Abstract
Industrial manufacturing of bioproducts, especially bioethanol, can benefit from high-temperature fermentation, which requires the use of thermotolerant yeast strains. Mitochondrial activity in yeast is closely related to its overall metabolism. However, the mitochondrial respiratory changes in response to adaptive thermotolerance are still poorly understood and have been rarely utilized for developing thermotolerant yeast cell factories. Here, adaptive evolution and transcriptional sequencing, as well as whole-genome-level gene knockout, were used to obtain a thermotolerant strain of Saccharomyces cerevisiae. Furthermore, thermotolerance and bioethanol production efficiency of the engineered strain were examined. Physiological evaluation showed the boosted fermentation capacity and suppressed mitochondrial respiratory activity in the thermotolerant strain. The improved fermentation produced an increased supply of adenosine triphosphate required for more active energy-consuming pathways. Transcriptome analysis revealed significant changes in the expression of the genes involved in the mitochondrial respiratory chain. Evaluation of mitochondria-associated gene knockout confirmed that ADK1, DOC1, or MET7 were the key factors for the adaptive evolution of thermotolerance in the engineered yeast strain. Intriguingly, overexpression of DOC1 with TEF1 promoter regulation led to a 10.1% increase in ethanol production at 42 °C. The relationships between thermotolerance, mitochondrial activity, and respiration were explored, and a thermotolerant yeast strain was developed by altering the expression of mitochondrial respiration-related genes. This study provides a better understanding on the physiological mechanism of adaptive evolution of thermotolerance in yeast.
Keywords: Saccharomyces cerevisiae, Adaptive evolution, Transcriptional sequencing, Mitochondrial respiratory, Thermotolerance
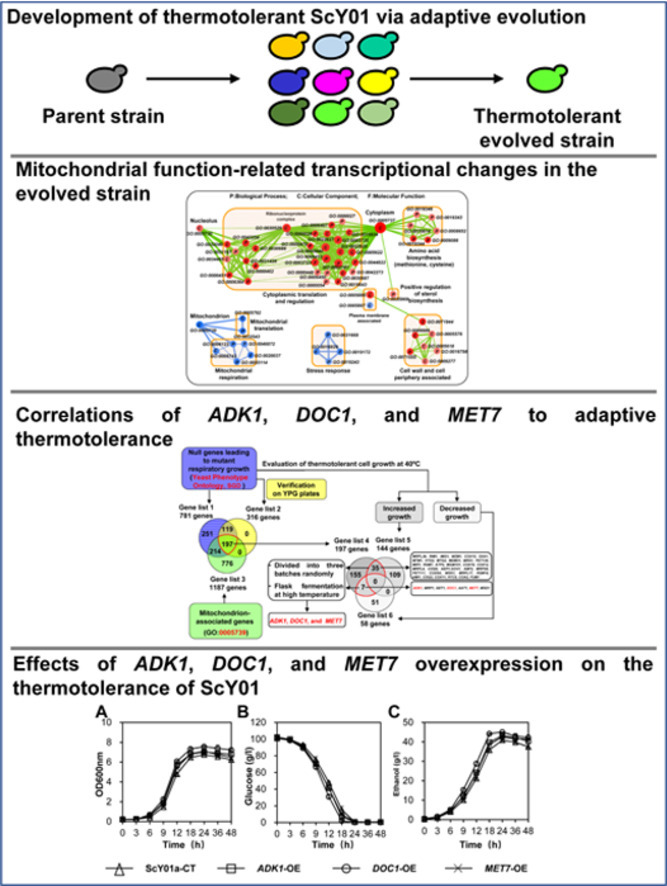
Graphical abstract
1. Introduction
Saccharomyces cerevisiae (S. cerevisiae) is widely used for producing various biofuels, biochemicals, and natural products, particularly bioethanol [1,2]. Usually, yeast cells grow optimally within a relatively narrow temperature range. However, even though deviations from this temperature range interrupt the cell structure and function, cells can tolerate these deviations to some extent via rapid physiological responses and adaptations [3]. Similar to many other eukaryotes [4], S. cerevisiae has evolved overlapping yet distinct cellular responses and adaptations to different aspects of high-temperature stress [5,6]. These responses include innate thermotolerance, acquired thermotolerance, thermotolerance to moderately high temperatures, and adaptive evolved thermotolerance. Adaptive laboratory evolution is garnering attention for the development of thermotolerant strains [7] that show elevated thermotolerance, which is one of the key desirable properties of S. cerevisiae for its industrial applications. High-temperature fermentation using thermotolerant S. cerevisiae can be advantageous for reducing the cost and increasing the productivity of industrial processes [8]. However, thermotolerance is a complex trait that is regulated by many genes and the complex interactions among them [9], [10], [11].
Our previous proteomic analysis showed the up-regulated expression of most glycolytic enzymes in S. cerevisiae during prolonged thermal stress, whereas the enzymes involved in the Tricarboxylic Acid Cycle, glycogen and glycerol biosynthesis, and pentose phosphate pathway were largely down-regulated [12,13]. This implies that cellular and mitochondrial metabolism reprogramming may play a key role in the thermotolerance exhibited by yeast during long-term thermal stress [14]. Sterol biosynthesis, cell periphery, and the cAMP-dependent protein kinase A signaling pathway were found to be crucial for the evolved thermotolerant strains. Surprisingly, some of these strains showed decreased or no respiratory growth in the presence of non-fermentable carbon sources, such as ethanol and glycerol [15,16]. These results imply that mitochondrial metabolism may play an important role in the adaptive evolution of thermotolerance in yeast. Mitochondria produce energy to provide adenosine triphosphate (ATP) for various biological processes and supply important metabolites for yeast cell growth and proliferation, as well as for stress adaptation [17,18]. However, mitochondrial functions, especially mitochondrial respiration, involved in the adaptive evolution of thermotolerance in yeast are not understood well.
Here, an evolved thermotolerant ScY01 strain was obtained through adaptive laboratory evolution. In previous studies, we investigated and compared the mechanisms of prolonged thermotolerance and transient heat shock response in the evolved ScY01 and the parental ScY by conducting proteomic analysis [12,13]. In this study, the underlying mechanisms controlling the physiological adaptations of the mitochondria were investigated. We further explored novel genes to develop more thermotolerant strains of yeast for industrial applications. Transcriptome analysis was used to determine the transcriptional variations between the evolved strain and the parent strain. The results of this study provide novel insights into mitochondrial functions, especially respiration, in response to adaptive evolution of thermotolerance in yeast.
2. Materials and methods
2.1. Plasmid construction
All the plasmids and primers used in the study are listed in Table 1 and Table S1, respectively. The ClonExpress® II One Step Cloning Kit (Vazyme Biotech Co., Ltd, Nanjing, China) based on the homologous recombination technology was used to construct the plasmids by following the instruction manual. The constructed plasmids were sequenced by the Beijing Genomics Institute (Beijing, China).
Table 1.
S. cerevisiae strains used in this study.
| Strains and plasmids | Description | Reference or source |
|---|---|---|
| S. cerevisiae strains | ||
| BY4743 knockout collection | The knockout strain collection of heterozygous diploid strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0) | EUROSCARF, Frankfurt, Germany |
| ScY | A diploid S. cerevisiae strain, | Lab collection |
| ScY01 | Evolved thermotolerant strain from ScY, diploid | This study |
| ScYa | MATa haploid strain derived from ScY | This study |
| ScY01a | MATa haploid strain derived from ScY01 | This study |
| ScYa (adk1∆) | ScY01a, MATa, adk1::KanMX | This study |
| ScYa (doc1∆) | ScY01a, MATa, doc1::KanMX | This study |
| ScYa (met7∆) | ScY01a, MATa, met7::KanMX | This study |
| ScY01a (adk1∆) | ScY01a, MATa, adk1::KanMX | This study |
| ScY01a (doc1∆) | ScY01a, MATa, doc1::KanMX | This study |
| ScY01a (met7∆) | ScY01a, MATa, met7::KanMX | This study |
| ScY01a-CT | ScY01a, MATa, harboring p315-KanMx-PTEF1-T8 | This study |
| ADK1-OE | ScY01a, MATa, harboring p315-KanMx-PTEF1-ADK1 | This study |
| DOC1-OE | ScY01a, MATa, harboring p315-KanMx-PTEF1-DOC1 | This study |
| MET7-OE | ScY01a, MATa, harboring p315-KanMx-PTEF1-MET7 | This study |
| Plasmid | ||
| pCAS | Kan+, 2-micron, KanMX | [59] |
| p315-PTEF1-T8 | Amp+, LEU2, ARS/CEN origin, containing the TEF1 promoter and the Tsynth8 terminator | [19] |
| p315-KanMX-PTEF1-T8 | p315-PTEF1-T8 containing KanMX cassette | This study |
| p315-KanMX-PTEF1-ADK1 | p315-KanMX-PTEF1-T8 expressing ADK1 gene | This study |
| p315-KanMX- PTEF1-DOC1 | p315-KanMX-PTEF1-T8 expressing DOC1 gene | This study |
| p315-KanMX-PTEF1-MET7 | p315-KanMX-PTEF1-T8 expressing MET7 gene | This study |
First, the KanMX cassette was amplified using the pCAS plasmid and subcloned into the p315-TEF1-T8 plasmid to finally generate the p315-KanMX-PTEF1-T8 plasmid. To determine the effects of ADK1, DOC1, and MET7 on the high-temperature fermentation efficiency, the open reading boxes of ADK1, DOC1, and MET7 were amplified from S. cerevisiae S288c genomic DNA and inserted into the p315-KanMX-PTEF1-T8 plasmid [19] to produce p315-KanMX-PTEF1-ADK1, p315-KanMX-DOC1, and p315-KanMX-PTEF1-MET7, respectively. The four constructed plasmids (p315-KanMX-PTEF1-T8, p315-KanMX-PTEF1-ADK1, p315-KanMX- PTEF1-DOC1, and p315-KanMX-PTEF1-MET7) were transferred into the evolved ScY01 strain using the standard lithium acetate method [20].
