Highlights
-
•
The Q291A mutant of Xltr1p accelerated the utilization of mannose as compared to the wild-type transporter.
-
•
The Y301W mutant lost the galactose utilization capacity but retained glucose utilization capacity.
-
•
Computer-assisted protein engineering facilitates the screening of substrate-specific hexose transporters.
Keywords: Hexose transporter, Glucose, Galactose, Mannose, Transmembrane α-helix 7, Sugar transport, Computer-aided screening
Abstract
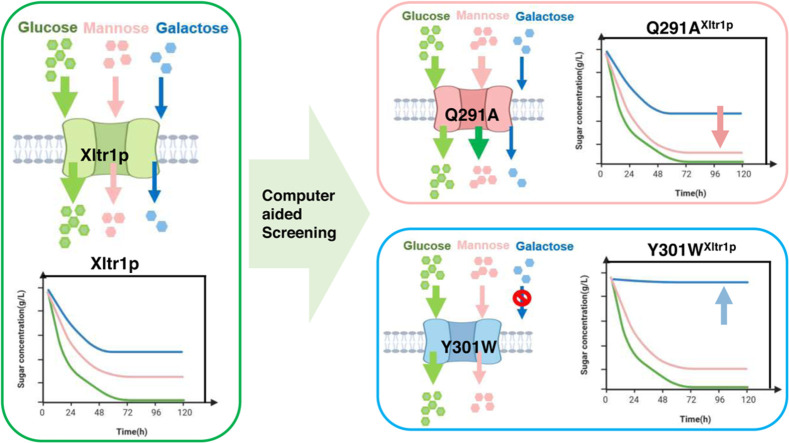
This research identified four amino acid residues (Leu174, Asn297, Tyr301, and Gln291) that contribute to substrate recognition by the high-affinity glucose transporter Xltr1p from Trichoderma reesei. Potential hotspots affecting substrate specificity were selected through homology modeling, evolutionary conservation analyses, and substrate-docking modeling of Xltr1p. Variants carrying mutations at these hotspots were subsequently obtained via in silico screening. Replacement of Leu174 or Asn297 in Xltr1p with alanine resulted in loss of hexose transport activity, indicating that Leu174 and Asn297 play essential roles in hexose transport. The Y301W variant exhibited accelerated mannose transport, but lost galactose transport capacity, and mutation of Gln291 to alanine greatly accelerated mannose transport. These results suggest that amino acids located in transmembrane α-helix 7 (Asn297, Tyr301, and Gln291) play critical roles in substrate recognition by the hexose transporter Xltr1p. Our results will help expand the potential applications of this transporter and provide insights into the mechanisms underlying its function and specificity.
Graphical abstract
1. Introduction
Synthetic biology is an emerging interdisciplinary field that has led to significant advances in diverse research disciplines [1,2]. Microbial consortia form efficient biosynthetic systems owing to their robustness, flexibility, and high product yields compared to monomicrobial systems [[2], [3], [4]]. Such consortia have increasingly garnered significant attention in synthetic biology research [5,6]. The construction of strains exhibiting preferences for different carbon sources eliminates carbon catabolic repression and substrate competition [3,7], facilitating stable maintenance of such systems. Relative ease of population ratio regulation in a microbial consortium would facilitate regulation of global metabolism [3,[8], [9], [10], [11]].
Sugar transport is the first step in carbohydrate metabolism in vivo. Sugar transporters located in cell membranes often serve as sentinels that regulate the transport of various sugars into the cells. Strains expressing different specific transporters can utilize specific carbon sources. Sugar transporters are widely distributed in nature and are of three main types: 1) ATP-binding cassette (ABC) transporters, which are the predominant sugar transporters among bacteria [12,13]; 2) the carbohydrate phosphotransferase system (PTS), which has been identified only in bacteria [14,15]; and 3) the major facilitator superfamily (MFS), which are the main family of transporters conserved among bacteria, archaea, and eukaryotes [16,17]. Saccharomyces cerevisiae is a well-established eukaryote model system that has been widely employed in synthetic biology as a chassis cell for the synthesis of a variety of high-value products [18,19]. Twenty hexose transporters are encoded in S. cerevisiae, including hexose transporters 1–17 (Hxt1–17), galactose transporter 2 (Gal2), two glucose sensors, sucrose non-fermenting 3 (Snf3), and restored glucose transport (Rgt2) [20,21]; of these, Hxt1–7 are mainly responsible for glucose transport. Based on their affinity for glucose, transporters are classified as high- (Hxt6, Hxt7, and Gal2), intermediate- (Hxt2, Hxt4, and Hxt5), or low-affinity transporters, all of which can transport a wide range of sugars.
The use of mixed carbon sources is highly beneficial for controlling bacterial growth ratios. However, homologous [22,23] and heterologous [24] expression of several transporters has revealed their potential to transport multiple sugars, although all of them exhibit innate sensitivity to competitive inhibition by glucose [25]. In fact, a hexose transporter that is insensitive to competitive inhibition by glucose has not been identified [26]. Exploration of the mechanisms underlying substrate recognition by these transporters is expected to greatly facilitate the development of specific transporters.
Several studies have elucidated mechanisms underlying substrate recognition by these transporters. Rojas et al. (2021) identified amino acid residues in Gal2 from S. cerevisiae (ScGal2) that are crucial for xylose transport. Tyr446, an aromatic amino acid in transmembrane segment 10 (TMS10) of ScGal2, is important for galactose transport, while Trp455 plays an important role in distinguishing galactose from glucose [[27], [28], [29]]. To date, mutants that exclusively transport galactose and not glucose have not been obtained. Tyr388 (corresponding to Trp455 of Gal2) in TM10 of rat glucose transporter 1 (Glut1) plays an important role in glucose transport [30]. Qiao et al. identified five residues (N321, N322, F325, G426, and P427) in transmembrane segment 7 (TMS7) and TMS10 regions of Hxt4 from Candida glycerinogenes (CgHxt4), which are essential for efficient glucose transport. Residues N321 and P427 likely play roles in glucose coordination and conformational flexibility, respectively [31]. Asn331 in TMS7 of Hxt2 plays a key role in determining substrate affinity, whereas Asp340 in TMS7 of Hxt7 is located at or in close proximity to the substrate-recognition site and plays a key role in high-affinity glucose transport by Hxt7 [32].
Xltr1p, a high-affinity glucose transporter from Trichoderma reesei that belongs to the MFS family, was identified in a previous study by our group [33]. Xltr1p exhibited the capacity to transport a wide range of hexose molecules and has been modified to obtain xylose transporter variants that escape competitive inhibition by glucose. These variants thus play an important role in the utilization of xylose-containing biomass. Despite these efforts, our understanding of the mechanisms underlying the function of this transporter remains limited. The goal of obtaining sugar transporters with varying capacities for transporting different hexose molecules necessitates exploration of the mechanisms underlying the recognition of different substrates.
