Abstract
Terpenoids are widely used as medicines, flavors, and biofuels. However, the use of these natural products is largely restricted by their low abundance in native plants. Fortunately, heterologous biosynthesis of terpenoids in microorganisms offers an alternative and sustainable approach for efficient production. Various genome-editing technologies have been developed for microbial strain construction. Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated protein 9 (Cas9) is the most commonly used system owing to its outstanding efficiency and convenience in genome editing. In this review, the basic principles of CRISPR-Cas9 systems are briefly introduced and their applications in engineering bacteria for the production of plant-derived terpenoids are summarized. The aim of this review is to provide an overview of the current developments of CRISPR-Cas9-based genome-editing technologies in bacterial engineering, concluding with perspectives on the challenges and opportunities of these technologies.
Keywords: CRISPR-Cas9, Genome editing, Terpenoids, Metabolic engineering, Bacteria
Graphical abstract
1. Introduction
Terpenoids, predominantly isolated from plants, are valuable natural treasures and have been widely used as flavors, fragrances, and pharmaceuticals [1]. However, the applications of terpenoids, such as the anticancer agent paclitaxel, antimalarial agent artemisinin, and the sugar substitute steviol, are severely impeded by their low abundance in plants [2,3]. In this respect, heterologous biosynthesis in different microorganisms and plants offers an alternative and sustainable approach to producing these highly valued products. Notably, microorganisms have distinct advantages over plants, such as faster growth and lower costs, as exemplified by the prokaryotic microorganisms Escherichia coli [4], Bacillus subtilis [5], and Corynebacterium glutamicum [6], as well as the eukaryotic microorganisms Saccharomyces cerevisiae [7], Pichia pastoris [8], and Yarrowia lipolytica [9]. Eukaryotic microorganisms have multiple organelles that facilitate compartmentalized biosynthesis and exhibit high tolerance to harsh industrial conditions [[10], [11], [12]]. Nonetheless, eukaryotic microorganisms face several insurmountable challenges including relatively slow growth, few selection markers, and easy contamination. As prokaryotic microorganisms are uncomplicated cells with a simple design, they possess many obvious advantages, including fast growth, diverse genetic tools, and simple maintenance, which enable their extensive application in metabolic engineering [13,14].
Reconstruction of the biosynthetic pathways of target compounds in heterologous cells is the most critical step in establishing an efficient chassis. Generally, plasmid expression is a common strategy for reconstruction and optimization; however, genes expressed in plasmids are prone to instability [15]. Concomitantly, the high-frequency use of antibiotics accelerates the antibiotic resistance crisis and increases experimental costs [16]. Therefore, the integration of genes into the genome is a more stable approach for gene expression and results in a lower growth burden. To integrate biosynthetic pathways into microbial genomes, various genome-editing tools have been developed such as homologous recombination (HR) [17], zinc finger nucleases (ZFNs) [18], and transcription activator-like effector nucleases (TALENs) [19]. Unfortunately, problems such as high cost and time consumption restrict their application, as reviewed elsewhere [20,21]. Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated protein 9 (Cas9) system has emerged with outstanding efficiency and convenience in genome editing and has become the predominant tool for various genetic modifications [[22], [23], [24]]. In this review, we briefly introduce the principle of the CRISPR-Cas9 system and provide an overview of its development and application in bacterial engineering for the production of diverse terpenoids. Additionally, challenges and perspectives regarding CRISPR-Cas9 systems for bacterial engineering are discussed.
2. The principles of CRISPR-Cas9 systems
Initially, CRISPR-Cas systems were identified as adaptive immune systems in prokaryotes. The immune response consists of three stages: (i) adaptation, which involves the incorporation of a spacer into the host genome, (ii) expression and maturation, which involves CRISPR RNA (crRNA) transcription and processing, and (iii) interference, in which the target genetic element is destroyed by crRNA-Cas protein effector complexes [[25], [26], [27]]. Based on the differences in Cas protein composition and sequence divergence among the effector complexes, CRISPR-Cas systems are classified into two classes (Class 1 and Class 2) and six types (type I–VI) (reviewed in [28,29]). In addition, a candidate type VII system has been proposed [30]. The CRISPR-Cas9 system is a type II adaptive immunity system and the most widely applied CRISPR-Cas system for genetic modifications because of its simplicity and high efficiency [[31], [32], [33]].
The CRISPR-Cas9 system used for genome editing consists of a guide RNA (gRNA) for target-specific recognition and a Cas9 protein to introduce double-stranded DNA breaks (DSBs) [[34], [35], [36], [37], [38]] (Fig. 1A). gRNA is a short synthetic RNA composed of a scaffold sequence for Cas9 binding and a 20 nt sequence for target sequence recognition [29,32]. gRNA can direct Cas9 to the genome where a protospacer adjacent motif (PAM) is located immediately downstream of the target site. The Cas9 protein, another fundamental component of the CRISPR-Cas9 system, has RNA-dependent endonuclease activity that introduces DSBs into the genome [39]. DSBs are mainly repaired by two endogenous cellular DNA repair pathways: non-homologous end joining (NHEJ) and HR [39,40] (Fig. 1A). Notably, the PAM sequence varies among different Cas9 proteins, and the canonical PAM sequence (i.e., NGG) is associated with the widely used Cas9 nuclease from Streptococcus pyogenes (SpCas9) [41,42]. In addition to genome editing, the CRISPR-Cas9 system has been modified to regulate gene expression at the transcriptional level. For instance, deactivated Cas9 (dCas9) generated by introducing point mutations (i.e. H840A and D10A) loses nuclease activity yet retains the ability for sequence-specific DNA binding. The dCas9 protein in complex with gRNA has been used alone to perform CRISPR interference (CRISPRi). Specifically, it targets the promoter to block the binding of RNA polymerase (RNAP), or the coding sequence region to block the elongation of RNAP (Fig. 1B-C). Alternatively, the fusion of dCas9 with a transcriptional repressor can also be used for CRISPRi [43] (Fig. 1D). In contrast, dCas9 fused with a transcriptional activator can perform CRISPR activation (CRISPRa) [44] (Fig. 1E).
Fig. 1.
Schematic diagram of CRISPR-Cas9-based technologies. (A) CRISPR-Cas9-mediated genome editing. Cas9 protein forms complex with gRNA, which will recognize the target site of genomic DNA, creating a double-stranded DNA break (DSB) upstream of the PAM sequence. The DSB can be repaired by HR or NHEJ, where HR introduces precise genome editing including gene insertion, replacement, and deletion, while NHEJ results in small insertion and deletion into the genome. (B) dCas9 based CRISPRi binding to promoter. The dCas9/gRNA complex targets the promoter of the target gene, and sterically blocks the binding of RNA polymerase (RNAP). (C) dCas9 based CRISPRi binding to coding sequence region. The dCas9/gRNA complex targets the coding sequence of the target gene, and blocks the elongation of RNAP by physical collision. (D) CRISPRi-mediated genetic interference. A fusion of dCas9 with a transcription repressor can be used to down-regulate transcription. (E) CRISPRa-mediated genetic activation. A fusion of dCas9 with a transcription activator can be used to upregulate transcription.
