Abstract
Mutations in splicing factor 3B subunit 1 (SF3B1) frequently occur in patients with chronic lymphocytic leukemia (CLL) and myelodysplastic syndromes (MDSs). These mutations have different effects on the disease prognosis with beneficial effect in MDS and worse prognosis in CLL patients. A full-length transcriptome approach can expand our knowledge on SF3B1 mutation effects on RNA splicing and its contribution to patient survival and treatment options. We applied long-read transcriptome sequencing (LRTS) to 44 MDS and CLL patients, as well as two pairs of isogenic cell lines with and without SF3B1 mutations, and found >60% of novel isoforms. Splicing alterations were largely shared between cancer types and specifically affected the usage of introns and 3′ splice sites. Our data highlighted a constrained window at canonical 3′ splice sites in which dynamic splice-site switches occurred in SF3B1-mutated patients. Using transcriptome-wide RNA-binding maps and molecular dynamics simulations, we showed multimodal SF3B1 binding at 3′ splice sites and predicted reduced RNA binding at the second binding pocket of SF3B1K700E. Our work presents the hitherto most-complete LRTS study of the SF3B1 mutation in CLL and MDS and provides a resource to study aberrant splicing in cancer. Moreover, we showed that different disease prognosises result most likely from the different cell types expanded during carcinogenesis rather than different mechanisms of action of the mutated SF3B1. These results have important implications for understanding the role of SF3B1 mutations in hematological malignancies and other related diseases.
Splicing is a fundamental step in eukaryotic gene expression in which noncoding introns are removed from premessenger RNA (pre-mRNA) transcripts and exons are joined to form mature mRNAs. This intricate process is often disrupted in cancer, either by mutations in spliceosomal genes or by other mechanisms that affect normal splicing function (Quesada et al. 2012; Seiler et al. 2018; Shiozawa et al. 2018; Yang et al. 2022; Bradley and Anczuków 2023). In turn, aberrant splicing can lead to changes in the composition of expressed isoforms and the formation of new isoforms that alter the encoded proteins and can have far-reaching consequences for cellular function. One striking example of splicing alterations in cancer are mutations in the gene encoding the splicing factor 3B subunit 1 (SF3B1) that have divergent ramifications for treatment efficiency and prognosis (Papaemmanuil et al. 2011; Rossi et al. 2011). Somatic SF3B1 mutations are frequently found in myelodysplastic syndrome (MDS; 20%), chronic lymphocytic leukemia (CLL; 15%), acute myeloid leukemia (3%), uveal melanoma (20%), cutaneous melanoma (4%), and prostate cancer (1%) and in 2% of all breast, pancreatic, and lung cancers (Bland et al. 2023).
In CLL patients, SF3B1 mutations are typically subclonal and have been linked to disease progression and shorter survival (Wan and Wu 2013; Landau et al. 2015). On the other hand, SF3B1 mutations in MDS patients have been associated with specific disease phenotypes that show erythroid dysplasia with ring sideroblasts and ineffective erythropoiesis (Malcovati et al. 2015). Unlike in CLL patients, a positive effect of the SF3B1 mutation on survival has been observed in almost all groups of MDS patients, except those with excess blasts, for whom no significant effect has been observed (Malcovati et al. 2020). However, so far there is no explanation for the divergent ramifications of SF3B1 mutations in CLL and MDS pathology.
Pre-mRNA splicing is a highly dynamic process, and the spliceosome undergoes several structural changes from the E to the A, B, and C complex during splicing. SF3B1 is part of the spliceosome and plays a critical role in 3′ splice-site usage. As a subunit of the U2 small nuclear ribonucleoprotein complex (snRNP), SF3B1 is UV cross-linked with the pre-mRNA on both sides of the branch-point (BP) adenosine in the A-complex, at nucleotide positions −6 and +5 (Gozani et al. 1996, 1998). The most common mutations in SF3B1 accumulate in the Huntington, Elongation Factor 3, PR65/A, TOR (HEAT) domain at its C terminus (Supplemental Fig. S1). The HEAT domain consists of 20 nonidentical HEAT repeats that form the RNA-binding interface (Cretu et al. 2016). These mutations are predicted to impact the N-terminal domain involved in complex formation with other splicing factors (Canbezdi et al. 2021). Mutations in SURP and G-patch domain containing 1 (SUGP1) mimic the splice alterations of mutant SF3B1 (Liu et al. 2020; Alsafadi et al. 2021), and mutations in DHX15 partially recapitulate the splicing alterations of mutant SF3B1 (Zhang et al. 2022). Both proteins have been shown to bind less to mutated SF3B1 (Zhang et al. 2019).
Previous studies based on short-read RNA sequencing (RNA-seq) have reported alternative 3′ splice-site usage (3′AS) and intron retention (IR) as the most prominent splicing alterations in CLL and MDS patients with mutated SF3B1 (DeBoever et al. 2015; Wang et al. 2016; Kesarwani et al. 2017; Shiozawa et al. 2018; Tang et al. 2020). The alternative 3′ splice sites (referred to as AG′) that were preferably used upon SF3B1 mutation are enriched at ∼20 nucleotides (nt) upstream of the canonical splice sites (AG) (DeBoever et al. 2015; Obeng et al. 2016; Wang et al. 2016; Tang et al. 2020). This strong positional constraint suggested that the mutations impacted SF3B1 binding and BP recognition upstream of the 3′ splice sites. Additionally, it was proposed that the mutation promotes the usage of otherwise inaccessible AG′ within the RNA secondary structure (Kesarwani et al. 2017). Despite these hypotheses, the exact mechanism of the effect of mutations in SF3B1 is still not resolved.
Here, we aimed to comprehensively characterize the effects of SF3B1 mutations in cancer using long-read transcriptome sequencing (LRTS) and combined complementary data derived from MDS and CLL patients with isogenic cell lines.
Results
Long-read RNA-seq expands patient transcriptome landscapes in divergent biological contexts
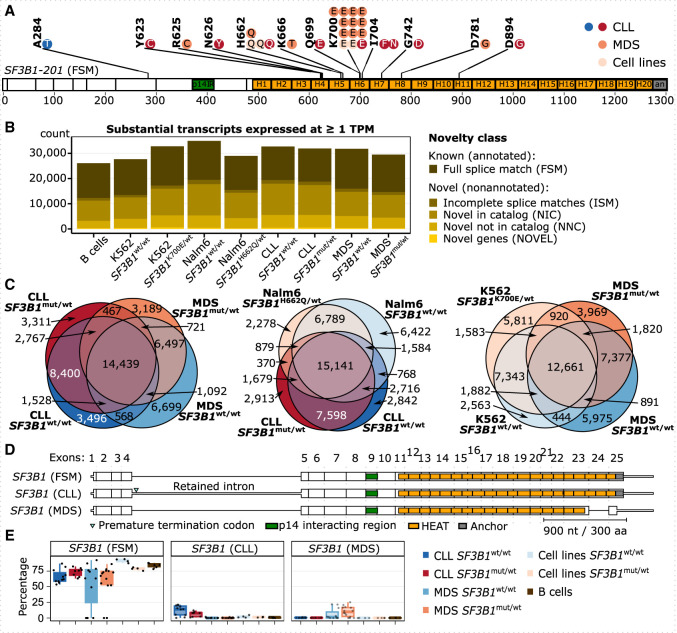
To investigate the effect of SF3B1 mutations on splicing, we characterized the transcriptomes of three data sets: CLL patients, MDS patients with ring sideroblasts, and isogenic cell lines with or without somatic SF3B1 mutations (Supplemental Fig. S2). In brief, we collected CLL cells or whole-blood samples from 19 CLL and 25 MDS patients, including eight CLL patients and 14 MDS patients with mutations in the SF3B1 HEAT repeat domain (Fig. 1A). These were complemented by two isogenic leukemia cell line pairs (K562 and Nalm6), both with SF3B1wt/wt and SF3B1mut/wt. K562 cells originated from a patient with chronic myeloid leukemia (CML), and the SF3B1mut/wt cells carried K700E the mutation, whereas the Nalm6 cells originated from a patient with B cell acute lymphoblastic leukemia (B-ALL), and the SF3B1mut/wt cells carried the H662Q mutation. As controls, we further included B cells from six healthy donors (Supplemental Table S1). The RNA expression level of mutated SF3B1 ranged from 14% to 52% (43% on average) in patients, 43% in K562, and 29% in the Nalm6 cell line (Supplemental Table S1). To detect complete transcript isoforms, we performed LRTS using Iso-Seq (Pacific Biosciences). We reached a mean of 582,135 full-length nonchimeric reads per sample, cumulating in a total of 33,763,806 reads with an average length of 2721 bp (Supplemental Table S1). Only 9% of the reads were potentially affected by technology-specific technical artifacts (Supplemental Figs. S3, S4; Cocquet et al. 2006).
Figure 1.
Long-read sequencing of CLL and MDS patient samples discovers novel isoforms. (A) Distribution of SF3B1 mutations in CLL and MDS patient samples used for Iso-Seq; each dot represents a mutated sample. One CLL patient is marked twice owing to two mutations (I704N and D894G). Note that the A284T mutation is outside the HEAT repeat domains and thus was grouped as a wild-type sample, also according to further analysis described below. SF3B1 is shown as the major isoform expressed, with the full splice match (FSM) to annotated isoform 201. (B) The number of substantial transcripts identified in each group and expressed at the level of at least one transcript per million (TPM) colored by the category of isoform novelty: with FSM, with incomplete splice matches (ISMs), with combinations of annotated splice junctions (novel in catalog [NIC]), with at least one novel splice site (novel not in catalog [NNC]), or from novel genes (NOVEL) (Tardaguila et al. 2018). (C) Venn diagrams showing the overlap between isoforms from B expressed at one or more TPM in each group. (D) SF3B1 isoforms expressed at >10% relative expression level. (E) Relative expression levels of SF3B1 isoforms from D.
In total, we identified 89,659 substantially expressed transcripts that contributed to at least 1% to a gene's total expression and were covered by at least five full-length reads (Supplemental Fig. S5). Almost one-third of these reads (28,261; 31.5%) were classified as full splice matches (FSMs) to annotated isoforms. Moreover, 58,168 (64.9%) represented novel isoforms that only partially overlapped with gene annotations, and 3230 (3.6%) reads originated from nonannotated, novel genes (Supplemental Fig. S6). Even for transcripts expressed at one or more transcript per million (TPM), the novel isoforms consisted of more than half of all transcripts detected (Fig. 1B). A large fraction of isoforms was shared by the different patient cohorts and isogenic cell lines, with a larger overlap of expressed isoforms between SF3B1mut/wt and SF3B1wt/wt of the same data set than between data sets (Fig. 1C; Supplemental Fig. S7).
The SF3B1 gene has multiple isoforms annotated and was indeed expressed in several isoforms in both samples with or without SF3B1 mutations (Fig. 1D; Supplemental Fig. S8). Although the most frequently expressed SF3B1-FSM isoform (∼70% of SF3B1 transcripts) fully corresponded to the annotated isoform, two shorter novel isoforms showed a disease-specific expression almost exclusively in either CLL or MDS patients. These contributed ∼10% each to the gene's overall expression, irrespective of the SF3B1 mutational status (Fig. 1E). The CLL-specific isoform (SF3B1-CLL) showed retention of the fourth intron, which introduced a premature termination codon (PTC) and likely targeted the isoform for nonsense-mediated mRNA decay (NMD). In the MDS-specific isoform (SF3B1-MDS), the penultimate exon was skipped and induced a frameshift that probably resulted in an NMD-resistant isoform that encoded for a C-terminally truncated protein devoid of HEAT repeats 18–20 and the anchor domain. In addition to the divergent splicing pattern, SF3B1 also showed three times higher expression in CLL compared with MDS patients, whereas its levels were reduced in MDS patients compared with B cells from healthy donors (Supplemental Fig. S8).
Overall, our results demonstrated a high transcriptome information content in the patient cohorts, which was dominated by a large number of novel transcripts.