In the present study, four amino acid residues (Leu174, Asp297, Tyr301, and Gln291) that contribute to substrate recognition by Xltr1p were identified. First, sequence comparison was performed and evolutionary conservation of Xltr1p was investigated using CulstalW and Consurf. Second, 3-dimensional (3D) structures of Xltr1p were obtained via homology modeling. Finally, docking studies were carried out between Xltr1p and the different hexose sugars, followed by identification of key amino acid residues for substrate binding. Based on our in silico docking analyses, the glucose, galactose, and mannose utilization capacities of S. cerevisiae EBYVW.4000 strain harboring each variant were examined. Among these, mutant Q291A exhibited an increased rate of mannose utilization compared to the strain expressing wild-type (WT) transporter. The strain expressing the Y301W variant retained its glucose utilization capacity, exhibited accelerated mannose utilization, and lost detectable galactose utilization capacity. This study facilitates the development of strategies to expand the potential applications of specific transporters and provides insights into mechanisms underlying substrate recognition by transporters.
2. Materials and methods
2.1. Chemicals, plasmids, and bacterial strain
Glucose, galactose, and mannose were purchased from Coolaber Co. Ltd. (Beijing, China). A KOD-Plus-Mutagenesis Kit was purchased from Toyobo Co., Ltd. (Osaka, Japan), and an ABclonal MultiF Seamless Assembly Kit was obtained from ABclonal Technology Co., Ltd. (Wuhan, China). The plasmid pJFE3 was purchased from Invitrogen (Carlsbad, CA, USA). Escherichia coli DH5α was purchased from Tsingke Biotechnology Co., Ltd. Primers were synthesized by Ruibiotech Co., Ltd. (Beijing, China). Phanta®Max Super-Fidelity DNA Polymerase was purchased from Vazyme Biotech Co., Ltd. (Nanjing, China). The other chemicals employed in this study were of analytical grade and obtained from commercial sources, unless otherwise noted.
2.2. Gene constructs and site-directed mutagenesis
The gene encoding Xltr1p (GenBank ID: XP_006966515) was amplified from T. reesei QM6a (ATCC 13631) using primers harboring homologous arms and ligated into the pJFE3 vector to obtain the recombinant plasmid pJFE3-Xltr1p. Mutants were generated using a KOD-Plus-Mutagenesis Kit with pJFE3-Xltr1p as template. The plasmids and strains used in this study are shown in Table S1, and the primers used are listed in Table S2.
2.3. Strains, media, and culture conditions
E. coli DH5α clones harboring recombinant plasmids were cultured in Luria broth containing 10 μg mL−1 ampicillin at 37 °C. S. cerevisiae EBY.VW4000, a strain devoid of all genes encoding hexose transporters, was used to investigate various transporters [34]. The strain EBY.VW4000 was cultured in YPM as described previously [33]. Recombinant plasmids were introduced into S. cerevisiae EBY.VW4000 to generate various recombinant yeast strains as previously reported [35,36]. Media glucose, galactose, and mannose depletion was estimated to investigate the effects of specific amino acid residues on hexose transport. Recombinant yeast strains were pre-cultured in a synthetic defined (SD) medium containing maltose (2 % w/v) for 12 h [37]. Cells were then washed thrice with cold double-distilled water and harvested. Harvested cells were inoculated into SD medium containing glucose, galactose, or mannose (2 % w/v) at 30 °C to obtain an initial optical density of 0.2 at 600 nm (OD600nm). Samples were withdrawn at specified time points (0, 12, 24, 48, 72, 96, and 120 h) to estimate residual concentrations of the carbon source and to measure OD600nm.
2.4. Determination of glucose, galactose, and mannose
The withdrawn samples were centrifuged at 13,000 × g for 2 min, and supernatants were filtered through 0.22-µm membrane filters. Concentrations of glucose, galactose, and mannose were determined using a high-performance liquid chromatography instrument equipped with a refractive index detector (RI detector L2490, HITACHI, Japan) and a Shodex SP-0810 column (300 × 8 mm, Showa Denko K.K., Tokyo, Japan). Doubly-distilled water was used as the mobile phase and samples were separated at 80 °C at a flow rate of 1.0 mL min−1. Respective glucose, galactose, and mannose retention times were 8.64, 9.92, and 11.04 min.
2.5. Homology modeling and evolutionary conservation analysis of Xltr1p
Homology modeling of Xltr1p was carried out using SWISS-MODEL (https://swissmodel.expasy.org/), MODELLER (https://salilab.org/modeller/), AI-lab (https://drug.ai.tencent.com/cn), and PHYRE2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) software. XylEp (PDB ID: 4GBY) and Arabidopsis thaliana STP10 (AtSTP10; PDB ID: 6H7D) were used as templates. The evolutionary conservation of the primary structure of Xltr1p was predicted using Consurf (https://predictprotein.org/).
2.6. Substrate docking and ligand–protein interaction analyses, and prediction of transmembrane helices in Xltr1p
Docking between transporters and d-glucose (PubChem CID: 5793), d-mannose (PubChem CID: 18950), and d-galactose (PubChem CID: 6036) was performed using Schrödinger Suites 2021-4 (https://newsite.schrodinger.com/). Substrate docking and interaction analyses were performed using Ligplot software (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/). Transmembrane helix prediction was conducted using PHYRE2 and DeepTMHMM (https://dtu.biolib.com/DeepTMHMM) tools.