2.2. S. cerevisiae strains, adaptive laboratory evolution, and culture conditions
All the S. cerevisiae strains used in this study are listed in Table 1. The diploid strain, ScY, was used as the parent strain for the adaptive evolution of thermotolerance in the laboratory. The evolution experiment was conducted in 50-mL shake flasks containing a yeast peptone (YP) medium (10 mL) with 200 g/L glucose at 200 rpm. The YP medium (pH 5.5) consisted of yeast extract (10 g/L) and peptone (20 g/L). The culture temperature was gradually increased from 37 to 39 °C and finally to 40 °C over the course of the evolution experiment. The cultured yeast cells were transferred into a fresh medium every 24 h, with 0.5 initial optical density at 600 nm (OD600). The OD600 of the culture was recorded before each serial transfer. After every five transfers, culture samples were collected and stored at −80 °C in a 15% (v/v) glycerol solution. The strain obtained from the last culture at 40 °C was named ScY01. The ScY01 strain was then assessed for fermentation efficiency and thermal stability at 40 °C and for other analyses. For RNA sequencing and real-time quantitative polymerase chain reaction (PCR), ScY and ScY01 cells were grown in the YP medium containing 200 g/L glucose at 40 °C and 200 rpm for 14–16 h (up to the early-log phase). After incubation, the yeast cells were harvested by applying centrifugation in Falcon tubes precooled in liquid N2 (5 min, 3220 × g). The supernatants were discarded and the pellets were stored at −80 °C until further use.
BY4743 and its knockout (KO) strains (Table 1), annotated for increased, decreased, or no respiratory growth in the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/), were purchased from EUROSCARF (Frankfurt, Germany) [21]. Strains ScYa and ScY01a were haploids of mating-type a, derived from ScY and ScY01, respectively. The KO method used for the key genes in ScYa and ScY01a was based on PCR amplification and one-step gene replacement [22]. The primers used for KO and detection are listed in Table S1.
2.3. RNA sequencing and differential gene expression (DEG) analysis
RNA libraries were prepared and sequenced with an Illumina Genome Analyzer IIx (Majorbio, Shanghai, China) using 36-bp single-end sequencing. Approximately 16 million clean reads corresponding to approximately 48-fold coverage of the reference genome were generated for each library. DEG in the evolved strain, ScY01, with respect to the parental strain, ScY, was analyzed by using a downstream analysis pipeline based on the reference genome of strain S288c (sequence assembly version R64) [23]. The clean reads were aligned to the reference transcriptome by using the Bowtie (version 1.1.1) software [24]. Subsequently, transcript quantification was carried out from RSEM mappings [25]. The MARS method implemented in the DEGseq R package was used for the DEG analysis [26]. Significant DEGs were then sorted by applying a p-value cutoff of 0.001 or less and an absolute fold-change threshold of 2.0 or more. Significantly upregulated and downregulated genes (Dataset S1) were analyzed for enrichment in gene ontology (GO) terms using DAVID Bioinformatics Resources 6.7 [27] (Dataset S2). Enrichment maps of ontology categories (clustering with significance of p-values <0.05 and Benjamini FDR < 0.5) were generated by using the Enrichment Map 2.0.1 software plug-in of Cytoscape 3.4.0 [28]. Transcription profile data and significant DEGs are provided in the supplementary material (Dataset S1) and the Gene Expression Omnibus (accession no. GSE96829).
2.4. Detection of ATP concentration in whole cells and mitochondria
ScY and ScY01 strains were grown in a YP medium containing 200 g/L glucose at 40 °C and 200 rpm for 16 h to the early log phase. The ATP levels in whole cells and mitochondria were quantified by using a method described in a previous study [29], with some minor modifications. To determine the ATP concentrations from whole cells and mitochondria, 10 OD600 and 50 OD600 cells were harvested, respectively, in Falcon tubes by performing centrifugation (5 min, at 3220 × g). The harvested cells were subsequently digested with zymolyase 20T (SilGreen), according to the protocol described in the manual. To separate mitochondria, the homogenized spheroplasts were subjected to differential centrifugation as described in an earlier study [30]. ATP concentration was measured using an ATP assay kit (Beyotime) in SpectraMax M5 (Molecular Devices) against ATP standard solutions, according to the manufacturer's protocol.
2.5. Assessing mutant respiratory growth and thermotolerance of mitochondria-associated single gene KO strains
The correlations among cell thermotolerance, mitochondrial function, and mutant respiratory growth was further examined. To achieve this objective, BY4743 and its KO strains were verified and evaluated for their growth capacities on a YP plate using 2% (v/v) glycerol as a single carbon source at 30 °C by setting up spot assays. The growth of strains was also evaluated in a liquid YP medium containing 100 g/L glucose at 40 °C using 96-well plates. The whole procedure is illustrated in Fig. 4. The cell growth of the KO mutants on the glycerol plates was confirmed by spot assays (Fig. S3). The dot density was measured by employing the NIH ImageJ software (https://imagej.nih.gov/ij/) according to the provided instructions (Dataset S3). The gene candidates whose KO mutants were verified for differential respiratory growth and thermotolerance were further selected as the mitochondria-associated genes (GO Term: mitochondrion, Cellular Component, GO: 0005739, SGD). The effects of these genes on the fermentation capacities of the mutants at the elevated temperature (40 °C) were evaluated. The fermentation capacities of the last selected KO mutants and their equivalent ScYa and ScY01a mutants were tested at the normal (30 °C and 35 °C) and elevated (40 °C) temperatures. Fermentation experiments were conducted using a 100-mL YP medium containing 100 g/L glucose in 250-mL flasks at 200 rpm. The thermotolerance and fermentation capacities of the ADK1-, DOC1-, and MET7-overexpressing mutants were evaluated at 40 °C.
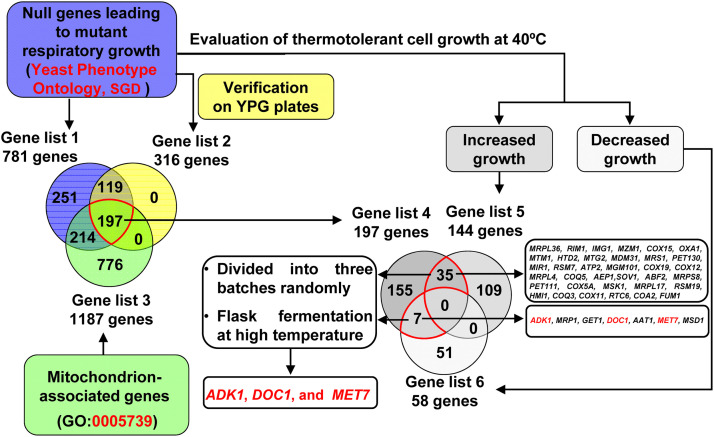
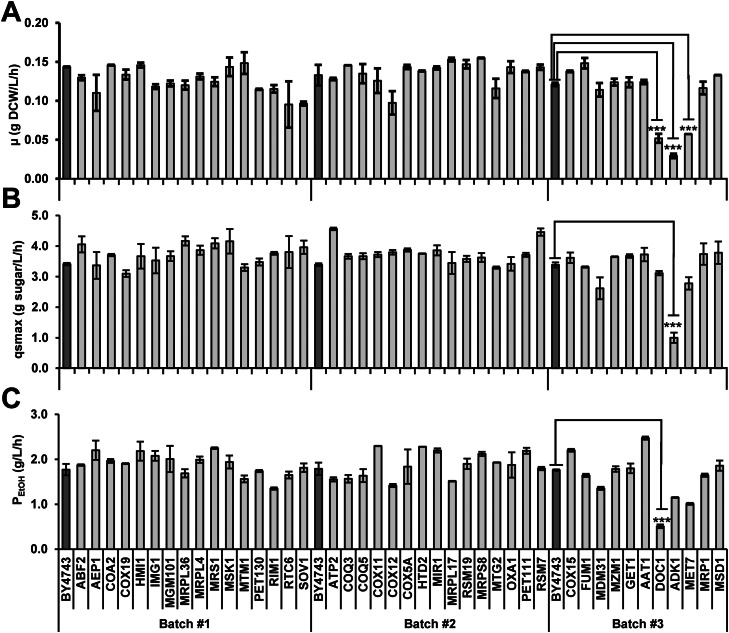
Fig. 4.