Patients and cell lines with SF3B1 mutations show similar splicing defects
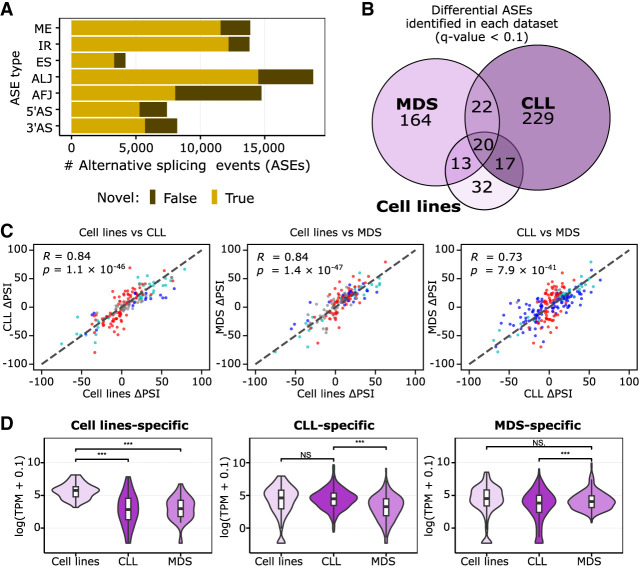
To investigate the transcriptome diversity at the splice-site level, we used the recently developed IsoTools software (Lienhard et al. 2023) to identify alternative splicing events (ASEs) in the transcripts expressed. IsoTools distinguishes exon skipping (ES), IR, mutually exclusive exons (MEs), and 5′ and 3′ alternative splice-site (5′AS and 3′AS) events, as well as alternative first and last junctions (AFJs and ALJs). Using a cut-off of 100 or more reads, ASEs were quantified as the proportion of reads supporting the ASE in relation to the sum of reads for all transcript isoforms, referred to as percentage spliced-in index (PSI). This threshold is motivated from extensive testing to optimize the detection of true splicing events while minimizing false positives and also ensures a sufficient number of individual samples (five or more) supporting the vast majority of ASEs (Supplemental Fig. S9). Across all samples, we discovered 80,995 ASEs in 9746 genes, for which the less-expressed ASEs made up for at least 10% of the reads. For 75% of these events, at least one of the alternatives was not annotated (novel event) (Fig. 2A).
Figure 2.
SF3B1 mutation effect is independent of the biological background, but its manifestation depends on the transcriptomic profile. (A) The number of alternative splicing events (ASEs) identified with Iso-Seq separated by splicing event type in all groups investigated, differentiated by the novelty class. (B) Overlap between significantly altered ASEs in samples with the SF3B1 mutation identified in the three data sets used (cell lines, CLL patients, or MDS patients). (C) Correlation of isoform usage measured by the difference in PSI from all events listed in B; namely, events called significant in at least one of the three data sets are shown. The colors of the dots correspond to significance reached only in one set: blue indicates x-axis only; red, y-axis only; light blue, both; and gray, none (i.e., called significant in a data set absent from the graph). Pearson correlation coefficient (R) and associated P-value (P) are given. (D) Violin plots with boxplots show the distribution of expression values of the genes with data set–specific ASEs from C. Significant differences are marked with as follows: (***) paired, two-tailed Student's t-test P-value < 0.001, (**) P-value < 0.01, (*) P-value < 0.05, (N.S.) not significant with P-value ≥ 0.05.
Next, we used IsoTools (Lienhard et al. 2023) to detect significant differences in splicing associated with SF3B1 mutations. Because SF3B1 mutations have been reported to convey either beneficial (MDS) or disadvantageous (CLL) effects on patient survival (Papaemmanuil et al. 2011; Rossi et al. 2011), we first tested for differential splicing in SF3B1mut/wt versus SF3B1wt/wt samples, separately in each data set. We detected 82, 288, and 219 ASEs in the isogenic cell lines, CLL, and MDS patients, respectively (adjusted P-value [Q-value] with false-discovery rate [FDR] <10%) (Supplemental Table S2; Benjamini and Hochberg 1995). Although we observed only a moderate overlap of the identified events between the data sets (Fig. 2B; Supplemental Fig. S10), to our surprise, the correlation of the PSI changes for the ASEs identified was high (Fig. 2C), indicating a common mutational effect. We found that the genes altered by the disease-specific ASE were generally expressed significantly higher in the corresponding group of patients (Fig. 2D; Supplemental Fig. S11), and out of the union of 531 genes with an ASE, 149 were relatively higher in MDS and 155 were higher in the CLL samples by at least twofold (FDR < 1%) based on the Iso-Seq data. Overall, about two-thirds of ASEs called in the MDS or CLL data set separately were significantly differentially expressed (FDR < 5%) when comparing MDS to CLL (Supplemental Fig. S12; Supplemental Table S3).
Our findings suggested that although SF3B1 mutations introduced shared splicing effects in both CLL and MDS patients, the divergence in the disease outcome could be attributed to the differential transcriptomic profiles. Specifically, the mutation seemed to exert its most potent effects on genes that were already dominantly expressed in each disease.
SF3B1 mutations affect 3′AS usage and IR
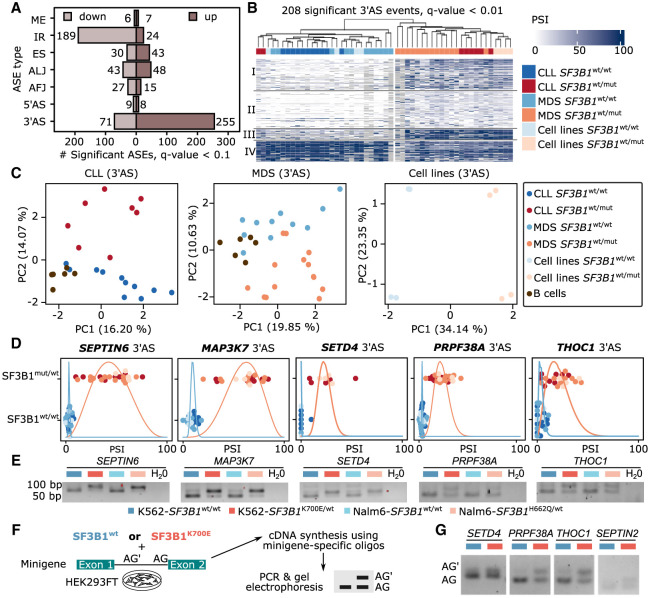
Because the individual analyses for patients and isogenic cell lines indicated a common effect of SF3B1 mutations, we combined all SF3B1mut/wt and SF3B1wt/wt samples to increase the statistical power for an overall estimation of the SF3B1 mutational impact. In total, we identified 775 differential splicing events in 530 different genes (Fig. 3A; Supplemental Table S4). As reported previously (Darman et al. 2015; DeBoever et al. 2015; Wang et al. 2016; Kesarwani et al. 2017; Shiozawa et al. 2018; Tang et al. 2020), the splicing changes upon SF3B1 mutation were strongly enriched for 3′AS (326, 42%) and IR (213, 27%) events, which together accounted for more than two-thirds of the significant changes. The majority of IR events showed decreased IR (89%), whereas the majority of 3′AS events (78%) showed higher PSI values, corresponding to longer exons in SF3B1mut/wt (Fig. 3B,C; Supplemental Fig. S13; Supplemental Table S4). Consistent with the common mutational effect, the regulated 3′AS events showed a uniform response across the cohorts and clustered SF3B1mut/wt and SF3B1wt/wt samples (Fig. 3B).
Figure 3.
SF3B1 mutation increases 3′ alternative splice site (3′AS) usage and decreases intron retention. (A) Number of differential ASEs with shorter or longer variant expression (percentage spliced index [PSI]) in SF3B1mut/wt versus SF3B1wt/wt samples. (B) Highly significantly altered (Q-value < 0.01) 3′ASs clearly separate samples by SF3B1 mutations in leukemia cell lines as well as CLL and MDS patients based on the longer variant PSI values showing four clusters described in Supplemental Figure S19. (C) Principal component (PC) analysis, based on the isoform usage of 3′ASs, clearly separates CLL and MDS patients, as well as cell lines, according to the SF3B1 mutational status. (D) Swarm plots showing the distribution of the isoform usage (PSI) among groups with or without SF3B1 mutation. (E) Validation of the differential splicing associated with SF3B1 mutation with RT-PCR experiments in isogenic K562 and Nalm6 cell lines. (F) Minigene assays workflow. HEK293T cells were cotransfected with minigenes and either SF3B1wt or SF3B1K700E for 48 h. RNA was extracted and used for amplification of splicing products with minigene-specific primers. (G) The results from the minigene assay from F. The lower band in the agarose gel corresponds to the usage of the canonical AG, the upper band to the upstream AG′.
Upon closer inspection, we found that two CLL-SF3B1mut/wt patient samples with mutations in the HEAT domain clustered with the SF3B1wt/wt samples when a subset of ASEs was used (208 ASEs with Q-value < 0.01). One of these patients carried a rare SF3B1 mutation, Q699E, that had so far been reported only once, in a single patient with bladder urothelial carcinoma included in the TCGA Pan-Cancer Atlas (according to cBioPortal [https://www.cbioportal.org/] accessed on October 16, 2023). Moreover, overexpression of the SF3B1-Q699H construct in the HEK293FT cells did not lead to any aberrant splicing, suggesting that this mutation is weakly pathogenic (Darman et al. 2015). The second patient carried two mutations: D894G in the HEAT repeat 11 (allele frequency [AF] = 51%) and I704N in the HEAT repeat 6 (AF = 18%) (Supplemental Table S1). Although the mutations in these two patients might have a weaker effect on the global splicing, we decided to keep the samples in our analysis to allow for more biological variance in the statistical analysis and detect stronger signals. For all other mutations, the position within the SF3B1 HEAT domain had no discernible influence on the clustering, indicating that the different mutations impair SF3B1 similarly (Fig. 3B). In fact, using general splicing information (PSI values), irrespective of regulation, we found that 3′AS events, but no other type of ASEs, clearly differentiated the samples based on the SF3B1 mutational status in an unsupervised principal component analysis (PCA) performed on each data set (Fig. 3C; Supplemental Figs. S14, S15), underlining the predominant effect of SF3B1 mutation on 3′AS events.
In addition to many new differential ASEs, previously published discoveries could be confirmed, including for example, five from 35 3′AS events (in SEPTIN2, ERGIC3, RHNO1, FDPS, and SNRPN) identified in CLL based on Nanopore sequencing (Tang et al. 2020), a 3′AS in SEPTIN6 reported in MDS-SF3B1mut/wt patients (Supplemental Fig. S16; Dolatshad et al. 2016), and 13 3′AS events (BCL2L1, COASY, DPH5, DYNLL1, EI24, ERGIC3, MED6, METTL5, SERBP1, SKIV2L, TMEM14C, ZBED5, and ZDHHC16) that were consistently found in CLL, MDS, and uveal melanoma patients (Pellagatti et al. 2018; Inoue et al. 2019). Moreover, we confirmed 13 from 32 (Zhou et al. 2020) and eight from 11 genes (Liu et al. 2020) reported as aberrantly spliced in either MDS or CLL patients.
The LRTS data opened the possibility to assess the splicing alterations in the context of complete transcript isoforms. Generally, we found that the effect of the SF3B1 mutation on splicing did not influence the choice of transcript start or end sites or the probability of other splicing events of the same gene. This means that the effect was local, and the resulting alternative transcript corresponded to the canonical transcript, except for the single alternative event. This is exemplified by the SF3B1mut/wt-induced inclusion of the poison cassette exon (PCE) in the BRD9 gene that introduces a PTC (Supplemental Figs. S17, S18; Inoue et al. 2019). Of note, the full-length reads allowed us not only to confirm differential splicing of this PCE but also to locate it to a specific isoform that has not been annotated yet (NIC class). This long-read-derived novel isoform otherwise resembled the canonical BRD9 isoform, whereas the annotated PTC isoforms were presumed to also harbor an alternative first exon and additional splicing alterations. Such incomplete and incorrect isoform annotations are likely to cause problems in quantifying transcriptome changes, especially when using short-read sequencing data.
Notably, we found multiple splicing factors among the genes affected by SF3B1 mutations. Overrepresentation analysis revealed 18 from the spliceosome pathway (KEGG: 03040, Q-value = 2.8 × 10−3) (Supplemental Table S4). This indicated a broad impact of SF3B1 mutations on the general splicing machinery, which could potentially lead to secondary effects on splicing. Taking a closer look at the 208 highly significant 3′AS events (Q-value < 0.01), we identified four clusters based on PSI values detected in SF3B1mut/wt and SF3B1wt/wt (Fig. 3B; Supplemental Fig. S19). The cluster II with low PSI values in SF3B1mut/wt and no expression in SF3B1wt/wt was enriched in spliceosome (Q-value = 0.0106) and cell cycle genes (GO: 0007049 Q-value = 2.8 × 10−4) (Supplemental Fig. S20).
