Abstract
Myxobacteria are famous for their capacity for social behavior and natural product biosynthesis. The unique sociality of myxobacteria is not only an intriguing scientific topic but also the main limiting factor for their manipulation. After more than half a century of research, a series of genetic techniques for myxobacteria have been developed, rendering these mysterious bacteria manipulable. Here, we review the advances in genetic manipulation of myxobacteria, with a particular focus on the exploitation of secondary metabolism. We emphasize the necessity and urgency of constructing the myxobacterial chassis for synthetic biology research and the exploitation of untapped secondary metabolism.
Keywords: Myxobacteria, Myxococcus xanthus, Genetic manipulation, Secondary metabolism, Epothilone
Graphical abstract
1. Introduction
Although previously misidentified as fungi in the early 20th century, myxobacteria, a type of bacteria, have been regarded as mysterious organisms due to their social behaviors. These include swarm-gliding movements, morphogenesis of multicellular fruiting body structures, and wolf-like predation, which are rare in bacteria but common in eukaryotes. Myxobacteria are thus used as amenable model organisms in the study of prokaryotic multicellularity and cell–cell communication to investigate the involved molecular mechanisms and their significance in evolution and ecology [1], [2], [3], [4]. In addition, myxobacteria are a prolific source of novel natural compounds with great potential for discovering new drugs with chemical diversity and uncommon mechanisms of action [5], [6], [7]. Myxobacteria typically have a large genome with numerous genes devoted to the biosynthesis of secondary metabolites; however, most of these genes have yet to be identified [8,9]. They are a group of predatory microorganisms that are ecologically applicable in biocontrol; for instance, a Corallococcus sp. was recently shown to attack pathogenic fungi by lysing cells with β-1,6-glucanase (GluM), as well as to regulate soil microbial community and control cucumber fusarium wilt [3,10]. Myxococcus xanthus DK1622 has also been shown to inhibit the growth of certain plant-pathogenic fungi using a toxin-immune protein system [11]. Thus, myxobacteria are relevant to both fundamental and applied researches. However, the unique sociality of myxobacteria is not only an intriguing scientific topic but also a limiting factor for their manipulation.
Myxobacteria typically grow slowly and rarely form colonies from a single cell [1]. It is, therefore, difficult to isolate myxobacterial strains, although they are among the most abundant and ubiquitous bacteria (accounting for 2% of the total OTUs) in soils worldwide [12]. Myxobacteria are one of the least cultured bacteria, and many unclassified myxobacteria exist [13]. The lack of genetic tools severely hampered research on cultured myxobacteria, particularly for non-model strains. Compared to common model strains such as Escherichia coli, myxobacteria lack efficient genetic manipulation techniques and selection markers, making it difficult to construct, screen, and purify mutant strains. Nevertheless, after more than half a century of research, several genetic tools have been developed and optimized in myxobacteria, primarily in the model species Myxococcus xanthus. These effective technologies facilitate myxobacteria research and exploitation. In this review, we provide a brief description of myxobacteria and their secondary metabolism and summarize the genetic tools developed in myxobacteria and their applications in the study of natural products, particularly epothilones. We propose myxobacterial chassis for synthetic biology research and genetic tools in non-model myxobacteria for untapped secondary metabolism.
2. Social and developmental biology of the myxobacteria
Myxobacteria are a group of gliding gram-negative bacteria phylogenetically related to the Delta-proteobacteria order Myxococcales. Myxococcales contains three validly published suborders, Cystobacterineae, Sorangiineae, and Nannocystineae, which are subdivided into 10 families, 29 genera, and 58 species [14]. Recently, the Myxococcales was reclassified as the Myxococcota phylum based on 120 conserved single-copy marker genes and rRNA genes [15].
Myxobacteria secrete enzymes capable of degrading various biological macromolecules or living microbial cells [16]. Myxobacteria are traditionally divided into two groups based on their feeding habits: bacteriolytic myxobacteria, which degrade living microorganisms, and cellulolytic myxobacteria, which decompose cellulose and dead cells. The feeding pattern is still the basis for myxobacteria isolation techniques [17]. To date, most cultured myxobacteria are bacteriolytic, and only two genera, Sorangium and Byssovorax [14,18], comprise the cellulolytic myxobacteria. Notably, some recently identified myxobacteria, including Vulgatibacter incomptus, Labilithrix luteola, and Simulacricoccus ruber, neither utilize living bacterial cells nor hydrolyze cellulose [19,20].
The motility of myxobacteria is driven by two distinct mechanisms: social motility (S motility), cells moving in large clusters mediated by the extending and retracting of polar type IV pili, and adventurous motility (A motility), cells moving as individuals driven by laterally embedded motility engines [2,21]. The A- and S-motilities collaborate to permit frequent encounters with prey, thereby facilitating efficient predation [16,22]. Under conditions of starvation, most myxobacteria can develop fruiting body structures: the developing cells migrate into aggregation centers and climb on top of each other to form fruiting bodies, where parts of cells inside cysts differentiate into environmentally resistant myxospores [23]. Notably, while myxospores of the model strain Myxococcus xanthus DK1622 can germinate from single cells when cultured in appropriate media, the germination of many other myxobacterial strains, particularly new isolates, is normally cell density-dependent even in these 'appropriate' media, and the fruiting body structures may ensure sufficient cell numbers for germination [17]. Notably, some recently discovered myxobacteria, such as Anaeromyxobacter [24], V. incomptus, L. luteola [19], S. ruber [20], and Polyangium aurulentum [25], are incapable of fruiting, and the non-fruiting myxobacteria are probably widespread in the environment, representing most of the uncultured myxobacteria in nature [26].
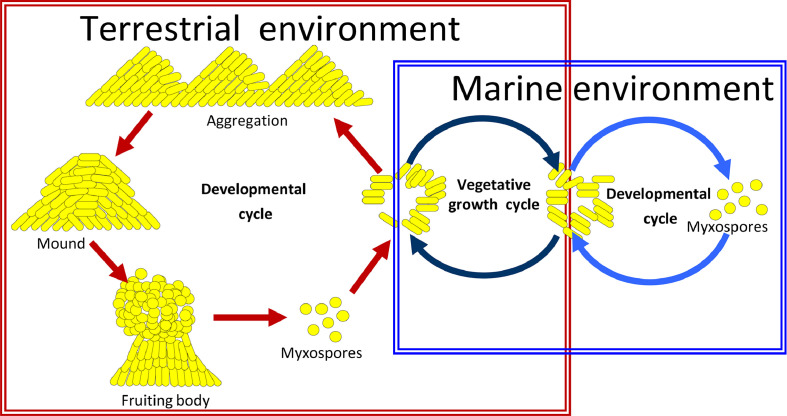
Although appealing, the multicellular social characteristics of myxobacteria are the primary factors limiting their manipulation. Li et al. reported a simple method for isolating salt-tolerant myxobacteria from marine samples. These salt-tolerant myxobacteria could grow but could not produce fruiting bodies at high salt concentrations [27]. Further research revealed that salt-tolerant Myxococcus fulvus HW-1 rapidly adapted to different living environments: cells grew normally and formed complete fruiting body structures in terrestrial environments (with low concentrations of seawater or salts); cells became shorter and formed rudimentary structures or even simple cell mounds as the seawater concentration increased [28], [29], [30], [31], [32]. Fig. 1 depicts the living pattern shift of HW-1 in response to salinity. The observed changes in fruiting body formation ability and cell density-dependent growth of salt-tolerant Myxococcus strains are indicative of ecological adaptation strategies of myxobacterial social behaviors to marine environments. Moreover, myxobacteria could be isolated and genetically modified by utilizing their simple living pattern in specific environments.
Fig. 1.
A schematic diagram showing the life pattern shift of halotolerant Myxococcus fulvus HW-1 between the marine and terrestrial conditions. The marine environment is saline, like seawater, whereas the terrestrial environment is relatively low in salt. In both cases, M. fulvus HW-1 grows (vegetative growth cycle) under eutrophic conditions and develops into fruiting bodies or myxospores (developmental cycle) with exhausted nutrition.
3. Diversity of secondary metabolism in myxobacteria
Microorganisms, particularly medicinal resource microorganisms rich in biosynthetic pathways of secondary metabolites, are consistently among the most promising sources for developing new drugs or lead compounds. Myxobacteria are a relatively new group of medicinal microorganisms with complex structures and diverse action mechanisms [33], whereas actinobacteria have been extensively screened for antibiotics for more than half a century. 109 distinct core structures and more than 600 derivatives have been determined from the normal screening of approximately 7500 identified myxobacterial strains; the genus Sorangium produces the most abundant secondary metabolites, accounting for 48.4% of the total products found in myxobacteria, followed by Myxococcus (20.7%), Chondromycetes (10.3%), Stigmatella (6.9%) and Polyangium (5.2%) [6].
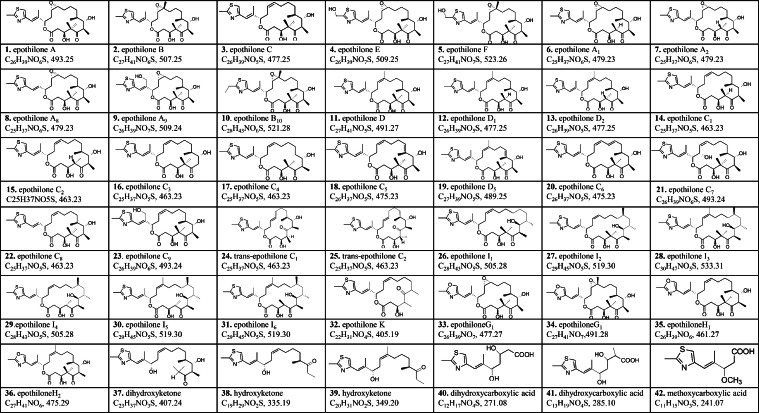
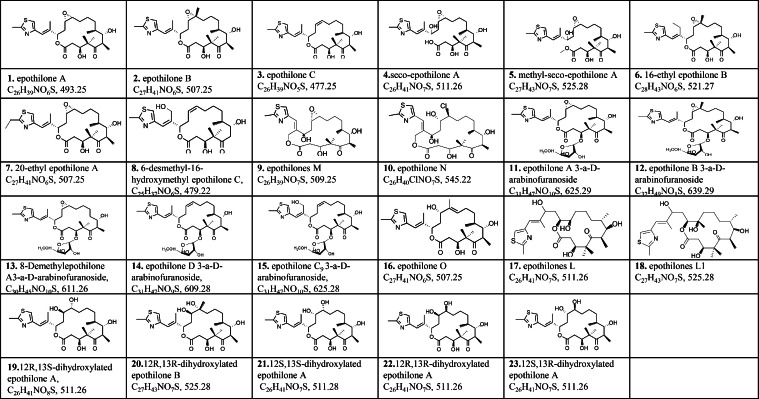
Myxobacterial bioactive compounds have various chemical structures, but most are the derivatives of products synthesized by polyketide synthase (PKS), non-ribosomal peptide synthase (NRPS), or hybrid PKS–NRPS systems [34]. The stepwise synthesis of polyketides, non-ribosomal peptides, and their hybrids involves the assembly of acyl-CoA thioester (in the case of polyketides) or amino acids (proteinogenic and non-proteinogenic in the case of non-ribosomal peptides). Diverse modifications may occur during assembly or after the release of a compound, resulting in products with complex chemical structures. Thus, a fundamental core structure can generate numerous analogs. More than 70 naturally occurring structural variants of soraphen were once discovered in S. cellulosum So ce26 [35]. Myxobacterial metabolites are also strain-specific; in S. cellulosum So ce90/B2, 37 analogs of epothilone have been isolated [36,37]. Li and colleagues identified 23 epothilone analogs from strain So0157-2 [38], [39], [40], [41], [42]. Only epothilones A, B, and C are present in So ce90/B2, whereas the others are unique (Fig. 2, Fig. 3).
Fig. 2.
Epothilone analogs identified in S. cellulosum So0157-2.
Fig. 3.
Epothilone analogs revealed in S. cellulosum So ce90/B2.
Moreover, myxobacterial products frequently exhibit diverse and novel action mechanisms, making them promising as new drugs for various applications, including antifungal, antibacterial, cytotoxic, and antiviral activities [43]. For example, bacterial RNA polymerase inhibitors such as thiolutin [44], streptolydigin [45], and holomycin [46] inhibit RNA synthesis in a manner distinct from that of the clinical drug rifamycin. Since they have no cross-resistance with rifamycin, these compounds have the potential to combat rifamycin resistance [47]. Epothilones, a type of microtubule stabilizer that mimics paclitaxel [36], exhibit potent anti-paclitaxel-resistant tumor activity [48]. Two epothilone varieties, Ixabepilone [49] and Utidelone [50] have been approved for use as antitumor drugs against advanced breast cancer in clinical settings. Epothilones are also promising in treating neurodegenerative disorders [51,52]. Several excellent reviews summarize the bioactivities and mechanisms of action of myxobacterial secondary metabolites, such as inhibitions of respiration, protein or nucleic acid synthesis, interferences of the protein phosphorylation system, electron transport, cell division, lipid synthesis, and carbohydrate metabolism [6,33,43,53].
Myxobacterial metabolites are highly specific. Althiomycin [54], saframycin [55], and pyrrolnitrin [56] are the only chemical core structures from myxobacteria that have been identified in other microbial resources. Similarly, approximately 40% of myxobacteria-derived compounds represent novel chemical structures that act on novel targets distinct from those imposed by compounds derived from Actinomycetes, Bacillus, or Pseudomonas [43]. The specificity of myxobacterial metabolites represents a significant opportunity for the discovery of new drugs.
A single myxobacteria species or strain is capable of producing numerous active secondary metabolites with distinct structural scaffolds. S. cellulosum strains have been found to contain a wide variety of compound structures, including macrolides (such as disorazol, chivosazol, soraphen, and epothilone), aromatics (such as jerangolid), quinones (such as sorangiolid), and polyenes (such as ratjadon and ambruticin) [6]. At least nine types of active natural products have been identified in Chondromyces crocatus Cm c5, including ajudazols, chondrochlorens, dialkylpyrazines, and thuggacins [7]. In the model strain M. xanthus DK1622, nine compounds with different types of structure have been found, of which Dkxanthenes, myxoprincomides, and homospermidin lipids are presently unique in DK1622, while myxothiazol, an inhibitor of mitochondrial respiration, has also been identified in Stigmatella aurantiaca [57], M. fulvus [58], and Angiococcus disciformis [59], and myxolamids are similarly produced in M. fulvus, M. xanthus, S. aurantiaca, and C. coralloides [34]. Systematic characterization of a single myxobacterial species or strain requires dogged efforts using the strategy of “one strain many compounds” (OSMAC) [60]. Examining strains from new genera rather than additional representatives within the same genus [61] increases the likelihood of discovering novel metabolites, but screening myxobacterial isolates from different environments could also reveal new active natural products such as nicotinic myxochelins [62], myxadazoles [63], and archangiumide [64].
With the advent of the genomic era, studies on the genome and the exploitation of secondary metabolite biosynthetic gene clusters (smBGCs) with undefined functions have gained attention as a novel strategy for expanding the range and diversity of bacterial secondary metabolites. According to the published genome information in the NCBI database, 91 cultured myxobacterial strains have been sequenced thus far. Except for a few small genomes of approximately 5 Mb [19,25,65], myxobacterial genomes range between 9 and 16 Mb in size. For example, the genomes of Minicystis rosea DSM 24000 [66] and S. cellulosum So0157-2 [67] are 16.04 Mb and 14.78 Mb, respectively, making them the two largest known prokaryotic genomes, surpassing many eukaryotic genomes such as Saccharomyces cerevisiae (12.16 Mb). A total of 42 myxobacteria have been fully sequenced, and their smBGCs have been analyzed using antiSMASH. As shown in Table 1, myxobacterial genomes contain more than 20 smBGCs, with the exception of the smallest genomes. In a specific strain of myxobacteria, such as the best-characterized M. xanthus DK1622, only nine of the 24 smBGCs and their products have been identified. Myxobacteria still contain a large number of metabolites awaiting further exploitation. Furthermore, considering the large number of uncultured myxobacteria and their unidentified secondary metabolic genes, the bioactive compounds reported to date from myxobacteria are merely the tip of the iceberg. Although omics-based research will reveal ever-increasing synthetic potentials of metabolites in myxobacteria [68], a thorough exploration of the potentials of drugs or leading compounds requires the development of efficient genetic tools for myxobacteria.