3. Results and discussion
3.1. Computer-aided protein engineering
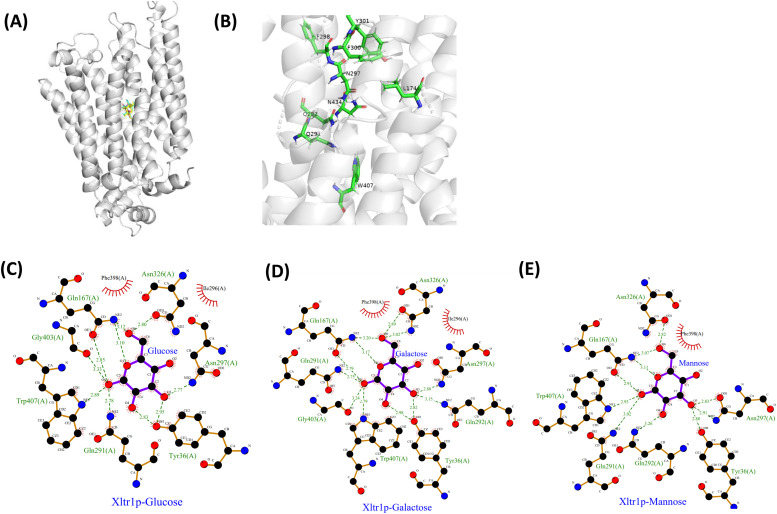
We first compared the amino acid sequence of Xltr1p with those of other sugar transporters using ClustalW (Fig. 1A). Additional sequence comparisons are shown in Fig. S1. The evolutionary conservation of amino acids was predicted using Consurf [38] (Fig. 1B). A high degree of evolutionary conservation was observed for the following residues, suggesting that they are likely to play key roles in sugar transport: aspartic acid (D) 39, arginine (R) 132, glutamine (Q) 288, glutamine (Q)291, glutamine (Q)292, phenylalanine (F) 298, isoleucine (I) 299, phenylalanine (F) 300, tyrosine (Y) 301, tyrosine (Y) 302, asparagine (N) 326, tryptophan (W) 407, and asparagine (N) 434 (Fig. 1A, B). Subsequently, XylEp (PDB ID: 4GBY) and AtSTP10 (PDB ID: 6H7D) were used as templates for homology modeling of Xltr1p using SWISS-MODEL [39], MODELLER [40], and PHYRE2 [41]. In addition, AI-lab [42] was employed for ab initio prediction-based modeling of Xltr1p. The four models built via different methods revealed similar characteristics, as shown in Fig. S2. Docking studies between Xltr1p and d-glucose, d-mannose, or d-galactose ligands were conducted to identify amino acid residues that play direct roles in hexose transport by Xltr1p (Fig. 2A). The structures of the three sugar molecules are shown in Fig. S3. Nine residues in Xltr1p (L174, Q291, Q292, N297, F298, F300, Y301, W407, and N434; Fig. 2B) were selected following analysis of the ligand–protein docking models using Ligplot software (Fig. 2C, D, E). These residues are located within 5-Å of the sugar molecules and interact with the latter via hydrogen bonds or hydrophobic interactions (Fig. 2C, D, E).
Fig. 1.
Amino acid conservation analysis of the sugar transporter protein Xltr1p. (A) Amino acid sequence comparison of Xltr1p with other hexose transporter proteins (GAT1 from Neurospora crassa, XP_963898.1; NcHXT-1 from Neurospora crassa, XP_965499.1; NcHXT-2 from Neurospora crassa, XP_959563; ScSUT3 from saccharomyces cerevisiae, XP_001386019.2; KlHgt1 from Kluyveromyces lactis, XP_451484.1; TrStr1 from Trichoderma reesei, XP_006967934.1; TrStr2 from Trichoderma reesei, XP_006967659.1; TrStr3 from Trichoderma reesei, EGR49690.1; Xltr1p from Trichoderma reesei, XP_006966515.1; ScGAL2 from saccharomyces cerevisiae, NP_013182.1). (B) Prediction of the evolutionary conservation of Xltr1p amino acid sequences using Consurf (https://predictprotein.org/).
Fig. 2.
Substrate–protein docking analyses. (A) Homology modeling of Xltr1p based on the crystal structure of XylEp using SWISS-MODEL, followed by docking analysis with the substrates d-glucose (blue), d-mannose (magenta), and d-galactose (yellow) using Schrödinger software. (B) Display of the Xltr1p amino acid residues selected for mutation analysis. Analysis of the interactions between Xltr1p and d-glucose (C), d-galactose (D), or d-mannose (E) using Ligplot software.
