Abstract
The 5′ leader of tobacco etch virus (TEV) genomic RNA directs efficient translation from the naturally uncapped viral mRNA. Two distinct regions within the TEV 143-nucleotide leader confer cap-independent translation in vivo even when present in the intercistronic region of a discistronic mRNA, indicating that the TEV leader contains an internal ribosome entry site (IRES). In this study, the requirements for TEV IRES activity were investigated. The TEV IRES enhanced translation of monocistronic or dicistronic mRNAs in vitro under competitive conditions, i.e., at high RNA concentration or in lysate partially depleted of eukaryotic initiation factor 4F (eIF4F) and eIFiso4F, the two cap binding complexes in plants. The translational advantage conferred by the TEV IRES under these conditions was lost when the lysate reduced in eIF4F and eIFiso4F was supplemented with eIF4F (or, to a lesser extent, eIFiso4F) but not when supplemented with eIF4E, eIFiso4E, eIF4A, or eIF4B. eIF4G, the large subunit of eIF4F, was responsible for the competitive advantage conferred by the TEV IRES. TEV IRES activity was enhanced moderately by the poly(A)-binding protein. These observations suggest that the TEV IRES directs cap-independent translation through a mechanism that involves eIF4G specifically.
During translation initiation, the 40S subunit of the ribosome binds to an mRNA and scans to the initiation codon, where it assembles with the 60S subunit to form the 80S ribosome, which is competent to carry out translation of the coding region. Numerous eukaryotic initiation factors (eIFs) assist in each step during the initiation process. Prior to 40S ribosomal subunit binding, eIF4E, the small subunit of eIF4F, binds to the cap structure (m7GpppN, where N represents any nucleotide) present at the 5′ terminus of most eukaryotic mRNAs. eIF4G, the large subunit of eIF4F, recruits several additional initiation factors including eIF4A, which is required to remove secondary structure within the 5′ leader sequence that would otherwise inhibit the scanning of the 40S ribosomal subunit, eIF3, which promotes 40S ribosomal subunit binding to the mRNA, and the poly(A)-binding protein (PABP), which stabilizes eIF4F binding to the 5′ cap (16, 21, 28, 29, 30, 45). The N-terminal domain of eIF4G is responsible for binding eIF4E and PABP, the middle domain binds eIF3 and eIF4A, and, in mammalian eIF4G, the C-terminal domain binds a second molecule of eIF4A as well as Mnk1, a mitogen-activated protein kinase responsible for phosphorylating eIF4E (15, 16, 39). Consequently, eIF4G functions as an adapter that recruits many of the factors involved in stimulating 40S ribosomal subunit binding to an mRNA. Two related but highly distinct eIF4G proteins are expressed in plants, animals, and yeast (7, 11, 12). The two plant eIF4G proteins, referred to as eIF4G and eIFiso4G, differ in size (165 and 86 kDa, respectively) and have 30% identity (K. Browning, personal communication), whereas mammalian eIF4GI and eIF4GII have 46% identity (12) and yeast eIF4G1 and eIF4G2 have 53% identity (11). How these two distinct forms of eIF4G differ functionally has not been thoroughly investigated.
The poly(A) tail serves as the binding site for PABP, which assists in the assembly of the initiation complex through a physical interaction with eIF4G, an interaction that is conserved in animals, plants, and yeast (16, 26, 28, 37, 42). An interaction between PABP and eIF4B, a factor that assists in the function of eIF4F, was also demonstrated in plants (26, 28) and was later confirmed in mammals (8). The interaction between PABP and eIF4G or between PABP and eIF4B increases the poly(A)-binding activity of PABP by over 1 order of a magnitude by reducing its rate of disassociation (26, 28) and increasing the affinity of eIF4F for the 5′ cap structure by 40-fold (45). eIF4G and eIF4B not only individually increase the binding affinity of PABP for poly(A) RNA but also together exert a synergistic effect on PABP activity (28), which indicates that the physical interaction between all three proteins serves to stabilize their association with their respective binding sites to increase their function during translation initiation.
Because they serve as binding sites for eIF4F and PABP, the 5′ cap and poly(A) tail are critical in recruiting translational machinery, and, as a consequence, virtually all mRNAs are capped and polyadenylated. However, some exceptions exist: several animal and plant viral RNAs naturally lack a 5′ cap and/or poly(A) tail. Poliovirus and encephalomyocarditis virus (EMCV) are two examples of animal picornaviruses whose genomic mRNA lacks a 5′ cap structure. Instead, these mRNAs possess a long, structured 5′ leader sequence that contains an internal ribosome entry site (IRES) to which the 40S ribosomal subunit is recruited (17, 33). For EMCV, eIF4G binds to the IRES, which in turn promotes internal binding of the 40S ribosomal subunit through its interaction with eIF3 (34, 35).
Tobacco etch virus (TEV) is a potyvirus, a member of the picornavirus supergroup of positive-strand RNA viruses, which infects plants. Like that of EMCV and poliovirus, the genomic RNA of TEV is a polyadenylated mRNA that naturally lacks a 5′ cap structure but that is nevertheless efficiently translated. The TEV 5′ leader is sufficient to confer cap-independent translation to an mRNA (9, 10) and is functionally analogous to a cap in that it interacts with the poly(A) tail to promote efficient translation (10). Two centrally located cap-independent regulatory elements within the 143-base TEV 5′ leader are required to direct cap-independent translation, and both are required to interact functionally with the poly(A) tail to promote optimal translation (31). Although, the TEV IRES evolved to function as part of the leader sequence of a monocistronic mRNA, it also promoted translation of the 5′-distal (i.e., second) cistron of a dicistronic mRNA in vivo when the IRES was present in the intercistronic region; these observations indicate that the TEV 5′ leader functions as an IRES.
The mechanism underlying IRES-mediated cap-independent translation has not been investigated for TEV or any other plant virus. In this study, we identified initiation factors that are required for TEV IRES activity. Wheat germ lysate in which the endogenous levels of eIF4F and eIFiso4F were reduced (eIF4F/eIF4F-reduced lysate) recapitulated the TEV IRES-mediated enhancement of cap-independent translation in in vitro translation assays. The translational advantage conferred by the TEV IRES under these conditions was lost when the depleted lysate was supplemented with eIF4F (or, to a lesser extent, eIFiso4F) but not when it was supplemented with eIF4E, eIFiso4E, eIF4A, or eIF4B. eIF4G, the large subunit of eIF4F, was responsible for the competitive advantage conferred by the TEV IRES. TEV IRES activity was enhanced moderately by PABP. These observations suggest that the TEV IRES directs cap-independent translation through a mechanism that involves eIF4G specifically.
MATERIALS AND METHODS
mRNA constructs and in vitro RNA synthesis.
The monocistronic (TEV-luc-A50 and Con144-luc-A50) and dicistronic (GUS-SL-TEV-luc-A50 and GUS-SL-Con144-luc-A50) luciferase constructs that contain the 144-nucleotide (nt) TEV leader sequence or control (i.e., 144- or 134-nt) sequences and that terminate in a poly(A)50 tract have been described previously (31). Dicistronic constructs contained the uidA gene, encoding β-glucuronidase (GUS), as the 5′-proximal cistron and luc as the 5′-distal cistron. Control constructs were designed to contain a 60% AT-rich sequence that was the same length as the TEV 5′ leader. The free energy (ΔG) calculated by the fold algorithm for the control sequence leader is −11.5 kcal/mol, which is approximately equal to the free energy of −10.7 kcal/mol of the 5′ leader of the TEV-luc-A50 mRNA construct (48). A 24-bp stem-loop structure (ΔG = −42.9 kcal/mol) introduced upstream of the TEV and control sequences in the dicistronic constructs was described previously (31).
Following linearization downstream of the poly(A)50 tract, the DNA concentration was quantitated spectrophotometrically and brought to 0.5 mg/ml. In vitro transcription was carried out as described previously (47) using a solution containing 40 mM Tris-HCl, pH 7.5; 6 mM MgCl2; 100 μg of bovine serum albumin/ml; 0.5 mM (each) ATP, CTP, UTP, and GTP; 10 mM dithiothreitol; 0.3 U of RNasin (Promega)/μl, and 0.5 U of T7 RNA polymerase/μl. All constructs used terminated in a poly(A)50 tail. Capped RNAs were synthesized using 3 μg of template in the reaction mixture described above except that GTP was used at 160 μM and 1 mM m7GpppG was included. Under these conditions more than 95% of the mRNA is capped.