Scheme of screening mitochondrion-associated gene knockout (KO) strains with mutant respiratory growth and differential thermotolerant cell growth in BY4743 background. All the genes used in Venn Diagrams were detailed in Dataset S3. Gene list 1 includes 781 genes whose KOs are annotated to lead to mutant respiratory growth (Yeast Phenotype Ontology, Observable: respiratory growth, SGD). After respiratory growth on YPG plates was verfied (YP medium containing 2% glycerol, Fig. S3), gene list 2 was generated from 316 KOs showing 50% decreased respiratory growth. Gene list 3 includes 1187 genes, which are annotated as mitochondrion-associated genes at SGD (GO: 0005739). Thus, gene list 4 includes those mitochondrion-associated gene whose KOs were confirmed to have decreased respiratory growth. In parallel, these 781 KOs were evaluated for thermotolerant cell growth in a liquid YP medium containing 20 g/L glucose at 40 °C. Gene lists 5 and 6 were generated from 144 KOs showing 50% increased cell growth and 58 KOs showing 50% decreased cell growth at 40 °C in contrast to the BY4743 wild-type strain, respectively. Finally, the mitochondrion-associated KOs with verified decreased respiratory cell growth and differential thermotolerant cell growth (35 KOs plus 7 KOs, gene names were listed on the right) were subjected to flask fermentation at high temperatures for further verification and evaluation. The 42 mutants were divided into three batches randomly, and BY4743 was used as the control strain in each batch.
2.6. Analytical methods and calculation of fermentation parameters
Cell growth was indirectly measured by recording OD600 using a SpectraMax M2 (Molecular Devices, USA). Glucose and ethanol concentrations were monitored by using high-performance liquid chromatography (Agilent Technologies, USA) with a refractive index detector and a BioRad Aminex HPX–87H Column (300 mm × 7.8 mm) (Bio–Rad Laboratories Inc., USA). The column was eluted with 0.01 N of H2SO4 at a flow rate of 0.6 mL/min at 63 °C. The fermentation parameters, including the maximum growth rate (µmax), maximum glucose consumption rate (qsmax), and ethanol productivity (PEtOH), corresponding to the fermentation profiles were calculated using Origin 8 (Originlab®, USA) as previously reported [31].
3. Results
3.1. Development of thermotolerant ScY01 via adaptive evolution
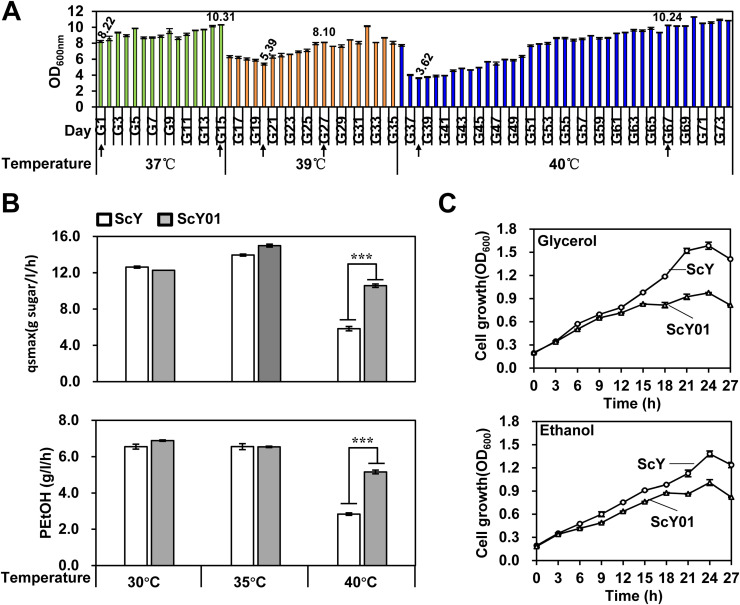
To improve the thermotolerance of S. cerevisiae and understand its origin, a diploid strain ScY was chosen for adaptive laboratory evolution by gradually increasing the temperature (Fig. 1(A)). The ScY strain is a derivative of the Ethanol Red industrial yeast [13]. During the serial transfers of the cultures grown at 37 °C for 15 d, no dramatic changes were observed in the cell growth. However, for the increased growth temperature of 39 °C, decreased cell growth was observed for the strains during earlier few transfers. The lowest OD600 of 5.39 was measured on the 20th day. Afterwards, cell growth gradually increased, with the increased OD600 of more than 8.00. On the 36th day, the growth temperature was increased to 40 °C. Although the culture showed similar growth (OD600 of 7.73) during the first transfer, severe growth inhibition was observed for the second and third transfers, with the lowest OD600 of 3.62. Afterwards, cell growth of the serial transfer cultures slowly increased and required a longer time (approximately 29 d) to surpass the OD600 of 10.00, which was similar to the cell growth at 37 °C. These observations suggested the possible enrichment of the cells adapted to the high growth temperature in the evolving cultures. Therefore, after continuous serial transfers for another 7 d at 40 °C, a strain was isolated from the last serial transfer culture on the 74th day and named ScY01. The thermal stability of the isolated ScY01 strain was then evaluated. ScY01 cell populations were serially transferred every 24 h for 20 d at normal temperature. Afterwards, the maximum glucose consumption rate (qsmax) and the fermentation capacity (PEtOH) were evaluated in the culture samples collected every five transfers. As expected, qsmax and PEtOH were found to be similar among the serially transferred cell populations of ScY01. However, the qsmax and PEtOH were clearly different from the corresponding parameters of the parent ScY strain (Fig. S1). This suggested that the ScY01 strain acquired robust physiological traits of adaptive thermotolerance.
Fig. 1.
Adaptive laboratory evolution and characterization of thermotolerant cell populations. (A) Cell growth (OD600) records of the evolving cultures during the adaptive laboratory evolution process. ScY strain was used for serial transfers every 24 h at increasing growth temperatures. The cultures showing the lowest OD600 at each temperature and reaching the average high OD600 are indicated by arrows. Data represent the mean and standard error of duplicate cultures at each condition. (B) Fermentation properties of the parental strain, ScY, and the evolved thermotolerant strain, ScY01, at different temperatures (30 °C, 35 °C, and 40 °C) using a YP medium with 200 g/L glucose. Maximum glucose consumption rates and ethanol productivities were calculated according to the fermentation profiles at each condition described in the Methods section. Data represent the mean and standard error of duplicate cultures at each condition. An initial OD600 of 0.5 was used. Statistical analysis was performed using one-way ANOVA (with strains as the factor) followed by Tukey's multiple-comparison posttest (***p < 0.001). (C) Growth profiles of ScY and ScY01 on 2% (v/v) glycerol (upper panel) or ethanol (bottom panel) at 40 °C. Data represent the mean and standard error of triplicate cultures at each condition. An initial OD600 of 0.2 was used.
The fermentation efficiencies of the evolved thermotolerant ScY01 and the parent ScY strain were compared at different temperatures (Fig. 1(B)). The difference between the two strains was more pronounced at 40 °C than at the other temperatures (30 °C and 35 °C). At 40 °C, ScY01 exhibited approximately 2.2- and 1.6-fold increases in the glucose consumption rate and ethanol productivities, respectively, compared to the ScY strain. These results indicated that the thermotolerance and the fermentation capacity of the evolved ScY01 strain were considerably improved compared with those of its parent strain.
Furthermore, compared with ScY, the evolved ScY01 showed decreased respiratory growth in the presence of a non-fermentable carbon source (Fig. 1(C)). This observation is consistent with the findings reported by a previous study [16]. Overall, the adaptive evolution of thermotolerance was confirmed to contribute to the improved fermentation capacity and the decreased respiratory growth of yeast.
3.2. Mitochondrial function-related transcriptional changes in the evolved strain
To identify the mitochondrial function-related transcriptional changes, transcriptome analysis was performed using RNA-seq on the parental strain ScY and the evolved strain ScY01 grown at 40 °C. Transcription profiling revealed that compared with ScY, 294 genes were significantly up-regulated in ScY01, while 91 genes were significantly down-regulated (absolute fold changes ≥ 2.0 or more; p ≤ 0.001) (Dataset S1).