Table 1.
smBGCs in completely sequenced myxobacteria.
| Strain | Genome size (Mb) | (G + C)% | Accession | Number of smBGCs | Proportion of smBGCs in chromosome |
|---|---|---|---|---|---|
| Minicystis rosea DSM 24000 | 16.04 | 69.1 | NZ_CP016211.1 | 40 | 6.60% |
| Sorangium cellulosum So0157-2 | 14.78 | 72.1 | NC_021658.1 | 35 | 9.60% |
| Sorangium cellulosum So ce836 | 14.60 | 72.0 | NZ_CP012672.1 | 42 | 12.70% |
| Sorangium cellulosum So ce26 | 14.56 | 71.7 | NZ_CP012673.1 | 36 | 10.90% |
| Archangium violaceum SDU8 | 13.21 | 68.9 | NZ_CP069396.1 | 38 | 12.80% |
| Pyxidicoccus sp. SCPEA002 | 13.21 | 69.6 | NZ_CP071090.1 | 32 | 12.40% |
| Sorangium cellulosum So ce 56 | 13.03 | 71.4 | NC_010162.1 | 35 | 9.00% |
| Archangium violaceum SDU34 | 12.68 | 68.4 | NZ_CP069338.1 | 35 | 9.60% |
| Archangium gephyra DSM 2261 | 12.49 | 69.4 | NZ_CP011509.1 | 39 | 11.30% |
| Cystobacter fuscus DSM 52655 | 12.35 | 68.5 | NZ_CP022098.1 | 47 | 12.00% |
| Labilithrix luteola DSM 27648 | 12.19 | 66.1 | NZ_CP012333.1 | 12 | 3.10% |
| Chondromyces crocatus Cm c5 | 11.39 | 68.7 | NZ_CP012159.1 | 33 | 12.90% |
| Sorangium cellulosum So ceGT47 | 11.26 | 72.6 | NZ_CP012670.1 | 27 | 8.50% |
| Myxococcus xanthus 124B02 | 11.05 | 70.0 | CP006003.1 | 29 | 9.60% |
| Myxococcus stipitatus DSM 14675 | 10.35 | 69.2 | NC_020126.1 | 25 | 13.70% |
| Sandaracinus amylolyticus DSM 53668 | 10.33 | 72.0 | NZ_CP011125.1 | 20 | 5.60% |
| Stigmatella aurantiaca DW4/3-1 | 10.26 | 67.5 | NC_014623.1 | 35 | 12.10% |
| Corallococcus coralloides DSM 2259 | 10.08 | 69.9 | NC_017030.1 | 31 | 13.10% |
| Melittangium boletus DSM 14713 | 9.91 | 68.4 | NZ_CP022163.1 | 33 | 11.80% |
| Corallococcus coralloides B035 | 9.59 | 70.3 | NZ_CP034669.1 | 32 | 15.30% |
| Myxococcus hansupus contaminant ex DSM 436 | 9.49 | 69.2 | NZ_CP012109.1 | 28 | 12.40% |
| Haliangium ochraceum DSM 14365 | 9.45 | 69.5 | NC_013440.1 | 21 | 8.10% |
| Corallococcus sp. EGB | 9.43 | 70.4 | CP079946.1 | 32 | 14.60% |
| Myxococcus xanthus KF4.3.9c1 | 9.43 | 68.9 | NZ_CP017172.1 | 21 | 11.10% |
| Myxococcus xanthus DZ2 | 9.36 | 68.9 | NZ_CP070500.1 | 22 | 12.50% |
| Myxococcus xanthus ATCC 27925 | 9.35 | 68.9 | NZ_CP080534.1 | 22 | 12.60% |
| Myxococcus xanthus MC3.5.9c15 | 9.32 | 69.0 | NZ_CP017174.1 | 22 | 11.50% |
| Myxococcus xanthus MC3.3.5c16 | 9.32 | 69.0 | NZ_CP017173.1 | 22 | 11.50% |
| Myxococcus xanthus GH3.5.6c2 | 9.32 | 69.0 | NZ_CP017169.1 | 21 | 11.20% |
| Myxococcus xanthus GH5.1.9c20 | 9.26 | 69.0 | NZ_CP017170.1 | 21 | 11.30% |
| Myxococcus xanthus R31 | 9.25 | 68.9 | NZ_CP068048.1 | 21 | 11.90% |
| Myxococcus xanthus DK1622::pDPO-Mxn116-Pvan-Tpase | 9.20 | 68.9 | NZ_CP065375.1 | 23 | 13.70% |
| Myxococcus xanthus DK 1622 | 9.14 | 68.9 | NC_008095.1 | 22 | 12.80% |
| Myxococcus fulvus HW-1 | 9.00 | 70.6 | NC_015711.1 | 23 | 11.40% |
| Corallococcus macrosporus DSM 14697 | 8.97 | 70.6 | NZ_CP022203.1 | 22 | 10.80% |
| Myxococcus xanthus KF3.2.8c11 | 8.95 | 69.0 | NZ_CP017171.1 | 24 | 13.50% |
| Anaeromyxobacter sp. Fw109-5 | 5.28 | 73.5 | NC_009675.1 | 6 | 2.40% |
| Anaeromyxobacter sp. K | 5.06 | 74.8 | NC_011145.1 | 6 | 3.40% |
| Anaeromyxobacter dehalogenans 2CP-1 | 5.03 | 74.7 | NC_011891.1 | 6 | 3.40% |
| Anaeromyxobacter dehalogenans 2CP-C | 5.01 | 74.9 | NC_007760.1 | 6 | 3.40% |
| Vulgatibacter incomptus DSM 27710 | 4.35 | 68.9 | NZ_CP012332.1 | 3 | 2.30% |
4. Genetic tools in myxobacteria
Exploitation of myxobacteria lags far behind that of other medicinal microorganisms, such as Actinomycetes and Bacillus, due to their slow and aggregate growth, complex multicellular behavior, and especially lack of efficient genetic methods. In conjunction with the genetic and biological research conducted on myxobacteria for more than half a century, several genetic manipulation techniques and tools have been developed, particularly for the model organism M. xanthus. The advantages and disadvantages of each of these tools are summarized in Table 2.
Table 2.
Summary of the major advantages and limitations of genetic manipulation methods in myxobacteria.
| Genetic manipulation method | Major components | Advantages | Limitations | Reference |
|---|---|---|---|---|
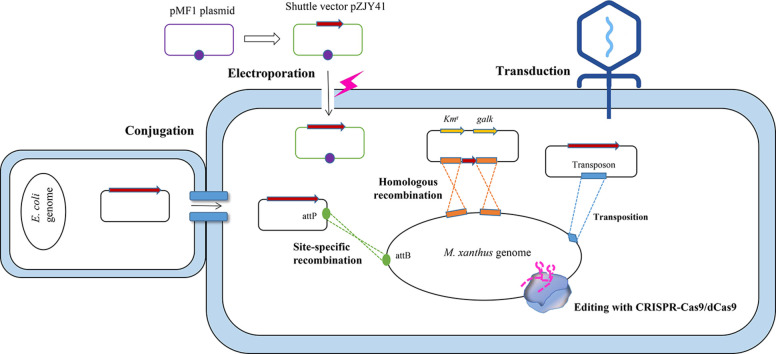
| Transduction | Phages | Applicable to any host that can be infected by phages | Phage-dependent operations are cumbersome and time-consuming | [70], [71], [72], [73] |
| Transformation (electroporation) | Plasmids | Efficient transformation of large smBGCs | Not suitable for agglomerate myxobacteria | [81], [82], [83] |
| Conjugation | Mobilizable plasmid | The transfer process is stable and gentle | Inefficient and cumbersome | [101], [102], [103], [104], [105] |
| Homologous recombination | Homologous arms and homologous recombinases | Directed editing | The accuracy and efficiency are affected by homologous sequences | [111,113] |
| Site-specific recombination | Intergrase and integration sites of phage Mx8 | Directed editing | The integration sites are limited, and integration efficiency decreases with the length of integrated fragments | [84,123,124] |
| Transposition | Transpon (Tn5, mariner) | Efficient insertion | Random insertion | [[136,140] |
| Plasmid | Shuttle vector pZJY41 derived from pMF1 | Stable multi-copy expression of target genes | There is only one kind of shuttle plasmid applicable to limited hosts | [79,162] |
| CRISPR-Cas9 system | Cas9 protein and sgRNA | Directed editing or activation of target genes | The simplicity and efficiency need to be optimized; the endogenous CRISPR-Cas system remains to be developed | [96,159,160] |
4.1. Gene delivery systems in myxobacteria
In bacteria, DNA is transported primarily through three mechanisms: transduction, transformation, and conjugation [69], and myxobacteria have demonstrated maneuverability with these gene delivery methods. The use of phage-dependent transduction for genetic modification of myxobacteria was once widespread but has since fallen out of favor due to its cumbersome nature and the advent of more effective techniques. Conjugation is predominantly utilized in S. cellulosum, but the operation method is strain-specific and lacks commonality. Transformation (electroporation rather than heat-shock transformation) stands out for its high efficiency in transporting DNA, particularly large fragments, and has become the primary method of gene delivery in myxobacteria.
4.1.1. Transduction
Transduction refers to the phage-mediated transfer of DNA from a donor cell to a recipient. E. coli phage P1 was the first phage to successfully introduce foreign DNA into an M. xanthus strain [70]. It allows the transfer of the transposon Tn5, which integrates randomly into the recipient chromosome to produce mutations [71,72]. In 1966, Burchard and Dworkin reported the first myxophage Mx1 from M. xanthus [73]. More than 35 myxophages have been isolated and classified into five serological groups: Mx1, Mx4, Mx8, Mx9, and Mxα [74]. Several generalized transduction phages have been derived [75], [76], [77], and Mx8 and Mx4 are capable of packaging up to approximately 50 kb of DNA [75]. Most of these studies on myxophages were conducted between 30–50 years ago, and myxophage-based transduction has been gradually phased out as more effective techniques have been developed. However, myxophage-derived integrases are still commonly used in myxobacteria gene editing, as will be discussed in detail in the “Site-specific recombination” section.
4.1.2. Transformation
Natural transformation is a significant way for bacteria to acquire genetic material in nature and a significant evolutionary force [78]. Shuttle plasmid pZJY41, which is derived from the autonomously replicating plasmid pMF1 of M. fulvus 124B02 [79], resulted in a successful transformation from E. coli to M. xanthus [80], according to a study conducted on M. xanthus. Exopolysaccharide (EPS), as an extracellular barrier, and restriction-modification system, as an inner barrier, both prevent the uptake of foreign DNA fragments, resulting in low efficiency of natural transformation, according to the authors.
Compared to natural transformation, electroporation employs transient high-voltage electrical pulses to induce the temporary formation of water channels or membrane pores on the cell membrane, allowing extracellular DNA to enter the cell. Electroporation has been developed as a fast, simple, and efficient technique for bacteria, yeast, plant cells, and mammalian cell lines [81]. Electrotransformation was first used in 1989 to introduce a plasmid containing 14 kb of M. xanthus DK101 genome DNA into development-deficient M. xanthus cells, producing fruiting transformants [82]. Subsequently, electrotransformation was optimized and widely adopted, eventually becoming the most prevalent method for transferring foreign DNA into M. xanthus cells [83]. For illustrative purposes, we highlighted several recent studies. With electroporation, the plasmids carrying target DNA of various sizes are efficiently transferred into myxobacteria, including smBGCs spanning tens or even a hundred kb [84]. In order to heterologously express myxobacterial secondary metabolites, the entire smBGC is assembled in E. coli cells using Red/ET recombineering technology or derivative technologies [87]. The smBGC was cloned based on the recombination of genomic library plasmids or even directly captured, modified precisely in E. coli, and then electroporated into M. xanthus or other hosts for heterologous expression. Combining advanced DNA recombineering techniques with an efficient electroporation technique allows for the exploration of secondary metabolite pathways in myxobacteria. Numerous known or unknown gene clusters from various myxobacteria were cloned and electroporated into heterologous hosts, resulting in the expression and identification of desired products such as epothilone [88], myxothiazol [89], myxochromide [88], disorazol [90], cystobactamid [91], myxopyronins [92], and myxarylin [93].
In addition to the model myxobacterium M. xanthus, the electroporation technique was also applied to S. aurantiaca DW4/3-1 [94], S. cellulosum So ce12 [95], So ce M4 [96,97], and A. disciformis An d48 [98]. One of the challenges of genetic manipulation in myxobacteria is the agglomerate growth characteristic, which severely restricts the uptake of external DNA as well as the screening and purification of mutant strains. Constructing dispersed-growing strains through mutation or domestication of wild-type strains effectively circumvents the genetic manipulation barrier. For instance, Zhang et al. obtained a completely dispersed growth mutant UV684 of M. fulvus HW-1 via ultraviolet mutagenesis. The mutant exhibits the same fruiting body formation and S-motility as the parent strain [99]. When the plasmid pMiniHimar1 was electrotransformed into UV684, the efficiency increased three to fivefold compared to HW-1. Notably, the model strain M. xanthus DK1622 is also a mutant of the original strain FB and is capable of developing fruiting bodies on solid surfaces as well as growing dispersedly in liquid, thereby facilitating numerous genetic manipulations.
4.1.3. Conjugation
Conjugation is a well-understood process in which a donor bacterium transfers DNA to a recipient bacterium [100]. It is linked to the type IV secretion system (T4SS), where the pilus appendage acts as a needle, propelling relaxase across membrane barriers into the recipient, and the conjugative plasmid is replicated using the rolling-circle replication mechanism and pumped into the recipient cytoplasm with the available T4SS transport conduit [101]. Although tedious, conjugation is a gentle and stable method for DNA transfer. Subsequently, using a mobilization plasmid, the recombinant conjugative plasmid carrying the target gene can be transferred from an E. coli donor strain to a myxobacterial recipient strain, where it can replicate autonomously or be incorporated into the genome.
The conjugation system has been utilized effectively in M. xanthus [102], S. aurantiaca [103], and S. cellulosum [104,105] to characterize the biosynthetic genes for secondary metabolites such as soraphen, epothilone, and chivosazole. It was reported that the conjugation efficiency of S. cellulosum increased as the length of the homologous arm increased [105]. Julien et al. cloned a mariner-based transposon into the conjugation plasmid in 2003, causing the plasmid to randomly insert into the genome of So ce90 following conjugation[107]. This conjugation system was applicable to So ce90 but not to other Sorangium strains, e.g., So ce12. Kopp et al. developed a method for DNA transfer from E. coli to So ce56 and So ce12 via conjugation using biparental or triparental mating and obtained transposon mutants with an inactive chivosazole biosynthetic gene cluster[108]. In addition to establishing the conjugative system in So0157-2 and So02007-3, Xia et al. found that the addition of low-dose antibiotics to the conjugation medium significantly increased conjugation efficiency [107]. Generally, the conjugation system developed for an S. cellulosum strain may be inapplicable to others, including strains of the same species [106], and no universal conjugation system has yet been established for different S. cellulosum strains, let alone other myxobacteria. The extraction of bioactive natural products from Sorangium strains might depend more on the heterologous expression of smBGCs than on the genetic performance of native producers.
4.2. Gene editing tools in myxobacteria
After transfer, the stable existence of extracellular DNA in host cells is dependent upon either autonomous plasmid replication or integration into the host genome. Homologous recombination between the host genome and homologous arms allows suicide plasmid or homologous fragment integration [108]. Typically derived from phages, integrases catalyze the site-specific recombination of foreign DNA with the genome at attachment sites [109]. Unlike site-specific insertion based on recombination and integration, during transposition, exogenous DNA is randomly inserted into the genome [112]. For myxobacteria, unless an autonomously replicating plasmid is available, gene editing studies in myxobacteria are carried out primarily via integration mediated by recombinases or integrases[79]. Myxobacteria have been introduced to the burgeoning clustered regularly interspaced short palindromic repeats (CRISPR) gene editing technology in recent years.
4.2.1. Homologous recombination
Through homologous recombination, a suicide plasmid carrying a fragment homologous to the host chromosome will be incorporated into the chromosome once introduced. Cho and Zusman once created a plasmid library containing approximately 500-bp random DNA fragments of M. xanthus. These fragments were electroporated into wild-type M. xanthus cells and integrated into the chromosome by homologous recombination, thereby producing a random pool of M. xanthus insertion mutants [111]. The screening of insertion mutants revealed two essential genetic regions for the production of EPS: the EPS synthesis region and the EPS-associated region [112]. The integration will result from the functional inactivation of the inserted gene or functional complementation of a gene carried by the plasmid. In order to avoid affecting the expression of in situ genes, it is necessary to choose an appropriate recombination site. For instance, the 3′ end of the dev operon was once chosen as the recombination site for the heterologous expression of the epothilone gene cluster in M. xanthus [85].
