovirus-K in human populations. PLoS Comput Biol 15: e1006564. 10.1371/journal.pcbi.1006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, Lu S, Lucas JK, Monlong J, Abel HJ, et al. 2023. A draft human pangenome reference. Nature 617: 312–324. 10.1038/s41586-023-05896-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Eichler EE. 2020. Long-read human genome sequencing and its applications. Nat Rev Genet 21: 597–614. 10.1038/s41576-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335: 823–828. 10.1126/science.1215040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Sola S, Moure JC, Moreto M, Espinosa A. 2021. Fast gap-affine pairwise alignment using the wavefront algorithm. Bioinformatics 37: 456–463. 10.1093/bioinformatics/btaa777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Ashizawa T. 1993. Spinocerebellar ataxia type 10. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [PubMed] [Google Scholar]
- Matsuura T, Fang P, Pearson CE, Jayakar P, Ashizawa T, Roa BB, Nelson DL. 2006. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am J Hum Genet 78: 125–129. 10.1086/498654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Sulovari A, Wang T, Loucks H, Hoekzema K, Munson KM, Lewis AP, Fuerte EPA, Paschal CR, Walsh T, et al. 2021. Targeted long-read sequencing identifies missing disease-causing variation. Am J Hum Genet 108: 1436–1449. 10.1016/j.ajhg.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morato Torres CA, Zafar F, Tsai Y-C, Vazquez JP, Gallagher MD, McLaughlin I, Hong K, Lai J, Lee J, Chirino-Perez A, et al. 2022. ATTCT and ATTCC repeat expansions in the ATXN10 gene affect disease penetrance of spinocerebellar ataxia type 10. HGG Adv 3: 100137. 10.1016/j.xhgg.2022.100137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA. 1993. Williams syndrome. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. 2022. The complete sequence of a human genome. Science 376: 44–53. 10.1126/science.abj6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Bhetariya PJ, Brown J, Kravitz SN, Marth G, Jensen RL, Bronner MP, Underhill HR, Quinlan AR. 2020. Somalier: rapid relatedness estimation for cancer and germline studies using efficient genome sketches. Genome Med 12: 62. 10.1186/s13073-020-00761-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin S, Ashizawa T, Teive HAG, Arruda WO, Fang P, Gao R, White MC, Werneck LC, Roa B. 2007. Reduced penetrance in a Brazilian family with spinocerebellar ataxia type 10. Arch Neurol 64: 591–594. 10.1001/archneur.64.4.591 [DOI] [PubMed] [Google Scholar]
- Reis ALM, Rapadas M, Hammond JM, Gamaarachchi H, Stevanovski I, Ayuputeri Kumaheri M, Chintalaphani SR, Dissanayake DSB, Siggs OM, Hewitt AW, et al. 2023. The landscape of genomic structural variation in indigenous Australians. Nature 624: 602–610. 10.1038/s41586-023-06842-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Gu B, Chaisson MJP. 2023. vamos: variable-number tandem repeats annotation using efficient motif sets. Genome Biol 24: 175. 10.1186/s13059-023-03010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal HM, Harbison MD, Netchine I. 1993. Silver-Russell syndrome. In GeneReviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [PubMed] [Google Scholar]
- Schloissnig S, Pani S, Rodriguez-Martin B, Ebler J, Hain C, Tsapalou V, Söylev A, Hüther P, Ashraf H, Prodanov T, et al. 2024. Long-read sequencing and structural variant characterization in 1,019 samples from the 1000 genomes project. bioRxiv 10.1101/2024.04.18.590093 [DOI] [Google Scholar]
- Scriba CK, Beecroft SJ, Clayton JS, Cortese A, Sullivan R, Yau WY, Dominik N, Rodrigues M, Walker E, Dyer Z, et al. 2020. A novel RFC1 repeat motif (ACAGG) in two Asia-Pacific CANVAS families. Brain 143: 2904–2910. 10.1093/brain/awaa263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafin K, Pesout T, Lorig-Roach R, Haukness M, Olsen HE, Bosworth C, Armstrong J, Tigyi K, Maurer N, Koren S, et al. 2020. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol 38: 1044–1053. 10.1038/s41587-020-0503-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafin K, Pesout T, Chang P-C, Nattestad M, Kolesnikov A, Goel S, Baid G, Kolmogorov M, Eizenga JM, Miga KH, et al. 2021. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat Methods 18: 1322–1332. 10.1038/s41592-021-01299-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman C, Kalish JM, Weksberg R. 1993. Beckwith-Wiedemann syndrome. In Genereviews® (ed. Adam MP, et al. ), University of Washington, Seattle. [Google Scholar]