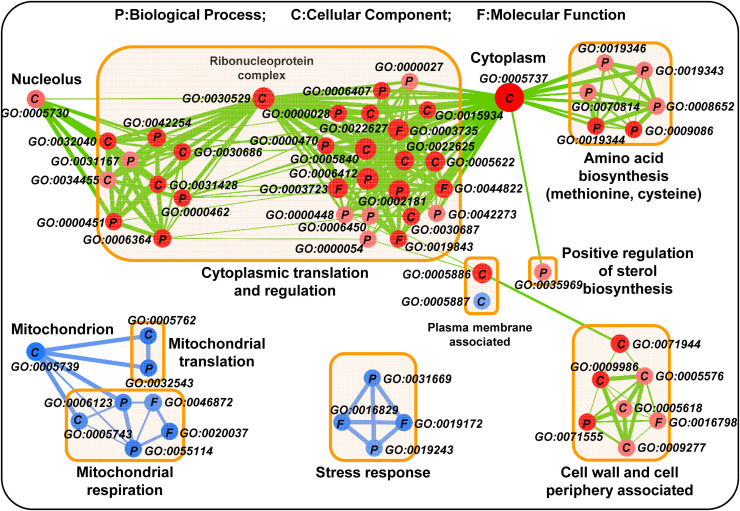
These differentially expressed genes were further used for functional clustering analysis to predict the changes in the metabolic activity, especially the mitochondrial activity in the evolved thermotolerant yeast cells (Fig. 2). A major cluster of the up-regulated genes in ScY01 were found to be involved in cytoplasmic translation, mainly including ribosome biogenesis, rRNA processing and translation regulation (Fig. 2, Fig. S2, Dataset S2). The remaining up-regulated enrichment gene clusters included the genes associated with cell wall and cell periphery function, and three transcription factors (i.e., SUT1, SUT2 and UPC2) involved in sterol biosynthesis regulation. By contrast, fewer genes were found to be down-regulated in ScY01, and were enriched in two main distinct clusters. The first cluster was associated with mitochondrial function, including mitochondrial translation and respiration, implying reduced mitochondria-dependent ATP production in ScY01 (Fig. 2). Interestingly, the second cluster was related to cellular responses to nutrient levels. This cluster included: (i) chaperone-encoding genes, such as HSP32, HSP33 and SNO4, required for transcriptional reprogramming during the diauxic shift and for survival during the stationary growth phase [32]; and (ii) genes that are repressed by glucose and induced prior to diauxic shift, such as ICL1, SNZ2, SNZ3 and ARO10 [33]. These results indicated that ScY01 may have altered its transcriptional functions to suppress the mitochondrial activity and gene expressions in response to glucose depletion, which resulted in the decreased respiratory growth (Fig. 1(C)) and impaired diauxic shift. This phenomenon has also been observed previously for other evolved thermotolerant strains [34].
Fig. 2.
Enrichment clustering of differentially expressed genes from RNA-seq analysis. Evolved strain ScY01 and wild-type strain ScY were compared for differential expression of genes when grown at 40 °C. Gene lists were compiled for significantly (p < 0.001) upregulated >2-fold or significantly downregulated <2-fold differentially expressed genes. The red color represents upregulation, whereas the blue color represents downregulation. For each enriched GO term with permissive p-value (<0.05) and FDR Q-value (<0.5) cutoffs, the following information is displayed: the ontology to which the GO term belongs (P = Biological Process; C = Cellular Component; F = Molecular Function). The GO term number was noted next to its GO term. Darker shades represent a relatively higher confidence of the enrichment score. Larger node sizes represent relatively larger numbers of the differentially regulated genes associated with the given ontology category as compared to the full gene. A smaller distance between the nodes denotes a higher degree of relationship between ontology categories, whereas thicker edge lines denote a relatively higher degree of similarity between the category nodes in terms of the degree of overlap between the specific gene sets that they are associated with. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
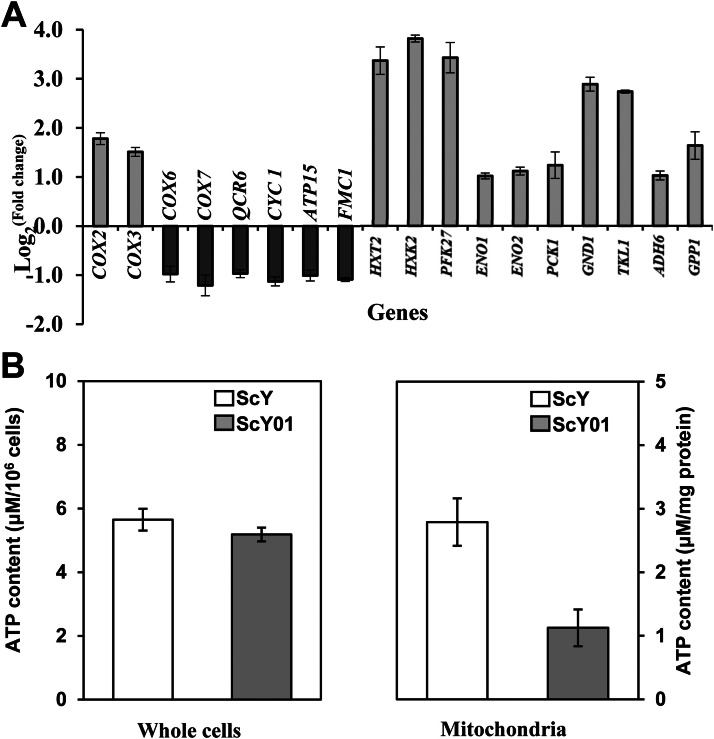
Transcriptome analysis revealed that the energy-consuming pathways were more active in ScY01 than in ScY, whereas ATP production relying on mitochondrial activity was found to be hampered in the ScY01 strain. This suggests that decreased respiration may result in the shortage of ATP and enhance other energy metabolism pathways. Therefore, significantly differentially expressed genes involved in mitochondrial respiration and glycolysis were evaluated (Fig. 3(A)). First, compared with ScY, increased expressions of the mitochondrial genes COX2 and COX3 encoding subunits of the respiratory chain complex IV were observed in ScY01. By contrast, decreased expressions of the nuclear genes COX6 and COX7 encoding another two subunits of the respiratory chain complex IV were observed in ScY01. Furthermore, expressions of the nuclear genes QCR6 and CYC1 encoding mitochondrial respiratory chain enzymes as well as ATP15 and FMC1 involved in the biosynthesis and assembly of mitochondrial F1F0 ATP synthase also decreased in ScY01.
Fig. 3.
(A) RNA-seq analysis of gene expression involved in mitochondrial respiration and glycolysis, and (B) ATP content in mitochondria and whole cells of ScY and ScY01. Fold changes were calculated by comparing gene expression levels in ScY01 with those in ScY. Error bars represent the standard deviation obtained from the six biological replicates.
To further investigate the mitochondria-function-related changes, we measured the ATP concentrations in a mitochondrion and in whole cells. Indeed, on the one hand, mitochondrial ATP concentration in ScY was 147.91% higher than that of ScY01. On the other hand, ATP concentration in whole cells was only 9.07% higher in ScY than in ScY01 (Fig. 3(B)). In addition, transcriptome analysis revealed that the expression of glycolytic enzyme genes, such as HXT2 (glucose transporter gene), HXK2 (predominant hexokinase gene), PFK27 (6-phosphofructo-2-kinase gene), ENO1 and ENO2 (enolase genes), increased in ScY01 (Fig. 3(A)). Additionally, PCK1 encoding a key enzyme in gluconeogenesis, GND1 and TKL1 involved in pentose phosphate pathway, GPP1 involved in glycerol biosynthesis, and ADH6 encoding alcohol dehydrogenase were also found to be up-regulated in ScY01, in contrast with ScY (Fig. 3(A)). These results preliminarily suggest that the evolved ScY01 may adjust the activities of mitochondrial respiration and boost other pathways, such as the glycolytic pathway. However, further research is required to confirm this hypothesis.