3. The development of CRISPR-Cas9-based genome-editing technologies in bacteria
3.1. In E. coli
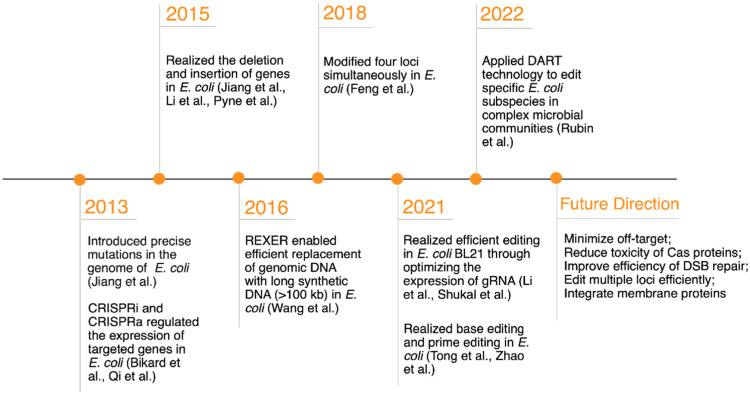
Among all types of bacteria, E. coli is the most widely investigated in terms of its application in synthetic biology, owing to its simple genetic background, fast growth and low cost, which constitutes a model organism for developing CRISPR-Cas9-based genome-editing technologies [[45], [46], [47]]. Considerable progress has been made in the application of CRISPR-Cas9 systems in E. coli (Fig. 2). Sapranauskas et al. found that the heterologous CRISPR-Cas9 system could be functionally transplanted into E. coli to provide immunity against relevant plasmids and phages [48]. Jiang et al. were the first to successfully generate precise site-directed mutations in the E. coli genome using a dual RNA and Cas9-based CRISPR system [49]. Subsequently, researchers managed to delete or insert intact genes in E. coli using the CRISPR-Cas9 system [4,50,51]. For example, Jiang et al. developed a two-plasmid-based CRISPR-Cas9 system, in combination with an effective phage-encoded λ-red recombination system to accomplish both gene insertion and deletion in E. coli [50]. Furthermore, as mentioned above, researchers have repurposed CRISPR systems for CRISPRi and CRISPRa to repress and activate target gene expression in E. coli using dCas9 [52]. Cas9 nickase (nCas9) was generated by introducing a D10A mutation that could only cut a single strand of DNA. With nCas9, base-editing and prime-editing technologies have been established that can directly introduce point mutations into cellular DNA without inducing DSBs [53,54]. Despite great progress, many shortcomings of CRISPR-Cas9-based editing technologies remain, such as low efficiency in editing long DNA sequences, huge difficulty in multiplex genome editing, and restricted applications in certain strains.
Fig. 2.
Timeline of key developments of CRISPR-Cas9-based technologies in E. coli.
3.1.1. Editing long DNA sequences
Researchers have made intensive efforts to engineer large DNA fragments in E. coli. Li et al. accomplished the deletion of genomic sequences of up to 12 kb using double-stranded donor DNA as an editing template with more than 90% efficiency, which was higher than that of single-stranded DNA (less than 20% efficiency) [4]. This increased efficiency can be attributed to decreased mismatch repair correction during homologous recombination. Specifically, using single-stranded donor DNA for CRISPR editing introduces a change near the DNA replication fork, which triggers mismatch repair correction [55]. In contrast, double-stranded donor DNA is less likely to cause these errors, thus achieving higher efficiency [4]. By introducing a mutation in the upstream homology arm, Bassalo et al. inactivated the targeted PAM sequence; thus, Cas9 did not cut the same site repeatedly, which enabled single-step 10 kb metabolic pathway insertion [56]. Wang et al. developed the REXER (replicon excision for enhanced genome engineering through programmed recombination) by combining the CRISPR-Cas9 and λ-red recombination systems, enabling the efficient replacement of genomic DNA with long synthetic DNA (> 100 kb) in E. coli [57].
3.1.2. Multiplex genome editing
CRISPR-Cas9-based technologies have also been established to edit multiple loci simultaneously with minimal labor and time in E. coli [4,58,59]. For example, Liu et al. developed a genome-editing system coupling CRISPR-Cas9 with λ-red recombination to achieve rapid two-gene modification at once. The efficiency of the double-locus point mutation reached 88.0%, whereas the double-locus deletion/insertion efficiency was only 38.7% [58]. Li et al. developed a CRISPR-Cas9 system comprising only a single plasmid that could express multiple gRNAs targeting distinct loci. Using this system, they introduced codon replacements in three genes simultaneously with a 23.0% editing efficiency [4]. Feng et al. used a CRISPR-Cas9 assisted multiplex genome editing (CMGE) technique to express multiple gRNAs together, realizing a four-locus modification with a 31.7% editing efficiency [59].
3.1.3. Expanding the scope of strains
Efforts have been made to expand the scope of E. coli strains that can be edited using CRISPR-Cas9-based technologies. Among these, E. coli BL21 is a popular chassis for the production of natural products. However, genetic modification is difficult to achieve due to the leaky expression of gRNA, which causes plasmid curing and hampers CRISPR-Cas9-based genome editing [50]. To overcome this problem, the promoter driving gRNA expression was either replaced or removed directly to avoid leaky expression, leading to efficient genome editing in E. coli BL21 [60,61]. Rubin et al. developed DNA-editing all-in-one RNA-guided CRISPR-Cas transposase (DART) systems to edit specific E. coli subspecies in complex microbial communities. This method overcomes the limitations of traditional genome-editing systems by enabling genome editing without the isolation of individual species [62].
3.2. In other bacteria
In addition to E. coli, other bacteria can be genetically modified using CRISPR-Cas9-based technologies for the production of natural products, including Streptococcus pneumoniae [49], Tatumella citrea [50], B. subtilis [[63], [64], [65]], Streptomyces coelicolor [66], Saccharopolyspora erythraea [67], C. glutamicum [68], and others. The CRISPRi system has been applied to Methylorubrum extorquens [69] and C. glutamicum [70], and the CRISPRa system has been applied to B. subtilis and Pseudomonas putida [71,72].
García-Moyano et al. developed a plasmid-based CRISPR-Cas9 system using high-throughput fragment exchange cloning techniques, which enabled precise gene integration in B. subtilis [65]. Westbrook et al. directly integrated the cas9 gene and gRNA transcription cassette into the B. subtilis genome, which realized the chromosomal expression of cas9 and ensured gRNA stability. This versatile toolkit enables point mutations, gene insertions, and multiplex editing with high efficiency and is further extended to CRISPRi for transcriptional modulation [63]. In C. glutamicum, Yao et al. constructed a single-plasmid CRISPR-Cas9 system, in which all elements including the cas9 gene, gRNA, and homologous arms, were designed in one temperature-sensitive plasmid. Using this system, the genome-editing efficiency reached 95.7% [68].
4. The application of CRISPR-Cas9-based technologies in engineering bacteria for the production of terpenoids
Terpenoids represent the largest family of natural products with wide pharmaceutical and industrial applications; however, limited resources severely hinder their use [73]. Nowadays, microbial production of terpenoids in bacteria, especially in E. coli, provides a promising alternative for the large-scale acquisition of these valuable compounds [4]. The reconstruction of the biosynthetic pathways of target compounds in heterologous cells is critical for efficient biosynthesis. The CRISPR-Cas9 system is robust for the reconstruction and optimization of biosynthetic pathways. Here, we review the recent progress in the heterologous biosynthesis of diverse terpenoids in bacteria using CRISPR-Cas9-based technologies (Table 1).
Table 1.
