Abstract
Members of the Closteroviridae and Potyviridae families of the plant positive-strand RNA viruses encode one or two papain-like leader proteinases. In addition to a C-terminal proteolytic domain, each of these proteinases possesses a nonproteolytic N-terminal domain. We compared functions of the several leader proteinases using a gene swapping approach. The leader proteinase (L-Pro) of Beet yellows virus (BYV; a closterovirus) was replaced with L1 or L2 proteinases of Citrus tristeza virus (CTV; another closterovirus), P-Pro proteinase of Lettuce infectious yellows virus (LIYV; a crinivirus), and HC-Pro proteinase of Tobacco etch virus (a potyvirus). Each foreign proteinase efficiently processed the chimeric BYV polyprotein in vitro. However, only L1 and P-Pro, not L2 and HC-Pro, were able to rescue the amplification of the chimeric BYV variants. The combined expression of L1 and L2 resulted in an increased RNA accumulation compared to that of the parental BYV. Remarkably, this L1-L2 chimera exhibited reduced invasiveness and inability to move from cell to cell. Similar analyses of the BYV hybrids, in which only the papain-like domain of L-Pro was replaced with those derived from L1, L2, P-Pro, and HC-Pro, also revealed functional specialization of these domains. In subcellular-localization experiments, distinct patterns were observed for the leader proteinases of BYV, CTV, and LIYV. Taken together, these results demonstrated that, in addition to a common proteolytic activity, the leader proteinases of closteroviruses possess specialized functions in virus RNA amplification, virus invasion, and cell-to-cell movement. The phylogenetic analysis suggested that functionally distinct L1 and L2 of CTV originated by a gene duplication event.
Papain-like cysteine proteinases of the positive-strand RNA viruses are multifunctional proteins involved not only in polyprotein processing but also in genome amplification, virus pathogenicity and spread in the infected organism, and suppression of host defenses (13–15, 26, 31, 39, 40, 44). Two major classes of the viral papain-like proteinases include “main” proteinases, which are required for the processing of nonstructural polyproteins and RNA replication (3, 13, 18, 39) and “accessory” or “leader” proteinases, which are typically responsible for a single autocatalytic cleavage at their C termini (1, 4, 5, 8, 9, 13, 14, 31, 44). Representatives of the each of these classes are found among diverse taxa of positive-strand RNA viruses infecting plants, animals, and fungi.
Closteroviridae and Potyviridae are two large families of plant viruses that share the filamentous morphology of the virions but belong to evolutionarily distant lineages of the positive-strand RNA viruses, the Sindbis virus-like supergroup and the picornavirus-like supergroup, respectively (26). An ∼10-kb genome of a typical potyvirus codes for a single polyprotein, which is processed by the three proteinases (35). One of these is a helper component-proteinase (HC-Pro), a leader proteinase that is also required for efficient genome amplification, suppression of RNA silencing, virus transport inside infected plants, and aphid transmission (6, 29). The papain-like, proteolytically active domain of the HC-Pro is located in its C-terminal region (4), whereas the large N-terminal domain is implicated in all additional functions of HC-Pro (21–23, 29).
The 15- to 20-kb genomes of closteroviruses are among the largest and most complex of all the genomes of viruses infecting plants (11, 19). Two currently recognized genera of the family Closteroviridae are Closterovirus and Crinivirus, with Beet yellows virus (BYV) and Lettuce infectious yellows virus (LIYV) as the prototype members, respectively. Although the gene content varies from one virus to another, all closteroviruses share the strategy of gene expression. 5′-terminal open reading frame 1 (ORF 1; see Fig. 1) encodes a large polyprotein that functions in genome amplification (25, 33, 36). The N-terminal part of this polyprotein encompasses the leader proteinase (1), which is traditionally abbreviated L-Pro, in BYV (33) and P-Pro in LIYV (24). Some closteroviruses, such as Citrus tristeza virus (CTV), possess two tandemly organized leader proteinases, designated L1 and L2 (20). The 3′-terminal part of the closterovirus genome harbors from 7 to 10 ORFs, which are expressed via formation of a set of 3′-coterminal subgenomic mRNAs (sgRNAs) (16, 30). In LIYV and other members of the Crinivirus genus, the genome is split between two RNAs; RNA 1 encodes P-Pro and replicase, whereas RNA 2 specifies most of the LIYV sgRNAs (24). It was demonstrated that the release of the BYV L-Pro from the polyprotein is mediated by the autocatalytic, papain-like domain (1, 33). This release is essential for genome replication. The N-terminal, nonproteolytic domain of L-Pro is required for efficient accumulation of BYV RNAs; its elimination results in an ∼1,000-fold reduction in the RNA levels (32).
FIG. 1.
