Abstract
Circular RNA (circRNA) serves as a competitive endogenous RNA (ceRNA) that plays a pivotal role in the initiation and progression of breast cancer (BC). However, compared to other noncoding RNAs (ncRNAs), research on circRNA in BC is still in its infancy. Through the analysis of circRNA datasets in the GEO database, hsa_circ_0001314, which is upregulated in BC, was selected as the focus of this study. RT-qPCR analysis showed that hsa_circ_0001314 was significantly upregulated in BC tissues and cells. Subsequently, the biological functions of hsa_circ_0001314 in BC cells were examined through CCK-8, wound healing, transwell invasion, and flow cytometry analyses. The research demonstrated that knocking down the expression level of hsa_circ_0001314 significantly inhibited cell proliferation, migration, and invasion abilities while notably promoting cell apoptosis. Bioinformatics methods were used to predict downstream miRNAs and mRNAs that may interact with hsa_circ_0001314, constructing a ceRNA regulatory network related to hsa_circ_0001314. RT-qPCR confirmed that hsa_circ_0001314 functions as a sponge for hsa-miR-548aj-3p, competitively binding to hsa-miR-548aj-3p to activate the MAPK signaling pathway and regulate the expression of MAPK8 and MAP3K1. The findings uncover the potential of hsa_circ_0001314 as a novel prognostic biomarker and therapeutic target for BC patients.
Keywords: breast cancer, ceRNA, circRNA, GEO
1. Introduction
Breast cancer (BC) is the most frequent disease among women and the primary cause of cancer-related deaths [1]. Despite improvements in the prognosis and early detection of BC, its morbidity and mortality remain very high and pose significant challenges, primarily due to the complex pathogenesis, multiple molecular subtypes, and high heterogeneity [2]. To effectively treat BC, it is imperative to discover new molecular biomarkers and develop novel diagnostic and therapeutic approaches.
Circular RNAs (circRNAs), a novel type of endogenous ncRNAs, unlike traditional linear RNAs, are a class of single-stranded RNA molecules with a covalently closed circular structure, which is mainly produced by reverse splicing of precursor mRNA (pre-mRNA) exons and lacks a 5′-terminal cap and a 3′-terminal polyadenylic acid tail (Poly A) [3], which makes them relatively stable in cells and not easily degraded by exonucleases [4]. As early as 1976, circRNA was first discovered by the Sanger team [5]. But at first, circRNAs did not attract much attention from the academic community [6]. Until the 21st century, as RNA-seq technology evolved dramatically, circRNAs were discovered in large numbers [7, 8], and the functionality of circRNAs began to be discovered and studied in depth [9]. The widespread expression of circRNA in many species has been demonstrated in numerous studies such as viral [10] and mammalian [11]. Meanwhile, there was a strong correlation found between circRNA and the emergence of certain malignancies, such as gastric cancer [12], BC [13], hepatocellular carcinoma [14], colorectal cancer [15], and other illnesses. Currently, the diversification of early diagnosis methods for BC and the pluralism of prognosis parameters provide a new evidence base and methods to screen for BC at an early stage and the selection of neoadjuvant chemotherapy regimens. The etiology and pathogenesis of BC are complex because tumors are highly heterogeneous and have various molecular subtypes. Therefore, making use of bioinformatics methods to find the molecular targets of BC and studying the detection methods and therapeutic drugs related to the targets has become a key link in the field of BC research.
circRNAs' primary function is to control gene expression by serving as the sponge for microRNAs [5]. For example, by adsorbing with miR-217-5p, CircEZH2 can upregulate KLF5 expression, thereby accentuating the carcinogenesis and metastasis of BC [16]. hsa_circ_0025202 has been shown to regulate tumor progression in BC cells by acting as a miRNA sponge for miR-182-5p [17].
We are primarily using the ceRNA mechanism of circRNA in this study. Using R software, we obtain the DEcircRNAs by using the circRNA database in BC tissue from the Gene Expression Omnibus (GEO). Use the database to predict miRNAs and mRNAs downstream of circRNA. The mRNAs were enriched according to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and the network of the ceRNAs was constructed. Verify the hsa_circ_0001314 expression and biological function by using in vitro experiments. The molecular mechanism by which hsa_circ_0001314 can act as a sponge of hsa-miR-548aj-3p, bind to hsa-miR-548aj-3p, activate the MAPK signaling pathway, and regulate the expression of MAPK8 and MAP3K1 genes. This study was aimed at identifying a novel therapeutic target for BC by focusing on circRNA-based ceRNA action, which not only facilitates a better understanding of the development mechanism of BC but also has important implications for the development of BC therapies.
2. Methods
2.1. Data Collection
Download the circRNA expression profile (GSE182471) and miRNA expression profile (GSE143564) in the GEO database (https://www.ncbi.nlm.nih.gov/) [18]. Based on the GPL21825 platform, the GSE182471 is a circRNA database that includes five BC tissue samples and five adjacent tissue samples. The GSE143564 is a miRNA database based on the GPL21572 platform that includes three BC tissue samples and three adjacent tissue samples.
2.2. Identification of circRNA
R is used to read the clinical and raw data for GSE182471. The probe and the gene symbol are matched based on the annotation file within the platform. The DEcircRNAs analysis was performed with the R, and the DEcircRNAs were identified using the p < 0.050 and |log2(fold change)| > 2. The volcano plot of the DEcircRNAs was displayed by the ggplot2 package in R. The heatmap plot of the DEcircRNAs was performed by heatmap plotting tools in Hiplot Pro (https://hiplot.com.cn/).
2.3. Prediction of miRNA and mRNA of circRNA
The target miRNAs of DEcircRNA were predicted using circBank (http://www.circbank.cn) [19]. For the differentially expressed miRNA (DEmiRNA) analysis, the R limma package was utilized. The DEmiRNAs in the GSE143564 dataset and these target miRNAs were intersected using a Venn diagram, and intersection miRNAs were subsequently obtained. To predict the interactions of miRNA and mRNA, miRDB (https://mirdb.org) [20] and TargetScan (https://www.targetscan.org/vert_80/) [21] were used. Then, we took the intersection of mRNAs between the miRDB and TargetScan, which we called DEmRNAs.
2.4. Functional Enrichment Analyses
The functional enrichment analysis of mRNAs was performed with the DAVID database (https://david.ncifcrf.gov) [22]. Three fields were included in the biological process (BP) section of the GO analysis: BPs, cellular component (CC), and molecular function (MF), while KEGG signaling pathways were used to further explore the pathway of DEmRNAs.
2.5. The ceRNA Network Construction
Using GeneMania (http://www.genemania.org) [23], the gene–gene interaction (GGI) network was created. The STRING database (https://cn.string-db.org) [24] was used to retrieve the interaction pairs between target genes, and the CytoNCA tool in Cytoscape was utilized to obtain the network's hub genes. The content of the analysis is betweenness centrality. Additionally, the PPI network was displayed using Cytoscape [25]. Based on the hub genes, the ceRNA network was built.