The applications of CRISPR-Cas9-based technologies and other strategies in engineering bacteria for the heterologous production of terpenoids.
| Strain | Compound | Integrated genes | Over-expressed genes | Deleted genes | Repressed genes | Gene origin | Other strategiesa | Titer | Increased foldb | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| B. subtilis | amorphadiene | GFP-ADS, FPPS | / | / | HepS*, UppS* | Plant | / | 116 mg/L | 1.4-fold | [84] |
| B. subtilis | amorphadiene | GFP-ADS, FPPS | / | / | / | Plant | Overexpress MEP pathway genes in plasmid; Optimize the medium. | 416 mg/L | 20.0-fold | [85] |
| E. coli DH1 | bisabolene | HMGS, HMGR, MK, PMK, PMD, IDI, FPPS, AgBIS | / | / | / | Plant and microorganism | / | 435 μg/L | 5.0-fold | [76] |
| E. coli MG1655 | steviol | GGPPS, CPS, KS | DXS, DXR, IDI | GdhA* | / | Microorganism | / | 38 mg/L | 2.5-fold | [77] |
| E. coli BL21 (DE3) | crocin | CCD, ALD | / | / | / | Plant and microorganism | / | 4 mg/L | 3.6-fold | [78] |
| C. glutamicum | squalene | SQS | DXS, IDI | / | GapA*, GdhA*, GGPPS* | Microorganism | High-throughput fermentation; Culture condition optimization. | 106 mg/L | 5.0-fold | [70] |
| E. coli BL21 (DE3) | lycopene | / | / | AdhE*, LdhA*, PflB*, PoxB* or AckA-Pta* | / | Microorganism | / | 135 mg/L | 3.0-fold | [61] |
| E. coli BL21 (DE3) | lycopene | / | / | / | AAS, HMGS, HMGR, MK, PMK, PMD, IDI | Microorganism | / | 71 mg/L | 8.0-fold | [83] |
| E. coli W3110 | lycopene | GGPPS, CrtI, CrtE, CrtB | DXS, DXR, IDI | / | / | Microorganism | / | 9 mg/g | 4.4-fold | [79] |
| E. coli MG1655 | β-carotene | FPPS, GGPPS, CrtB, CrtI, | DXS, CMK, HDR, IDI, GalP | PtsHIcrr* | / | Microorganism | / | 2 g/L | 28.0-fold | [4] |
| E. coli ATCC 8739 | β-carotene | Almgs | / | / | / | Microorganism | / | 37 mg/L | 1.4-fold | [92] |
| E.coli ATCC 8739 | zeaxanthin | CrtZ | / | / | / | Microorganism | / | 451 μg/L | 1.3-fold | [93] |
| E.coli ATCC 8739 | astaxanthin | CrtW, CrtZ | / | / | / | Microorganism | / | 410 μg/L | 1.3-fold | [93] |
Note:
Host genes.
Strategies unrelated to CRISPR-Cas9-based technologies.
“Increased fold” means the increased fold of titers compared with the control.
4.1. Reconstruction and optimization of terpenoid biosynthetic pathways via CRISPR-Cas9-based technologies
4.1.1. Biosynthetic pathways of diverse terpenoids
The biosynthesis of plant-derived terpenoid compounds can be divided into three modules: central, upstream, and downstream pathways. The central pathway plays a pivotal role in metabolic regulation by generating essential precursors such as pyruvate, glyceraldehyde-3-phosphate (G3P), and acetyl-CoA to synthesize target compounds [73]. Two parallel upstream pathways, methylerythritol phosphate (MEP) and mevalonate (MVA) provide universal precursors: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). The MEP pathway starts with pyruvate and G3P, whereas the MVA pathway utilizes acetyl-CoA [73]. The downstream pathway is specialized for distinct target compounds regarding genes involved and generates various terpenoids, including monoterpenoids (C10), sesquiterpenoids (C15), diterpenoids (C20), triterpenoids (C30), and tetraterpenoids (C40) (Fig. 3) [74,75].
Fig. 3.
Schematic diagram of the central, upstream, and downstream pathways of terpenoid biosynthesis. All genes marked with blue were upregulated in E. coli; all genes marked with red were downregulated in E. coli. The arrows with solid line represent the single-step reaction, and the ones with dotted line represent the multi-step reaction. Terpenoid compounds through biosynthesis described in this review are shown in blue. The crossed arrows represent the deletion of genes. GalP, galactose permease; PtsHIcrr, phosphoenolpyruvate; PTS: carbohydrate phosphotransferase; GapA, glyeraldehyde-3-phosphate dehydrogenase; AdhE, alcohol dehydrogenase; LdhA, lactate dehydrogenase; PoxB, pyruvate oxidase; Pta, phosphate acetyltransferase; AckA, acetate kinase; PflB, pyruvate-formate lyase; GdhA, glutamate dehydrogenase; GltA, citrate synthase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase; CMK, 4-(cytidine-5’-diphospho)-2-C-methyl-D-erythritol kinase; MCS, 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl-diphosphate reductase; AAS, acetyl-CoA thiolase; HMGS, HMG-CoA synthase; HMGR, HMG-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; PMD, diphosphomevalonate decarboxylase; IDI, isopentenyl diphosphate isomerase; GPPS, GPP synthase; FPPS, FPP synthase; GGPPS, GGPP synthase; CrtB, phytoene synthase; CrtI, phytoene desaturase; CrtY, lycopene cyclase; CrtZ, 3,3′-hydroxylase; CrtW, 4,4′-oxygenase; CCD, carotenoid-cleaving dioxygenase; ALD, aldehyde dehydrogenase; ADS, amorphadiene synthase; BIS, bisabolene synthase; SQS, squalene synthase; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAH, kaurenoic acid 13-hydroxylase.
4.1.2. Reconstructing terpenoid biosynthetic pathways
The basic principle of engineering microorganisms for terpenoid production is to enhance the expression of biosynthetic pathway genes and suppress the expression of competitive pathway genes, which can decrease by-products and redistribute flux to target compounds. The CRISPR-Cas9 system has been used to tune the central pathway to boost the levels of related precursors in E. coli. For instance, Shukal et al. achieved increased levels of acetyl-CoA by knocking out the competitive branch pathway gene LdhA using the CRISPR-Cas9 system, leading to improved terpenoid production [61]. Li et al. improved the concentrations of both pyruvate and G3P by overexpressing GalP and knocking out PtsHIcrr using the CRISPR-Cas9 system, thus providing a more efficient strain for terpenoid production [4]. Therefore, modifying the central pathway is an effective strategy for optimizing the biosynthesis of terpenoid compounds.
Similarly, both the MEP and MVA pathways were individually optimized to afford a larger supply of IPP and DMAPP via CRISPR-Cas9-based technologies in bacteria. Given that prokaryotes inherently possess intact MEP pathways, Li et al. simply integrated extra copies of four pathway genes (i.e., DXS, CMK, HDR, and IDI) to upregulate gene expression via CRISPR-Cas9 systems in E. coli MG1655, accomplishing a 3.0-fold improvement in β-carotene production [4]. In contrast, the MVA pathway-associated genes, which are not inherently present in bacteria, are usually integrated exogenously. For instance, genes such as HMGS, HMGR, MK, PMK, PMD, and IDI were integrated into the E. coli DH1 genome, enabling a 5.0-fold increase in terpenoid production [76]. Overall, the genes involved in the MEP and MVA pathways are integrated into the genomes of engineered bacteria to boost terpenoid production, indicating their significant role in improving terpenoid biosynthesis.
In addition, the downstream pathway genes are generally distinct and non-native to bacteria, and need to be integrated first to reconstruct the biosynthetic pathways of specific terpenoids. For example, Moon et al. integrated the ent-kaurene pathway genes (i.e., GGPPS, CPS, and KS) along with the MEP pathway genes (i.e., DXS, DXR, and IDI) into the E. coli MG1655 genome, which enables the production of ent-kaurene in combination with the 5′UTR engineering strategy [77]. Wang et al. integrated the carotenoid-cleaving dioxygenase gene (CCD) and aldehyde dehydrogenase gene (ALD) into a modified zeaxanthin-producing strain, completing the pathway reconstruction of crocetin [78]. Another example is the reconstruction of the lycopene pathway in E. coli W3110 by integrating essential pathway genes (i.e., CrtE, CrtB, and CrtI) into its genome via the CRISPR-Cas9 system, resulting in the heterologous production of lycopene with an appealing yield [79]. Thus, the integration of downstream pathway genes is a key step in the reconstruction of terpenoid biosynthetic pathways. In summary, CRISPR-Cas9-based genome-editing technologies have enabled the heterologous production of terpenoids by integrating pathway genes and deleting branch pathway genes, thereby laying the foundation for the further optimization of these pathways.
