Abstract
Objective:
To investigate the impact of global and local genetic ancestry and neighborhood socioeconomic status (nSES), on breast cancer (BC) subtype, and gene expression.
Summary of Background Data:
Higher rates of aggressive BC subtypes (TNBC) and worse overall BC survival are seen in black women [Hispanic (HB) and non-Hispanic (NHB)] and women from low nSES. However, the complex relationship between genetic ancestry, nSES, and BC subtype etiology remains unknown.
Methods:
Genomic analysis was performed on the peripheral blood from a cohort of 308 stage I-IV non-Hispanic White (NHW), Hispanic White (HW), HB and NHB women with BC. Patient and tumor characteristics were collected. Global and local ancestral estimates were calculated. Multinomial logistic regression was performed to determine associations between age, stage, genetic ancestry, and nSES on rates of TNBC compared to ER+/HER2−, ER+/HER2+, and ER−/HER2+ disease.
Results:
Among 308 women, we identified a significant association between increasing West African (WA) ancestry and odds of TNBC (OR 1.06,95%CI 1.001-1.126, p=0.046) as well as an inverse relationship between higher nSES and TNBC (OR 0.343,95%CI 0.151-0.781, p=0.011). WA ancestry remained significantly associated with TNBC when adjusting for patient age and tumor stage, but not when adjusting for nSES (OR 1.049, 95%CI-0.987-1.116, p=0.120). Local ancestry analysis revealed nSES-independent enriched WA ancestral segment centered at chr2:42004914 (p=3.70x10−5) in patients with TNBC.
Conclusions:
In this translational epidemiologic study of genetic ancestry and nSES on BC subtype, we discovered associations between increasing WA ancestry, low nSES, and higher rates of TNBC compared to other BC subtypes. Moreover, on admixture mapping, specific chromosomal segments were associated with WA ancestry and TNBC, independent of nSES. However, on multinomial logistic regression adjusting for WA ancestry, women from low nSES were more likely to have TNBC, independent of genetic ancestry. These findings highlight the complex nature of TNBC and the importance of studying potential gene-environment interactions as drivers of TNBC.
Keywords: Breast cancer, genetic ancestry, neighborhood socioeconomic status, health care disparities
Mini Abstract
In this translational epidemiologic study of genetic ancestry and neighborhood socioeconomic status (nSES) on breast cancer subtype, we found that increasing West African ancestry and low nSES is associated with higher rates of TNBC. On multinomial logistic regression adjusting for WA ancestry, women from a high nSES had one-third the odds of having TNBC compared to women from a low nSES. On admixture mapping, specific chromosomal segments were associated with WA ancestry and TNBC, independent of nSES. These findings highlight the importance of studying both genetic ancestry associated and gene-environment interactions as drivers of TNBC.
INTRODUCTION
Racial/ethnic minority and socioeconomically disadvantaged populations continue to experience a disproportionate burden of breast cancer mortality1. Earlier onset, more advanced stage at diagnosis, and aggressive tumor subtype (triple negative breast cancer (TNBC)) are some of the characteristic features of breast cancer in Black and Hispanic women, representing one of the most significant examples of racial/ethnic differences in oncology2, 3. This has been extended further as we recently identified intra-ethnic racial differences, showing Hispanic Black (HB) women had lower rates of TNBC compared to non-Hispanic Black (NHB), however still had higher rates of TNBC when compared to their Hispanic White (NH) and non-Hispanic White (NHW) counterparts3. These inter-racial and intra-ethnic disparities seen throughout oncology have prompted questions regarding the role of genetic ancestry in prognosis and tumor biology4-6.
Genetic ancestry reflects population history, providing background information about genetic variation that is essential to determine genomic associations with diseases. Recent work has shown that genetic ancestry is associated with breast cancer characteristics7-9. Specifically, in a study by Yuan et al5, African ancestry compared with European ancestry was associated with higher levels of chromosomal instability with more TP53 mutations and fewer PIK3CA mutations, which may in turn be leading to more aggressive tumor biology and worse survival. However, the absolute number of samples from racial/ethnic minorities was limited10.
Racial/ethnic minorities are also more likely to live in disadvantaged neighborhoods and are exposed to considerably higher levels neighborhood-level social stressors which studies have hypothesized may contribute to the development of aggressive breast cancer tumor subtypes and higher mortality11, 12. Analysis of these underlying gene-environment interactions are complex and difficult to study but are critical to dismantle racial/ethnic and socioeconomic disparities in breast cancer.
Given there are no publicly available cancer genetic ancestry databases with neighborhood-level socioeconomic annotation5, this study fills this critical knowledge gap by integrating neighborhood socioeconomic status (nSES) and genetic ancestry to analyze their impact on breast cancer subtype, specifically rates of TNBC. In doing so, we leveraged the socioeconomic diversity and admixed genetic ancestry of our Miami-Dade County population, a melting pot of Latin America and the Caribbean.
METHODS
Patient samples and variables of interest
Patient samples were collected at the University of Miami-Sylvester Comprehensive Cancer Center (SCCC) and its sister safety-net hospital Jackson Memorial Hospital between January 1, 2017-January 1, 2021 under an Institutional Review Board approved prospective study and performed in adherence to the tenets of the Declaration of Helsinki. Patients with stages I-IV invasive ductal or lobular carcinoma were included in the analysis. Patients with other malignancies of the breast (i.e. sarcoma, lymphoma) were excluded in the analysis. Patient and tumor characteristics were collected from electronic medical records and deidentified for further analysis. For each individual, demographic information (age, sex, race, ethnicity, neighborhood-level income), clinical information (body-mass index, tobacco use, alcohol use), and tumor characteristics [stage, grade (unknown, well differentiated, moderately differentiated, and poorly differentiated/anaplastic), and estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) status). The AJCC 7th edition guidelines were used to determine the clinical stage at the time of diagnosis. Tumors were also grouped into four subtypes based on hormone receptor status: ER+/HER−, ER+/HER2+, ER−/HER2+, and TNBC (ER−/HER2−).
Self-identified race and ethnicity (SIRE) were grouped as NHW, HW, NHB, and HB. The other group accounted for only 3.8% of the population and included Asians or unknown. Median income was calculated based on the patient’s home zip code, ranging from $19,138–$154,868 for the entire population. The population was divided into three groups representing those at the poverty line in Miami-Dade County (nSES 1: <$35,600), those above the poverty line to the median income (nSES 2: $35,600-55,600), and those with income greater than the median in Miami-Dade County (nSES 3: > $55,600).