Protein purification and Western analysis.
Wheat PABP (27), eIF4F and eIFiso4F (7), eIF4B (6), eIF4A (25), and recombinant eIFiso4G and eIFiso4E (44) were purified as described previously. The purification of eIF4G and eIF4E will be described elsewhere. Purified wheat eIF4F used in these studies contained eIF4G and eIF4E. Similarly, purified wheat eIFiso4F contained eIFiso4G and eIFiso4E. eIF4A, the third subunit of mammalian eIF4F, was not present in purified plant eIF4F and eIFiso4F.
Protein from control and depleted wheat germ lysate was resolved using standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the protein was transferred to a 0.22-μm-pore-size nitrocellulose membrane by electroblotting. Following transfer, the nitrocellulose membranes were blocked in 5% milk–0.01% thimerosal in TPBS (0.1% Tween 20, 13.7 mM NaCl, 0.27 mM KCl, 1 mM Na2HPO4, 0.14 mM KH2PO4), followed by incubation with primary antibodies diluted typically 1:1,000 to 1:2,000 in TPBS with 1% milk for 1.5 h. The blots were then washed twice with TPBS and incubated with goat anti-rabbit horseradish peroxidase-conjugated antibodies (Southern Biotechnology Associates, Inc.) diluted 1:10,000 for 1 h. The blots were washed twice with TPBS, and the signal was detected typically after between 1 and 15 min using chemiluminescence (Amersham Corp.).
In vitro translation assays.
Two hundred microliters of wheat germ extract (Promega) was added to 300 μl of m7GTP-Sepharose (Pharmacia) or 100 μl of poly(A)-agarose (Sigma), and the mixture was incubated with rotation at 4°C for 30 min. The lysate was collected by centrifugation (800 × g for 1 min) through a spin column (Promega) and used immediately. Depletion of eIF4G, eIF4E, eIFiso4G, eIFiso4E, eIF4A, eIF4B, eIF3, and PABP was verified by Western analysis following resolution of the extract by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. mRNA constructs were translated using complete or depleted wheat germ lysate as described by the manufacturer (Promega) except that all amino acids were unlabeled. The lysates were supplemented with recombinant initiation factors or factors purified from wheat germ extract as indicated in each experiment. The reaction mixtures were incubated for 3 h at 22°C, and 2-μl aliquots were assayed for luciferase activity. Each mRNA construct was translated in triplicate, and the average value and standard deviation for each construct are reported.
Lysate was assayed in luciferase assay buffer (25 mM Tricine [pH 8]–5 mM MgCl2–0.1 mM EDTA supplemented with 33.3 mM dithiothreitol, 270 μM coenzyme A, and 500 μM ATP) following injection of 0.5 mM luciferin using a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
RESULTS
Recapitulation of TEV 5′-leader-mediated enhancement of cap-independent translation in vitro.
The TEV 5′ leader was shown previously to enhance cap-independent translation in vivo but not in vitro (31), suggesting the possibility that a factor required for the function of the TEV leader might be lacking in the wheat germ lysate. However, wheat germ lysate is highly message dependent because of a low concentration of endogenous transcript and a high level of unengaged translational machinery. As a consequence, those features that increase the competitiveness of an mRNA, such as the TEV 5′ leader, would not be expected to confer a translational advantage under the noncompetitive conditions that prevail in normal lysate. Competitive translation might be achieved by increasing the concentration of mRNA to a level that exceeds the translational capacity of the lysate or by removing the excess capacity of those factors most important in facilitating translation initiation. Both of these approaches were examined to determine whether wheat germ lysate was competent to support the TEV 5′-leader-mediated enhancement of cap-independent translation in vitro.
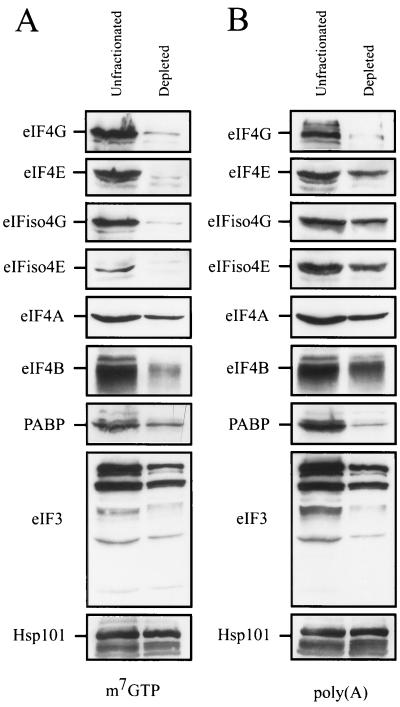
Because eIF4F (in which eIF4G and eIF4E are subunits) and eIFiso4F (in which eIFiso4G and eIFiso4E are subunits) bind m7GTP, the level of both factors in wheat germ lysate could be readily reduced through their binding to m7GTP-Sepharose. Western analysis confirmed that the levels of eIF4E and eIF4G and the levels of eIFiso4E and eIFiso4G were reduced by 90 to 95% (Fig. 1). The levels of eIF4A, eIF4B, and eIF3, factors known to associate with eIF4G proteins, were also reduced, as was that of PABP, which also physically interacts with eIF4G and eIFiso4G (26, 28). No reduction in the level of Hsp101, a control protein, was observed (Fig. 1).
FIG. 1.
Depletion of eIF4F and eIFiso4F from wheat germ lysate. Wheat germ lysate was incubated with m7GTP-Sepharose (A) or poly(A)-Sepharose (B) for 30 min. Western analysis was performed to determine the levels of eIF4G, eIF4E, eIFiso4G, eIFiso4E, eIF4A, eIF4B, eIF3, and PABP relative to that of unfractionated lysate. Western analysis of heat shock protein Hsp101 was performed as a control.
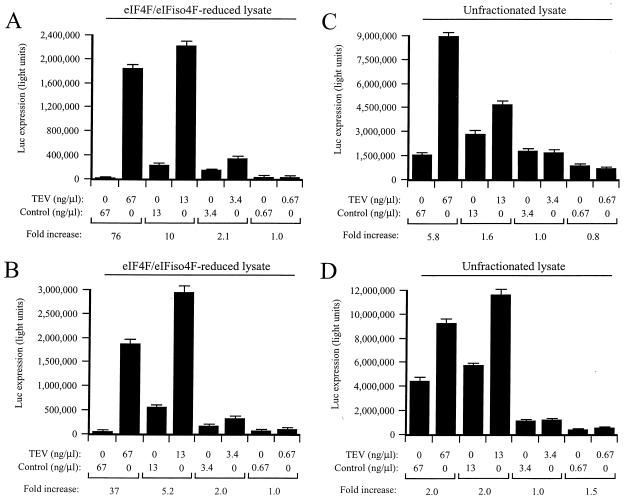
To examine whether cap-independent translation would be stimulated by the TEV 5′ leader in eIF4F/eIFiso4F-reduced lysate, luc mRNA with the TEV 5′ leader (i.e., TEV-luc-A50) or with a control leader of similar length (i.e., Con144-luc-A50) was translated. The constructs were translated as capped or uncapped mRNAs, and the extent to which the reporter mRNA was translated was determined by measuring luciferase activity. Translation from uncapped TEV-luc-A50 mRNA was 76-fold greater than that from uncapped control mRNA when the lysate was programmed with a high level of RNA (Fig. 2A). The degree to which the TEV 5′ leader stimulated cap-independent translation decreased with the decrease in RNA concentration. The same uncapped mRNAs were translated in unfractionated lysate using the same range of RNA concentrations (Fig. 2C). TEV-luc-A50 mRNA was translated only 5.8-fold higher than that of the control at the highest RNA concentration tested, and the translational advantage conferred by the TEV 5′ leader was lost at lower RNA concentrations. The addition of a 5′ cap to the TEV and control mRNAs reduced the ability of the TEV 5′ leader to promote translation by about twofold in the eIF4F/eIFiso4F-reduced and unfractionated lysates (Fig. 2B and D, respectively). These data suggest that the TEV 5′ leader can promote cap-independent translation in vitro but does so most under competitive conditions, e.g., when a high concentration of RNA is used and when the endogenous level of eIF4F and eIFiso4F has been reduced. The data also suggest that the presence of a 5′ cap, the binding site of eIF4F or eIFiso4F, reduces the translational advantage conferred by the TEV 5′ leader, suggesting functional redundancy. These observations are in good agreement with those made during in vivo translation (31).