To independently validate the detected splicing changes, we performed short-read RNA-seq on the isogenic cell line pairs and on 27 CLL patient samples, which included the same 19 CLL samples used for Iso-Seq, and collected publicly available RNA-seq data from 398 MDS patients (Supplemental Table S1; Supplemental Fig. S21). Although we detected more events using rMATS (Supplemental Fig. S22, bottom; Supplemental Table S4; Shen et al. 2014), we observed a high and significant (P-values < 0.001) correlation of PSI values in the ASEs detected with IsoTools (Lienhard et al. 2023) and rMATS (Shen et al. 2014) on the same cell lines (Pearson correlation coefficient R = 0.840, P-value = 2.47 × 10−17). The same held true for the samples from CLL and MDS patients (R = 0.794 with P-value = 2.95 × 10−36 and R = 0.800 with P-value = 3.60 × 10−24, respectively) (Supplemental Fig. S23).
As an orthogonal approach, we employed semiquantitative reverse-transcription PCR (RT-PCR) to test 15 differential 3′AS events in the isogenic cell line pairs (Supplemental Fig. S24; Supplemental Table S5). From these 15 tested, 12 (80%) clearly showed an increase in alternatively spliced isoform expression in the SF3B1mut/wt conditions. The strongest effects were observed for 3′AS events in MAP3K7, SEPTIN6, and SETD4, which showed an almost complete switch to the alternative splicing variant (Fig. 3D,E). We additionally performed splicing reporter assays using minigenes for six 3′AS events in HEK293T cells (Fig. 3F). Indeed, we observed differential 3′AS usage upon ectopic SF3B1K700E expression in four out of six minigenes tested (SETD4, PRPF38A, THOC1, and SEPTIN2) (Fig. 3G), supporting that the effect of the SF3B1 mutation persists in an unrelated cell line. In the two remaining cases (TPP2 and BRCA1), the usage of the upstream AG′ was already low in the validation assays using the K562 and Nalm6 cell line pairs (Supplemental Fig. S24).
Overall, these results supported a common effect of SF3B1 mutations in different biological backgrounds and confirmed their predominant impact on 3′AS usage and IR.
Computational prediction of protein function and stability of individual splicing isoforms
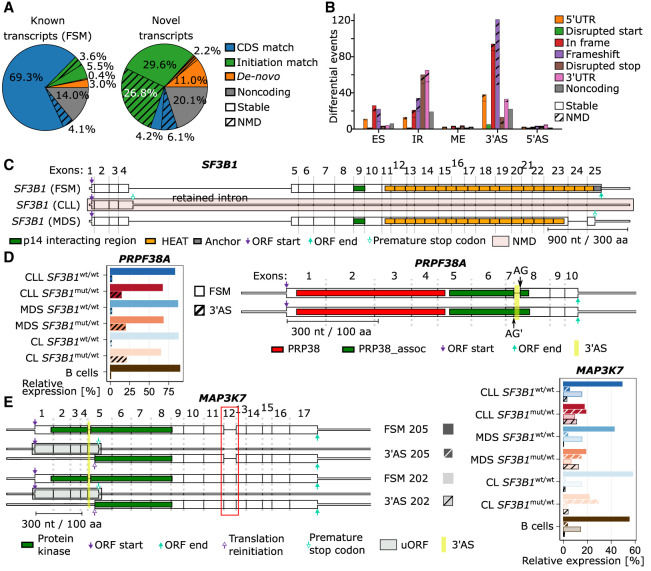
Further on, we used the LRTS information on full-length transcripts to predict the potential coding sequences (CDSs) of the transcripts (Supplemental Methods). Among the 28,261 known transcripts (FSM), we found that 73.4% had a matching reference CDS (Fig. 4A). In contrast, the majority of the 58,168 novel transcripts (89.7%) did not match a CDS and either lacked an open reading frame (ORF; 20.1%), initiated from an unannotated start codon (13.2%), or began at an annotated initiation site but deviated from the reference CDS owing to alternative splicing (56.4%). Additionally, we observed that 35.1% of the expressed novel transcripts were likely to be targeted by NMD compared with only 8.2% among the known transcripts (Fig. 4A).
Figure 4.
SF3B1 mutation results in altered mRNAs potentially translated into modified proteins. (A) Coding potential of the known and novel isoforms identified divided by CDS similarity to annotated isoforms and NMD prediction. (B) Effect of SF3B1mut-associated ASEs on the protein-coding potential. (C) SF3B1 isoforms detected in this study with CLL- or MDS-specific isoforms. (D) The PRPF38A isoform expression levels (left) and structure with Pfam domains indicated (right). Highlighted in yellow is the SF3B1mut-associated 3′AS that may influence the protein function. (E) Schematic of major MAP3K7 isoforms (left) with the protein kinase domain showed as green boxes. The SF3B1mut-associated 3′AS is highlighted in yellow, and ORF start/end are indicated by arrows. Highlighted in light green are predicted upstream ORFs (uORFs). Red box highlights the additional exon 12 in the isoform 202, which is absent in the isoform 205. Expression of each isoform is shown on the right. The expression of isoforms with 3′AS event is shown as striped bars.
We next determined how SF3B1 mutation-induced alternative splicing impacts the coding potential and the function of the proteins. To this end, we classified ASEs into categories based on their relative location and the impact on the CDS: 5′ UTR, disrupted start codon, in-frame, frameshift, disrupted stop codon, and 3′ UTR. Of the 326 events featuring 3′AS, the majority led to either frameshift modifications (121 events) or in-frame changes (94 events) in the CDS (Fig. 4B). In total, we identified 274 ASEs predicted to yield stable alternative proteins that are not predicted to undergo NMD.
We further examined more thoroughly the functional consequences of the two novel SF3B1 isoforms, one predominantly expressed in CLL and one in MDS (Fig. 1D,E). In the SF3B1-CLL transcript, the fourth intron was retained, which contained a PTC that shortened the CDS to 465 nt. Our prediction showed a strong signal for NMD owing to the presence of 19 downstream exon–exon junctions. In contrast, in the SF3B1-MDS transcript, the penultimate exon was skipped and a frameshift was introduced and, subsequently, a PTC. However, because this PTC was located within the last exon, the SF3B1-MDS transcript was unlikely to be targeted by NMD and should result in a protein product missing its C-terminal section, that is, HEAT domains 18–20 and the terminal anchor domain (Fig. 4C).
To further investigate the functional outcomes of the altered proteins, we examined the impact on protein domain levels by aligning Pfam (Mistry et al. 2021) domains to the predicted protein sequences. For 57 of the ASEs, we found at least one Pfam domain that overlapped the divergent part of the protein sequence, indicating partially altered protein functions (Fig. 4D; Supplemental Table S4), and we found evidence that some aberrantly spliced transcripts may induce a translations reinitiation event (Fig. 4E; Supplemental Fig. S25). We show this as an example for MAP3K7, a frequently described gene with an ASE in SF3B1mut/wt (DeBoever et al. 2015; Wang et al. 2016; Shiozawa et al. 2018).
The effect of SF3B1 mutations depends on the distance and sequence context of 3′ASs
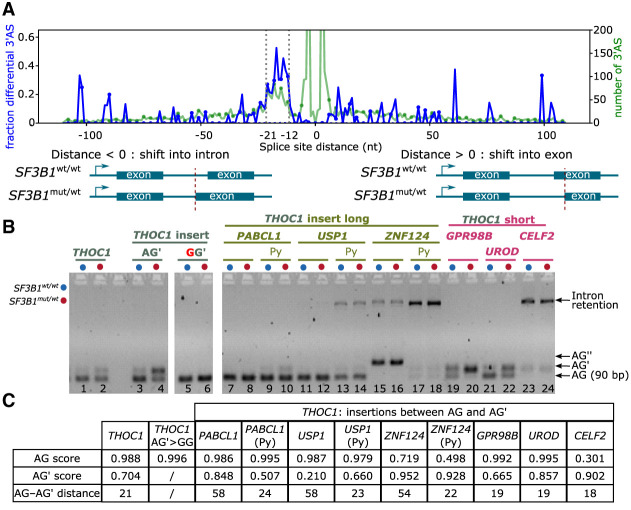
When we plotted the fraction of differential 3′AS against the splice-site differences, we noticed, consistent with previous findings, that the alternative splice sites of differential 3′AS events were enriched within 12–21 nt upstream of the canonical splice site (AG) mainly used in the SF3B1wt/wt samples (Fig. 5A; DeBoever et al. 2015; Obeng et al. 2016; Wang et al. 2016; Kesarwani et al. 2017; Tang et al. 2020). Within this range, 30.8% of 3′ASs were significantly differentially used in SF3B1mut/wt compared with 1.8% outside this range. To investigate the significance of the AG′–AG distance and sequence context for the differential splicing event, we constructed a minigene assay with a part of the THOC1 transcript harboring the significantly affected 3′AS. The insert was then modified by replacing the 21 nt fragment between the AG′ and AG of THOC1 with 45 nt to 50 nt AG′–AG fragments from alternatively but nondifferentially spliced introns (PABCL1, USP1, ZNF124) (Supplemental Fig. S26). As a control, we mutated AG′ to GG′ to disrupt any alternative splicing. (Fig. 5B). These experiments suggested that increasing the AG′–AG distance was sufficient to remove the 3′AS from SF3B1 regulation. We also found that specific sequences, such as the BP region upstream of AG′, were responsible for the differential splicing between SF3B1wt- and SF3B1K700E-expressing cells and confirmed that a strong AG is required for AG′ usage (Fig. 5C; for details, please see Supplemental Notes; Darman et al. 2015).
Figure 5.
SF3B1 mutations promote upstream 3′ASs and partially dependent on the sequence context. (A) 3′AS distance distribution. Negative distances indicate an alternative was located upstream, and positive values indicate an alternative located downstream, leading to a shorter exon. Blue line represents proportion; green, the total number of 3′ASs. Dotted vertical lines indicate the enriched region of 12–21 nt upstream of the canonical AG. (B) Minigene assays with long (45–50 bases) AG– AG′ inserts, shortened inserts containing ∼20 nt directly upstream the AG including the polypyrimidine (Py) tract, and short (15–20 nt) AG–AG′ inserts from nondifferentially alternatively spliced 3′AS events. The chosen events without differential splicing detected with SF3B1 mutation were from PAPCL1, USP1, and ZNF124 (AG′–AG distance >50 nt) as well as GPR98B, UROD, and CELF2 (AG′–AG distance <20 nt). (C) Table showing splice-site strength for AG and AG′ calculated with SpliceRover (Zuallaert et al. 2018).
Our results confirmed that mutations in SF3B1 primarily affected proximal AG′, but the AG′–AG distance did not seem to be the sole factor required for the usage of AG′. Moreover, we did not find any motif enriched at this position that could indicate a binding of another protein potentially disrupting or competing with SF3B1. We therefore speculated that SF3B1 binding at the sites with ASE may be altered in patients carrying SF3B1 mutation.
K700E mutation may lead to destabilization of SF3B1-mRNA binding
To scrutinize the effect of the most common SF3B1 mutation, K700E, we performed molecular dynamics (MD) simulations of 20 transcripts with a 3′AS within 50 nt distance, including 14 transcripts that were differentially spliced between SF3B1mut/wt and SF3B1wt/wt and six nondifferentially spliced transcripts (Supplemental Methods). For each transcript, we performed four replicas of 200 ns MD simulations of the mRNA with (1) the BP of the downstream AG (BP) bound to SF3B1wt, (2) the BP of the upstream AG (BP′) bound to SF3B1wt, (3) the BP bound to the SF3B1K700E mutant, and (4) the BP′ bound to the SF3B1K700E mutant. There were no differences in BP binding between all transcript–protein combinations in the first binding pocket (Supplemental Fig. S27A–C), whereas the frequency of the interaction in the second binding pocket decreased in SF3B1K700E owing to repulsive interactions of like-charged atoms (Supplemental Methods; Supplemental Fig. S27A,D,E).