3.3. Correlations of ADK1, DOC1, and MET7 to adaptive thermotolerance
The evolved ScY01 exhibited decreased respiratory growth in the presence of non-fermentable carbon sources. Moreover, the evolved strain was found to be capable of resisting high-temperature stress by adjusting mitochondrial function. Therefore, the genes related to respiratory defects and mitochondrial function were further evaluated. The KO collections of laboratory strain BY4743 were employed. In this library, 781 gene null strains (Dataset S3, Gene list 1) showing mutated respiratory growth have been annotated in Yeast Phenotype Ontology in SGD (Fig. 4). After their growth on glycerol plates at 40 °C was verified, respiratory growth of KO mutants were evaluated and compared with the wild type strain BY4743. 316 KOs (Dataset S3, Gene list 2) showed a decrease of approximately 50% in the respiratory growth on glycerol relative to the wild type strain BY4743 (Fig. S3, Dataset S3). Among these KOs, 197 KOs were annotated in Yeast Phenotype Ontology in SGD, and contained mitochondria-associated genes (Dataset S3, Gene list 4). In parallel, the thermotolerance of 781 respiratory growth mutant strains were evaluated at 40 °C (Dataset S3). Compared with the wild type strain BY4743, 144 KOs (Dataset S3, Gene list 5) showed 50% increase in the cell growth at 40 °C, whereas 58 KOs showed 50% decrease in the cell growth. Among these KOs, 42 KOs (Fig. 4) were found to be mitochondria-associated KOs with verified decreased respiratory cell growth and differential thermotolerance. These 42 KOs were further subjected to shake-flask fermentation at 40 °C. However, none of these KOs exhibited significant improvement in the fermentation capacity at high temperature (Fig. 5). Nevertheless, the results confirmed that the KO of ADK1, DOC1, and MET7 genes resulted in a significant suppression of cell growth at 40 °C. In addition, DOC1 KO led to a significant decrease in the fermentation rates (qsmax and PEtOH). All of these observations suggested that the decreased respiratory growth of thermotolerant strains might be the effect of adaptive thermotolerance.
Fig. 5.
Shake-flask fermentation evaluation of the mitochondrion-associated KOs with verified decreased respiratory cell growth and differential thermotolerant cell growth. Fermentations were performed using a YP medium containing 100 g/L glucose at 40 °C. An initial OD of 0.2 was used. Maximum glucose consumption rates and ethanol productivities were calculated according to the fermentation profiles at each condition described in the Methods section. Data represent the mean and standard error of duplicate cultures at each condition. Statistical analysis was performed using one-way ANOVA (with strains as the factor) followed by Tukey's multiple-comparison posttest (***p < 0.001).
ADK1, DOC1, and MET7 KOs showed obvious inhibitory effects on the growth of BY4743 at high temperature. This suggests that these three genes may be required for thermotolerant cell growth of yeast. Thus, these three genes were also knocked out in the haploid strains of ScY and ScY01. This allowed a comprehensive comparison of the effects of these genes on the fermentation capacities of different strains at different temperatures (Fig. 6).
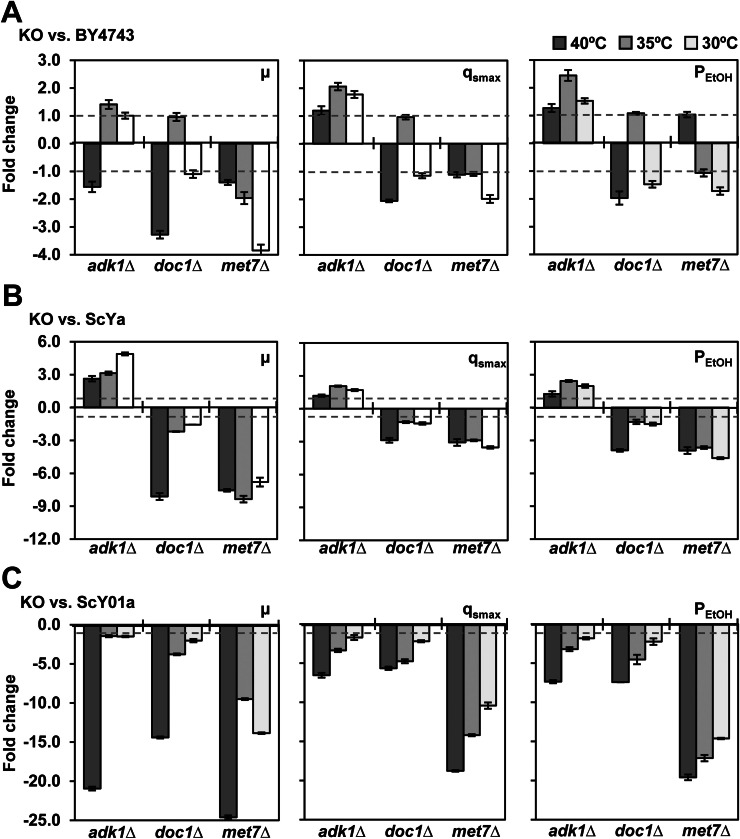
Fig. 6.
Different effects of ADK1, DOC1, and MET7 KO on fermentation capacities of (A) BY4743, (B) ScYa, and (C) ScY01a. Fermentations were performed using a YP medium containing 100 g/L glucose at 40 °C, 35 °C, and 30 °C. An initial OD of 0.2 was used. Maximum glucose consumption rates and ethanol productivities were calculated according to the fermentation profiles at each condition described in the Methods section and compared with those of the corresponding wild type strains. Data represent the mean and standard error of duplicate cultures at each condition.
ADK1 encodes the adenylate kinase enzyme required for purine metabolism [35]. The KO of this gene resulted in reduced cell growth of BY4743 at high temperature (40 °C), but not at the mild (35 °C) and normal (30 °C) temperatures (Fig. 6(A)). Notably, ADK1 KO stimulated glucose consumption and ethanol accumulation at 35 °C and 30 °C, which was consistent with the results of a previously reported study [36]. For haploid ScYa (MATa) derived from ScY, ADK1 KO surprisingly exhibited positive effects on cell growth, glucose consumption and ethanol accumulation at all studied temperatures (Fig. 6(B)). However, the effects at 40 °C were less favorable than those at 35 °C and 30 °C. Interestingly, for ScY01a (MATa) i.e., haploid derived from ScY01, ADK1 KO resulted in the loss of cell growth at 40 °C, along with severely decreased glucose consumption and ethanol production (Fig. 6(C)). Furthermore, ADK1 KO also inhibited the fermentation capacity of ScY01a at 35 °C and 30 °C. However, the inhibitory effects of ADK1 KO were considerably less severe at 30 °C and 35 °C than that at 40 °C. These results indicated the greater dependence of strain ScY01a (compared to strain ScYa) on the function of ADK1, especially at high temperature.
DOC1 encodes a processivity factor required for the ubiquitous activity of the anaphase promoting complex [37]. The KO of DOC1 led to significant inhibition of cell growth and glucose fermentation capacity of BY4743 at 40 °C, but not at 35 °C and 30 °C (Fig. 6(A)). Similar trends were also observed for the ScYa (Fig. 6(B)) and ScY01a (Fig. 6(C)) strains. Certainly, with the increase in temperature, the inhibitory effects of DOC1 KO in ScY01a became stronger than those in ScYa. This suggests that DOC1 function might be more crucial for the evolved ScY01a strain than for ScYa. Furthermore, DOC1 might be essential for thermotolerant glucose fermentation, regardless of the employed yeast strain.
MET7 encodes the folylpolyglutamate synthetase enzyme required for methionine synthesis and for maintenance of mitochondrial DNA [38]. The KO of this gene caused a significant decrease in the cell growth and glucose fermentation capacities of BY4743 at 30 °C relative to those at 35 °C and 40 °C (Fig. 6(A)). MET7 KO in ScYa led to severe inhibition of cell growth and glucose fermentation at all temperatures, resulting in only 37–44% glucose consumption at 48 h and similar inhibitory extents (Fig. 6(B)). In contrast with BY4743, MET7 KO in ScY01a led to a more severe inhibition of cell growth and glucose fermentation at 40 °C than at normal temperatures (Fig. 6(C)). Furthermore, the glucose consumption at 48 h was 81%, 59%, and 15% at 30 °C, 35 °C, and 40 °C, respectively. The contrasting results for ScYa and ScY01a suggested that MET7 function was more crucial for the evolved thermotolerant strain ScY01a.
3.4. Effects of ADK1, DOC1, and MET7 overexpression on the thermotolerance of ScY01
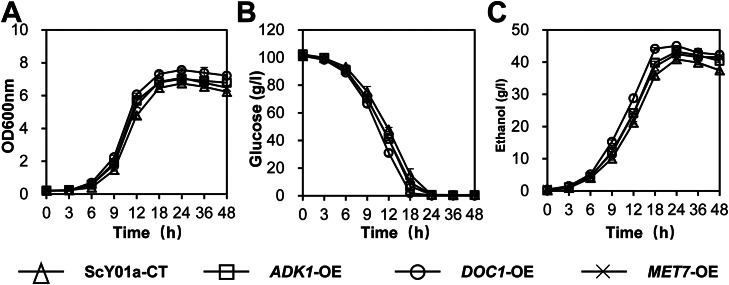
As the deletion of ADK1, DOC1, and MET7 resulted in reduced thermotolerance of ScY01a, these genes were overexpressed in the evolved ScY01a to confirm their impact on high-temperature fermentation. The strong promoter TEF1 and synthetic terminator T8 were employed for continuous overexpression of ADK1, DOC1, and MET7 and generation of ADK1-OE, DOC1-OE, MET7-OE mutants, correspondingly. ScY01a strains containing p315-KanMx-TEF1-T8 were used as negative control and named ScY01a-CT.