FIG. 2.
The TEV IRES confers a translational advantage in vitro under competitive conditions. eIF4F/eIFiso4F-reduced (A and B) and unfractionated (C and D) wheat germ lysate was programmed with Con144-luc-A50 (i.e., control) or TEV-luc-A50 (i.e., TEV) mRNAs. The constructs were translated as uncapped (A and C) or capped (B and D) mRNAs at the indicated concentrations. The degree to which each mRNA was translated was determined by luciferase assays. Luciferase activity is indicated as the average (from 2 μl of lysate) of three translation reactions with the standard deviation for each construct shown. The degree to which the presence of the TEV 5′ leader increased translation relative to the control (i.e., fold increase) is indicated below each pair of mRNAs for each concentration tested.
When present in the intercistronic region of a discistronic mRNA, the TEV 5′ leader sequence can promote translation from the second cistron in vivo (31), indicating that it possesses IRES activity. To examine whether the TEV IRES can function in vitro, dicistronic mRNAs were constructed such that the TEV 5′ leader sequence or the control sequence of similar length (the same sequence as that present in the 5′ leader of the Con144-luc-A50 construct) was introduced into the intercistronic region upstream of the 5′-distal luc cistron. The GUS gene served as the 5′-proximal cistron in both constructs, and a 24-bp stem-loop structure (ΔG = −42.9 kcal/mol) was introduced upstream of the intercistronic region to prevent ribosome scanning from the 5′ terminus. The resulting uncapped mRNAs, i.e., GUS-SL-TEV-luc-A50 and GUS-SL-Con144-luc-A50, were translated in the eIF4F/eIFiso4F-reduced lysate using a range of RNA concentrations. The TEV IRES stimulated translation from the luc cistron as a function of the RNA concentration: up to a 73-fold increase in translation from the luc cistron was observed at the highest concentration tested, whereas little stimulation was observed at the lowest concentration (Fig. 3). These data demonstrate that the TEV 5′ leader sequence has IRES activity in vitro that is revealed under conditions of competitive translation.
FIG. 3.
The TEV IRES directs internal initiation in vitro under competitive conditions. eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with uncapped control (i.e., GUS-SL-Con144-luc-A50) or uncapped TEV IRES-containing (i.e., GUS-SL-TEV-luc-A50) dicistronic constructs at the concentrations indicated below the histograms. Luciferase activity is reported as the average (from 2 μl of lysate) of three translation reactions with the standard deviation for each construct shown. The degree to which the presence of the TEV 5′ leader increased translation relative to the control (i.e., fold increase) is indicated below each pair of mRNAs for each concentration tested.
TEV IRES activity requires eIF4G.
Because the TEV IRES functionally replaces the 5′ cap that normally serves as the binding site for eIF4F or eIFiso4F, we examined whether either of these factors was necessary for TEV IRES activity. The observation that the translation of an mRNA in eIF4F/eIFiso4F-reduced lysate improves substantially if the mRNA contains the TEV IRES suggests that this sequence promotes the recruitment of a factor that is required for translation initiation but whose availability is limited. Consequently, restoring the factor in question to the lysate would be expected to increase expression from the mRNA lacking the TEV IRES to an extent greater than that from the mRNA containing the TEV IRES, thereby reducing the translational advantage conferred by the TEV IRES to a level similar to that observed in unfractionated lysate, as seen in Fig. 2C.
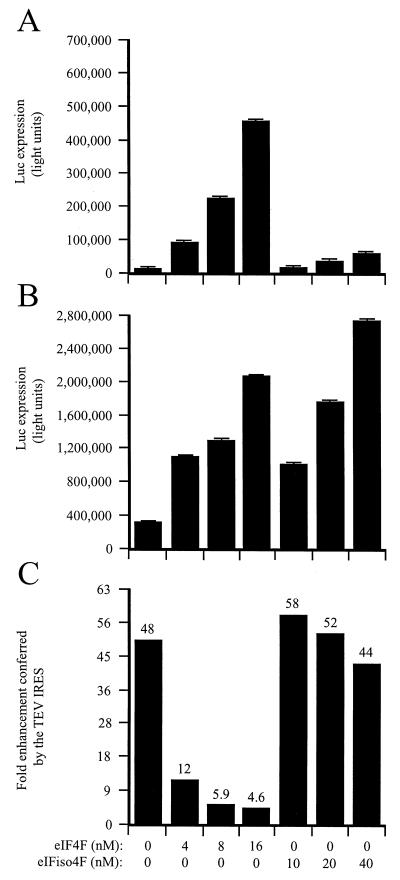
Therefore, to assay the requirement for eIF4F or eIFiso4F for TEV IRES function, the same uncapped, discistronic mRNA constructs used in Fig. 3 were translated in eIF4F/eIFiso4F-reduced lysate supplemented with either factor and their effect on the translation from each mRNA and the translational advantage conferred by the TEV IRES were determined. eIF4F and eIFiso4F purified from wheat do not contain eIF4A; therefore, the purified eIF4F used in these studies contained eIF4E and eIF4G whereas the purified eIFiso4F contained eIFiso4E and eIFiso4G. In eIF4F/eIFiso4F-reduced lysate, the TEV IRES increased translation from the 5′-distal luc cistron in GUS-SL-TEV-luc-A50 mRNA 48-fold relative to that from the GUS-SL-Con144-luc-A50 control mRNA (Fig. 4C). The addition of eIF4F increased expression from both the control (Fig. 4A) and TEV (Fig. 4B) constructs; however, expression from the control construct increased disproportionately as competition for eIF4F was relieved. Consequently, the translational advantage conferred by the TEV IRES was reduced in a dose-dependent manner (Fig. 4C). In contrast, although supplementation with eIFiso4F increased expression from both mRNAs, it did so equally for both (Fig. 4A and B) and therefore had little effect on TEV IRES activity (Fig. 4C). These data suggest that the TEV IRES recruits eIF4F when the factor is present in limited amounts but that the translational advantage conferred by the TEV IRES is lost when the concentration of eIF4F is no longer limiting.
FIG. 4.
eIF4F but not eIFiso4F mediates TEV IRES function in a dicistronic mRNA. eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with uncapped GUS-SL-Con144-luc-A50 mRNA (A) or uncapped GUS-SL-TEV-luc-A50 mRNA (B) and was supplemented with the indicated amounts of eIF4F or eIFiso4F. eIF4A was included at a ratio of 1:70 (for eIF4F assays) or 1:30 (for eIFiso4F assays). eIF4B was included at a ratio of 1:4 (for eIF4F assays) or 1:1.6 (for eIFiso4F assays). Each mRNA construct was translated in triplicate, and the average value and standard deviation for each construct are reported. Luciferase expression is indicated as light units from 2 μl of translation lysate. (C) Degree to which the TEV IRES stimulated translation relative to the control construct.