To investigate whether this decrease in interactions is associated with an increase in mRNA mobility, we computed the per-residue root mean square fluctuations (RMSF) of the mRNA nucleobases. Indeed, the mobility of nucleobases significantly increased for nucleobases in the 3′ direction after the K700 binding site for SF3B1K700E compared with SF3B1wt, when the BP′ was bound to SF3B1 (Supplemental Figs. S27F, S28–S32).
Taken together, these results indicated that the K700E mutation did not lead to differences in the BP recognition. However, the mutation led to significant differences in SF3B1-mRNA contacts within the second mRNA-binding pocket at least for the 20 mRNAs tested with an upstream alternative AG within 50 nt.
SF3B1 shows multimodal binding at 3′ splice sites
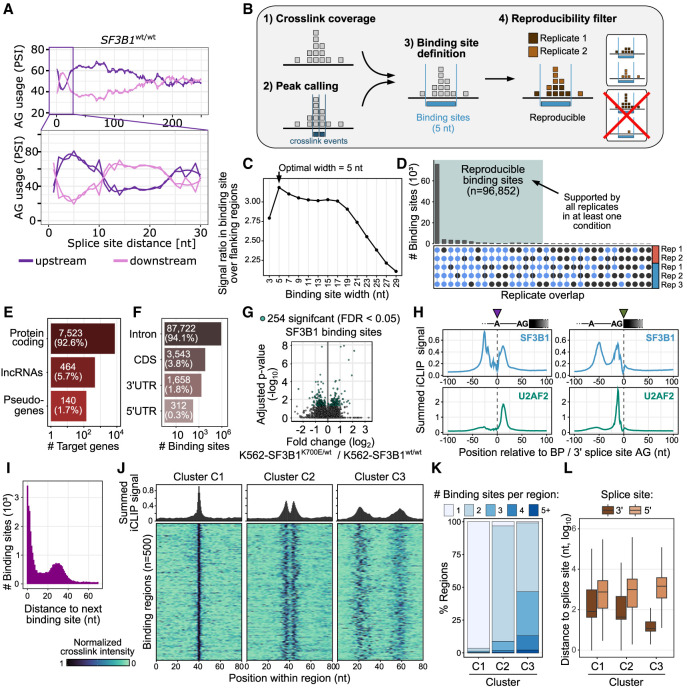
Our splicing analyses showed that mutations in SF3B1 resulted in the activation of alternative splice sites 12 to 21 nt upstream of the canonical AG (Figs. 5A, 6A). To understand how SF3B1 recognizes these sites, we performed individual-nucleotide resolution UV cross-linking and immunoprecipitation (iCLIP) to map SF3B1 binding sites throughout the transcriptome (König et al. 2010). After UV cross-linking, we immunoprecipitated SF3B1 from K562-SF3B1wt/wt and K562-SF3B1K700E/wt cells, yielding together more than 100 million SF3B1 cross-link events (Fig. 6B; Supplemental Table S1). To facilitate direct comparisons, we randomly subsampled the sequencing reads to adjust the library size of the replicates (see Methods). Based on the merged iCLIP data, we identified 96,852 SF3B1 reproducible binding sites with an optimal width of 5 nt (Fig. 6C,D). The binding sites occurred in 8127 genes, with the vast majority being protein-coding genes (93%). As expected, within the protein-coding transcripts, SF3B1 mostly bound introns (94%, Fig. 6E,F).
Figure 6.
Multimodal SF3B1 binding. (A) The SF3B1 choice of AG within a narrow window of 12–21 nt is strongly affected by the alternative splice-site distance. The AG usage is shown as the PSI as a function of the splice-site distance in SF3B1wt/wt. The rolling mean across 20 nt and the smoothed (loess method) trend line are shown for upstream (violet) and downstream (pink) AG. (B) Schematic workflow of processing iCLIP reads and calling SF3B1 binding sites. (C) Defining optimal site binding width. A binding site width of 5 nt optimally captures the SF3B1 cross-link events. Dot plot shows average ratio of cross-link events within binding sites of increasing widths (x-axis) over the mean background signal in flanking windows of the same size, indicating how much more signal occurs within the binding sites compared with their immediate surrounding. (D) SF3B1 binding sites reproducibility across replicates. Upper panel shows overlaps of supported binding sites in the replicates, with threshold for sufficient coverage individually adjusted to the signal depth in each replicate (Busch et al. 2020). (E) Gene classes targeted by SF3B1 based on iCLIP. (F) Transcript regions of protein-coding genes targeted by the SF3B1 based on iCLIP. (G) Differential SF3B1 binding sites in K562-SF3B1K700E/wt versus K562-SF3B1wt/wt. (H) Metaprofiles of SF3B1 (top) and U2AF2 (bottom) binding centered at branch-point adenosine (left) and 3′ splice-site AG (right). (I) Distribution of distances between neighboring binding sites. (J) Density plot and heat map showing SF3B1 patterns in regions from three cluster types identified in Supplemental Figure S35. (K) Distribution of the number of binding sites per region for each of the three clusters. (L) Distribution of the distance between SF3B1 binding region and closest splice site.
Because K562-SF3B1K700E/wt cells expressed both wild-type and mutated SF3B1 protein and both variants were recognized by the anti-SF3B1 antibody that specifically binds to the SF3B1 N terminus, we tested for differences in the SF3B1 binding between K562-SF3B1K700E/wt and K562-SF3B1wt/wt. Consistent with a recent study (Porter et al. 2021), the K700E mutation did not generally impair RNA binding (Supplemental Fig. S33). Moreover, at the level of binding sites, we detected only minor differences between K562-SF3B1K700E/wt and K562-SF3B1wt/wt (Fig. 6G). Thus, although subtle differences may have been masked by the overlay of both protein variants in the heterozygous cells, the K700E mutation does not obviously change the global RNA-binding behavior of SF3B1. However, local changes as predicted with the MD simulations might be too dynamic to be caught by the global iCLIP analysis.
Next, we examined SF3B1 binding at 3′ splice sites. Using metaprofiles, we detected two prominent peaks of SF3B1 binding (Fig. 6H). The two peaks were centered at about −50 nt and −10 nt upstream of the canonical 3′ splice site and surrounded the BP (Fig. 6H). Visual inspection indicated multiple SF3B1 binding sites within each peak (Supplemental Fig. S34). When centering the metaprofiles to the predicted BP adenosine, SF3B1 binds ∼25 nt upstream of and directly downstream from the predicted BP adenosine (Fig. 6H). The binding peak at −10 nt of the canonical 3′splice site, coincided with the Py-tract region bound by U2AF2 (Zarnack et al. 2013). To test this, we performed iCLIP experiments with U2AF2 in K562-SF3B1wt/wt cells, which confirmed that the −10 nt SF3B1 peak overlapped with U2AF2 binding (Fig. 6H). Together, these observations indicated that SF3B1 binds at both sites of the BP and that the binding site directly downstream from the BP encompasses the Py-tract.
Consistent with the two peaks of SF3B1 binding at 3′ splice sites, we found that SF3B1 binding sites frequently occurred at distances of ∼30 nt to each other (Fig. 6I). To globally classify the SF3B1 binding pattern, we merged adjacent binding sites into equal-sized binding regions (80 nt, 56,224 regions) (Supplemental Table S6) and performed unsupervised uniform manifold approximation and projection (UMAP) followed by density-based applications with noise (DBSCAN). This yielded three distinct clusters: Cluster C1 harbored mostly isolated binding sites (35,907 regions); cluster C2 included two closely spaced binding sites (5635 regions); and cluster C3 showed a more complex arrangement of three to four binding sites with wider spacing (13,847 regions) (Fig. 6J,K; Supplemental Fig. S35). The latter were located closest to 3′ splice sites (Fig. 6L), suggesting that multiple SF3B1 binding sites assemble into complex binding patterns at 3′ splice sites. We then investigated the differences in binding between K562-SF3B1K700E/wt and K562-SF3B1wt/wt solely in the C3 cluster. We noticed that K562-SF3B1K700E/wt showed a slight decrease in the left peak, which was more distal to the canonical AG (Supplemental Fig. S36). Within cluster C3, we noticed a slight decrease in binding of K562-SF3B1K700E/wt compared with K562-SF3B1wt/wt specifically in the left peak, which was more proximal to the canonical AG (Supplemental Fig. S36). This suggested that the K700E mutation led to change in the complex arrangement of SF3B1 binding sites at 3′ splice sites, preferentially affecting AG-proximal binding.
Altogether, our SF3B1 iCLIP data showed that SF3B1 adopted a multimodal mode of binding at 3′ splice sites, with two major peaks of SF3B1 binding that surround the BP. The peaks include multiple binding sites, which may reflect the dynamic binding rearrangements during the splicing process. The strong enrichment of this binding pattern at 3′ splice sites suggested that the defined arrangement of binding is required for SF3B1's function in splicing.
SF3B1 alternates within a small window of alternative splice-site distances, and the K700E mutation leads to the use of the proximal upstream AG
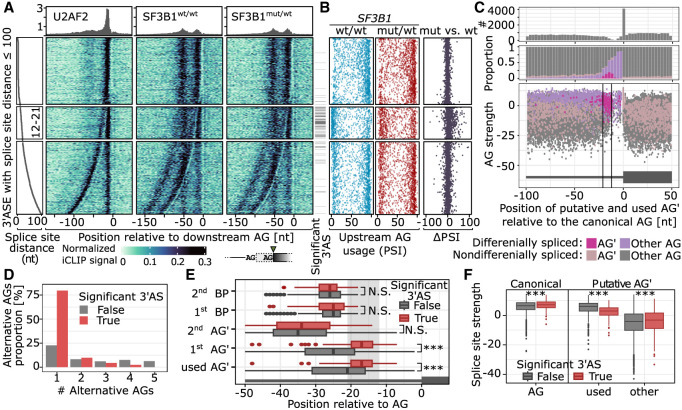
We and others (Darman et al. 2015; DeBoever et al. 2015; Alsafadi et al. 2016; Zhang et al. 2019) found that 3′ splice sites are particularly sensitive to SF3B1 mutations when they are directly preceded by an 3′AS. To test how this relates to binding, we overlaid the iCLIP data with the splicing quantifications from the same isogenic cell lines (K562-SF3B1wt/wt and K562-SF3B1K700E/wt). As shown above, the SF3B1 mutations showed most prominent effects on 3′ASs 12–21 nt upstream of the canonical 3′ splice site (Fig. 5A). At this distance, the upstream AG was at the border of the proximal peak of SF3B1 binding (Fig. 7A). Moving further away, the effects subsided drastically as soon as the upstream AG emerged from the proximal peak (Fig. 7B).
Figure 7.
SF3B1 promotes alternative proximal AG′ usage. (A) U2AF2 and SF3B1 binding to pre-mRNA based on the iCLIP signal. U2AF2 iCLIP was performed using K562-SF3B1wt/wt cells, and SF3B1 iCLIP was performed on K562-SF3B1wt/wt and K562-SF3B1K700E/wt. On the left panel, the splice-site distance is shown, followed by the iCLIP signal aligned to the more downstream AG used. The significance of the differential 3′AS usage between K562-SF3B1wt/wt and K562-SF3B1K700E/wt is shown as annotation bar on the right side of the iCLIP signal heatmap. (B) For every 3′AS from A, the upstream AG PSI value is denoted for SF3B1wt/wt (blue, left), SF3B1mut/wt (red, center), and the difference between SF3B1mut/wt and SF3B1wt/wt (black, right). (C) Distribution of AG occurrence (top), proportion of significantly alternatively used AG′ (middle), and AG′ scores among introns with significant (pink) or nonsignificant (violet) difference in usage between SF3B1mut/wt and SF3B1wt/wt (bottom). (D) Number of alternative AGs (AG′s) among regions with multiple AG′s. AG′s within 6 nt distance from AG were removed to avoid NAGNAG acceptor sites (Hiller et al. 2004). (E) Distance of AG′s and BPs from AGs among nonsignificant (gray) and significant (red) differentially splicing events. (F) Splice-site strength of canonical and alternative AGs as calculated with MaxEnt score (Yeo and Burge 2003). (A–F) Only 3′ASs with the following features were used: (1) placed chromosome scaffold, (2) classical AGs, (3) an intron became shorter in the mutant (the use of upstream alternative AG′), and (4) overlap with an iCLIP cross-link. The more used AG in SF3B1wt/wt was set as canonical AG.
