Highlights
-
•
The proposed selective cultivation method using P. elgii supernatant is effective in isolating bacterial strains from environmental samples.
-
•
Isolation of 35 bacterial strains with antibacterial activity from the soil microbiota.
-
•
Genomic analysis revealed several gene clusters of biotechnological relevance.
-
•
Most gene clusters identified in K003 strain showed low (<11 %) or no similarity to already known compounds, suggesting the potential production of new bioactive compounds.
Keywords: Prospecting, Supernatant, Antibacterial activity
Abstract
The biotechnological industry faces a crucial demand for novel bioactive compounds, particularly antimicrobial agents, to address the rising challenge of bacterial resistance to current available antibiotics. Traditional strategies for cultivating naturally occurring microorganisms often limit the discovery of novel antimicrobial producers. This study presents a protocol for targeted selection of bacterial strains using the supernatant of Paenibacillus elgii, which produces abundant signal molecules and antimicrobial peptides. Soil samples were inoculated in these enriched culture media to selectively cultivate bacteria resistant to the supernatant, indicating their potential to produce similar compounds. The bacterial strains isolated through this method were assessed for their antibacterial activity. In addition, the functional annotation of the genome of one of these strains revealed several gene clusters of biotechnological interest. This study highlights the effectiveness of using this approach for selective cultivation of microorganisms with potential for biotechnological applications.
Graphical Abstract
1. Introduction
The advent of molecular techniques, such as PCR and next generation sequencing, led to a diminished emphasis on bacterial cultivation in the 1970s [1]. The wealth of genomic insights derived from these techniques shifted attention away from traditional culture-based methods. However, these methods are a crucial tool for microbiologists since both culture-dependent and independent methods are complementary [2]. Bacterial DNA sequencing unveils interesting potential physiological traits of microorganisms guiding direct cultivation efforts [3]. The reciprocal relationship between genomic and cultivation techniques enriches our further understanding of bacterial ecology. Despite their challenges, microbial cultures allow not only well-annotated microbial genomes for predictions used in genomic and metagenomic analyses, but also enable deep investigation of their metabolic and biotechnological potential [[1], [4], [5], [6]].
One of the primary industrial demands for bioactive compounds involves the production of antimicrobial molecules, driven by the need for alternatives to existing market products to combat the growing resistance observed in various bacteria [7]. Many antimicrobials are produced by bacteria as a result of microbial competition or communication in their natural environments [8]. However, a significant obstacle in the study of prokaryotes and discovering novel microbial compounds is the inability to cultivate most of the environmental diversity under laboratory conditions [6].
Certain bacterial groups require specific growth conditions, indicating the necessity for refining existing strategies for microbial cultivation and developing novel optimized techniques to access the uncultivated fraction of microorganisms for biotechnological applications. Potential improvements in culture methods encompass use of specialized culture media and antimicrobials to selectively promote growth [9]. Additionally, further advancements may involve the implementation of co-cultures, diffusion chambers, membrane systems, and bioreactors [[10], [11], [12], [13]]. Thus, specific enrichment and suplementation strategies can be tailored to build substantially larger collections of microorganisms with industrial relevance [14].
In their natural environment, bacteria thrive within communities and utilize sophisticated communication networks driven by metabolic changes that impact basal behavioral processes such as reproduction, sporulation, biofilm formation, and antimicrobial expression [15]. A particular type of chemical interaction is the production and detection of signaling compounds. Signaling molecules play a central role in microbial growth, as certain microorganisms struggle to survive under in vitro conditions where intercellular communication, known as quorum sensing, may be absent [16].
Among the known signaling molecules, acylhomoserine lactones represent the most well-understood compounds that are used by many Gram-negative bacteria [17,18]. In constrast, autoinductive peptides are involved in intercellular communication in Gram-positive bacteria and exhibit a dual function by acting as both antimicrobial agents and signaling molecules [19]. Quorum sensing systems also regulate the biosynthesis of siderophores, which also stimulate bacterial growth [20]. Remarkably, Paenibacillus elgii is known to produce peptides, self-inducing lactones, and siderophores, all of which are signaling molecules associated with bacterial growth stimulation [21,22].
Antimicrobial producing organisms are expected to contain self-resistance strategies against their own bioactive molecules. Typically, antibiotic-producing bacteria carry resistance genes as a protection mechanism against their antimicrobial products [23]. Therefore, in this study, we used a bacterial culture methodology that leverages a P. elgii supernatant as a selection mechanism for cultivating bacterial strains with biotechnological relevance. Furthermore, a strain obtained through this methodology was thoroughly characterized in order to confirm the effectiveness of the method proposed herein for isolating antimicrobial producing microbes.
2. Material and methods
2.1. Recovery of P. elgii supernatants
Supernatants of P. elgii cultures were obtained following the inoculation of a single P. elgii AC13 colony from LB agar into LB broth (Sigma-Aldrich, USA). The culture was grown at 28°C and 200 rpm, and then incubated at 4°C to induce bacterial sporulation. Subsequently, the culture was centrifuged at 5,000 × g and 4°C for 10 minutes, and the resulting supernatant was filtered using a 0.22 µm membrane filter. The absence of P. elgii AC13 cells was confirmed by plating 100 µL of the filtered supernatant onto LB agar plates following incubation for 48 hours at 28°C.
2.2. Isolation of resistant bacterial strains
Soil samples from the Brazilian Cerrado biome (Federal District, Brazil) were collected and suspended in Middlebrook media following a previously described protocol [24]. These suspensions were diluted with fresh media at the ratio of 1:20 and inoculated onto R2A agar plates (BD Difco, USA) supplemented with 0.3, 0.6, 1, or 2 mL.L−1 of P. elgii supernatant, and then incubated at room temperature for seven days. Resulting colonies were transferred to fresh R2A agar plates until pure cultures were obtained. The isolated resistant bacteria were stored at -80°C in 15 % glycerol (Sigma-Aldrich, USA) for further analysis [24].
2.3. Antimicrobial activity of resistant bacterial strains
The antimicrobial activity of soil bacteria was assessed using a modified version of the method described by Brady and collaborators [25], employing soft agar overlay assay. Each bacterial isolate was inoculated was centrally inoculated onto R2A agar plates and incubated at room temperature for six days. Subsequently, a soft R2A agar overlay previously inoculated with Bacillus subtilis, Escherichia coli, or Pseudomonas aeruginosa was poured over the individual isolates to evaluate the formation of growth inhibition zones after up to 48 hours of incubation at room temperature. A strain named K003, potentially identified as Methylobacterium radiotolerans, was selected for further analysis due to its ability to inhibit the growth of all tested strains. All experiments were conducted in biological and technical triplicates.
2.4. DNA extraction and 16S rRNA analysis of resistant bacterial strains
GenElute Bacterial Genomic DNA kit (Sigma-Aldrich, USA) was used to purify the total DNA from bacterial isolates following the manufacturer's instructions. DNA samples were analyzed by electrophoresis in 1 % agarose gel (Sigma-Aldrich, USA) and quantified using the Qubit dsDNA BR Quant-it assay kit (Invitrogen, USA) on a Qubit Fluorometer (Invitrogen, USA). Then, 16S rRNA gene amplification was performed using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGAC-3′) [26], and the resultant amplicons were purified using the Gene Jet PCR Purification Kit (Thermo Scientific, USA). Additionally, the purified 16S rRNA genes were also cloned into E. coli to enable precise identification [24]. Bidirectional sequencing of the 16S rRNA genes was performed on a Sanger platform (Macrogen, South Korea) to allow for the identification of bacterial sequences against the GenBank database (NCBI, NIH, USA). 16S rRNA marker genes from the strain K003 and other 11 representative sequences of valid species of the genus Methylobacterium were aligned using the Clustal W tool [27] implemented in Mega 11 software [28] to construct a similarity cladogram for the phylogenetic analysis [29]. The Maximum Likelihood (ML) method was used with 1000 sample repetitions (bootstrap) [30].
2.5. Whole-genome sequencing of Methylobacterium sp. K003
Genomic sequencing of Methylobacterium sp. K003 was conducted using a dual approach, which consisted of (i) short-read paired-end technology on the Illumina NextSeq 500 platform (Macrogen, South Korea) and (ii) long-read sequencing technology on the Oxford Nanopore Technologies platform (Oxford Nanopore Technologies, United Kingdom). Resultant long-read sequences supported the complete genome assembly from Illumina short-read sequences [[31], [32]]. Both DNA libraries were prepared according to the manufacturer's instructions, and genomic assembly and metrics were achieved following a previously established protocol [24]. Additionally, a visual representation of the circular genome and its coding regions was obtained using the Artemis software [33].
2.6. Genomic and pangenomic analysis of Methylobacterium sp. K003
First, the average nucleotide index (ANI) and digital DNA-DNA hybridization (dDDH) values related to Methylobacterium sp. K003 were calculated to perform taxonomic analysis using the JSpeciesWS v.3.9.1 [34] and the GGDC v.3.0 [35] servers, respectively. The genomes of the strain K003 and its reference strain – M. radiotolerans JCM 2831 – were aligned using the Plot Nucmer tool of the MUMmer v.3.1 toolkit [36], and then the ggbio package was used to generate a circular visualization of the resultant alignments [37].
Orthologous genes from the genomic sequences of twelve strains – including Methylobacterium sp. K003 and Enterovirga rhinocerotis YIM 100770 as the outgroup – were concatenated to construct a similarity cladogram through M1CR0B1AL1Z3R [38]. In addition, the genomes of K003 and its 10 closest strains according to dDDH analysis were annotated using Prokka v.1.14.6 [39]. These annotated genomes were then subjected to pangenome analysis using Roary v.3.13.0 [40,41], with a minimum of 80 % identity as the threshold to group genes encoding complete protein sequences into the core genome [42]. Based on these results, similarities in gene numbers between the strains were analyzed using the OrthoVenn 3 platform [43]. Phylogeny and gene distribution were further assessed by constructing a phylogenetic tree using Phandango [44].
2.7. Biotechnological potential of Methylobacterium sp. K003
Gene functions related to Methylobacterium sp. K003 were identified and classified into Clusters of Orthologous Groups (COGs) using eggNOG-mapper v.2 [45] and the KEGG database [46]. Subsequently, AntiSMASH v.6.1.1 was employed to identify secondary metabolism related genes [47] and CAMPR4 was used to predict antimicrobial peptides (AMPs) [48] [49]. In addition, peptide sequences were extracted from the annotated genomes using FaBox tools v.1.61 [50] to search for biologically active peptides (BAPs). Sequences with less than 100 amino acid residues were filtered and selected, and Peptide Ranker was used to identify BAP sequences. Sequences with a probability of over 0.5 were chosen for functional prediction. AntiCP [51], AntiFP [52], and AntiAngioPred [53] software were used to predict anticancer, antifungal, and anti-inflammatory functions, respectively.
Potential antibiotic resistance-related genes and CRISPR loci were inferred using the (i) resistance gene identifier (RGI) [54] and (ii) CRISPRCasFinder [55]. The similarity of biosynthetic gene clusters (BCGs) between K003 and its reference strain were explored using BiG-SCAPE with a distance cutoff of 0.3 [56] based on the AntiSMASH results [47,24]. This software was also used to compare the non-ribosomal peptide synthetase (NRPS)- and polyketide synthases (PKS)-related BCGs of both K003 and P. elgii AC13 strains.