4.1.3. Optimizing the terpenoid biosynthetic pathways
The promoter [80] and coding sequence (CDS) [81] are potential target sites for genomic modification using the CRISPR-Cas9 system to regulate target gene expression in bacteria. Promoters can profoundly impact the level of gene expression by affecting transcription initiation rates; thus, it is feasible to enhance gene expression using stronger promoters. Alonso-Gutierrez et al. achieved a 5.0-fold increase in terpenoid production in E. coli DH1 by simply replacing native promoters with stronger T7 promoters using the CRISPR-Cas9 system [76]. CDS can also be modified to fine-tune gene expression via codon optimization. As reported, replacement of the native GltA gene with a rare codon containing GltA caused the downregulation of GltA expression in E. coli, which repressed the transformation of acetyl-CoA to citrate, thus redistributing the metabolic flux to other downstream compounds of acetyl-CoA origin [82]. Undoubtedly, all the above-mentioned elements provide many target sites for modification by CRISPR-Cas9 systems to improve terpenoid production.
In addition to the CRISPR-Cas9 system, CRISPRi has emerged as a promising tool for modulating the expression of pathway genes. Kim et al. tuned the MVA pathway using CRISPRi in E. coli BL21 (DE3), where the promoters and internal regions of key genes (i.e., AAS, HMGS, HMGR, MK, PMK, PMD, and IDI) were targeted to interfere with gene transcription, resulting in an increased lycopene yield [83]. Park et al. repressed the expression of GapA, GdhA, and GGPPS, which are distributed in different biosynthetic modules, to achieve combinatorial gene repression via CRISPRi in C. glutamicum. Correspondingly, squalene yield increased [70]. Thus, CRISPRi has proven to be a powerful tool for the heterologous production of terpenoids by tuning gene expression. CRISPRa is also a potential tool for regulating gene expression and has been used to enhance the mevalonate yield in P. putida [71]. Nonetheless, CRISPRa has not yet been applied to bacterial terpenoid production although it holds great application potential.
4.2. Engineering bacteria for the production of diverse terpenoids via CRISPR-Cas9-based technologies
Plant terpenoids, including amorphadiene, lycopene, β-carotene, etc., have been investigated thoroughly in terms of their production in heterologous hosts via CRISPR-Cas9-based technologies. These representative examples provide valuable references for the heterologous biosynthesis of additional terpenoids in microorganisms.
4.2.1. Sesquiterpenoids
Sesquiterpenoids derived from the C15 linear precursor farnesyl diphosphate (FPP) exhibit versatile pharmacological activities. (Sesqui)terpenes are non-oxygenated (sesqui)terpenoids, which are hydrocarbon skeletons generally produced by terpene synthases. Currently, the heterologous expression of sesquiterpenoids in bacteria using CRISPR-Cas9 systems has already been realized for certain compounds, as described in detail below.
Amorphadiene, the key precursor of the antimalarial agent artemisinin, is a well-recognized sesquiterpene. Song et al. developed a single plasmid-based CRISPR-Cas9 system to edit the genome of B. subtilis, making it capable of heterologously producing amorphadienes [84]. Specifically, the central pathway was regulated by the introduction of strong promoters for genes in the Krebs cycle. Green fluorescent protein (GFP) was fused with amorphadiene synthase (ADS) to improve its solubility, and the fused GFP-ADS and FPP synthase genes (FPPS) were integrated into the B. subtilis genome. Additionally, the ADS gene was mutated to improve protein performance, and the branched pathway genes (i.e., HepS and UppS) were attenuated to ensure sufficient FPP supply. All these strategies collaboratively increased amorphadiene production from 81 mg/L to 116 mg/L [84]. By episomally overexpressing key MEP pathway genes and optimizing the medium, the titer of amorphadiene was further increased to 416 mg/L [85].
Bisabolene is a sesquiterpene with anti-cancer activity [86]. Alonso-Gutierrez et al. integrated foreign MVA pathway genes (i.e., HMGS, HMGR, MK, PMK, PMD, and IDI) and the bisabolene synthase gene (BIS) into the E. coli DH1 genome, completing the necessary pathway reconstruction. Additionally, they replaced the native promoters of the MVA pathway genes with stronger T7 promoters using the CRISPR-Cas9 system and achieved a 5.0-fold increase in bisabolene production compared to that of the original strain [76]. In conclusion, the CRISPR-Cas9 system is successfully used to edit genes in all three modules to boost the heterologous production of sesquiterpenes. Although the titers of the products are still low, they lay the foundation for future bacterial engineering to produce oxidized sesquiterpenoids.
4.2.2. Diterpenoids
Diterpenoids are a class of compounds derived from the C20 linear precursor geranylgeranyl diphosphate (GGPP). Steviol, the key precursor of the steviol glycosides used as natural sweeteners, is a well-studied diterpenoid. [87]. Taking the CRISPR-Cas9-based genome-editing technologies, Moon et al. redesigned and reconstructed the steviol biosynthetic pathway in E. coli MG1655. In particular, they first integrated the key MEP pathway genes (i.e., DXS, DXR, and IDI) and the ent-kaurene pathway genes (i.e., GGPPS, CPS, and KS) into the bacterial genome. In combination with the 5′UTR engineering strategy, they achieved a 624 mg/L titer of ent-kaurene [77]. Moreover, the glutamate dehydrogenase-encoding gene GdhA was deleted to increase the cellular NADPH/NADP+ ratio, which presumably favored ent-kaurene oxidase (KO) and kaurenoic acid 13-hydroxylase (KAH). Ultimately, the titer of ent-kaurenoic acid increased from 32 mg/L to 41 mg/L, and the titer of steviol also increased from 5 mg/L to 38 mg/L [77].
Crocetin, an important diterpenoid with anti-apoptotic activity, is derived from carotenoids. Wang et al. integrated the carotenoid-cleaving dioxygenase gene CCD and aldehyde dehydrogenase gene ALD into a modified zeaxanthin-producing strain, completing the biosynthetic pathway reconstruction of crocetin. Furthermore, the strength of different promoters in regulating the expression of the integrated CCD and ALD genes was evaluated, and the highest titer of crocetin (4 mg/L) was obtained under the control of promoter M1-46 [78]. Although the yield of crocetin was still low, this research made a useful exploration and laid the foundation for subsequent production improvements.
Many important diterpenoids have a common diterpene skeleton; for instance, ent-kaurene is a general precursor of gibberellins. The construction of a strain that generates a diterpene skeleton with high efficiency can ultimately increase the diterpenoid yield. With the discovery of numerous diterpenoid synthases, it is promising to expand the application of CRISPR-Cas9 systems to the synthesis of more diterpenoids in the near future.
4.2.3. Triterpenoids
Triterpenoids are C30 terpenoids derived from the common precursor squalene, which is generated by the condensation of two FPP molecules by squalene synthase (SQS). Squalene has also become a popular target for scientists to synthesize [88,89]. The CRISPRi system has been utilized in C. glutamicum to increase squalene yield. Combinatorial metabolic engineering strategies for precursor rebalancing, redox balancing, and blocking competing pathways for IPP availability were applied by repressing target genes. In particular, the genes GapA, GdhA, and GGPPS were down-regulated by CRISPRi, so that the metabolic flux was redirected to favor the production of squalene. Moreover, the genes DXS, IDI, and SQS were overexpressed to enhance squalene production. Combined with high-throughput fermentation and culture condition optimization, the highest squalene production reached 105 mg/L, representing a 5.0-fold increase over that of the parental strain [70]. Squalene, a common precursor of triterpenoids, has attracted increasing attention, and oxidative compounds derived from squalene, such as amyrin, friedelin, and lupeol, have great application potential owing to their versatile properties. Efficient heterologous production of squalene will pave the way for the sustainable production of oxidized triterpenoids.