The abovementioned nine amino acid residues of Xltr1p were substituted with alanine (A), lysine (L), phenylalanine (F), and tryptophan (W) using site-directed mutagenesis to obtain the following 33 variants: L174A, L174F, L174W, Q291A, Q291L, Q291F, Q291W, Q292A, Q292L, Q292F, Q292W, N297A, N297L, N297F, N297W, F298A, F298L, F298W, F300A, F300L, F300F, F300W, Y301A, Y301L, Y301F, Y301W, W407A, W407L, W407F, N434A, N434L, N434F, and N434W. The 3D structures of the variants were obtained using UCSF Chimera software (https://www.cgl.ucsf.edu/chimera/), and used for substrate docking analysis of each of the variant models with d-glucose, d-galactose, or d-mannose; the scores obtained for the molecular docking analyses are shown in Table S3. The binding energies of the N297A, L174A, Q291A, or Y301W variants with the ligands, as estimated by substrate docking analyses, were lower than −7 kcal mol−1, while the ligand–protein affinities for these variants were higher than those of the other variants. Thus, the Xltr1p variants N297A, L174A, Q291A, and Y301W were used in further investigations.
3.2. Detection of d-glucose, d-galactose, and d-mannose utilization by S. cerevisiae EBY.VW4000 harboring Xltr1p variants
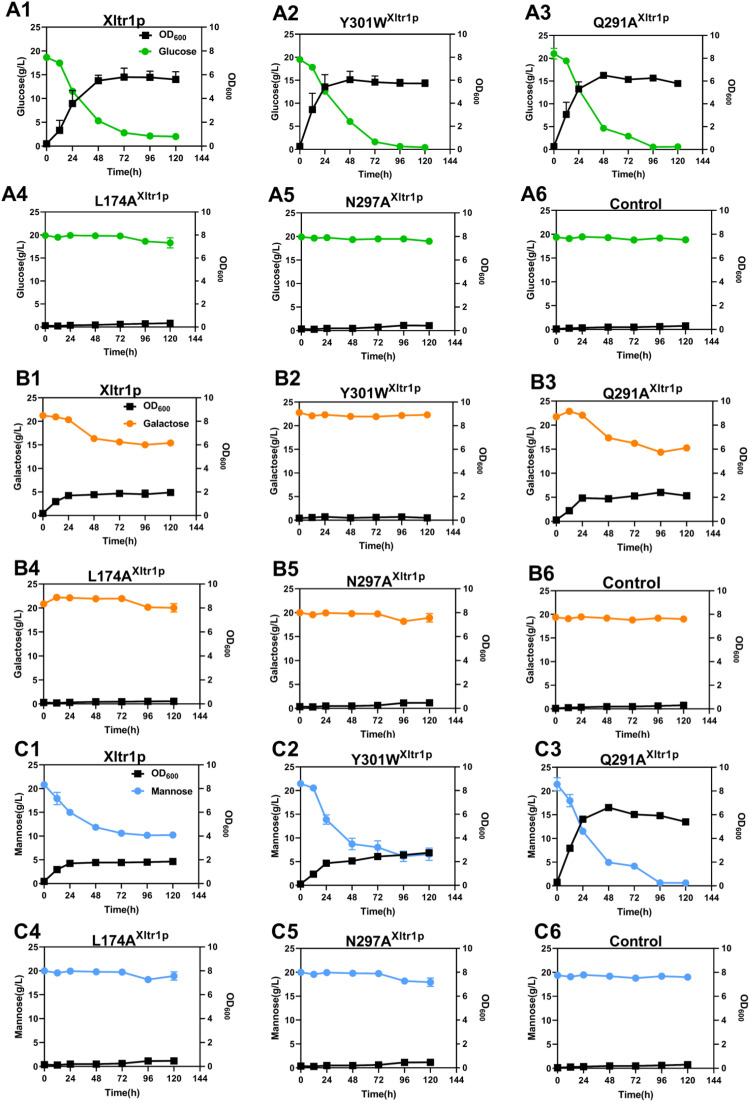
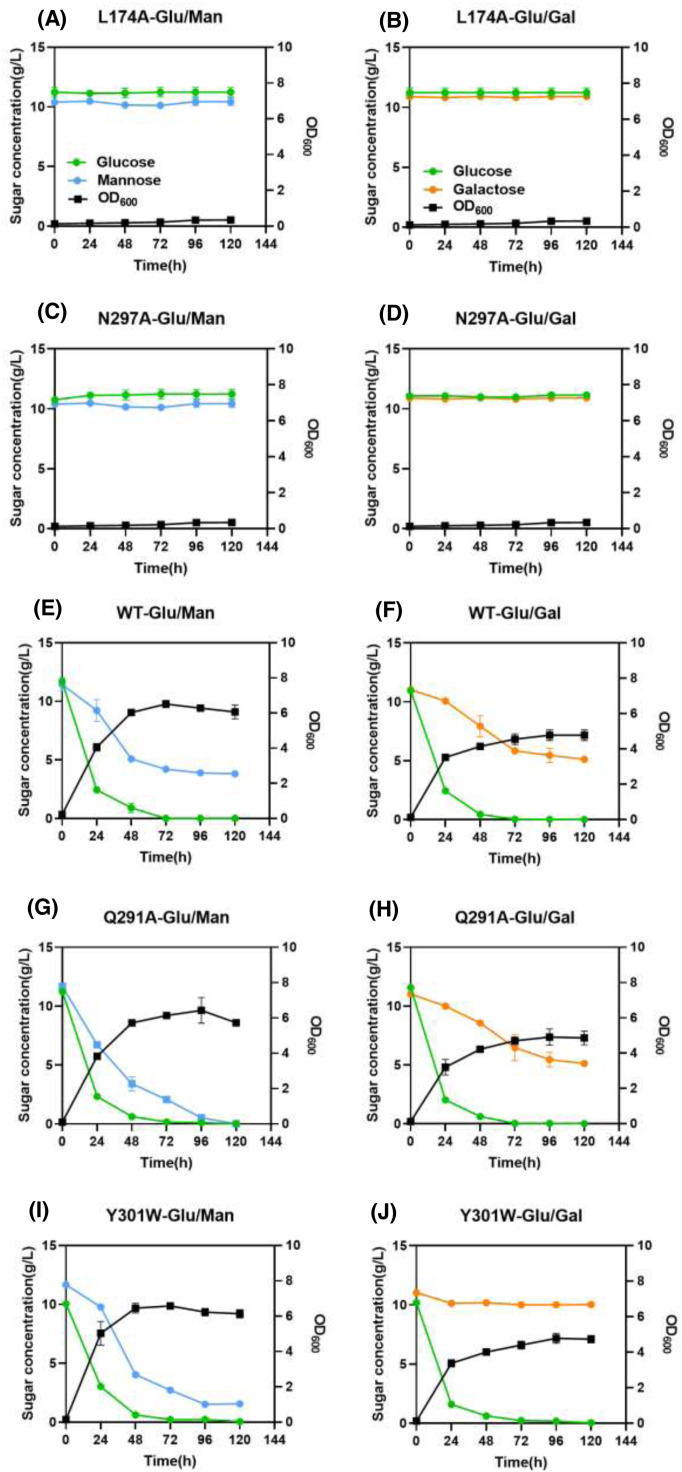
The utilization of hexose sugars by S. cerevisiae EBY.VW4000 harboring the Xltr1p variants L174A, Q291A, Y301W, and N297A or WT Xltr1p (henceforth referred to as strains L174AXltr1p, Q291AXltr1p, Y301WXltr1p, N297AXltr1p, and WTXltr1p, respectively) was evaluated (Fig. 3). Strains L174AXltr1p and N297AXltr1p lost the ability to utilize d-glucose, d-galactose, and d-mannose (Fig. 3 panels A4, B4, C4, A5, B5, C5). Interestingly, strain Y301WXltr1p lost the ability to utilize d-galactose but utilized d-mannose at an accelerated rate (Fig. 3 panels B2 and C2), whereas the rate of d-glucose utilization (Fig. 3 panel A2) remained equivalent to that of WTXltr1p. The Q291AXltr1p strain retained uncompromised ability to utilize d-glucose and d-galactose (Fig. 3 panels A3 and B3), while utilizing d-mannose at a significantly accelerated rate (Fig. 3 panel C3) compared to WTXltr1p (Fig. 3 panel C1).
Fig. 3.
Estimation of d-glucose (A), d-galactose (B), and d-mannose (C) utilization by S. cerevisiae EBY.VW4000 harboring WT Xltr1p and its variants Y301W, Q291A, L174A, and N297A. All experiments were performed using three biological replicates, with the strain harboring the plasmid pJFE3 without Xltr1p representing the negative control.