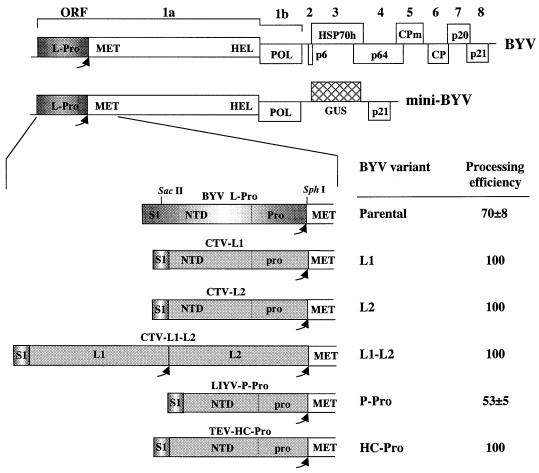
Diagrams of the BYV genome (top) and the cDNA clone of a mini-BYV variant tagged by insertion of the GUS gene. BYV ORFs 1 to 8 encode L-Pro, a replicase that harbors methyltransferase (MET), RNA helicase (HEL) and RNA polymerase (POL) domains, a 6-kDa protein (p6), an HSP70 homolog (HSP70 h), a 64-kDa protein (p64), a minor capsid protein (CPm), a major capsid protein (CP), a 20-kDa protein (p20), and a 21-kDa protein (p21). Arrows, self-processing sites for the viral leader proteinases. An expanded diagram of the BYV L-Pro-coding region and its chimeric variants is shown below. SacII and SphI endonuclease restriction sites engineered to facilitate generation of the replacement mutants are shown. S1, a short region of L-Pro coding sequence (32); NTD, N-terminal, nonproteolytic domains; Pro and pro, papain-like, proteolytic domains of BYV L-Pro and foreign proteinases, respectively.
In this study we conducted comparative analyses of the leader proteinases of plant viruses using a gene swapping approach and computer-assisted phylogenetic reconstructions. Our results indicate that the nonconserved N-terminal domains of the leader proteinases provide several distinct functions, which may vary from one proteinase to another. Moreover, conserved papain-like domains of the leader proteinases also exhibited an unexpected degree of functional specialization. In addition to being involved in autocatalytic processing, these domains were implicated in activation of genome amplification, virus invasion, and virus movement from cell to cell.
MATERIALS AND METHODS
Generation of the chimeric BYV variants.
To generate a cassette for replacing the BYV L-Pro-coding region with regions encoding foreign leader proteinases, two restriction endonuclease sites were engineered into pBYV-GUS-p21 (16) (see Fig. 1). One of these sites, a SacII site, was introduced as mutation A2, described earlier (32). The second site, an SphI site, was engineered downstream from the two glycine codons that specify a scissile dipeptide using oligonucleotide primer GG-Sph (5′-CGT.TTC.ATC.GGC-GGC.ATG.CAA.GAA.GAA.GCT.CCT.G; dots separate codons; a hyphen separates two glycine codons; the SphI site is in boldface). This modification resulted in replacement of valine and glutamic acid codons with methionine and glutamine codons, respectively. The regions encoding tobacco etch virus (TEV) proteinase HC-Pro, CTV proteinases L1 and L2, a combination of L1 and L2, and LIYV proteinase P-Pro were amplified by PCR using biologically active cDNA clones of TEV (10), CTV (36), and LIYV (25) as templates. The SacII and SphI sites flanking the resulting cDNA fragments were introduced concomitant with PCR and used to clone these fragments into mini-BYV (Fig. 1). The chimeric leader proteinases shown in Fig. 2 possessed authentic N-terminal domains of BYV L-Pro (encoded by the sequence up to nucleotide [nt] 1443) and papain-like proteinase domains derived from TEV (encoded by nt 1957 to 2436), CTV L1 (encoded by nt 1114 to 1563) and L2 (encoded by nt 2593 to 3039), and LIYV (encoded by nt 956 to 1336). The engineering of the corresponding chimeric variants of mini-BYV was done as described above, except that the SacII site corresponded to mutation A12 rather than A2 (32). The four chimeric BYV variants shown in Table 3 were subcloned into pBYV-GFP, the BYV variant tagged via insertion of the green fluorescent protein (GFP) gene (34). To this end, the NheI-EagI fragment of pBYV-GFP was replaced with corresponding chimeric cDNA fragments derived from mini-BYV variants.
FIG. 2.
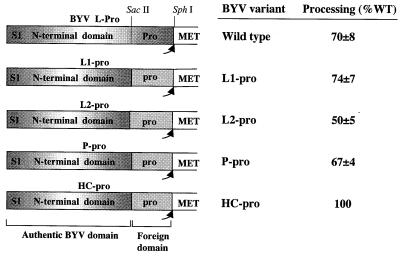
Diagrams of the chimeric variants in which the authentic N-terminal domain of BYV L-Pro (Pro) was fused with the foreign proteinase domains derived from CTV L1 or L2, LIYV P-Pro, and TEV HC-Pro (each designated pro). SacII and SphI endonuclease restriction sites engineered to facilitate generation of the chimeric variants are shown. WT, wild type.
TABLE 3.
Specific infectivity and cell-to-cell movement of the chimeric BYV variants in the leaves of C. perfoliata
| BYV-GFP variant | No. of foci per leaf | Mean diam (cells) |
|---|---|---|
| Parent | 12.4 ± 3.5 | 4.3 ± 1.8 |
| CTV-L1 | 0.5 ± 0.2 | 1 |
| CTV-L1-L2 | 0.8 ± 0.3 | 1 |
| LIYV-P-Pro | 0.6 ± 0.2 | 1 |
| CTV-L1-pro | 0.9 ± 0.3 | 1 |
Analysis of the mutant phenotypes in vitro and in vivo.