Genotyping
Peripheral blood was collected at time of surgery for each patient. Buffy coat DNA was extracted using the QIAamp DNA mini kit. DNA was genotyped using the Illumina Expanded Multi-Ethnic Global Array (MEGAex) beadchip. Autoconvert 2.0 was used to convert raw .idat files to .gtc which were then converted to variant call file (VCF) format. Quality control (QC) was performed, genotypes with call rates < 97% and/or incorrect sex identification were excluded from the study.
Global and local ancestry estimation
Global and local ancestry estimation was performed as previously described4. Reference populations from the 1000 genomes project phase 3 data set were used to estimate European (EU), West African (WA), and East Asian (EA) global ancestry13. Global ancestral estimation for Native American (NAT) ancestry was performed using samples representing 52 indigenous groups14-16. Study patient genotypes were merged with the global ancestry reference samples using vcftools17. Variants that were non-biallelic or that were not called in at least one individual were removed from the analysis. Population structure was estimated using ADMIXTURE18 (K=4) and PCA analysis using TRACE19, 20.
Study patient genotypes were merged with local ancestry reference samples and processed using vcftools, filtering out variants that were non-biallelic or that were missing in at least one individual. After pruning, a total of >400K variants were used for local ancestry estimation. Common variants were phased using Beagle 5.021, 22. Since no patients were known to be related, pedigree information was not used for phasing. Local ancestry was inferred across the autosome for each sample by discriminative modeling with random forests using RFMix23. With a minimum node size of 5, PopPhased option was used. Posterior probabilities of local ancestry were used to generate karyograms24.
Statistical Analysis
Descriptive statistics were calculated for patient, clinical, and tumor characteristics using mean, standard deviation, median, and interquartile rage Q1–Q3 for continuous data and frequencies (percentage) for categorical data. Univariate analysis using ANOVA, Kruskal–Wallis, and Chi-squared analysis as appropriate. Multinomial logistic regressions were used for model building of covariates including the proportion of WA, NAT, and EA ancestries with respect to EU ancestry, age, stage, and income bracket. Admixture mapping of local ancestral calls was performed using a multinomial regression on tumor subtype correcting for the proportion of WA, NAT, and EA ancestries with respect to EU ancestry, age, stage, and income bracket. A significance threshold of 3.65x10−4 was determined based on the mean number of ancestral switches across the autosome for our cohort. Manhattan plots were generated using CMplot25.
RESULTS
Patient and Tumor Characteristics Based on Self-Identified Race and Ethnicity (SIRE)
This study included 308 patients with breast cancer, of which 46 (14.9%) were NHW, 192 (62.3%) were HW, 11 (3.6%) were HB, and 47 (15.3%) were NHB (Table 1). The median age at diagnosis was lower in both HB and NHB (52 and 51 years, respectively) when compared to NHW and HW (55 and 55 years, respectively) patients. There was a significant difference in nSES across our cohort (p=0.011). There were no significant differences across the cohort for tumor stage (p=0.386) or grade (p=0.116). Although there was no significant difference across the cohort for tumor subtype (p=0.104) based on SIRE.
Table 1.
Sociodemographics and clinical features in 308 patients with breast cancer by self-identified race and ethnicity
| Factor | Non- Hispanic White |
Hispanic White |
Hispanic Black |
Non- Hispanic Black |
p- value |
|
|---|---|---|---|---|---|---|
| N=46 | N=192 | N=11 | N=47 | |||
| Age (years) | Mean (SD) | 56.59 (12.9) | 55.4 (11.7) | 48.64 (13.5) | 52.2 (10.6) | 0.077 |
| Neighborhood-Level Income (nSES) | <$35600 | 5 (10.9%) | 43 (22.4%) | 5 (45.5%) | 16 (34.0%) | 0.011 |
| $35600-$55600 | 16 (34.8%) | 84 (43.8%) | 5 (45.5%) | 18 (38.3%) | ||