2.8. Phenotypic characterization of Methylobacterium sp. K003
An in silico phenotypic prediction was performed to compare the phenotypes of Methylobacterium sp. K003 and its closest strains using Traitar [[57], [58], [59]]. Comparative phenotypic and physiological analysis between M. radiotolerans K003 and M. radiotolerans JCM 2831, obtained from the Brazilian Collection of Environmental and Industrial Microorganisms (CBMAI), were conducted to evaluate their potential singularities and unique features. Initially, K003 and JCM 2831 strains were analyzed using light microscopy with a DM750 microscope (Leica Microsystems, Germany) after Gram staining [60]. Optimal growth temperature, pH, and salt tolerance related to K003 and JCM 2831 were then assessed following a previously established protocol [24].
Biochemical tests were performed on cultures of strains K003 and JCM 2831 grown on R2A agar (BD Difco, USA) at 30°C for 48 hours. Potential catalase activity was determined following the protocol of Tindall and collaborators [61], whereas the oxidase activity was assessed using Oxidase Strips 40560 (Millipore Sigma, USA). Fatty acid profiling was carried out using the fatty acid methyl ester (FAME) isolation method [62] followed by gas chromatography (Agilent 7890A, Agilent, USA) equipped with the software Sherlock Microbial Identification System v.6.2 and the method library RTSBA6 [63]. Finally, the antibiotic susceptibility of these strains was assessed using the disk diffusion method [64] (Table 1).
Table 1.
Antibiotics and concentrations tested during the antibiotic susceptibility assay of Methylobacterium sp. K003 and Methylobacterium radiotolerans JCM 2831.
| Antibiotic | Concentration (μg/disc) |
|---|---|
| Amikacin | 30 |
| Amoxicillin/Clavulanic acid | 20/10 |
| Ampicillin | 10 |
| Aztreonam | 30 |
| Cephalexin | 30 |
| Cefazolin | 30 |
| Cefepime | 30 |
| Ceftazidime | 30 |
| Ceftriaxone | 30 |
| Ciprofloxacin | 5 |
| Clindamycin | 2 |
| Chloramphenicol | 30 |
| Doxycycline | 30 |
| Erythromycin | 15 |
| Gentamicin | 10 |
| Imipenem | 10 |
| Meropenem | 10 |
| Norfloxacin | 10 |
| Ofloxacin | 5 |
| Oxacillin | 1 |
| Penicillin G | 10 |
| Piperacillin + Tazobactam | 110 |
| Polymyxin B | 300 |
| Sulfonamide | 300 |
| Tetracycline | 30 |
| Vancomycin | 30 |
3. Results
3.1. Effects of P. elgii supernatant on the isolation of resistant bacterial strains
P. elgii and its supernatant exhibited strong antibacterial activity against B. subtilis (Fig. S1). The use of this supernatant was effective in stimulating selective microbial growth. Notably, the concentration of P. elgii supernatant revealed a relevant impact on the growth of microorganisms in the culture media inoculated with soil samples. The results indicated variations in bacterial growth at different supernatant concentrations, with significantly higher growth observed at lower concentrations. Consequently, as the concentration of the supernatant increased, the supplemented culture media became more selective (Fig. S2). Among the bacterial strains cultivated with P. elgii supernatant, 35 showed antibacterial activity [24]. Most of these strains were isolated from a supernatant concentration of 1000 mL.L−1, 11 strains from a concentration of 600 mL.L−1, and only three strains from the lowest concentration of 300 mL.L−1 (Tab. S1). The isolated strains were then identified by analyzing their sequenced 16S rRNA gene (Table 2), which revealed the presence of several genera, including Paenibacillus, Pseudomonas, Enterobacter, Kitasatospora, Sphingomonas, Xanthobacter, Burkholderia, and Methylobacterium.
Table 2.
Identification of the bacterial strains isolated from cultures supplemented with the supernatant of P. elgii after 16S rRNA genes sequencing.
| Strain | Access number | Description | Max Score | Total Score | Query Coverage | Max Ident |
|---|---|---|---|---|---|---|
| A1 | HQ236048.1 | P. elgii | 167 | 167 | 59 % | 92 % |
| A2 | JX174235.1 | Enterobacter sp. | 302 | 302 | 96 % | 95 % |
| A3 | AB739006.1 | P. ehimensis | 623 | 623 | 33 % | 92 % |
| A4 | AB739006.1 | P. ehimensis | 645 | 645 | 26 % | 98 % |
| A5 | HQ236085.1 | P. elgii | 1831 | 1831 | 98 % | 95 % |
| A6 | HM063032.1 | Paenibacillus sp. | 1943 | 1943 | 99 % | 97 % |
| A7 | AB739006.1 | P. ehimensis | 287 | 287 | 96 % | 93 % |
| A8 | AB739006.1 | P. ehimensis | 248 | 248 | 95 % | 90 % |
| A9 | HQ236085.1 | P. elgii | 474 | 474 | 98 % | 97 % |
| A10 | JQ735955.1 | P. elgii | 808 | 808 | 68 % | 98 % |
| A11 | HQ236048.1 | P. elgii | 1034 | 1034 | 99 % | 99 % |
| A12 | JX436405.1 | Pseudomonas sp. | 350 | 350 | 96 % | 99 % |
| B1 | JF701960.1 | Burkholderia sp. | 512 | 512 | 36 % | 92 % |
| B2 | GQ895737.1 | Sphingomonas sp. | 760 | 760 | 34 % | 95 % |
| B3 | JX090597.1 | P. aeruginosa | 22292 | 22292 | 99 % | 99 % |
| B4 | AF247494.1 | Burkholderia sp. | 642 | 642 | 97 % | 95 % |
| B5 | GU518381.1 | Uncultured bacterium | 115 | 115 | 83 % | 80 % |
| B6 | JX090597.1 | P. aeruginosa | 1000 | 1000 | 98 % | 98 % |
| B7 | EU139032.1 | Kitasatospora sp. | 263 | 263 | 94 % | 92 % |
| B8 | HQ651053.1 | Pseudomonas sp. | 1018 | 1018 | 98 % | 99 % |
| B9 | AF247494.1 | Burkholderia sp. | 388 | 388 | 48 % | 97 % |
| B10 | FR872408.1 | Burkholderia sp. | 329 | 329 | 90 % | 97 % |
| B11 | DQ095878.1 | P. aeruginosa | 176 | 176 | 64 % | 91 % |
| B12 | GU191167.1 | Burkholderia sp. | 294 | 294 | 88 % | 97 % |
| C1 | JX436403.1 | Pseudomonas sp. | 675 | 675 | 67 % | 96 % |
| C2 | JX436403.1 | Pseudomonas sp. | 497 | 675 | 63 % | 94 % |
| C3 | JX178938.1 | Xanthobacter sp. | 425 | 425 | 27 % | 95 % |
| C4 | HM771642.1 | P. aeruginosa | 990 | 990 | 99 % | 93 % |
| C5 | EU912440.1 | Methylobacterium sp. | 453 | 453 | 66 % | 88 % |
| K003 | EU912444.1 | Methylobacterium sp. | 193 | 193 | 73 % | 90 % |
| C7 | JX436403.1 | Pseudomonas sp. | 545 | 545 | 65 % | 94 % |
| C8 | JQ659918.1 | P. aeruginosa | 156 | 156 | 94 % | 81 % |
| C10 | JX436405.1 | Pseudomonas sp. | 279 | 279 | 98 % | 92 % |
| C11 | GU447238.1 | P. aeruginosa | 2017 | 2017 | 95 % | 96 % |
| C12 | AF247494.1 | Burkholderia sp. | 887 | 887 | 97 % | 95 % |
3.2. Antimicrobial activity of resistant bacterial strains
All cultivated strains were subjected to an overlay assay to evaluate antibacterial activity against B. subtilis, and they exhibited inhibition clear zones after incubation (Tab. S2; Fig. S3). Out of the 35 strains, five were selected for further testing against the Gram-negative bacteria E. coli and P. aeruginosa. The selection included at least one representative strain from each identified genus, except for Paenibacillus, Pseudomonas, and Enterobacter. The first was excluded for being related to P. elgii, and the second for being one of the seeded bacteria in the agar overlayassay. Enterobacter was also filtered out based on its reported lower biotechnological potential when compared to the other isolated strains. All tested strains showed antibacterial activity against E. coli, whereas only two showed activities against P. aeruginosa. A strain classified as Methylobacterium – Methylobacterium sp. K003 – was selected for genomic analysis based on the reported potential to promote plant growth [65], resistance against radiation [66], and antimicrobial and antioxidative properties [67,68].