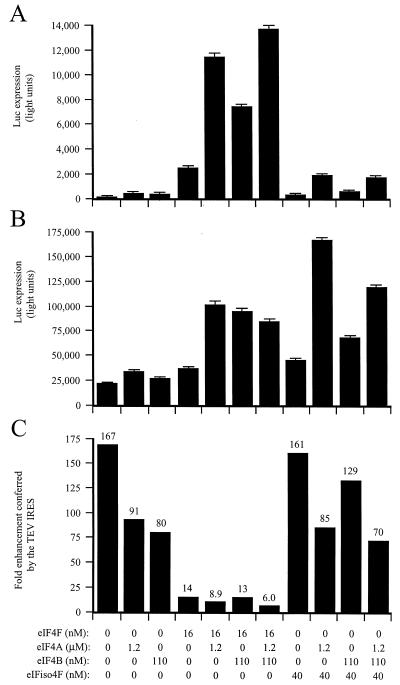
eIF4A and eIF4B were included in the above assays as they increase the activity of eIF4F and eIFiso4F. To examine whether they are required for TEV IRES activity, the TEV and control dicistronic mRNAs were translated in eIF4F/eIFiso4F-reduced lysate supplemented with either factor individually or with both factors in combination (Fig. 5). Supplementation of the depleted lysate with 16 nM eIF4F (which lacks eIF4A) increased expression from the TEV and control mRNAs but did so preferentially for the control mRNA. Consequently, the addition of eIF4F reduced the translational advantage conferred by the TEV IRES from a level of a 167-fold enhancement of translation (in the absence of added eIF4F) to just 14-fold enhancement (in the presence of added eIF4F), demonstrating that eIF4F alone is sufficient to relieve the translational advantage conferred by the TEV IRES. In contrast, addition of 40 nM eIFiso4F (which lacks eIF4A) did not reduce the translational advantage conferred by the TEV IRES (Fig. 5C). Addition of either eIF4A or eIF4B increased translation from both mRNAs but, as with eIF4F, they increased translation from the control mRNA disproportionately. Thus, addition of eIF4A or eIF4B alone reduced the translational advantage conferred by the TEV IRES by approximately twofold and the combination of eIF4A or eIF4B with eIF4F reduced the translational advantage conferred by the TEV IRES to a level of just sixfold enhancement of translation (Fig. 5C). Any combination of eIF4A, eIF4B, and eIFiso4F had only a moderate effect on the translational advantage conferred by the TEV IRES. These results suggest that eIF4A or eIF4B plays a moderate role in the function of the TEV IRES. However, the degree to which they are required cannot be fully assessed as a moderate level of each factor remains in the lysate following binding to m7GTP-Sepharose (Fig. 1).
FIG. 5.
eIF4A and eIF4B enhance eIF4F-mediated TEV IRES function. eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with uncapped GUS-SL-Con144-luc-A50 mRNA (A) or uncapped GUS-SL-TEV-luc-A50 mRNA (B) and was supplemented with the indicated amounts of initiation factors. Each mRNA construct was translated in triplicate, and the average value and standard deviation for each construct are reported. Luciferase expression is indicated as light units from 2 μl of translation lysate. (C) Degree to which the TEV IRES stimulated translation relative to the control construct.
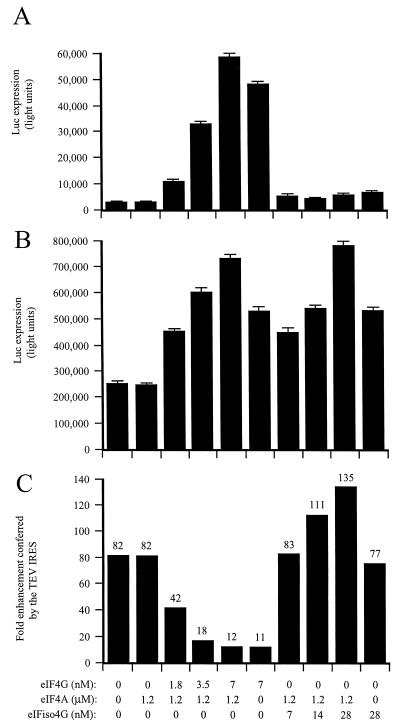
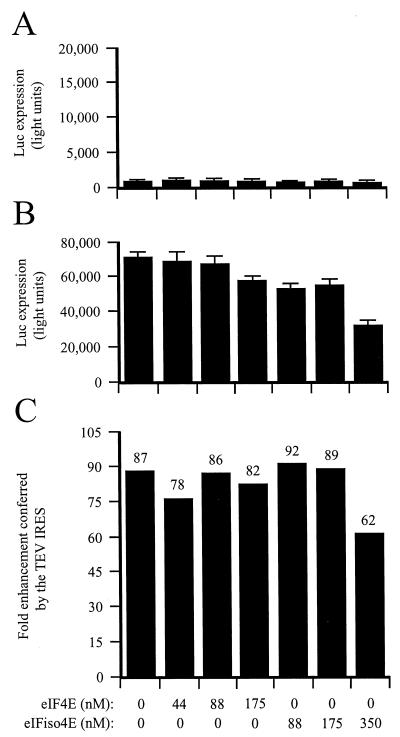
To determine which subunit of eIF4F, i.e., eIF4E or eIF4G, is required for TEV IRES function, the TEV and control dicistronic mRNAs were translated in eIF4F/eIFiso4F-reduced lysate supplemented with recombinant eIF4E or eIF4G. Addition of eIF4G reduced the translational advantage conferred by the TEV IRES from a level of an 82-fold enhancement of translation (in the absence of added eIF4G) to just 11-fold enhancement (in the presence of added eIF4G) (Fig. 6C), demonstrating that eIF4G alone (i.e., in the absence of eIF4E) is sufficient for TEV IRES function. In contrast, the addition of recombinant eIFiso4G had little effect on TEV IRES function (Fig. 6C), a result that is in good agreement with the lack of effect on TEV IRES activity exhibited by eIFiso4F (Fig. 4C). Supplementation of the lysate with recombinant eIF4E or eIFiso4E also had little effect on TEV IRES function (Fig. 7C), although both factors inhibited expression from both the control and TEV-containing mRNAs somewhat at a high molar concentration (Fig. 7A and B).
FIG. 6.
eIF4G is the subunit of eIF4F that mediates TEV IRES function. eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with uncapped GUS-SL-Con144-luc-A50 mRNA (A) or uncapped GUS-SL-TEV-luc-A50 mRNA (B) and was supplemented with the indicated amounts of eIF4G, eIFiso4G, and eIF4A. Each mRNA construct was translated in triplicate, and the average value and standard deviation for each construct are reported. Luciferase expression is indicated as light units from 2 μl of translation lysate. (C) Degree to which the TEV IRES stimulated translation relative to the control construct.
FIG. 7.
eIF4E and eIFiso4E do not mediate TEV IRES function. eIF4F/eIFiso4F-reduced wheat germ lysate was programmed with uncapped GUS-SL-Con144-luc-A50 mRNA (A) or uncapped GUS-SL-TEV-luc-A50 mRNA (B) and was supplemented with the indicated amounts of eIF4E or eIFiso4E. Each mRNA construct was translated in triplicate, and the average value and standard deviation for each construct are reported. Luciferase expression is indicated as light units from 2 μl of translation lysate. (C) Degree to which the TEV IRES stimulated translation relative to the control construct.
Exogenous TEV IRES RNA sequesters eIF4G.
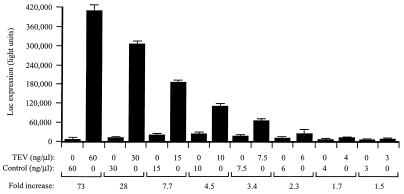
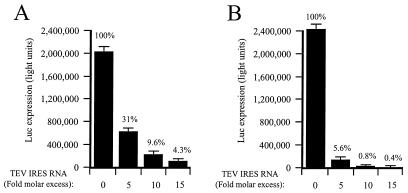
If the TEV 5′ leader confers a competitive translational advantage to an mRNA by virtue of its greater ability to recruit a general translation initiation factor such as eIF4G, the presence of the TEV IRES RNA in trans in a translation lysate would be expected to result in the sequestration of the factor and in a reduction in the translation of all mRNA constructs but should affect most those mRNAs least competent to recruit the factor. To test this prediction, the effect of TEV IRES RNA supplied in trans on the translation of 0.67/μl ng of uncapped TEV-luc-A50 and uncapped Con144-luc-A50 mRNAs in unfractionated lysate was examined. In unfractionated lysate, the TEV-containing and control mRNAs were translated to similar extents in the absence of exogenous TEV IRES RNA (Fig. 8), results that are in good agreement with those presented in Fig. 2C for these mRNAs at the same concentration. Addition of TEV IRES RNA reduced expression from both mRNAs, but translation from Con144-luc-A50 mRNA was reduced up to 10-fold more by the TEV IRES RNA than was translation from TEV-luc-A50 mRNA (Fig. 8). These data indicate that the TEV IRES RNA sequesters a factor that is required for the translation of mRNAs whether or not they contain the TEV 5′ leader. Moreover, the disproportionate reduction in translation observed for the control mRNA following addition of the TEV IRES RNA and sequestration of the factor suggests that an uncapped mRNA without the TEV 5′ leader competes less efficiently for the factor.
FIG. 8.