2.6. Expression Level and Survival Analysis of mRNAs
The online analytic system of the assistance for clinical bioinformatics, which is based on the TCGA database, examined the expression of hub genes in patients with BC. The UALCAN database analyzes the survival of hub genes in BC patients to explore whether the hub genes affect the survival rate of BC patients.
2.7. Tissue Sample Collection
From the Third Affiliated Hospital of Qiqihar Medical University, a total of 15 BC samples and 9 adjacent breast tissue samples were obtained. The study was approved by the Ethics Committee of Qiqihar Medical University, and informed consent was obtained from all patients.
2.8. Cell Culture and Transfection
Normal human mammary epithelial cells (MCF-10A) and human BC cells (MCF-7 and MDA-MB-231) were supplied by the Chinese Academy of Sciences Cell Bank (Shanghai, China). Fetal bovine serum (FBS, TBDscience, China) and penicillin–streptomycin solution (Biosharp, China) were added to RPMI 1640 (Procell, China) to cultivate the MCF-7 cells. MCF-10A and MDA-MB-231 were cultured using MCF-10A cell-specific medium (Procell, China) and DMEM high glucose medium (Procell, China), respectively. All cells were grown at 5% CO2 and 37°C. Si-hsa_circ_0001314-1, si-hsa_circ_0001314-2, and hsa-miR-548aj-3p for RNA interference in BC cells were designed and synthesized by GenePharma (Shanghai, China). Si-hsa_circ_0001314-1, si-hsa_circ_0001314-2, hsa-miR-548aj-3p, and negative control (NC) were transfected into MCF-7 cells. Then, the seeding of cells in six-well culture plates and transfecting with siRNA and NC using liposomal 6000 (Beyotime, China). Table 1 contains a list of the siRNA and miRNA mimic synthesis sequences.
Table 1.
Sequences of the synthesis.
| Target name | Sequences (5′-3′) | |
|---|---|---|
| Negative control | Sense | UUCUCCGAACGUGUCACGUTT |
| Antisense | ACGUGACACGUUCGGAGAATT | |
| si-hsa_circ_0001314-1 | Sense | GGAACUAGGCAGGAAUAUCTT |
| Antisense | GAUAUUCCUGCCUAGUUCCTT | |
| si-hsa_circ_0001314-2 | Sense | CUAGGCAGGAAUAUCUUAUTT |
| Antisense | AUAAGAUAUUCCUGCCUAGTT | |
| hsa-miR-548aj-3p | Sense | UAAAAACUGCAAUUACUUUUA |
| Antisense | AAAGUAAUUGCAGUUUUUAUU |
2.9. RT-qPCR
From tissue samples and cells, total RNA was extracted using TRIzol reagent (Tiangen, China), and reverse transcription of cDNA was performed using FastKing gDNA Dispelling RT SuperMix (Tiangen, China). RT-qPCR studies were conducted using Talent qPCR PreMix (Tiangen, China). Reference genes for GAPDH and U6 were used. The 2−ΔΔCT method was employed to calculate the relative expression levels. Specific primer sequences refer to Table 2. To investigate the association between circRNA and miRNA (hsa_circ_0001314 and hsa-miR-548aj-3p), Pearson's correlation coefficients were employed.
Table 2.
Sequences of primers for RT-qPCR.
| Primer name | Primer sequences (5′-3′) |
|---|---|
| hsa_circ_0001314 | F: GAAACTAAGGAACTAGGCAGG |
| R: CAAGCCAGGTATCTTGTGAATG | |
| GAPDH | F: AGAAGGCTGGGGCTCATTTG |
| R: GCAGGAGGCATTGCTGATGAT | |
| hsa-miR-548aj-3p | F: CGCGCGTAAAAACTGCAATTA |
| R: AGTGCAGGGTCCGAGGTATT | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| KRAS | F: TAGGCAAGAGTGCCTTGACG |
| R: CCCTCCCCAGTCCTCATGTA | |
| MAP3K1 | F: AGGTCGCACAGTGAAATCAG |
| R: GTTTCCTCAGGGCTATATGGTG | |
| MAPK8 | F: GACGCCTTATGTAGTGACTCGC |
| R: TCCTGGAAAGAGGATTTTGTGGC |
2.10. Cell Counting Kit-8 (CCK-8) Proliferation Assay
The transfected BC cells will continue to be cultured under appropriate conditions for 24, 48, or 72 h. Then, add 10 μL of CCK-8 reagent (Biosharp, China) to each well following the kit instructions. Using a microplate reader, the absorbance of the cells was determined at 450 nm after they had been incubated for 1 h at 37°C.
2.11. Wound Healing Assay
In six-well plates, transfected MCF-7 cells were cultivated until 80% confluence was attained. Using a 200 μL sterile pipette tip, the cell surface was scraped. After PBS washes to remove cell debris, followed by culture at 37°C with fresh medium. Cells were visualized and photographed using a microscope (Olympus, Japan) at 0, 24, and 48 h. ImageJ was used to estimate scratch areas and calculate the relative migration rate.
2.12. Transwell Invasion Assay
Matrigel (ABW, China)-covered upper chamber was seeded with cells. The lower chamber was filled with a medium containing 10% FBS, and it was incubated for 48 h. Four percent paraformaldehyde is used to fix cells for 15 min, and 0.1% crystal violet is used to dye them for 10 min. Then, a cotton swab was used to wipe the top chamber. Cells were visualized and counted using a microscope.
2.13. Cell Apoptosis Assay
Apoptosis was assessed using the V-FITC/PI Apoptosis Assay Kit (Biosharp, China). Following trypsinization, the cells were resuspended in 250 μL of binding buffer after being washed with cold PBS. Add 5 μL of Annexin V-FITC and 10 μL of PI, then incubate for 10 min at room temperature. Apoptosis was detected using a FACScalibur flow cytometer (BD Biosciences, United States).
2.14. Statistical Analysis
Triplicates of each experiment were run. ImageJ software and GraphPad Prism 9.5.1 software were used for statistical analysis of image production and experimental data. A T-test was used for univariate variable comparison, and variance analysis was used for data analysis between multiple groups. A possibility of p < 0.050 reflects a significant difference.
3. Results
3.1. Differentially Expressed circRNAs
The datasets of GSE182471 and GSE143564 downloaded from the GEO database have been standardized (Figures 1(a) and 1(b)). Following the analysis, 142 DEcircRNAs were discovered, four of which had downregulation and 138 of which had upregulation. As demonstrated by the DEcircRNAs' heatmaps and volcano plot in Figures 1(c) and 1(d), one of the circRNAs that is most significantly upregulated in BC tissues is hsa_circ_001831. Finally, hsa_circ_001831 was regarded as the study's final DEcircRNA. The hsa_circ_001831 is converted into a universal name hsa_circ_0001314 based on circBase [26]. The structure pattern diagram of hsa_circ_0001314 (Figure 1(e)) was shown by CircPrimer 2.0 [27].
Figure 1.