| >$55600 | 22 (47.8%) | 62 (32.3%) | 1 (9.1%) | 9 (19.1%) | ||
| Unknown | 3 (6.5%) | 3 (1.6%) | 0 (0%) | 4 (8.5%) | ||
| Tobacco Use | Never Smoker | 26 (56.5%) | 129 (67.2%) | 8 (72.7%) | 36 (76.6%) | 0.108 |
| Current Smoker | 1 (2.2%) | 14 (7.3%) | 1 (9.1%) | 3 (6.4%) | ||
| Former Smoker | 19 (41.3%) | 45 (23.4%) | 1 (9.1%) | 8 (17.0%) | ||
| Unknown | 0 (0%) | 4 (2.1%) | 1 (9.1%) | 0 (0%) | ||
| Alcohol Use | None | 13 (28.3%) | 141 (73.4%) | 9 (81.8%) | 33 (70.2%) | <0.001 |
| Current Use | 33 (71.7%) | 46 (24.0%) | 1 (9.1%) | 14 (29.8%) | ||
| Former Use | 0 (0%) | 1 (0.5%) | 0 (0%) | 0 (0%) | ||
| Unknown | 0 (0%) | 4 (2.1%) | 1 (9.1%) | 0 (0%) | ||
| Body Mass Index (BMI) | Underweight (<18.5) | 0 (0%) | 2 (1.0%) | 0 (0%) | 0 (0%) | <0.001 |
| Normal weight (18.5–24.9) | 15 (32.6%) | 38 (19.8%) | 5 (45.5%) | 5 (10.6%) | ||
| Overweight (25–29.9) | 12 (26.1%) | 61 (31.8%) | 3 (27.3%) | 8 (17.0%) | ||
| Class I Obesity (30–34.9) | 8 (17.4%) | 38 (19.8%) | 0 (0%) | 9 (19.1%) | ||
| Class II Obesity (35–39.9) | 5 (10.9%) | 12 (6.3%) | 2 (18.2%) | 8 (17.0%) | ||
| Class III Obesity (40+) | 0 (0%) | 6 (3.1%) | 0 (0%) | 10 (21.3%) | ||
| Unknown | 6 (13.0%) | 35 (18.2%) | 1 (9.1%) | 7 (14.9%) | ||
| Stage | 1 | 20 (43.5%) | 59 (30.7%) | 2 (18.2%) | 12 (25.5%) | 0.386 |
| 2 | 20 (43.5%) | 78 (40.6%) | 6 (54.5%) | 18 (38.3%) | ||
| 3 | 5 (10.9%) | 50 (26%) | 3 (27.3%) | 16 (34.0%) | ||
| 4 | 1 (2.2%) | 5 (2.6%) | 0 (0%) | 1 (2.1%) | ||
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Grade | Well-Differentiated | 10 (21.7%) | 29 (15.1%) | 0 | 5 (10.6%) | 0.116 |
| Moderately Differentiated | 24 (52.2%) | 82 (42.7%) | 4 (36.4%) | 16 (34.0%) | ||
| Poorly Differentiated / Anaplastic | 11 (23.9%) | 74 (38.5%) | 6 (54.5%) | 23 (48.9%) | ||
| Unknown | 1 (2.2%) | 7 (3.6%) | 1 (9.1%) | 3 (6.4%) | ||
| Tumor Subtype | ER+/HER2− | 35 (76.1 %) | 120 (62.5%) | 5 (45.5%) | 29 (61.7%) | 0.104 |
| ER+/HER2+ | 7 (15.2%) | 27 (14.1%) | 1 (9.1%) | 3 (6.4%) | ||
| ER−/HER2+ | 3 (6.5%) | 12 (6.3%) | 1 (9.1%) | 3 (6.4%) | ||
| ER−/HER2− | 1 (2.2%) | 32 (16.7%) | 4 (36.4%) | 12 (25.5%) | ||
| Unknown | 0 (0%) | 1 (0.5%) | 0 (0%) | 0 (0%) |
Patient and Tumor Characteristics Based on Global Genetic Ancestry
To estimate the global ancestral proportions of this cohort, patients were genotyped and compared to WA, EA, EU, and NAT reference populations. The breast cancer cohort exhibited considerable diverse population structure (Figure 1A). NHW and NHB patients clustered toward EU and WA reference populations, respectively, however HW and HB patients were more spread out due to contributions of different ancestries (Figure 1B). Mean EU ancestry by SIRE categories was 87.9% for NHW patients, 73.2% for HW patients, 36.8% for HB patients, and 12.7% for NHB patients (Figure 1C and Table 2). An inverse relationship along SIRE groups was observed where WA ancestry decreased from a mean of 85.4%, 53.9%, 9.5%, and 3.9% for NHB, HB, HW, and NHW patients, respectively. There were significant differences with respect to EU (p<0.001), WA (p<0.001), NAT (p<0.001), and EA (p=0.002), ancestries across the cohort. HW and HB patients had a higher percentage of NAT ancestry (15.5% and 8.0%, respectively) when compared to the non-Hispanic patients.
Figure 1. Population structure of patients with invasive breast cancer.
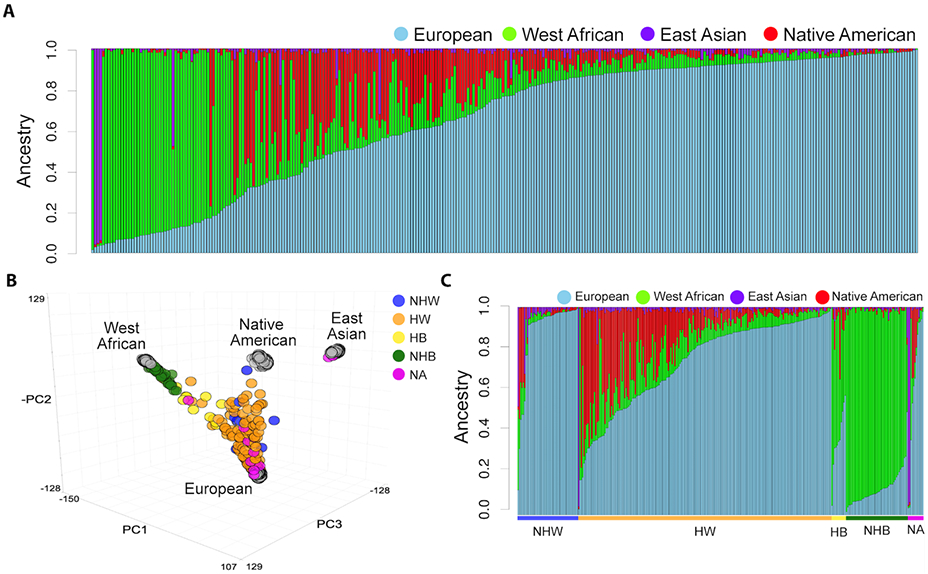
(A) Stacked barplot of ancestral percentages after unsupervised clustering using ADMIXTURE analysis of our cohort assuming K=4 ancestral clusters. Each bar represents one individual and the height of each segment represents the respective ancestral percentage. (B) Principal component analysis (PCA) based global ancestral genotypes across the cohort (colored circles) and the global ancestry reference panel (grey circles). (C) Stacked barplot of ancestral percentages grouped by self-reported race/ethnicity category. Non-Hispanic white (NHW), Hispanic white (HW), non-Hispanic black (NHB), Hispanic black (HB), and not available (NA).
Table 2.