TEV IRES RNA inhibits translation when present in trans. Unfractionated wheat germ lysate was programmed with uncapped TEV-luc-A50 (A) or uncapped Con144-luc-A50 (B) mRNAs at a concentration of 0.67 ng/μl. TEV IRES RNA was added in trans to each lysate at the indicated molar ratio, and the degree to which each mRNA was translated was determined by luciferase assays. Luciferase activity is indicated as the average (from 2 μl of lysate) of three translation reactions, with the standard deviation for each construct shown. The degree to which an mRNA was translated relative to expression from the mRNA in lysate containing no TEV IRES RNA is indicated as a percentage.
The TEV IRES is active in lysate depleted of PABP.
Because PABP interacts with eIF4G and eIFiso4G (16, 26, 28, 37, 42), recruits eIF4G to an mRNA in the absence of a cap or functional eIF4E (the cap-binding subunit of eIF4F) (38, 43), and promotes translational initiation both in cis and in trans (32), we examined whether reducing the concentration of PABP present in lysate would affect the extent to which the TEV 5′ leader sequence functions to stimulate cap-independent translation. Lysate was depleted of PABP following incubation with poly(A)-agarose. The reduction in PABP was confirmed by Western blotting (Fig. 1). Because eIF4G, eIFiso4G, and eIF4B also bind poly(A) RNA, albeit with a lower affinity than does PABP, and because PABP interacts with eIF4G, eIFiso4G, and eIF4B, which in turn interact with eIF4A and eIF3, the levels of these initiation factors were expected to be reduced following incubation with poly(A)-agarose; this was also confirmed by Western analysis (Fig. 1). No reduction in the control protein, Hsp101, was observed (Fig. 1).
To examine the activity of the TEV 5′ leader in the PABP-reduced lysate, monocistronic TEV-luc-A50 and the control construct, Con144-luc-A50, were translated over a range of RNA concentrations (Fig. 9A). Translation from uncapped TEV-luc-A50 mRNA was up to 95-fold greater than that from uncapped control mRNA when the lysate was programmed with a high level of RNA (Fig. 9A). As was observed in eIF4F/eIFiso4F-reduced lysate, the degree to which the TEV 5′ leader stimulated cap-independent translation decreased with a reduction in RNA concentration. The TEV and control dicistronic mRNA constructs used in Fig. 3 were then translated in the PABP-reduced lysate to determine whether internal initiation could be promoted by the TEV IRES. In the PABP-reduced lysate, the TEV IRES increased internal initiation up to 79-fold relative to that for the control mRNA (Fig. 9B). These data suggest that a reduction in PABP (as well as the partial reduction in eIF4F and eIFiso4F as shown in Fig. 1) increases the extent to which the TEV IRES promotes translation.
FIG. 9.
The TEV IRES confers a translational advantage to monocistronic and dicistronic mRNAs in vitro under competitive conditions. PABP-reduced wheat germ lysate was programmed with uncapped Con144-luc-A50 (i.e., control) or uncapped TEV-luc-A50 (i.e., TEV) mRNAs (A) and with uncapped control (GUS-SL-Con144-luc-A50) or TEV IRES-containing (GUS-SL-TEV-luc-A50) dicistronic mRNAs (B) at the concentrations indicated, and the degree to which each mRNA was translated was determined by luciferase assays. Luciferase activity is indicated as the average (from 2 μl of lysate) of three translation reactions, with the standard deviation for each construct shown. The degree to which the presence of the TEV IRES increased translation relative to that for the control (i.e., fold increase) is indicated below each pair of mRNAs for each concentration tested.
eIF4F is sequestered by TEV IRES RNA present in trans.
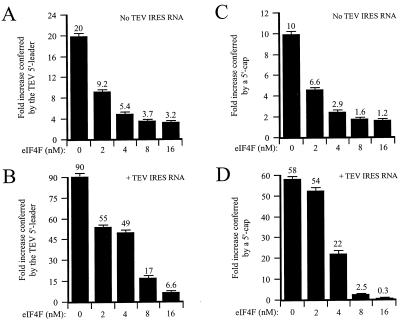
To determine whether the translational advantage conferred by the TEV 5′ leader in the PABP-reduced lysate was a result of competition for eIF4F, monocistronic TEV-luc-A50 mRNA and the control, Con144-luc-A50 mRNA, were translated in PABP-reduced (and eIF4G/eIFiso4G-reduced) lysate in a series of reactions in which the concentration of eIF4F was raised by adding purified eIF4F. The ratio of expression from the TEV-containing mRNA to that from the control mRNA (i.e., fold increase in expression conferred by the TEV IRES when present as the 5′ leader) was calculated and displayed as the histograms in Fig. 10A. Without the addition of purified eIF4F, the presence of the TEV-IRES increased translation 20-fold (Fig. 10A, left histogram). Addition of eIF4F reduced the translational advantage conferred by the TEV IRES to just 3.2-fold enhancement at 16 nM eIF4F (Fig. 10A). This was a result of a preferential increase in translation from the control construct (data not shown). Translation of capped and uncapped Con144-luc-A50 mRNAs in the PABP-reduced lysate was performed in parallel, yielding similar results for the function of the 5′ cap (Fig. 10C). The presence of the 5′ cap increased translation 10-fold (Fig. 10C, left histogram). Addition of eIF4F reduced the translational advantage conferred by the cap to just a 1.2-fold enhancement at 16 nM eIF4F (Fig. 10C), which resulted from a preferential increase in the translation of the control construct (data not shown).
FIG. 10.
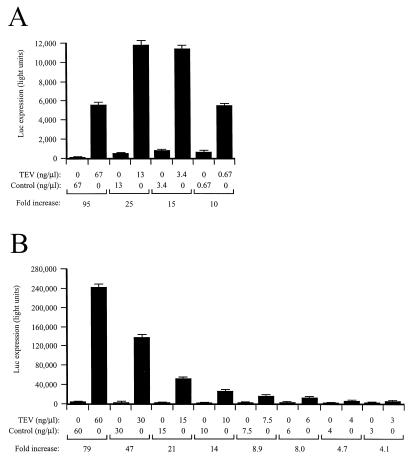
eIF4F is sequestered by TEV IRES RNA supplied in trans. (A and B) PABP-reduced wheat germ lysate was programmed with uncapped Con144-luc-A50 or uncapped TEV-luc-A50 mRNA in the absence (A) or presence (B) of a fivefold molar excess of TEV IRES RNA supplied in trans. Purified eIF4F was added in increasing amounts to the reaction mixtures. The histograms show the ratios of expression from the TEV-containing mRNA to that from the control mRNA (i.e., fold increases in expression conferred by the TEV IRES when present as the 5′ leader). (C and D) PABP-reduced wheat germ lysate was programmed with uncapped Con144-luc-A50 or capped Con144-luc-A50 mRNA in the absence (C) or presence (D) of a fivefold molar excess of TEV IRES RNA supplied in trans. Purified eIF4F was added in increasing amounts to the reaction mixtures. The histograms show the ratios of expression from the capped mRNA to that from uncapped mRNA (i.e., fold increase in expression conferred by the 5′ cap). eIF4A was included at a ratio of 1:70 and eIF4B was included at a ratio of 1:4 for the reactions in all panels.
To determine whether eIF4F was sequestered when TEV IRES RNA was supplied in trans, uncapped, monocistronic TEV-luc-A50 and control mRNAs were translated in PABP-reduced lysate in the presence of TEV IRES RNA supplied in trans. To determine whether eIF4F was sequestered by the TEV IRES RNA, increasing amounts of purified eIF4F were added to the lysate. The ratios of expression from the TEV-containing mRNA to that from the control mRNA were calculated and displayed as the histograms in Fig. 10B. Without the addition of purified eIF4F, TEV IRES RNA supplied in trans in a 5-fold molar excess relative to the test mRNAs resulted in a level of translation from the TEV-luc-A50 mRNA that was 90-fold greater than that from the control mRNA (Fig. 10B, left histogram). This is substantially higher than the level of TEV IRES-mediated stimulation observed when no TEV IRES RNA was added in trans (Fig. 10A) and was due to a preferential reduction in translation from the control mRNA (data not shown), as was seen in Fig. 8. The increase in the ratio of expression from the TEV-containing mRNA relative to that from the control mRNA in the presence of TEV IRES RNA is consistent with the sequestration of a general translation factor by the TEV IRES for which the TEV-containing mRNA, but not the control mRNA, can effectively compete. Addition of eIF4F reduced the ratio of the expression from TEV-luc-A50 mRNA to that from control mRNA from the 90-fold observed in the absence of added eIF4F to just 6.6-fold in the presence of 16 nM eIF4F (Fig. 10B), which was a greater reduction (90-fold reduced to 6.6-fold) than that observed for lysate not containing the TEV IRES in trans (20-fold reduced to 3.2-fold; Fig. 10A).