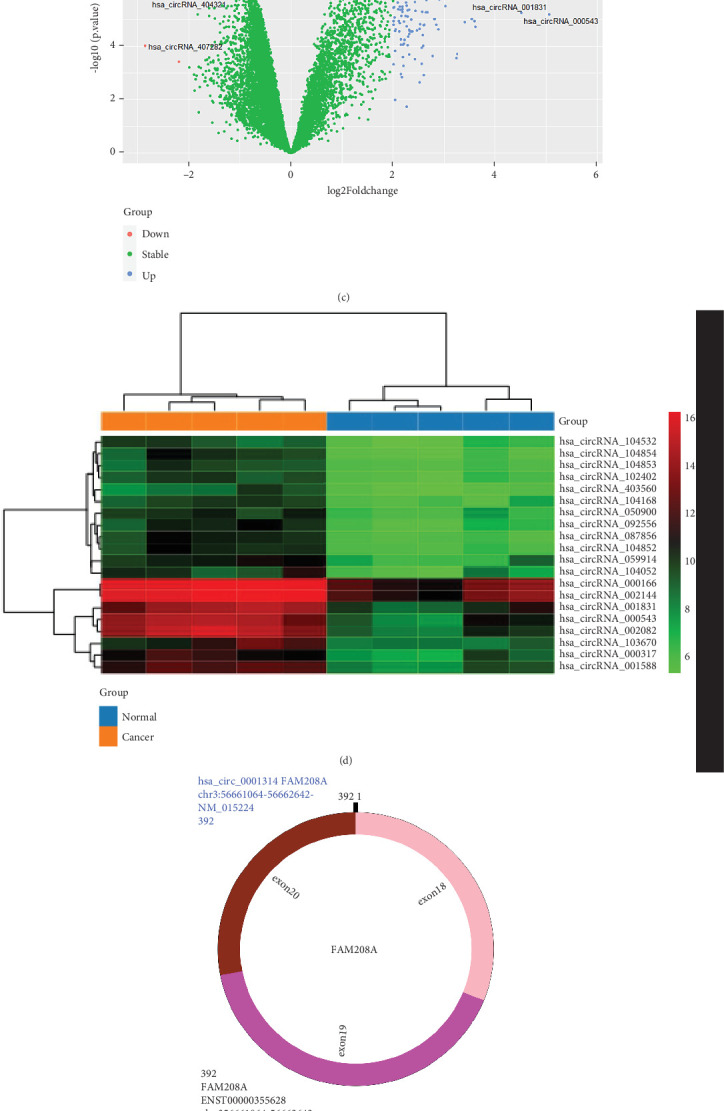
Identification of the DEcircRNAs, DEmiRNAs, and DEmRNAs. (a) Normalization of circRNA dataset. (b) Normalization of miRNA dataset. (c) Volcano plot of DEcircRNAs. (d) Heatmap of DEcircRNAs. (e) Structural patterns of hsa_circ_0001314. (f) Venn diagram of the intersection between target miRNAs and the DEmiRNA in the GSE143564 dataset. (g) Venn diagram of the intersection between target mRNAs from the miRDB and TargetScan predictions. (h) The binding site of hsa_circ_0001314 with hsa-miR-548aj-3p.
3.2. Prediction of miRNA and mRNA of circRNA
Through analysis and screening of the GEO database (GSE143564), a total of 4422 DEmiRNAs were obtained. Through predictions generated by the circBank database, 25 miRNAs were identified as binding to hsa_circ_0001314. Subsequently, as shown in Figure 1(f), a Venn diagram was used to intersect the 4422 DEmiRNAs with the 25 target miRNAs, yielding 23 intersecting miRNAs. Among them, hsa-miR-548aj-3p (p = 2.41e − 03) was significantly downregulated in BC tissues. CircMir displayed the binding site of hsa_circ_0001314 with hsa-miR-548aj-3p (Figure 1(h)). Therefore, hsa-miR-548aj-3p was selected as the downstream miRNA of hsa_circ_0001314 for subsequent research.
Using miRDB and TargetScan, the interactions between miRNA and mRNA were predicted. Then, as illustrated in Figure 1(g), the DEmRNAs of the miRDB and the TargetScan were intersected using the Venn diagram, yielding 360 intersection mRNAs.
3.3. GO and KEGG Pathway Enrichment Analyses
GO and KEGG analyses were performed on the 360 intersecting mRNAs to assess the role of the DEmRNAs in the network (Figure 2). The findings of the GO analysis demonstrated that the regulation of transcription from the RNA Polymerase II promoter and the negative regulation of transcription from the RNA Polymerase II promoter were the primary areas of enrichment for changes in BPs of DEmRNAs. The nucleus, cytoplasm, and nucleoplasm showed the greatest enrichment in changes in the CC of DEmRNAs. Protein binding and metal ion binding exhibited a high enrichment in the MF changes of DEmRNAs. The DEmRNAs were primarily abundant in the pathways related to cancer, the MAPK signaling system, the PI3K-Akt signaling pathway, the signaling pathways regulating pluripotency of stem cells, and the Ras signaling pathway, according to KEGG pathway analysis. Most of the pathways identified above are closely correlated with cancer progression. After analysis, MAPK signaling was the most significant number of enriched genes. The MAPK signaling pathway is one of the fundamental pathways of mammalian cells, as demonstrated by numerous research studies, and is closely related to important physiological activities such as cell proliferation, differentiation, apoptosis, and angiogenesis. Therefore, subsequent research will concentrate on the genes that are enriched in the MAPK signaling pathway. The KEGG analysis of partial results is listed in Table 3.
Figure 2.