The 3′AS events within the critical window showed a distinct behavior already in wild-type K562-SF3B1wt/wt cells. If the distance between 3′ potential splice sites was either <12 nt or >21 nt, splicing predominantly occurred at the upstream AG in the vast majority of cases, indicating that the spliceosome generally favored upstream AG usage. However, within the critical window, this pattern was inverted, such that the upstream AG at these distances was generally outdone by the canonical downstream AG. This indicated that in the presence of two AGs within 12–21 nt, the downstream AG is predominately used for splicing. In line with this notion, we found that upstream AGs were generally depleted from this region (Fig. 7C, top). Moreover, upstream AG still falling into this window showed basal usage already in wild-type cells in >50% of cases and were predominantly activated upon SF3B1 mutation (Fig. 7C, middle), which could be facilitated by less stable binding of SF3B1K700E to the pre-mRNA, as suggested by the MD simulations.
Based on these observations, we hypothesized that diminished SF3B1K700E/wt binding to the pre-mRNA results in increased upstream AG usage within a critical window that may particularly impair splicing fidelity. Indeed, SF3B1 mutation preferentially affected the first (nearest) upstream AG (Fig. 7D), which lay significantly closer to the 3′ splice site compared with nondifferential 3′AS (Fig. 7E). In contrast, the BP of the differential spliced 3′AS events neither moved closer to the 3′ splice site nor differed in its predicted strengths (Fig. 7E; Supplemental Fig. S37), indicating that the positioning of the upstream AG rather than the BP was the primary determinant for the observed effects. The differentially used upstream AG had slightly lower predicted splice-site strengths (MaxEnt score) (Yeo and Burge 2003) compared with nondifferential 3′AS. The downstream AGs were considerably stronger than unused upstream AGs (Fig. 7C, lower panel). This was accompanied by higher splice-site strengths of the canonical 3′ splice sites, indicating that a strong canonical 3′ splice site was required to support differentially spliced 3′AS events (Fig. 7F).
Taken together, we propose that SF3B1 binding often directly overlaps with alternative AG′ in a constrained window upstream of 3′ splice sites, thereby using downstream 3′ splice sites in wild-type conditions. In the presence of SF3B1 mutations, SF3B1 cannot properly bind the Py-tract of the downstream canonical AG when a strong upstream AG′ lies within a short distance, leading to increased alternative splicing with the use of an alternative BP (Supplemental Fig. S38). This is partly explained by changes in the second binding pocket of SF3B1, as predicted by MD simulations, that destabilize the SF3B1–mRNA interaction. Thus, although mutated SF3B1 still binds to both sides of the BP in this scenario, the changed SF3B1 protein structure reduces usage of the downstream AG, resulting in widespread splicing defects.
Discussion
Alternative splicing plays a critical role in generating transcriptome diversity, and its dysregulation has been linked to various diseases, including cancer. Yet, complex transcriptome studies are challenging to perform with classical RNA-seq owing to the frequent ambiguity in mapping short reads and the difficulties in identifying novel isoforms (Rehrauer et al. 2013; Hooper 2014; Zhang et al. 2017). Of note, our capacity to study transcriptomes and ASEs has expanded with the recent advancements in long-read sequencing technologies. To the best of our knowledge, up to now only a few studies applied long-read Oxford Nanopore sequencing to analyze splice-site alterations mediated by mutated SF3B1 in CLL (bulk or single cell) (Tang et al. 2020; Peng et al. 2024) and MDS (single-cell) (Cortés-López et al. 2023).
Here, we used long-read Iso-Seq (Pacific Biosciences [PacBio]) sequencing of 44 patients to investigate the impact of SF3B1 mutations on alternative splicing. Our LRTS analysis revealed a wide variety of transcripts, with more than two-thirds unannotated, or even more than 3000 transcripts from novel genes. This highlighted the importance of long-read sequencing for comprehensive transcriptome profiling, particularly for detecting novel splice variants and ASEs. Our results supported previous findings that SF3B1 mutations specifically alter the usage of 3′ASs and IR (Darman et al. 2015; DeBoever et al. 2015; Alsafadi et al. 2016). Importantly, using a comprehensive setup of two cohorts of CLL and MDS patients, complemented by two isogenic cell line pairs, we were able to substantially expand the catalog of differentially spliced 3′AS to a total of 326 3′AS events in 266 genes (Supplemental Table S4). We observed similar effects in both patient cohorts and the cell lines studied, indicating a common effect of SF3B1 mutation on splicing. The clinical differences and prognosis of the CLL and MDS patients with SF3B1 mutations most likely depend on the specific gene expression profiles and thus their different relevances of splicing alterations.
Our results revealed that SF3B1 mutations affect genes involved in the major mRNA splicing pathway, indicating a broader impact on the splicing machinery. This may trigger secondary effects on splicing, potentially leading to altered transcriptomes and disease phenotypes. In particular and in concordance with previous studies, we observed an enrichment of the 3′AS events 12–21 nt upstream of the canonical 3′ splice sites (Darman et al. 2015; DeBoever et al. 2015; Alsafadi et al. 2016). This region is typically depleted of alternative AG dinucleotides. Why AGs are depleted in the critical region remains unclear. One possibility is that they are removed by purifying selection and might have an evolutionary advantage. However, a subset of introns still contains AGs within this critical region leading to 3′AS. Why some AGs remain present in the critical region is also unclear. We and others (Darman et al. 2015) observed that in these cases, the canonical AG is often stronger than at other 3′ splice sites, indicating that a strong 3′ splice site may be required to tolerate an upstream AG within the critical window.
Our minigene assays did not confirm that these 3′AS events depend solely on the distance between canonical and alternative AGs. Coinciding with the critical range of AG′–AG distances of 12–21 nt, we observed a bimodal binding of SF3B1 surrounding the BP, whereby the BP often coincided or was near the upstream AG′. Thereby, SF3B1 binding may shield the upstream AG from recognition during splicing as was proposed by Kesarwani et al. (2017). We did not detect obvious global changes in SF3B1K700E binding, which might be owing to a temporary effect and/or binding affinity of the SF3B1K700E that might not be caught by iCLIP analyses. However, our MD simulations predict that the number of contacts between the mRNA and residue 700 of SF3B1 resulted in increased mobility of the mRNA at both the canonical and alternative 3′ splice sites in SF3B1K700E.
We confirmed with minigene assays that SF3B1mut uses an alternative BP that leads to 3′AS usage in SF3B1mut/wt cells (Darman et al. 2015; Alsafadi et al. 2016). However, transcriptome-wide studies revealed that about one-third of all human exons have multiple BPs (Mercer et al. 2015; Pineda and Bradley 2018), which argues against the hypothesis that mutated SF3B1 always prefers usage of an alternative BP. Consistently, we did not observe any strong alterations in the binding of SF3B1K700E/wt to mRNAs, although a slight increase of the peak at the Py-tract directly upstream of the 3′ splice site was observed (Supplemental Fig. S39). Although we cannot exclude that we partially coprecipitated U2AF2, we and others did not find obvious changes in U2AF2 binding to SF3B1 in immunoprecipitations (Alsafadi et al. 2016; Cretu et al. 2016). Furthermore, U2AF2 binds to the Py-tract only during early stages of the splicing process and is released during transition to the activated B complex (Agafonov et al. 2011). In contrast, SF3B1 is assembled into the spliceosome later, and subsequently replaces U2AF2 in the activated B complex and binds to the Py-tract with its binding pocket consisting of HEAT domains 3–7, which harbor most of the mutational hotspots (Schmitzová et al. 2023). Therefore, the downstream peak observed in our SF3B1 iCLIP experiments most likely corresponds to SF3B1 binding.
The differential splicing observed in SF3B1mut/wt may result from a weakened binding of SF3B1mut-containing spliceosomes. There might be an additional effect of decreased binding of DDX46/PRP5, a kinase involved in proofreading of the pre-mRNA branch site (Tang et al. 2016; Carrocci et al. 2017; Zhang et al. 2021; Zhao et al. 2022). Other splicing proteins that have been shown to bind less to SF3B1mut are DDX42 (Zhao et al. 2022), DHX15 (Zhang et al. 2024), and SUGP1 (Zhang et al. 2019, 2023). DDX42 and DDX46 have been shown to sequentially occupy the RNA-binding pocket consisting of HEAT repeats 3–7 during early steps of the splicing process (Zhang et al. 2021, 2024; Yang et al. 2023).
Besides gaining additional insight into the SF3B1 splicing mechanism, we also explored the splicing and expression alterations identified through the Iso-Seq approach in CLL and MDS. Apart from identification of large numbers of new differentially spliced genes, we were able to specifically map the toxic exon of BRD9 to its isoform and predict its amino acid composition. We also identified SF3B1 isoforms specifically more present in MDS or CLL, albeit their expression levels were at ∼10%. The SF3B1-CLL transcript is predicted to undergo NMD, whereas the SF3B1-MDS is predicted to miss its C-terminal part. This shortened protein might rescue part of the detrimental effect of the mutated SF3B1, leading to a slightly favorable prognosis. SF3B1 overexpression in CLL B cells with respect to normal B cells has been reported before (Wan and Wu 2013). Possible reasons for this increase might be: (1) regulatory mechanisms, such as alterations in transcription factors or epigenetic changes; (2) oncogenic pathways involving growth factors or cytokines; or (3) feedback mechanisms (Huang et al. 2011). We would speculate that owing to the NMD-sensitive SF3B1-CLL transcript, the cell increases the transcription of the regular version of the SF3B1 transcript to compensate for this loss. This is not necessary in the case of MDS, because the SF3B1-MDS transcript is functional and only misses its C-terminal part. This would explain how the SF3B1-CLL isoform under NMD is related to the overexpression of SF3B1 in CLL.
This overexpression can now be brought in context with the worse prognosis of mutated SF3B1 in CLL, because in the presence of the mutation, this overexpression leads to a more massive dysregulation of alternative splicing caused by SF3B1 in CLL (compared to MDS), which in turn disrupts multiple pathways and lowers survival of the patients. Indeed, if we further investigate the disease-specific ASEs in mutated versus wild-type SF3B1 patients (Fig. 2B; Supplemental Table S2), we observed that the overall number of ASEs is fairly similar (CLL 288, MDS 219) with 1.31-fold increase in ASEs in CLL, but there was a drastic (×2.75) increase of CLL-specific IR events (CLL 77, MDS 28) (Supplemental Fig. S40A). The ASEs break down to 69 (CLL) and 27 (MDS) unique genes and were largely different with only six genes in common (Supplemental Fig. S40B). The pathways affected by CLL-IRs were related to mRNA splicing machinery, oncogenic signaling pathways, and immune pathways that might affect the survival of the patients (Supplemental Fig. S40C). Thus, we would argue that the overexpression of SF3B1 in CLL compared with MDS leads to elevated splicing effects, in particular introns, that target a different, more signaling-related panel of genes with multiple cellular functions that promote tumorigenesis and reduce survival. However, further functional analyses will show the impact of these altered SF3B1 proteins and if they influence MDS and CLL pathomechanisms.
Another example, which is frequently reported to be differentially spliced within SF3B1 mutated cancers, is MAP3K7. We were able to show that mutations in SF3B1 led to reduced expression of longer isoforms and increased expression of isoforms to a shortened protein kinase domain, likely impacting its function. Thus, with these data at hand, it is possible to not only identify splicing events but also map them to their cognate isoform and thus provide information on the resulting protein composition. This is a fundamental information for understanding splicing data and to gain insight into pathomechanisms underlying CLL and MDS.
Our study provides new insights into the mechanism by which SF3B1 mutations affect splicing regulation, as well as the potential consequences on protein function. Our findings highlight the importance of long-read sequencing for investigating differential alternative splicing usage and splicing factor function. These results have implications for understanding the role of SF3B1 mutations in hematological malignancies and other diseases and may be used in the future to predict new approaches for targeted therapies for these conditions.
Methods
Ethics approval
The study was approved by the ethics committee of the University of Cologne (Ethikvotum 11-319 from December 11, 2011, with an amendment from June 7, 2016) and the ethics committee of the University of Düsseldorf (Ethikvotum 3768, amendment from October 24, 2018). Informed consent has been obtained from all patients involved.