3.3. Phylogenetic, genomic, and pangenomic analyses of Methylobacterium sp. K003
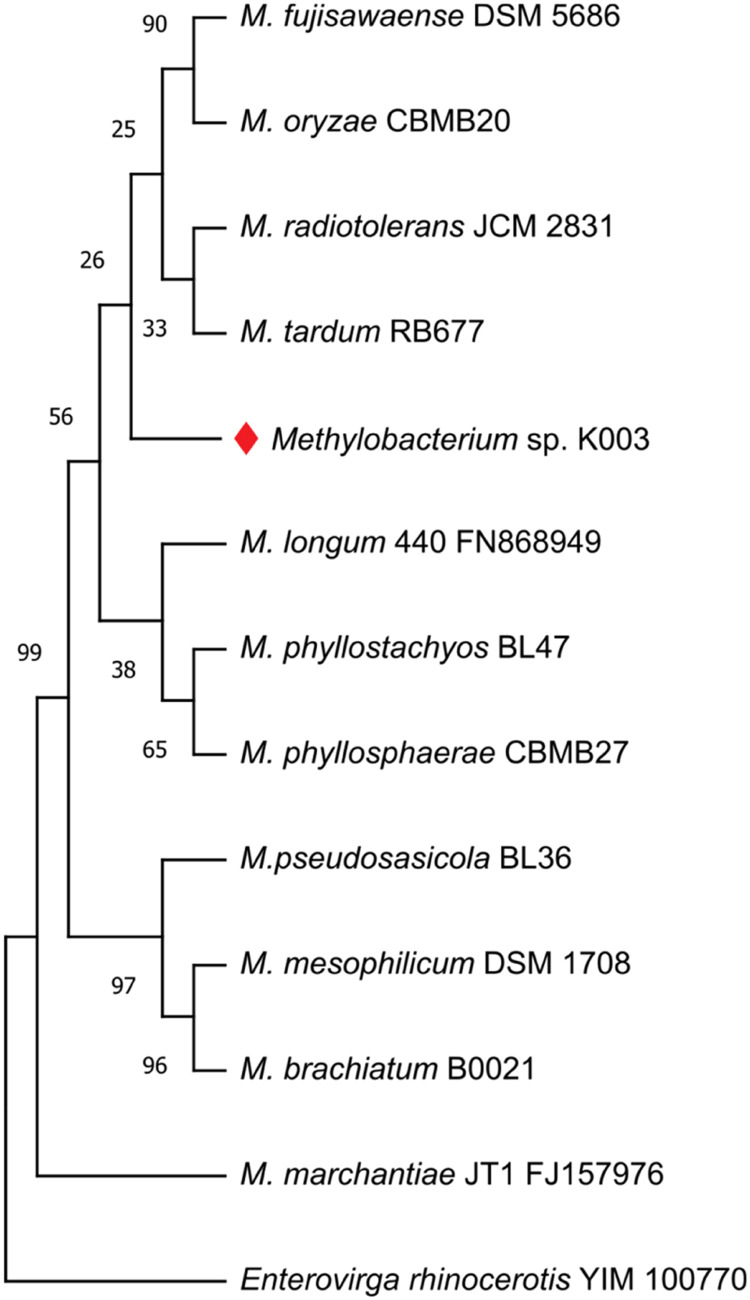
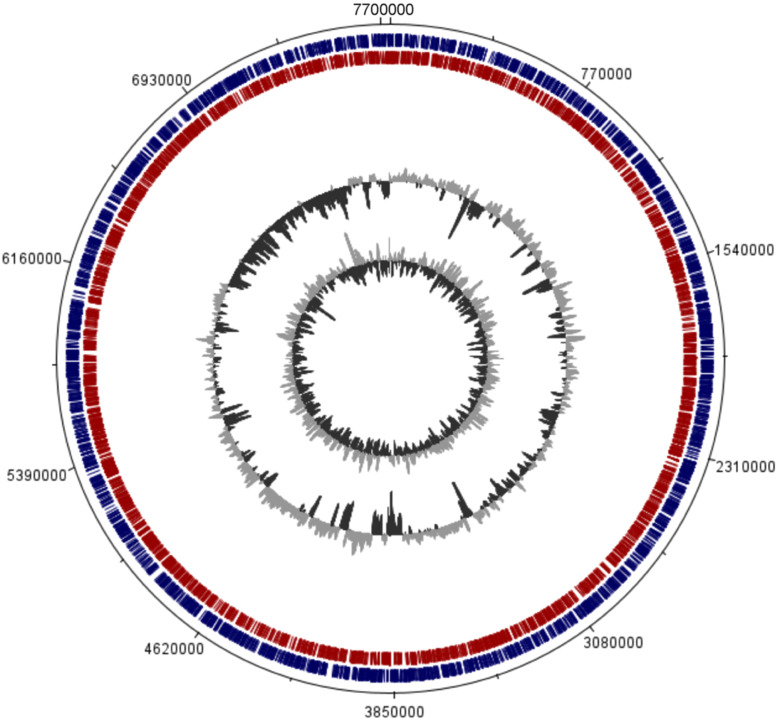
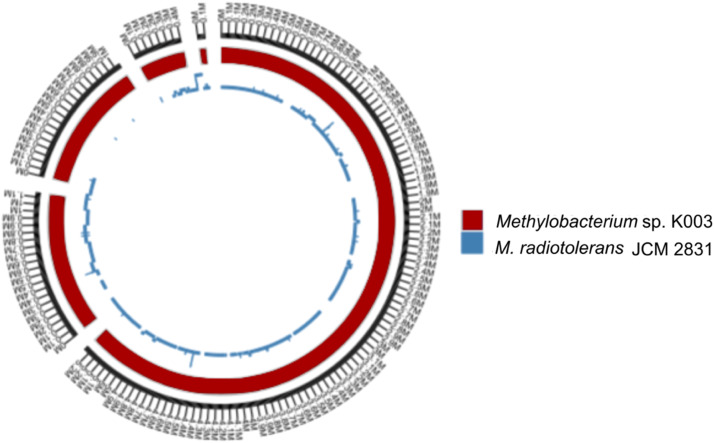
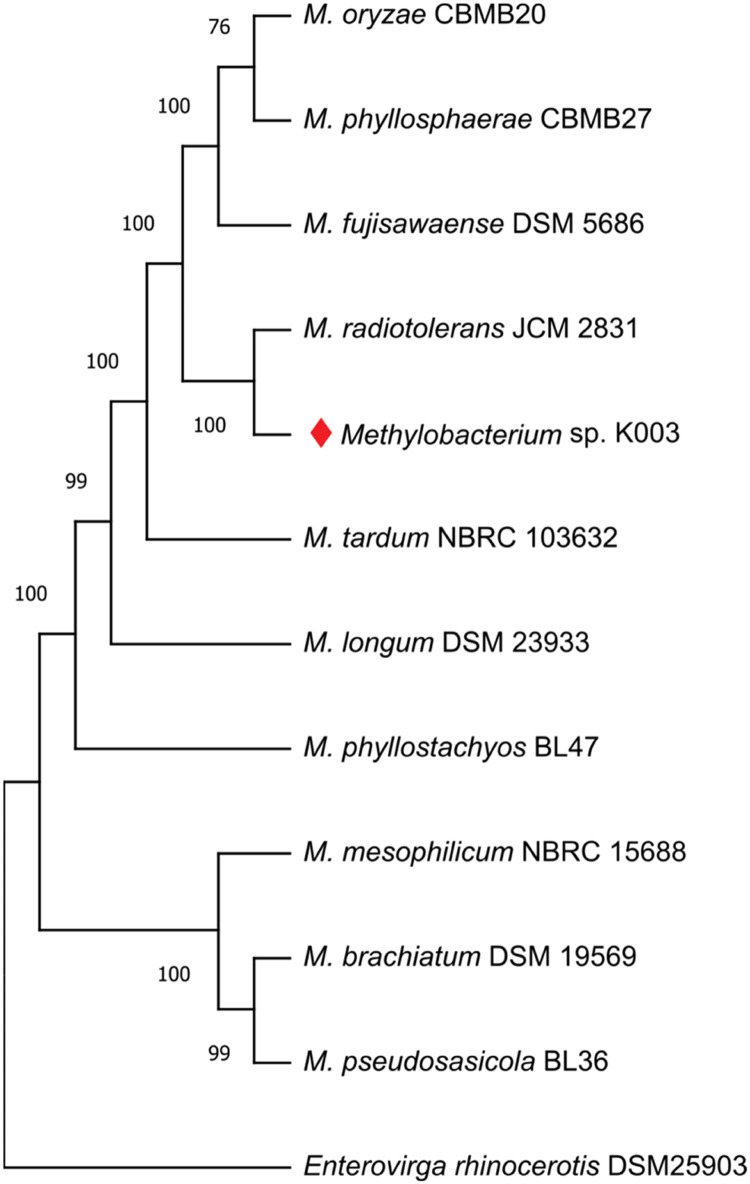
The identification of Methylobacterium sp. K003 was carried out using 16S rRNA gene sequence analysis, which also revealed its close relationship to the strain M. radiotolerans JCM 2831 with 99.88 % similarity. A similarity cladogram was then constructed using the 16S rRNA gene sequences of the Methylobacterium sp. K003 strain, other Methylobacterium strains, and Enterovirga rhinoceotis YIM 100770 as the outgroup (Fig. 1). Genome assembly of Methylobacterium sp. K003 resulted in a total size of 7,735,228 bp across five contigs and a GC content of 70.84 % (Table 3; Fig. 2). Genomic dDDH analysis indicated that M. radiotolerans JCM 2831 is the closest strain to K003, with a dDDH value of 90.3–94 % and ANI of 97.51 % (Fig. 3).
Fig. 1.
Similarity cladogram of 16S rRNA genes of Methylobacterium sp. K003 and other valid Methylobacterium strains. The evolutionary position of Methylobacterium sp. K003 was assessed using the Maximum-Likelihood (ML) method with 1000 sampling repetitions (Bootstrap).
Table 3.
Genomic assembly of Methylobacterium sp. K003 performed using long- and short-read sequencing.
| Assembly Data | |
|---|---|
| Contigs | 5 |
| CDS | 7292 |
| rRNA | 9 |
| Repeat region | 0 |
| tRNA | 73 |
| TmRNA | 2 |
| GC content (%) | 70,84 |
| Contamination (%) | 2,46 |
| Completeness (%) | 100 |
| Coverage | 400x |
| L50 | 1 |
| N50 (pb) | 5241738 |
| Total Size (bp) | 7735228 |
| Major contig | 5241738 |
Fig. 2.
Circular map of the genome assembly of Methylobacterium sp. K003, showing the coding regions and the GG content. Outside to center: forward CDS (dark blue), reverse CDS (red), GC content, and GC slope (dark gray and light gray).
Fig. 3.
Genomic alignment of Methylobacterium sp. K003 (red) and Methylobacterium radiotolerans JCM 2831 (blue), showing regions with a minimum of 1000 nucleotides and 90 % identity. The outer band represents the size scale of the contigs, whereas blue bands with a two-layer depth indicate overlapping sequences present in both strains.
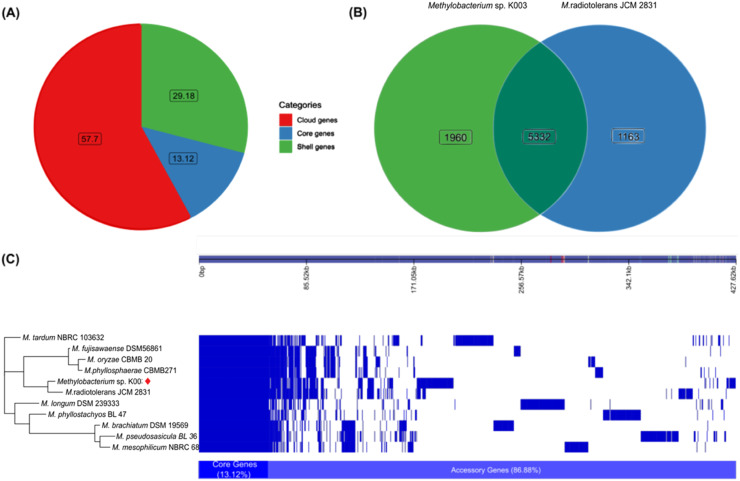
To assess the phylogenetic relationships within the genus Methylobacterium, the genomes of the twelve analyzed strains underwent orthologous gene set analysis. A total of 233 core genes were recovered and used to construct a maximum likelihood phylogenetic tree (Fig. 4). A total of 21,379 gene families constituted the K003 pangenome, resulting in 2,806 (13.12 %) core genes, classified as accessory genes with rare genes representing 57.7 % (Fig. 5A). Comparison between the genomes of K003 and JCM 2831 showed a total of 8,455 genes, with 5,332 genes (63.06 %) shared between both strains (Fig. 5B). Analysis of the pangenome facilitated the determination of the presence or absence of genes in each genome. The phylogenetic tree highlighted a core set of genes shared by all twelve genomes, as well as distinct clades corresponding to the analysis of core orthologous genes. Notably, Methylobacterium sp. K003 and its closest strain, M. radiotolerans JCM 2831, clustered together within the same clade (Fig. 5C).
Fig. 4.
Maximum-likelihood phylogenetic tree of Methylobacterium strains based on 233 core orthologous genes concatenated from genomic sequences. The dataset included 12 Methylobacterium strains, including Methylobacterium sp. K003 and its closely related strains, with Enterovirga rhinocerotis YIM 100770 as the outgroup.
Fig. 5.
Pangenomic and phylogenetic analysis of Methylobacterium sp. K003. (A) Pangenome distribution illustrating three gene categories across genomes: core genes (present in 99 % to 100 % of the genomes), shell genes (present in 15 % to 95 % of the genomes), and cloud genes (present in 0 % to 15 % of the genomes). (B) Venn diagram showing the number of genes common between Methylobacterium sp. K003 and its closest strain Methylobacterium radiotolerans JCM 2831. (C) Maximum-likelihood phylogenetic tree based on the presence and absence of genes from Methylobacterium sp. K003 pangenome. Blue bar represents the pangenome, with each line corresponding to a strain and each column indicating gene variation. Gene presence is depicted in a descending frequency, with empty spaces representing gene absence. Genome comparison includes Methylobacterium sp. K003 and its closest genomes, using a minimum blastp identity of 80 % for grouping genes into the core genomes.