4.2.4. Tetraterpenoids
Tetraterpenoids, derived from the condensation of two GGPP molecules, are C40 terpenoids, which contain a series of bioactive compounds, such as lycopene, β-carotene, and zeaxanthin. Lycopene is utilized as a pigment and food ingredient due to its antioxidant activity. Shukal et al. modified the central pathway by individually deleting acetyl-CoA-associated genes, including AdhE, LdhA, PflB, PoxB, and AckA-Pta in E. coli BL21 (DE3) using the CRISPR-Cas9 system. The results showed that the LdhA knockout strain increased acetyl-CoA availability, leading to an improved lycopene yield [61]. Kim et al. successfully tuned the MVA pathway using CRISPRi in E. coli BL21 (DE3), in which key genes (i.e., AAS, HMGS, HMGR, MK, PMK, PMD, and IDI) were interfered with to minimize their leaky expression at the seed culture stage, where their metabolites were toxic and hindered bacterial growth. Accordingly, interference was removed during the main flask culture to restore MVA pathway expression. Using this strategy, lycopene production was improved to 71 mg/L, an 8.0-fold increase [83]. In another example, the MEP pathway was modulated to enhance the precursor supply by introducing an extra copy of rate-limiting enzyme genes (i.e., DXS, DXR, and IDI) into the E. coli W3110 genome using CRISPR-Cas9 systems. Additionally, the integration of downstream pathway genes (i.e., CrtE, CrtB, and CrtI) resulted in an efficient lycopene-producing strain with a final yield of 9 mg/g [79].
Another attractive tetraterpene β-carotene, the downstream product of lycopene catalyzed by CrtY, possesses various bioactivities [90]. To realize its efficient production in microorganisms, Li et al. increased the intracellular concentrations of pyruvate and G3P by overexpressing GalP and knocking out PtsHIcrr in E. coli MG1655, which significantly increased the β-carotene titer by 1.7-fold compared to the original strain [4]. To increase the supply of IPP and DMAPP, Li et al. simultaneously overexpressed four MEP pathway genes (i.e., DXS, CMK, HDR, and IDI), obtaining a 28.0-fold increased titer of β-carotene (2 g/L) compared with the control [4]. As the membrane compartment is important to accumulate the hydrophobic β-carotene [91], Wu et al. integrated Almgs encoding monoglucosyldiacylglycerol synthase into the E. coli ATCC 8739 genome, whose expression was further tuned by using the optimum promoter M1-37. These modification strategies using CRISPR-Cas9 systems significantly enhanced the storage capacity of the cell chassis for β-carotene, promoting the titer of β-carotene from 27 mg/L to 37 mg/L [92]. The above studies prove that CRISPR-Cas9-based technologies can greatly enhance the yield of β-carotene through integrating relevant key genes.
Other high-value-added tetraterpenoids, such as zeaxanthin (a pigment) and astaxanthin (an antioxidant), have also been synthesized by microorganisms. Xie et al. developed a novel molecular device for fusing location tags with dCas9 (Cas9-Lag) in E. coli ATCC 8739. Cas9-Lag localized the CrtZ expression cassette to the membrane, and the zeaxanthin titer was increased to 451 μg/L, which was 1.3-fold higher than that of the control strain. Likewise, by localizing the combined cassette expressing CrtW and CrtZ via Cas9-Lag, the astaxanthin titer reached 410 μg/L, which was 1.3-fold higher than that of the control strain [93].
In short, CRISPR-Cas9-based technologies have been extensively developed and applied to tune the metabolic pathways of tetraterpenoids in bacteria compared to the above-mentioned terpenoids of other types. The highest titer of β-carotene reached gram scale, however, the yield of its additionally oxidized products (i.e., zeaxanthin and astaxanthin) dropped drastically, thereby requiring profound optimization in future research.
5. Conclusion
Plants provide a treasury of bioactive metabolites such as alkaloids, phenolic acids, and terpenoids, which have been widely utilized in the pharmaceutical, industrial, and agricultural industries. Nevertheless, as represented by terpenoids in this review, these plant-derived compounds are usually limited in nature and have difficulty meeting market demand. Fortunately, synthetic biology techniques have emerged in a timely manner, providing an alternative strategy for the sustainable production of these low natural content yet high value-added chemicals. Accordingly, scientists have made tremendous efforts to produce these compounds heterologously in numerous types of chassis cells, including bacteria, yeast, and tobacco. Among them, bacteria stand out owing to their fast growth, low cost, and simple genetic background, and have attracted significant attention from scientists.
Integrating intact biosynthetic pathways of target compounds into bacterial hosts is essential for achieving heterologous biosynthesis. Among the available genome-editing systems, newly emerged CRISPR-Cas9-based genome-editing technologies have rapidly become the dominant tools for microbial genome editing because of their simplicity and efficiency [94]. As briefly overviewed here, current CRISPR-Cas9 systems allow the efficient insertion and deletion of large DNA fragments of over 10 kb at the chromosomal level, making it theoretically operable to integrate exogenous target genes into bacterial genomes. Besides, CRISPR-Cas9 systems have also been developed to tune the expression of target genes. Notably, CRISPRi and CRISPRa were used to repress and activate gene expression, respectively, which enabled the exquisite control of relevant genes and greatly improved the yield of target compounds [95]. To our knowledge, the progress of CRISPRa lags behind that of CRISPRi, owing to the paucity of effective transcriptional activators [96], which remains to be further elucidated.
In this review, we focus on the current progress in the application of CRISPR-Cas9-based genome-editing technologies in engineering bacteria for the heterologous production of plant-derived terpenoids. Using representative examples, we have introduced how CRISPR-Cas9-based technologies have been adopted to reconstruct and optimize the biosynthetic pathways of each terpenoid product by interfering with all three biosynthetic modules (i.e., the central, upstream, and downstream pathways). Many research groups have obtained genetically stable bacterial strains that afford satisfactory yield of various terpenoids, ranging from sesquiterpenoids to tetraterpenoids.
6. Discussion and future perspectives
Although considerable progress has been made in the application of CRISPR-Cas9 systems in bacteria, numerous challenges and problems associated with CRISPR-Cas9 systems need to be addressed to achieve heterologous biosynthesis of these valued compounds on an industrial scale. First, bacterial strains are intrinsically unique and exhibit distinct features when chromosomally edited; thus, low efficiency or even total failure of regular CRISPR-Cas9 systems is often observed for certain strains. To solve this problem, a more robust and generalized editing platform is required that ideally possesses the following advantages: minimal off-target genome editing, reduced toxicity of Cas9 proteins to bacterial hosts, and improved homologous recombination efficiency.
A series of studies have been conducted and significant progress has been made. First, off-target effects can be reduced by replacing regular gRNAs with RNA-DNA hybrids, with the latter exhibiting higher specificity than the former [97]. Alternatively, nCas9 was developed to cut one DNA strand, and two gRNAs for directing two copies of nCas9 were used to decrease off-target effects [98]. To reduce Cas9 toxicity, the expression of cas9 has been strictly regulated [99], and endogenous CRISPR systems have been identified and engineered to ensure the successful implementation of CRISPR systems in bacteria [100]. As reported, a series of endogenous CRISPR systems have been found and applied in E. coli [101,102], Clostridium tyrobutyricum [103], Clostridium pasteurianum [104], Lactobacillus crispatus [105], Zymomonas mobilis [106], and others [107,108]. To improve the efficiency of DSB repair in bacteria, the λ-red system was coupled with the CRISPR-Cas9 system [56], while increasing the length of the homologous sequence is also a viable strategy to enhance recombination efficiency.
Although solutions to existing issues are emerging, CRISPR-Cas9 systems have a number of challenges that require further research. Most current CRISPR-Cas9 systems can simultaneously target only a single site. Reconstructing a biosynthetic pathway usually involves editing bulky genes; thus, it is time-consuming to obtain recombinant strains. Many studies have focused on modifying CRISPR-Cas9 systems to edit multiple loci simultaneously [4,58,59], however, the current efficiency of multiplex editing remains far from satisfactory and can be improved by expressing multiple gRNAs under an appropriate promoter [109]. Furthermore, prokaryotic bacteria require systematic optimization to cope with the demands of operating such intricate metabolic pathways. For instance, cytochrome P450 (CYP450) is essential for generating oxygenated terpenoids, yet no CYP450 genes have been integrated into bacteria via the CRISPR-Cas9 system, which can otherwise exert biocatalytic activity episomally in bacteria [110]. With improvements of current CRISPR-Cas9-based genome-editing technologies, problems are expected to be solved.