The plasmids containing cloned BYV cDNAs were linearized using XbaI (for in vitro analysis) or SmaI (for in vivo experiments) and transcribed using SP6 RNA polymerase (33). To assess the proteolytic activity of the leader proteinases, the resulting capped-RNA transcripts were translated using wheat germ extracts (Promega) and [35S]methionine (Amersham/Pharmacia Biotech) according to the manufacturer's protocol. After 1 h of incubation at 25°C, the labeled translation products were separated by polyacrylamide gel electrophoresis, and the radioactivity in the bands corresponding to processed and unprocessed products was quantified using a PhosphorImager (Molecular Dynamics) and the ImageQuant, version 5, software package. This radioactivity was normalized to the number of methionine residues present in each product and used to calculate the efficiency of proteolysis as described previously (33). The results represent means and standard deviations from four independent reactions. The mini-BYV variants were further characterized using protoplasts isolated from a suspension culture of Nicotiana tabacum cells (12) or from the leaves of Nicotiana benthamiana (36). Protoplasts were harvested at 4 days posttransfection and used to measure the β-glucuronidase (GUS) activity (10). Each recombinant variant was characterized in four independent transfections. The pBYV-GFP-based variants were manually inoculated to leaves of Claytonia perfoliata or N. benthamiana, and the number and diameter of the resulting fluorescent infection foci were determined at 8 days postinoculation (34). At least four independent inoculation experiments, each involving six leaves, were conducted for each variant. The total numbers of infection foci counted were ∼300 for the parental BYV-GFP and ∼20 for the chimeric viruses.
Subcellular localization of the leader proteinases.
The expression cassette encompassing the duplicated cauliflower mosaic virus 35S promoter, the TEV leader, the poly(A) signal (5), and the coding sequence for GFP was cloned into binary vector pCB302 (42). The genes encoding GUS, L-Pro, L1 and L2, and P-Pro were amplified by PCR using the full-length cDNA clones of TEV-GUS (10), BYV (34), CTV (36), and LIYV (25), respectively, with the concomitant addition of the AvrII and XbaI sites into their 5′- and 3′-terminal regions. The translation stop codons were introduced into amplified genes upstream from the XbaI sites. The modified genes were cloned downstream from the GFP gene using the AvrII and XbaI sites. The resulting plasmids were transformed into Agrobacterium tumefaciens strain EHA 105, and used for transient protein expression via infiltration of the bacteria to the leaves of N. benthamiana (28). GFP fluorescence was detected at 2 days postinfiltration using a confocal laser scanning microscope (Leica; TCS 4D). For GFP imaging, a 488/568-nm excitation beam generated by a krypton-argon laser was used with an RSP580 beam splitter and BP-fluorescein isothiocyanate emission filter.
Phylogenetic analysis.
The multiple alignments of amino acid sequences were generated using the Macaw program (37). The Gibbs sampler option of Macaw was used to detect the blocks with highest sequence similarity. The Phylip package (J. Felsenstein, PHYLIP, Phylogeny Inference Package, version 3.5c, Department of Genetics, University of Washington, Seattle, 1993) was used for construction of the phylogenetic trees. One hundred bootstrap replicates were obtained using the SEQBOOT program; the trees were built using the neighbor-joining algorithm (NEIGHBOR program) or maximum-likelihood algorithm (KITSCH program).
RESULTS
To facilitate the generation and characterization of interviral hybrids, we used a cDNA clone of a mini-BYV variant that was tagged by insertion of the bacterial GUS gene (16). This reporter gene replaced six BYV ORFs that are nonessential for the genome amplification (Fig. 1) and provided a convenient and sensitive marker for quantification of the amplification and expression of the viral genome. It was demonstrated that accumulation of the GUS activity strictly correlates with accumulation of the viral RNAs (32).
Replacement of the BYV L-Pro with heterologous leader proteinases.
Each of the cDNA fragments encoding the foreign proteinase replaced almost the entire BYV L-Pro-coding region (Fig. 1). The 70-codon part of this region designated S1 was retained because it contains an RNA element that is crucial for BYV RNA amplification (32). Hence, each of the foreign leader proteinases expressed by chimeric BYV variants possessed an N-terminal extension. Two artificial restriction endonuclease sites were introduced immediately downstream from the S1 region and from the last L-Pro codon to accommodate the foreign inserts (see Materials and Methods). The possible effect of corresponding mutations on the amplification and expression of the BYV genome was assessed using transfection of the corresponding RNA transcript into tobacco protoplasts. It was found that the resulting double mutant amplified to a level similar to that of the parental BYV variant (data not shown).
In addition to replacing the BYV L-Pro with the CTV L1 or L2, the LIYV P-Pro, or the TEV HC-Pro, we engineered a mini-BYV variant containing both of the CTV proteinases to mimic the tandem organization of the L1-L2 region of the CTV polyprotein (Fig. 1). The processing of chimeric polyproteins was examined in the in vitro translation system. As in previous studies (1, 33), the authentic BYV L-Pro was able to process ∼70% of the translation product after 1 h of incubation in the cell-free system. In contrast, the processing of the chimeric translation products by the CTV L1 and L2 and the TEV HC-Pro was essentially complete. For LIYV P-Pro, processing efficiency was somewhat lower than that for the parental BYV variant (Fig. 1).
The genome amplification of the chimeric mini-BYV variants was examined using transfection of two types of protoplasts: suspension culture protoplasts derived from N. tabacum and N. benthamiana leaf protoplasts. Replacement of the BYV L-Pro with each of the CTV leader proteinases resulted in two conspicuously distinct phenotypes. L1 was able to partially replace the L-Pro function in genome amplification, whereas L2 failed to do so (Table 1). However, the replacement hybrid expressing the combination of L1 and L2 reproduced more efficiently than one expressing L1 only. In N. benthamiana protoplasts, this L1-L2 variant reproducibly outperformed the parental BYV variant expressing the authentic L-Pro.
TABLE 1.