3.4. Biotechnological potential of Methylobacterium sp. K003
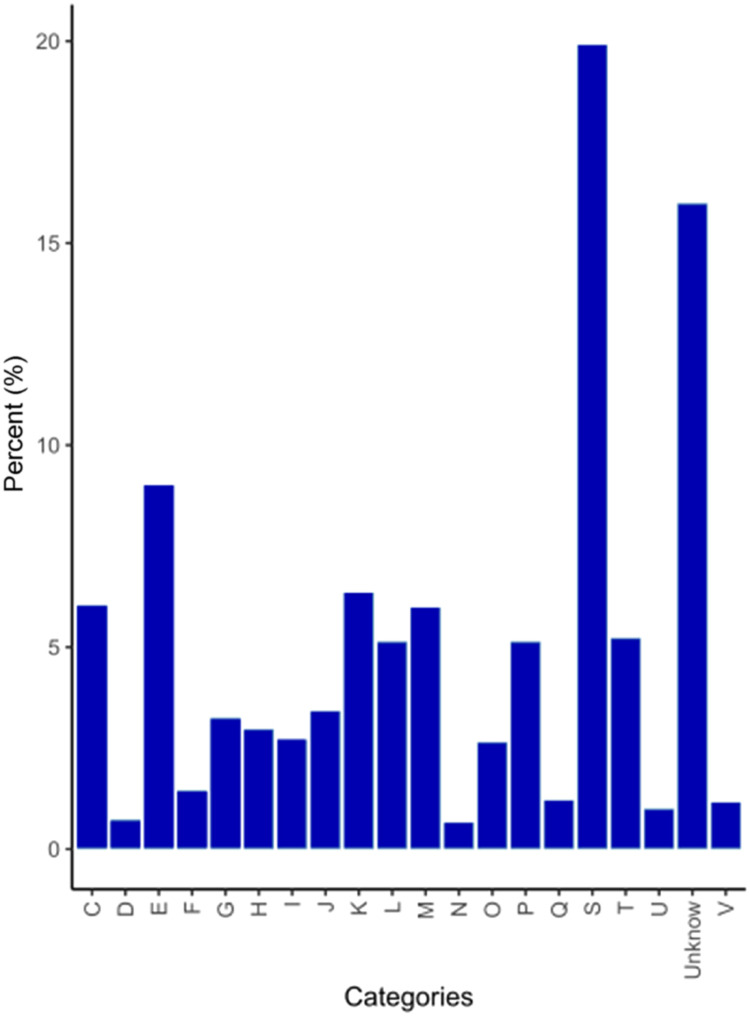
The genome of Methylobacterium sp. K003 was annotated and the predicted proteins were successfully classified into COG categories (Fig. 6). From the proteins encoded in the Methylobacterium sp. K003 genome, 34 % were assigned to unknown function (19 %) or to any of the COG categories (15 %). Other COG categories corresponded to amino acid metabolism (9 %), transcription (6 %), and energy production and conversion (6 %), which were the most abundant functions identified in the genome. The least abundant functions were related to cell cycle (0.7 %) and cell motility (0.6 %).
Fig. 6.
Distribution of encoded proteins in the genome of Methylobacterium sp. K003 across their respective COG categories.
Four genes associated with the resistance-nodulation-cell division (RND) efflux pumps were identified in both K003 and JCM 2831 genomes, all of which were related to fluoroquinolone and tetracycline resistance with similarities ranging from 41 % to 71 %. A total of 40 AMPs were identified in the K003 genome with probability scores ranging from 0.68 to 0.75 [49]. In addition, it was also detected in the genome of K003 two CRISPR matrices with 11 and 19 spacers, and a Type IIIA locus lacking CRISPR arrays. However, no complete CRISPR-cas systems were detected. Accordingly, two CRISPR arrays with 1 and 6 spacers were identified in the genome of JCM 2831 with no cas genes found.
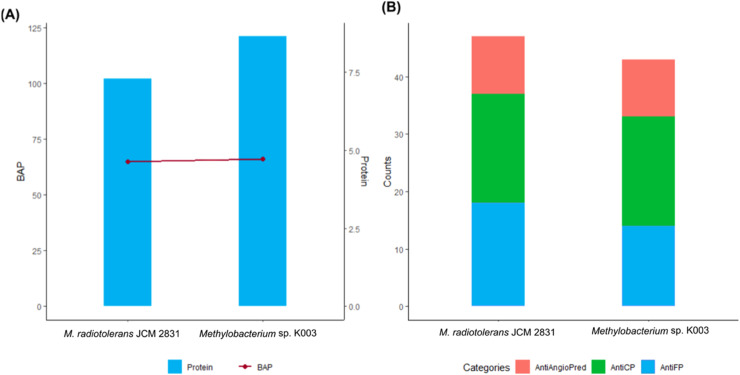
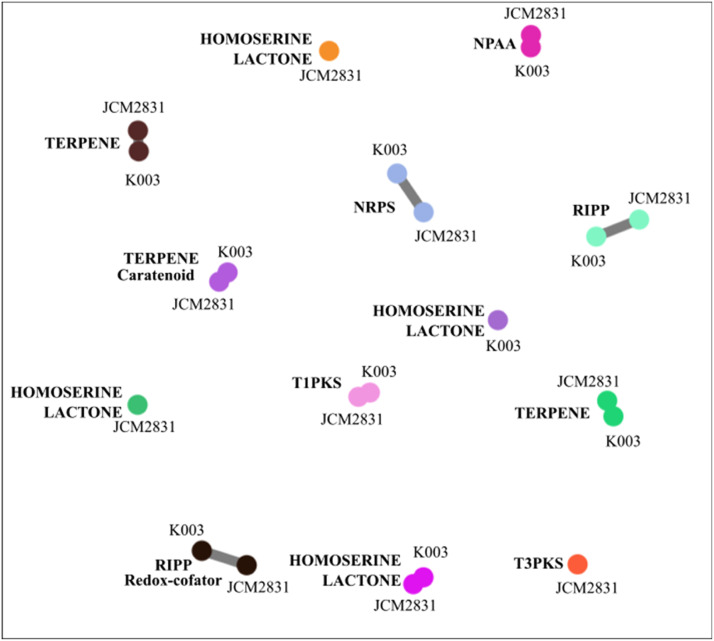
The functional annotation of genomes revealed 10 gene clusters in the K003 genome (Table 4), and most of these were associated with the production of antibacterial compounds. Overall, K003 and JCM 2831 gene clusters were highly similar, except for the absence of T3PKS genes and one less cluster of Hserlactone in K003. A total of 121 peptides were identified in the strain K003, whereas 102 were found in the strain JCM 2831. Among these peptides, 66 and 65, respectively, were classified as BAPs (Fig. 7A). Specifically, Methylobacterium sp. K003 showed 14 BAPs classified as antifungal, 19 as anticancer, 10 as anti-inflammatory, and 23 showed no predicted biological function. In contrast, the strain JCM 2831 showed 18 peptides classified as antifungal, 19 as anticancer, 10 as anti-inflammatory, and 18 with no predicted biological function (Fig. 7B).
Table 4.
Biosynthetic gene clusters of secondary metabolites from the genome of Methylobacterium sp. K003 compared to its reference strain Methylobacterium radiotolerans JCM 2831.
| Secondary metabolites | ||
|---|---|---|
| No. of clusters | ||
| Type | K003 | JCM 2831 |
| Hserlactone | 2 | 3 |
| NAPAA | 1 | 1 |
| NRPS | 1 | 1 |
| Redox-cofator | 1 | 1 |
| RIPP | 1 | 1 |
| T1PKS | 1 | 1 |
| T3PKS | 0 | 1 |
| Terpenoid | 3 | 3 |
Fig. 7.
Peptide distribution and functional prediction in the genomes of Methylobacterium sp. K003 and Methylobacterium radiotolerans JCM 2831. (A) Total number of peptides with fewer than 100 residues and their predicted biological function in each strain (BAP). (B) Predicted biological functions of biologically active peptides (BAP) in each strain, with a focus on anti-inflammatory (AntiAngioPred), anticancer (AntiCP), and antifungal (AntiFP) activities.
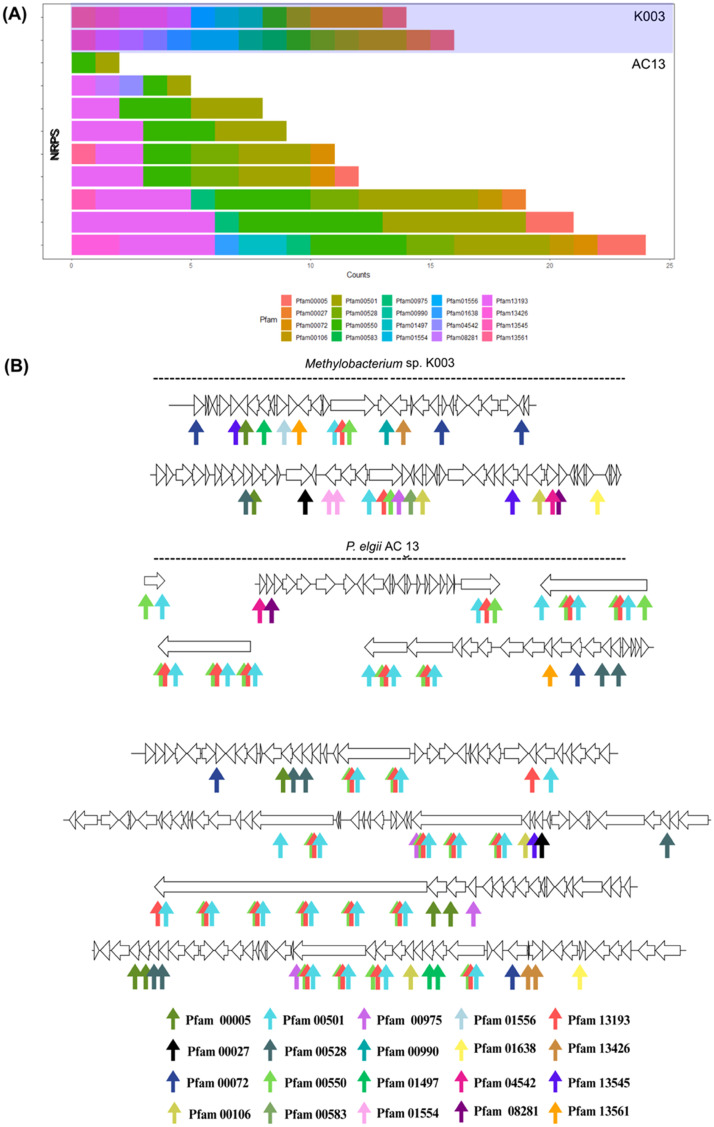
BGC analysis identified 10 families related to the strain K003 and 12 to the strain JCM 2831. Genes associated with similar products in both strains include three terpene-, one NRPS-, one homoserine lactone-, one non-proteinogenic amino acid (NPAA)-, two RIPP-, and one T1PKS-related genes. One homoserine lactone gene cluster from the strain K003 did not show similarity to the homoserine lactone clusters found in the strain JCM 2831 (Fig. 8). A total of 24 BCGs were classified as NRPS in the strain K003 (2 BCGs) and in P. elgii AC13 (22 BCGs). These BCGs comprise 325 different Pfam domains. Of these Pfam domains, 226 were related to a single BCG and, therefore, were disregarded. From the remaining 99 Pfam domains, 20 were found in both the NRPS of K003 and of P. elgii AC13 strains. The most abundant domains corresponded to the AMP-binding enzyme, phosphopantetheine-binding, and AMP-binding enzyme C-terminal domain (Fig. 9A). These domains were found in all BCGs of both K003 and P. elgii AC13 strains, except for a smaller BCG found exclusively in the strain AC13 (Fig. 9B). Additionally, three PKS-NRPS hybrids and three other PKS were found in the strain AC13, but none were detected in the strain K003.
Fig. 8.
Graphical representation of the similarity between biosynthetic gene cluster (BCG) families from Methylobacterium sp. K003 and Methylobacterium radiotolerans JCM 2831. Each link represents similar gene clusters between the analyzed strains. Singletons, presented in isolated form, are specific clusters unique to each strain, indicating no similarities between the strains.