Compared to ScY01a-CT, the fermentation capacity of ADK1-OE, DOC1-OE, and MET7-OE mutants at 40 °C increased to varying extents. The cell growth and ethanol production of ADK1-OE were increased by 4.2% and 5.7%, respectively, whereas for MET7-OE, the cell growth and ethanol production increased by 5.0%, and 4.0%, respectively (Fig. 7). Similarly, overexpression of DOC1 significantly improved the high-temperature fermentation performance of ScY01a. In particular, the DOC1-OE mutant displayed a 10.1% increase in the ethanol production (Fig. 7). These results confirmed that overexpression of the overexpressing mitochondrial respiratory-related genes identified in this study can successfully improve ethanol production.
Fig. 7.
Different effects of ADK1, DOC1, and MET7 overexpression on fermentation capacities of ScY01a. (A) The cell growth, (B) glucose consumption, and (C) ethanol production of ADK1, DOC1, and MET7 overexpressed mutants and the control strains were evaluated. Fermentations were performed using a YP medium containing 100 g/L glucose at 40 °C. An initial OD of 0.2 was used. ScY01a-CT: ScY01a containing blank plasmid. ADK1-OE, DOC1-OE, and MET7-OE: overexpressing ADK1, DOC1, or MET7 gene in ScY01a. Blue column: cell growth; yellow column: ethanol production; gray column: glucose consumption. Data represent the mean and standard error of duplicate cultures at each condition. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Overall, this study indicated that mitochondrial respiration might be a response to the adaptive evolution of thermotolerance, and engineering of mitochondrial respiration-related genes is an effective strategy for improving high-temperature fermentation capacity and ethanol production.
4. Discussion
Much evidence has been found for the distinct adaptation mechanisms of S. cerevisiae in response to long-term thermal stress that differ from the mechanisms of heat shock response to short-term thermal stress [6,39,40]. Recently, adaptive laboratory evolution experiments have been conducted to enhance the thermotolerance of S. cerevisiae, which is a desired property for its industrial applications [7,41,42]. Meanwhile, the underlying mechanisms of adaptive evolved thermotolerance are gradually being uncovered. Interestingly, cellular adjustments and reprogramming in the evolved thermotolerant strains were found to be associated with mitochondrial functions and respiratory growth capacity [16,43,44]. These reports suggested that the adjustment of mitochondrial functions might be one of the driving forces for the long-term adaptation of yeast strains to high-temperature stress. In this work, the physiological and transcriptomic changes in the evolved thermotolerant strain relative to the parent strain were systematically evaluated. This study provided more detailed information about the molecular changes that determine the mitochondrial function and revealed the key factors involved in the regulation of mitochondrial functions during the adaptive evolution of thermotolerance in the evolved strain.
Compared to the parent strain, the evolved thermotolerant strain obtained in this study showed significant physiological changes, including increased glucose fermentation capacity and decreased respiratory growth (Fig. 1(B) and (C)), and these changes were consistent with the results obtained in the previous studies [16,34]. Similarly, some previously reported transcriptomic changes in the evolved stress tolerant strains were also observed in the thermotolerant strain evolved in this study. For instance, the previously found up-regulated expression of the gene involved in sterol biosynthesis [34] and up-regulated expressions of the translation-related and amino acid biosynthetic genes were consistent with the results of this study [45] (Fig. 2). In addition, some unique mitochondria-associated transcriptome changes were revealed in the evolved thermotolerant strain examined in this study. The expression levels of the genes associated with mitochondrial translation and respiration significantly decreased in the evolved thermotolerant strain in contrast with the parent strain. This suggested that mitochondrial activity might be strongly suppressed in the evolved thermotolerant strain.
An earlier study reported that thermotolerance of yeast depends strongly on the ATP metabolism [46]. The ability of yeast to withstand high temperatures is significantly influenced by mitochondrial metabolism, particularly by the state of the electron transport chain [43]. Li et al. [44] found that mitochondria-related genes contribute to the evolution of thermotolerance in yeast. Caspeta and Nielsen [47] reported that evolution of thermotolerance is associated with the loss of the respiration capacity of yeast. Rikhvanov et al. [48] reported higher tolerance of the respiration-deficient yeast mutant to high temperature, compared to the wild-type strain. Previous studies have mainly focused on transient heat shock. However, in this study, the changes in the mitochondrial function-related genes in response to adaptive thermotolerance were investigated. While no novel relationships were observed between thermotolerance and reduced mitochondrial electron transport chain functions, the results of this study confirmed the previous findings to some extent.
Previous studies have reported an increase in the reactive oxygen species (ROS) during growth at non-optimal temperatures [49,50]. Respiration is known to generate considerably more ROS than the fermentative metabolism in yeasts. Therefore, the decreased respiration activity in the evolved strain is speculated to be a protective mechanism for preventing excessive generation of ROS, thereby avoiding excessive oxidative stress. In addition, the evolved ScY01 strain may boost other metabolic pathways, such as glycolysis, to generate abundant ATP as a response against high-temperature stress. We preliminarily found that glycolytic-associated genes showed increased expression in the evolved strain, which was consistent with its increased glucose fermentation capacity (Fig. 4). However, the evolved strain seemed to require more ATPs to fuel amino acid and protein biosynthesis and to maintain high thermotolerance. The inability of the cells to obtain enough ATP from mitochondria promoted greater production of ATP by glycolytic flux. To further confirm the changes in the Embden-Meyerhof-Parnas pathway fluxes in the evolved ScY01, flux balance analysis or the use 13C labelled fluxes are required. Mitochondria are known to regulate the expressions of nuclear genes as well as the cellular functions [51]. Remarkably, the significant impacts of genomic and transcriptomic changes associated with mitochondrial functions were inversely related to the regulation of the cytochrome c oxidase genes. Up-regulated expressions of mitochondrial genes (i.e., COX2 and COX3) were accompanied by the down-regulated expressions of nuclear genes. These contradictory regulation trends may be related to mitochondrial retrograde regulation. However, the detailed mechanism remains unclear and requires further exploration.
Based on the assessment of thermotolerance of mitochondria-associated KO strains, three mitochondria-associated genes, which may play critical roles in determining the yeast cell response and adaptation to long-term thermal stress, were identified. Adenylate kinase, encoded by ADK1, is a pivotal enzyme involved in energetic and adenylic nucleotide metabolisms. In previous work, an ADK1 KO strain derived from the BY4742 laboratory strain showed increased glucose consumption and ethanol accumulation [36]. This finding was consistent with the observations related to the wild type strain ScYa and the laboratory strain BY4743 examined in this study (Fig. 6(A) and (B)). However, ADK1 KO resulted in a significant decrease in the glucose fermentation capacity of the evolved thermotolerant strain ScY01a, especially at high temperature (Fig. 6(C)). Several previous studies have indicated that ADK1 null mutants showed increased heat sensitivity [52,53]. In a previous study, an ADK1 null mutant displayed slow growth and inability to utilize non-fermentable carbon sources, such as glycerol [54]. In the ADK1 KO mutant of BY4742 [36], ATP and ADP accumulations decreased, while AMP accumulation increased, inducing further glucose consumption to sustain ATP levels. Furthermore, genes encoding mitochondrial enzymes were also found to be up-regulated in the ADK1 KO mutant strain of BY4742, resulting in the improved respiration capacity and higher oxygen consumption. These findings suggest that ADK1 may play an important role in the long-term thermal response of the evolved thermotolerant strain by maintaining the adenine nucleotide pools of ATP, ADP, and AMP, as well as by reducing the oxygen consumption to avoid cell damage caused by ROS. However, the detailed mechanism of these functions remains to be determined. Both DOC1 and MET7 have been found to be essential for the maintenance of mitochondrial DNA [38,55]. A large-scale survey showed that DOC1 deletion may lead to increased heat sensitivity [52,56]. The DOC1 and MET7 null mutants showed no respiratory growth in presence of a non-fermentable carbon source, especially when glycerol was the sole carbon source [54,55,57]. According to the results of this study (Fig. 6), both DOC1 and MET7 KOs severely decreased the cell growth and glucose fermentation capacities of the BY4743, ScYa, and ScY01a strains. This implies that mitochondrial genome maintenance is essential for the cellular response of yeast strains to long-term thermal stress. Notably, MET7 KO resulted in a more severe inhibition of cell growth and glucose fermentation capacity of the evolved strain ScY01a (Fig. 6(C)), compared to the wild type strain ScYa (Fig. 6(B)). Altogether, these observations indicate that MET7 may play a vital role in the adaptation of yeast cells to long-term thermal stress.