Notably, CRISPR-Cas9 systems have undergone continuous development for new applications. The CRISPR-nCas9 mediated DNA base editing and prime editing enable precise nucleotide substitutions without inducing DSBs, as reviewed elsewhere [111]. Meanwhile, the artificial intelligence-aided protein engineering efforts have created enzymes with high efficiency, providing fundamental elements for the biosynthesis of natural products. High-throughput sequencing technology promotes a deeper systematic understanding of various hosts. All these advances make it conceivable that a longer list of natural products, beyond the terpenoids described here, can be heterologously produced via a synthetic biology approach in a larger variety of microbial hosts. We can even imagine that rapidly emerging automation technologies will eventually realize labor-free heterologous production of all desired chemicals, thus solving the long-lasting contradiction of short supply and growing demand.
CRediT authorship contribution statement
Xin Sun: Writing – review & editing, Writing – original draft, Conceptualization. Haobin Zhang: Writing – review & editing. Yuping Jia: Writing – review & editing, Conceptualization. Jingyi Li: Writing – review & editing, Writing – original draft, Conceptualization. Meirong Jia: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Beijing Nova Program (Z211100002121004), CAMS Innovation Fund for Medical Sciences (CIFMS 2021-I2M-1-029, CIFMS 2022-I2M-2-002), and Nonprofit Central Research Institute Fund of the Chinese Academy of Medical Sciences (2021-RC350-003).
Contributor Information
Jingyi Li, Email: lijingyi@imm.ac.cn.
Meirong Jia, Email: jiameirong@imm.ac.cn.
References
- 1.Gershenzon J., Dudareva N. The function of terpene natural products in the natural world. Nat. Chem. Biol. 2007;3:408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 2.Mewalal R., Rai D.K., Kainer D., Chen F., Külheim C., Peter G.F., Tuskan G.A. Plant-derived terpenes: a feedstock for specialty biofuels. Trends Biotechnol. 2017;35:227–240. doi: 10.1016/j.tibtech.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Wilson B.A.P., Thornburg C.C., Henrich C.J., Grkovic T., O'Keefe B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020;37:893–918. doi: 10.1039/c9np00068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Lin Z., Huang C., Zhang Y., Wang Z., Tang Y.J., Chen T., Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Westbrook A.W., Ren X., Oh J., Moo-Young M., Chou C.P. Metabolic engineering to enhance heterologous production of hyaluronic acid in Bacillus subtilis. Metab. Eng. 2018;47:401–413. doi: 10.1016/j.ymben.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Li M., Chen J., He K., Su C., Wu Y., Tan T. Corynebacterium glutamicum cell factory design for the efficient production of cis, cis-muconic acid. Metab. Eng. 2024;82:225–237. doi: 10.1016/j.ymben.2024.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Cao X., Yu W., Chen Y., Yang S., Zhao Z.K., Nielsen J., Luan H., Zhou Y.J. Engineering yeast for high-level production of diterpenoid sclareol. Metab. Eng. 2023;75:19–28. doi: 10.1016/j.ymben.2022.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Cai P., Wu X., Deng J., Gao L., Shen Y., Yao L., Zhou Y.J. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris. Proc. Natl. Acad. Sci. U S A. 2022;119 doi: 10.1073/pnas.2201711119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y., Liu N., Greisen P., Li J., Qiao K., Huang S., Stephanopoulos G. Removal of lycopene substrate inhibition enables high carotenoid productivity in Yarrowia lipolytica. Nat. Commun. 2022;13:572. doi: 10.1038/s41467-022-28277-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong K.K., Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol. Life Sci. 2012;69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lian J., Zhao H. Recent advances in biosynthesis of fatty acids derived products in Saccharomyces cerevisiae via enhanced supply of precursor metabolites. J. Ind. Microbiol. Biotechnol. 2015;42:437–451. doi: 10.1007/s10295-014-1518-0. [DOI] [PubMed] [Google Scholar]
- 12.Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keasling J.D. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 14.Yim H., Haselbeck R., Niu W., Pujol-Baxley C., Burgard A., Boldt J., Khandurina J., Trawick J.D., Osterhout R.E., Stephen R., Estadilla J., Teisan S., Schreyer H.B., Andrae S., Yang T.H., Lee S.Y., Burk M.J., Van Dien S. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 15.Summers D.K. The kinetics of plasmid loss. Trends Biotechnol. 1991;9:273–278. doi: 10.1016/0167-7799(91)90089-z. [DOI] [PubMed] [Google Scholar]
- 16.Sengupta M., Austin S. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 2011;79:2502–2509. doi: 10.1128/IAI.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smithies O., Gregg R.G., Boggs S.S., Koralewski M.A., Kucherlapati R.S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 18.Geurts A.M., Cost G.J., Freyvert Y., Zeitler B., Miller J.C., Choi V.M., Jenkins S.S., Wood A., Cui X., Meng X., Vincent A., Lam S., Michalkiewicz M., Schilling R., Foeckler J., Kalloway S., Weiler H., Ménoret S., Anegon I., Davis G.D., Zhang L., Rebar E.J., Gregory P.D., Urnov F.D., Jacob H.J., Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tesson L., Usal C., Ménoret S., Leung E., Niles B.J., Remy S., Santiago Y., Vincent A.I., Meng X., Zhang L., Gregory P.D., Anegon I., Cost G.J. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 20.Lambert J.M., Bongers R.S., Kleerebezem M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 2007;73:1126–1135. doi: 10.1128/AEM.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M.M., Wang Y., Ang E.L., Zhao H. Engineering microbial hosts for production of bacterial natural products. Nat. Prod. Rep. 2016;33:963–987. doi: 10.1039/c6np00017g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontrelli S., Chiu T.Y., Lan E.I., Chen F.Y., Chang P., Liao J.C. Escherichia coli as a host for metabolic engineering. Metab. Eng. 2018;50:16–46. doi: 10.1016/j.ymben.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Becker J., Rohles C.M., Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 2018;50:122–141. doi: 10.1016/j.ymben.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Hille F., Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin E.V., Makarova K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2019;374 doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 28.Wang J.Y., Pausch P., Doudna J.A. Structural biology of CRISPR-Cas immunity and genome editing enzymes. Nat. Rev. Microbiol. 2022;20:641–656. doi: 10.1038/s41579-022-00739-4. [DOI] [PubMed] [Google Scholar]
- 29.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., Moineau S., Mojica F.J.M., Scott D., Shah S.A., Siksnys V., Terns M.P., Venclovas C., White M.F., Yakunin A.F., Yan W., Zhang F., Garrett R.A., Backofen R., van der Oost J., Barrangou R., Koonin E.V. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altae-Tran H., Kannan S., Suberski A.J., Mears K.S., Demircioglu F.E., Moeller L., Kocalar S., Oshiro R., Makarova K.S., Macrae R.K., Koonin E.V., Zhang F. Uncovering the functional diversity of rare CRISPR-Cas systems with deep terascale clustering. Science. 2023;382:1910. doi: 10.1126/science.adi1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westra E.R., Buckling A., Fineran P.C. CRISPR-Cas systems: beyond adaptive immunity. Nat. Rev. Microbiol. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 32.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson R.A., Martin C., Nemudryi A.A., Wiedenheft B. CRISPR RNA-guided autonomous delivery of Cas9. Nat. Struct. Mol. Biol. 2019;26:14–24. doi: 10.1038/s41594-018-0173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 35.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters J.M., Silvis M.R., Zhao D., Hawkins J.S., Gross C.A., Qi L.S. Bacterial CRISPR: accomplishments and prospects. Curr. Opin. Microbiol. 2015;27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y., Fu Y. Class 2 CRISPR/Cas: an expanding biotechnology toolbox for and beyond genome editing. Cell Biosci. 2018;8:59. doi: 10.1186/s13578-018-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong Y., Weber T., Lee S.Y. CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 2019;36:1262–1280. doi: 10.1039/c8np00089a. [DOI] [PubMed] [Google Scholar]