Fig. 9.
Similarities between biosynthetic gene clusters (BCGs) found in Methylobacterium sp. K003 and Paenibacillus elgii AC13. (A) The total number of Pfam domains per BCG shared between the analyzed strains. (B) Graphical representation of similar BCGs between K003 and AC13 strains.
3.5. Phenotypic characterization of Methylobacterium sp. K003
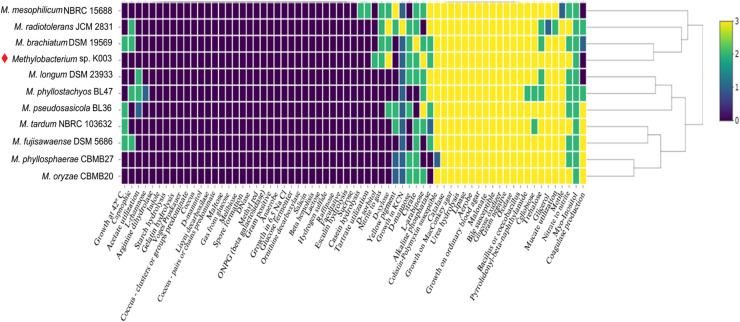
Phenotypic prediction of Methylobacterium sp. K003 revealed that the strain was positive for D-sorbitol, in contrast to its closest strains (Fig. 10). Despite its similarities with M. radiotolerans JCM 2831, the strain K003 tested negative for capnophilic and coagulase production, and positive for D-sorbitol and L-arabinose. Indeed, M. brachiatum DSM 19569 was found to be the closest strain to the strain K003 based on the phenotypes. However, Methylobacterium sp. K003 tested negative for growth at 42°C as well as capnophilic and coagulase production, as opposed to M. brachiatum DSM 19569.
Fig. 10.
Comparative analysis of phenotypic predictions from the genome assembly of Methylobacterium sp. K003. The comparison includes genomes of the closest strains according to dDDH, as available in the NCBI database. Scoring system: a value of zero indicates a negative phenotype, whereas values of 1 and 2 correspond to positive phenotypes in the phypat and phypat+PGL predictors, respectively. A value of 3 indicates a positive phenotype in both predictors.
Methylobacterium sp. K003 and reference strain exhibit aerobic growth within a temperature range of 15–40°C when cultivated in NB medium with NaCl concentrations of 0–2 % (w/v) and 0–3 %, respectively. It also grows under the pH range of 5–9, whereas the strain JCM 2831 grows under a broader pH range of 4–9. The optimal growth conditions for both strains were observed at 30°C, pH 8, and 0 % (w/v) NaCl (Table 5), and both strains tested positive for catalase and oxidase.
Table 5.
Comparative analysis between the phenotypes of Methylobacterium sp. K003 and Methylobacterium radiotolerans JCM 2831 strain, where (+) represents the presence and (–) the absence.
| Growth range (optimum) | K003 | JCM 2831 |
|---|---|---|
| Temperature (°C) | 15-40 (30) | 15-40 (30) |
| pH | 5-9 (8) | 4-9 (8) |
| NaCl (%) | 0-2 (0) | 0-3 (0) |
| Catalase | + | + |
| Oxidase | + | + |
The most abundant fatty acid found in the strains K003 and JCM 2831 was Summed Feature 8, corresponding to over 85 % of the total content of fatty acids in both strains. The least abundant fatty acid was Summed Feature 2 (0.74 %) in the strain K003 and C12:0 (0 .67 %) in the strain JCM 2831. Overall, the resultant fatty acid profiles showed clear differences between JCM 2831 and K003 strains due to the absence of C12:0 and 18:0 3OH in the latter (Table 6). Out of the 26 antibiotics tested for antimicrobial susceptibility, both strains were resistant to amoxicillin/clavulanic acid, aztreonam, cefazolin, cefepime, clindamycin, meropenem, oxacillin, polymyxin B, and sulfonamide. In addition, the strain K003 also showed resistance to vancomycin (Table 7).
Table 6.
Fatty acids profile (%) of Methylobacterium sp. K003 compared to its reference strain Methylobacterium radiotolerans JCM 2831. Peaks with imperfect separation were collectively reported as "Summed Features" with specific components identified: Summed Feature 2 (putative 12:0 aldehyde), Summed Feature 3 (C16:1 ω6c and/or C16:1 ω7c), and Summed Feature 8 (C18:1 ω7c and/or C18:1 ω6c). Absence of a particular fatty acid is denoted by (−).
| Fatty acid | Name | Types of fatty acid | K003 (%) | JCM 2831 (%) |
|---|---|---|---|---|
| C12:0 | Dodecanoic acid | Medium chain Saturated | - | 0.67 |
| C16:0 | Hexadecanoic acid | Long chain Saturated | 2.37 | 2.81 |
| C18:0 | Octadecanoic acid | Long chain Saturated | 5.75 | 5.59 |
| 18:0 3OH | 3-Hydroxyoctadecanoic acid | Long chain Saturated with a hydroxyl | - | 0.84 |
| Summed Feature 2 | - | - | 0.74 | 0.87 |
| Summed Feature 3 | - | - | 0.76 | 0.90 |
| Summed Feature 8 | - | - | 90.39 | 88.33 |
Table 7.
Antibiotic susceptibility assay of the isolated bacterial strains using the disc diffusion method [64], where (+) indicates susceptibility and (−) indicates resistance.
| Anibiotic (μg per disc) | K003 | JCM 2831 |
|---|---|---|
| Amikacin (30) | + | + |
| Amoxicillin/Clavulanic acid (20/10) | - | - |
| Ampicillin (10) | + | + |
| Aztreonam (30) | - | - |
| Cephalexin (30) | + | + |
| Cefazolin (30) | - | - |
| Cefepime (30) | - | - |
| Ceftazidime (30) | + | + |
| Ceftriaxone (30) | + | + |
| Ciprofloxacin (5) | + | + |
| Clindamycin (2) | - | - |
| Chloramphenicol (30) | + | + |
| Doxycycline (30) | + | + |
| Erythromycin (15) | + | + |
| Gentamicin (10) | + | + |
| Imipenem (10) | + | + |
| Meropenem (10) | - | - |
| Norfloxacin (10) | + | + |
| Ofloxacin (5) | + | + |
| Oxacillin (1) | - | - |
| Penicillin G (10) | + | + |
| Piperacillin + Tazobactam (110) | + | + |
| Polymyxin B (300) | - | - |
| Sulfonamide (300) | - | - |
| Tetracycline (30) | + | + |
| Vancomycin (30) | - | + |
4. Discussion
For years, the scientific community considered microorganisms as self-sufficient individual entities incapable of communicating or organizing themselves in groups [69]. However, it is now known that bacteria and other prokaryotes have communication systems that encompass different processes involving the production, release, and detection of chemical compounds called autoinducers. These are small molecules similar to hormones that trigger a variety of cellular responses and enable individual microorganisms to assess the cell density of a local population and coordinate gene regulation in a population [70].
Cell-to-cell communication systems (i.e., quorum sensing) involve density-dependent formation of autoinducing acylhomoserine lactone (AHL) molecules or short peptides that control light production, expression of virulence factors, swarming, biofilm formation, cell aggregation, and genetic competence [18]. Known metabolic interactions also involve compounds participating in central metabolism, such as siderophores [20].
Most soil microorganisms resist cultivation under laboratory conditions, leading to an enormous gap in the assessment of its biochemical diversity. To unlock this metabolic potential, it is necessary to simulate the biochemical context found in the natural environments of the microbial communities. Isolating novel environmental microorganisms allows us to access the still untapped source of metabolic pathways and natural products [71]. In traditional culture methods, bacteria are normally grown in pure cultures with a high availability of nutrients and resources, which contrasts to their complex and competitive natural habitat [72]. Complex environments, such as soil, comprise a broad diversity of microorganisms [73] that modify the microenvironmental conditions, thus facilitating the growth of other microorganisms by sharing metabolites and essential compounds for growth [74]. These molecules include those that play a role in quorum sensing, bacterial fitness, and community shapping [75], whereas the absence of other community members in pure cultures reduces the potential for microbial recovery [76].
In this scenario, some bacteria can only grow under laboratory conditions when in co-culture with another member of the microbial community [10]. Co-culturing can be achieved by directly culturing the microorganism and a helper strain together or by using supernatants of helper strains [71,77]. The use of P. elgii supernatant enriches the culture with inducing molecules and antimicrobial compounds that can mimic naturally occurring chemical-ecological relationships. This may enhance microbial recovery from environmental samples [78], and thus has emerged as an effective and targeted approach to finding new bioactive compounds with activity against the pathogens for which new antibiotics are needed [79].
The supernatant of P. elgii is rich in siderophores and self-inducing lactones, as well as peptides with both self-inducing and antibacterial properties [22]. This may explain the growth stimulus at different concentrations of P. elgii supernatants, the diversity within the genus cultivated, and the selective growth for strains with biotechnological potential. The positive results obtained from antibacterial tests conducted on both the supernatant of P. elgii and the strains obtained through the approach presented herein demonstrate the protocol's efficacy in selecting strains with activity similar to that of the supernatant.
Indeed, by using enriched cultures with P. elgii supernatant, 35 fast-growing cultivable bacterial strains were successfully isolated and identified, most of which were from genera known to have significant biotechnological potential. These include Paenibacillus, Pseudomonas, Enterobacter, Kitasatospora, Sphingomonas, Xanthobacter, Burkholderia, and Methylobacterium. A Methylobacterium strain was characterized to confirm the efficacy of this methodology in selecting microorganisms with potential biotechnological applications.
Species within the genus Sphingomonas exhibit a spectrum of properties, ranging from the bioremediation of environmental contamination to the production of highly beneficial phytohormones. The antibacterial activity of Sphingomonas was confirmed by Akinsanya et al. (2017) [80] against several pathogens, including S. typhimurium, P. vulgaris, K. pneumoniae, E. coli, S. aureus, S. pyogenes, B. cereus, and C. albicans. Recent studies have shown an intriguing role of Sphingomonas species in the degradation of organometallic compounds [81]. Morevover, endophytic bacteria belonging to the genus Xanthobacter are known to promote plant growth. Xanthobacter exhibits promising potential as biofertilizer, as a control of weeds that compete with plant cultivation, and as an useful biological tool for the bioremediation of water and soils polluted by chemical agents [82]. The Kitasatospora genus is renowned for producing novel secondary metabolites with diverse structures, that exhibit activity against eukaryotic organisms [83]. Addittionally, it demonstrates effectiveness against both Gram-positive and Gram-negative pathogens [84] and showcases resistance to various heavy metals [85].
Traditionally recognized for its pathogenic role, the Burkholderia genus shows a broader spectrum of environmental interactions, suggesting multiple beneficial properties. These bacteria are increasingly acknowledged for their involvement in bioremediation, biological control, and substantial biotechnological potential, particularly as producers of antimicrobial agents [[86], [87], [88]]. Although the genus Enterobacter is known as a phytopathogen [89], it has been recognized for its effectiveness as a bioinoculant for rice under alkaline salinity stress [90] and its ability to degrade polyethylene [91]. Similarly, Pseudomonas – another genus with significant biotechnological and health impact – has representatives identified as plant growth promoters [92], bioremediation agents [93], and producers of various natural compounds, including rhamnolipids, terpenoids, polyketides, and non-ribosomal peptides [94]. Furthermore, Pseudomonas species are known for their ability to produce biopolymers [95,96].