Similar observations were made for the effect on 5′ cap function when capped and uncapped Con144-luc-A50 mRNAs were translated in the PABP-reduced lysate in the presence or absence of TEV IRES RNA in trans. The ratio of translation expression from capped mRNA to that from uncapped mRNA was calculated and displayed as the histograms in Fig. 10C and D. Addition of TEV IRES RNA in trans increased the extent to which the 5′ cap enhanced translation from 10-fold in the absence of TEV IRES RNA (Fig. 10C, left histogram) to 58-fold in the presence of TEV IRES RNA (Fig. 10D, left histogram). The increase in cap dependency resulted from a preferential inhibition of translation from the uncapped mRNA (data not shown). This observation suggests that the TEV IRES RNA and the 5′ cap compete for the same factor and that an uncapped mRNA competes only poorly for the factor. When TEV IRES RNA was present in trans, addition of eIF4F reduced the ratio of translation expression from capped mRNA to that from uncapped mRNA from the 58-fold observed in the absence of added eIF4F to 0.3-fold in the presence of 16 nM eIF4F (Fig. 10D), which was a greater reduction than that observed for lysate not containing the TEV IRES in trans (10-fold reduced to 1.2-fold; Fig. 10C). These data demonstrate that the effect of TEV IRES RNA supplied in trans has similar effects on cap- and TEV IRES-mediated translation in that it increased the translational advantage conferred by each and that their advantage was relieved by the addition of eIF4F. Consequently, the TEV IRES and the cap are functionally similar in that each confers a translational advantage to an mRNA under competitive translation conditions which is relieved when eIF4F is no longer limiting.
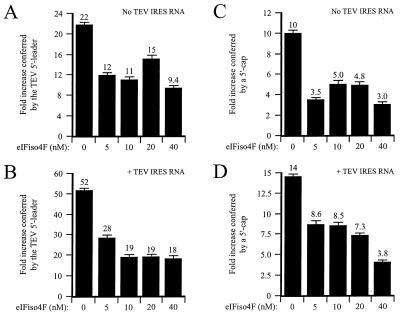
In contrast to eIF4F, eIFiso4F did not affect the function of the TEV IRES when this sequence was present in the intercistronic region of a dicistronic mRNA as shown in Fig. 4. However, eIFiso4F is more 5′ end dependent than is eIF4F in its functional interaction with mRNAs (9a). To determine whether eIFiso4F was sequestered when TEV IRES RNA was supplied in trans, the same translation assays described for Fig. 10 were carried out using eIFiso4F instead of eIF4F for the supplementation. As in the previous experiment, the presence of TEV IRES RNA in trans increased the translational advantage exhibited by the TEV-luc-A50 mRNA 52-fold relative to that for the control mRNA (Fig. 11B), which was greater than that observed in the same lysate lacking TEV IRES RNA in trans (Fig. 11A, left histogram). When TEV IRES RNA was present in trans, addition of eIFiso4F reduced the ratio of the expression of TEV-luc-A50 mRNA to that of control mRNA from the 52-fold observed in the absence of added eIFiso4F to 18-fold in the presence of 40 nM eIFiso4F (Fig. 11B), which is a degree of reduction similar to that observed for lysate not containing the TEV IRES in trans (22-fold reduced to 9.4-fold; Fig. 11A). These data suggest that when the TEV IRES is present in a 5′-proximal position, eIFiso4F can partially relieve the translational advantage that it confers to an mRNA; however, it is not as effective as eIF4F in this regard. Similar results were obtained when the ratio of the expression of capped mRNA to that of uncapped mRNA was examined (Fig. 11C and D).
FIG. 11.
eIFiso4F is sequestered by the TEV IRES supplied in trans. (A and B) PABP-reduced wheat germ lysate was programmed with uncapped Con144-luc-A50 or uncapped TEV-luc-A50 mRNA in the absence (A) or presence (B) of a fivefold molar excess of TEV IRES RNA supplied in trans. Purified eIFiso4F was added in increasing amounts to the reaction mixtures. The histograms show the ratios of expression from the TEV-containing mRNA to that from the control mRNA (i.e., fold increases in expression conferred by the TEV IRES when present as the 5′ leader). (C and D) PABP-reduced wheat germ lysate was programmed with uncapped Con144-luc-A50 or capped Con144-luc-A50 mRNA in the absence (C) or presence (D) of a fivefold molar excess of TEV IRES RNA supplied in trans. Purified eIFiso4F was added in increasing amounts to the reaction mixtures. The histograms show the ratios of expression from the capped mRNA to that from uncapped mRNA (i.e., fold increases in expression conferred by the 5′ cap). eIF4A was included at a ratio of 1:30 and eIF4B was included at a ratio of 1:1.6 for the reactions in all panels.
PABP stimulates TEV IRES function.
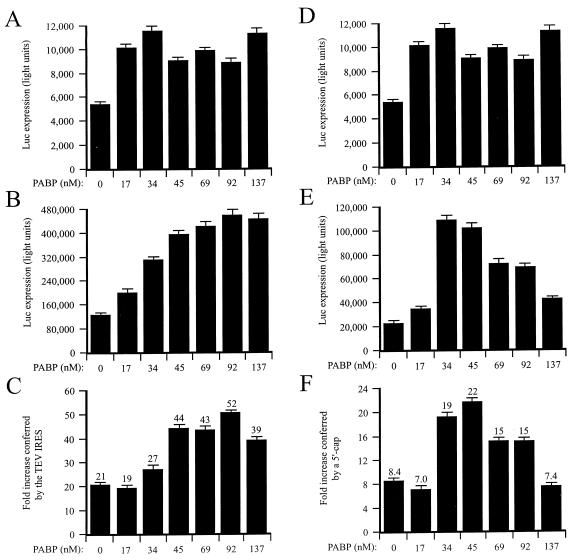
Although the TEV IRES can promote cap-independent translation of a nonpolyadenylated mRNA, the presence of a poly(A) tail enhances its function (10, 31), an observation suggesting a functional interaction between the TEV IRES and PABP. To examine whether the presence of PABP might affect TEV 5′ leader activity, monocistronic TEV-luc-A50 and control mRNAs were translated in the PABP-reduced lysate supplemented with increasing amounts of purified PABP (Fig. 12). In the absence of PABP, translation from uncapped, monocistronic TEV-luc-A50 mRNA was 21-fold greater than that from the uncapped, control mRNA (Fig. 12C). The addition of PABP increased translation from the control (Fig. 12A) and TEV-containing (Fig. 12B) mRNAs; however, translation from the TEV-luc-A50 mRNA benefited most from the increased availability of PABP. Consequently, the degree to which the TEV IRES enhanced translation increased to 52-fold in the presence of 92 nM PABP (Fig. 12C). Similar observations were made for the function of the 5′ cap: capped Con144-luc-A50 mRNA was translated 8.4-fold greater than uncapped Con144-luc-A50 mRNA in the PABP-reduced lysate. Addition of PABP increased translation from uncapped (Fig. 12D) and capped (Fig. 12E) mRNAs but did so most from the capped construct, thereby increasing the translational advantage conferred by the cap such that the 5′ cap stimulated translation 19- to 22-fold in the presence of 34 to 45 nM PABP (Fig. 12F). These data illustrate that PABP can stimulate the function of the TEV IRES as it does for a 5′ cap. Because PABP stabilizes the binding of eIF4F to a 5′ cap (45), PABP is expected to increase the function of the cap in promoting translation. The stimulatory effect of PABP on the function of the TEV IRES may also be a consequence of PABP stabilizing the functional interaction between eIF4F and the TEV leader sequence. Note that at the higher concentrations of PABP (e.g., 137 nM), the stimulation of translation from the capped mRNA afforded by PABP was less than that observed at lower PABP concentrations (e.g., 34 to 45 nM), which was not observed for the uncapped mRNAs. The lower stimulation of capped mRNA translation may represent PABP that is in excess (relative to the binding capacity of the input mRNA), which may compete with the bound PABP for other factors (such as eIF4F) needed for translation, resulting in an apparently lower level of stimulation. The fact that the mRNA containing the TEV IRES is not similarly affected suggests that it may very efficiently recruit eIF4F, thus rendering the mRNA less susceptible to the effect of excess PABP.