Summary of global genetic ancestry for self-identified race and ethnicity
| Factor | Non-Hispanic White |
Hispanic White |
Hispanic Black |
Non- Hispanic Black |
All | p-value | |
|---|---|---|---|---|---|---|---|
| N=46 | N=192 | N=11 | N=47 | N=308 | |||
| European Ancestry (%) | Mean (SD) | 87.9 (19.9) | 73.2 (21.4) | 36.8 (14.9) | 12.7 (8.2) | 64.5 (31.1) | <0.001 |
| Median | 95.7 | 82.9 | 35.3 | 10.6 | 76.7 | ||
| Minimum - Maximum | 12.0 - 99.4 | 2.8 - 99.3 | 4.5 - 57.5 | 1.6 - 49.6 | 1.6 - 99.4 | ||
| 25th-75th Percentile | 92.6 - 97.6 | 58.1 - 90.4 | 32.2 - 49.8 | 7.0 - 14.8 | 38.1 - 91.7 | ||
| West African Ancestry (%) | Mean (SD) | 3.9 (7.1) | 9.5 (10.3) | 53.9 (19.3) | 85.4 (10.2) | 21.8 (30.3) | <0.001 |
| Median | 1.3 | 6.6 | 54.3 | 88.1 | 6.98 | ||
| Minimum - Maximum | 0 - 38.7 | 0 - 82.6 | 29.3 - 95.4 | 32.4 - 97.7 | 0 - 97.8 | ||
| 25th-75th Percentile | 0 - 3.8 | 3.8 - 11.3 | 36.2 - 62.4 | 82.1 - 92.1 | 3.6 - 21.4 | ||
| Native American Ancestry (%) | Mean (SD) | 4.8 (11.6) | 15.5 (17.5) | 8.0 (9.4) | 0.8 (1.0) | 11.2 (15.9) | <0.001 |
| Median | 1.4 | 7.1 | 4.7 | 0.4 | 3.3 | ||
| Minimum - Maximum | 0 - 53.3 | 0 - 77.2 | 0.1 - 33.2 | 0 - 3.4 | 0 - 77.2 | ||
| 25th-75th Percentile | 0.2 - 2.5 | 2.4 - 26.5 | 2.3 - 11 | 0 - 1.5 | 0.9 - 14.5 | ||
| East Asian Ancestry (%) | Mean (SD) | 3.4 (8.4) | 1.8 (7.0) | 1.3 (2.0) | 1.0 (2.3) | 2.5 (9.9) | 0.002 |
| Median | 1 | 1.1 | 0.7 | 0.5 | 0.9 | ||
| Minimum - Maximum | 0 - 47.6 | 0 - 95.9 | 0 - 6.8 | 0 - 14.8 | 0 - 95.9 | ||
| 25th-75th Percentile | 0 - 2.8 | 0.5 - 1.8 | 0 - 1.4 | 0 - 1.1 | 0.3 - 1.8 |
Increased WA ancestry was associated with various demographic variables including earlier age of diagnosis (p=0.018) and lower income (p<0.001) (Table 3). The inverse relationship was seen for EU ancestry. WA ancestry was also associated with differences in clinical characteristics such as stage (p=0.039), grade (p=0.0083), and subtype (p=0.012) (Table 3 and Figure 2), which was not observed when using SIRE.
Table 3.
Associations between sociodemographic and clinical features with global genetic ancestry
| European Ancestry |
West African Ancestry |
Native American Ancestry |
East Asian Ancestry |
||
|---|---|---|---|---|---|
| Age (years) | Correlation Coefficient | 0.124 | −0.135 | −0.014 | 0.073 |
| p-value | 0.03 | 0.018 | 0.805 | 0.198 | |
| nSES Bracket | p-value | <0.001 | <0.001 | 0.095 | 0.098 |
| Tobacco Use | p-value | 0.061 | 0.081 | 0.349 | 0.176 |
| Alcohol Use | p-value | 0.009 | 0.054 | 0.109 | 0.732 |
| BMI | p-value | <0.001 | 0.003 | 0.01 | 0.361 |
| Stage | p-value | 0.001 | 0.039 | 0.376 | 0.167 |
| Grade | p-value | 0.012 | 0.008 | 0.618 | 0.225 |
| Tumor Subtype | p-value | 0.008 | 0.012 | 0.234 | 0.302 |
Figure 2. Association between tumor characteristics and global ancestry.
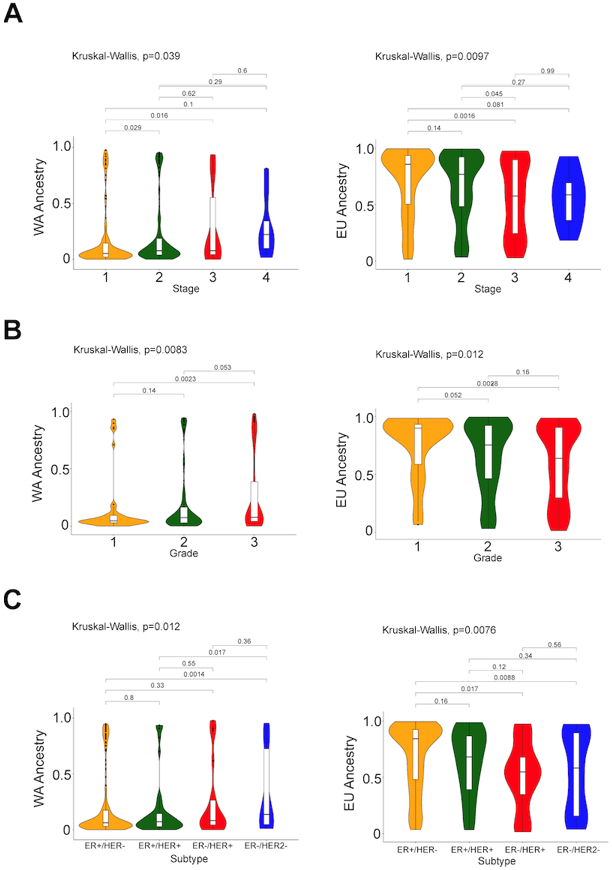
Violin plots of ancestral percentages [(European (EU) and West African(WA)] of patients stratified by (A) stage, (B) grade, and (C) subtype
Multinomial logistic regression with respect to tumor subtype revealed a significant association between increasing WA ancestry and TNBC (OR 1.06,95%CI 1.001-1.126, p=0.046) when adjusting for age and stage (Table 4). When adjusting for nSES, WA was no longer significantly associated (OR 1.049, 95%CI 0.987-1.116, p=0.120), however there was a significant inverse association between higher nSES and TNBC (OR 0.366, 95%CI 0.158-0.848, p=0.019).
Table 4.