GUS activity in N. tabacum and N. benthamiana protoplasts transfected with the chimeric BYV variants harboring full-size, heterologous leader proteinases
| BYV variant | Mean % of mini-BYV activity ± SD for:
|
|
|---|---|---|
| N. tabacum | N. benthamiana | |
| Parent | 100 | 100 |
| CTV-L1 | 10 ± 2 | 29 ± 5 |
| CTV-L2 | <0.001 | <0.001 |
| CTV-L1-L2 | 19 ± 5 | 185 ± 25 |
| LIYV-P-Pro | 2 ± 1 | 34 ± 6 |
| TEV HC-Pro | <0.001 | <0.001 |
The functional profile of the replacement chimera harboring LIYV P-Pro resembled that of L1: P-Pro supported the amplification of the chimeric genome much more efficiently in N. benthamiana protoplasts than in N. tabacum protoplasts (Table 1). In contrast, the TEV HC-Pro was unable to functionally substitute for the BYV L-Pro in either of the protoplast systems. These results revealed a high degree of functional specialization of the closterovirus leader proteinases and indicated that CTV L1 and LIYV P-Pro, but not CTV L2 or TEV HC-Pro, can provide functions required for the efficient amplification of the chimeric mini-BYV genome. The strikingly better performance of all three viable replacement variants in N. benthamiana protoplasts than in N. tabacum protoplasts suggested that the function of leader proteinases is affected by species-specific host factors.
Functional specialization of the papain-like proteolytic domains.
To determine if the sole function of the papain-like domains of the closteroviral and potyviral leader proteinases is autoprocessing, we engineered a series of chimeric viruses in which only the C-terminal proteinase domain of L-Pro was replaced with the homologous domains derived from L1, L2, P-Pro, and HC-Pro (Fig. 2). If autoprocessing is the sole function, the papain-like domains of different viruses would be functionally equivalent to each other. In vitro assays revealed that the proteolytic activities of the chimeric proteinases harboring papain-like domains of L1 and P-Pro were not significantly different from that of the parental variant (Fig. 2). The activity of the L2 chimera was somewhat lower, whereas the TEV proteinase domain processed ∼100% of the polyprotein.
The in vivo experiments indicated that only the CTV proteinase domains derived from L1 and L2 were capable of supporting limited genome amplification of the corresponding chimeric variants (Table 2). None of the variants that harbored LIYV or TEV proteinase domains were viable. Comparison of the data in Fig. 2 and Table 2 reveals no apparent correlation between the levels of proteolytic activity and genome amplification of the chimeric variants. It should be emphasized that, unlike what was found for chimeras expressing the full-size L1, L2, P-Pro, and HC-Pro, reproduction of those expressing only the corresponding proteinase domains did not depend on the source of protoplasts (compare Tables 1 and 2). It can be concluded that, in addition to proteolytic processing, the homologous, papain-like proteolytic domains of the closterovirus leader proteinases have additional specialized functions required for efficient genome amplification.
TABLE 2.
GUS activity in N. tabacum and N. benthamiana protoplasts transfected with the chimeric BYV variants harboring heterologous proteinase domains
| BYV variant | Mean % of mini-BYV activity ± SD for:
|
|
|---|---|---|
| N. tabacum | N. benthamiana | |
| Parent | 100 | 100 |
| CTV-L1-pro | 11 ± 3 | 14 ± 2 |
| CTV-L2-pro | 2 ± 1 | 3 ± 1 |
| LIYV-P-pro | <0.001 | <0.001 |
| TEV HC-pro | <0.001 | <0.001 |
Invasiveness and cell-to-cell movement of the hybrid viruses.
To examine the phenotypes of the chimeric viruses in intact plants, we employed the BYV variant tagged via insertion of the reporter gene encoding GFP (BYV-GFP). A subset of viable chimeric variants that accumulated in N. benthamiana protoplasts to the levels of 14 to 185% of the wild-type level (Tables 2 and 3) was engineered into BYV-GFP. Other variants were not included in this analysis, because the very low (<3%) levels of their amplification would not allow confident identification of infected cells by observation of the GFP fluorescence. The corresponding RNA transcripts were mechanically inoculated to the leaves of C. perfoliata. The multicellular infection foci formed by BYV-GFP can be easily detected and measured using the fluorescence microscope. At 8 days postinoculation, the parental virus produced on average 12 infection foci per leaf; the mean diameter of the foci was ∼4 cells (Table 3). In contrast, numbers of the infection foci produced by the chimeric variants were reduced by factors of from 14 to 25. Moreover, all of these foci were unicellular (Table 3). Importantly, a similar phenotype of infection was observed when only the proteinase domain of L-Pro was replaced with that of CTV L1.
Although C. perfoliata is susceptible to infection by BYV via local lesions, it is not a reported host for CTV or LIYV. Because of that, this plant species could impose host-specific constraints on the functions of CTV and LIYV proteinases. To determine if the invasiveness and intercellular translocation of the chimeric viruses can be improved in a more permissive host, we inoculated leaves of N. benthamiana with a BYV-GFP variant expressing CTV L1 and L2. As shown above, the leaf protoplasts derived from this plant species supported very efficient amplification of the L1-L2 chimera (Table 1). In accord with the earlier work (34), the average number of infection foci found on N. benthamiana leaves was much less than that on the C. perfoliata leaves. The parental BYV-GFP and its L1-L2 replacement chimera produced 0.94 ± 0.3 and 0.83 ± 0.2 foci per leaf, respectively. The mean diameters of the infection foci were 4.3 ± 2.1 cells for BYV-GFP and 1 cell for the L1-L2 chimera. Thus, the specific infectivity (invasiveness), but not the cell-to-cell movement, of the L1-L2 chimera was improved in N. benthamiana compared to C. perfoliata.