Mitochondrial fusion was previously reported to increase the copy number of mitochondrial DNA by facilitating ROS-triggered, recombination-mediated DNA replication [58]. Furthermore, mitochondrial fusion causes the dissociation of respiratory complex III–IV, followed by disassembly of complex IV [58]. This suggests that mitochondrial fusion may induce mitochondrial retrograde signaling to down-regulate the gene expression of complex IV during adaptation to high temperature. The underlying mechanisms of the roles of the mitochondrial morphology and complex IV in mitochondrial retrograde regulation of evolved thermotolerant strains need to be explored further.
In this study, the respiratory growth of the thermotolerant ScY01 and its parent strain ScY was initially assessed at 40 °C using a non-fermentable carbon source. In the presence of glycerol or ethanol, ScY01 exhibited slower cell growth than ScY, indicating low respiratory growth of ScY01. Thus, decreased respiratory growth was preliminarily assumed to contribute to the adaptive evolution of thermotolerance in the evolved ScY01 strain. Subsequently, transcriptional analysis was performed for the thermotolerant ScY01 and its parent strain ScY, cultured in a very sugary medium. Both strains were subjected to the same culture conditions. To better understand the impact of respiration on the thermotolerance of the yeast strain, RNA sequencing must be carried out in future work under non-fermentable conditions. In this study, the respiratory growth capacity of the KO collection of laboratory strains BY4743 (with mutated respiratory-associated genes) were further evaluated on glycerol plates and in a YP medium with a lower glucose concentration (100 g/L) at 40 °C. In particular, the KO of ADK1, DOC1, and MET7 lowered the cell growth and fermentation rates at 40 °C. Overall, this study confirms that adjusting the respiratory activity in yeast strains may be advantageous for the adaptive evolution of thermotolerance. However, the relationship between thermotolerance, mitochondrial activity, and respiration requires further investigation.
Although adaptive laboratory evolution has been effectively used to develop thermotolerant S. cerevisiae strains, the driving force for the adaptive evolution of thermotolerance has not been well understood. In the present study, the transcriptomic changes in the evolved thermotolerant strain were systematically evaluated and compared with the wild type strain. The results of the RNA sequencing analysis revealed that the evolved strain activated energy-consuming pathways including amino acid and protein biosynthetic pathways, and suppressed mitochondrial activities including translation and respiration. As a result, glucose fermentation produced more energy to meet the energy demands due to enhanced amino acid and protein biosynthesis. Overall, the results suggest that functional adjustment of mitochondrial respiration may be the driving force of adaptive evolution of thermotolerance in S. cerevisiae strain, thus allowing the evolved strain to maintain improved fermentation performance during prolonged thermal stress. This study provided a better understanding on the adaptive evolution of thermotolerance, which may be helpful for developing novel S. cerevisiae strains with improved thermotolerance for industrial applications.
Data Availability Statement
The RNA sequencing raw data are deposited in the NCBI Gene Expression Omnibus (GEO) under the accession number GSE96829. The S. cerevisiae S288c genome as a reference can be accessed at https://www.ncbi.nlm.nih.gov/assembly/GCF_000146045.2/.
CRediT authorship contribution statement
Xianni Qi: Investigation, Methodology, Writing – original draft. Zhen Wang: Investigation, Methodology, Writing – original draft, Writing – review & editing, Funding acquisition. Yuping Lin: Supervision, Data curation, Writing – original draft. Yufeng Guo: Methodology, Visualization, Formal analysis. Zongjie Dai: Supervision. Qinhong Wang: Supervision, Funding acquisition, Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2021YFC2103300), Research Equipment Program of Chinese Academy of Sciences (YJKYYQ20170023), National Natural Science Foundation of China (32071423, 31470214, and 32200067), Natural Science Foundation of Hebei Province (C2020204013), and Development Program Projects of Hebei Province (22322905D). Qinhong Wang and Yuping Lin were supported by Tianjin Industrial Synthetic Biology Innovation Team.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.engmic.2023.100108.
Appendix. Supplementary materials
Fig. S1. Thermal stability evaluation of ScY01, the evolved thermotolerant S. cerevisiae strain.
Fig. S2. Visualization of significantly differentially expressed genes in Cytoscape.
Fig. S3. Spot assay of respiratory growth mutants on glycerol plates.
Table S1. Primers used in this study.
Dataset S1. Transcriptome profiles and differential gene expression in ScY and ScY01.
Dataset S2. DAVID Gene Ontology enrichment of up-regulated and down-regulated genes.
Dataset S3. Verification and high-temperature growth evaluation of respiratory growth mutants and gene lists in Venn Diagrams of Fig. 2.
References
- 1.Nielsen J. Yeast systems biology: model organism and cell factory. Biotechnol. J. 2019;14 doi: 10.1002/biot.201800421. [DOI] [PubMed] [Google Scholar]
- 2.Wang S., Zhao F., Yang M., et al. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of valuable chemicals. Crit. Rev. Biotechnol. 2023:1–28. doi: 10.1080/07388551.2022.2153008. [DOI] [PubMed] [Google Scholar]
- 3.Verghese J., Abrams J., Wang Y., et al. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter K., Haslbeck M., Buchner J. The heat shock response: life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Kainth A.S., Chowdhary S., Pincus D., et al. Primordial super-enhancers: heat shock-induced chromatin organization in yeast. Trends Cell Biol. 2021;31:801–813. doi: 10.1016/j.tcb.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan Y., Qi X., Lin Y., et al. A hierarchical transcriptional regulatory network required for long-term thermal stress tolerance in an industrial Saccharomyces cerevisiae strain. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.826238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salas-Navarrete P.C., de Oca Miranda A.I.M., Martinez A., et al. Evolutionary and reverse engineering to increase Saccharomyces cerevisiae tolerance to acetic acid, acidic pH, and high temperature. Appl. Microbiol. Biotechnol. 2022;106:383–399. doi: 10.1007/s00253-021-11730-z. [DOI] [PubMed] [Google Scholar]
- 8.Boonchuay P., Techapun C., Leksawasdi N., et al. Bioethanol production from cellulose-rich corncob residue by the thermotolerant Saccharomyces cerevisiae TC-5. J. Fungi (Basel) 2021;7:547. doi: 10.3390/jof7070547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao L., Liu Y., Sun H., et al. Advances in mechanisms and modifications for rendering yeast thermotolerance. J. Biosci. Bioeng. 2016;121:599–606. doi: 10.1016/j.jbiosc.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Qi Q., Lin Y., et al. QTL analysis reveals genomic variants linked to high-temperature fermentation performance in the industrial yeast. Biotechnol. Biofuels. 2019;12:59. doi: 10.1186/s13068-019-1398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phong H.X., Klanrit P., Dung N.T.P., et al. High-temperature ethanol fermentation from pineapple waste hydrolysate and gene expression analysis of thermotolerant yeast Saccharomyces cerevisiae. Sci. Rep. 2022;12:13965. doi: 10.1038/s41598-022-18212-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao W., Duan X., Lin Y., et al. Distinct proteome remodeling of industrial Saccharomyces cerevisiae in response to prolonged thermal stress or transient heat shock. J. Proteome Res. 2018;17:1812–1825. doi: 10.1021/acs.jproteome.7b00842. [DOI] [PubMed] [Google Scholar]
- 13.Shui W., Xiong Y., Xiao W., et al. understanding the mechanism of thermotolerance distinct from heat shock response through proteomic analysis of industrial strains of Saccharomyces cerevisiae. Mol. Cell Proteom. 2015;14:1885–1897. doi: 10.1074/mcp.M114.045781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspeta L., Coronel J., Montes de Oca A., et al. Engineering high-gravity fermentations for ethanol production at elevated temperature with Saccharomyces cerevisiae. Biotechnol. Bioeng. 2019;116:2587–2597. doi: 10.1002/bit.27103. [DOI] [PubMed] [Google Scholar]