Multinomial regression models of individual-level and contextual-level measures associated with breast cancer subtype
| Models | Subtype* | Model Outputs |
Age | Stage** | Ancestry | nSES*** | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| II | III | IV | African / European |
Native American / European |
East Asian / European |
2 | 3 | ||||
| Age + Stage | ER+/HER2+ | OR | 0.998 | 2.519 | 3.522 | 2.036 | |||||
| P | 0.873 | 0.041 | 0.013 | 0.539 | |||||||
| 95% CI | 0.968-1.027 | 1.04-6.104 | 1.3-9.535 | 0.211-19.668 | |||||||
| ER−/HER2+ | OR | 0.763 | 3.019 | 9.125 | 17.116 | ||||||
| P | 0.238 | 0.185 | 0.007 | 0.010 | |||||||
| 95% CI | 0.931-1.018 | 0.588-15.487 | 1.835-45.422 | 1.95-150.355 | |||||||
| TNBC | OR | 1.014 | 5.669 | 9.855 | 0.000 | ||||||
| P | 0.307 | 0.001 | <0.001 | 0.986 | |||||||
| 95% CI | 0.987-1.042 | 2.067-15.534 | 3.35-28.991 | 0- >100 | |||||||
| Age + Stage + nSES | ER+/HER2+ | OR | 0.998 | 2.497 | 3.655 | 1.904 | 2.440 | 1.723 | |||
| P | 0.871 | 0.043 | 0.011 | 0.581 | 0.129 | 0.379 | |||||
| 95% CI | 0.969-1.027 | 1.027-6.068 | 1.347-9.954 | 0.194-18.728 | 0.771-7.721 | 0.589-2.974 | |||||
| ER−/HER2+ | OR | 0.972 | 2.983 | 8.390 | 21.073 | 0.264 | 0.665 | ||||
| P | 0.223 | 0.193 | 0.010 | 0.008 | 0.053 | 0.483 | |||||
| 95% CI | 0.93-1.017 | 0.576-15.456 | 1.667-42.224 | 2.223-199.737 | 0.069-1.016 | 0.213-2.079 | |||||
| TNBC | OR | 1.017 | 6.038 | 9.507 | 0.000 | 0.348 | 0.343 | ||||
| P | 0.229 | 0.001 | <0.001 | 0.982 | 0.011 | 0.011 | |||||
| 95% CI | 0.989-1.046 | 1.08-16.81 | 3.177-28.446 | 0- >100 | 0.154-0.789 | 0.151-0.781 | |||||
| Age + Stage + Ancestry | ER+/HER2+ | OR | 0.996 | 2.524 | 3.327 | 2.036 | 0.992 | 1.338 | 1.069 | ||
| P | 0.080 | 0.041 | 0.020 | 0.540 | 0.873 | 0.382 | 0.228 | ||||
| 95% CI | 0.967-1.026 | 1.037-6.147 | 1.204-9.198 | 0.21-19.767 | 0.897-1.097 | 0.696-2.57 | 0.959-1.194 | ||||
| ER−/HER2+ | OR | 0.969 | 3.736 | 10.299 | 26.076 | 1.100 | 1.533 | 1.120 | |||
| P | 0.193 | 0.150 | 0.011 | 0.006 | 0.006 | 0.301 | 0.052 | ||||
| 95% CI | 0.924-1.016 | 0.621-22.466 | 1.707-62.178 | 2.54-268.003 | 1.027-1.177 | 0.682-3.442 | 0.999-1.256 | ||||
| TNBC | OR | 1.015 | 5.795 | 10.054 | 0.000 | 1.062 | 1.057 | 0.997 | |||
| P | 0.278 | 0.001 | <0.001 | 0.984 | 0.046 | 0.873 | 0.976 | ||||
| 95% CI | 0.988-1.043 | 2.071-16.216 | 3.33-30.326 | 0- >100 | 1.001-1.126 | 0.537-2.077 | 0.811-1.225 | ||||
| Age + Stage + Ancestry + nSES | ER+/HER2+ | OR | 0.995 | 2.489 | 3.391 | 1.954 | 0.999 | 1.539 | 1.067 | 2.829 | 2.059 |
| P | 0.752 | 0.046 | 0.019 | 0.566 | 0.994 | 0.208 | 0.239 | 0.090 | 0.251 | ||
| 95% CI | 0.966-1.106 | 1.017-6.092 | 1.218-9.44 | 0.199-19.221 | 0.903-1.106 | 0.787-3.007 | 0.958-1.19 | 0.85-9.507 | 0.601-7.05 | ||
| ER−/HER2+ | OR | 0.969 | 3.490 | 9.459 | 29.964 | 1.091 | 1.275 | 1.115 | 0.320 | 0.736 | |
| P | 0.191 | 0.173 | 0.015 | 0.005 | 0.015 | 0.581 | 0.067 | 0.118 | 0.627 | ||
| 95% CI | 0.923-1.016 | 0.577-21.115 | 1.56-57.34 | 2.779-323.112 | 1.017-1.169 | 0.537-3.028 | 0.992-1.255 | 0.077-1.332 | 0.215-2.524 | ||
| TNBC | OR | 1.018 | 6.135 | 9.905 | 0.000 | 1.049 | 0.896 | 1.006 | 0.366 | 0.366 | |
| P | 0.213 | 0.001 | <0.001 | 0.980 | 0.120 | 0.755 | 0.951 | 0.019 | 0.019 | ||
| 95% CI | 0.99-1.046 | 2.166-17.392 | 3.235-30.356 | 0- >100 | 0.987-1.116 | 0.448-1.793 | 0.82-1.236 | 0.158-0.848 | 0.158-0.850 | ||
Reference ER+/HER2−
Reference Stage 1
Reference nSES 1: nSES 1: <$35,600, nSES 2: $35,600-55,600, nSES 3: > $55,600
TNBC and Local Genetic Ancestry
Ancestry was estimated locally at each single nucleotide polymorphism (SNP) with respect to our four ancestral reference groups. There was rich diversity of ancestral blocks throughout the autosome, especially in our HW and HB patients (Figure 3A). Admixture mapping, through a multinomial logistic regression model correcting for stage, nSES, and age, was performed at each variant. Our analysis revealed nSES-independent enriched WA ancestral segments (chr2:40114470-47026542), where segment chr2:42004851-42329329 (p=3.71x10−5) exhibited the most significant enrichment in patients with TNBC (Figure 3B). Additionally, increased NAT ancestry along chr17:79328964-80456621, with a peak enrichment at chr17:79693136 (p=1.72x10−4), was associated with TNBC.
Figure 3. Admixture and fine mapping.