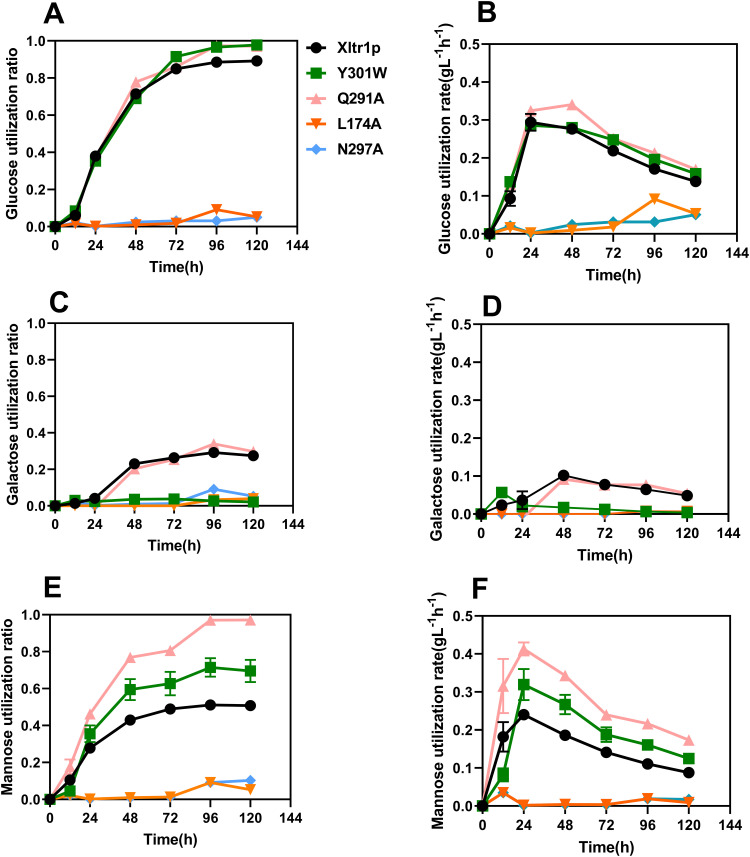
The hexose utilization capacity of the strains harboring Xltr1p mutants was subsequently assessed. The sugar utilization capacity of these strains was used to infer the transport capacity of the corresponding transporter variants. As shown in Fig. 4, WTXltr1p exhibited hexose utilization capacity in the following order: d-glucose > d-mannose > d-galactose. The sugar utilization ratio represents the sugar utilization capacity, calculated as follows: amount of sugar consumed (g L−1)/total sugar added (g L−1). The d-glucose utilization ratios of the strains Y301WXltr1p, Q291AXltr1p, and WTXltr1p were 0.97, 0.97, and 0.89, respectively, at 120 h (Fig. 4A). The respective corresponding d-glucose utilization rates were 0.158, 0.169, and 0.138 g L−1 h−1 at 120 h (Fig. 4B). Therefore, the Y301WXltr1p and Q291AXltr1p variants were inferred to possess stronger glucose transport capacity than WTXltr1p. The respective d-galactose utilization ratios of the strains Y301WXltr1p, Q291AXltr1p, and WTXltr1p were 0.021, 0.297, and 0.270 at 120 h (Fig. 4C). The respective corresponding d-galactose utilization rates were 0.004, 0.054, and 0.048 g L−1 h−1 at 120 h (Fig. 4D). The utilization rate of d-galactose by the strain expressing the Q291A mutant was 12.5 % higher than that expressing the WT transporter, possibly owing to the slightly improved transport of d-galactose by strain Q291AXltr1p compared to WTXltr1p. The d-galactose utilization rate of the Y301W mutant decreased by 91.7 % compared to that of the WT transporter, possibly due to the loss of d-galactose transport capacity by strain Y301WXltr1p. The respective d-mannose utilization ratio of the strains Y301WXltr1p, Q291AXltr1p, and WTXltr1p were 0.690, 0.970, and 0.500 at 120 h (Fig. 4E), and the respective corresponding d-mannose utilization rates were 0.124, 0.173, and 0.087 g L−1 h−1 at 120 h (Fig. 4F). The d-mannose utilization rates corresponding to the mutants Q291A and Y301W were 98.8 % and 42.5 % higher than those of the WT transporter, respectively, which could be explained by the assumption that d-mannose transport was significantly accelerated in strains Q291AXltr1p and Y301WXltr1p.
Fig. 4.
Comparison of d-glucose (A, B), d-galactose (C, D), and d-mannose (E, F) utilization ratios (A, C, E) and rates (B, D, F) by S. cerevisiae EBY.VW4000 harboring WT Xltr1p and its variants. All experiments were performed using three biological replicates. The sugar utilization ratio represents sugar utilization capacity, calculated as follows: amount of sugar consumed (g L−1)/total sugar added (g L−1). The sugar utilization rate was calculated as follows: amount of sugar consumed (g L−1)/time (h).
We performed mixed-sugar experiments under the following conditions: 1) a mixture of 10 g/L glucose and 10 g/L mannose and 2) a mixture of 10 g/L glucose and 10 g/L galactose. The utilization of mixed sugars by strains L174AXltr1p, Q291AXltr1p, Y301WXltr1p, N297AXltr1p, and WTXltr1p was investigated using galactose or mannose mixed with glucose as the carbon source in the medium. The results are shown in Fig. 5. Strains L174AXltr1p and N297AXltr1p lost their ability to utilize glucose, mannose, and galactose. Glucose was completely consumed by strains Q291AXltr1p and Y301WXltr1p in 72 h. The respective d-galactose utilization ratios of the strains Q291AXltr1p and Y301WXltr1p were 0.53 and 0.09 at 120 h. The respective d-mannose utilization ratios of strains Q291AXltr1p and Y301WXltr1p were 0.99 and 0.86 at 120 h. As mentioned above, the mannose transport capacity of Q291AXltr1p was significantly higher than that of WTXltr1p, and the galactose transport capacity of Y301WXltr1p was weakened.
Fig. 5.
Growth and sugar utilization of strains L174AXltr1p(A-B), N297AXltr1p (C-D), WTXltr1p (E-F), Q291AXltr1p (G-H) and Y301WXltr1p (I-J) in the presence of mixed sugar. All experiments were performed using three biological replicates.
3.3. Structure analyses
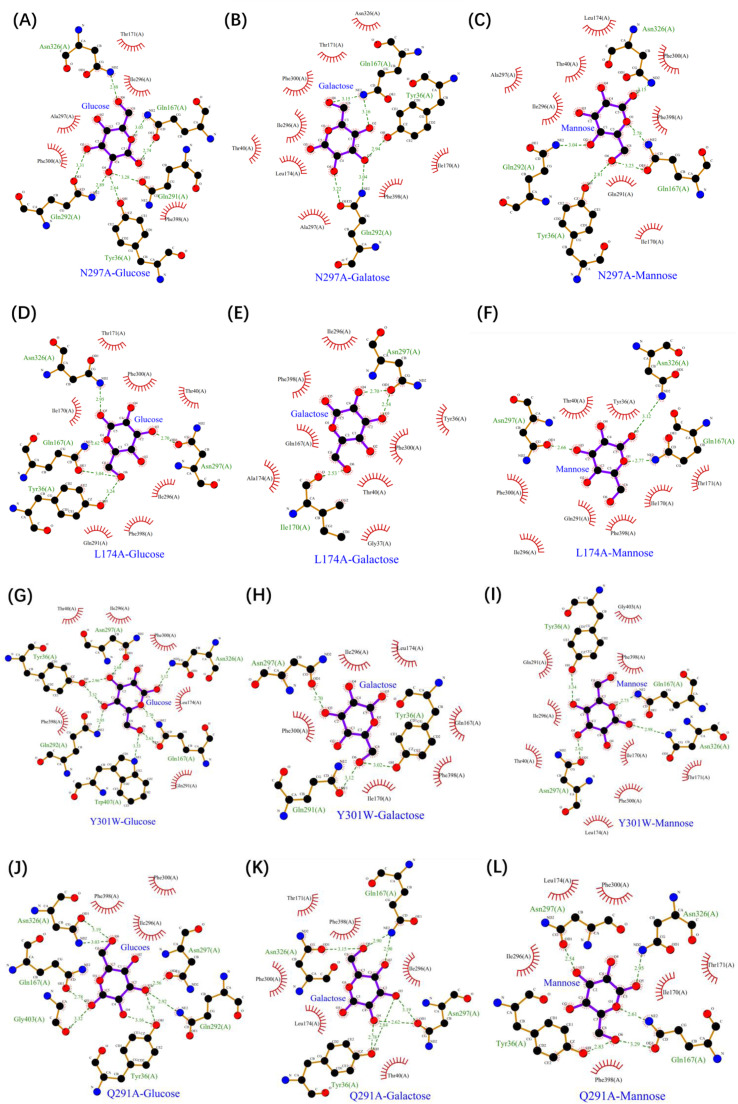
These results prompted us to explore the mechanisms governing hexose transport by Xltr1p. LigPlot software was used to analyze interactions between transporters and sugar molecules (Fig. 6). In WT Xltr1p, a hydrogen bond was observed between N297 and d-glucose, d-galactose, or d-mannose molecules, which was lost following mutation of the asparagine residue to alanine (Fig. 6A–C). Residue N297 was thus inferred to play a critical role in hexose transport by Xltr1p, consistent with the observed loss of hexose transport capacity in the N297A mutant transporter (Fig. 3 panels A5, B5, C5).