Based on homologous recombination, a knockout system based on the pBJ113 plasmid [113] containing the kanamycin resistance (Kmr)-galactosidase kinase (galK) gene cassette (KG) has been developed and is widely used for traceless gene editing in M. xanthus [114], [115], [116], [117]. Kmr is used as a positive selection marker for the first round of homologous recombination, and galK from E. coli makes cells sensitive to 1% galactose for isolating mutants that have lost galK in the second round of homologous recombination. If the second recombination occurs between the same homologous arms as the initial recombination, the strain will revert to its wild-type state; otherwise, the target gene is deleted completely. This double-crossover method was once used to determine the necessity of groEL2, but not groEL1, for the biosynthesis of the secondary metabolite myxovirescin in M. xanthus DK1622 [117,118]. Similar methods have been used to identify transcriptional regulators of secondary metabolites, primarily in M. xanthus, including specific regulators for the biosynthesis of epothilone [119] or carotenoid [120], as well as some global regulators [121,122]. Knockout of epoF, the last gene of the epothilone gene cluster, caused the conversion of epothilones A and B to epothilones C and D, indicating that the gene is responsible for the formation of the epoxide at C12-C13 [85].
Exploring the biosynthetic pathways of smBGC via the traceless mutation is also very encouraging. Because there are numerous similar smBGCs in a myxobacterial strain and highly similar sequences in smBGCs such as PKS and NRPS, homologous recombination-based gene editing is unavoidably time-consuming and inefficient. Moreover, using the two-round schedule in M. xanthus, homologous recombination, mutant screening, and purification of a traceless knockout strain typically require at least 1 month.
4.2.2. Site-specific recombination
In M. xanthus, the site-specific recombination system mediated by a myxophage-derived recombinase is also effective in enhancing genetic performance. Mx8 has been completely sequenced and is the myxophage that is best understood. Mx8 integrates into the host genome as a stable prophage via site-specific recombination between the phage attP site and attB1 or attB2, two neighboring attachment sites in the host chromosome [123]. Julien et al. characterized the integrase gene and attP site in Mx8 and the insertion sites in M. xanthus chromosome [124]. A lacZ reporter gene integrated at the Mx9 attB site demonstrated a higher transcriptional level than the same integration at the Mx8 attB site. Approximately 300-fold more site-specific recombination occurred at the chromosomal attB site than at the native lonD locus in M. xanthus [125]. In M. xanthus, the phage attachment sites facilitate the efficient and stable expression of exogenous genes or additional copies of existing genes.
Salmi et al. cloned the attP site and the integrase coding gene into the E. coli plasmid pBGS18, constructing the plasmid pPLH343, which cannot replicate in M. xanthus cells but can insert into the attB site of the M. xanthus genome [126,127]. Similar plasmids such as pSWU19 (Kmr), pSWU30 (Tetracycline-resistant, Tetr), and pCK T7A1 att (Kmr) were constructed and are currently widely used in M. xanthus for expression and functional verification of genes associated with primary metabolism [128], motility [2,129], predation [130], development [131], and self-recognition [4]. Gross et al. modified and chemically synthesized the cystobactamid biosynthetic gene cluster (about 58 kb) of Cystobacter velatus Cbv34 in fragments, which were then assembled and integrated into the M. xanthus DK1622 genome via the Mx8 integrase [91]. According to Zhu et al., however, the recombination efficiency of the Mx8 attB site decreased sharply as the integration fragment size increased, and site-specific recombination at the attB site is generally not preferred for the expression of smBGCs larger than 50 kb [84].
In addition to efficiently transporting large smBGCs, these suicide plasmids effectively identify genes involved in the synthesis of natural products by complementing or overexpressing target genes. Using the Pck-T7A1-att plasmid, Volz et al. identified two bacterial enhancer-binding proteins (bEBPs) in M. xanthus that functioned as direct regulators to activate or inhibit various secondary metabolites [121]. Subsequently, using plasmid pSWU30 to overexpress the esi gene upstream of the epothilone gene cluster, Yue et al. determined that this gene encodes a transcription factor that inhibits the transcription of the epothilone gene cluster specifically [119]. Integrated expression based on Mx8 site-specific integration remains one of the most widely used and efficient methods for gene expression in myxobacteria.
4.2.3. Transposition
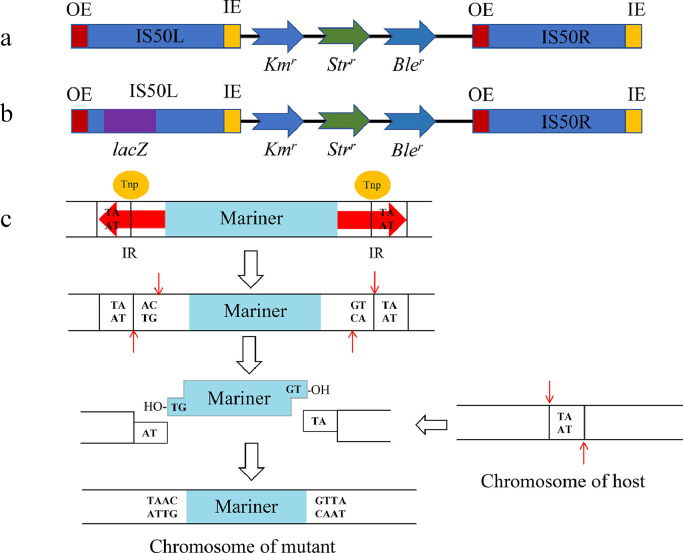
Transposons are genetic elements that can “jump” to various genome locations within a cell. Tn5 is an E. coli transposon made up of a sequence encoding three antibiotic resistance genes (kanamycin, bleomycin, and streptomycin) and a Tn5 transposase Tnp gene, flanked by two inverted insertion sequences, IS50L and IS50R (Fig. 4a) [132]. Each of the two IS50 sequences has a pair of 19-bp reverse sequences at both ends, outside ends (OEs) and inside ends (IEs). Tnp recognizes and cleaves the OE sequence to liberate Tn5, which then binds and inserts randomly into the host genome [133]. The random insertion characteristic with the antibiotic selection marker is convenient for subsequent phenotypic screening, genetic mapping, or clonal identification, making the Tn5 system a highly productive DNA transposon [134].
Fig. 4.
Transposons widely used in myxobacteria. a, Tn5 transposon; b, mutant Tn5-lacZ transposon; c, Mariner transposon and insertion mechanism. IS50L and IS50R are two inverted insertion sequences flanking a transposase gene; OE and IE, 19-bp reverse sequence recognized by transposase at both ends of IS50L and IS50R. Tnp, transposase; IR, Tnp-recognized inverted repeat sequence. A thin red arrow slicing through Tnp. Kmr, kanamycin-resistant gene; Strr, streptomycin-resistant gene; Bler, bleomycin-resistant gene; lacZ, galactosidase gene.
Tn5 is the first transposable element used in myxobacteria, and it continues to be widely employed in the construction of insertion mutants in M. xanthus. Initially, Tn5 was introduced into M. xanthus via phage transfection. Following the transduction of the E. coli phage P1::Tn5 into M. xanthus, Tn5 was randomly inserted into the genome, producing stable kanamycin-resistant mutants [71]. Once Tn5, along with the antibiotic resistance gene, is integrated into the host genome, the likelihood of this element being transposed to other sites is less than 1/1000 [135]. Therefore, P1::Tn5 is predisposed to generate mutants with a single Tn5 insertion, which facilitates the identification of mutant genes responsible for phenotypic changes. Based on Tn5, Sasakawa & Yoshikawa constructed a series of Tn5 transposon variants by replacing the selection marker with various antibiotic resistance genes (including resistance genes against tetracycline, chloramphenicol, gentamicin, trimethoprim, streptomycin, and ampicillin). These variants provide more options for various scenes[138]. Additionally, the authors created a temperature-sensitive suicide plasmid R388 as a vector for transporting Tn5 elements into various hosts. Kroos et al. replaced a portion of IS50L with the lacZ gene lacking a promoter and constructed the Tn5-lacZ cassette [76] (refer to Fig. 4b). When P1::Tn5-lacZ is transduced into M. xanthus, lacZ will be co-transcribed with the inserted gene if the Tn5-lacZ gene was inserted into a transcription unit on the host chromosome in the same orientation, which can be detected by β-galactosidase activity [137], [138], [139], [140].
The Mini–Himar element (magellan-4) is a derivative of the mariner transposon superfamily from eukaryotes and is frequently used in myxobacteria [141]. The Mini–Himar element integrates into the chromosome through a straightforward mechanism of cut-and-paste. The transposase cleaves DNA at the inverted repeat (IR) sequences that mark each transposon end and then insert the excised transposon at the dinucleotide TA site (Fig. 4c) [142]. In contrast to other transposons, the mariner's transposition is dependent solely on the self-transposase. This allows the mariner to be transferred horizontally between species, making it an attractive tool for genetic engineering [143]. Youderian et al. (2003) reported that Mini–Himar is active in M. xanthus and generates more extensive insertions than bacterial transposons, including Tn5 [140]. The authors identified 30 new genes required for A motility using the protocol. This transposon has been utilized since then to analyze and identify genes in M. xanthus [144,145]. The mariner-based transposon was also developed and introduced into S. cellulosum strains via conjugation; similarly, the transposon could randomly insert into the chromosomes of So ce90 and So ce12 [146].
Random insertion makes Tn5 and its variants useful for identifying and validating the coding or regulatory genes for secondary metabolites. Varon et al. reported that the transposition of Tn5-lacZ and TnV (another Tn5 derivative containing the replication origin of plasmid pSC101) into M. xanthus resulted in 8,381 kanamycin-resistant mutants, of which 24 mutants were nonproducers, and three produced higher levels of TA than the parent strain [147]. There was a co-transduction between kanamycin resistance and the altered TA phenotype in most mutant strains. The gene cluster for the biosynthesis of the heterocyclic quinone antibiotic saframycin Mx1 was inactivated and tagged by Tn5 insertions in M. xanthus DM504/15, and the tagged genes were cloned and identified, thereby identifying the genomic region involved in saframycin biosynthesis [148]. Isolation and genetic analysis of M. xanthus mutants generated by Tn5 insertions helped identify the key genetic loci involved in light-induced carotenogenesis and elucidate the molecular mechanisms underlying the light-dependent signaling and regulation of the transcriptional response leading to carotenoid biosynthesis [120,149].
In addition, since transposition-mediated gene integration is not constrained by fragment size, transposable elements are a useful tool for the transfer of large smBGCs. For example, Fu et al. achieved heterologous expression of epothilone by introducing the epothilone biosynthesis gene cluster derived from S. cellulosum So ce90 into M. xanthus DK1622 with the aid of the Mini–Himar element[88]. The biosynthetic gene cluster of disorazol, a macrocyclic polyketide produced by S. cellulosum So ce12, was identified by transposon mutagenesis with Mini–Himar [95] and cloned in a bacterial artificial chromosome library on the basis of which the 58-kb core gene cluster was reconstituted by Red/ET recombineering and expressed in M. xanthus DK1622 [90]. Zhu et al. also utilized the Mini–Himar method to randomly insert the epothilone gene cluster of S. cellulosum So0157-2 into M. xanthus DZ2, and obtained heterologous expression mutants of epothilone with varying production capacities [84]. They discovered that the transposon efficiently integrated large fragments into the host genome and that the insertion sites substantially affected the expression of foreign genes.
4.2.4. Expression vectors
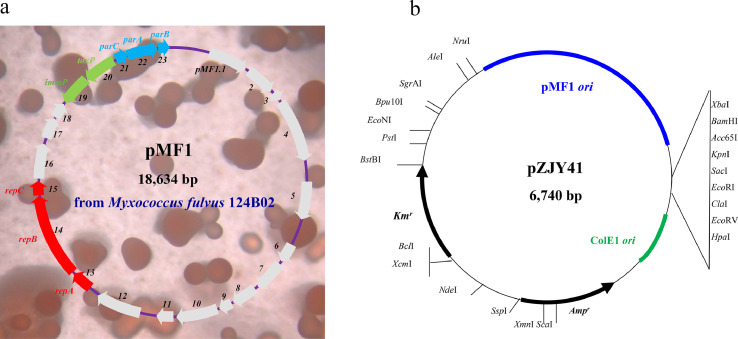
All of the abovementioned plasmids used in the genetic manipulation of myxobacteria must integrate into the host genome via homologous recombination, transposition, or site-specific integration after entering the host, and the self-replicating plasmid from myxobacteria was not reported until 2008. Zhao et al. screened 150 myxobacterial strains and reported pMF1, the first circular plasmid capable of self-replication, from M. fulvus 124B02 [79] (Fig. 5a). This plasmid is 18,634 bp long, contains a G+C content of 68.7%, containing 23 open-reading frames. In the repABC operon consisting of pMF1.13-pMF1.15 (Fig. 5a), pMF1.14 encodes an essential replication initiation protein with abundant disordered regions but devoid of DNA/protein binding motifs [79,150]. It is hypothesized that the extremely restricted host range of the pMF1 replicon in Myxococcus cells is due to its unmatched characteristics. Further research revealed that the pMF1.21-pMF1.23 operon (labeled parCAB in Fig. 5a) was responsible for the partitioning of pMF1 [151] and that the natural expression of parC positioned upstream of parA was essential for the plasmid inheritance in Myxococcus cells [152]. In addition, pMF1.20 and pMF1.19 encode a pair of nuclease toxins and immune protein that function as a post-segregational killing system, which has been proposed to aid in the maintenance of plasmid PMF1 in M. fulvus cells [153]. The other predicted genes in pMF1 either have homologs in sequenced myxobacterial genomes or lack homology to any known sequence [154].
Fig. 5.
pMF1 plasmid from M. fulvus 124B02 (a) and the shuttle vector pZJY41 (b). parCAB, par operon for the partitioning of pMF1, related genes were highlighted with blue arrows; repABC, operon encoding the replication proteins, related genes were highlighted with red arrows; taxP and immP, a pair of nuclease toxins and immune protein expressed by pMF1, were highlighted with corresponding green and blue arrows, respectively. Other genes in pMF1 with uncertain functions were labeled with gray arrows. pMF1 ori, a replicon of pMF1, responsible for the replication of pZJY41 in M. xanthus; ColE1 ori, replicon responsible for the replication of pZJY41 in E. coli.
The Myxococcus plasmid pMF1, which replicates autonomously, provides a new tool for genetic manipulation. Based on the pMF1 replicon, vectors such as pZJY41 and pZJY156 were designed to shuttle expression between M. xanthus and E. coli [79]. The plasmid pZJY41 contains the pMF1 ori, the high-copy-number ColE1 ori, two antibiotic resistance genes as selection markers, and a region with multiple clone sites (MCS) (Fig. 5b). The target gene cloned in the MCS of pZJY41 will express 10–20 copies of the plasmid in M. xanthus DK1622, resulting in overexpression [155], [156], [157], [158]. In addition to the expression and identification of targeted genes, pZJY41 is also used to express small guide RNA (sgRNA) to mediate gene editing with CRISPR and CRISPR-associated protein 9 (Cas9) system [159] or to promote the transcription of epothilone biosynthesis with the sgRNA recruited RNAP in M. xanthus [160,161].
In 2021, Panter et al. reported the presence of the second self-replicating plasmid pSa001 in Sandaracinus sp. MSr10575 [162]. The 209.7-kb pSa001 contained a large PKS–NRPS hybrid gene cluster that was activated by homologous recombination mediated exchange of the native promoter and produced antitumor sandarazols. The replication and division mechanisms of this plasmid require additional research.
4.2.5. CRISPR-Cas system
Numerous prokaryotic organisms employ CRISPR-Cas systems to repel mobile genetic elements [163]. The discovery of the CRISPR-Cas system and its transformation into a potent gene editing tool have revolutionized the field of molecular biology [164], [165], [166]. The widespread use of the CRISPR-Cas9 system in both prokaryotes and eukaryotes is due to its simplicity and flexibility. This system provides a robust and multiplexable platform for genome editing, transcriptional perturbation, epigenetic modulation, and genome imaging [167,168]. CRISPR-Cas9-based genome engineering has also been utilized for the discovery of natural products in various bacteria, fungi, and plants [169].