Taken together, these results clearly indicated that the replacement of the authentic BYV L-Pro with proteinases derived from other members of the family Closteroviridae resulted in a dramatic decrease in invasiveness of the chimeric RNA transcripts. Moreover, these chimeric viruses completely lost the ability to move from cell to cell.
Subcellular localization of the GFP-leader proteinase fusion proteins.
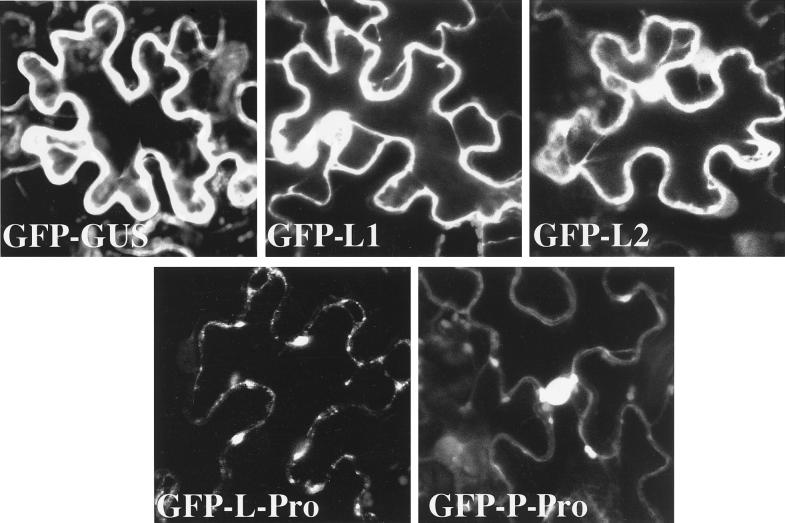
To determine if the closterovirus leader proteinases possess signals for targeting to specific cellular compartments, we employed Agrobacterium-mediated, transient expression of these proteins in the leaves of N. benthamiana. Each of the ORFs encoding BYV L-Pro, CTV L1 and L2, and LIYV P-Pro was fused in frame with the 3′ terminus of the GFP ORF. An analogous GFP-GUS fusion was used as a control because neither of these reporter proteins possesses specific targeting signals. Furthermore, the molecular mass of GFP-GUS (∼95 kDa) is similar to that of GFP–L-Pro (∼93 kDa) and other tested fusion products. Figure 3 shows that the green fluorescence of GFP-GUS was uniformly distributed in the cytosol, although a fraction of the product in some cells was localized to nuclei (not shown). A similar pattern of subcellular localization was observed for the CTV L1 and L2 fusion products, whereas GFP–L-Pro formed distinct cytoplasmic inclusion bodies (Fig. 3). In contrast, very little of the GFP–P-Pro was detected in the cytosol; most of the fluorescence was confined to the nuclei (Fig. 3).
FIG. 3.
Subcellular localization of the GUS (control) and viral leader proteinases fused to the GFP reporter. White indicates GFP-specific fluorescence, whereas small gray bodies are the autofluorescent chloroplasts.
Phylogenetic analysis of the closterovirus papain-like proteinases.
We examined the evolutionary relationships of the leader proteinases from BYV, CTV, LIYV, and two additional members of the family Closteroviridae, Grapevine leafroll-associated virus 2 (GLRaV-2) (43) and Little cherry virus (17). The multiple alignment of corresponding amino acid sequences clearly revealed the conserved C-terminal domain that possessed signature motifs common to the viral papain-like proteinases (Fig. 4). In contrast, extensive comparisons of the N-terminal, nonproteolytic domains revealed no amino acid motifs common to all included leader proteinases. Only a limited conservation in the regions located upstream from the proteinase domains was observed in subsets of leader proteinases (e.g., CTV L1 and L2; data not shown).
FIG. 4.
Multiple alignment of the amino acid sequences of the papain-like proteinase domains encoded in diverse representatives of the family Closteroviridae. GLR, GLRaV-2 (43); LChV, Little cherry virus (17). Invariant residues are in bold face, and blocks of conserved residues are capitalized. ∗, cysteine residue predicted to directly attack the scissile peptide bond and the histidine residue also likely to participate in catalysis; arrow and gap, scissile bond between a glycine and another small residue (glycine, alanine, or serine).
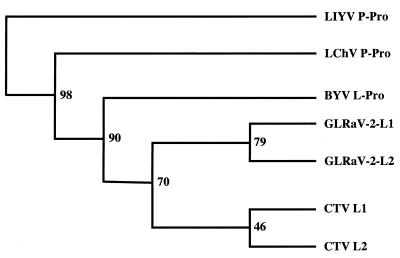
The phylogenetic trees of the papain-like domains based on the neighbor-joining algorithm (Fig. 5) and maximum-likelihood algorithm (not shown) were very similar. The proteinase of LIYV was selected as an outgroup in accordance with the distinct genome organization and sequence relationships of the LIYV replicational proteins (19). Notably, specific affinities between the L1 and L2 proteinase domains in both CTV and GLRaV-2 were observed. These pairs of proteinases appear to be more closely related to each other than to homologs present in other family members. This tree topology suggests intragenomic duplication as a likely scenario for the origin of the tandem genes encoding L1 and L2 of CTV and GLRaV-2.
FIG. 5.
Dendrogram illustrating phylogenetic relations of the conserved, papain-like domains of closterovirus proteinases. The numbers indicate the results of the bootstrap analysis. The proteinase domain of the LIYV was used as an outgroup. Although the bootstrap value corresponding to the grouping of the CTV L1 and L2 is rather low, the affinity of these leader proteinases was further supported by the presence of conserved amino acid sequence motifs upstream from the proteinase domains (not shown). LChV, Little cherry virus.