Fig. 6.
Analyses of interactions between the Xltr1p mutants N297A (A–C), L174A (D–F), Y301W (G–I), and Q291A (J–L) and d-glucose, d-galactose, or d-mannose using Ligplot software.
The L174A variant also lost its capacity to transport d-glucose, d-mannose, and d-galactose, which was attributed to a significant reduction in hydrogen-bonding interactions between the L174A variant and the sugar molecules, as evidenced by comparing the ligand–protein interaction diagrams for WT and L174A Xltr1p (Figs. 6D–F, 2C–E). In WT Xltr1p, Q167, N326, G403, W407, Q291, Y36, and N297 residues form hydrogen bonds with d-glucose, as opposed to only Y36, Q167, N326, and N297 residues in the L174A variant, maintaining hydrogen bonds with d-glucose. The Q167, N326, G403, W407, Q291, Q292, Y36, and N297 residues of WT Xltr1p form hydrogen bonds with d-galactose, as opposed to only T170 and N297 residues in the L174A variant forming hydrogen bonds with d-galactose. The Q167, N326, W407, Q291, Q292, Y36, and N297 residues of WT Xltr1p form hydrogen-bonds with d-mannose, while only N297, Q167, and N326 residues of the L174A variant form hydrogen bonds with d-mannose. These results support the important role of L174 in governing the interactions of adjacent amino acids with sugar molecules, such that its mutation results in the loss of hexose transport capacity.
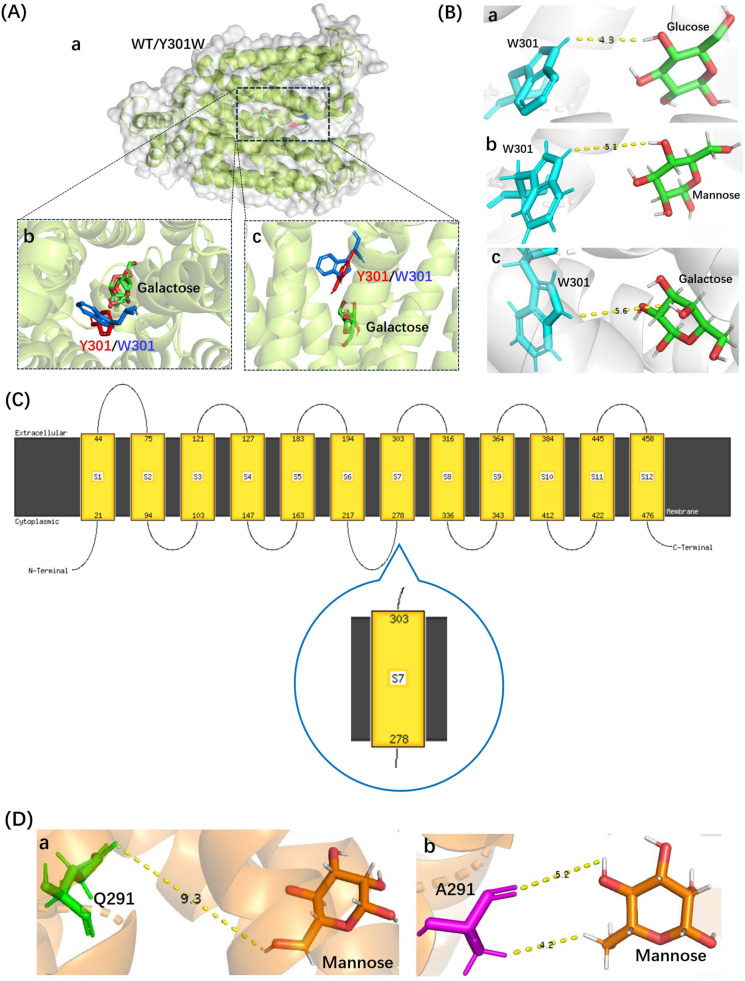
The Y301W variant of Xltr1p exhibited a loss of galactose transport capacity, consistent with the observed weaker interactions of the Y301W mutant with sugar molecules than those of WT Xltr1p (Fig. 6G–I). An analysis of the Xltr1p protein structure revealed that the benzene ring of the indole component of tryptophan 301 in the mutant protein was spatially altered to lie almost perpendicular to the pore, relative to tyrosine 301 in the WT transporter (Fig. 7A). The rotated conformation of this mutated residue was hypothesized to create a spatial barrier to d-galactose transport, resulting in loss of galactose transport capacity in the Y301W variant. The Y301W variant lost its galactose transport capacity while retaining higher glucose and mannose transport capacities. Analysis of the conformations of the three sugar molecules revealed that the configuration of the fourth carbon atom of galactose (S-configuration) differed significantly from those of the other two sugar molecules (R-configuration; Fig. S3). The distances between the tryptophan residue at site 301 in the Y301W variant and the C4 sites of d-glucose, d-mannose, and d-galactose were found to be 4.3, 5.1, and 5.6 Å, respectively (Fig. 7B). The weak interaction of Y301W with galactose was thus hypothesized to result in loss of galactose transport capacity. Prediction of the transmembrane helices of Xltr1p using DeepTMHMM (Fig. S4) and PHYRE2 (Fig. 7C) revealed that amino acid residue 301 is located in TM7, which has been previously reported to play an important role in hexose transport [31,32].