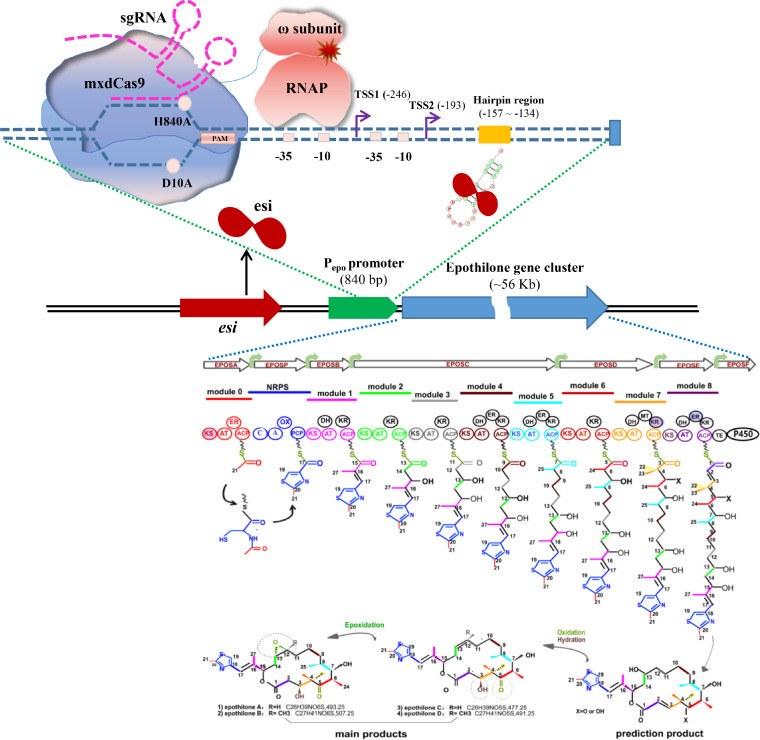
In 2018, Peng et al. constructed the CRISPR-based activation system in myxobacteria to promote the biosynthesis of secondary metabolites for the first time [160]. A codon-optimized, nuclease-dead Cas9 (dCas9) was fused with various activator proteins and introduced into the epothilone-producing producer M. xanthus ZE9. With the expression of sgRNA targeting the promoter of the epothilone operon, the dCas9-activator proteins recruited RNAP to promote the transcription of the epothilone genes, resulting in a 1.5-fold increase in epothilone yield. This CRISPR-dCas9 system was then used to individually or collectively activate internal promoters in the epothilone operon, revealing the multiple interferences between promoters that altered the transcriptional profile of operon genes and the production of epothilones [161]. Similarly, Ye et al. introduced the CRISPR-dCas9 system into the S. cellulosum strain So ce M4 to activate the transcription of the epothilone gene cluster using VP4 as the activator [97]. It was also determined that the CRISPR-Cas9 system is effective for the deletion of large genome fragments in M. xanthus DK1622 [159]. According to the protocol, the Cas9 gene and sgRNA were integrated into the host chromosome via site-specific recombination. A temporally high expression strategy was employed to reduce cell damage caused by high Cas9 expression, and a tRNA-sgRNA-tRNA chimeric structure was designed to ensure correct targeting by sgRNA sequence. Additionally, 14% of the 92-kb smBGC for myxalamids was deleted using the strategies, demonstrating the application potentials of CRISPR-Cas9 in the genome rational design and deep modification of M. xanthus.
In spite of its applicability, the CRISPR-Cas9 system in M. xanthus requires further optimization to make its operation easier and more efficient. For instance, independently introduced sgRNA, not in conjunction with Cas9, will make it possible to edit different sites or simultaneously implement multi-site editing. The introduction of a non-homologous end-joining system (NHEJ) into M. xanthus may omit the need for homologous arm introduction. Notably, M. xanthus DK1622 possesses three endogenous CRISPR-Cas systems, MxI-B, I-C, and III-B [170], which are being identified and developed as effective gene editing tools (unpublished). For the discovery of natural products in myxobacteria, ever-more-effective CRISPR-Cas systems are anticipated to be utilized.
5. Engineering of secondary metabolism in myxobacteria
The social characteristics, indistinct genetic backgrounds, and limited genetic manipulation techniques make it very difficult to directly modify smBGCs in their original myxobacterial producers. Heterologous expression is an alternative solution to this issue.M. xanthus DK1622 is the model strain of myxobacteria and is an ideal host for studying and expressing the secondary metabolites of myxobacteria, which are typically expressed poorly in other microbial hosts [171]. Among numerous myxobacterial metabolites, the heterologous expression of epothilones is currently the most thoroughly investigated. Epothilones are 16-member macrolides that were first isolated from S. cellulosum So ce90 and exhibit antifungal and cytotoxic properties [36,172]. Epothilones function as antineoplastics similarly to paclitaxel. They bind to microtubules to prevent the formation of spindles during mitosis, causing cancer cells to undergo apoptosis [173,174]. Moreover, they have therapeutic potential in the treatment of neurodegeneration and nervous system injury [51,52]. In myxobacteria research, heterologous expression and biosynthesis regulation of epothilones are among the hottest topics due to the low productivity and limited modification options of the original producers.
5.1. Biosynthesis and regulation of epothilone
In 2000, the biosynthetic gene cluster and biosynthetic pathway of epothilones were identified and deduced [175,176]. As depicted in Fig. 6, the 56-kb gene cluster consists of six multifunctional genes in the same transcription direction (epoA, epoP, epoB, epoC, epoD, and epoE) and a P450 gene epoF (also epoK in some publications), encoding five PKS (EPOSA, EPOSB, EPOSC, EPOSD, and EPOSE), an NRPS (EPOSP) and a cytochrome P450 epoxidation enzyme (EPOSF) for the elongation and processing of epothilones. Each PKS protein is comprised of one or more modules containing discrete domains, including the indispensable β-ketoacyl synthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) domains, and the dispensable modification domains such as ketoreductase (KR), dehydratase (DH), enoyl reductase (ER), methyltransferase (MT) or thioesterase (TE), whereas EPOSP consists of condensation domain (C), a peptidyl carrier protein (PCP), adenylation domain (A), and an oxidation domain (OX). The synthetic pathway begins with the synthesis of a thiazole ring, followed by eight steps for the polyketide chain extension, the cyclic release of the polyketide chain, and the epoxidation of epothilone C/D to epothilone A/B [85].
Fig. 6.
The gene cluster and biosynthesis pathway of epothilone (modified from reference 178), as well as investigations into its regulatory mechanisms. KS denotes the β-ketoacyl synthase domain; AT, the acyltransferase domain; ACP, the acyl carrier protein domain; KR, the ketoreductase domain; DH, the dehydratase domain; ER, the enoyl reductase domain; MT, the methyltransferase domain; TE, the thioesterase domain; C, the condensation domain; PCP, peptidyl carrier protein; A, adenylation domain; OX, oxidation domain. The shaded domains are inactive in the biosynthesis pathway. TSS, transcriptional start site; the base position of the TSSs and hairpin region was defined with reference to the translation start site as +1. esi, negative transcriptional regulator bing to the hairpin region of Pepo. H840A and D10A were two mutant amino acids in dCas9. Green arrows indicate the internal promoters of the epothilone gene cluster.
Zhu et al. characterized the promoters of epothilone gene clusters from various producers and discovered that the expression of all operons is regulated by a dual promoter Pepo (with two −35 and −10 regions and two distinct TSSs), a hairpin structure, and the distal non-coding region between the promoter and operon [177] (Fig. 6). Simple replacement of epothilone promoter Pepo with strong endogenous promoters (PpilA, PgroEL) did not improve epothilone synthesis in M. xanthus, most likely due to inconsistency in the transcriptional temporal modes between nominated promoters and those of secondary metabolite synthesis [178]. Nonetheless, the multi-copied Pepo tandem substantially increased the transcription level of epothilone genes, resulting in a remarkable increase in epothilone yield. A gene located directly upstream of epoA in S. cellulosum So0157-2 encodes the negative transcriptional regulator Esi, which binds to the hairpin region and inhibits the transcription of epothilone genes; inactivation of esi in So0157-2 or in heterologous host ZE10 increased the yield of epothilones by 4.1-fold and 38%, respectively [119] (Fig. 6). This is the first transcriptional regulator of epothilone biosynthesis that has been identified. In addition to the promoter in front of the epothilone gene operon (Pepo), multiple internal promoters were identified within the operon. Activation of the Pepo and/or internal promoters with the CRISPR-dCas9 activation system promoted the epothilone synthesis, demonstrating the complex regulation of epothilone expression [160,161] (Fig. 6). Notably, aided by high-throughput evaluation of mutants, the epothilone production in S. cellulosum So0157-2 could be significantly enhanced by a combination of UV mutation, recursive protoplast fusion, and genome shuffling [179]. However, the underlying mechanism remains unclear. Additional research into the transcriptional regulatory mechanism will inform the genetic modification of producing strains to optimize epothilone production.
5.2. Heterologous expression of epothilone biosynthetic gene cluster
Tang et al. (2000) introduced the epothilone biosynthesis gene cluster from S. cellulosum SMP44 into Streptomyces coelicolor CH999, and epothilone levels reached 50–100 µg/L [180]. In vivo knockout of epoF and in vitro assays with recombinant epoF expressed in E. coli demonstrated the function of EPOSF. Other Streptomyces strains, such as S. venezuelae DHS2001 [181] and S. lividans ZX7 [182], were also used to express epothilone with various applicable promoters, but yields were either undetectable or extremely low. In S. venezuelae, overexpression of pikD, a positive regulatory gene of the endogenous pikromycin biosynthetic gene cluster, increased epothilone production. Two adjacent genes of the epothilone gene cluster, orf3 and orf14, were identified as putative transport genes because their deletion resulted in a threefold decrease in epothilone D production. Also redesigned and synthesized was the entire gene cluster, which expressed as soluble proteins in E. coli K431-37-2A and produced trace amounts of epothilones C and D (< 1 µg/L) [183]. In addition, cells expressing the epoC-epoD-epoE genes synthesized epothilones C and D when fed a thioester of the normal substrate for epoC, demonstrating the composability of the epothilone synthetic pathway. Bian et al. (2017) expressed the entire epothilone gene cluster from S. cellulosum So ce90 in Schlegelella brevitalea DSM 7029. Host-specific medium optimization and genetic modification (introduction of the biosynthetic pathway for precursors and overexpression of rare tRNA genes) significantly increased epothilone production in S. brevitalea [184]. The epothilone gene cluster of S. cellulosum So0157-2 was similarly expressed in S. brevitalea DSM 7029, resulting in an initial yield of 80 µg/L, which was increased by 7.5-fold through the reconstruction of the methylmalonyl-CoA biosynthetic pathway to improve precursor supply [185]. The authors reassembled the mega epothilone gene cluster using BioBricks and SSRTA techniques, thereby creating an artificial pathway in which each gene was transcribed with an independent promoter to regulate the pathway's equilibrium and maximize its activity. Further addition of sucrose debottlenecked the DSM 7029 strain, which improved epothilone production by enhancing the potentials of the optimally reassembled gene cluster; the epothilone yield finally reached 82 mg/L.
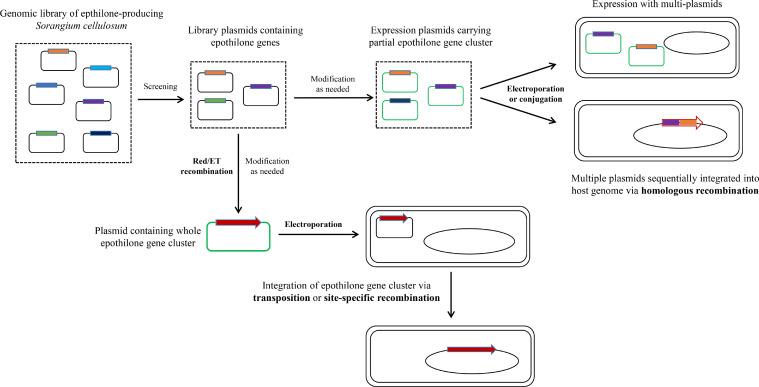
To date, the epothilone gene cluster has been expressed in various hosts (genetic modifications of the epothilone gene cluster for heterologous expression are depicted in Fig. 7), among which the model species M. xanthus possesses the most promising properties (Table 3). Julien et al. inserted the epothilone gene cluster into the M. xanthus DZ1 genome by means of homologous recombination. The initial yield of epothilone A and epothilone B in mutant strains was ∼100 µg/L, whereas the removal of epoF increased the yield of epothilone D to 200–400 µg/L [85]. Using Red/ET recombination, Fu et al. stitched the full-length epothilone gene cluster into a single plasmid carrying the magellan-4 transposon, which mediated the integration of epothilone gene cluster into the genomes of M. xanthus DK1622 and Pseudomonas putida FG2005, respectively [88]. This study demonstrated that transposons can efficiently deliver large transgenes to heterologous hosts. The mutants derived from M. xanthus DK1622 synthesized ∼100 µg/L of epothilones, whereas none was detected in the P. putida mutants. The epothilone gene cluster of S. cellulosum So0157-2 was introduced into various M. xanthus (M. xanthus DZ2, DK1622, and SW504) by a mariner transposon, and allopatric integrations in genome significantly altered host transcriptomes, resulting in a range of epothilone production capacities (0.1–1000 µg/L) [84]. The 1 mg/L production represents the highest initial yield of heterologous expression of epothilones, and the different yields resulting from genomic position effects suggest that by screening more mutants, more productive strains can be anticipated.
Fig. 7.
Genetic modification of the epothilone gene cluster for heterologous expression. Library plasmids containing partial epothilone gene cluster are screened from the genomic library of epothilone-producing Sorangium cellulosum. After customized modifications (such replacement of suitable promoter and addition of necessary components like transposon), plasmid expression is introduced into the host for extranuclear or integrated expression. Alternatively, the library plasmids are modified with Red/ET to assemble the entire epothilone gene cluster, and the resulting expression plasmids are introduced into the host and then integrated into the genome through transposition or site-specific recombination. Red arrow, whole epothilone gene cluster; colored rectangles, various epothilone gene cluster components; black rounded rectangle, skeleton of common plasmid; green rounded rectangle, skeleton of expression plasmid; black oval, host genome.
Table 3.
Heterologous expression of the epothilone biosynthetic gene cluster in different hosts.
| Expression host | Origin of gene cluster | Expression form | Initial yield | Optimized method | Optimized yield | Reference |
|---|---|---|---|---|---|---|
| S. coelicolor CH999 | S. cellulosum SMP44 | Expressed by plasmids | 50–100 µg/L | - | - | [180] |
| S. venezuelae DHS2001 | S. cellulosum So ce90 | Expressed by plasmids | 0.1 µg/L | Knockout of epoD | 0.4 µg/L | [181] |
| E. coli K431-37-2A | Chemical synthesized | Expressed by plasmids | < 1 µg/L | - | - | [183] |
| P. putida FG2005 | S. cellulosum So ce90 | Integrated via transposition | Undetectable | - | - | [86] |
| M. xanthus DZ1 | S. cellulosum So ce90 | Integrated via homologous recombination | 200–400 µg/L | Addition of resin and methyl oleate | 85 mg/L (Fed-batch fermentation) | [112,188,189] |
| M. xanthus DK1622 | S. cellulosum So ce90 | Integrated via transposition | 100 µg/L | - | - | [86] |
| M. xanthus DK1622 | S. cellulosum So ce90 | Integrated via site-specific recombination | 100 µg/L | - | - | [187] |
| M. xanthus DK1622, DZ2, SW504 | S. cellulosum So0157-2 | Integrated via transposition | 0.1–1000 µg/L | Addition of resin and methyl oleate; knockout of negative transcriptional regulator; CRISPR-activation | 28 mg/L | [84,119,160,178] |
| S. brevitalea DSM 7029 | S. cellulosum So ce90 | Integrated via transposition | 4 µg/L | Medium Optimization; enhancement of precursor supply; overexpression of rare tRNA genes | 307 µg/L | [184] |
| S. brevitalea DSM 7029 | S. cellulosum So0157-2 | Integrated via site-specific recombination | 80 µg/L | Enhancement of precursor supply; reassembling of gene cluster; debottlenecking of the cell autolysis | 82 mg/L | [185] |
In general, compared to other heterologous expression hosts, M. xanthus exhibited superior productivity of epothilones, and heterologous expression of epothilones in M. xanthus resulted in relatively higher initial yields. M. xanthus, unlike E. coli and Burkholderia, produces diverse PKS/NRPS secondary metabolites, such as myxovirescin, myxalamid, DKxanthene, and myxochromide, which are constructed similarly from malonyl-CoA or methylmalonyl-CoA [34]. Engineering of pathway-specific regulators [121] or enzymes responsible for posttranslational activation of natural product assembly lines may increase the total amount of the abovementioned compounds [186], which implies that the metabolic pathways of M. xanthus can provide abundant precursors for the synthesis of epothilones without special metabolic pathway modification. However, despite the fact that competition for precursor monomers was not regarded as the primary constraint on epothilone production [187], engineering or feeding monomers will increase production yields. In 2014, Osswald et al. redesigned and reassembled the epothilone gene cluster by codon optimization and introduction of unique restriction sites to permit pathway assembly and future interchangeability of modular building blocks from the epothilone megasynthetase [187] and the successful heterologous production of epothilones unambiguously demonstrated the functionality of the artificial pathway in M. xanthus. This research promises future engineering of epothilone biosynthesis and production optimization through a highly adaptable assembly strategy.