FIG. 12.
PABP stimulated TEV IRES function. PABP-reduced wheat germ lysate was programmed with uncapped GUS-SL-Con144-luc-A50 (A) or uncapped GUS-SL-TEV-luc-A50 (B) mRNA and supplemented with the indicated amounts of PABP. Each mRNA construct was translated in triplicate, and the average value and standard deviation for each construct are reported. Luciferase expression is indicated as light units from 2 μl of translation lysate. (C) Degree to which the TEV IRES stimulated translation relative to that for the control construct (i.e., fold increase). (D and E) PABP-reduced wheat germ lysate was also programmed with uncapped (D) or capped (E) GUS-SL-Con144-luc-A50 mRNA and supplemented with the indicated amounts of PABP. (F) Degree to which the 5′ cap stimulated translation relative to that for the control construct (i.e., fold increase).
DISCUSSION
TEV, like animal picornaviruses, naturally lacks a 5′ cap, the binding site for eIF4F. Despite what would appear to be a handicap, the genomic mRNA of TEV is efficiently translated by means of a 5′ leader sequence that recruits the translational machinery in the absence of a cap. In addition to the 5′ leader, a virus-encoded protein (Vpg) that is covalently attached to the 5′ terminus of TEV or turnip mosaic virus (a potyvirus similar to TEV) genomic RNA has been shown to interact with eIF4E or eIFiso4E, respectively (41a, 46) although the consequence of this for viral translation remains unknown. The TEV 5′ leader does not require Vpg to confer cap-independent translation, as it enhances translation in the absence of any viral protein. In addition to conferring cap-independent translation, the TEV 5′ leader promotes internal initiation (31), an observation demonstrating that IRES elements are present in plant viral mRNAs and that plant ribosomes, like those of animals, are capable of internal initiation. In this study, we investigated which initiation factors are required for TEV IRES function and found that eIF4G plays as central a role in the function of the TEV IRES as it does for the IRES elements of animal picornaviruses, such as EMCV, foot-and-mouth disease virus, and Theiler murine encephalomyelitis virus (19, 20, 34, 35, 36).
The EMCV IRES promotes 40S ribosomal subunit binding, in part by the ability of eIF4G to bind to a defined region within the IRES (20, 34, 35). The binding of eIF4G to the IRES is considered a prerequisite for the recruitment of eIF3, which in turn promotes 40S ribosomal subunit binding. Interestingly, infection by several picornaviruses, such as poliovirus, results in cleavage of mammalian eIF4GI and eIF4GII (13). Cleavage of eIF4G removes the N-terminally located eIF4E-binding site, inhibiting the participation of the cleaved eIF4G in cap-dependent translation. However, the C-terminal region of eIF4G, which is released by the cleavage event, contains the binding sites for eIF3 and eIF4A and is sufficient for EMCV IRES function (35). As little as the central one-third of eIF4G has been shown to be sufficient for specific binding to the EMCV IRES (35). However, cleavage of eIF4G is not essential for it to mediate translation from the an IRES. Indeed, no cleavage of eIF4G occurs following infection by hepatitis A virus (HAV) or EMCV (reviewed in reference 3), and the IRES present in HAV requires intact eIF4G for its function (4). Whether TEV employs a strategy similar to that of poliovirus, in which eIF4G is targeted for cleavage following infection, or one more similar to that of EMCV or HAV, in which eIF4G is not cleaved, remains to be determined. In several aspects, the TEV IRES differs substantially from those of animal picornaviruses in that it is shorter and less structured and contains no AUG triplets upstream of the initiation codon. This might suggest that the functional interactions between the plant IRES and the translational machinery are simpler than those required in animal cells. However, the region of the EMCV IRES that is responsible for eIF4G binding (34, 35) is similar in length to the TEV IRES (31). The greater structural complexity of the picornavirus IRESs may be involved in aspects of the viral life cycle in ways that have not yet been elucidated.
The TEV IRES promoted cap-independent translation and internal initiation in wheat germ lysate, demonstrating that the IRES is capable of functioning in vitro. However, IRES function was observed only at higher mRNA concentrations (>13 ng/μl) or when the endogenous levels of eIF4F and eIFiso4F in the lysate had been reduced. These results indicate that the TEV IRES confers a translational advantage only under conditions of competitive translation. Similar observations were made for the 5′ cap: the stimulatory effect of a cap was greatest at high mRNA concentrations (9a) or in lysate in which the level of the translational initiation machinery had been reduced. It should be noted that the depletion of eIF4F and eIFiso4F from the lysate was not complete, as a low level of each factor remained following depletion (Fig. 1), which provided the basis for the increase in cap dependency observed for the depleted lysate (Fig. 10C and 11C). These results suggest that the TEV IRES efficiently recruited, either directly or indirectly, a trans-acting factor(s) involved in general translation which is present in excess in unfractionated lysate but which is limiting in depleted lysate. When the factor is present in excess, those elements such as a 5′ cap or the TEV IRES are not able to confer a translational advantage to an mRNA. Therefore, the translational advantage conferred by a 5′ cap or the TEV IRES under conditions of competitive translation where the factor is present in limiting amounts would be increasingly reduced as the concentration of the limiting factor is increased. By supplementing depleted lysate with initiation factors alone or in combination, the eIF4G subunit of eIF4F was identified as the factor responsible for TEV IRES function. eIF4G is a subunit of eIF4F the latter of which exhibited an effect on TEV IRES activity similar to that observed for eIF4G, suggesting that the viral mRNA may utilize eIF4G either alone or when present as part of eIF4F. Little or no effect on TEV IRES function was observed when eIF4A or eIF4B was tested either alone or in combination, suggesting that the effect exhibited by eIF4G was specific to that factor. However, eIF4A and eIF4B increased the effectiveness of eIF4G, particularly when eIF4G was present as part of eIF4F. eIF4A and eIF4B have been shown to stimulate the RNA-dependent ATPase and ATP-dependent RNA-unwinding activities of eIF4F (1, 2, 5, 14, 18, 22, 23, 24, 40, 41). The binding of mammalian eIF4G to the EMCV IRES was strongly stimulated by eIF4A and eIF4B, and both were required for the formation of the 48S initiation complex (34, 35), observations that are consistent with the ability of plant eIF4A and eIF4B to increase the effectiveness of eIF4G in mediating TEV IRES function during translation. The observation that the addition of exogenous eIF4A stimulated, but was not essential for, the eIF4G-mediated function of the TEV IRES may be due to the level of eIF4A that remained in lysates following depletion of eIF4F and eIFiso4F or PABP.
Two eIF4G proteins are expressed in plants, animals, and yeast and are only 30, 46, and 53% conserved within these groups, respectively (K. Browning, personal communication; 11, 12), suggesting the possibility that they have functionally specialized. In plants, eIF4G (as part of eIF4F) exhibits a greater ability to facilitate translation from nonstandard mRNAs, i.e., those that contain secondary structure proximal to the 5′ cap, those that lack a cap, and those that contain more than one cistron (9a). In contrast, eIFiso4G (as part of eIFiso4F) functions optimally in supporting translation from capped, monocistronic mRNAs. These observations may explain why eIF4F and not eIFiso4F mediated TEV IRES function when this sequence was present in the intercistronic region of a dicistronic mRNA. When the IRES was present as the 5′ leader, supplementation with eIFiso4F partially reduced the translational advantage conferred by the TEV sequence although to a much lesser extent than that observed for eIF4F, particularly when the molar concentration of each factor is considered. When this is done, eIF4F was approximately 1 order of magnitude more effective in mediating TEV IRES function than was eIFiso4F when the IRES was present as the 5′ leader of an uncapped mRNA. These observations suggest that the translational strategy of the TEV IRES may have evolved to exploit the greater ability of eIF4F to promote internal initiation and cap-independent translation. The observation that the TEV IRES can functionally discriminate between the two divergent plant eIF4G proteins, at least in vitro, raises the possibility that animal picornavirus IRES elements might exhibit a preference for one of the two eIF4G proteins that are expressed in animals.