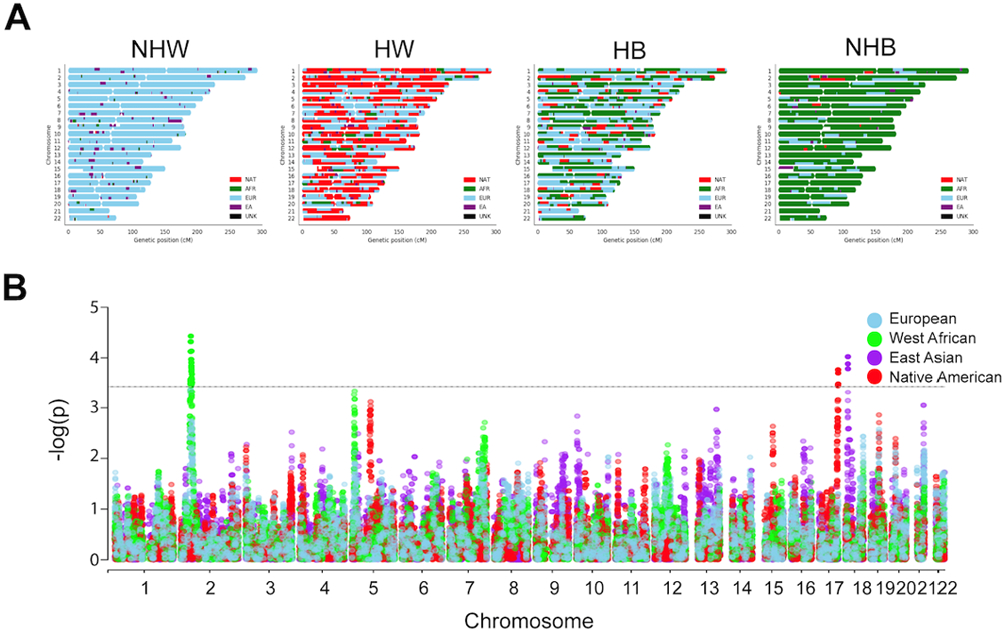
(A) Representative karyograms, after local ancestry estimation, for patients identifying as non-Hispanic white (NHW), Hispanic white (HW), Hispanic black (HB), or non-Hispanic black (NHB). European (EU), West African (WA), East Asian (EA), Native American (NAT), and unknown (UNK). (B) Manhattan plot of ancestral segments associated with triple negative breast cancer (TNBC) representing results of multinomial regression and admixture mapping. X-axis, chromosome position; Y-axis, −log10 (p value) after multinomial regression for tumor subtype and local ancestry at each variant, correcting for age, stage, nSES, and the proportion of WA, NAT, and EA ancestries with respect to EU ancestry. Each dot represents an ancestral segment tested in the association test. Horizontal dashed lines represent significance threshold, p < 3.7x10−4.
To further investigate these enriched regions, we analyzed expression quantitative trait loci (eQTL) data from the PanCanQTL26 database, which provides cis and trans eQTL data for 33 cancer types from The Cancer Genome Atlas (TCGA). Within the NAT enriched region (chr17:79328964-80456621) 15 cis and 0 trans eQTLs were identified specific for the breast cancer (BRCA) cohort. Additionally, the expression of 2 unique genes were impacted: TSPAN10 (FDR=5.79x1016) and ARL16 (1.95x10−12). Within the WA enriched region (chr2:42004851-42329329) 1845 cis and 40 trans eQTLs were identified specific for the BRCA cohort. These variants impacted the expression of 38 unique genes (egenes). Of those, in cis, COX7AR’s expression was the most significantly impacted (FDR=2.62x10−26). In trans, SLC46A3’s (FDR=6.6x10−4) and COX7AR’s (FDR=0.002) expression were most significantly impacted. Enrichr27-29 pathway analysis was performed on the 38 egenes. Interestingly, these genes were associated with an impact on IL6 signaling (q=0.012), ERBB2 and Her2 pathway signaling (q=0.029). Pathway analysis of the clinvar database investigating 11 trans-egenes revealed disease association with non-Hodgkin’s lymphoma (q=0.008), breast neoplasms (q=0.014) familial breast neoplasms (q=0.014).
DISCUSSION
This novel translational epidemiologic study revealed associations between genetic ancestry and nSES on TNBC. Both WA ancestry and nSES were independently associated with TNBC; however, on the multinomial logistic regression when nSES and WA ancestry were both added in our model, only nSES remained significant, suggesting potential gene-environment interactions. Moreover, admixture mapping of WA ancestry was associated with TNBC, independent of nSES, at specific regions along the autosome. These regions of WA ancestral origin may predispose individuals to this highly aggressive tumor subtype, reflecting tumor subtype trends typically observed in self-identified racial/ethnic (SIRE) groups. Our findings underscore the importance of integrating precise genomic measures and contextual-level measures to identify molecular alterations that can be used to improve disparities through ancestrally and socioeconomically-calibrated patient stratification, prognostication, and development of novel therapies in patients with TNBC.
Moving Beyond Self-Identified Race and Ethnicity (SIRE)
Self-identified Black and Hispanic women with breast cancer, when compared to their White counterparts, classically exhibit earlier onset, more advanced stage at diagnosis, and TNBC. Numerous studies have suggested that inequality of screening, treatment, and follow up lead to much of this mortality disparity. However, recent papers, including our current study, suggest biologically and clinically relevant differences in the tumors from women with WA ancestry compared to EU ancestry5, 8
SIRE has been a metric classically characterized in most publications describing patient demographics. As the prevalence of admixture increases throughout the United States, the more difficult it will be to fit individuals into racial “boxes”. The 2020 Census data revealed a 276% increase in the multiracial population and a 23% increase in the Hispanic or Latino population. As racial groups become increasingly ancestrally heterogeneous, more precise genomic measures are needed in order to appropriately stratify patients4.
In this study we found significant differences in tumor characteristics (e.g., later stage, higher grade, and TNBC) between those with increasing WA genetic ancestry compared to increasing EU genetic ancestry; however, these differences were not seen using SIRE. These results show that genetic ancestry may be a more sensitive measure of differences in tumor characteristics than SIRE. This may also be in part due to the large number of admixed individuals analyzed in the study. As a result, SIRE may be unreliable or misinterpreted, leading to misclassification and the generation of unreliable results in diverse racial/ethnic populations30.