Resin XAD-16 can absorb epothilone products [36], whereas methyl oleate increases cell density and epothilone yield [85]. Combining. XAD-16, a suitable carbon source, and the fed-batch process, the yield of epothilone D in M. xanthus increased 140-fold from an initial titer of 0.16 mg/L [188] to 85 mg/L after 22 days of fed-batch cultivation [189]. In addition, the enhancement of epothilone gene transcription by tandem Pepo promoters [178] or CRISPR-dCas9 activation [160,161] significantly boosted epothilone production. Thus, there is good reason to believe that the epothilone yield in the heterologous host M. xanthus will be significantly enhanced by optimizing culture conditions, combining different approaches, and modifying the producer strain comprehensively.
6. Construction of myxobacterial chassis
Due to the lack of genetic manipulation techniques in most myxobacteria, heterologous expression of smBGCs is an efficient means of exploiting the biosynthetic potentials of myxobacteria, including functional elucidation of cryptic gene clusters and optimization of product yields [190]. However, a high yield is typically not guaranteed when a biosynthetic gene cluster is isolated and transferred to a heterologous host. Different chassis of cyanobacteria [191], actinobacteria [192], filamentous fungi [193], and yeast [194,195] have been successfully constructed and utilized for the heterologous expression of natural active products. However, their performance in expressing smBGCs from myxobacteria was typically poor [196]. In order to achieve efficient heterologous expression and improve product supply for further exploitation of smBGCs from myxobacteria, it is highly desirable to establish an efficient and versatile microbial production platform. Considering issues such as promoter recognition, precursor supply, and self-resistance, we conclude that phylogenetic relatedness is crucial for successful heterologous expression. M. xanthus DK1622 is a preferred heterologous host of secondary metabolites from myxobacteria not only due to its phylogenetically close relationship to the native producers but also some advantageous features. The DK1622 strain grows dispersedly in a liquid medium with a shorter generation time (3–4 h) compared to many other myxobacteria, as well as available fermentation knowledge [92]. As a promising producer of natural products, M. xanthus DK1622 is characterized by the availability of common biosynthetic precursors [197] and broad spectrum phosphopantetheinyl transferases required for posttranslational activation of PKS/NRPS megasynthetases [186]. Additionally, the genetic manipulation methods developed for myxobacteria are mostly applicable in M. xanthus DK1622, making it easier to redesign and renovate the strain as an ideal chassis host [198].
M. xanthus DK1622 is frequently used to heterologously express exogenous smBGCs [7]. The 9.14-Mb genome of DK1622 contains numerous secondary metabolic genes, regulatory genes, and transfer elements, all of which contribute to the intricate regulatory network [8,121]. This, however, is not conducive to the construction of stable, high yield engineering strains and can easily result in an unstable and unstable strain state [199], [200], [201]. Screening the M. xanthus DK1622 genome revealed at least 24 smBGCs (Table 4), which are primarily clustered in two regions (4.4–5.8 Mb and 1.5–3.5 Mb), but only nine gene clusters of their products have been identified [7]. The characterized and unidentified secondary metabolites are predominantly PKS/NRPS compounds, which may compete for precursors and energy with the synthesis of the target product. Therefore, the endogenous smBGCs should be deleted to simplify the autochthonous metabolic background and the complexity of the chromosomes. For instance, the myxochromides A gene cluster, one of the most abundant compound families produced by M. xanthus DK1622 under standard cultivation conditions, was deleted by homologous recombination and replaced with a tetracycline resistance gene, resulting in the mutant strain ΔmchA-tet. The mutant was once used as a host for parallel expression studies involving myxobacterial products such as myxopyronin [92], myxochromides [202], vioprolides [203], and argyrins [89]. Similarly, using the CRISPR-Cas9 system, some smBGCs were deleted in an epothilone-producing M. xanthus strain to examine their effects on epothilone production [159]. Regarding mobile elements, IslandPath-DIMBO predicts the existence of 19 genomic islands (GIs), which account for 4.5% of the genome sequence length (Table 4). Moreover, the deletion of 12 of the 19 GIs had no effect on cellular growth (unpublished), but the deletion of multiple GI genes is a time-consuming process.
Table 4.
smBGCs and GIs in M. xanthus DK1622.
| smBGC | Start | End | Length (bp) | Type | Products |
|---|---|---|---|---|---|
| cluster 1 | 1015682 | 1031120 | 15438 | Terpene | Carotenoid |
| cluster 2 | 1490426 | 1537319 | 46893 | NRPS | |
| cluster 3 | 1771438 | 1813469 | 42031 | NRPS | |
| cluster 4 | 1863446 | 1927180 | 63734 | NRPS | |
| cluster 5 | 2778046 | 2800369 | 22323 | Thioamitides | |
| cluster 6 | 3235611 | 3284053 | 48442 | NRPS/Type I PKS | |
| cluster 7 | 3318246 | 3340534 | 22288 | Lanthipeptide-claaa-II | |
| cluster 8 | 4013494 | 4058514 | 45020 | Type I PKS | Myxalamid |
| cluster 9 | 4135538 | 4145774 | 10236 | RiPP | |
| cluster 10 | 4202141 | 4363714 | 161573 | NRPS/Type I PKS | Myxochelin |
| cluster 11 | 4432172 | 4452468 | 20296 | RRE-containing | |
| cluster 12 | 4481594 | 4563638 | 82044 | NRPS/Type I PKS | Myxoprincomide |
| cluster 13 | 4736846 | 4822556 | 85710 | PKS/NRPS | Myxovericin |
| cluster 14 | 4885909 | 4923967 | 38058 | NRPS/Type I PKS | |
| cluster 15 | 4977639 | 5044926 | 67287 | NRPS/Type I PKS | Myxochromide |
| cluster 16 | 5105157 | 5116326 | 11169 | RiPP | |
| cluster 17 | 5235101 | 5311872 | 76771 | NRPS/Type I PKS | Dkxanthene |
| cluster 18 | 5387898 | 5488693 | 100795 | NRPS/Type I PKS | |
| cluster 19 | 5585364 | 5671886 | 86522 | NRPS/Type I PKS | |
| cluster 20 | 5715538 | 5794912 | 79374 | NRPS/RiPP | |
| cluster 21 | 6157224 | 6205340 | 48116 | Thiopeptide/RiPP | |
| cluster 22 | 7706986 | 7729253 | 22267 | Terpene | Cittilin |
| cluster 23 | 7855962 | 7897173 | 41211 | Ladderane/lanthipeptide-class-II | |
| cluster 24 | 8147550 | 8188632 | 41082 | T3PKS | Alkylpyrone |
| GI1 | 148705 | 177189 | 28485 | Containing mobile element | |
| GI2 | 660595 | 676929 | 16335 | Containing mobile element | |
| GI3 | 1402684 | 1448711 | 46028 | Containing mobile element | |
| GI4 | 1543126 | 1569486 | 26361 | Containing mobile element | |
| GI5 | 2013057 | 2037294 | 24238 | Containing mobile element | |
| GI6 | 2080781 | 2095849 | 15069 | Containing mobile element | |
| GI7 | 2107749 | 2127300 | 19552 | Containing mobile element | |
| GI8 | 2179596 | 2204689 | 25094 | Containing mobile element | |
| GI9 | 2216216 | 2242401 | 26186 | Adjacent to tRNA gene | |
| GI10 | 2427372 | 2460798 | 33427 | Containing mobile element | |
| GI11 | 3290237 | 3299422 | 9186 | Adjacent to tRNA gene | |
| GI12 | 4192698 | 4217512 | 24815 | Adjacent to tRNA gene | |
| GI13 | 5059847 | 5063412 | 3566 | Containing mobile element | |
| GI14 | 5066197 | 5081299 | 15103 | Containing mobile element | |
| GI15 | 5532695 | 5545491 | 12797 | Adjacent to tRNA gene | |
| GI16 | 6077090 | 6109421 | 32332 | Adjacent to tRNA gene | |
| GI17 | 6493076 | 6508978 | 15903 | Adjacent to tRNA gene | |
| GI18 | 7763756 | 7767905 | 4150 | Containing mobile element | |
| GI19 | 9048513 | 9050527 | 2015 | Containing mobile element |
RiPP, ribosomally synthesized and posttranslationally modified peptides. The GIs that were successfully deleted in M. xanthus DK1622 are underlined.
Genome reduction based on the deletion of non-essential genes, such as competing endogenous smBGCs and mobile elements, will simplify metabolic profiles and increase the genetic stability of chassis cells. Numerous examples indicate that a reduced genome leads to improved growth characteristics (including smaller cell size, reduced cell autolysis, and more dispersed growth), increased electroporation efficiency, and enhanced heterologous protein and secondary metabolite production [204], [205], [206], [207], [208]. However, some secondary metabolites that appear to be non-essential may play unknown roles in cell growth, and in some cases, deletion of genomic regions predicted to be non-essential may hinder cell growth [205,209]. Hence, rational construction of genome-reduced myxobacteria chassis with M. xanthus DK1622 necessitates an in-depth analysis of dispensable genetic elements and sequential knockout verification of non-essential gene elements, which may be demanding but essential.
In addition to conventional genetic operations such as homologous recombination, site-specific recombination mediated by phage transposase, transposition, and plasmid expression system, new genome editing tools for efficient deletion of large genomic regions and reduction of metabolic background have been developed. Yang et al. develop efficient genome engineering systems for large fragment deletion in M. xanthus DK1622 based on the CRISPR-Cas9 system and Cre/loxP-mediated recombination and successfully delete 83-466 kb genomic regions [159,210]. In addition, recently reported endogenous strong constitutive promoters in M. xanthus DK1622 provide a crucial synthetic toolkit for efficient gene expression [211]. With the advancement of genome editing tools, a genetically accessible M. xanthus DK1622 chassis for the efficient heterologous expression of unexplored secondary metabolites from myxobacteria is highly anticipated.
7. Conclusion and perspectives
The methods of genetic manipulation in myxobacteria and their applications in the exploitation of myxobacterial natural products are summarized in this article. As a rich source of structurally diverse secondary metabolites with intriguing biological activities, myxobacteria exhibit a high potential for the discovery of new drugs with high chemical diversity and uncommon mechanisms of action. The exceptionally large genome sizes and abundance of smBGCs indicate vast untapped natural product resources. Traditional recombination- or transposition-based genetic manipulation, in addition to CRISPR-Cas and Red/ET techniques, enables the operation of myxobacteria and the identification of novel myxobacterial secondary metabolites. As stated previously, the CRISPR-Cas9 system must be optimized prior to its efficient application in M. xanthus DK1622, such as the flexible substitution of sgRNA. The NHEJ system is recommended for further development of the CRISPR-Cas system in M. xanthus DK1622 because the homologous recombination repair after shearing of Cas9 is dependent on the introduction of homologous arms, which prolongs the cycle of engineering mutant strains. Similarly, Cre/loxP-mediated recombination requires the introduction of two loxP sites in the same orientation; a similar issue exists in the Cre/loxP system. Moreover, similar to the Rec/ET system in E. coli, the host-specific Redα/Redβ recombinase has been identified and used for highly efficient genome editing in S. brevitalea DSM 7029 [205,212]. Therefore, we anticipate that myxobacteria-specific homologous recombinases will develop more efficient genetic manipulation techniques in M. xanthus.
From high-abundance molecules that could be fractionated from grown cultures to massive transcriptionally silent or cryptic smBGCs, we also consider the necessity and maneuverability of constructing myxobacterial chassis for expression and yield optimization of smBGCs. Classic chassis cells such as E. coli and Streptomyces are incapable of expressing smBGCs from myxobacteria, necessitating the use of myxobacterial chassis cells. As the most well-studied model strain, M. xanthus DK1622 is the preferred candidate. The increasingly sophisticated genetic manipulation techniques established in myxobacteria, combined with the flourishing omics tools (genomics, transcriptome, metabolomics, and proteomics), should enable rational modification of M. xanthus DK1622 as an ideal chassis. The numerous endogenous smBGCs and GIs are favored as deletion targets for genome reduction. With a simplified host, the optimal transcription and translation of smBGCs are required for the efficient expression of the target product. The establishment of an enriched promoter library has expanded the synthetic biology toolkit available for M. xanthus DK1622 [211]. The identification of secondary metabolism regulatory factors, particularly global regulatory factors such as ROK-like proteins [122], will provide potent tools for regulating the transcription of target gene clusters. Considering genetic modification at the translational level, ribosome engineering merits further study in M. xanthus DK1622 to activate the silent gene clusters or optimize the production of known products, just as in Streptomyces [213]. Although the mechanism is not well understood, some molecular chaperones, such as groEL in M. xanthus DK1622 [214], seem to regulate secondary metabolism at a non-transcriptional level, making them important regulatory factors in the myxobacteria chassis.
In the future, natural product mining in myxobacteria is anticipated to be accelerated by the cloning or refactoring of biosynthetic pathways coupled with systematic heterologous expression in optimized chassis cells.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Given his role as Editorial Board Member, Dr. Yuezhong Li, had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to Dr. Linquan Bai.
Acknowledgements
This work was financially supported by the National Key Research and Development Programs of China (2018YFA0900400, 2018YFA0901704 and 2021YFC2101000) and the Natural Science Foundation of Shandong Province (ZR2019BC041).
Contributor Information
Xinjing Yue, Email: xjy2018@sdu.edu.cn.
Duohong Sheng, Email: dhsheng@sdu.edu.cn.
Li Zhuo, Email: zhuoli39@163.com.
Yue-Zhong Li, Email: lilab@sdu.edu.cn.