Depletion of PABP increased the cap dependency of the lysate. It should be noted that depletion of PABP resulted in a reduction in the level of eIF4F and eIFiso4F, presumably through their interaction with PABP, as observed previously (26). PABP at a concentration of 34 to 45 nM stimulated cap function, whereas concentrations higher or lower than this reduced the competitive advantage that a cap conferred to an mRNA (Fig. 12F). These data suggest that PABP at an appropriate concentration is required for full function of the cap. Similarly, TEV IRES function was stimulated by 45 to 137 nM PABP (Fig. 12C), data indicating that although the TEV IRES can function when the concentration of PABP is low, PABP at an appropriate concentration is necessary for full TEV IRES function. This result is in good agreement with our previous observation that, although the TEV IRES can enhance translation from a poly(A)− mRNA, its function is stimulated by the presence of a poly(A) tail (10). Analysis of the TEV IRES had revealed two sequence elements required for cap-independent translation (31). The combinatorial effect of the two elements on cap-independent translation was multiplicative and dependent on the poly(A) tail. This observation and the fact that PABP was required for full IRES function in vitro suggest that PABP may assist in the recruitment of eIF4G to the IRES. This is analogous to the effect that the interaction of PABP has on the ability of the 5′ cap to recruit eIF4F. PABP interacts with eIF4G (16, 26, 28, 37, 42), which results in an increase in the affinity of eIF4F for the 5′ cap structure as well as an increase in the binding affinity of PABP for poly(A) RNA (28, 45), suggesting that the physical interaction between PABP and eIF4F serves to stabilize their binding to their respective binding sites. Whether eIF4G binds directly to the TEV IRES or requires other initiation factors such as eIF4A and eIF4B, as is the case for the mammalian EMCV IRES (34, 35), or PABP remains to be determined. It is also possible that recruitment of eIF4G to the TEV IRES requires the assistance of a trans-acting factor similar to the requirement for polypyrimidine tract-binding protein (PTB) for Theilers' murine encephalomyelitis virus or the requirement for PTB and the IRES trans-acting factor 45 (ITAF45) protein for foot-and-mouth disease virus (36). These trans-acting factors may promote the binding of eIF4G directly or act as RNA chaperones that maintain the structure of the IRES in a conformation that facilitates optimal eIF4G binding. Identification of those proteins that bind to the TEV IRES will be necessary to determine which picornavirus translational strategy this plant member of the supergroup has adopted for its own expression.
ACKNOWLEDGMENTS
This work was supported by grants from the USDA (NRICGP 99-35301-7866 and 00-35301-9086).
I thank Karen Browning for the purified initiation factors and initiation factor antiserum used in this study and Christian Caldwell for technical assistance.
REFERENCES
- 1.Abramson R D, Browning K S, Dever T E, Lawson T G, Thach R E, Ravel J M, Merrick W C. Initiation factors that bind mRNA. A comparison of mammalian factors with wheat germ factors. J Biol Chem. 1988;263:5462–5467. [PubMed] [Google Scholar]
- 2.Altmann M, Muller P P, Wittmer B, Ruchti F, Lanker S, Trachsel H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham G J, Jackson R J. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 4.Borman A M, Kean K M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- 5.Browning K S, Lax S R, Ravel J M. Identification of two messenger RNA cap binding proteins in wheat germ. Evidence that the 28-kDa subunit of eIF-4B and the 26-kDa subunit of eIF-4F are antigenically distinct polypeptides. J Biol Chem. 1987;262:11228–11232. [PubMed] [Google Scholar]
- 6.Browning K S, Maia D M, Lax S R, Ravel J M. Identification of a new protein synthesis initiation factor from wheat germ. J Biol Chem. 1987;262:538–541. [PubMed] [Google Scholar]
- 7.Browning K S, Webster C, Roberts J K, Ravel J M. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem. 1992;267:10096–10100. [PubMed] [Google Scholar]
- 8.Bushell M, Wood W, Carpenter G, Pain V M, Morley S J, Clemens M J. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J Biol Chem. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- 9.Carrington J C, Freed D D. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Gallie D R, Browning K S. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem. 2001;276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- 10.Gallie D R, Tanguay R L, Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are synergistic regulators of translation. Gene. 1995;165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 11.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Trachsel H, Sonenberg N. Tif4631 and Tif4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor-4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grifo J A, Abramson R D, Satler C A, Merrick W C. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 15.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang S K, Krausslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaramillo M, Dever T E, Merrick W C, Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991;11:5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolupaeva V G, Hellen C U, Shatsky I N. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 20.Kolupaeva V G, Pestova T V, Hellen C U, Shatsky I N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 21.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 22.Lawson T G, Lee K A, Maimone M M, Abramson R D, Dever T E, Merrick W C, Thach R E. Dissociation of double-stranded polynucleotide helical structures by eukaryotic initiation factors, as revealed by a novel assay. Biochemistry. 1989;28:4729–4734. doi: 10.1021/bi00437a033. [DOI] [PubMed] [Google Scholar]
- 23.Lax S, Browning K S, Maia D M, Ravel J M. ATPase activities of wheat germ initiation factors 4A, 4B, and 4F. J Biol Chem. 1986;261:15632–15636. [PubMed] [Google Scholar]
- 24.Lax S, Fritz W, Browning K, Ravel J. Isolation and characterization of factors from wheat germ that exhibit eukaryotic initiation factor 4B activity and overcome 7-methylguanosine 5′-triphosphate inhibition of polypeptide synthesis. Proc Natl Acad Sci USA. 1985;82:330–333. doi: 10.1073/pnas.82.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lax S R, Lauer S J, Browning K S, Ravel J M. Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol. 1986;118:109–128. doi: 10.1016/0076-6879(86)18068-2. [DOI] [PubMed] [Google Scholar]
- 26.Le H, Browning K S, Gallie D R. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem. 2000;275:17452–17462. doi: 10.1074/jbc.M001186200. [DOI] [PubMed] [Google Scholar]
- 27.Le H, Chang S-C, Tanguay R L, Gallie D R. The wheat poly(A)-binding protein functionally complements pab1 in yeast. Eur J Biochem. 1997;243:350–357. doi: 10.1111/j.1432-1033.1997.0350a.x. [DOI] [PubMed] [Google Scholar]
- 28.Le H, Tanguay R L, Balasta M L, Wei C-C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 29.Metz A M, Browning K S. Mutational analysis of the functional domains of the large subunit of the isozyme form of wheat initiation factor eIF4F. J Biol Chem. 1996;271:31033–31036. doi: 10.1074/jbc.271.49.31033. [DOI] [PubMed] [Google Scholar]
- 30.Neff C L, Sachs A B. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol Cell Biol. 1999;19:5557–5564. doi: 10.1128/mcb.19.8.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niepel M, Gallie D R. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J Virol. 1999;73:9080–9088. doi: 10.1128/jvi.73.11.9080-9088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otero L J, Ashe M P, Sachs A B. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 34.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilipenko E V, Pestova T V, Kolupaeva V G, Khitrina E V, Poperechnaya A N, Agol V I, Hellen C U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 37.Piron P, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 39.Pyronnet S, Imataka H, Gingras A C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray B K, Lawson T G, Kramer J C, Cladaras M H, Grifo J A, Abramson R D, Merrick W C, Thach R E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 41.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Schaad M C, Anderberg R J, Carrington J C. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology. 2000;273:300–306. doi: 10.1006/viro.2000.0416. [DOI] [PubMed] [Google Scholar]
- 42.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 43.Tarun S Z, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Heerden A, Browning K S. Expression in Escherichia coli of the two subunits of the isozyme form of wheat germ protein synthesis initiation factor 4F. Purification of the subunits and formation of an enzymatically active complex. J Biol Chem. 1994;269:17454–17457. [PubMed] [Google Scholar]
- 45.Wei C-C, Balasta M L, Ren J, Goss D J. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry. 1998;37:1910–1916. doi: 10.1021/bi9724570. [DOI] [PubMed] [Google Scholar]
- 46.Wittmann S, Chatel H, Fortin M G, Laliberte J F. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology. 1997;234:84–92. doi: 10.1006/viro.1997.8634. [DOI] [PubMed] [Google Scholar]
- 47.Yisraeli J K, Melton D A. Synthesis of long, capped transcripts in vitro by SP6 and T7 RNA polymerases. Methods Enzymol. 1989;180:42–50. doi: 10.1016/0076-6879(89)80090-4. [DOI] [PubMed] [Google Scholar]
- 48.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]