- 15.Satomura A., Miura N., Kuroda K., et al. Reconstruction of thermotolerant yeast by one-point mutation identified through whole-genome analyses of adaptively-evolved strains. Sci. Rep. 2016;6:23157. doi: 10.1038/srep23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace-Salinas V., Brink D.P., Ahren D., et al. Cell periphery-related proteins as major genomic targets behind the adaptive evolution of an industrial Saccharomyces cerevisiae strain to combined heat and hydrolysate stress. BMC Genom. 2015;16:514. doi: 10.1186/s12864-015-1737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R., Reichert A.S. Common principles and specific mechanisms of mitophagy from yeast to humans. Int. J. Mol. Sci. 2021;22:4363. doi: 10.3390/ijms22094363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zdralevic M., Guaragnella N., Antonacci L., et al. Yeast as a tool to study signaling pathways in mitochondrial stress response and cytoprotection. ScientificWorldJournal. 2012;2012 doi: 10.1100/2012/912147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Lin Y., Dai Z., et al. Modulating DNA repair pathways to diversify genomic alterations in Saccharomyces cerevisiae. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02326-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiestl R.H., Gietz R.D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 21.Winzeler E.A., Shoemaker D.D., Astromoff A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 22.Guldener U., Heck S., Fielder T., et al. A new efficient gene disruption cassette for repeated use in budding yeast. Nucl. Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas B.J., Papanicolaou A., Yassour M., et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Trapnell C., Pop M., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Feng Z., WANG X., et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 27.Huang D.A.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Merico D., Isserlin R., Bader G.D. Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Methods Mol. Biol. 2011;781:257–277. doi: 10.1007/978-1-61779-276-2_12. [DOI] [PubMed] [Google Scholar]
- 29.Choi J.S., Lee C.K. Maintenance of cellular ATP level by caloric restriction correlates chronological survival of budding yeast. Biochem. Biophys. Res. Commun. 2013;439:126–131. doi: 10.1016/j.bbrc.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Gergondey R., Garcia C., Marchand C.H., et al. Modulation of the specific glutathionylation of mitochondrial proteins in the yeast Saccharomyces cerevisiae under basal and stress conditions. Biochem. J. 2017;474:1175–1193. doi: 10.1042/BCJ20160927. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y., Chomvong K., Acosta-Sampson L., et al. Leveraging transcription factors to speed cellobiose fermentation by Saccharomyces cerevisiae. Biotechnol. Biofuels. 2014;7:126. doi: 10.1186/s13068-014-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller-Fleming L., Antas P., Pais T.F., et al. Yeast DJ-1 superfamily members are required for diauxic-shift reprogramming and cell survival in stationary phase. Proc. Natl. Acad. Sci. USA. 2014;111:7012–7017. doi: 10.1073/pnas.1319221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padilla P.A., Fuge E.K., Crawford M.E., et al. The highly conserved, coregulated SNO and SNZ gene families in Saccharomyces cerevisiae respond to nutrient limitation. J. Bacteriol. 1998;180:5718–5726. doi: 10.1128/jb.180.21.5718-5726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caspeta L., Chen Y., Ghiaci P., et al. Biofuels. Altered sterol composition renders yeast thermotolerant. Science. 2014;346:75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- 35.Jayachandran C., Palanisamy A.B., Sankaranarayanan M. Cofactor engineering improved CALB production in Pichia pastoris through heterologous expression of NADH oxidase and adenylate kinase. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauthier S., Coulpier F., Jourdren L., et al. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Mol. Microbiol. 2008;68:1583–1594. doi: 10.1111/j.1365-2958.2008.06261.x. [DOI] [PubMed] [Google Scholar]
- 37.Qin L., Mizrak A., Guimaraes D., et al. The pseudosubstrate inhibitor Acm1 inhibits the anaphase-promoting complex/cyclosome by combining high-affinity activator binding with disruption of Doc1/Apc10 function. J. Biol. Chem. 2019;294:17249–17261. doi: 10.1074/jbc.RA119.009468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt T.T., Sharma S., Reyes G.X., et al. Inactivation of folylpolyglutamate synthetase Met7 results in genome instability driven by an increased dUTP/dTTP ratio. Nucl. Acids Res. 2020;48:264–277. doi: 10.1093/nar/gkz1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magalhaes R.S.S., Popova B., Braus G.H., et al. The trehalose protective mechanism during thermal stress in Saccharomyces cerevisiae: the roles of Ath1 and Agt1. FEMS Yeast Res. 2018;18 doi: 10.1093/femsyr/foy066. [DOI] [PubMed] [Google Scholar]
- 40.Gibney P.A., Lu C., Caudy A.A., et al. Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. Proc. Natl. Acad. Sci. USA. 2013;110:E4393–E4402. doi: 10.1073/pnas.1318100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C.J., Lu M.Y., Chang Y.W., et al. Experimental evolution of yeast for high-temperature tolerance. Mol. Biol. Evol. 2018;35:1823–1839. doi: 10.1093/molbev/msy077. [DOI] [PubMed] [Google Scholar]
- 42.Randez-Gil F., Prieto J.A., Rodriguez-Puchades A., et al. Myriocin-induced adaptive laboratory evolution of an industrial strain of Saccharomyces cerevisiae reveals its potential to remodel lipid composition and heat tolerance. Microb. Biotechnol. 2020;13:1066–1081. doi: 10.1111/1751-7915.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mejia-Barajas J.A., Martinez-Mora J.A., Salgado-Garciglia R., et al. Electron transport chain in a thermotolerant yeast. J. Bioenerg. Biomembr. 2017;49:195–203. doi: 10.1007/s10863-017-9696-x. [DOI] [PubMed] [Google Scholar]
- 44.Li X.C., Peris D., Hittinger C.T., et al. Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci. Adv. 2019;5:eaav1848. doi: 10.1126/sciadv.aav1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinel D., Colatriano D., Jiang H., et al. Deconstructing the genetic basis of spent sulphite liquor tolerance using deep sequencing of genome-shuffled yeast. Biotechnol. Biofuels. 2015;8:53. doi: 10.1186/s13068-015-0241-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piper P.W. Molecular events associated with acquisition of heat tolerance by the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1993;11:339–355. doi: 10.1111/j.1574-6976.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 47.Caspeta L., Nielsen J. Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. mBio. 2015;6:e00431. doi: 10.1128/mBio.00431-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rikhvanov E.G., Varakina N.N., Rusaleva T.M., et al. [Heat shock-induced changes in the respiration of the yeast Saccharomyces cerevisiae] Mikrobiologiia. 2001;70:531–535. [PubMed] [Google Scholar]
- 49.Pyatrikas D.V., Fedoseeva I.V., Varakina N.N., et al. Relation between cell death progression, reactive oxygen species production and mitochondrial membrane potential in fermenting Saccharomyces cerevisiae cells under heat-shock conditions. FEMS Microbiol. Lett. 2015;362:fnv082. doi: 10.1093/femsle/fnv082. [DOI] [PubMed] [Google Scholar]
- 50.Fedoseeva I.V., Pyatrikas D.V., Stepanov A.V., et al. The role of flavin-containing enzymes in mitochondrial membrane hyperpolarization and ROS production in respiring Saccharomyces cerevisiae cells under heat-shock conditions. Sci. Rep. 2017;7:2586. doi: 10.1038/s41598-017-02736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parikh V.S., Morgan M.M., Scott R., et al. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- 52.Sinha H., David L., Pascon R.C., et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics. 2008;180:1661–1670. doi: 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng X., Xu Z., Wang J., et al. ATP-dependent pre-replicative complex assembly is facilitated by Adk1p in budding yeast. J. Biol. Chem. 2010;285:29974–29980. doi: 10.1074/jbc.M110.161455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dimmer K.S., Fritz S., Fuchs F., et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merz S., Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10:R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruiz-Roig C., Vieitez C., Posas F., et al. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 2010;76:1049–1062. doi: 10.1111/j.1365-2958.2010.07167.x. [DOI] [PubMed] [Google Scholar]
- 57.Stenger M., Le D.T., Klecker T., et al. Systematic analysis of nuclear gene function in respiratory growth and expression of the mitochondrial genome in S. cerevisiae. Microb. Cell. 2020;7:234–249. doi: 10.15698/mic2020.09.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hori A., Yoshida M., Ling F. Mitochondrial fusion increases the mitochondrial DNA copy number in budding yeast. Genes Cells. 2011;16:527–544. doi: 10.1111/j.1365-2443.2011.01504.x. [DOI] [PubMed] [Google Scholar]
- 59.Ryan O.W., Skerker J.M., Maurer M.J., et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Thermal stability evaluation of ScY01, the evolved thermotolerant S. cerevisiae strain.
Fig. S2. Visualization of significantly differentially expressed genes in Cytoscape.
Fig. S3. Spot assay of respiratory growth mutants on glycerol plates.
Table S1. Primers used in this study.
Dataset S1. Transcriptome profiles and differential gene expression in ScY and ScY01.
Dataset S2. DAVID Gene Ontology enrichment of up-regulated and down-regulated genes.
Dataset S3. Verification and high-temperature growth evaluation of respiratory growth mutants and gene lists in Venn Diagrams of Fig. 2.
Data Availability Statement
The RNA sequencing raw data are deposited in the NCBI Gene Expression Omnibus (GEO) under the accession number GSE96829. The S. cerevisiae S288c genome as a reference can be accessed at https://www.ncbi.nlm.nih.gov/assembly/GCF_000146045.2/.