- Smolka M, Paulin LF, Grochowski CM, Horner DW, Mahmoud M, Behera S, Kalef-Ezra E, Gandhi M, Hong K, Pehlivan D, et al. 2024. Detection of mosaic and population-level structural variants with Sniffles2. Nat Biotechnol 42: 1571–1580. 10.1038/s41587-023-02024-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8: 90. 10.1186/1742-4690-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulovari A, Li R, Audano PA, Porubsky D, Vollger MR, Logsdon GA, Human Genome Structural Variation Consortium, Warren WC, Pollen AA, Chaisson MJP, et al. 2019. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc Natl Acad Sci 116: 23243–23253. 10.1073/pnas.1912175116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanudisastro HA, Deveson IW, Dashnow H, MacArthur DG. 2024. Sequencing and characterizing short tandem repeats in the human genome. Nat Rev Genet 25: 460–475. 10.1038/s41576-024-00692-3 [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Chhetri SB, Tassia MG, Biddanda A, Yan SM, Wojcik GL, Battle A, McCoy RC. 2024. Sources of gene expression variation in a globally diverse human cohort. Nature 632: 122–130. 10.1038/s41586-024-07708-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretina K, Park ES, Maminska A, MacMicking JD. 2019. Interferon-induced guanylate-binding proteins: guardians of host defense in health and disease. J Exp Med 216: 482–500. 10.1084/jem.20182031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twesigomwe D, Drögemöller BI, Wright GEB, Adebamowo C, Agongo G, Boua PR, Matshaba M, Paximadis M, Ramsay M, Simo G, et al. 2023. Characterization of CYP2D6 pharmacogenetic variation in sub-Saharan African populations. Clin Pharmacol Ther 113: 643–659. 10.1002/cpt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twesigomwe D, Drögemöller BI, Wright GEB, Adebamowo C, Agongo G, Boua PR, Matshaba M, Paximadis M, Ramsay M, Simo G, et al. 2024. Characterization of CYP2B6 and CYP2A6 pharmacogenetic variation in sub-Saharan African populations. Clin Pharmacol Ther 115: 576–594. 10.1002/cpt.3124 [DOI] [PubMed] [Google Scholar]
- Wagner J, Olson ND, Harris L, Khan Z, Farek J, Mahmoud M, Stankovic A, Kovacevic V, Yoo B, Miller N, et al. 2022. Benchmarking challenging small variants with linked and long reads. Cell Genomics 2: 100128. 10.1016/j.xgen.2022.100128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. 2021. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol 39: 1348–1365. 10.1038/s41587-021-01108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Antonacci-Fulton L, Howe K, Lawson HA, Lucas JK, Phillippy AM, Popejoy AB, Asri M, Carson C, Chaisson MJP, et al. 2022. The Human Pangenome Project: a global resource to map genomic diversity. Nature 604: 437–446. 10.1038/s41586-022-04601-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman A, D'haene E, Baetens M, Yankee TN, Winchester EW, Glidden N, Roets E, Van Dorpe J, Janssens S, Miller DE, et al. 2024. A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development. Nat Commun 15: 136. 10.1038/s41467-023-44506-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik MH, Reuter CM, Marwaha S, Mahmoud M, Duyzend MH, Barseghyan H, Yuan B, Boone PM, Groopman EE, Délot EC, et al. 2023. Beyond the exome: what's next in diagnostic testing for Mendelian conditions. Am J Hum Genet 110: 1229–1248. 10.1016/j.ajhg.2023.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Metzger GA, Padilla Del Valle R, Delgadillo Rubalcaba D, McLaughlin RN. 2024. Evolutionary insights from profiling LINE-1 activity at allelic resolution in a single human genome. EMBO J 43: 112–131. 10.1038/s44318-023-00007-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalusky MP, Miller DE. 2024. Methylation Operation Wizard (MeOW): identification of differentially methylated regions in long-read sequencing data. arXiv 10.48550/arXiv.2402.17182 [DOI] [Google Scholar]
- Zanger UM, Schwab M. 2013. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138: 103–141. 10.1016/j.pharmthera.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Zhao X, Collins RL, Lee W-P, Weber AM, Jun Y, Zhu Q, Weisburd B, Huang Y, Audano PA, Wang H, et al. 2021. Expectations and blind spots for structural variation detection from long-read assemblies and short-read genome sequencing technologies. Am J Hum Genet 108: 919–928. 10.1016/j.ajhg.2021.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Li S, Su J, Leung AW-S, Lam T-W, Luo R. 2022. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Nat Comput Sci 2: 797–803. 10.1038/s43588-022-00387-x [DOI] [PubMed] [Google Scholar]
- Zook JM, Hansen NF, Olson ND, Chapman L, Mullikin JC, Xiao C, Sherry S, Koren S, Phillippy AM, Boutros PC, et al. 2020. A robust benchmark for detection of germline large deletions and insertions. Nat Biotechnol 38: 1347–1355. 10.1038/s41587-020-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.