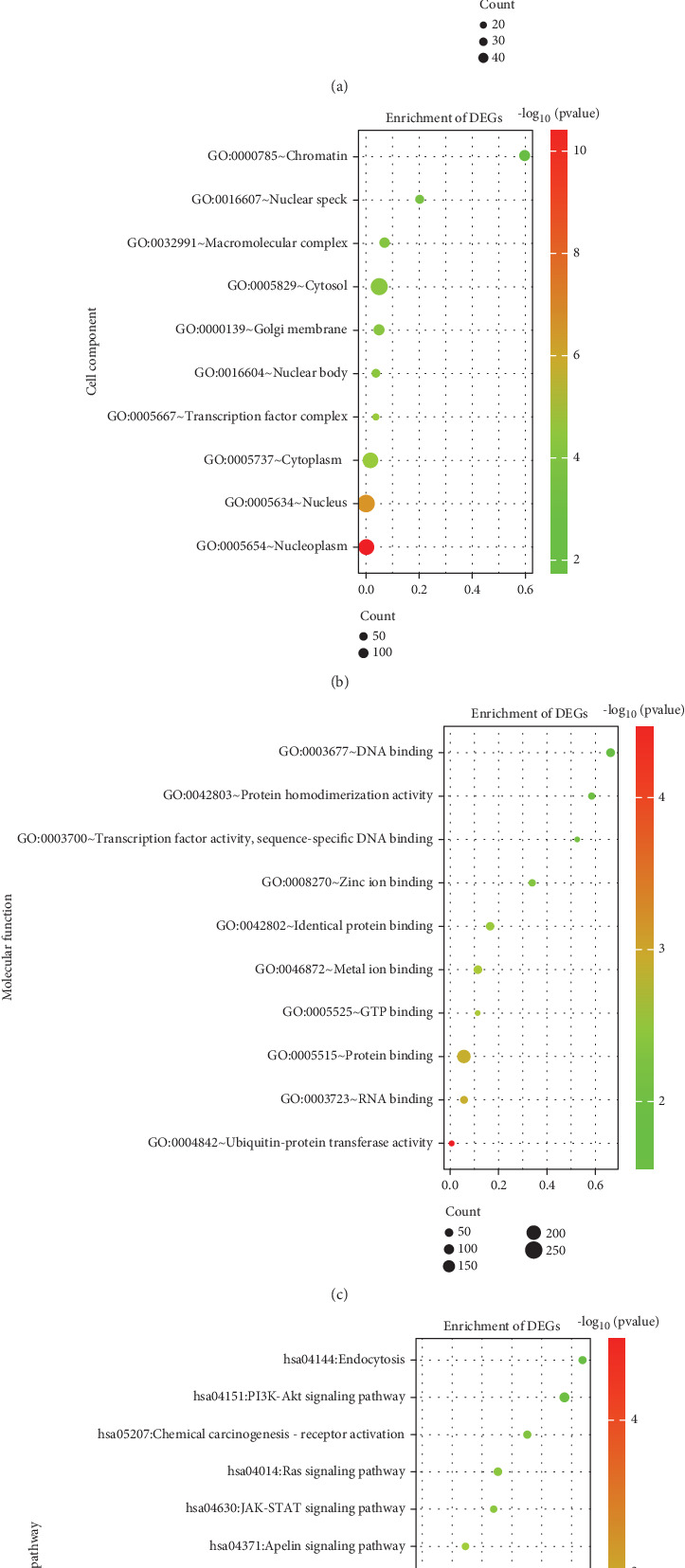
GO and KEGG pathway enrichment analyses. (a–c) The dot plot of Top 10 GO analysis. (d) The dot plot of Top 10 KEGG pathway analysis.
Table 3.
KEGG analysis of partial results.
| Terms | Count | p value | Genes |
|---|---|---|---|
| hsa05200: Pathways in cancer | 21 | 4.52e − 03 | SMAD2, GSK3B, SMAD4, HDAC2, CHUK, GADD45A, IL15, DLL1, RASGRP1, EPOR, GNAI1, AGT, MAPK8, CCND1, GNG7, COL4A5, KRAS, VHL, JAK2, SOS2, and FGFR2 |
| hsa04010: MAPK signaling pathway | 15 | 2.25e − 03 | MEF2C, MAP3K1, CHUK, GADD45A, CACNA2D1, RRAS2, RASGRP1, RAP1B, MAPK8, RASA2, MKNK2, KRAS, LAMTOR3, SOS2, and FGFR2 |
| hsa04151: PI3K-Akt signaling pathway | 13 | 5.15e − 02 | GSK3B, CHUK, YWHAZ, EPOR, CCND1, PPP2R5E, GNG7, COL4A5, KRAS, JAK2, SOS2, FGFR2, and MCL1 |
| hsa04014: Ras signaling pathway | 11 | 1.79e − 02 | RAP1B, RAB5B, MAPK8, CHUK, GNG7, RASA2, RRAS2, KRAS, RASGRP1, SOS2, and FGFR2 |
| hsa05224: Breast cancer | 7 | 7.44e − 02 | GSK3B, CCND1, GADD45A, TNFSF11, KRAS, DLL1, and SOS2 |
3.4. Construction of ceRNA Network
The GeneMania database was used to construct the GGI network of the mRNAs (Figure 3(a)). These genes were closely related to genetic interactions, pathways, physical interactions, coexpression, colocalization, shared protein domains, etc. The STRING database and Cytoscape were used to construct the PPI network to perform a topological analysis of the network (Figure 3(b)). PPI network results showed that the Top 5 hub genes were KRAS, RASGRP1, MAPK8, MAP3K1, and SOS2 (Table 4).
Figure 3.
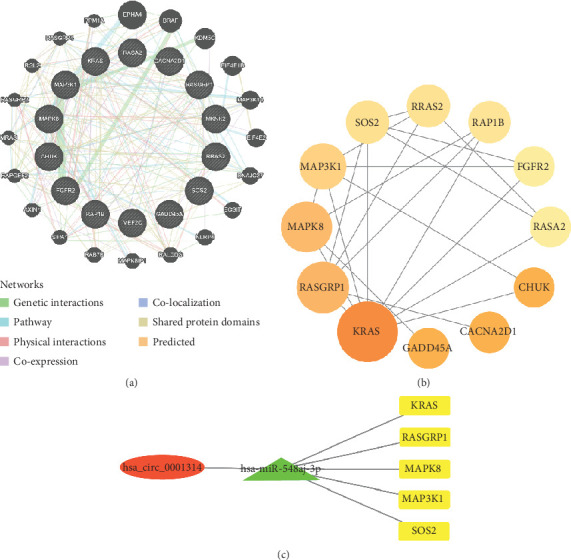
Construction of the network. (a) The GGI network of mRNAs. (b) The PPI network of mRNAs. (c) Cytoscape software was applied to create a visualization of the ceRNA network, which included one circRNA (hsa_circ_0001314), one miRNA (hsa-miR-548aj-3p), and five mRNAs (KRAS, RASGRP1, MAPK8, MAP3K1, and SOS2) (red: circRNA; green: miRNA; yellow: mRNAs).
Table 4.
The table shows the genes ranked by BC.
| Nodes | Betweenness | |
|---|---|---|
| 1 | KRAS | 41.0 |
| 2 | RASGRP1 | 23.5 |
| 3 | MAPK8 | 21.0 |
| 4 | MAP3K1 | 12.5 |
| 5 | SOS2 | 5.8333335 |
| 6 | RRAS2 | 5.0 |
| 7 | RAP1B | 4.0 |
| 8 | FGFR2 | 0.6666667 |
| 9 | RASA2 | 0.5 |
| 10 | GADD45A | 0.0 |
| 11 | CHUK | 0.0 |
| 12 | CACNA2D1 | 0.0 |
The hub genes were chosen to form a circRNA-miRNA-mRNA regulatory network for subsequent investigation. Then, a ceRNA network was built (Figure 3(c)), which included one circRNA (hsa_circ_0001314), one miRNA (hsa-miR-548aj-3p), and five mRNAs (KRAS, RASGRP1, MAPK8, MAP3K1, and SOS2).
3.5. The Expression and Survival Analysis of mRNAs
The expression of five hub genes in BC patients was analyzed (Figure 4). The findings demonstrated that the gene expression of KRAS (p = 1.1e − 149), MAP3K1 (p = 5.7e − 29), MAPK8 (p = 5e − 04), RASGRP1 (p = 2.9e − 111), and SOS2 (p = 7.8e − 31) was considerably upregulated in BC as compared to the normal tissue.
Figure 4.