DISCUSSION
The leader proteinases of the positive-strand RNA viruses from the families Potyviridae and Closteroviridae have similar plans of organization: they possess a variable N-terminal domain and a conserved C-terminal papain-like domain. In addition to being involved in polyprotein processing, these proteinases were implicated in efficient genome amplification (21, 22, 32, 33), and it has been suggested that they have similar functional profiles (12). To test this possibility, we replaced the L-Pro of BYV, a closterovirus, with HC-Pro of TEV, a potyvirus. Although the chimeric polyprotein was efficiently processed, the hybrid BYV was nonviable, suggesting that the functions of L-Pro and HC-Pro in genome amplification are mechanistically different. It was proposed recently that the HC-Pro role in genome amplification is mediated by the suppression of RNA silencing (6). The BYV L-Pro, however, lacks detectable silencing suppression activity (J. Reed, K. D. Kasschau, J. C. Carrington, and V. V. Dolja, unpublished data).
We further asked whether the functional specialization of L-Pro and HC-Pro is provided solely by their unrelated N-terminal domains. A chimeric protein in which the N-terminal domain of the BYV L-Pro was fused to the papain-like domain of the TEV HC-Pro failed to support genome amplification of BYV, suggesting that, despite their homology, the papain-like domains of the closteroviruses and potyviruses are functionally distinct. It should be noted that functional differences between L-Pro and HC-Pro cannot be attributed to differences in the host ranges of BYV and TEV since these viruses readily infect N. benthamiana and have several other common hosts. We can also exclude the possibility that the expression of the TEV proteinase domain or insertion of the corresponding RNA exerted an inhibitory effect on BYV amplification. Indeed, this domain was expressed from several locations within the BYV genome without affecting the viability of the resulting hybrid variants (16).
To examine functional specialization of the closterovirus leader proteinases, we replaced BYV genes with the corresponding genes of CTV and LIYV, which belong to two distinct evolutionary lineages within the family Closteroviridae. Among these viruses, BYV and LIYV possess only one leader proteinase, whereas CTV possesses two, L1 and L2. Phylogenetic analysis of the proteinase domains (Fig. 5) suggested that the corresponding gene tandem in CTV evolved via a duplication event. It was not, however, known if L1 and L2 are functionally distinct and, if so, which of them is more similar to the leader proteinases of BYV and LIYV. The ability of the CTV L1 and LIYV P-Pro to substitute for the BYV L-Pro in genome amplification indicated that these three leader proteinases belong to the same functional class. In contrast, failure of the CTV L2 to support amplification of the chimeric genome suggested that L2 function had diverged from that of L1, L-Pro, and P-Pro. This assumption was further confirmed by a phenotype of the BYV chimera that expressed both L1 and L2. This chimera amplified almost twice as efficiently as the original BYV, suggesting the synergistic mode of action for L1 and L2, and providing a remarkable example of a hybrid virus that outperformed its parent.
The transient-expression experiments revealed distinct patterns of subcellular localization of the leader proteinases fused with the GFP reporter. Most of the CTV L1 and L2 proteinases were uniformly distributed in the cytoplasm and nucleus, whereas the LIYV P-Pro almost exclusively localized to nuclei. In contrast, the BYV L-Pro was observed predominantly in cytoplasmic inclusion bodies. The possibility that the localization of the leader proteinases in the context of the virus-infected cell might be different from that observed in the transient-expression experiments cannot be excluded. Nevertheless, our results suggest that the leader proteinases of CTV, BYV, and LIYV possess distinct intrinsic signals for interaction with the cell environment and may function in different compartments of the infected cell.
Duplication and functional divergence of the leader proteinase genes are not unique to closteroviruses. A tandem arrangement of the leader proteinases is found among several animal viruses from the order Nidovirales (9, 38, 41, 44). Although Closteroviridae and Nidovirales are phylogenetically dissimilar, they are the most complex positive-strand RNA viruses of plants and animals, respectively (26, 44). Apparently independent duplication of the leader proteinases in these viruses may be interpreted as one of the means to facilitate evolution of the larger and more-complex genomes. In accord with this speculation, acquisition of the second leader proteinase gene in the ∼20-kb CTV genome is accompanied by three additional genes that have no homologs in the otherwise closely related ∼15-kb BYV genome.
The gene swapping experiments revealed an unexpected degree of functional specialization of the papain-like domains of the closterovirus proteinases. Each of these domains efficiently processed the chimeric polyprotein. However, the papain-like domains of the CTV L1 and L2 supported relatively low levels of BYV genome amplification, whereas the corresponding domain of the LIYV P-Pro was completely nonfunctional (Fig. 2 and Table 2). Although the mechanistic basis for this specialization is unknown, it seems possible that the proper function of the leader proteinases requires structural compatibility between the N-terminal and C-terminal domains.
Perhaps the most important outcome of this work is a better understanding of the multifunctional nature of the closterovirus proteinases. In addition to the primary role in the autocatalytic processing, each of the studied four proteinases functions in activation of genome amplification. Because the L1, L1-L2, and P-Pro chimeras amplified to severalfold-higher levels in the N. benthamiana protoplasts than in N. tabacum protoplasts (Table 1), this activation appears to work in a host-specific manner. Furthermore, at least L-Pro is critical for the ability of BYV to establish infection in the initially inoculated cells (virus invasiveness) and to translocate from cell to cell (Table 3).