References
- 1.Dawid W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 2000;24(4):403–427. doi: 10.1111/j.1574-6976.2000.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 2.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye X, Li Z, Luo X, Wang W, Li Y, Li R, et al. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome. 2020;8(1):49. doi: 10.1186/s40168-020-00824-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong Y, Zhang Z, Liu Y, Zhou XW, Anwar MN, Li ZS, et al. A nuclease-toxin and immunity system for kin discrimination in Myxococcus xanthus. Environ. Microbiol. 2018;20(7):2552–2567. doi: 10.1111/1462-2920.14282. [DOI] [PubMed] [Google Scholar]
- 5.Wenzel SC, Müller R. The biosynthetic potential of myxobacteria and their impact on drug discovery. Curr. Opin. Drug Discov. Develop. 2009;12(2):220–230. [PubMed] [Google Scholar]
- 6.Herrmann J, Abou Fayad A, Müller R. Natural products from myxobacteria: novel metabolites and bioactivities. Nat. Product Reports. 2017;34(2):135–160. doi: 10.1039/c6np00106h. [DOI] [PubMed] [Google Scholar]
- 7.Bader CD, Panter F, Müller R. In depth natural product discovery - myxobacterial strains that provided multiple secondary metabolites. Biotechnol. Adv. 2020;39 doi: 10.1016/j.biotechadv.2019.107480. [DOI] [PubMed] [Google Scholar]
- 8.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 2006;103(41):15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissman KJ, Müller R. A brief tour of myxobacterial secondary metabolism. Bioorg. Med. Chem. 2009;17(6):2121–2136. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Ye X, Liu M, Xia C, Zhang L, Luo X, et al. A novel outer membrane beta-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J. 2019;13(9):2223–2235. doi: 10.1038/s41396-019-0424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wang J, Zhang Z, Wang F, Gong Y, Sheng DH, et al. Two PAAR proteins with different C-terminal extended domains have distinct ecological functions in Myxococcus xanthus. Appl. Environ. Microbiol. 2021;87(9) doi: 10.1128/AEM.00080-21. pp.e00080-00081-e00080-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-Gonzalez A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359(6373):320–325. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 13.Wang JJ, Wang JN, Wu SG, Zhang Z, Li YZ. Global geographic diversity and distribution of the myxobacteria. Microbiol. Spectrum. 2021;9(1) doi: 10.1128/Spectrum.00012-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr KI, Wolf C, Nubel U, Szafranska AK, Steglich M, Hennessen F, et al. A polyphasic approach leads to seven new species of the cellulose-decomposing genus Sorangium, Sorangium ambruticinum sp nov., Sorangium arenae sp nov., Sorangium bulgaricum sp nov., Sorangium dawidii sp nov., Sorangium kenyense sp nov., Sorangium orientale sp nov and Sorangium reichenbachii sp nov. Int. J. Syst. Evol. Microbiol. 2018;68(11):3576–3586. doi: 10.1099/ijsem.0.003034. [DOI] [PubMed] [Google Scholar]
- 15.Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, Wagner M, et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020;70(11):5972–6016. doi: 10.1099/ijsem.0.004213. [DOI] [PubMed] [Google Scholar]
- 16.Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. Fems Microbiol. Rev. 2009;33(5):942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimkets LJ, Reichenbach H, Dworkin M. In: The Prokaryotes. Hedderich R, Whitman WB, editors. Springer-Verlag; New York: 1992. The myxobacteria; pp. 3416–3487. [DOI] [Google Scholar]
- 18.Reichenbach H, Lang E, Schumann P, Sproer C. Byssovorax cruenta gen. nov., sp. nov., nom. rev., a cellulose-degrading myxobacterium: rediscovery of 'Myxococcus cruentus' Thaxter 1897. Int. J. Syst. Evol. Microbiol. 2006;56(Pt 10):2357–2363. doi: 10.1099/ijs.0.63628-0. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto E, Muramatsu H, Nagai K. Vulgatibacter incomptus gen. nov., sp. nov. and Labilithrix luteola gen. nov., sp. nov., two myxobacteria isolated from soil in Yakushima Island, and the description of Vulgatibacteraceae fam. nov., Labilitrichaceae fam. nov. and Anaeromyxobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 10):3360–3368. doi: 10.1099/ijs.0.063198-0. [DOI] [PubMed] [Google Scholar]
- 20.Garcia R, Müller R. Simulacricoccus ruber gen. nov., sp nov., a microaerotolerant, non-fruiting, myxospore-forming soil myxobacterium and emended description of the family Myxococcaceae. Int. J. Syst. Evol. Microbiol. 2018;68(10):3101–3110. doi: 10.1099/ijsem.0.002936. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher D, Sogaard-Andersen L. Regulation of cell polarity in motility and cell division in Myxococcus xanthus. Ann. Rev. Microbiol. 2017;71(71):61–78. doi: 10.1146/annurev-micro-102215-095415. [DOI] [PubMed] [Google Scholar]
- 22.Thiery S, Kaimer C. The predation strategy of Myxococcus xanthus. Front. Microbiol. 2020;10:2. doi: 10.3389/fmicb.2020.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nature Rev. Microbiol. 2007;5(11):862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 24.Sanford RA, Cole JR, Tiedje JM. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 2002;68(2):893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Ran Q, Du X, Wu S, Wang J, Sheng D, et al. Two new Polyangium species, P. aurulentum sp. nov. and P. jinanense sp. nov., isolated from a soil sample. Syst. Appl. Microbiol. 2021;44(6) doi: 10.1016/j.syapm.2021.126274. [DOI] [PubMed] [Google Scholar]
- 26.Jiang DM, Wu ZH, Zhao JY, Li YZ. Fruiting and non-fruiting myxobacteria: a phylogenetic perspective of cultured and uncultured members of this group. Mol. Phylogenet. Evol. 2007;44(2):545–552. doi: 10.1016/j.ympev.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Li YZ, Hu W, Zhang YQ, Qiu Z, Zhang Y, Wu BH. A simple method to isolate salt-tolerant myxobacteria from marine samples. J. Microbiol. Methods. 2002;50(2):205–209. doi: 10.1016/s0167-7012(02)00029-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang B, Hu W, Liu H, Zhang CY, Zhao JY, Jiang DM, et al. Adaptation of salt-tolerant Myxococcus strains and their motility systems to the ocean conditions. Microb. Ecol. 2007;54(1):43–51. doi: 10.1007/s00248-006-9169-y. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YQ, Li YZ, Wang B, Wu ZH, Zhang CY, Gong X, et al. Characteristics and living patterns of marine myxobacterial isolates. Appl. Environ. Microbiol. 2005;71(6):3331–3336. doi: 10.1128/Aem.71.6.3331-3336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang CY, Cai K, Liu H, Zhang Y, Pan HW, Wang B, et al. New locus important for Myxococcus social motility and development. J. Bacteriol. 2007;189(21):7937–7941. doi: 10.1128/JB.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan HW, Liu H, Liu T, Li CY, Li ZF, Cai K, et al. Seawater-regulated genes for two-component systems and outer membrane proteins in myxococcus. J. Bacteriol. 2009;191(7):2102–2111. doi: 10.1128/JB.01556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan HW, Tan ZG, Liu H, Li ZF, Zhang CY, Li CY, et al. Hdsp, a horizontally transferred gene required for social behavior and halotolerance in salt-tolerant Myxococcus fulvus HW-1. ISME J. 2010;4(10):1282–1289. doi: 10.1038/ismej.2010.52. [DOI] [PubMed] [Google Scholar]
- 33.Landwehr W, Wolf C, Wink J. Actinobacteria and Myxobacteria-Two of the most important bacterial resources for novel antibiotics. Curr. Top. Microbiol. Immunol. 2016;398:273–302. doi: 10.1007/82_2016_503. [DOI] [PubMed] [Google Scholar]
- 34.Wenzel SC, Müller R. Myxobacteria-'microbial factories' for the production of bioactive secondary metabolites. Mol. Biosyst. 2009;5(6):567–574. doi: 10.1039/b901287g. [DOI] [PubMed] [Google Scholar]
- 35.Naini A, Sasse F, Bronstrup M. The intriguing chemistry and biology of soraphens. Nat. Prod. Rep. 2019;36(10):1394–1411. doi: 10.1039/c9np00008a. [DOI] [PubMed] [Google Scholar]
- 36.Gerth K, Bedorf N, Hofle G, Irschik H, Reichenbach H. Epothilons A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). Production, physico-chemical and biological properties. J. Antibiot. (Tokyo) 1996;49(6):560–563. doi: 10.7164/antibiotics.49.560. [DOI] [PubMed] [Google Scholar]
- 37.Hardt IH, Steinmetz H, Gerth K, Sasse F, Reichenbach H, Hofle G. New natural epothilones from Sorangium cellulosum, strains So ce90/B2 and So ce90/D13: isolation, structure elucidation, and SAR studies. J. Nat. Prod. 2001;64(7):847–856. doi: 10.1021/np000629f. [DOI] [PubMed] [Google Scholar]
- 38.Lu C, Liu X, Li Y, Shen Y. Two 18-membered epothilones from Sorangium cellulosum So0157-2. J. Antibiot. (Tokyo) 2010;63(9):571–574. doi: 10.1038/ja.2010.81. [DOI] [PubMed] [Google Scholar]
- 39.Zhang YJ, Deng AW, Zhang H, Xi FY, Ying LP, Wang JD, et al. Epothilone O, a new member of this family from Sorangium cellulosum strain So0157-2. J. Antibiot. (Tokyo) 2013;66(5):285–286. doi: 10.1038/ja.2012.121. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Zhang H, Ying L, Wang C, Jiang N, Zhou Y, et al. Five new epothilone metabolites from Sorangium cellulosum strain So0157-2. J. Antibiot. (Tokyo) 2009;62(9):483–487. doi: 10.1038/ja.2009.55. [DOI] [PubMed] [Google Scholar]
- 41.Wang JD, Jiang N, Zhang H, Ying LP, Wang CX, Xiang WS, et al. New epothilone congeners from Sorangium cellulosum strain So0157-2. Nat. Prod. Res. 2011;25(18):1707–1712. doi: 10.1080/14786419.2011.553719. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L, Li PF, Lu CH, Li SG, Shen YM, Li YZ. Glycosylation and production characteristics of epothilones in alkali-tolerant Sorangium cellulosum strain So0157-2. J. Microbiol. 2010;48(4):438–444. doi: 10.1007/s12275-010-0048-3. [DOI] [PubMed] [Google Scholar]
- 43.Bhat MA, Mishra AK, Bhat MA, Banday MI, Bashir O, Rather IA, et al. Myxobacteria as a source of new bioactive compounds: a perspective study. Pharmaceutics. 2021;13(8):1265. doi: 10.3390/pharmaceutics13081265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tipper DJ. Inhibition of yeast ribonucleic-acid polymerases by thiolutin. J. Bacteriol. 1973;116(1):245–256. doi: 10.1128/Jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuske S, Sarafianos SG, Wang XY, Hudson B, Sineva E, Mukhopadhyay J, et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122(4):541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122(3):351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 47.O'Neill AJ, Huovinen T, Fishwick CWG, Chopra I. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob. Agents Chemother. 2006;50(1):298–309. doi: 10.1128/Aac.50.1.298-309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodin S, Kane MP, Rubin EH. Epothilones: mechanism of action and biologic activity. J. Clin. Oncol. 2004;22(10):2015–2025. doi: 10.1200/Jco.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Peterson JK, Tucker C, Favours E, Cheshire PJ, Creech J, Billups CA, et al. In vivo evaluation of ixabepilone (BMS247550), a novel epothilone B derivative, against pediatric cancer models. Clin Cancer Res. 2005;11(19):6950–6958. doi: 10.1158/1078-0432.Ccr-05-0740. [DOI] [PubMed] [Google Scholar]
- 50.Xu B, Sun T, Zhang Q, Zhang P, Yuan Z, Jiang Z, et al. Efficacy of utidelone plus capecitabine versus capecitabine for heavily pretreated, anthracycline- and taxane-refractory metastatic breast cancer: final analysis of overall survival in a phase III randomised controlled trial. Ann. Oncol. 2021;32(2):218–228. doi: 10.1016/j.annonc.2020.10.600. [DOI] [PubMed] [Google Scholar]
- 51.De Vos KJ, Grierson AJ, Ackerley S, Miller CC. Role of axonal transport in neurodegenerative diseases. Annu. Rev. Neurosci. 2008;31:151–173. doi: 10.1146/annurev.neuro.31.061307.090711. [DOI] [PubMed] [Google Scholar]
- 52.Zhang B, Carroll J, Trojanowski JQ, Yao Y, Iba M, Potuzak JS, et al. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J. Neurosci. 2012;32(11):3601–3611. doi: 10.1523/JNEUROSCI.4922-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissman KJ, Müller R. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat. Prod. Rep. 2010;27(9):1276–1295. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 54.Gaspari F, Paitan Y, Mainini M, Losi D, Ron EZ, Marinelli F. Myxobacteria isolated in Israel as potential source of new anti-infectives. J. Appl. Microbiol. 2005;98(2):429–439. doi: 10.1111/j.1365-2672.2004.02477.x. [DOI] [PubMed] [Google Scholar]
- 55.Nelson JT, Lee J, Sims JW, Schmidt EW. Characterization of SafC, a catechol 4-O-methyltransferase involved in saframycin biosynthesis. Appl. Environ. Microbiol. 2007;73(11):3575–3580. doi: 10.1128/Aem.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Yi SX, Zang Y, Yao Q, Zhu HH. The Predatory Myxobacterium Citreicoccus inhibens gen. nov. sp. nov. showed antifungal activity and bacteriolytic property against phytopathogens. Microorganisms. 2021;9(10):2137. doi: 10.3390/microorganisms9102137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silakowski B, Schairer HU, Ehret H, Kunze B, Weinig S, Nordsiek G, et al. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 1999;274(52):37391–37399. doi: 10.1074/jbc.274.52.37391. [DOI] [PubMed] [Google Scholar]
- 58.Gerth K, Irschik H, Reichenbach H, Trowitzsch W. Myxothiazol, an antibiotic from Myxococcus fulvus (myxobacterales). I. Cultivation, isolation, physico-chemical and biological properties. J. Antibiot. (Tokyo) 1980;33(12):1474–1479. doi: 10.7164/antibiotics.33.1474. [DOI] [PubMed] [Google Scholar]
- 59.Sandmann A, Frank B, Müller R. A transposon-based strategy to scale up myxothiazol production in myxobacterial cell factories. J. Biotechnol. 2008;135(3):255–261. doi: 10.1016/j.jbiotec.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 2002;3(7):619–627. doi: 10.1002/1439-7633(20020703)3:7<619::Aid-Cbic619>3.0.Co;2-9. [DOI] [PubMed] [Google Scholar]
- 61.Hoffmann T, Krug D, Bozkurt N, Duddela S, Jansen R, Garcia R, et al. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018;9(1):803. doi: 10.1038/s41467-018-03184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang DG, Niu L, Lin ZM, Wang JJ, Gao DF, Sui HY, et al. The discovery and biosynthesis of nicotinic myxochelins from an Archangium sp. SDU34. J. Nat. Prod. 2021;84(10):2744–2748. doi: 10.1021/acs.jnatprod.1c00524. [DOI] [PubMed] [Google Scholar]
- 63.Li YL, Zhuo L, Li XB, Zhu YQ, Wu SG, Shen T, et al. Myxadazoles, myxobacterium-derived isoxazole-benzimidazole hybrids with cardiovascular activities. Angewandte Chemie-Int. Ed. 2021;60(47):24748. doi: 10.1002/anie.202113164. 24748. [DOI] [PubMed] [Google Scholar]
- 64.Hu JQ, Wang JJ, Li YL, Zhuo L, Zhang A, Sui HY, et al. Combining NMR-based metabolic profiling and genome mining for the accelerated discovery of Archangiumide, an allenic macrolide from the myxobacterium Archangium violaceum SDU8. Organic Lett. 2021;23(6):2114–2119. doi: 10.1021/acs.orglett.1c00265. [DOI] [PubMed] [Google Scholar]
- 65.Hwang C, Copeland A, Lucas S, Lapidus A, Barry K, Glavina Del Rio T, et al. Complete genome sequence of Anaeromyxobacter sp. Fw109-5, an anaerobic, metal-reducing bacterium isolated from a contaminated subsurface environment. Genome Announc. 2015;3(1):e01449. doi: 10.1128/genomeA.01449-14. 01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pal S, Sharma G, Subramanian S. Complete genome sequence and identification of polyunsaturated fatty acid biosynthesis genes of the myxobacterium Minicystis rosea DSM 24000(T) BMC Genomics. 2021;22(1):655. doi: 10.1186/s12864-021-07955-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han K, Li ZF, Peng R, Zhu LP, Zhou T, Wang LG, et al. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci. Reports. 2013;3:2101. doi: 10.1038/srep02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baltz RH. Genome mining for drug discovery: progress at the front end. J. Ind. Microbiol. Biotechnol. 2021;48(9-10) doi: 10.1093/jimb/kuab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310(5753):1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaiser D, Dworkin M. Gene transfer to myxobacterium by Escherichia coli phage P1. Science. 1975;187(4177):653–654. doi: 10.1126/science.803710. [DOI] [PubMed] [Google Scholar]