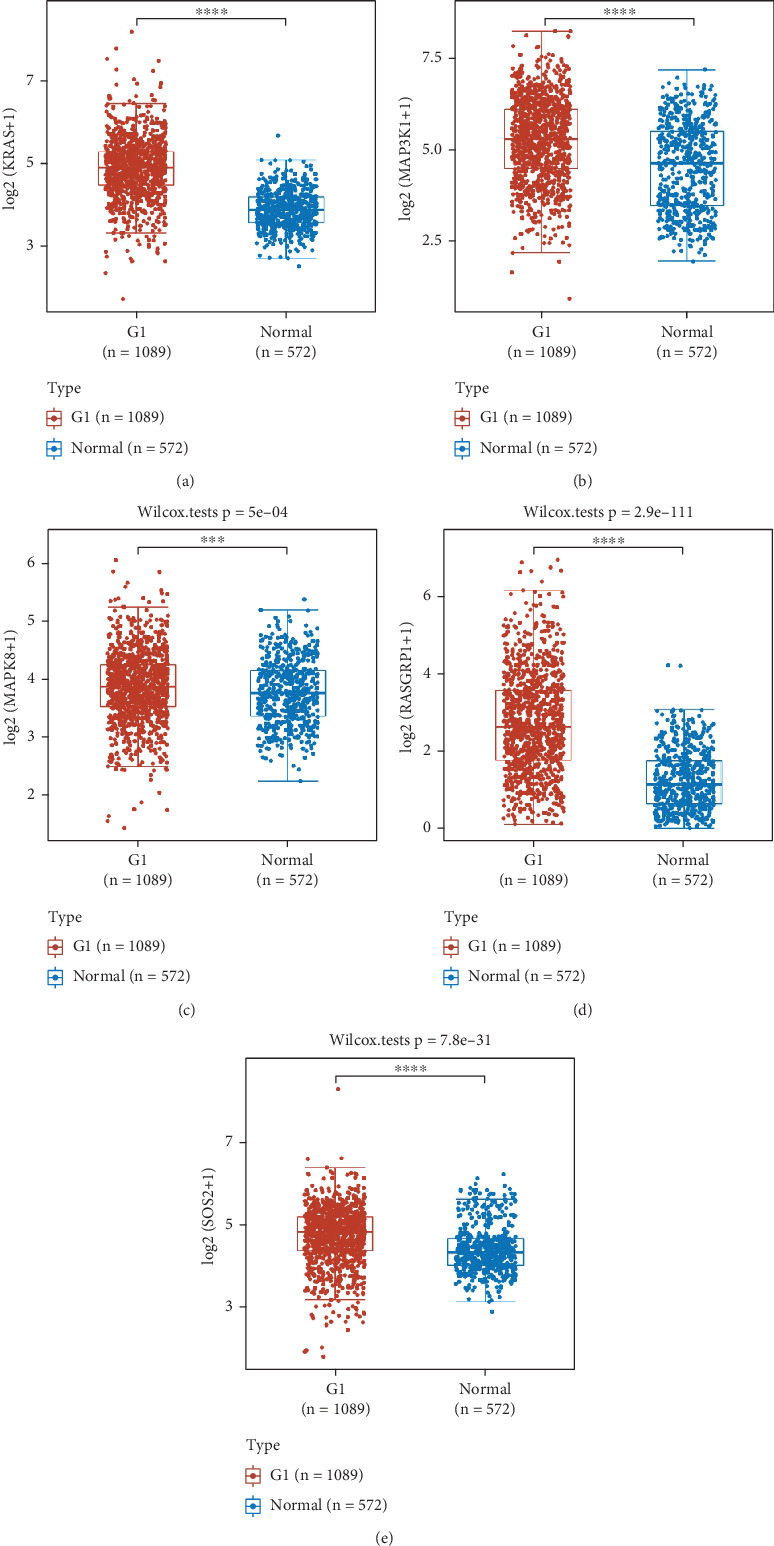
The expression of five hub genes in BC patients was analyzed (the G1 group was BC tissue, and the normal group was normal breast tissue; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). (a) KRAS; (b) MAP3K1; (c) MAPK8; (d) RASGRP1; (e) SOS2.
To further explore whether the hub genes influence the prognostic survival in BC patients, the survival rate of the five hub genes in BC patients was analyzed using UALCAN. By survival analysis, the expression of KRAS, MAP3K1, and MAPK8 genes correlated with survival rate, while BC patients with low expression of KRAS (p < 0.0001), MAP3K1 (p < 0.0001), and MAPK8 (p = 0.00064) had a significantly higher survival rate than those with high expression, while the expression of RASGRP1 gene (p = 0.260) and SOS2 gene (p = 0.210) showed no significant relationship with survival rate (Figure 5).
Figure 5.
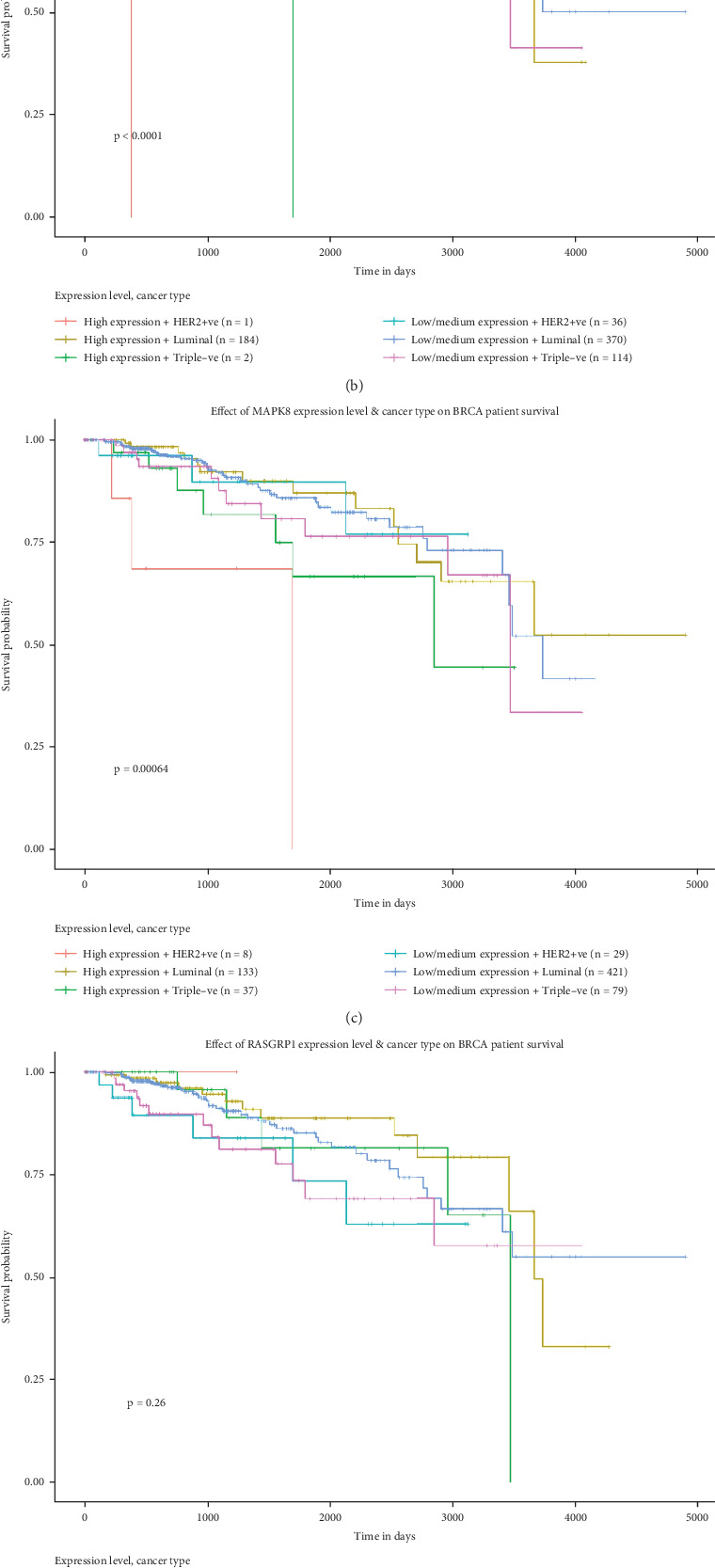
The survival rate of the five hub genes in BC patients was analyzed. (a) KRAS; (b) MAP3K1; (c) MAPK8; (d) RASGRP1; (e) SOS2.