Cell lines and patients’ samples
The isogenic cell line pairs, K562-SF3B1K700E/wt and its parental K562-SF3B1wt/wt (RRID:CVCL_0004), as well as Nalm6-SF3B1H662Q/wt and its parental Nalm6-SF3B1wt/wt, were obtained from Horizon Discovery (HD181-012, HD115-110). Because homozygous SF3B1 mutations were reported to be lethal (Lee et al. 2016), we used heterozygous cell lines. The K700E mutation is the most frequent SF3B1 mutation reported in CLL and MDS, and H662Q mutation is also frequently reported (Rossi et al. 2011; Quesada et al. 2012; Wan and Wu 2013). The SF3B1-mutated cell lines were described previously (Darman et al. 2015). HEK293-FT (RRID: CVCL_6911) was purchased from Thermo Fisher Scientific (R70007). Information on cell line authentication and cell growth conditions is provided in the Supplemental Methods.
CLL and B cell samples were obtained from the CLL-Biobank Cologne. IGHV mutational status was determined as previously described (Rosenquist et al. 2017). Peripheral blood B cells were isolated via negative selection using RosetteSep immunodensity-based cell separation (Stemcell Technologies). The purity of CLL/ B cells was analyzed by flow cytometry and revealed that ≥90% cells coexpressed CD5/CD19.
Specimens from MDS with ring sideroblast (MDS-RS) patients were obtained from the MDS Biobank of the University Clinic Düsseldorf. Either RNA or cells were obtained from the Biobank. If cells were obtained, RNA was isolated using the NucleoSpin RNA kit (Macerey Nagel). RNA quality was accessed by RNA ScreenTape analysis (Agilent) or a Bioanalyzer (Agilent).
Clinical information on the patients is summarized in Supplemental Table S1.
Plasmids
Plasmids pCMV-3Tag-1A-SF3B1wt and pCMV-3Tag-1A-SF3B1K700E (Alsafadi et al. 2016) were designed by Angelos Constantinou (Department of Molecular Bases of Human Diseases, IGH-Institute of Human Genetics) and kindly provided by Marc-Henri Stern, Institut Curie. Plasmids pcDNA3.1-FLAG-SF3B1-WT and pcDNA3.1-FLAG-hSF3B1-K700E (Kesarwani et al. 2017) were obtained from Addgene (82576 and 82577). The human full-length SF3B1 sequence has been previously reported to be impossible to clone into bacteria (Wang et al. 1998; Yokoi et al. 2011). Therefore, the plasmids consisted of synthetic sequences, codon-optimized for expression in bacteria (Alsafadi et al. 2016; Kesarwani et al. 2017). For the minigene constructs, the intron and parts/complete adjacent upstream and downstream exons were PCR-amplified from K562 genomic DNA using Phusion Hot Start Flex DNA Polymerase (New England Biolabs) and cloned by the Hot Fusion (Fu et al. 2014) method into the BamHI restriction site of pcDNA3 (Invitrogen; https://www.addgene.org/vector-database/2092/). The ORF of the exons was left intact. The oligonucleotides used for cloning of the constructs are listed in Supplemental Table S5. Mutations and insertions were introduced by site-directed mutagenesis using the Q5 site-directed mutagenesis kit (New England Biolabs). Oligonucleotides for site-directed mutagenesis were designed using the NEBaseChanger version 1.3.3 (New England Biolabs) and are listed in Supplemental Table S5. All constructs were verified by Sanger sequencing (Microsynth Seqlab).
cDNA synthesis and validation of the splicing alterations
Transfections and RNA isolation were performed following standard procedures with PEI Max (PolyScience 24765 1) and a NucleoSpin RNA mini kit (Macherey Nagel 740955.250). An amount of 500 ng total RNA was reverse-transcribed using SuperScript II (Thermo Fisher Scientific 18064014) and hexamer oligonucleotides for the cDNA synthesis from K562 and Nalm6 RNA. For the minigene assays, 500 ng RNA was reverse-transcribed using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific 18090010) with the plasmid-specific BGH-rev oligo in 20 µL. Subsequently, RNA in DNA–RNA hybrids was digested by RNase H incubation. For RT-PCR, we used 1 µL of cDNA, Taq DNA polymerase, recombinant (Thermo Fisher Scientific 10342020), and specific oligonucleotides (Supplemental Table S5) in a volume of 25 µL. The PCR ran for 35 PCR cycles. PCR products were separated on a 3%–4% TAE-agarose gel.
PacBio Iso-Seq library preparation and sequencing
For the cDNA synthesis, we used oligo(dT) oligonucleotides and the TeloPrime Full-Length cDNA Amplification Kit V2 (Lexogen) to ensure the amplification of full-length mRNAs that contained a cap structure. Barcoded primers were used in the cDNA amplification step to enable multiplexing before library preparation. To enrich for slightly larger cDNAs, we adjusted the magnetic bead concentration in the bead clean-up after cDNA amplification. Subsequent library preparation was performed with the SMRTbell Express Template Prep Kit 2.0 (PacBio).
In total, 58 libraries were sequenced on the PacBio Sequel II platform, by multiplexing four samples per 8 million SMRT Cell at the Genomics and Transcriptomics Laboratory, the production site of the West German Genome Center in Düsseldorf (Heinrich Heine Universität) (Supplemental Table S1).
Processing of PacBio Iso-Seq data
Preprocessing of raw Iso-Seq sequencing data was performed with IsoSeq (https://github.com/PacificBiosciences/IsoSeq) software version 3.4, using the recommended parameters. In brief, we used the ccs tool to call circular consensus sequences by clustering and collapsing steps, lima to remove primers and adapters, and isoseq refine to demultiplex samples and filter out reads not featuring poly(A) sequences. This resulted more than 33 million aligned full-length nonchimeric (flnc) poly(A) HiFi reads (100,302–1,258,653 per library, average 582,135) (Supplemental Table S1), with an average length of 2721 bp. At this sequencing depth, transcript isoforms expressed at one TPM are expected to be sequenced by at least 25 reads and two TPM isoforms by at least 50 reads, with >95% probability (Supplemental Fig. S1).
According to the base quality values, 58 samples had an error rate of <1% in at least 99.7% of the HiFi reads, and only four samples had higher error rates, with 96.7% to 96.9% reads with <1% error rate. Overall, the quality of the reads was high, with only 9% of reads potentially affected by technology-specific technical artifacts (Supplemental Fig. S2; Cocquet et al. 2006).
The flnc reads were converted to FASTQ using SAMtools v.1.18 (Li and Durbin 2009) and, without an additional clustering step, aligned to the human genome GRCh38.p13 using minimap2 (Li 2018) version 2.22 with the preset parameters for high-quality spliced reads (-ax splice:hq). For each sample, at least 99.85% of the reads were mapped to the genome, and at least 91.9% were uniquely mapped. Samples for which we sequenced more than one library were merged using SAMtools v.1.18 (Li and Durbin 2009) after the mapping step. Sequencing and mapping statistics per sample are detailed in Supplemental Table S1.
SF3B1 mutation calling was done with BCFtools v.1.13 (Danecek et al. 2021) mpileup and SnpEff v.5.1d (Cingolani et al. 2012).
Further analysis of Iso-Seq data was performed in Python v3, using IsoTools (Lienhard et al. 2023) version 0.2.8. In brief, aligned reads were imported and compared with the human reference annotation version 36 from GENCODE (Frankish et al. 2021) to call, annotate, classify, and quantify transcripts, using IsoTools’ add_sample_from_bam function.
For exploratory analysis, ASEs were detected using IsoTools’ alternative_splicing_events function. For each sample, individual events were quantified by PSI values, that is, the number of reads supporting transcripts that include additional exonic sequence over all transcripts spanning that event. Based on these PSI values, PCA plots for different alternative splicing categories were computed using IsoTools’ plot_embedding function.
Differential splicing events between SF3B1mut/wt and SF3B1wt/wt CLL, MDS, and cell line samples were computed with the IsoTools altsplice_test function, using the betabinomial likelihood ratio test. This test models the variability within the tested groups with a beta-binomial mixture distribution, a binomial distribution in which the probability parameter, p, of the binomial distribution B(n,p) follows a beta distribution, Beta(a, b). The test compares the group-wise coverage of the splicing event with the total coverage.
Here, BB(k|α, β, n) is the probability mass function of the beta-binomial distribution, and are maximum likelihood estimates for the parameters. The maximum log-likelihood parameters are determined numerically by a quasi-Newton optimization method (LM-BFGS from SciPy) (Virtanen et al. 2020). Under the null hypothesis (i.e., no differential splicing), the test statistic is X2 distributed with two degrees of freedom.
This formulation allows for considering within-group variability in a similar manner as tests based on negative binomial distribution for RNA-seq data, which is crucial for heterogeneous samples such as individual cancer patients. To be tested, we required the events to be covered by at least 10 reads in at least four samples per group, as well as the minor alternative to be covered by at least 5% of the total reads (test = “betabinom_lr”, min_n = 10, min_sa = 4, min_alt_fraction = 0.05). Because of the limited number of samples, we did not include any covariates in the model analysis. We did not observe any bias toward highly expressed genes (Supplemental Fig. S41).
All 3′ ASEs (including those that were not differentially expressed between SF3B1mut/wt and SF3B1wt/wt) were exported for further analysis (Supplemental Table S4).
Differential expression analysis on the Iso-Seq read counts was performed with DESeq2 (Love et al. 2014).
Estimation of BP position and splice-site strength score
For all 3′ ASEs, we used the R (R Core Team 2017) Bioconductor package branchpointer (Signal et al. 2018) to predict BP probabilities for both canonical and alternative splice sites. We used the position with the highest BP probability as the predicted BP.
The strength of the 3′ splice sites for the minigene constructs was calculated with SpliceRover (Zuallaert et al. 2018; http://bioit2.irc.ugent.be/rover/splicerover, accessed July 23, 2023, using the model “human acceptors”).
Transcript coding potential
For a detailed description of the methods, please see Supplemental Notes.
Illumina RNA-seq library preparation and sequencing
RNA from K562 and Nalm6 cells was isolated using the NucleoSpin RNA mini kit (Macherey Nagel) followed by DNase-digestion with the DNase I set (Zymo Research E1010) and clean-up using the NucleoSpin RNA clean-up mini kit (Macherey-Nagel 740948.50). RNA quality was surveyed using the RNA ScreenTape system (Agilent), and the RNA integrity number (RIN) was 10 for all samples. CLL cells from 19 CLL patients used for Iso-Seq and an additional eight patients (including four patients with SF3B1 mutation) were stored in RNA later, and RNA was isolated using the RNeasy mini kit (Qiagen) followed by DNase digestion using the DNase I amplification grade kit (Invitrogen) and a clean-up using RNeasy MinElute columns (Qiagen). RNA-seq libraries for the cell lines and the CLL RNA were prepared using the TruSeq Stranded Total RNA Sample Prep Kit (Illumina) according to the manufacturer's protocol. In brief, 2 µg of total RNA was depleted for ribosomal RNA using a Ribo-Zero rRNA Removal Kit (Illumina), followed by random primed cDNA synthesis. Sequencing libraries were run at the sequencing core unit of the Max-Planck Institute for Molecular Genetics on a HiSeq 2500 (Illumina) using 50 bp paired-end reads.
Processing of Illumina RNA-seq data
We collected publicly available MDS RNA-seq data from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) using the following criteria: “MDS” in the study description, paired-end Illumina reads, available FASTQ files, human blood cell samples, and at least five samples per study. After merging technical replicates, our data set consisted of 263 samples from SF3B1wt/wt MDS patients and 135 SF3B1mut/wt patients. All study and sample IDs are listed in Supplemental Table S1.
Illumina RNA-seq reads were aligned to the human reference genome GRCh38.p13 using STAR aligner version 2.7.6a (Dobin et al. 2013), with provided GFF annotation from GENCODE release 36, including annotation of nonchromosomal scaffolds. ASEs were called and quantified using rMATS (v4.1.1) (Shen et al. 2014).