Fig. 7.
Structural analyses of the Y301W and Q291A mutants of Xltr1p. (A) Substrate docking analysis of Xltr1p (WT or Y301W) with d-galactose. a. Overall structures of WT (gray) and Y301W (green) Xltr1p with docked d-galactose, b. Vertical view, and c. Front view, of Y301 and W301 residues (before [in red] and after [in blue] mutation, respectively). (B) Schematic representation of the interaction between the W301 residue of Xltr1p Y301W and d-glucose (a), d-mannose (b), or d-galactose (c). (C) Predictions of transmembrane helices using PHYRE2. Predictions using DeepTMHMM are shown in Fig. S4. (D) Schematic representation of the interactions between Q291 of WT Xltr1p (a) or A291 of Q291A (b) with d-mannose.
The Q291A mutation greatly accelerated d-mannose transport. Similar analyses revealed that when glutamine (Q291) was mutated to alanine (A), more hydrophobic interactions were formed between the alanine and mannose molecules (Fig. 6L). We thus speculated that the mutation of glutamine (Q) to the small side-chain residue alanine (A) favored the passage of mannose. Conformational analyses of the three sugar molecules revealed that the configuration of the second carbon atom of d-mannose (S-configuration) differed greatly from those of the other two sugar molecules (R-configuration; Fig. S3). The spatial distance and orientation of the d-mannose molecule with respect to residue 291 of Xltr1p were altered by the mutation, as shown (Fig. 7D). The Q291A mutation was hypothesized to affect interactions of the surrounding amino acids with the d-mannose molecule, as shown (Figs. 2E, 6L, and 7D), resulting in easier passage of molecules with a configuration corresponding to that of d-mannose. Prediction of transmembrane helices revealed that residues N297, Q291, and Y301 are located in TM7. These amino acid residues have been found to play an important role in sugar transport, in agreement with previous reports [31,32].
4. Conclusions
The current study successfully identified four amino acid residues (L174, N297, Q291, and Y301) that play key roles in the transport of glucose, galactose, and mannose by Xltr1p. Computer-assisted screening followed by experimental validation revealed a loss of hexose transport capacity in Xltr1p mutants L174A and N297A, revealing an important role for residues L174 and N297 in hexose transport by Xltr1p. The utilization rate of d-mannose by strain Q291AXltr1p was 98 % higher than that of WTXltr1p, prompting speculation that the mannose transporter capacity of the Q291A variant was higher than that of WT Xltr1p, which is expected to impact utilization of mixed carbon sources. The Y301W mutant, compared to WT Xltr1p, retained glucose utilization capacity and exhibited accelerated mannose utilization, but lost its capacity to utilize galactose. This observation, in turn, is expected to permit improved selectivity and specificity for glucose and galactose in S. cerevisiae strains harboring the mutant transporter. The transporter modification strategy employed in the current study can be exploited as a reference for the protein engineering of specific hexose transporters.
CRediT authorship contribution statement
Wei Ma: Writing – review & editing, Writing – original draft, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Shiyu Yuan: Supervision, Formal analysis. Zixian Wang: Supervision, Formal analysis. Kangle Niu: Supervision, Methodology. Fengyi Li: Writing – original draft, Supervision. Lulu Liu: Supervision, Formal analysis. Lijuan Han: Supervision, Methodology. Xu Fang: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Dr. Xiangmei Ren from State Key Laboratory of Microbial Technology of Shandong University for help and guidance in the high-performance liquid chromatography. We thank Prof. Dr Eckhard Boles from the Institutfür Molekulare Biowissenschaf-ten Goethe-Universität Frankfurt for kindly providing the EBY.VW4000 strain. This work was supported by National Key R&D Program of China (No. 2018YFA0901700), National Natural Science Foundation of China (No. 32271526).
Chemical compounds studied in this article
d-Glucose (PubChem CID:5793), d-Mannose (PubChem CID:18950), d-Galactose (PubChem CID:6036).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.engmic.2024.100151.
Appendix. Supplementary materials
Data Availability Statement
The original/source data and resources are available from the correspondence author Xu Fang (fangxu@sdu.edu.cn) on request.
References
- 1.Bittihn P., Din M.O., Tsimring L.S., Hasty J. Rational engineering of synthetic microbial systems: from single cells to consortia. Curr. Opin. Microbiol. 2018;45:92–99. doi: 10.1016/j.mib.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarty N.S., Ledesma-Amaro R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 2019;37(2):181–197. doi: 10.1016/j.tibtech.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song H., Ding M.Z., Jia X.Q., Ma Q., Yuan Y.J. Synthetic microbial consortia: from systematic analysis to construction and applications. Chem. Soc. Rev. 2014;43(20):6954–6981. doi: 10.1039/C4CS00114A. [DOI] [PubMed] [Google Scholar]
- 4.Said S.B., Tecon R., Borer B., Or D. The engineering of spatially linked microbial consortia–potential and perspectives. Curr. Opin. Biotechnol. 2020;62:137–145. doi: 10.1016/j.copbio.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns N.I., Blazejewski T., Gomes A.L., Wang H.H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 2016;31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao B., Sabnis R., Costantini T., Jinkerson R., Sun Q. A peek in the micro-sized world: a review of design principles, engineering tools, and applications of engineered microbial community. Biochem. Soc. Trans. 2020;48(2):399–409. doi: 10.1042/BST20190172. [DOI] [PubMed] [Google Scholar]
- 7.Song H., Payne S., Tan C., You L. Programming microbial population dynamics by engineered cell–cell communication. Biotechnol. J. 2011;6(7):837–849. doi: 10.1002/biot.201100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang J.S. Engineering multicellular traits in synthetic microbial populations. Curr. Opin. Chem. Biol. 2012;16(3–4):370–378. doi: 10.1016/j.cbpa.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Bacchus W., Fussenegger M. Engineering of synthetic intercellular communication systems. Metab. Eng. 2013;16:33–41. doi: 10.1016/j.ymben.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Brenner K., You L., Arnold F.H. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26(9):483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Wintermute E.H., Silver P.A. Dynamics in the mixed microbial concourse. Genes Dev. 2010;24(23):2603–2614. doi: 10.1101/gad.1985210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarr P.T., Tarling E.J., Bojanic D.D., Edwards P.A., Baldán Á. Emerging new paradigms for ABCG transporters. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids. 2009;1791(7):584–593. doi: 10.1016/j.bbalip.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locher K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016;23(6):487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 14.Lengeler J.W., Titgemeyer F., Vogler A.P., Wohrl B.M. Structures and homologies of carbohydrate: phosphotransferase system (PTS) proteins. Philo. Trans. R. Soc. Lond. B, Biol. Sci. 1990;326(1236):489–504. doi: 10.1098/rstb.1990.0027. [DOI] [PubMed] [Google Scholar]