3.6. hsa_circ_0001314 Is Upregulated in BC
To confirm the differential expression of hsa_circ_0001314 in BC, the expression of hsa_circ_0001314 in BC tissue and normal breast tissue, human BC cells (MCF-7 and MDA-MB-231), and normal human breast cells (MCF-10A) was detected by RT-qPCR (Figure 6). The results were consistent with bioinformatics predictions. hsa_circ_0001314 was highly expressed in BC tissues (p = 0.0028, p < 0.010) and BC cells (p = 0.0069, p < 0.010; p = 0.0053, p < 0.010), especially in MCF-7 cells, so MCF-7 cells were chosen for further investigations.
Figure 6.
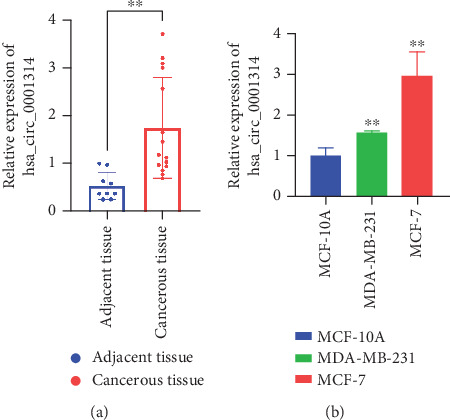
The level of hsa_circ_0001314 in BC tissue and cell is significantly increased. (a) RT-qPCR revealed that the level of hsa_circ_0001314 in BC tissues was higher than that in adjacent tissues (∗∗p < 0.010). (b) Compared with MCF-10A, the expression of hsa_circ_0001314 in BC cells (MCF-7 and MDA-MB-231) was increased significantly (∗∗p < 0.010).
3.7. hsa_circ_0001314 Knockdown Suppressed BC Cell Proliferation, Migration, Invasion, and Promoted Apoptosis
Reduce the expression of hsa_circ_0001314 in MCF-7 to investigate the impact of hsa_circ_0001314 on the biological function of BC cells. After 12 h of transfection, the distribution of intracellular fluorescence signals in MCF-7 was examined using fluorescence microscopy (Figure 7(a)). RT-qPCR was used to detect the expression of hsa_circ_0001314 and verify the transfection efficiency. The outcomes demonstrated that, in comparison to the NC, the expression levels of hsa_circ_0001314 in MCF-7 transfected with si-hsa_circ_0001314-1 and si-hsa_circ_0001314-2 were lower. In addition, the expression level of hsa_circ_0001314 in MCF-7 transfected with si-hsa_circ_0001314-2 was lower than that of transfected with si-hsa_circ_0001314-1; that is, si-hsa_circ_0001314-2 had higher interference efficiency. The si-hsa_circ_001314-2 with better transfection efficiency was chosen for further investigation (Figure 7(b)).
Figure 7.
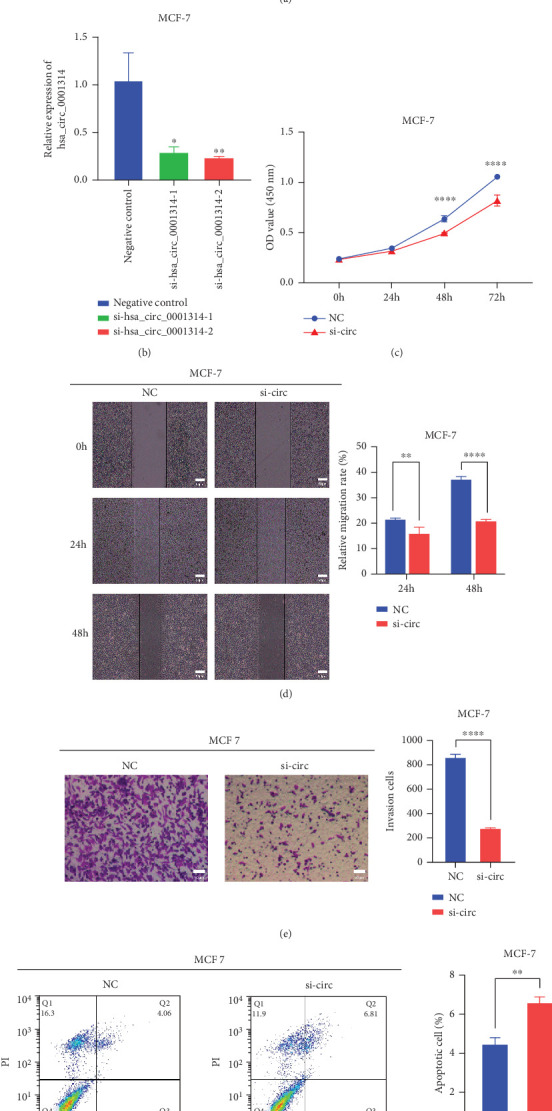
Effect of hsa_circ_0001314 on the proliferative and metastatic capacity of BC cells. (a) Fluorescence signals of the transfected cells were observed under a fluorescence microscope. (b) The siRNAs of hsa_circ_0001314 were transfected to BC cells. Compared with si-hsa_circ_0001314-1, si-hsa_circ_0001314-2 had higher interference efficiency (∗p < 0.050, ∗∗p < 0.010). (c) CCK-8 assay revealed that the knockdown of hsa_circ_001314 significantly reduced the proliferation of BC cells (∗∗∗∗p < 0.0001). (d) Wound healing assay revealed that the knockdown of hsa_circ_001314 dramatically inhibited the migration of BC cells (∗∗p < 0.010, ∗∗∗∗p < 0.0001). (e) Transwell assay revealed that the knockdown of hsa_circ_001314 dramatically inhibited the invasion of BC cells (∗∗∗∗p < 0.0001). (f) Flow cytometry assay revealed that the knockdown of hsa_circ_001314 significantly induced cell apoptosis in BC cells (∗∗p < 0.010).