Our findings of increasing WA ancestry among women who are NHB and HB can also be extended to prior studies by Goel et al3 which uncovered tumor differences exist between SIRE groups. Specifically, they identified intra-ethnic differences by Black race between Hispanic Blacks and Hispanic Whites. Hispanic Blacks had more aggressive tumor characteristics (e.g., later stage at diagnosis, higher grade, TNBC) compared to Hispanic Whites. More striking, breast cancer-specific survival outcomes were worse among Hispanic Black women compared to Hispanic White women, but better among Hispanic Black women compared to non-Hispanic Black women. Extending the results of this current study suggest that an increase in WA ancestry may drive more aggressive tumor biology (TNBC) and worse breast cancer-specific survival among NHB and HB compared to NHW and HW. The sooner that the direct etiologies of disease susceptibility (e.g., TNBC) and aggressiveness are identified, the better our understanding of intervenable factors to mitigate population-based disparities using more accurate biomarkers such as genetic ancestry instead of SIRE. Thus, these emerging data suggest that biological features associated with genetic ancestry may both inform our understanding of aggressive disease and provide pathways to precision screening and treatment approaches.
Integration of nSES and Genetic Ancestry Identifies Potential Gene-Environment Interactions
In addition to disparities in more aggressive breast cancer tumor characteristics (e.g., later stage, higher grade, and TNBC) associated with increasing WA ancestry, we would be remiss to assume that these disparities in exist in a vacuum. To adequately understand the impact of WA and EU ancestry on breast cancer disparities, we need to evaluate contextual-level measures as well such as nSES. 5, 31
By integrating genetic ancestry and nSES in evaluation of breast cancer subtype, we discovered that both WA ancestry and nSES are independently associated with TNBC. More striking, on the fully adjusted multinomial logistic regression with age, stage, WA ancestry, and nSES, the associated between low nSES and higher rates of TNBC compared to other breast cancer subtypes remained statistically significant. This suggests potential gene-environment interactions associated with disadvantaged neighborhoods as drivers of TNBC.
The Ecosocial theory of disease distribution posits that health disparities may arise due to social, ecological, political and/or historical exposures within a neighborhood33. Recent studies have identified disparities in rates of TNBC based on extremes of residential economic segregation, with higher rates of TNBC compared to ER+/HER2− in the most economically disadvantaged neighborhoods, even independent of race11, 32. These race-independent disparities, due in part by living in low nSES, may reflect the downstream effects of negative health behaviors (poor diet, limited physical activity), lack of healthcare access, and chronic stress33. Poor diet and limited physical activity may lead to the development of obesity and diabetes which have been associated with an increased risk of developing TNBC through the dysregulation of cell cycle regulation and cell proliferation signaling pathways34-36. Synergistically, stress of living in a disadvantaged environment has been thought to shape tumor biology by altering gene expression and impacting inflammatory or immune response systems37. Newman et al38 also highlights that the allostatic load (“wear and tear on the body”) from social inequality and epigenetic alterations need to be considered in studying complexity of studying breast cancer disparities. Collectively, our findings that nSES, independent of increasing WA ancestry, is also associated with TNBC suggests, within the background of these prior genomic frameworks, a potential biological mechanism through which nSES may impact differences in breast cancer subtype. As a result, we emphasize the importance of contextual-level annotation of genetic ancestry studies5,6.
These findings also bring to light a “chicken or egg” situation--are women of WA ancestry more likely to inherit genetic components of geographically defined WA ancestry that are associated with hereditary susceptibility for TNBC (“nature”) verses is the Black phenotype associated with increasing WA ancestry sorting women into disadvantaged neighborhoods as a result of structural racism from historical redlining and driving TNBC etiology through gene-environment interactions (“nurture”). Or, more likely, a combination of both “nature” and “nurture” are at play and associated with TNBC.
Novel West African Ancestry SNPs associated with TNBC Independent of nSES
To study this point further, we performed admixture mapping to identify genomic loci of ancestral origin that are associated with TNBC. We found WA and NAT genomic loci. independent of nSES that are associated with TNBC. These loci may predispose women to TNBC, which may be one reason this subtype is seen at a higher proportion in Black and Hispanic women when compared to their White counterparts. Of these genomic segments WA enrichment was most significant with TNBC. Our results identified 38 genes whose expression in breast cancer were altered by variants found in the segments enriched with WA ancestry. These genes were associated with ERBB2 and Her2 pathway signaling. Additionally, expression of Cox subunit VIIa polypeptide 2-like protein (COX7AR) was impacted most significantly by multiple variants found in these enriched segments, which is known to facilitate human breast cancer malignancy39. These results suggest that there may be an ancestrally driven predisposition of the development of TNBC.
Our findings need to be evaluated while considering the study limitations, which include a case-only, two-institution study in South Florida, which may not be generalizable to other health systems caring for similar populations. Nevertheless, these two-institutions consist of an NCI designated cancer center and safety-net hospital which reflects two diverse racial/ethnic and socioeconomic populations. Furthermore, the small sample size of 47 NHB and 11 Hispanic Blacks (12 and 4 with TNBC, respectively) limits statistical power in disentangling neighborhood disadvantage from WA ancestry and its association with TNBC. Future studies need to expand on our findings to further understand the association between neighborhood disadvantage and genetic ancestry on rates of TNBC. Nevertheless, the racial/ethnic and economic mix of our population in one of the most diverse counties in the US, which likely reflects the future demographics of the US, particularly the Sun Belt40.
Overall, this prospective study evaluates the impact of both genetic ancestry and nSES on rates of breast cancer subtype among an admixed population. Given evidence in the literature that one’s social and environmental exposures and individual experiences may lead to epigenetic changes, influencing a woman’s risk for developing breast cancer, we extend this further to also influencing a woman’s risk for developing a specific breast cancer subtype. Quantifying these alterations may provide insight into the extent nSES has impacted an individual over time. Future studies must integrate nSES, genetic ancestry, and additional molecular features associated with breast cancer subtype to improve outcomes in historically marginalized individuals. Utilizing this translational approach will allow for improved methods of patient risk stratification, cancer control interventions in the community setting, and precision oncology treatment strategies for patients. It also presents an exciting opportunity to further investigate novel SNP biomarkers of nSES, WA ancestry, and TNBC.