Mutations in SF3B1 were called as for Iso-Seq described above.
iCLIP experiments
iCLIP of K562-SF3B1wt/wt (three replicates) and K562-SF3B1K700E/wt (two replicates) was performed as described previously (Sutandy et al. 2016). To this end, exponentially growing K562 cells were pelleted, washed once with PBS, and 10 × 106 were subjected to UV cross-linking at 400 mJ/cm2 at 254 nm in 6 mL PBS in a 10 cm petri dish on ice. Cross-linked cells were scraped from the dish, collected by centrifugation for 2 min at 500g at 4°C, snap frozen in liquid nitrogen and stored at −80°C. About 3 × 106 cells were immunoprecipitated using 10 µg of a monoclonal SF3B1 antibody (clone 16, MBL D221-3) or 10 µg of the monoclonal U2AF2 antibody (U4759, Sigma-Aldrich).
iCLIP data processing
Initial quality control was done using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) before and after quality filtering. All reads having at least one position with a sequencing quality <10 in the barcode area (positions 1–3, 4–7, 8–9) were removed. Demultiplexing and adapter trimming were done on quality-filtered data using Flexbar (Roehr et al. 2017). No mismatches were allowed during demultiplexing, whereas an error rate of 0.1 was accepted when trimming the adapter at the right end of the reads. Furthermore, a minimal overlap of 1 bp between the reads and adapter was required, and only trimmed reads with a minimal length of 15 bp (24 bp including the barcode) were kept for further analysis. Barcodes of remaining reads were trimmed (but kept as additional information with the reads). Trimmed reads were then mapped to genome assembly version GRCh38 using STAR (v. 2.6.1b) (Dobin et al. 2013) with 4% mismatched bases allowed and turned-off soft-clipping on the 5′-end, as well as GENCODE gene annotation v31 (Frankish et al. 2019). Although technical duplicates were removed with UMI-tools (Smith et al. 2017) with the unique method, all real duplicate reads were kept. Then, we checked the cross-link quality with iCLIPro (Hauer et al. 2015).
To facilitate comparisons, the cross-link events, that is, reads after duplicate removal, of the replicates were randomly subsampled to the size of the smallest replicate (n = 13,892,358; replicate 2 of SF3B1K700E/wt) (Supplemental Table S1).
Definition and classification of SF3B1 binding sites
Binding sites were identified from the merged cross-link events of SF3Bwt/wt and SF3B1K700E/wt as described previously (Supplemental Fig. S29; Busch et al. 2020). For this, the cross-link events of all five replicates were combined and subjected to peak calling with PureCLIP (version 1.3.1) (Krakau et al. 2017) with default parameters. The PureCLIP-called sites (Psites) were filtered by first removing 5% of the Psites with the lowest score associated and then keeping only the top 20% of Psites within each gene annotated (GENCODE release 36, GRCh38; only annotations with a gene support level of one or two and transcript support level from one to three). The Psites were then merged into binding sites using the R/Bioconductor package BindingSiteFinder (version 1.0.3) (https://bioconductor.org/packages/release/bioc/html/BindingSiteFinder.html), using the following options: width of 5 nt (bsSize = 5); two or more Psites (minWidth = 2, minClSites = 1) and one or more cross-link position within each binding site (minCrosslinks = 1). In brief, Psites closer than 5 nt were merged into regions, and isolated Psites were discarded. Within each region, binding site centers were iteratively placed at the position with the most cross-link events and extended by 2 nt on both sides. Binding site centers were required to harbor the maximum cross-link signal within the binding site. The optimal binding site width of 5 nt was determined by an evaluation of the ratio of cross-link events within binding sites of increasing width over the mean background signal in flanking windows of the same size (Supplemental Fig. S30). Next, binding sites that were not supported by all replicates in at least one condition (SF3B1wt/wt or SF3B1K700E/wt) were filtered out (Supplemental Fig. S31). The threshold for sufficient coverage in a replicate was determined using the fifth percentile and a lower boundary of two cross-link events as described by Busch et al. (2020). Finally, binding sites were assigned to target genes using GENCODE annotation (release 36, GRCh38; filtered as above) as described by Busch et al. (2020). In total, this procedure identified 96,852 SF3B1 binding sites in 8127 genes.
To classify distinct SF3B1 binding patterns in introns, bound regions were defined by merging intronic binding sites within a distance <55 nt and resizing the obtained regions to 81 nt around the center, resulting in 56,224 regions harboring 87,199 binding sites. Following the approaches suggested previously (Heyl and Backofen 2021), we used unsupervised clustering to separate the cross-link patterns in the bound regions. For this, the cross-link coverage (sum of all replicates) was subjected to min–max normalization (Tarantola 2008) within each window (i.e., scaling such that the lowest and highest number of cross-link events are set to zero and one, respectively), followed by spline-smoothing using the smooth.spline function (R package stats, version 4.1.0) with lambda 0.2 (spar = 0.2) and inflated dimensions (dim = 150). This changed the shape of the matrix from A × B (56,224 × 81) to A × B′ (56,244 × 150), where A is the number of regions, B is the nucleotide positions, and B′ is the inflated nucleotide positions. The matrix A × B′ of normalized and smoothed cross-link coverages was then subjected to dimension reduction using uniform manifold approximation and projection (UMAP) (McInnes et al. 2018) with the umap function (package umap, version 0.2.7) with the parameters n_epochs = 5000, n_components = 2, min_dist = 0.01, and n_neighbors = 5 (Supplemental Fig. S35A). The UMAP results were assigned to clusters using density-based clustering of applications with noise (DBSCAN) (Ester et al. 1996) with the dbscan function (R package dbscan, version 1.1, eps = 0.3) (Hahsler et al. 2019), with a minimum number of 150 points per cluster (MinPts = 150), yielding three clusters: C1 (n = 35,907 regions), C2 (n = 5635), and C3 (n = 13,847). Bound regions in cluster C0 (n = 835) were deemed as outliers that could not be assigned to any of the fitted density centers and were excluded from further analysis. Bound regions in cluster C3 (wide pattern) were smoothed more finely (spar = 0.1, dim = 500) and then subjected to a second round of UMAP dimension reduction (parameters as above) and DBSCAN clustering (MinPts = 60, eps = 0.23), yielding subclusters 0–33 (Supplemental Fig. S35B). Cluster numbering is based on the increasing distance between the two modes in the arrangement of binding sites, calculated on the summed and smoothed coverages within each cluster using the locmodes function (R package multimode, version 1.5) (Supplemental Fig. S35C; Ameijeiras-Alonso et al. 2021).
Data access
Cell lines’ and patients’ transcriptome raw FASTQ files or PacBio CCS unaligned BAM files generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA1037338 or to the European Genome-Phenome Archive (https://ega-archive.org/) under accession number EGAS50000000053, respectively. iCLIP raw FASTQ data and processed files have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE247658. Patient data access will be controlled by the data access committee at the Institute for Translational Epigenetics at the University Hospital Cologne, University of Cologne, Cologne, Germany.
The complete Iso-Seq/iCLIP data analysis code is available as a Jupyter Notebook at GitHub (https://github.com/ZarnackGroup/go_long2023), Zenodo (https://doi.org/10.5281/zenodo.12597946), and as Supplemental Code.
Supplemental Material
Acknowledgments
We acknowledge the IMB Genomics Core Facility and its NextSeq 500 sequencer (funded by the Deutsche Forschungsgemeinschaft [DFG, German Research Foundation] INST 247/870-1 FUGG). We thank Angelos Constantinou (IGH-Institute of Human Genetics, France) and Marc-Henri Stern (Institut Curie, Paris, France) for sharing the SF3B1 plasmids, Anke Busch (IMB Mainz, Germany) for iCLIP data preprocessing, and Elena Wasserburger-Zichel (University Hospital Cologne, Germany) and Bernd Timmermann (Sequencing Core Unit, Max Planck Institute for Molecular Genetics, Berlin, Germany) for their technical assistance. We furthermore thank the Regional Computing Center of the University of Cologne (RRZK) for providing computing time on the DFG-funded (funding number INST 216/512/1FUGG) high-performance computing (HPC) system CHEOPS as well as IT support. In addition, we acknowledge computational support of the Center for Information and Media Technology, especially the HPC team at the Heinrich Heine University, as well as the computing time provided by the John von Neumann Institute for Computing on the supercomputer JUWELS at the Jülich Supercomputing Centre (user IDs: VSK33, DNAzyme). This work was supported by the DFG Research Infrastructure West German Genome Center (407493903) as part of the Next Generation Sequencing Competence Network (project 423957469). High-throughput sequencing was carried out at the West German Genome Center, and production sites in Cologne and Düsseldorf. The study was funded by the German Research Foundation: KFO286-RP8/SCHW1605/1-1, SCHW1605/4-1 (GO-LONG), SFB1399 and SFB1530 to M.R.S., KFO-286-RP6 to M.H., KFO-286-CP to C.D.H., SFB1530 to M.H., the Volkswagen Stiftung Lichtenberg program to M.R.S., the Center for Molecular Medicine Cologne, CMMC (A12 to M.R.S.), and the EU Horizon 2021 LongTREC (no. 101072892) to R.H.
Authors contributions: M.R.S., C.G., R.H., K.Z., H.G., N.G., M.H., and Ju.K. designed the study. H.H., L.B., A.K., K.B., K.K., Je.K., and C.D.H. acquired the data. A.P., M.L., M.B., Je.K., H.G., C.G., R.H., K.Z., and M.R.S. analyzed and interpreted the data. A.P., M.L., C.G., R.H., K.Z., and M.R.S. drafted and wrote the manuscript. All authors have read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279327.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
M.H. received honoraria (speakers bureau and/or advisory board) from Roche, Janssen, and Abbvie, as well as research support from Roche, Janssen, Abbvie, Astra Zeneca, and Beigene.