- 71.Kuner JM, Kaiser D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc. Natl. Acad. Sci. U. S. A. 1981;78(1):425–429. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimkets LJ, Gill RE, Kaiser D. Developmental cell-interactions in Myxococcus xanthus and the Spoc Locus. Proc. Nat. Acad. Sci. U.S.A.-Biol. Sci. 1983;80(5):1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burchard RP, Dworkin M. A Bacteriophage for Myxococcus xanthus - isolation characterization and relation of infectivity to host morphogenesis. J. Bacteriol. 1966;91(3):1203. doi: 10.1128/Jb.91.3.1305-1313.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vasse M, Wielgoss S. Bacteriophages of Myxococcus xanthus, a social bacterium. Viruses-Basel. 2018;10(7):374. doi: 10.3390/v10070374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaiser D. Genetic systems in myxobacteria. Methods Enzymol. 1991;204:357–372. doi: 10.1016/0076-6879(91)04018-j. [DOI] [PubMed] [Google Scholar]
- 76.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Develop. Biol. 1986;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 77.Bowden MG, Kaplan HB. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 1998;30(2):275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 78.Johnsborg O, Eldholm V, Havarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 2007;158(10):767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 79.Zhao JY, Zhong L, Shen MJ, Xia ZJ, Cheng QX, Sun X, et al. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl. Environ. Microbiol. 2008;74(7):1980–1987. doi: 10.1128/Aem.02143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J, Hu W, Lux R, He X, Li Y, Shi W. Natural transformation of Myxococcus xanthus. J. Bacteriol. 2011;193(9):2122–2132. doi: 10.1128/JB.00041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P, Nagarajan A, Uchil PD. DNA transfection by electroporation. Cold Spring Harb. Protoc. 2019;2019(7):559–563. doi: 10.1101/pdb.prot095471. [DOI] [PubMed] [Google Scholar]
- 82.Kuspa A, Kaiser D. Genes required for developmental signalling in Myxococcus xanthus: three asg loci. J. Bacteriol. 1989;171(5):2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murphy KA, Garza AG. In: Myxobacteria: Multicellularity and Differentiation. Whitworth DE, editor. ASM Press; Washington, D.C: 2007. Genetic Tools for studying Myxococcus xanthus biology; pp. 491–501. [Google Scholar]
- 84.Zhu LP, Yue XJ, Han K, Li ZF, Zheng LS, Yi XN, et al. Allopatric integrations selectively change host transcriptomes, leading to varied expression efficiencies of exotic genes in Myxococcus xanthus. Microb. Cell Fact. 2015;14:105. doi: 10.1186/s12934-015-0294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Julien B, Shah S. Heterologous expression of epothilone biosynthetic genes in Myxococcus xanthus. Antimicrob. Agents Chemotherapy. 2002;46(9):2772–2778. doi: 10.1128/Aac.46.9.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perlova O, Fu J, Kuhlmann S, Krug D, Stewart AF, Zhang Y, et al. Reconstitution of the myxothiazol biosynthetic gene cluster by Red/ET recombination and heterologous expression in Myxococcus xanthus. Appl. Environ. Microbiol. 2006;72(12):7485–7494. doi: 10.1128/AEM.01503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abbasi MN, Fu J, Bian X, Wang H, Zhang Y, Li A. Recombineering for Genetic Engineering of Natural Product Biosynthetic Pathways. Trends Biotechnol. 2020;38(7):715–728. doi: 10.1016/j.tibtech.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 88.Fu J, Wenzel SC, Perlova O, Wang J, Gross F, Tang Z, et al. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic. Acids. Res. 2008;36(17):e113. doi: 10.1093/nar/gkn499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pogorevc D, Tang Y, Hoffmann M, Zipf G, Bernauer HS, Popoff A, et al. Biosynthesis and heterologous production of argyrins. ACS Synth. Biol. 2019;8(5):1121–1133. doi: 10.1021/acssynbio.9b00023. [DOI] [PubMed] [Google Scholar]
- 90.Tu Q, Herrmann J, Hu S, Raju R, Bian X, Zhang Y, et al. Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via Red/ET recombineering. Sci. Rep. 2016;6:21066. doi: 10.1038/srep21066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gross S, Schnell B, Haack PA, Auerbach D, Müller R. In vivo and in vitro reconstitution of unique key steps in cystobactamid antibiotic biosynthesis. Nat. Commun. 2021;12(1):1696. doi: 10.1038/s41467-021-21848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sucipto H, Pogorevc D, Luxenburger E, Wenzel SC, Müller R. Heterologous production of myxobacterial alpha-pyrone antibiotics in Myxococcus xanthus. Metab. Eng. 2017;44:160–170. doi: 10.1016/j.ymben.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 93.Hug JJ, Frank NA, Walt C, Senica P, Panter F, Müller R. Genome-guided discovery of the first myxobacterial biarylitide myxarylin reveals distinct C-N biaryl crosslinking in RiPP biosynthesis. Molecules. 2021;26(24):7483. doi: 10.3390/molecules26247483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stamm I, Leclerque A, Plaga W. Purification of cold-shock-like proteins from Stigmatella aurantiaca - molecular cloning and characterization of the cspA gene. Arch. Microbiol. 1999;172(3):175–181. doi: 10.1007/s002030050757. [DOI] [PubMed] [Google Scholar]
- 95.Kopp M, Irschik H, Pradella S, Müller R. Production of the tubulin destabilizer disorazol in Sorangium cellulosum: biosynthetic machinery and regulatory genes. ChemBioChem. 2005;6(7):1277–1286. doi: 10.1002/cbic.200400459. [DOI] [PubMed] [Google Scholar]
- 96.Ye W, Zhang W, Chen Y, Li H, Li S, Pan Q, et al. A new approach for improving epothilone B yield in Sorangium cellulosum by the introduction of vgb epoF genes. J. Ind. Microbiol. Biotechnol. 2016;43(5):641–650. doi: 10.1007/s10295-016-1735-9. [DOI] [PubMed] [Google Scholar]
- 97.Ye W, Liu T, Zhu M, Zhang W, Huang Z, Li S, et al. An easy and efficient strategy for the enhancement of epothilone production mediated by TALE-TF and CRISPR/dcas9 systems in Sorangium cellulosum. Front. Bioeng. Biotechnol. 2019;7:334. doi: 10.3389/fbioe.2019.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sandmann A, Sasse F, Müller R. Identification and analysis of the core biosynthetic machinery of tubulysin, a potent cytotoxin with potential anticancer activity. Chem. Biol. 2004;11(8):1071–1079. doi: 10.1016/j.chembiol.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 99.Zhang CY, Cai K, Wu ZH, Li YZ. Improving cellular properties for genetic manipulation by dispersed growing mutagenesis in Myxococcus fulvus HW-1. Curr. Microbiol. 2010;60(6):393–399. doi: 10.1007/s00284-009-9554-0. [DOI] [PubMed] [Google Scholar]
- 100.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 2010;34(1):18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 101.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz, Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002;45(1):1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 102.Breton AM, Jaoua S, Guespin-Michel J. Transfer of plasmid RP4 to Myxococcus xanthus and evidence for its integration into the chromosome. J. Bacteriol. 1985;161(2):523–528. doi: 10.1128/jb.161.2.523-528.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Glomp I, Saulnier P, Guespinmichel J, Schairer HU. Transfer of incp plasmids into Stigmatella aurantiaca leading to insertional mutants affected in spore development. Mol. General Genet. 1988;214(2):213–217. doi: 10.1007/Bf00337713. [DOI] [PubMed] [Google Scholar]
- 104.Jaoua S, Neff S, Schupp T. Transfer of mobilizable plasmids to Sorangium cellosum and evidence for their integration into the chromosome. Plasmid. 1992;28(2):157–165. doi: 10.1016/0147-619x(92)90046-D. [DOI] [PubMed] [Google Scholar]
- 105.Pradella S, Hans A, Sproer C, Reichenbach H, Gerth K, Beyer S. Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch. Microbiol. 2002;178(6):484–492. doi: 10.1007/s00203-002-0479-2. [DOI] [PubMed] [Google Scholar]
- 106.Kopp M, Irschik H, Gross F, Perlova O, Sandmann A, Gerth K, et al. Critical variations of conjugational DNA transfer into secondary metabolite multiproducing Sorangium cellulosum strains So ce12 and So ce56: development of a mariner-based transposon mutagenesis system. J. Biotechnol. 2004;107(1):29–40. doi: 10.1016/j.jbiotec.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 107.Xia ZJ, Wang J, Hu W, Liu H, Gao XZ, Wu ZH, et al. Improving conjugation efficacy of Sorangium cellulosum by the addition of dual selection antibiotics. J. Ind. Microbiol. Biotechnol. 2008;35(10):1157–1163. doi: 10.1007/s10295-008-0395-9. [DOI] [PubMed] [Google Scholar]
- 108.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 109.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 110.Hickman AB, Dyda F. Mechanisms of DNA transposition. Microbiol. Spectr. 2015;3(2) doi: 10.1128/microbiolspec.MDNA3-0034-2014. MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cho K, Zusman DR. Sporulation timing in Myxococcus xanthus is controlled by the espAB locus. Mol. Microbiol. 1999;34(4):714–725. doi: 10.1046/j.1365-2958.1999.01633.x. [DOI] [PubMed] [Google Scholar]
- 112.Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, et al. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 2005;55(1):206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- 113.Ueki T, Inouye S, Inouye M. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene. 1996;183(1-2):153–157. doi: 10.1016/s0378-1119(96)00546-x. [DOI] [PubMed] [Google Scholar]
- 114.Julien B, Kaiser AD, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U S A. 2000;97(16):9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peng R, Chen JH, Feng WW, Zhang Z, Yin J, Li ZS, et al. Error-prone DnaE2 balances the genome mutation rates in Myxococcus xanthus DK1622. Front. Microbiol. 2017;8:122. doi: 10.3389/fmicb.2017.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pan Z, Zhang Z, Zhuo L, Wan TY, Li YZ. Bioinformatic and functional characterization of Hsp70s in Myxococcus xanthus. mSphere. 2021;6(3):00305–00321. doi: 10.1128/mSphere.00305-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhuo L, Wan TY, Pan Z, Wang JN, Sheng DH, Li YZ. A dual-functional orphan response regulator negatively controls the differential transcription of duplicate groELs and plays a global regulatory role in Myxococcus. mSystems. 2022;7(2) doi: 10.1128/msystems.01056-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Y, Li X, Zhang W, Zhou X, Li YZ. The groEL2 gene, but not groEL1, is required for biosynthesis of the secondary metabolite myxovirescin in Myxococcus xanthus DK1622. Microbiology. 2014;160(Pt 3):488–495. doi: 10.1099/mic.0.065862-0. [DOI] [PubMed] [Google Scholar]
- 119.Yue XJ, Cui XW, Zhang Z, Peng R, Zhang P, Li ZF, et al. A bacterial negative transcription regulator binding on an inverted repeat in the promoter for epothilone biosynthesis. Microbial. Cell Factories. 2017;16:92. doi: 10.1186/s12934-017-0706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Padmanabhan S, Monera-Girona AJ, Perez-Castano R, Bastida-Martinez E, Pajares-Martinez E, Bernal-Bernal D, et al. Light-triggered carotenogenesis in Myxococcus xanthus: new paradigms in photosensory signaling, transduction and gene regulation. Microorganisms. 2021;9(5):1067. doi: 10.3390/microorganisms9051067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Volz C, Kegler C, Müller R. Enhancer binding proteins act as hetero-oligomers and link secondary metabolite production to myxococcal development, motility, and predation. Chem. Biol. 2012;19(11):1447–1459. doi: 10.1016/j.chembiol.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 122.Izzat S, Rachid S, Ajdidi A, El-Nakady YA, Liu XX, Ye BC, et al. The ROK like protein of Myxococcus xanthus DK1622 acts as a pleiotropic transcriptional regulator for secondary metabolism. J. Biotechnol. 2020;311:25–34. doi: 10.1016/j.jbiotec.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 123.Tojo N, Sanmiya K, Sugawara H, Inouye S, Komano T. Integration of bacteriophage Mx8 into the Myxococcus xanthus chromosome causes a structural alteration at the C-terminal region of the IntP protein. J. Bacteriol. 1996;178(14):4004–4011. doi: 10.1128/jb.178.14.4004-4011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Julien B. Characterization of the integrase gene and attachment site for the Myxococcus xanthus bacteriophage Mx9. J. Bacteriol. 2003;185(21):6325–6330. doi: 10.1128/JB.185.21.6325-6330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tojo N, Komano T. The IntP C-terminal segment is not required for excision of bacteriophage Mx8 from the Myxococcus xanthus chromosome. J. Bacteriol. 2003;185(7):2187–2193. doi: 10.1128/JB.185.7.2187-2193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Magrini V, Storms ML, Youderian P. Site-specific recombination of temperate Myxococcus xanthus phage Mx8: regulation of integrase activity by reversible, covalent modification. J. Bacteriol. 1999;181(13):4062–4070. doi: 10.1128/Jb.181.13.4062-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Salmi D, Magrini V, Hartzell PL, Youderian P. Genetic determinants of immunity and integration of temperate Myxococcus xanthus phage Mx8. J. Bacteriol. 1998;180(3):614–621. doi: 10.1128/JB.180.3.614-621.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bode HB, Ring MW, Schwar G, Altmeyer MO, Kegler C, Jose IR, et al. Identification of additional players in the alternative biosynthesis pathway to isovaleryl-CoA in the Myxobacterium Myxococcus xanthus. ChemBioChem. 2009;10(1):128–140. doi: 10.1002/cbic.200800219. [DOI] [PubMed] [Google Scholar]
- 129.Müller S, Shen H, Hofmann D, Schairer HU, Kirby JR. Integration into the phage attachment site, attB, impairs multicellular differentiation in Stigmatella aurantiaca. J. Bacteriol. 2006;188(5):1701–1709. doi: 10.1128/Jb.188.5.1701-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Y, Zhang WY, Zhang Z, Li J, Li ZF, Tan ZG, et al. Mechanisms involved in the functional divergence of duplicated GroEL chaperonins in Myxococcus xanthus DK1622. PLos Genet. 2013;9(2) doi: 10.1371/journal.pgen.1003306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Müller FD, Treuner-Lange A, Heider J, Huntley SM, Higgs PI. Global transcriptome analysis of spore formation in Myxococcus xanthus reveals a locus necessary for cell differentiation. BMC Genomics. 2010;11:264. doi: 10.1186/1471-2164-11-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blot M, Heitman J, Arber W. Tn5-mediated bleomycin resistance in Escherichia coli requires the expression of host genes. Mol. Microbiol. 1993;8(6):1017–1024. doi: 10.1111/j.1365-2958.1993.tb01646.x. [DOI] [PubMed] [Google Scholar]
- 133.Li N, Jin K, Bai Y, Fu H, Liu L, Liu B. Tn5 transposase applied in genomics research. Int. J. Mol. Sci. 2020;21(21):8329. doi: 10.3390/ijms21218329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reznikoff WS. Transposon Tn5. Annu. Rev. Genet. 2008;42:269–286. doi: 10.1146/annurev.genet.42.110807.091656. [DOI] [PubMed] [Google Scholar]
- 135.Avery L, Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol. Gen. Genet. 1983;191(1):99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- 136.Sasakawa C, Yoshikawa M. A series of Tn5 variants with various drug-resistance markers and suicide vector for transposon mutagenesis. Gene. 1987;56(2-3):283–288. doi: 10.1016/0378-1119(87)90145-4. [DOI] [PubMed] [Google Scholar]
- 137.Balsalobre JM, Ruiz-Vazquez RM, Murillo FJ. Light induction of gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 1987;84(8):2359–2362. doi: 10.1073/pnas.84.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Perez-Burgos M, Garcia-Romero I, Jung J, Valvano MA, Sogaard-Andersen L. Identification of the lipopolysaccharide O-antigen biosynthesis priming enzyme and the O-antigen ligase in Myxococcus xanthus: critical role of LPS O-antigen in motility and development. Mol. Microbiol. 2019;112(4):1178–1198. doi: 10.1111/mmi.14354. [DOI] [PubMed] [Google Scholar]
- 139.Sydney N, Swain MT, So JMT, Hoiczyk E, Tucker NP, Whitworth DE. The genetics of prey susceptibility to myxobacterial predation: a review, including an investigation into Pseudomonas aeruginosa mutations affecting predation by Myxococcus xanthus. Microb. Physiol. 2021;31(2):57–66. doi: 10.1159/000515546. [DOI] [PubMed] [Google Scholar]
- 140.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 2003;49(2):555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 141.Tellier M, Bouuaert CC, Chalmers R. Mariner and the ITm superfamily of transposons. Microbiol. Spectr. 2015;3(2) doi: 10.1128/microbiolspec.MDNA3-0033-2014. MDNA3-0033-2014. [DOI] [PubMed] [Google Scholar]