CONCLUSIONS
This integrative study illustrates that global and local genetic ancestry and nSES are independently associated with TNBC, suggesting potential gene-environment interactions. This study is among the first to estimate ancestral proportions in a large cohort of admixed individuals with breast cancer. Our results reflect the genomic diversity of these patients and the changing racial/ethnic landscape of the United States in the years to come. Integrating genetic ancestry and contextual-level measures can result in novel tests for patient stratification and prognostication. Overall, this study reveals a need for larger genomic studies of admixed racial/ethnic minorities with breast cancer, that comprehensively integrate social and environmental factors in which patients live in order to reach health equity.
Sources of support:
Supported by grants from the National Institutes of Health (K12CA226330, to N.G. F31CA243426, to D.A.R) and American Surgical Association Fellowship Award (N.G.)
Footnotes
Competing Interests:
J.W.H is an inventor of intellectual property related to gene expression profiling in uveal melanoma. He is a paid consultant for Castle Biosciences, licensee of this intellectual property, and he receives royalties from its commercialization. All remaining authors declare no competing financial interests.
REFERENCES
- 1.Kelly KN, Hernandez A, Yadegarynia S, et al. Overcoming disparities: Multidisciplinary breast cancer care at a public safety net hospital. Breast Cancer Res Treat 2021; 187(1):197–206. [DOI] [PubMed] [Google Scholar]
- 2.Daly B, Olopade OI. A perfect storm: How tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J Clin 2015; 65(3):221–38. [DOI] [PubMed] [Google Scholar]
- 3.Goel N, Yadegarynia S, Lubarsky M, et al. Racial and Ethnic Disparities in Breast Cancer Survival: Emergence of a Clinically Distinct Hispanic Black Population. Ann Surg 2021; 274(3):e269–e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez DA, Sanchez MI, Decatur CL, et al. Impact of Genetic Ancestry on Prognostic Biomarkers in Uveal Melanoma. Cancers (Basel) 2020; 12(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan J, Hu Z, Mahal BA, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell 2018; 34(4):549–560 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrot-Zhang J, Soca-Chafre G, Patterson N, et al. Genetic Ancestry Contributes to Somatic Mutations in Lung Cancers from Admixed Latin American Populations. Cancer Discov 2021; 11(3):591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fejerman L, John EM, Huntsman S, et al. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res 2008; 68(23):9723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huo D, Hu H, Rhie SK, et al. Comparison of Breast Cancer Molecular Features and Survival by African and European Ancestry in The Cancer Genome Atlas. JAMA Oncol 2017; 3(12):1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer JR, Ruiz-Narvaez EA, Rotimi CN, et al. Genetic susceptibility loci for subtypes of breast cancer in an African American population. Cancer Epidemiol Biomarkers Prev 2013; 22(1):127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spratt DE, Chan T, Waldron L, et al. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol 2016; 2(8):1070–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goel N, Westrick AC, Bailey ZD, et al. Structural Racism and Breast Cancer-Specific Survival: Impact of Economic and Racial Residential Segregation. Ann Surg 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bemanian A, Beyer KM. Measures Matter: The Local Exposure/Isolation (LEx/Is) Metrics and Relationships between Local-Level Segregation and Breast Cancer Survival. Cancer Epidemiol Biomarkers Prev 2017; 26(4):516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015; 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reich D, Patterson N, Campbell D, et al. Reconstructing Native American population history. Nature 2012; 488(7411):370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science 2002; 298(5602):2381–5. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg NA, Mahajan S, Ramachandran S, et al. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet 2005; 1(6):e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics 2011; 27(15):2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19(9):1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Zhan X, Bragg-Gresham J, et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet 2014; 46(4):409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Zhan X, Liang L, et al. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet 2015; 96(6):926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browning BL, Zhou Y, Browning SR. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am J Hum Genet 2018; 103(3):338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81(5):1084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maples BK, Gravel S, Kenny EE, et al. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 2013; 93(2):278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin ER, Tunc I, Liu Z, et al. Properties of global- and local-ancestry adjustments in genetic association tests in admixed populations. Genet Epidemiol 2018; 42(2):214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin L, Zhang H, Tang Z, et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated tool for Genome-Wide Association Study. Genomics Proteomics Bioinformatics 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong J, Mei S, Liu C, et al. PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res 2018; 46(D1):D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44(W1):W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Z, Bailey A, Kuleshov MV, et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 2021; 1(3):e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez SL, Kelsey JL, Glaser SL, et al. Inconsistencies between self-reported ethnicity and ethnicity recorded in a health maintenance organization. Ann Epidemiol 2005; 15(1):71–9. [DOI] [PubMed] [Google Scholar]
- 31.Carrot-Zhang J, Chambwe N, Damrauer JS, et al. Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer. Cancer Cell 2020; 37(5):639–654 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krieger N, Singh N, Waterman PD. Metrics for monitoring cancer inequities: residential segregation, the Index of Concentration at the Extremes (ICE), and breast cancer estrogen receptor status (USA, 1992-2012). Cancer Causes Control 2016; 27(9):1139–51. [DOI] [PubMed] [Google Scholar]
- 33.Krieger N. Methods for the scientific study of discrimination and health: an ecosocial approach. Am J Public Health 2012; 102(5):936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vargas-Hernandez VM, Vargas-Aguilar V, Moreno-Eutimio MA, et al. Metabolic syndrome in breast cancer. Gland Surg 2013; 2(2):80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis AA, Kaklamani VG. Metabolic syndrome and triple-negative breast cancer: a new paradigm. Int J Breast Cancer 2012; 2012:809291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiti B, Kundranda MN, Spiro TP, et al. The association of metabolic syndrome with triple-negative breast cancer. Breast Cancer Res Treat 2010; 121(2):479–83. [DOI] [PubMed] [Google Scholar]
- 37.Linnenbringer E, Gehlert S, Geronimus AT. Black-White Disparities in Breast Cancer Subtype: The Intersection of Socially Patterned Stress and Genetic Expression. AIMS Public Health 2017; 4(5):526–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman LA, Kaljee LM. Health Disparities and Triple-Negative Breast Cancer in African American Women: A Review. JAMA Surg 2017; 152(5):485–493. [DOI] [PubMed] [Google Scholar]
- 39.Zhang K, Wang G, Zhang X, et al. COX7AR is a Stress-inducible Mitochondrial COX Subunit that Promotes Breast Cancer Malignancy. Sci Rep 2016; 6:31742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bureau USC, QuickFacts Florida. Available at: https://www.census.gov/quickfacts/fact/table/FL,US/PST045219. Accessed May 5, 2021.