References
- Agafonov DE, Deckert J, Wolf E, Odenwälder P, Bessonov S, Will CL, Urlaub H, Lührmann R. 2011. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol 31: 2667–2682. 10.1128/MCB.05266-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, et al. 2016. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun 7: 10615. 10.1038/ncomms10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsafadi S, Dayot S, Tarin M, Houy A, Bellanger D, Cornella M, Wassef M, Waterfall JJ, Lehnert E, Roman-Roman S, et al. 2021. Genetic alterations of SUGP1 mimic mutant-SF3B1 splice pattern in lung adenocarcinoma and other cancers. Oncogene 40: 85–96. 10.1038/s41388-020-01507-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameijeiras-Alonso J, Crujeiras RM, Rodriguez-Casal A. 2021. Multimode: an R package for mode assessment. J Stat Softw 97: 1–32. 10.18637/jss.v097.i09 [DOI] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bland P, Saville H, Wai PT, Curnow L, Muirhead G, Nieminuszczy J, Ravindran N, John MB, Hedayat S, Barker HE, et al. 2023. SF3B1 hotspot mutations confer sensitivity to PARP inhibition by eliciting a defective replication stress response. Nat Genet 55: 1311–1323. 10.1038/s41588-023-01460-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Anczuków O. 2023. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer 23: 135–155. 10.1038/s41568-022-00541-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Brüggemann M, Ebersberger S, Zarnack K. 2020. iCLIP data analysis: a complete pipeline from sequencing reads to RBP binding sites. Methods 178: 49–62. 10.1016/j.ymeth.2019.11.008 [DOI] [PubMed] [Google Scholar]
- Canbezdi C, Tarin M, Houy A, Bellanger D, Popova T, Stern M-H, Roman-Roman S, Alsafadi S. 2021. Functional and conformational impact of cancer-associated SF3B1 mutations depends on the position and the charge of amino acid substitution. Comput Struct Biotechnol J 19: 1361–1370. 10.1016/j.csbj.2021.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. 2017. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res 45: 4837–4852. 10.1093/nar/gkw1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Chong A, Zhang G, Veitia RA. 2006. Reverse transcriptase template switching and false alternative transcripts. Genomics 88: 127–131. 10.1016/j.ygeno.2005.12.013 [DOI] [PubMed] [Google Scholar]
- Cortés-López M, Chamely P, Hawkins AG, Stanley RF, Swett AD, Ganesan S, Mouhieddine TH, Dai X, Kluegel L, Chen C, et al. 2023. Single-cell multi-omics defines the cell-type-specific impact of splicing aberrations in human hematopoietic clonal outgrowths. Cell Stem Cell 30: 1262–1281.e8. 10.1016/j.stem.2023.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretu C, Schmitzová J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Lührmann R, Pena V. 2016. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol Cell 64: 307–319. 10.1016/j.molcel.2016.08.036 [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience 10: giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al. 2015. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep 13: 1033–1045. 10.1016/j.celrep.2015.09.053 [DOI] [PubMed] [Google Scholar]
- DeBoever C, Ghia EM, Shepard PJ, Rassenti L, Barrett CL, Jepsen K, Jamieson CHM, Carson D, Kipps TJ, Frazer KA. 2015. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput Biol 11: e1004105. 10.1371/journal.pcbi.1004105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V, Roy S, Scifo L, Armstrong RN, Shaw J, et al. 2016. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 30: 2322–2331. 10.1038/leu.2016.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, Xu X. 1996. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, pp. 226–231. AAAI Press. [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland JE, Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I, et al. 2021. GENCODE 2021. Nucleic Acids Res 49: D916–D923. 10.1093/nar/gkaa1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Donovan WP, Shikapwashya-Hasser O, Ye X, Cole RH. 2014. Hot fusion: an efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS One 9: e115318. 10.1371/journal.pone.0115318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 10: 233–243. 10.1101/gad.10.2.233 [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760. 10.1128/MCB.18.8.4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahsler M, Piekenbrock M, Doran D. 2019. dbscan: fast density-based clustering with R. J Stat Softw 91: 1–30. 10.18637/jss.v091.i01 [DOI] [Google Scholar]
- Hauer C, Curk T, Anders S, Schwarzl T, Alleaume A-M, Sieber J, Hollerer I, Bhuvanagiri M, Huber W, Hentze MW, et al. 2015. Improved binding site assignment by high-resolution mapping of RNA–protein interactions using iCLIP. Nat Commun 6: 7921. 10.1038/ncomms8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl F, Backofen R. 2021. StoatyDive: evaluation and classification of peak profiles for sequencing data. GigaScience 10: giab045. 10.1093/gigascience/giab045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet 36: 1255–1257. 10.1038/ng1469 [DOI] [PubMed] [Google Scholar]
- Hooper JE. 2014. A survey of software for genome-wide discovery of differential splicing in RNA-seq data. Hum Genomics 8: 3. 10.1186/1479-7364-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lou C-H, Chan W, Shum EY, Shao A, Stone E, Karam R, Song H-W, Wilkinson MF. 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 43: 950–961. 10.1016/j.molcel.2011.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Chew G-L, Liu B, Michel BC, Pangallo J, D'Avino AR, Hitchman T, North K, Lee SC-W, Bitner L, et al. 2019. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574: 432–436. 10.1038/s41586-019-1646-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani AK, Ramirez O, Gupta AK, Yang X, Murthy T, Minella AC, Pillai MM. 2017. Cancer-associated SF3B1 mutants recognize otherwise inaccessible cryptic 3′ splice sites within RNA secondary structures. Oncogene 36: 1123–1133. 10.1038/onc.2016.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakau S, Richard H, Marsico A. 2017. PureCLIP: capturing target-specific protein–RNA interaction footprints from single-nucleotide CLIP-seq data. Genome Biol 18: 240. 10.1186/s13059-017-1364-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, et al. 2015. Mutations driving CLL and their evolution in progression and relapse. Nature 526: 525–530. 10.1038/nature15395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC-W, Dvinge H, Kim E, Cho H, Micol J-B, Chung YR, Durham BH, Yoshimi A, Kim YJ, Thomas M, et al. 2016. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med 22: 672–678. 10.1038/nm.4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard M, van den Beucken T, Timmermann B, Hochradel M, Börno S, Caiment F, Vingron M, Herwig R. 2023. IsoTools: a flexible workflow for long-read transcriptome sequencing analysis. Bioinformatics 39: btad364. 10.1093/bioinformatics/btad364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yoshimi A, Wang J, Cho H, Chun-Wei Lee S, Ki M, Bitner L, Chu T, Shah H, Liu B, et al. 2020. Mutations in the RNA splicing factor SF3B1 promote tumorigenesis through MYC stabilization. Cancer Discov 10: 806–821. 10.1158/2159-8290.CD-19-1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jädersten M, Jansson M, Elena C, Gallì A, Walldin G, Della Porta MG, et al. 2015. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 126: 233–241. 10.1182/blood-2015-03-633537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, Bowen DT, Campbell PJ, Ebert BL, Fenaux P, et al. 2020. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the international working group for the prognosis of MDS. Blood 136: 157–170. 10.1182/blood.2020004850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, Großberger L. 2018. UMAP: Uniform Manifold Approximation and Projection. J Open Source Softw 3: 861. 10.21105/joss.00861 [DOI] [Google Scholar]
- Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. 2015. Genome-wide discovery of human splicing branchpoints. Genome Res 25: 290–303. 10.1101/gr.182899.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49: D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, Schneider RK, Lord AM, Wang L, Gambe RG, et al. 2016. Physiologic expression of Sf3b1K700E causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30: 404–417. 10.1016/j.ccell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, et al. 2011. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 365: 1384–1395. 10.1056/NEJMoa1103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellagatti A, Armstrong RN, Steeples V, Sharma E, Repapi E, Singh S, Sanchi A, Radujkovic A, Horn P, Dolatshad H, et al. 2018. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood 132: 1225–1240. 10.1182/blood-2018-04-843771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Jabbari JS, Tian L, Chua CC, Anstee NS, Amin N, Wei AH, Davidson NM, Roberts AW, Huang DCS, et al. 2024. Single-cell rapid capture hybridization sequencing (scRaCH-seq) to reliably detect isoform usage and coding mutations in targeted genes at a single-cell level. bioRxiv 10.1101/2024.01.30.577942 [DOI]
- Pineda JMB, Bradley RK. 2018. Most human introns are recognized via multiple and tissue-specific branchpoints. Genes Dev 32: 577–591. 10.1101/gad.312058.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DF, Miao W, Yang X, Goda GA, Ji AL, Donohue LKH, Aleman MM, Dominguez D, Khavari PA. 2021. easyCLIP analysis of RNA-protein interactions incorporating absolute quantification. Nat Commun 12: 1569. 10.1038/s41467-021-21623-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. 2012. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44: 47–52. 10.1038/ng.1032 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rehrauer H, Opitz L, Tan G, Sieverling L, Schlapbach R. 2013. Blind spots of quantitative RNA-seq: the limits for assessing abundance, differential expression, and isoform switching. BMC Bioinformatics 14: 370. 10.1186/1471-2105-14-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehr JT, Dieterich C, Reinert K. 2017. Flexbar 3.0: SIMD and multicore parallelization. Bioinformatics 33: 2941–2942. 10.1093/bioinformatics/btx330 [DOI] [PubMed] [Google Scholar]
- Rosenquist R, Ghia P, Hadzidimitriou A, Sutton LA, Agathangelidis A, Baliakas P, Darzentas N, Giudicelli V, Lefranc MP, Langerak AW, et al. 2017. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: updated ERIC recommendations. Leukemia 31: 1477–1481. 10.1038/leu.2017.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, Fangazio M, Vaisitti T, Monti S, Chiaretti S, et al. 2011. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 118: 6904–6908. 10.1182/blood-2011-08-373159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzová J, Cretu C, Dienemann C, Urlaub H, Pena V. 2023. Structural basis of catalytic activation in human splicing. Nature 617: 842–850. 10.1038/s41586-023-06049-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, Smith PG, Buonamici S, Yu L, Caesar-Johnson SJ, et al. 2018. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep 23: 282–296.e4. 10.1016/j.celrep.2018.01.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci 111: E5593–LP-E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa Y, Malcovati L, Gallì A, Sato-Otsubo A, Kataoka K, Sato Y, Watatani Y, Suzuki H, Yoshizato T, Yoshida K, et al. 2018. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun 9: 3649. 10.1038/s41467-018-06063-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signal B, Gloss BS, Dinger ME, Mercer TR. 2018. Machine learning annotation of human branchpoints. Bioinformatics 34: 920–927. 10.1093/bioinformatics/btx688 [DOI] [PubMed] [Google Scholar]
- Smith T, Heger A, Sudbery I. 2017. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res 27: 491–499. 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutandy FXR, Hildebrandt A, König J. 2016. Profiling the binding sites of RNA-binding proteins with nucleotide resolution using iCLIP. In Post-transcriptional gene regulation (ed. Dassi E), pp. 175–195. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, Gupta V, Moldón A, Xu Y-Z, Query CC. 2016. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev 30: 2710–2723. 10.1101/gad.291872.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AD, Soulette CM, van Baren MJ, Hart K, Hrabeta-Robinson E, Wu CJ, Brooks AN. 2020. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat Commun 11: 1438. 10.1038/s41467-020-15171-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantola S. 2008. European innovation scoreboard: strategies to measure country progress over time. JRC Scientific and Technical Reports, EUR 23526 EN. JRC46943. OPOCE, Luxembourg. https://publications.jrc.ec.europa.eu/repository/handle/JRC46943.
- Tardaguila M, de la Fuente L, Marti C, Pereira C, Pardo-Palacios FJ, del Risco H, Ferrell M, Mellado M, Macchietto M, Verheggen K, et al. 2018. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res 28: 396–411. 10.1101/gr.222976.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17: 261–272. 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Wu CJ. 2013. SF3B1 mutations in chronic lymphocytic leukemia. Blood 121: 4627–4634. 10.1182/blood-2013-02-427641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev 12: 1409–1414. 10.1101/gad.12.10.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brooks AN, Fan J, Wan Y, Gambe R, Li S, Hergert S, Yin S, Freeman SS, Levin JZ, et al. 2016. Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell 30: 750–763. 10.1016/j.ccell.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Beutler B, Zhang D. 2022. Emerging roles of spliceosome in cancer and immunity. Protein Cell 13: 559–579. 10.1007/s13238-021-00856-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Bian T, Zhan X, Chen Z, Xing Z, Larsen NA, Zhang X, Shi Y. 2023. Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly. Nat Commun 14: 897. 10.1038/s41467-023-36489-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2003. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. In Proceedings of the seventh annual international conference on research in computational molecular biology, RECOMB ’03, pp. 322–331, Association for Computing Machinery, New York. [DOI] [PubMed] [Google Scholar]
- Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, et al. 2011. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J 278: 4870–4880. 10.1111/j.1742-4658.2011.08387.x [DOI] [PubMed] [Google Scholar]
- Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. 10.1016/j.cell.2012.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang B, Lin L-L, Zhao S. 2017. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 18: 583. 10.1186/s12864-017-4002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ali AM, Lieu YK, Liu Z, Gao J, Rabadan R, Raza A, Mukherjee S, Manley JL. 2019. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol Cell 76: 82–95.e7. 10.1016/j.molcel.2019.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Rigo N, Dybkov O, Fourmann J-B, Will CL, Kumar V, Urlaub H, Stark H, Lührmann R. 2021. Structural insights into how Prp5 proofreads the pre-mRNA branch site. Nature 596: 296–300. 10.1038/s41586-021-03789-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang J, Xu K, Xing P, Huang Y, Liu Z, Tong L, Manley JL. 2022. DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer. Proc Natl Acad Sci 119: e2216712119. 10.1073/pnas.2216712119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xie J, Huang J, Liu X, Xu R, Tholen J, Galej WP, Tong L, Manley JL, Liu Z. 2023. Characterization of the SF3B1–SUGP1 interface reveals how numerous cancer mutations cause mRNA missplicing. Genes Dev 37: 968–983. 10.1101/gad.351154.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhan X, Bian T, Yang F, Li P, Lu Y, Xing Z, Fan R, Zhang QC, Shi Y. 2024. Structural insights into branch site proofreading by human spliceosome. Nat Struct Mol Biol 31: 835–845. 10.1038/s41594-023-01188-0 [DOI] [PubMed] [Google Scholar]
- Zhao B, Li Z, Qian R, Liu G, Fan M, Liang Z, Hu X, Wan Y. 2022. Cancer-associated mutations in SF3B1 disrupt the interaction between SF3B1 and DDX42. J Biochem 172: 117–126. 10.1093/jb/mvac049 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Gong Q, Wang Y, Li M, Wang L, Ding H, Li P. 2020. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark Res 8: 38. 10.1186/s40364-020-00220-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuallaert J, Godin F, Kim M, Soete A, Saeys Y, De Neve W. 2018. SpliceRover: interpretable convolutional neural networks for improved splice site prediction. Bioinformatics 34: 4180–4188. 10.1093/bioinformatics/bty497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.