- 142.Dornan J, Grey H, Richardson JM. Structural role of the flanking DNA in mariner transposon excision. Nucleic. Acids. Res. 2015;43(4):2424–2432. doi: 10.1093/nar/gkv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, et al. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6(6):415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Youderian P, Hartzell PL. Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics. 2006;172(3):1397–1410. doi: 10.1534/genetics.105.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gong Y, Zhang Z, Zhou XW, Anwar MN, Hu XZ, Li ZS, et al. Competitive interactions between incompatible mutants of the social bacterium Myxococcus xanthus DK1622. Front. Microbiol. 2018;9:1200. doi: 10.3389/fmicb.2018.01200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Julien B, Fehd R. Development of a mariner-based transposon for use in Sorangium cellulosum. Appl. Environ. Microbiol. 2003;69(10):6299–6301. doi: 10.1128/Aem.69.10.6299-6301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Varon M, Fuchs N, Monosov M, Tolchinsky S, Rosenberg E. Mutation and mapping of genes involved in production of the antibiotic TA in Myxococcus xanthus. Antimicrob. Agents Chemother. 1992;36(10):2316–2321. doi: 10.1128/AAC.36.10.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pospiech A, Cluzel B, Bietenhader J, Schupp T. A new Myxococcus xanthus gene cluster for the biosynthesis of the antibiotic saframycin Mx1 encoding a peptide synthetase. Microbiology (Reading) 1995;141(Pt 8):1793–1803. doi: 10.1099/13500872-141-8-1793. [DOI] [PubMed] [Google Scholar]
- 149.Martinezlaborda A, Elias M, Ruizvazquez R, Murillo FJ. Insertions of Tn5 linked to mutations affecting carotenoid synthesis in Myxococcus xanthus. Mol. General Genetics. 1986;205(1):107–114. doi: 10.1007/Bf02428039. [DOI] [Google Scholar]
- 150.Feng J, Chen XJ, Sun X, Wang N, Li YZ. Characterization of the replication origin of the myxobacterial self-replicative plasmid pMF1. Plasmid. 2012;68(2):105–112. doi: 10.1016/j.plasmid.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 151.Sun X, Chen XJ, Feng J, Zhao JY, Li YZ. Characterization of the partitioning system of Myxococcus plasmid pMF1. PLoS One. 2011;6(12):e28122. doi: 10.1371/journal.pone.0028122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sheng D, Chen X, Li Y, Wang J, Zhuo L, Li Y. ParC, a new partitioning protein, is necessary for the active form of parA from Myxococcus pMF1 plasmid. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.623699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li YJ, Liu Y, Zhang Z, Chen XJ, Gong Y, Li YZ. A Post-segregational killing mechanism for maintaining plasmid PMF1 in its Myxococcus fulvus host. Front. Cell Infect. Microbiol. 2018;8:274. doi: 10.3389/fcimb.2018.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen XJ, Han K, Feng J, Zhuo L, Li YJ, Li YZ. The complete genome sequence and analysis of a plasmid-bearing myxobacterial strain Myxococcus fulvus 124B02 (M 206081) Stand. Genomic Sci. 2016;11:1. doi: 10.1186/s40793-015-0121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao JY, Xia ZJ, Sun X, Zhong L, Jiang DM, Liu H, et al. Cloning and characterization of an rRNA methyltransferase from Sorangium cellulosum. Biochem. Biophys. Res. Commun. 2008;370(1):140–144. doi: 10.1016/j.bbrc.2008.03.045. [DOI] [PubMed] [Google Scholar]
- 156.Tan ZG, Li HM, Pan HW, Zhou XW, Liu X, Luo NN, et al. Characterization of four type IV pilin homologues in Stigmatella aurantiaca DSM17044 by heterologous expression in Myxococcus xanthus. PLoS One. 2013;8(9):e75105. doi: 10.1371/journal.pone.0075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhuo L, Wang Y, Zhang Z, Li J, Zhang XH, Li YZ. Myxococcus xanthus DK1622 coordinates expressions of the duplicate groEL and single groES genes for synergistic functions of groELs and groES. Front. Microbiol. 2017;8:733. doi: 10.3389/fmicb.2017.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sheng DH, Wang YX, Qiu M, Zhao JY, Yue XJ, Li YZ. Functional division between the RecA1 and RecA2 proteins in Myxococcus xanthus. Front. Microbiol. 2020;11:140. doi: 10.3389/fmicb.2020.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang YJ, Wang Y, Li ZF, Gong Y, Zhang P, Hu WC, et al. Increasing on-target cleavage efficiency for CRISPR/Cas9-induced large fragment deletion in Myxococcus xanthus. Microb. Cell Factories. 2017;16:142. doi: 10.1186/s12934-017-0758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peng R, Wang Y, Feng WW, Yue XJ, Chen JH, Hu XZ, et al. CRISPR/dCas9-mediated transcriptional improvement of the biosynthetic gene cluster for the epothilone production in Myxococcus xanthus. Microbial Cell Factories. 2018;17:15. doi: 10.1186/s12934-018-0867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang Y, Yue XJ, Yuan SF, Hong Y, Hu WF, Li YZ. Internal promoters and their effects on the transcription of operon genes for epothilone production in Myxococcus xanthus. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.758561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Panter F, Bader CD, Müller R. The sandarazols are cryptic and structurally unique plasmid-encoded toxins from a rare Myxobacterium. Angewandte Chemie-Int. Ed. 2021;60(15):8081–8088. doi: 10.1002/anie.202014671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34(8):401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 164.Shivram H, Cress BF, Knott GJ, Doudna JA. Controlling and enhancing CRISPR systems. Nat. Chem. Biol. 2021;17(1):10–19. doi: 10.1038/s41589-020-00700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 167.Savic N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl. Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 168.Xu X, Qi LS. A CRISPR-dCas toolbox for genetic engineering and synthetic biology. J. Mol. Biol. 2019;431(1):34–47. doi: 10.1016/j.jmb.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 169.Tong Y, Weber T, Lee SY. CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 2019;36(9):1262–1280. doi: 10.1039/c8np00089a. [DOI] [PubMed] [Google Scholar]
- 170.Bernal-Bernal D, Abellon-Ruiz J, Iniesta AA, Pajares-Martinez E, Bastida-Martinez E, Fontes M, et al. Multifactorial control of the expression of a CRISPR-Cas system by an extracytoplasmic function sigma/anti-sigma pair and a global regulatory complex. Nucleic. Acids. Res. 2018;46(13):6726–6745. doi: 10.1093/nar/gky475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu J, Wang X, Dai G, Zhang Y, Bian X. Microbial chassis engineering drives heterologous production of complex secondary metabolites. Biotechnol. Adv. 2022;59 doi: 10.1016/j.biotechadv.2022.107966. [DOI] [PubMed] [Google Scholar]
- 172.Höfle G BN, Gerth K, Reichenbach H. Epothilones, Process for Preparing the Same and their use as Medicaments and as Plant Protecting Agents. WO, WO1993010121 A1.
- 173.Bollag DM, Mcqueney PA, Zhu J, Hensens O, Koupal L, Liesch J, et al. Epothilones, a new class of microtubule-stabilizing agents with a Taxol-like mechanism of action. Cancer Res. 1995;55(11):2325–2333. [PubMed] [Google Scholar]
- 174.Ibrahim NK. Ixabepilone: Overview of effectiveness, safety, and tolerability in metastatic breast cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.617874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Julien B, Shah S, Ziermann R, Goldman R, Katz L, Khosla C. Isolation and characterization of the epothilone biosynthetic gene cluster from Sorangium cellulosum. Gene. 2000;249(1-2):153–160. doi: 10.1016/s0378-1119(00)00149-9. [DOI] [PubMed] [Google Scholar]
- 176.Molnár I, Schupp T, Ono M, Zirkle R, Milnamow M, Nowak-Thompson B, et al. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 2000;7(2):97–109. doi: 10.1016/s1074-5521(00)00075-2. [DOI] [PubMed] [Google Scholar]
- 177.Zhu LP, Li ZF, Sun X, Li SG, Li YZ. Characteristics and activity analysis of epothilone operon promoters from Sorangium cellulosum strains in Escherichia coli. Appl. Microbiol. Biotechnol. 2013;97(15):6857–6866. doi: 10.1007/s00253-013-4830-0. [DOI] [PubMed] [Google Scholar]
- 178.Yue XJ, Cui XW, Zhang Z, Hu WF, Li ZF, Zhang YM, et al. Effects of transcriptional mode on promoter substitution and tandem engineering for the production of epothilones in Myxococcus xanthus. Appl. Microbiol. Biotechnol. 2018;102(13):5599–5610. doi: 10.1007/s00253-018-9023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gong GL, Sun X, Liu XL, Hu W, Cao WR, Liu H, et al. Mutation and a high-throughput screening method for improving the production of Epothilones of Sorangium. J. Ind. Microbiol. Biotechnol. 2007;34(9):615–623. doi: 10.1007/s10295-007-0236-2. [DOI] [PubMed] [Google Scholar]
- 180.Tang L, Shah S, Chung L, Carney J, Katz L, Khosla C, et al. Cloning and heterologous expression of the epothilone gene cluster. Science. 2000;287(5453):640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 181.Park SR, Park JW, Jung WS, Han AR, Ban YH, Kim EJ, et al. Heterologous production of epothilones B and D in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 2008;81(1):109–117. doi: 10.1007/s00253-008-1674-0. [DOI] [PubMed] [Google Scholar]
- 182.Zhang L, Zhao G, Ding X. Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination. Sci. Rep. 2011;1:141. doi: 10.1038/srep00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mutka SC, Carney JR, Liu Y, Kennedy J. Heterologous production of epothilone C and D in Escherichia coli. Biochemistry. 2006;45(4):1321–1330. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- 184.Bian XY, Tang B, Yu YC, Tu Q, Gross F, Wang HL, et al. Heterologous production and yield improvement of epothilones in Burkholderiales strain DSM 7029. Acs Chem. Biol. 2017;12(7):1805–1812. doi: 10.1021/acschembio.7b00097. [DOI] [PubMed] [Google Scholar]
- 185.Yu YC, Wang HM, Tang B, Liang JH, Zhang L, Wang HK, et al. Reassembly of the biosynthetic gene cluster enables high epothilone yield in engineered Schlegelella brevitalea. Acs Synth. Biol. 2020;9(8):2009–2022. doi: 10.1021/acssynbio.0c00100. [DOI] [PubMed] [Google Scholar]
- 186.Meiser P, Müller R. Two functionally redundant Sfp-type 4′-phosphopantetheinyl transferases differentially activate biosynthetic pathways in Myxococcus xanthus. ChemBioChem. 2008;9(10):1549–1553. doi: 10.1002/cbic.200800077. [DOI] [PubMed] [Google Scholar]
- 187.Osswald C, Zipf G, Schmidt G, Maier J, Bernauer HS, Müller R, et al. Modular construction of a functional artificial epothilone polyketide pathway. Acs Synth. Biol. 2014;3(10):759–772. doi: 10.1021/sb300080t. [DOI] [PubMed] [Google Scholar]
- 188.Lau J, Frykman S, Regentin R, Ou S, Tsuruta H, Licari P. Optimizing the heterologous production of epothilone D in Myxococcus xanthus. Biotechnol. Bioeng. 2002;78(3):280–288. doi: 10.1002/bit.10202. [DOI] [PubMed] [Google Scholar]
- 189.Frykman SA, Tsuruta H, Licari PJ. Assessment of fed-batch, semicontinuous, and continuous epothilone D production processes. Biotechnol. Prog. 2005;21(4):1102–1108. doi: 10.1021/bp050010. + [DOI] [PubMed] [Google Scholar]
- 190.Ongley SE, Bian X, Neilan BA, Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 2013;30(8):1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- 191.Luan G, Zhang S, Lu X. Engineering cyanobacteria chassis cells toward more efficient photosynthesis. Curr. Opin. Biotechnol. 2020;62:1–6. doi: 10.1016/j.copbio.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 192.Weber T, Charusanti P, Musiol-Kroll EM, Jiang XL, Tong YJ, Kim HU, et al. Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015;33(1):15–26. doi: 10.1016/j.tibtech.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 193.Yuan YJ, Cheng S, Bian GK, Yan P, Ma ZN, Dai W, et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi. Nature Catal. 2022;5(4) doi: 10.1038/s41929-022-00762-x. 277-+. doi. [DOI] [Google Scholar]
- 194.Rin Kim S, Kim SJ, Kim SK, Seo SO, Park S, Shin J, et al. Yeast metabolic engineering for carbon dioxide fixation and its application. Bioresour. Technol. 2022;346 doi: 10.1016/j.biortech.2021.126349. [DOI] [PubMed] [Google Scholar]
- 195.Chen R, Jia QD, Mu X, Hu B, Sun X, Deng ZX, et al. Systematic mining of fungal chimeric terpene synthases using an efficient precursor-providing yeast chassis. Proc. Nat. Acad. Sci. U.S.A. 2021;118(29) doi: 10.1073/pnas.2023247118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wenzel S.C., Müller R. In: Industrial Biotechnology: Microorganism. Christoph W, James CL, editors. Wiley-VCH Verlag GmbH & Co. KGaA; 2017. Host organisms: myxobacterium; pp. 453–485. [DOI] [Google Scholar]
- 197.Krug D, Zurek G, Revermann O, Vos M, Velicer GJ, Müller R. Discovering the hidden secondary metabolome of Myxococcus xanthus: a study of intraspecific diversity. Appl. Environ. Microbiol. 2008;74(10):3058–3068. doi: 10.1128/Aem.02863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Huo L, Hug JJ, Fu C, Bian X, Zhang Y, Müller R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019;36(10):1412–1436. doi: 10.1039/c8np00091c. [DOI] [PubMed] [Google Scholar]
- 199.Yu YN, Kleiner M, Velicer GJ. Spontaneous reversions of an evolutionary trait loss reveal regulators of a small RNA that controls multicellular development in myxobacteria. J. Bacteriol. 2016;198(23):3142–3151. doi: 10.1128/JB.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yu YTN, Yuan X, Velicer GJ. Adaptive Evolution of an sRNA that controls Myxococcus development. Science. 2010;328(5981):993. doi: 10.1126/science.1187200. 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Birch A, Hausler A, Hutter R. Genome rearrangement and genetic instability in Streptomyces spp. J. Bacteriol. 1990;172(8):4138–4142. doi: 10.1128/jb.172.8.4138-4142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yan F, Burgard C, Popoff A, Zaburannyi N, Zipf G, Maier J, et al. Synthetic biology approaches and combinatorial biosynthesis towards heterologous lipopeptide production. Chem. Sci. 2018;9(38):7510–7519. doi: 10.1039/c8sc02046a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yan F, Auerbach D, Chai Y, Keller L, Tu Q, Huttel S, et al. Biosynthesis and heterologous production of vioprolides: rational biosynthetic engineering and unprecedented 4-methylazetidinecarboxylic acid formation. Angew. Chem. Int. Ed Engl. 2018;57(28):8754–8759. doi: 10.1002/anie.201802479. [DOI] [PubMed] [Google Scholar]
- 204.Bu QT, Yu P, Wang J, Li ZY, Chen XA, Mao XM, et al. Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches. Microb. Cell Fact. 2019;18(1):16. doi: 10.1186/s12934-019-1055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu J, Zhou H, Yang Z, Wang X, Chen H, Zhong L, et al. Rational construction of genome-reduced Burkholderiales chassis facilitates efficient heterologous production of natural products from proteobacteria. Nat. Commun. 2021;12(1):4347. doi: 10.1038/s41467-021-24645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Posfai G, Plunkett G, 3rd, Feher T, Frisch D, Keil GM, Umenhoffer K, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312(5776):1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 207.Fan X, Zhang Y, Zhao F, Liu Y, Zhao Y, Wang S, et al. Genome reduction enhances production of polyhydroxyalkanoate and alginate oligosaccharide in Pseudomonas mendocina. Int. J. Biol. Macromol. 2020;163:2023–2031. doi: 10.1016/j.ijbiomac.2020.09.067. [DOI] [PubMed] [Google Scholar]
- 208.Martinez-Garcia E, Nikel PI, Aparicio T, de Lorenzo V. Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Fact. 2014;13:159. doi: 10.1186/s12934-014-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Choe D, Lee JH, Yoo M, Hwang S, Sung BH, Cho S, et al. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun. 2019;10(1):935. doi: 10.1038/s41467-019-08888-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yang YJ, Singh RP, Lan X, Zhang CS, Li YZ, Li YQ, et al. Genome editing in model strain Myxococcus xanthus DK1622 by a site-specific Cre/loxP recombination system. Biomolecules. 2018;8(4):137. doi: 10.3390/biom8040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hu WF, Niu L, Yue XJ, Zhu LL, Hu W, Li YZ, et al. Characterization of constitutive promoters for the elicitation of secondary metabolites in myxobacteria. ACS Synth Biol. 2021;10(11):2904–2909. doi: 10.1021/acssynbio.1c00444. [DOI] [PubMed] [Google Scholar]
- 212.Wang X, Liu J, Zheng W, Zhang Y, Bian X. Recombineering-mediated genome editing in Burkholderiales strains. Methods Mol. Biol. 2022;2479:21–36. doi: 10.1007/978-1-0716-2233-9_3. [DOI] [PubMed] [Google Scholar]
- 213.Ochi K. Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J. Antibiot. (Tokyo) 2017;70(1):25–40. doi: 10.1038/ja.2016.82. [DOI] [PubMed] [Google Scholar]
- 214.Wang Y, Li X, Zhang W, Zhou X, Li YZ. The groEL2 gene, but not groEL1, is required for biosynthesis of the secondary metabolite myxovirescin in Myxococcus xanthus DK1622. Microbiology (Reading) 2014;160(Pt 3):488–495. doi: 10.1099/mic.0.065862-0. [DOI] [PubMed] [Google Scholar]