- 41.Wang J., Teng Y., Zhang R., Wu Y., Lou L., Zou Y., Li M., Xie Z.R., Yan Y. Engineering a PAM-flexible SpdCas9 variant as a universal gene repressor. Nat. Commun. 2021;12:6916. doi: 10.1038/s41467-021-27290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Russa M.La, Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 43.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashemi A. CRISPR-Cas9/CRISPRi tools for cell factory construction in E. coli. World J. Microbiol. Biotechnol. 2020;36:96. doi: 10.1007/s11274-020-02872-9. [DOI] [PubMed] [Google Scholar]
- 46.Arroyo-Olarte R.D., Bravo Rodriguez R., Morales-Rios E. Genome editing in bacteria: CRISPR-Cas and beyond. Microorganisms. 2021;9:844. doi: 10.3390/microorganisms9040844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong H., Cui Y., Zhang D. CRISPR/Cas technologies and their applications in Escherichia coli. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.762676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pyne M.E., Moo-Young M., Chung D.A., Chou C.P. Coupling the CRISPR/Cas9 system with lambda Red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl. Environ. Microbiol. 2015;81:5103–5114. doi: 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qi D.D., Jin J., Liu D., Jia B., Yuan Y.J. In vitro and in vivo recombination of heterologous modules for improving biosynthesis of astaxanthin in yeast. Microb. Cell Fact. 2020;19:103. doi: 10.1186/s12934-020-01356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong Y., Jorgensen T.S., Whitford C.M., Weber T., Lee S.Y. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 2021;12:5206. doi: 10.1038/s41467-021-25541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao D., Li J., Li S., Xin X., Hu M., Price M.A., Rosser S.J., Bi C., Zhang X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 55.Costantino N., Court D.L. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. U S A. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bassalo M.C., Garst A.D., Halweg-Edwards A.L., Grau W.C., Domaille D.W., Mutalik V.K., Arkin A.P., Gill R.T. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth. Biol. 2016;5:561–568. doi: 10.1021/acssynbio.5b00187. [DOI] [PubMed] [Google Scholar]
- 57.Wang K., Fredens J., Brunner S.F., Kim S.H., Chia T., Chin J.W. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu H.Q., Hou G.F., Wang P., Guo G.Y., Wang Y., Yang N., Rehman M.N.U., Li C.L., Li Q., Zheng J.P., Zeng J.F., Li S.H. A double-locus scarless genome editing system in Escherichia coli. Biotechnol. Lett. 2020;42:1457–1465. doi: 10.1007/s10529-020-02856-7. [DOI] [PubMed] [Google Scholar]
- 59.Feng X., Zhao D., Zhang X., Ding X., Bi C. CRISPR/Cas9 assisted multiplex genome editing technique in Escherichia coli. Biotechnol. J. 2018;13 doi: 10.1002/biot.201700604. [DOI] [PubMed] [Google Scholar]
- 60.Li Q., Sun B., Chen J., Zhang Y., Jiang Y., Yang S. A modified pCas/pTargetF system for CRISPR-Cas9-assisted genome editing in Escherichia coli. Acta Biochim. Biophys. Sin. (Shanghai) 2021;53:620–627. doi: 10.1093/abbs/gmab036. [DOI] [PubMed] [Google Scholar]
- 61.Shukal S., Lim X.H., Zhang C., Chen X. Metabolic engineering of Escherichia coli BL21 strain using simplified CRISPR-Cas9 and asymmetric homology arms recombineering. Microb. Cell Fact. 2022;21:19. doi: 10.1186/s12934-022-01746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubin B.E., Diamond S., Cress B.F., Crits-Christoph A., Lou Y.C., Borges A.L., Shivram H., He C., Xu M., Zhou Z., Smith S.J., Rovinsky R., Smock D.C.J., Tang K., Owens T.K., Krishnappa N., Sachdeva R., Barrangou R., Deutschbauer A.M., Banfield J.F., Doudna J.A. Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 2022;7:34–47. doi: 10.1038/s41564-021-01014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westbrook A.W., Moo-Young M., Perry C.C. Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis. Appl. Environ. Microbiol. 2016;82:4876–4895. doi: 10.1128/AEM.01159-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang K., Duan X., Wu J. Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas9 system. Sci. Rep. 2016;6:27943. doi: 10.1038/srep27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia-Moyano A., Larsen O., Gaykawad S., Christakou E., Boccadoro C., Puntervoll P., Bjerga G.E.K. Fragment exchange plasmid tools for CRISPR/Cas9-mediated gene integration and protease production in Bacillus subtilis. Appl. Environ. Microbiol. 2021;87:e02090. doi: 10.1128/AEM.02090-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang H., Zheng G., Jiang W., Hu H., Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim. Biophys. Sin. 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 67.Mo J., Wang S., Zhang W., Li C., Deng Z., Zhang L., Qu X. Efficient editing DNA regions with high sequence identity in actinomycetal genomes by a CRISPR-Cas9 system. Synth. Syst. Biotechnol. 2019;4:86–91. doi: 10.1016/j.synbio.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao C., Hu X., Wang X. Construction and application of a CRISPR/Cas9-assisted genomic editing system for Corynebacterium glutamicum. AMB Express. 2021;11:70. doi: 10.1186/s13568-021-01231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mo X.H., Zhang H., Wang T.M., Zhang C., Zhang C., Xing X.H., Yang S. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis. Appl. Microbiol. Biotechnol. 2020;104:4515–4532. doi: 10.1007/s00253-020-10543-w. [DOI] [PubMed] [Google Scholar]
- 70.Park J., Yu B.J., Choi J.I., Woo H.M. Heterologous production of squalene from glucose in engineered Corynebacterium glutamicum using multiplex CRISPR interference and high-throughput fermentation. J. Agric. Food Chem. 2019;67:308–319. doi: 10.1021/acs.jafc.8b05818. [DOI] [PubMed] [Google Scholar]
- 71.Kiattisewee C., Dong C., Fontana J., Sugianto W., Peralta-Yahya P., Carothers J.M., Zalatan J.G. Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metab. Eng. 2021;66:283–295. doi: 10.1016/j.ymben.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 72.Liao C., Cui J., Gao M., Wang B., Ito K., Guo Y., Zhang B. Dual-sgRNA CRISPRa system for enhanced MK-7 production and salmonella infection mitigation in Bacillus subtilis natto applied to Caco-2 cells. J. Agric. Food Chem. 2024;72:4301–4316. doi: 10.1021/acs.jafc.3c08866. [DOI] [PubMed] [Google Scholar]
- 73.Wang C., Liwei M., Park J.B., Jeong S.H., Wei G., Wang Y., Kim S.W. Microbial platform for terpenoid production: Escherichia coli and yeast. Front. Microbiol. 2018;9:2460. doi: 10.3389/fmicb.2018.02460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C., Chen Q., Fan D., Li J., Wang G., Zhang P. Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol. Plant. 2016;9:195–204. doi: 10.1016/j.molp.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 75.Lichman B.R., Kamileen M.O., Titchiner G.R., Saalbach G., Stevenson C.E.M., Lawson D.M., O'Connor S.E. Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis. Nat. Chem. Biol. 2019;15:71–79. doi: 10.1038/s41589-018-0185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alonso-Gutierrez J., Koma D., Hu Q., Yang Y., Chan L.J.G., Petzold C.J., Adams P.D., Vickers C.E., Nielsen L.K., Keasling J.D., Lee T.S. Toward industrial production of isoprenoids in Escherichia coli: lessons learned from CRISPR-Cas9 based optimization of a chromosomally integrated mevalonate pathway. Biotechnol. Bioeng. 2018;115:1000–1013. doi: 10.1002/bit.26530. [DOI] [PubMed] [Google Scholar]