The results of this study encompass the characterization of the K003 strain and a draft of its genome, explored through genome mining. This integrative approach has yielded promising findings related to new antimicrobial metabolites. Despite being a previously described species, the K003 strain exhibits unique characteristics. While it shares similarities with its reference strain (JCM 2831) in terms of genome alignment, pangenome, and the diversity of biosynthetic clusters, it is distinguished from the JCM 2831 strain in several aspects. These include growth at pH 5, absence of specific fatty acids, vancomycin resistance and coagulase production, as well as presence of a Type IIIA case and both D-sorbitol and L-arabinose activities.
In comparison to a previous study on 12 strains of Methylobacterium [97], our findings align particularly well with the observation of their main fatty acid Summed Feature 8. Notably, our strains exhibited a higher tolerance to salinity levels, reaching 2 % and 3 % for K003 and JCM 2831 strains, respectively, as opposed to the 1 % salinity tolerance reported in the cited study. Regarding pH, all strains generally thrived within the pH range of 4-5 and 8. In a comprehensive study of 62 strains of Methylobacterium and Methylorubrum [98], a diverse range of their genomic characteristics was assessed. Specifically, the genome size of the strains varied from 4.4 to 8.8 Mb, encompassing a wide span of coding sequences (CDS) from 4171 to 8622, and GC content ranging from 65.9 % to 72.7 %. Indeed, the data obtained in this study indicate that the K003 strain aligns with the expected GC content (70.84 %) and stands among the strains with larger genomes, with a genome size of 7.7 Mb.
Although the in vitro tests indicated susceptibility of K003 and JCM 2831 strains to tetracycline, a detailed annotation of the results on the Antibiotic Resistance Gene Database (ARDB) revealed the presence of four RND antibiotic efflux pumps. These pumps, identified as potential contributors to antibiotic resistance, were associated not only with tetracycline but also with fluoroquinolones. Notably, the strain K003 was resistant to Polymyxin B, which was an expected result since it refers to a class of antibiotic lipopeptides traditionally produced by the genus Paenibacillus with structural similarity to pelgipeptins produced by P. elgii [99,21]. Polymyxins are lipopeptides with a wide range of activities, from antimicrobial to anticancer. Together with octapeptins, polypeptins, iturins, surfactins, fengycins, fusaricidins, and tridecaptins, they are produced through NRPS, of which its cluster is responsible for the biosynthetic pathway that was found in the strain K003 [99].
In addition, the high percentage (34 %) attributed to S and Unknown categories in the K003 genome underscores the existing limitations in understanding the proteome of these microorganisms [100]. This observation highlights a challenge in genome annotation, which relies on databases that primarily focus on previously described structures. Alessa et al. [98] found that the genomes of their 62 strains analyzed were significantly enriched in metabolism processes, which is consistent with our findings in the strain K003. Specifically, category E, related to amino acid metabolism, emerged as the most abundant among the described categories.
In the strain K003, the identification of CRISPR systems, which are a defense mechanism employed by prokaryotes against invaders like viruses and plasmids [101], holds significant biotechnological relevance. This system's DNA editing capabilities are particularly noteworthy and offer substantial potential for genetic engineering applications [102]. Many classes of natural products have also been identified, including terpenoids, alkaloids, polyketides, and non-ribosomal peptides [103]. Specific attention will be given to some of these classes, as their production-related gene clusters are present in the genome of the strain obtained in this study.
The NRPS cluster identified in the strain K003 is associated with enzyme complexes organized into specific amino acid modules. NRPS utilizes non-proteinogenic amino acids, expanding the possibilities beyond the standard 20 amino acids found in ribosomal peptides, resulting in several hundreds of potential structures [104]. The natural products synthesized by NRPS clusters often exhibit diverse properties, including antiviral, antitumor, or antibiotic activities against both human and plant pathogens [105]. This versatility has captured the interest of various industries, including biotechnology, pharmaceuticals, and agriculture [106].
NRPS are responsible for the production of the majority of lipopeptides from Paenibacillus, the genus from which the supernatant was used to isolate and grow the K003 strain. The primary mode of action of these lipopeptides tipically involves the disruption of the target cell membrane. Due to the inherent difficulty of these cells in reorganizing their membranes, the development of resistance to lipopeptides is suggested to be slow and limited. This is pivotal from the perspective of developing new drugs [99].
The identification of NRPS clusters in the K003 genome, responsible for pelgipeptin production – a non-ribosomal peptide derived from P. elgii [107] – suggests the potential production of analogous compounds in both the K003 and P. elgii strains. According to the AntiSMASH results, Paenibactin, which is a linear counterpart of pelgipeptins [108], showed 100 % similarity with an NRPS cluster of the strain P. elgii AC13, a cluster that in terms of Pfam domains showed similarities with the M. radiotolerans K003 clusters through BiG-SCAPE. These observations support the notion that bacteria resistant to specific antimicrobials can synthesize compounds similar to those antimicrobials.
PKS enzymes constitute a large group of natural products with great diversity and biological activities, such as antibiotics, antifungals, anticancer agents, cholesterol-lowering agents, and immunosuppressants [109]. They are classified into Types I, II, and III, but solely Type I was identified in the strain K003. The Type I PKS class, known as polyether ionophores, has been widely used as effective veterinary drugs and exhibits bioactivity, including antifungal, antiviral, antitumor, herbicidal, and anti-inflammatory activity. Polyether ionophores play a crucial role in inducing cell death in target cells as they are capable of transporting cations across plasma membranes, leading to depolarization and subsequent cell death. This ability becomes very interesting due to its antimicrobial activities [110].
The Hserlactone cluster in the K003 genome refers to a homoserine lactone and is associated with quorum sensing. In Gram-negative bacteria, acylated homoserine lactones (acyl-HSL) act as common signal molecules that regulate several target functions and are frequently implicated in interactions with the host. Quorum sensing plays a crucial role in the formation of biofilms in bacteria, including opportunistic human pathogens. Thus, if pathogens use acyl-HSL signals to induce virulence gene expression, these systems become unique targets for drug discovery [111].
Terpenes found in microorganisms have demonstrated potent antimicrobial properties [112]. In the genome of the strain K003, terpenoid clusters are evident and one of them was identified as a carotenoid, an important component for coping with hostile environments characterized by intense UV light exposure. This is particularly relevant for Methylobacterium sp., as they are known to be abundant and adapted to the phyllosphere [98,113]. The gene clusters identified in the strain K003, with the exception of a terpene recognized as a carotenoid, displayed minimal (<11 %) or no similarity to known compounds. This suggests the potential production of novel bioactive compounds by the isolated strain.
The NPAA found in the K003 genome consists of a cluster of polyamino acids similar to ε-poly-L-lysine (ε-PL). It is an antimicrobial peptide with activity against bacteria that cause spoilage and food poisoning [114]. Another large class of natural products comprises ribosomally synthesized and post-translationally modified peptides (RiPP). Their related gene clusters were also found in the K003 genome and result in the production of compounds with antimicrobial activity. Natural products that present post-translational modifications have diverse structures, which improves target recognition and, therefore, their chemical functionality [115]. Our findings related to the secondary metabolism of the strain K003 through AntiSMASH, together with the predicted biologically active peptides with antibacterial, anti-inflammatory, anti-cancer, and antifungal functions, indicate the biotechnological relevance of the strain K003 selected using the supernatant of P. elgii.
5. Conclusions
The methodology tested in this work demonstrated efficacy in isolating diverse genera of bacteria with biotechnological significance, including Paenibacillus, Pseudomonas, Enterobacter, Kitasatospora, Sphingomonas, Xanthobacter, Burkholderia, and Methylobacterium. All strains obtained using the supernatant of P. elgii exhibited antibacterial activity in overlay assays, consistant with the resistance to antimicrobials produced by P. elgii. Therefore, the use of supplemented cultures sets the stage for future investigations of relevant metabolic traits of cultured strains within an industrial perspective.
Genomic sequencing data confirmed that the strain K003 is classified as M. radiotolerans. However, the strain K003 showed unique features compared to its reference strain, includind differences in the fatty acid profile, resistance to vancomycin, distinct biosynthetic gene clusters, and predicted phenotypic traits. The draft genome of Methylobacterium sp. K003 provided a glimpse into the biosynthetic diversity available in strains selected using P. elgii supernatant. The identified gene clusters in the strain K003 is likely linked to the antibacterial activity observed in overlay assays. Indeed, similarities were found in NRPS of K003 and P. elgii AC13 strains, which are responsible for the production of pelgipeptin in P.elgii, supporting that bacteria resistant to certain antimicrobial peptides likely produce similar compounds. Furthermore, future endeavors focusing on identifying and isolating Methylobacterium sp. K003 metabolites can leverage genomic insights into the biosynthetic origins of its potential bioactive compounds. It is noteworthy that most of the biosynthetic gene clusters lacked similarity with published clusters, suggesting a novel source of pharmaceutically relevant products.
Data Availability Statement
The type strain is K003, which is isolated from the soil of the Brazilian savanna-like Cerrado biome, Brasilia, Brazil. The deposit of the K003 genome in the GenBank database is being processed under accession number JAZBNP000000000.
CRediT authorship contribution statement
I. C. Cunha-Ferreira: Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization. C. S. Vizzotto: Investigation, Formal analysis, Data curation, Conceptualization. T. D. Frederico: Formal analysis, Data curation. J. Peixoto: Writing – review & editing, Writing – original draft, Formal analysis, Data curation. L. S Carvalho: Writing – review & editing, Methodology. M. R. Tótola: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. R. H. Krüger: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors thank the Laboratório de Biotecnologia e Biodiversidade para o Meio Ambiente (Universidade Federal de Viçosa - UFV) for the infrastructure to carry out part of the experiments in this work. This study was funded by the National Council for Scientific and Technological Development (CNPq), the Coordination for the Improvement of Higher Education Personnel (CAPES), and the Federal District Research Support Foundation (FAP-DF).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.engmic.2024.100163.
Appendix. Supplementary materials
Table S1 Concentrations of Paenibacillus elgii supernatant used to isolate resistant strains from soil samples.
Table S2 Overlay assay of antibacterial activity for resistant strains isolated from cultures supplemented with the supernatant of Paenibacillus elgii. After the growth of the isolated strains, a soft-agar layer inoculated with Bacillus subtilis was poured onto the plate, and its growth was observed for up to 48 h at room temperature.
Fig. S1 Antibacterial activity of (A) Paenibacillus elgii (red arrow) and (B) its supernatant (blue arrow). Antibiotic production assays were conducted against Bacillus subtillis (yellow arrows).
Fig. S2 Cultures of resistant strains isolated from different concentrations of Paenibacillus elgii supernatant: (A) 300 µL.L−1, (B) 600 µL.L−1, (C) 1 mL.L−1, and (D) 2 mL.L−1. Strains were selected based on the presence of inhibition zones (red arrows).
Fig. S3 Overlay assay demonstrating the antibacterial activity of Methylobacterium sp. K003 against Bacillus subtillis.