The CCK-8 assay revealed that the knockdown of hsa_circ_001314 significantly reduced the proliferation of BC cells (Figure 7(c)). Wound healing and transwell assays revealed that knocking down hsa_circ_001314 dramatically inhibited the invasion and migration ability of BC cells (Figures 7(d) and 7(e)). Flow cytometry assay revealed that the knockdown of hsa_circ_001314 significantly induced cell apoptosis in BC cells (Figure 7(f)). These results demonstrated that knocking down hsa_circ_001314 inhibited BC cell proliferation, migration, and invasion while increasing apoptosis.
3.8. hsa_circ_0001314 Binds to hsa-miR-548aj-3p in BC and Regulate the Expression of MAPK8/MAP3K1
hsa-miR-548aj-3p expression in BC tissues was assessed by RT-qPCR. The findings illustrated that hsa-miR-548aj-3p was significantly downregulated (Figure 8(a)). Furthermore, hsa-miR-548aj-3p expression was found to be negatively linked with hsa_circ_0001314 expression in BC tissues (Figure 8(b)). These investigations preliminarily demonstrated that hsa_circ_0001314 could be bound to hsa-miR-548aj-3p in BC.
Figure 8.
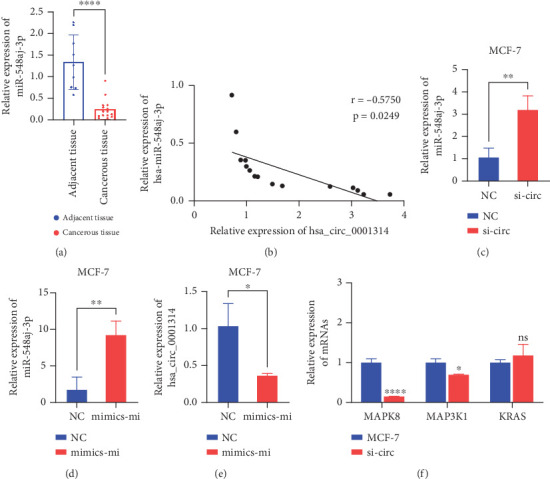
hsa_circ_0001314 binds to hsa-miR-548aj-3p in BC and regulates the expression of MAPK8/MAP3K1. (a) RT-qPCR revealed that the level of hsa-miR-548aj-3p in BC tissues was lower than that in adjacent tissues (∗∗∗∗p < 0.0001). (b) Correlation analysis indicated that the expression levels of hsa_circ_0001314 and hsa-miR-548aj-3p were negatively correlated in BC tissues (r = −0.5750, p = 0.0249). (c) After the knockdown of hsa_circ_0001314, the expression level of hsa-miR-548aj-3p was higher than that in the NC (∗∗p < 0.010). (d) Transfection efficiency was verified after transfection of hsa-miR-548aj-3p mimics in MCF-7 (∗∗p < 0.010). (e) After overexpression of hsa-miR-548aj-3p, the expression level of hsa_circ_0001314 was lower than that of NC (∗p < 0.050). (f) Compared with the expression in MCF-7, the expression of MAPK8 and MAP3K1 genes was significantly reduced after knockdown hsa_circ_0001314, and there was no significant change in KRAS (∗p < 0.050, ∗∗∗∗p < 0.0001; the ns indicates that the difference is not significant).
To further confirm that hsa-miR-548aj-3p is the miRNA bound by hsa_circ_0001314, si-hsa_circ_0001314-2 was transfected into MCF-7 cells to knock down the expression level of hsa_circ_0001314, and the expression of hsa-miR-548aj-3p was detected by RT-qPCR. The results showed that, after the knockdown of hsa_circ_0001314, the expression level of hsa-miR-548aj-3p was higher than that in the NC group (Figure 8(c)). Furthermore, hsa-miR-548aj-3p mimics were transfected into MCF-7 cells to overexpress hsa-miR-548aj-3p, with this group designated as the mimics-mi group. The transfection efficiency was verified using RT-qPCR. The results indicated that the expression level of hsa-miR-548aj-3p in the mimics-mi group was higher than that in the NC group, confirming successful transfection and suitability for subsequent experiments (Figure 8(d)). RT-qPCR was employed to detect the expression of hsa_circ_0001314. The results revealed that, compared to the NC group, overexpression of hsa-miR-548aj-3p led to a decrease in the expression level of hsa_circ_0001314 (Figure 8(e)). These findings demonstrate that the knockdown of hsa_circ_0001314 increases the expression of hsa-miR-548aj-3p, while overexpression of hsa-miR-548aj-3p decreases the expression of hsa_circ_0001314. In conclusion, hsa_circ_0001314 functions as a sponge for hsa-miR-548aj-3p, targeting and binding to it.
According to the prediction of the database, hsa-miR-548aj-3p can target KRAS, RASGRP1, MAPK8, MAP3K1, and SOS2 genes. Because the expression of MAPK8, MAP3K1, and KRAS genes is significantly upregulated in BC and is associated with the survival of BC patients, MAPK8, MAP3K1, and KRAS genes were selected for follow-up research. MCF-7 was transfected with si-hsa_circ_0001314-2 for knockdown of hsa_circ_0001314 expression, and the expression of MAPK8, MAP3K1, and KRAS genes was detected by RT-qPCR (Figure 8(f)). The results showed that compared with the expression in MCF-7, the expression of MAPK8 and MAP3K1 genes was significantly reduced after knockdown hsa_circ_0001314, and there was no significant change in KRAS. These suggested that hsa_circ_0001314 could regulate the expression of MAPK8 and MAP3K1.
4. Discussion
As one of the noncoding RNAs, the circRNA is mainly formed by the pre-mRNA through “reverse splicing,” which has a variety of functions and special properties [28]. circRNA is mostly found in the cytoplasm, and very little of it is found in the nucleus [29]. Compared to its linear counterparts, circRNA has been shown in certain studies to have longer half-lives [30]. Due to their high expression levels, structural stability, and diverse biological functions, circRNAs are increasingly attracting the attention of researchers. With the rapid advancements in sequencing and bioinformatics technologies, preliminary studies have revealed that circRNAs play crucial roles in carcinogenesis, metastasis, or apoptosis in BC, and potential new therapeutic targets for the treatment of this disease may be offered by them. However, the specific functions of most circRNAs and their molecular mechanisms in BC remain largely unexplored. Therefore, identifying novel circRNAs with biological functions would contribute to elucidating the mechanisms underlying the development of BC, providing a solid theoretical basis for the diagnosis and treatment of this disease.