- 77.Moon J.H., Lee K., Lee J.H., Lee P.C. Redesign and reconstruction of a steviol-biosynthetic pathway for enhanced production of steviol in Escherichia coli. Microb. Cell Fact. 2020;19:20. doi: 10.1186/s12934-020-1291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang W., He P., Zhao D., Ye L., Dai L., Zhang X., Sun Y., Zheng J., Bi C. Construction of Escherichia coli cell factories for crocin biosynthesis. Microb. Cell Fact. 2019;18:120. doi: 10.1186/s12934-019-1166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su B., Song D., Zhu H. Homology-dependent recombination of large synthetic pathways into E. coli genome via λ-Red and CRISPR/Cas9 dependent selection methodology. Microb. Cell Fact. 2020;19:108. doi: 10.1186/s12934-020-01360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yona A.H., Alm E.J., Gore J. Random sequences rapidly evolve into de novo promoters. Nat. Commun. 2018;9:1530. doi: 10.1038/s41467-018-04026-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nieuwkoop T., Claassens N.J., van der Oost J. Improved protein production and codon optimization analyses in Escherichia coli by bicistronic design. Microb. Biotechnol. 2019;12:173–179. doi: 10.1111/1751-7915.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu S.Z., Guo L.W., Zhao L.Y., Chen Z.Y., Huo Y.X. Metabolic engineering of E. coli for producing phloroglucinol from acetate. Appl. Microbiol. Biotechnol. 2020;104:7787–7799. doi: 10.1007/s00253-020-10591-2. [DOI] [PubMed] [Google Scholar]
- 83.Kim S.K., Han G.H., Seong W., Kim H., Kim S.W., Lee D.H., Lee S.G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016;38:228–240. doi: 10.1016/j.ymben.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 84.Song Y., He S., Abdallah II, Jopkiewicz A., Setroikromo R., van Merkerk R., Tepper P.G., Quax W.J. Engineering of multiple modules to improve amorphadiene production in Bacillus subtilis using CRISPR-Cas9. J. Agric. Food Chem. 2021;69:4785–4794. doi: 10.1021/acs.jafc.1c00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pramastya H., Xue D., Abdallah II, Setroikromo R., Quax W.J. High level production of amorphadiene using Bacillus subtilis as an optimized terpenoid cell factory. Nat. Biotechnol. 2021;60:159–167. doi: 10.1016/j.nbt.2020.10.007. [DOI] [PubMed] [Google Scholar]
- 86.Yeo S.K., Ali A.Y., Hayward O.A., Turnham D., Jackson T., Bowen I.D., Clarkson R. β-Bisabolene, a sesquiterpene from the essential oil extract of opoponax (Commiphora guidottii), exhibits cytotoxicity in breast cancer cell lines. Phytother. Res. 2016;30:418–425. doi: 10.1002/ptr.5543. [DOI] [PubMed] [Google Scholar]
- 87.Pól J., Hohnová B., Hyötyläinen T. Characterisation of Stevia rebaudiana by comprehensive two-dimensional liquid chromatography time-of-flight mass spectrometry. J. Chromatogr. A. 2007;1150:85–92. doi: 10.1016/j.chroma.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 88.Spanova M., Daum G. Squalene – biochemistry, molecular biology, process biotechnology, and applications. Eur. J. Lipid Sci. Technol. 2011;113:1299–1320. [Google Scholar]
- 89.Reddy L.H., Couvreur P. Squalene: a natural triterpene for use in disease management and therapy. Adv. Drug Deliv. Rev. 2009;61:1412–1426. doi: 10.1016/j.addr.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 90.Palozza P., Krinsky N.I. Antioxidant effects of carotenoids in vivo and in vitro: an overview. Meth. Enzymol. 1992;213:403–420. doi: 10.1016/0076-6879(92)13142-k. [DOI] [PubMed] [Google Scholar]
- 91.Ahrazem O., Rubio-Moraga A., Berman J., Capell T., Christou P., Zhu C., Gomez-Gomez L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016;209:650–663. doi: 10.1111/nph.13609. [DOI] [PubMed] [Google Scholar]
- 92.Wu T., Ye L., Zhao D., Li S., Li Q., Zhang B., Bi C., Zhang X. Membrane engineering - A novel strategy to enhance the production and accumulation of β-carotene in Escherichia coli. Metab. Eng. 2017;43:85–91. doi: 10.1016/j.ymben.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 93.Xie Q., Li S., Zhao D., Ye L., Li Q., Zhang X., Zhu L., Bi C. Manipulating the position of DNA expression cassettes using location tags fused to dCas9 (Cas9-Lag) to improve metabolic pathway efficiency. Microb. Cell Fact. 2020;19:229. doi: 10.1186/s12934-020-01496-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Donohoue P.D., Barrangou R., May A.P. Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol. 2018;36:134–146. doi: 10.1016/j.tibtech.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 95.Lian J., HamediRad M., Hu S., Zhao H. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 2017;8:1688. doi: 10.1038/s41467-017-01695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dong C., Fontana J., Patel A., Carothers J.M., Zalatan J.G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 2018;9:2489. doi: 10.1038/s41467-018-04901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin H., Song C.Q., Suresh S., Kwan S.Y., Wu Q., Walsh S., Ding J., Bogorad R.L., Zhu L.J., Wolfe S.A., Koteliansky V., Xue W., Langer R., Anderson D.G. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Biol. 2018;14:311–316. doi: 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J., Wang Y., Lu Y., Zheng P., Sun J., Ma Y. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb. Cell Fact. 2017;16:205. doi: 10.1186/s12934-017-0815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goh Y.J., Barrangou R. Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli. Curr. Opin. Biotechnol. 2019;56:163–171. doi: 10.1016/j.copbio.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 101.Chang Y., Su T., Qi Q., Liang Q. Easy regulation of metabolic flux in Escherichia coli using an endogenous type I-E CRISPR-Cas system. Microb. Cell Fact. 2016;15:195. doi: 10.1186/s12934-016-0594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luo M.L., Mullis A.S., Leenay R.T., Beisel C.L. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J., Zong W., Hong W., Zhang Z.T., Wang Y. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab. Eng. 2018;47:49–59. doi: 10.1016/j.ymben.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 104.Pyne M.E., Bruder M.R., Moo-Young M., Chung D.A., Chou C.P. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 2016;6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hidalgo-Cantabrana C., Goh Y.J., Pan M., Sanozky-Dawes R., Barrangou R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. U S A. 2019;116:15774–15783. doi: 10.1073/pnas.1905421116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng Y., Han J., Wang B., Hu X., Li R., Shen W., Ma X., Ma L., Yi L., Yang S., Peng W. Characterization and repurposing of the endogenous Type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 2019;47:11461–11475. doi: 10.1093/nar/gkz940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu Z., Li Y., Cao H., Si M., Zhang G., Woo P.C.Y., Yan A. A transferrable and integrative type I-F Cascade for heterologous genome editing and transcription modulation. Nucleic Acids Res. 2021;49:e94. doi: 10.1093/nar/gkab521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hao Y., Wang Q., Li J., Yang S., Zheng Y., Peng W. Double nicking by RNA-directed Cascade-nCas3 for high-efficiency large-scale genome engineering. Open Biol. 2022;12 doi: 10.1098/rsob.210241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao J., Fang H., Zhang D. Expanding application of CRISPR-Cas9 system in microorganisms. Synth. Syst. Biotechnol. 2020;5:269–276. doi: 10.1016/j.synbio.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu B., Zhao X., Wang E., Zhou J., Li J., Chen J., Du G. Efficient heterologous expression of cytochrome P450 enzymes in microorganisms for the biosynthesis of natural products. Crit. Rev. Biotechnol. 2023;43:227–241. doi: 10.1080/07388551.2022.2029344. [DOI] [PubMed] [Google Scholar]
- 111.Kantor A., McClements M.E., MacLaren R.E. CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 2020;21:6240. doi: 10.3390/ijms21176240. [DOI] [PMC free article] [PubMed] [Google Scholar]