The cell-to-cell movement of plant viruses proceeds through plasmodesmata and is activated by the movement proteins (27). In BYV, as many as five proteins that are encoded by a conserved gene block were implicated in virus movement. These proteins include three dedicated movement proteins (p6, HSP70 h, and p64) and two capsid proteins (2, 34). Since virion assembly is a prerequisite for BYV cell-to-cell movement (2), it was possible that the debilitated movement of the chimeric BYV-GFP variants was due to defective assembly. However, analysis of the chimeric virus progeny from the transfected protoplasts revealed normal virion assembly (data not shown). It should be noted that L-Pro by no means could be considered the movement protein since its primary functions are in proteolysis and virus genome amplification. Nevertheless, the fact that variant CTV-L1-L2 accumulates in N. benthamiana protoplasts almost twice as much RNA as the wild type (Table 2) yet does not move from cell to cell even in the leaves of N. benthamiana (Table 3) clearly indicates that L-Pro plays an essential role in cell-to-cell movement. This role, albeit indirect, suggests the need for coordination between the processes of genome amplification and virus translocation. Intriguingly, the leader proteinase of Foot-and-mouth disease virus, an aphtovirus, was recently implicated in virus spread within infected animals (7). Although the mechanisms of virus transport in plants and animals are different, this functional parallelism highlights the evolutionary plasticity of the viral papain-like proteinases that provide a structural platform for a variety of functions.
The mechanistic basis of the multifunctionality and specialization of the closterovirus leader proteinases is yet to be determined. These proteinases may act via cleavage of or via interaction with the particular viral or host target proteins. The host-dependent mode of activation of genome amplification and its role in virus invasiveness suggest that the intracellular targets of the closterovirus leader proteinases may include host factors. In conclusion, the gene swapping approach allowed us to reveal novel functions of the leader proteinases encoded in diverse representatives of the family Closteroviridae in genome amplification and virus invasiveness and spread. We also characterized the autonomous subcellular distribution and evolutionary relations of these leader proteinases and generated capable interviral hybrids. Further study of these hybrids will provide an insight into molecular mechanisms underlying multiple activities of leader proteinases and help to design more-efficient viral gene expression vectors.
ACKNOWLEDGMENTS
We thank T. Satyanarayana and S. Gowda for their help with N. benthamiana protoplasts and B. W. Falk for providing a cDNA clone of the LIYV.
This work was supported by grants from the U.S. Department of Agriculture (NRICGP 2001-35319-10875) and National Institutes of Health (R1GM53190B) to V.V.D.
REFERENCES
- 1.Agranovsky A A, Koonin E V, Boyko V P, Maiss E, Frotschl R, Lunina N A, Atabekov J G. Beet yellows closterovirus: complete genome structure and identification of a leader papain-like thiol protease. Virology. 1994;198:311–324. doi: 10.1006/viro.1994.1034. [DOI] [PubMed] [Google Scholar]
- 2.Alzhanova D V, Hagiwara Y, Peremyslov V V, Dolja V V. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology. 2000;268:192–200. doi: 10.1006/viro.1999.0155. [DOI] [PubMed] [Google Scholar]
- 3.Bransom K L, Dreher T W. Identification of the essential cysteine and histidine residues of the turnip yellow mosaic virus protease. Virology. 1994;198:148–154. doi: 10.1006/viro.1994.1017. [DOI] [PubMed] [Google Scholar]
- 4.Carrington J C, Cary S M, Parks T D, Dougherty W G. A second proteinase encoded by a plant potyvirus genome. EMBO J. 1989;8:365–370. doi: 10.1002/j.1460-2075.1989.tb03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington J C, Freed D D, Oh C-S. Expression of potyviral polyproteins in transgenic plants reveals three proteolytic activities required for complete processing. EMBO J. 1990;9:1347–1353. doi: 10.1002/j.1460-2075.1990.tb08249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrington J C, Kasschau K D, Johansen L K. Activation and suppression of RNA silencing by plant viruses. Virology. 2001;281:1–5. doi: 10.1006/viro.2000.0812. [DOI] [PubMed] [Google Scholar]
- 7.Chinsangaram J, Piccone M E, Grubman M J. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J Virol. 1999;73:9891–9898. doi: 10.1128/jvi.73.12.9891-9898.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven M G, Pawlyk D M, Choi G H, Nuss D L. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J Virol. 1993;67:6513–6521. doi: 10.1128/jvi.67.11.6513-6521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon J A, Faaberg K S, Meulenberg J J M, Wassenaar A L M, Plagemann P G W, Gorbalenya A E, Snijder E J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolja V V, McBride H J, Carrington J C. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase (GUS) into the viral polyprotein. Proc Natl Acad Sci USA. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolja V V, Karasev A V, Koonin E V. Molecular biology and evolution of closteroviruses: sophisticated build-up of large RNA genomes. Annu Rev Phytopathol. 1994;32:261–285. [Google Scholar]