References
- 1.Gutleben J., Loureiro C., Ramírez Romero L.A., Shetty S., Wijffels R.H., Smidt H., et al. Cultivation of bacteria from Aplysina aerophoba: effects of oxygen and nutrient gradients. Front. Microbiol. 2020;11:175. doi: 10.3389/fmicb.2020.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015;28(1):237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson L., Iqbal K.M., Simmons-Ehrhardt T., Bertino M.F., Shah M.R., et al. Customizable 3D printed diffusion chambers for studies of bacterial pathogen phenotypes in complex environments. J. Microbiol. Methods. 2019;162:8–15. doi: 10.1016/j.mimet.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Choi J., Yang F., Stepanauskas R., Cardenas E., Garoutte A., et al. Strategies to improve reference databases for soil microbiomes. ISME J. 2017;11:829–834. doi: 10.1038/ismej.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S., Moon C.D., Zheng N., Huws S., Zhao S., Wang J. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome. 2022;10:76. doi: 10.1186/s40168-022-01272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overmann J., Abt B., Sikorski J. Present and future of culturing bacteria. Annu. Rev. Microbiol. 2017;71:711–730. doi: 10.1146/annurev-micro-090816-093449. [DOI] [PubMed] [Google Scholar]
- 7.Laws M., Shaaban A., Rahman K.M. Antibiotic resistance breakers: current approaches and future directions. FEMS Microbiol. Rev. 2019;43(5):490–516. doi: 10.1093/femsre/fuz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand G.A., Raoult D., Dubourg G. Antibiotic discovery: history, methods and perspectives. Int. J. Antimicrob. Agents. 2019;53:371–382. doi: 10.1016/j.ijantimicag.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Overmann J. In: The Prokaryotes: Prokaryotic Biology and Symbiotic Associations. 4th edn. Rosenberg E., DeLong E.F., Stackebrandt E., Lory S., Thompson F., editors. Springer; New York: 2013. Principles of enrichment, isolation, cultivation, and preservation of bacteria; pp. 149–207. [Google Scholar]
- 10.Boilattabi N., Barrassi L., Bouanane-Darenfed A., La Scola B. Isolation and identification of Legionella spp. from hot spring water in Algeria by culture and molecular methods. J. Appl. Microbiol. 2021;130:1394–1400. doi: 10.1111/jam.14871. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary D.K., Khulan A., Kim J. Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 2017;9:6666. doi: 10.1038/s41598-019-43182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pascual J., García-López M., Carmona C., Sousa T.S., de Pedro N., et al. Pseudomonas soli sp. nov., a novel producer of xantholysin congeners. Syst. Appl. Microbiol. 2014;37:412–416. doi: 10.1016/j.syapm.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 13.van Dorst J.M., Hince G., Snape I., Ferrari B.C. Novel culturing techniques select for heterotrophs and hydrocarbon degraders in a subantarctic soil. Sci. Rep. 2016;6:36724. doi: 10.1038/srep36724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmann B.O., Van Lanem S.G., Baltz R.H. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014;41(2):175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y., Zou H., Li J., Song T., Lv W., et al. Impact of quorum sensing signaling molecules in gram-negative bacteria on host cells: current understanding and future perspectives. Gut. Microbes. 2022;14(1) doi: 10.1080/19490976.2022.2039048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols D., Lewis K., Orjala J., Mo S., Ortenberg R., O'Connor P., et al. Short peptide induces an "uncultivable" microorganism to grow in vitro. Appl. Environ. Microbiol. 2008;74(15):4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babenko L.M., Kosakivska I.V., Romanenko К.О. Molecular mechanisms of N-acyl homoserine lactone signals perception by plants. Cell. Biol. Int. 2022;46(4):523–534. doi: 10.1002/cbin.11749. [DOI] [PubMed] [Google Scholar]
- 18.Papenfort K., Bassler B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016;14(9):576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verbeke F., De Craemer S., Debunne N., Janssens Y., Wynendaele E., et al. Peptides as quorum sensing molecules: measurement techniques and obtained levels In Vitro and In Vivo. Front. Neurosci. 2017;11:183. doi: 10.3389/fnins.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soares E.V. Perspective on the biotechnological production of bacterial siderophores and their use. Appl. Microbiol. Biotechnol. 2022;106(11):3985–4004. doi: 10.1007/s00253-022-11995-y. [DOI] [PubMed] [Google Scholar]
- 21.da Costa R.A., Andrade I.E.P.C., Pinto O.H.B., de Souza B.B.P., Fulgêncio D.L.A., et al. A novel family of non-secreted tridecaptin lipopeptide produced by Paenibacillus elgii. Amino Acids. 2022;54(11):1477–1489. doi: 10.1007/s00726-022-03187-9. [DOI] [PubMed] [Google Scholar]
- 22.Ortega D.B., Costa R.A., Pires A.S., Araújo T.F., Araújo J.F., et al. Draft genome sequence of the antimicrobial-producing strain Paenibacillus elgii AC13. Genome Announc. 2018;6(26) doi: 10.1128/genomeA.00573-18. e00573-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak S., Xu Y., Nodwell J.R. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 2014;93(3):391–402. doi: 10.1111/mmi.12689. [DOI] [PubMed] [Google Scholar]
- 24.Cunha-Ferreira I.C., Vizzotto C.S., Freitas M.A.M., Peixoto J., Carvalho L.S., et al. Genomic and physiological characterization of Kitasatospora sp. nov., an actinobacterium with potential for biotechnological application isolated from Cerrado soil. Braz J Microbiol. 2024 doi: 10.1007/s42770-024-01324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady S.F. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc. 2007;2(5):1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 26.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420. [DOI] [PubMed] [Google Scholar]
- 31.Hughes G.M., Gang L., Murphy W.J., Higgins D.G., Teeling E.C. Using Illumina next generation sequencing technologies to sequence multigene families in de novo species. Mol Ecol Resour. 2013;13(3):510–521. doi: 10.1111/1755-0998.12087. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Zhao Y., Bollas A., Wang Y., Au K.F. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348–1365. doi: 10.1038/s41587-021-01108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32(6):929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier-Kolthoff J.P., Carbasse J.S., Peinado-Olarte R.L., Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022;50(D1):D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin T., Cook D., Lawrence M. Ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13(8):R77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avram O., Rapoport D., Portugez S., Pupko T. M1CR0B1AL1Z3R-a user-friendly web server for the analysis of large-scale microbial genomics data. Nucleic Acids Res. 2019;47:W88–W92. doi: 10.1093/nar/gkz423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Afgan E., Baker D., Van Den Beek M., Blankenberg D., Bouvier D., et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids. Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corretto E., Antonielli L., Sessitsch A., Compant S., Höfer C., et al. Complete genome sequence of the heavy metal resistant bacterium Agromyces aureus AR33T and comparison with related actinobacteria. Stand. Genomic Sci. 2017;12:2. doi: 10.1186/s40793-016-0217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J., Lu F., Luo Y., Bie L., Xu L., Wang Y. OrthoVenn3: anintegrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023;51(W1):W397–W403. doi: 10.1093/nar/gkad313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadfield J., Croucher N.J., Goater R.J., Abudahab K., Aanensen D.M., Harris S.R. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34(2):292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blin K., Shaw S., Kloosterman A.M., Charlop-Powers Z., van Wezel G.P., et al. AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49(W1):W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gawde U., Chakraborty S., Waghu F.H., Barai R.S., Khanderkar A., et al. CAMPR4: a database of natural and synthetic antimicrobial peptides. Nucleic Acids Res. 2023 doi: 10.1093/nar/gkac933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waghu F.H., Barai R.S., Gurung P., Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016;44:D1094–D1097. doi: 10.1093/nar/gkv1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villesen P. FaBox: an online toolbox for FASTA sequences. Mol. Ecol.Notes. 2007;7:965–968. doi: 10.1111/j.1471-8286.2007.01821.x. [DOI] [Google Scholar]
- 51.Agrawal P., Bhagat D., Mahalwal M., Sharma N., Raghava G.P.S. AntiCP 2.0: an updated model for predicting anticancer peptides. Brief. Bioinform. 2021;22(3) doi: 10.1093/bib/bbaa153. [DOI] [PubMed] [Google Scholar]
- 52.Agrawal P., Bhalla S., Chaudhary K., Kumar R., Sharma M., Raghava G.P.S. In silico approach for prediction of antifungal peptides. Front. Microbiol. 2018;9:323. doi: 10.3389/fmicb.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramaprasad A.S.E., Singh S., Gajendra R.P.S., Venkatesan S. AntiAngioPred: a server for prediction of anti-angiogenic peptides. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0136990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu B., Pop M. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46(W1):W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Navarro-Muñoz J.C., Selem-Mojica N., Mullowney M.W., Kautsar S.A., et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020;16(1):60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. 2010 Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weimann A., Mooren K., Frank J., Pope P.B., Bremges A., McHardy A.C. From genomes to phenotypes: traitar, the microbial trait analyzer. mSystem. 2016 doi: 10.1101/043315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy C.A., Beveridge T.J., Breznak J.A., Marzluf G.A., Schmidt T.M., Snyder L.R. Methods for general and molecular microbiology. Am. Soc. Microbiol. 2007 doi: 10.1128/9781555817497. [DOI] [Google Scholar]
- 61.Tindal B., Sikorski J., Smibert R., Krieg N. In: Methods for General and Molecular Microbiology. Reddy C., Berridge T., Breznak J., Marzluf G., Schmidr T., Snyder L., editors. ASM Press; Washington, DC: 2007. Phenotypic characterization and the principles of comparative systematics; pp. 330–393. [Google Scholar]
- 62.Ramaprasad E.V.V., Mahidhara G., Sasikala C., Ramana C.V. Rhodococcus electrodiphilus sp. nov., a marine electro active actinobacterium isolated from coral reef. Int. J. Syst. Evol. Microbiol. 2018;68(8):2644–2649. doi: 10.1099/ijsem.0.002895. [DOI] [PubMed] [Google Scholar]
- 63.Sasser, M. 2001. Technical Note # 101 identification of bacteria by gas chromatography of cellular fatty acids.