- 15.Postma P.W., Lengeler J.W., Jacobson G. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 1993;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pao S.S., Paulsen I.T., Saier M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S.C., Davejan P., Hendargo K.J., Javadi-Razaz I., Chou A., Yee D.C., Ghazi F., Lam K.J.K., Conn A.M., Madrigal A., Medrano-Soto A., Saier M.H., Jr. Expansion of the major facilitator superfamily (MFS) to include novel transporters as well as transmembrane-acting enzymes. Biochim. Biophys. Acta (BBA)-Biomembr. 2020;1862(9) doi: 10.1016/j.bbamem.2020.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao X., Shen B., Li M., Ye L., Yu H. Secretory production of tocotrienols in Saccharomyces cerevisiae. ACS Synth. Biol. 2022;11(2):788–799. doi: 10.1021/acssynbio.1c00484. [DOI] [PubMed] [Google Scholar]
- 19.Tippelt A., Nett M. Saccharomyces cerevisiae as host for the recombinant production of polyketides and nonribosomal peptides. Microb. Cell Fact. 2021;20(1):1–24. doi: 10.1186/s12934-021-01650-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anjos J., de Sousa H.R., Roca C., Cássio F., Luttik M., Pronk J.T., Gonçalves P. Fsy1, the sole hexose-proton transporter characterized in Saccharomyces yeasts, exhibits a variable fructose: H+ stoichiometry. Biochim. Biophys. Acta (BBA)-Biomembr. 2013;1828(2):201–207. doi: 10.1016/j.bbamem.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Ng T.S., Chew S.Y., Rangasamy P., Mohd Desa M.N., Sandai D., Chong P.P., Than L.T.L. SNF3 as high affinity glucose sensor and its function in supporting the viability of Candida glabrata under glucose-limited environment. Front. Microbiol. 2015;6(1334) doi: 10.3389/fmicb.2015.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamacher T., Becker J., Gárdonyi M., Hahn-Hagerdal B., Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148(9):2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- 23.Sedlak M., Ho N.W. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast. 2004;21(8):671–684. doi: 10.1002/yea.1060. [DOI] [PubMed] [Google Scholar]
- 24.Young E., Poucher A., Comer A., Bailey A., Alper H. Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl. Environ. Microbiol. 2011;77(10):3311–3319. doi: 10.1128/AEM.02651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas S.A.T., Schadeweg V., Kirchner F., Boles E., Oreb M. Identification of a glucose-insensitive variant of Gal2 from Saccharomyces cerevisiae exhibiting a high pentose transport capacity. Sci. Rep. 2021;11(1):24404. doi: 10.1038/s41598-021-03822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colabardini A.C., Ries L.N.A., Brown N.A., Dos Reis T.F., Savoldi M., Goldman M.H., Menino J.F., Rodrigues F., Goldman G.H. Functional characterization of a xylose transporter in Aspergillus nidulans. Biotechnol. Biofuels. 2014;7:46. doi: 10.1186/1754-6834-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishizawa K., Shimoda E., Kasahara M. Substrate recognition domain of the Gal2 galactose transporter in yeast Saccharomyces cerevisiae as revealed by chimeric galactose-glucose transporters. J. Biol. Chem. 1995;270(6):2423–2426. doi: 10.1074/jbc.270.6.2423. [DOI] [PubMed] [Google Scholar]
- 28.Kasahara M., Shimoda E., Maeda M. Amino acid residues responsible for galactose recognition in yeast Gal2 transporter. J. Biol. Chem. 1997;272(27):16721–16724. doi: 10.1074/jbc.272.27.16721. [DOI] [PubMed] [Google Scholar]
- 29.Kasahara M., Maeda M. Contribution to substrate recognition of two aromatic amino acid residues in putative transmembrane segment 10 of the yeast sugar transporters Gal2 and Hxt2. J. Biol. Chem. 1998;273(44):29106–29112. doi: 10.1074/jbc.273.44.29106. [DOI] [PubMed] [Google Scholar]
- 30.Kasahara T., Kasahara M. Tryptophan 388 in putative transmembrane segment 10 of the rat glucose transporter Glut1 is essential for glucose transport. J. Biol. Chem. 1998;273(44):29113–29117. doi: 10.1074/jbc.273.44.29113. [DOI] [PubMed] [Google Scholar]
- 31.Qiao Y., Li C., Lu X., Zong H., Zhuge B. Identification of key residues for efficient glucose transport by the hexose transporter CgHxt4 in high sugar fermentation yeast Candida glycerinogenes. Appl. Microbiol. Biotechnol. 2021;105:7295–7307. doi: 10.1007/s00253-021-11567-6. [DOI] [PubMed] [Google Scholar]
- 32.Kasahara T., Kasahara M. Identification of a key residue determining substrate affinity in the yeast glucose transporter Hxt7: a two-dimensional comprehensive study. J. Biol. Chem. 2010;285(34):26263–26268. doi: 10.1074/jbc.M110.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y., Shen Y., Gu L., Wang Z., Su N., Niu K., Guo W., Hou S., Bao X., Tian C., Fang X. Identification and characterization of an efficient d-xylose transporter in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020;68(9):2702–2710. doi: 10.1021/acs.jafc.9b07113. [DOI] [PubMed] [Google Scholar]
- 34.Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C.P., Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464(3):123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 35.Guo W., Huang Q., Feng Y., Tan T., Niu S., Hou S., Chen Z., Du Z., Shen Y., Fang X. Rewiring central carbon metabolism for tyrosol and salidroside production in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2020;117(8):2410–2419. doi: 10.1002/bit.27370. [DOI] [PubMed] [Google Scholar]
- 36.Guo W., Huang Q., Liu H., Hou S., Niu S., Jiang Y., Bao X., Shen Y., Fang X. Rational engineering of chorismate-related pathways in Saccharomyces cerevisiae for improving tyrosol production. Front. Bioeng. Biotechnol. 2019;7:152. doi: 10.3389/fbioe.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C., Bao X., Li Y., Jiao C., Hou J., Zhang Q., Zhang W., Liu W., Shen Y. Cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Metab. Eng. 2015;30:79–88. doi: 10.1016/j.ymben.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1):W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016;54(1):5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., Ye Z., Qiu J., Feng R., Li D., Hsieh C., Allcock J., Zhang S. A mutation-induced drug resistance database (MdrDB) Commun. Chem. 2023;6(1):123. doi: 10.1038/s42004-023-00920-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original/source data and resources are available from the correspondence author Xu Fang (fangxu@sdu.edu.cn) on request.