- 12.Dolja V V, Hong J, Keller K E, Martin R R, Peremyslov V V. Suppression of potyvirus infection by coexpressed closterovirus protein. Virology. 1997;234:243–252. doi: 10.1006/viro.1997.8660. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty W G, Semler B L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya A E, Koonin E V, Lai M M-C. Putative papain-related thiol proteases of positive-strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarne A, Tormo J, Kirchweger R, Pfistermueller D, Fita I, Skern T. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 1998;17:7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagiwara Y, Peremyslov V V, Dolja V V. Regulation of closterovirus gene expression examined by insertion of a self-processing reporter and by Northern hybridization. J Virol. 1999;73:7988–7993. doi: 10.1128/jvi.73.10.7988-7993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jelkman W, Fetchner B, Agranovsky A A. Complete genome structure and phylogenetic analysis of little cherry virus, a mealybug-transmissible closterovirus. J Gen Virol. 1997;78:2067–2071. doi: 10.1099/0022-1317-78-8-2067. [DOI] [PubMed] [Google Scholar]
- 18.Kadare G, Rozanov M, Haenni A-L. Expression of the turnip yellow mosaic virus proteinase in Escherichia coli and determination of the cleavage site within the 206 kDa protein. J Gen Virol. 1995;76:2853–2857. doi: 10.1099/0022-1317-76-11-2853. [DOI] [PubMed] [Google Scholar]
- 19.Karasev A V. Genetic diversity and evolution of closteroviruses. Annu Rev Phytopathol. 2000;38:293–324. doi: 10.1146/annurev.phyto.38.1.293. [DOI] [PubMed] [Google Scholar]
- 20.Karasev A V, Boyko V P, Gowda S, Nikolaeva O V, Hilf M E, Koonin E V, Niblett C L, Cline K, Gumpf D J, Lee R F, Garnsey S M, Lewandowski D J, Dawson W O. Complete sequence of the citrus tristeza virus RNA genome. Virology. 1995;208:511–520. doi: 10.1006/viro.1995.1182. [DOI] [PubMed] [Google Scholar]
- 21.Kasschau K D, Carrington J C. Requirement for HC-Pro processing during genome amplification of tobacco etch potyvirus. Virology. 1995;209:268–273. doi: 10.1006/viro.1995.1254. [DOI] [PubMed] [Google Scholar]
- 22.Kasschau K D, Cronin S, Carrington J C. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology. 1997;228:251–262. doi: 10.1006/viro.1996.8368. [DOI] [PubMed] [Google Scholar]
- 23.Kasschau K D, Carrington J C. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 24.Klaassen V A, Boeshore M, Koonin E V, Tian T, Falk B W. Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly transmitted, bipartite closterovirus. Virology. 1995;208:99–110. doi: 10.1006/viro.1995.1133. [DOI] [PubMed] [Google Scholar]
- 25.Klaassen V A, Mayhew D, Fisher D, Falk B W. In vitro transcripts from cloned cDNAs of the lettuce infectious yellows closterovirus bipartite genomic RNAs are competent for replication in Nicotiana benthamiana protoplasts. Virology. 1996;222:169–175. doi: 10.1006/viro.1996.0407. [DOI] [PubMed] [Google Scholar]
- 26.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 27.Lazarowitz S G, Beachy R N. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell. 1999;11:535–548. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llave C, Kaschau K D, Carrington J C. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maia I G, Haenni A, Bernardi F. Potyviral HC-Pro: a multifunctional protein. J Gen Virol. 1996;77:1335–1341. doi: 10.1099/0022-1317-77-7-1335. [DOI] [PubMed] [Google Scholar]
- 30.Navas-Castillo J, Albiach-Marti M R, Gowda S, Hilf M, Garnsey S M, Dawson W O. Kinetics of accumulation of citrus tristeza virus RNAs in host and non-host protoplasts. Virology. 1997;228:92–97. doi: 10.1006/viro.1996.8369. [DOI] [PubMed] [Google Scholar]
- 31.Nuss D L. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol Rev. 1992;56:561–576. doi: 10.1128/mr.56.4.561-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng C W, Dolja V V. Leader proteinase of the beet yellows closterovirus: mutation analysis of the function in genome amplification. J Virol. 2000;74:9766–9770. doi: 10.1128/jvi.74.20.9766-9770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peremyslov V V, Hagiwara Y, Dolja V V. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J Virol. 1998;72:5870–5876. doi: 10.1128/jvi.72.7.5870-5876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peremyslov V V, Hagiwara Y, Dolja V V. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc Natl Acad Sci USA. 1999;96:14771–14776. doi: 10.1073/pnas.96.26.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revers F, Le Gall O, Candresse T, Maule A J. New advances in understanding the molecular biology of plant/potyvirus interactions. Mol Plant-Microbe Interact. 1999;12:367–376. [Google Scholar]
- 36.Satyanarayana T, Gowda S, Boyko V P, Albiach-Marti M R, Mawassi M, Navas-Castillo J, Karasev A V, Dolja V, Hilf M E, Lewandowski D J, Moreno P, Bar-Joseph M, Garnsey S M, Dawson W O. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc Natl Acad Sci USA. 1999;96:7433–7438. doi: 10.1073/pnas.96.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 38.Snijder E J, Meulenberg J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 39.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki N, Chen B, Nuss D L. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J Virol. 1999;73:9478–9484. doi: 10.1128/jvi.73.11.9478-9484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tijms M A, van Dinten L C, Gorbalenya A E, Snijder E J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc Natl Acad Sci USA. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang C, Han P, Lutziger I, Wang K, Oliver D J. A mini binary vector series for plant transformation. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- 43.Zhu H-Y, Ling K-S, Goszczynski D E, McFerson J R, Gonsalves D. Nucleotide sequence and genome organization of grapevine leafroll-associated virus-2 are similar to beet yellows virus, the closterovirus type member. J Gen Virol. 1998;79:1289–1298. doi: 10.1099/0022-1317-79-5-1289. [DOI] [PubMed] [Google Scholar]
- 44.Ziebur J, Snijder E J, Gorbalenya A E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]