- 64.Bauer A.W., Kirby W.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 65.Grossi C.E.M., Fantino E., Serral F., Zawoznik M.S., Fernandez Do Porto D.A., Ulloa R.M. Methylobacterium sp. 2A is a plant growth-promoting rhizobacteria that has the potential to improve potato crop yield under adverse conditions. Front. Plant. Sci. 2020;11:71. doi: 10.3389/fpls.2020.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J., Chhetri G., Kim I., Kim M.K., Seo T. Methylobacterium durans sp. nov., a radiation-resistant bacterium isolated from gamma ray-irradiated soil. Antonie Van Leeuwenhoek. 2020;113(2):211–220. doi: 10.1007/s10482-019-01331-2. [DOI] [PubMed] [Google Scholar]
- 67.Photolo M.M., Mavumengwana V., Sitole L., Tlou M.G. Antimicrobial and antioxidant properties of a bacterial endophyte, Methylobacterium radiotolerans MAMP 4754, Isolated from Combretum erythrophyllum Seeds. Int. J. Microbiol. 2020;18:2020–9483670. doi: 10.1155/2020/9483670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S.S., Liu J.M., Sun J., Sun Y.F., Liu J.N., et al. Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different Dendrobium stems. Sci. Rep. 2019;9:10389. doi: 10.1038/s41598-019-46863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Njoroge J., Sperandio V. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol. Med. 2009;1(4):201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gray B., Hall P., Gresham H. Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors (Basel) 2013;13(4):5130–5166. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kapinusova G., Lopez Marin M.A., Uhlik O. Reaching unreachables: obstacles and successes of microbial cultivation and their reasons. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1089630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho A., Di Lonardo D.P., Bodelier P.L. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 2017;93(3) doi: 10.1093/femsec/fix006. [DOI] [PubMed] [Google Scholar]
- 73.Bahram M., Hildebrand F., Forslund S.K., Anderson J.L., Soudzilovskaia N.A., et al. Structure and function of the global topsoil microbiome. Nature. 2018;560(7717):233–237. doi: 10.1038/s41586-018-0386-6. [DOI] [PubMed] [Google Scholar]
- 74.Schink B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek. 2002;81(1-4):257–261. doi: 10.1023/a:1020579004534. [DOI] [PubMed] [Google Scholar]
- 75.Jacoby R., Peukert M., Succurro A., Koprivova A., Kopriva S. The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant. Sci. 2017;8:1617. doi: 10.3389/fpls.2017.01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pande S., Kost C. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol. 2017;25(5):349–361. doi: 10.1016/j.tim.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 77.Stewart E.J. Growing unculturable bacteria. J. Bacteriol. 2012;194(16):4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turan N.B., Chormey D.S., Büyükpınar C., Engin G.O., Bakirdere S. Quorum sensing: little talks for an effective bacterial coordination. Tr. A. Chem. 2017;91:1–11. doi: 10.1016/j.trac.2017.03.007. [DOI] [Google Scholar]
- 79.van Bergeijk D.A., Terlouw B.R., Medema M.H., van Wezel G.P. Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat. Rev. Microbiol. 2020;18(10):546–558. doi: 10.1038/s41579-020-0379-y. [DOI] [PubMed] [Google Scholar]
- 80.Akinsanya M.A., Goh J.K., Lim S.P., Ting A.S. Diversity, antimicrobial and antioxidant activities of culturable bacterial endophyte communities in Aloe vera. FEMS Microbiol. Lett. 2015;362(23):fnv184. doi: 10.1093/femsle/fnv184. [DOI] [PubMed] [Google Scholar]
- 81.Asaf S., Numan M., Khan A.L., Al-Harrasi A. Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020;40(2):138–152. doi: 10.1080/07388551.2019. [DOI] [PubMed] [Google Scholar]
- 82.Sánchez-Yañez J.M. Xanthobacter autotrophicus an endophytic beneficial bacterium for wheat and other plants: a short review. Curr. Tr. W. Res. pp. 2022;73 doi: 10.5772/intechopen.102066. [DOI] [Google Scholar]
- 83.Takahashi Y. Genus Kitasatospora, taxonomic features and diversity of secondary metabolites. J. Antibiot. 2017;70:506–513. doi: 10.1038/ja.2017.8. [DOI] [PubMed] [Google Scholar]
- 84.Zhu D., Xie C., Huang Y., Sun J., Zhang W. Description of Comamonas serinivorans sp. nov., isolated from wheat straw compost. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 12):4141–4146. doi: 10.1099/ijs.0.066688-0. [DOI] [PubMed] [Google Scholar]
- 85.Yun B.R., Malik A., Kim S.B. Genome based characterization of Kitasatospora sp. MMS16-BH015, a multiple heavy metal resistant soil actinobacterium with high antimicrobial potential. Gene. 2020;5:733–144379. doi: 10.1016/j.gene.2020.144379. [DOI] [PubMed] [Google Scholar]
- 86.Cain C.C., Henry A.T., Waldo R.H., Casida L.J., Falkinham J.O., Jr Identification and characteristics of a novel Burkholderia strain with broad-spectrum antimicrobial activity. Appl. Environ. Microbiol. 2000;66(9):4139–4141. doi: 10.1128/AEM.66.9.4139-4141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Depoorter E., Bull M.J., Peeters C., Coenye T., Vandamme P., Mahenthiralingam E. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016;100(12):5215–5229. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 88.Wang Y., Hoffmann J.P., Chou C.W., Höner Z.U., Bentrup K.H., et al. Burkholderia thailandensis outer membrane vesicles exert antimicrobial activity against drug-resistant and competitor microbial species. J. Microbiol. 2020;58(7):550–562. doi: 10.1007/s12275-020-0028-1. [DOI] [PubMed] [Google Scholar]
- 89.Singh R.P., Mishra S., Jha P., Raghuvanshi S., Jha P.N. Effect of inoculation of zinc-resistant bacterium Enterobacter ludwigii CDP-14 on growth, biochemical parameters and zinc uptake in wheat (Triticum aestivum L.) plant. Ecol. Eng. 2018;116:163–173. doi: 10.1016/j.ecoleng.2017.12.033. [DOI] [Google Scholar]
- 90.Sagar A., Sayyed R.Z., Ramteke P.W., Sharma S., Marraiki N., Elgorban A.M., Syed A. ACC deaminase and antioxidant enzymes producing halophilic Enterobacter sp. PR14 promotes the growth of rice and millets under salinity stress. Physiol. Mol. Biol. Plants. 2020;26(9):1847–1854. doi: 10.1007/s12298-020-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ren L., Men L., Zhang Z., Guan F., Tian J., et al. Biodegradation of Polyethylene by Enterobacter sp. D1 from the Guts of Wax Moth Galleria mellonella. Int. J. Environ. Res. Public. Health. 2019;16(11):1941. doi: 10.3390/ijerph16111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sah S., Krishnani S., Singh R. Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Curr. Res. Microb. Sci. 2021;2 doi: 10.1016/j.crmicr.2021.100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ojewumi M.E., Okeniyi J.O., Ikotun J.O., Okeniyi E.T., Ejemen V.A., Popoola A.P.I. Bioremediation: data on Pseudomonas aeruginosa effects on the bioremediation of crude oil polluted soil. Data Brief. 2018;19:101–113. doi: 10.1016/j.dib.2018.04.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Loeschcke A., Thies S. Pseudomonas putida a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 2015;99(15):6197–6214. doi: 10.1007/s00253-015-6745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mozejko-Ciesielska J., Szacherska K., Marciniak P. Pseudomonas Species as Producers of Eco-friendly Polyhydroxyalkanoates. J. Polym. Environ. 2019;27:1151–1166. doi: 10.1007/s10924-019-01422-1. [DOI] [Google Scholar]
- 96.Salvachúa D., Rydzak T., Auwae R., De Capite A., Black B.A., et al. Metabolic engineering of Pseudomonas putida for increased polyhydroxyalkanoate production from lignin. Microb. Biotechnol. 2020;13(1):290–298. doi: 10.1111/1751-7915.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knief C., Dengler V., Bodelier P.L., Vorholt J.A. Characterization of Methylobacterium strains isolated from the phyllosphere and description of Methylobacterium longum sp. nov. Antonie Van Leeuwenhoek. 2012;101(1):169–183. doi: 10.1007/s10482-011-9650-6. [DOI] [PubMed] [Google Scholar]
- 98.Alessa O., Ogura Y., Fujitani Y., Takami H., Hayashi T., Sahin N., Tani A. Comprehensive comparative genomics and phenotyping of Methylobacterium species. Front. Microbiol. 2021;6:12–740610. doi: 10.3389/fmicb.2021.740610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cochrane S.A., Vederas J.C. Lipopeptides from Bacillus and Paenibacillus spp.: a Gold Mine of Antibiotic Candidates. Med. Res. Rev. 2016;36(1):4–31. doi: 10.1002/med.21321. [DOI] [PubMed] [Google Scholar]
- 100.Galperin M.Y., Wolf Y.I., Makarova K.S., Vera Alvarez R., Landsman D., Koonin E.V. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021;49(D):D274–D281. doi: 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wimmer F., Beisel C.L. CRISPR-cas systems and the paradox of self-targeting spacers. Front. Microbiol. 2020;22(10):3078. doi: 10.3389/fmicb.2019.03078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sedeek K.E.M., Mahas A., Mahfouz M. Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 2019;10:114. doi: 10.3389/fpls.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ding Y., Ting J.P., Liu J., Al-Azzam S., Pandya P., Afshar S. Impact of non-proteinogenic amino acids in the discovery and development of peptide therapeutics. Amino Acids. 2020;52(9):1207–1226. doi: 10.1007/s00726-020-02890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang S., Liu Y., Liu W.Q., Neubauer P., Li J. The nonribosomal peptide valinomycin: from discovery to bioactivity and biosynthesis. Microorganisms. 2021;9(4):780. doi: 10.3390/microorganisms9040780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Biniarz P., Łukaszewicz M., Janek T. Screening concepts, characterization and structural analysis of microbial-derived bioactive lipopeptides: a review. Crit. Rev. Biotechnol. 2017;37(3):393–410. doi: 10.3109/07388551.2016.1163324. [DOI] [PubMed] [Google Scholar]
- 107.Qian C.D., Liu T.Z., Zhou S.L., Ding R., Zhao W.P., Li O., Wu X.C. Identification and functional analysis of gene cluster involvement in biosynthesis of the cyclic lipopeptide antibiotic pelgipeptin produced by Paenibacilluselgii. BMC Microbiol. 2012;8:12–197. doi: 10.1186/1471-2180-12-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Araújo T.F., Ortega D.B., da Costa R.A., Andrade I.E.P.C., Fulgêncio D.L.A., Mendonça M.L., et al. Co-occurrence of linear and cyclic pelgipeptins in broth cultures of Paenibacillus elgii AC13. Braz J Microbiol. 2021;52(4):1825–1833. doi: 10.1007/s42770-021-00597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H., Liu N., Xi L., Rong X., Ruan J., Huang Y. Genetic screening strategy for rapid access to polyether ionophore producers and products in actinomycetes. Appl. Environ. Microbiol. 2011;77(10):3433–3442. doi: 10.1128/AEM.02915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nivina A., Yuet K.P., Hsu J., Khosla C. Evolution and diversity of assembly-line polyketide synthases. Chem. Rev. 2019;119(24):12524–12547. doi: 10.1021/acs.chemrev.9b00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar L., Patel S.K.S., Kharga K., Kumar R., Kumar P., Pandohee J., et al. Molecular mechanisms and applications of N-Acyl homoserine lactone-mediated quorum sensing in bacteria. Molecules. 2022;27(21):7584. doi: 10.3390/molecules27217584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamada Y., Kuzuyama T., Komatsu M., Shin-Ya K., Omura S., et al. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA. 2015;112(3):857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoshida S., Hiradate S., Koitabashi M., Kamo T., Tsushima S. Phyllosphere Methylobacterium bacteria contain UVA-absorbing compounds. J. Photochem. Photobiol. 2017;167:168–175. doi: 10.1016/j.jphotobiol.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 114.Wang J., Gao C., Chen X., Liu L. Expanding the lysine industry: biotechnological production of l-lysine and its derivatives. Adv. Appl. Microbiol. 2021;115:1–33. doi: 10.1016/bs.aambs.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 115.Ongpipattanakul C., Desormeaux E.K., DiCaprio A., van der Donk W.A., Mitchell D.A., Nair S.K. Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides. Chem. Rev. 2022;122(18):14722–14814. doi: 10.1021/acs.chemrev.2c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Concentrations of Paenibacillus elgii supernatant used to isolate resistant strains from soil samples.
Table S2 Overlay assay of antibacterial activity for resistant strains isolated from cultures supplemented with the supernatant of Paenibacillus elgii. After the growth of the isolated strains, a soft-agar layer inoculated with Bacillus subtilis was poured onto the plate, and its growth was observed for up to 48 h at room temperature.
Fig. S1 Antibacterial activity of (A) Paenibacillus elgii (red arrow) and (B) its supernatant (blue arrow). Antibiotic production assays were conducted against Bacillus subtillis (yellow arrows).
Fig. S2 Cultures of resistant strains isolated from different concentrations of Paenibacillus elgii supernatant: (A) 300 µL.L−1, (B) 600 µL.L−1, (C) 1 mL.L−1, and (D) 2 mL.L−1. Strains were selected based on the presence of inhibition zones (red arrows).
Fig. S3 Overlay assay demonstrating the antibacterial activity of Methylobacterium sp. K003 against Bacillus subtillis.
Data Availability Statement
The type strain is K003, which is isolated from the soil of the Brazilian savanna-like Cerrado biome, Brasilia, Brazil. The deposit of the K003 genome in the GenBank database is being processed under accession number JAZBNP000000000.