The circRNA dataset (GSE182471) was used in this study to successfully screen for the hsa_circ_0001314, whose expression was upregulated in BC. hsa_circ_0001314 was validated to be highly expressed in BC tissues and cells by RT-qPCR, which is consistent with the predicted results from bioinformatics. Therefore, it is speculated that hsa_circ_0001314 may be involved in the onset and progression of BC, and its cell function and related mechanisms will be analyzed and verified. According to research, hsa_circ_0001314 expression knockdown can dramatically reduce BC cell proliferation, migration, and invasion while increasing apoptosis.
In addition, some recent studies have suggested that circRNA comprises characters with several binding sites of miRNA. Through the ceRNA mechanism, which binds to particular miRNAs to control gene expression, it can function as a sponge, such as circBACH2 (hsa_circ_0001625), which functions as an hsa-miR-944 sponge to enhance HNRNPC expression and stimulate BC cell proliferation [31]. The circ_0008039 regulates SKA3 by sponging miR-140-3p [32]. CircCD44 stimulates the growth of TNBC by focusing on the miR-502-5p-KRAS axis and C-myc [33]. Therefore, it is considered that hsa_circ_0001314 may also promote the malignant progression of BC through this mechanism. Subsequently, the regulatory mechanism of hsa_circ_0001314 in BC is investigated. Initially, bioinformatics analysis predicted that hsa_circ_0001314 targets and binds to hsa-miR-548aj-3p. It was confirmed by RT-qPCR that hsa-miR-548aj-3p is lowly expressed in BC tissues, which contrasts with the high expression of hsa_circ_0001314 in the BC tissues. Pearson's analysis revealed a significant negative correlation between the expression levels of hsa_circ_0001314 and hsa-miR-548aj-3p in BC tissues. Therefore, it can be tentatively concluded that hsa_circ_0001314 targets and binds to hsa-miR-548aj-3p. hsa_circ_0001314 was knocked down and hsa-miR-548aj-3p was overexpressed in MCF-7 cells, respectively, and the expression levels of hsa-miR-548aj-3p and hsa_circ_0001314 were detected by RT-qPCR. The results showed that knocking down hsa_circ_0001314 increased the expression of hsa-miR-548aj-3p while overexpressing hsa-miR-548aj-3p decreased the expression of hsa_circ_0001314. In summary, it is established that hsa_circ_0001314 functions as a sponge for hsa-miR-548aj-3p, enabling targeted binding to it.
Next, bioinformatics methods were employed to predict the potential target mRNAs of hsa-miR-548aj-3p, and functional enrichment analysis was further conducted on these predicted mRNAs. Analysis indicated that the majority of genes were enriched in the MAPK signaling pathway, which is one of the core pathways in mammalian cells and is closely associated with crucial physiological processes such as cell growth, differentiation, apoptosis, and angiogenesis. Therefore, genes enriched in the MAPK signaling pathway were selected as the main focus of subsequent research. Fifteen DEmRNAs enriched in the MAPK signaling pathway were selected for constructing both the GGI network and the PPI network. KRAS, RASGRP1, MAPK8, MAP3K1, and SOS2, which ranked in the Top 5 based on betweenness centrality, were identified as hub genes. These hub genes were then chosen to construct the ceRNA regulatory network involving hsa_circ_0001314-hsa-miR-548aj-3p-mRNAs. Bioinformatics analysis revealed that the KRAS, MAP3K1, and MAPK8 genes were significantly upregulated in BC and correlated with the prognosis and survival of BC patients, prompting their selection for further study. The expression of hsa_circ_0001314 was knocked down in MCF-7 cells, and the expression levels of KRAS, MAP3K1, and MAPK8 genes were detected by RT-qPCR. The results showed that, compared to their expression in MCF-7 cells, the expression of MAPK8 and MAP3K1 genes was significantly reduced after knocking down hsa_circ_0001314, while there was no significant change in KRAS gene expression. This suggests that hsa_circ_0001314 can regulate the expression of MAPK8 and MAP3K1 genes.
For the first time, hsa_circ_0001314 was discovered to be considerably overexpressed in BC tissues and cell lines in our investigation. Then, in vitro experiments confirmed that knocking down the expression level of hsa_circ_0001314 could significantly inhibit the proliferation, migration, and invasion of BC cells and promote the biological effect of apoptosis of BC cells. The molecular mechanism that hsa_circ_0001314 can act as a sponge of hsa-miR-548aj-3p and combine with hsa-miR-548aj-3p to activate MAPK signaling pathway, regulate the expression of MAPK8 and MAP3K1 genes, and exert biological effects. However, our study only preliminarily validated the interaction between hsa_circ_0001314 and hsa-miR-548aj-3p, which has some limitations. In the future, we will verify the interaction between hsa_circ_0001314 and hsa-miR-548aj-3p through a luciferase reporter assay to support the mechanism of ceRNA. Our main goal in the next phase of research is to verify the biological function of hsa_circ_0001314 through in vivo studies and further explore the role of hsa_circ_0001314 as a therapeutic target, hoping to improve the molecular mechanism of hsa_circ_0001314 in the occurrence and progression of BC.
5. Conclusions
In conclusion, hsa_circ_0001314 is highly expressed in BC tissues and cells. Significant inhibition of cell proliferation, migration, and invasion, as well as promotion of apoptosis, was achieved through knockdown of the hsa_circ_0001314 expression level, promoting apoptosis and constructing the ceRNA network for the hsa_circ_0001314-hsa-miR-548aj-3p-mRNAs. The molecular mechanism by which hsa_circ_0001314 can act as a sponge of hsa-miR-548aj-3p, bind to hsa-miR-548aj-3p, activate the MAPK signaling pathway, regulate the expression of MAPK8 and MAP3K1 genes, and play a biological role was preliminarily demonstrated. The research offers a new possible target for the identification and treatment of BC by elucidating the mechanism of action of hsa_circ_0001314 in BC for the first time.
Nomenclature
- BC
breast cancer
- BPs
biological processes
- CC
cellular component
- ceRNA
competitive endogenous RNA
- circRNA
circular RNA
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DEcircRNAs
differentially expressed circRNAs
- DEmiRNAs
differentially expressed miRNAs
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GEO
Gene Expression Omnibus
- GGI
gene–gene interaction
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MF
molecular function
- miRNA
microRNA
- mRNA
messenger RNA
- PPI
protein–protein interaction
- RT-qPCR
real-time quantitative PCR
- STRING
Search Tool for the Retrieval of Interacting Genes
- UALCAN
University of Alabama at Birmingham Cancer data analysis portal
Data Availability Statement
Our study used a public online database. The data can be accessed by the following websites: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182471, and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143564.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Yuge Ma: methodology, software, writing–original draft. Bing Zhang: visualization, formal analysis, data curation. Liping Wang: writing–review and editing, formal analysis. Wei Rong: supervision, resources. Ting Liu: conceptualization, project administration, funding acquisition.
Funding
Ting Liu (grant number QY2016B-24), Yuge Ma (grant number QYYCX2022-10), and Liping Wang (grant number 2020-KYYWF-0005) received fund.
References
- 1.Sung H., Ferlay J., Siegel R. L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer Journal for Clinicians . 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg O., Bray F., Coleman M. P., et al. The global burden of women’s cancers: a grand challenge in global health. The Lancet . 2017;389(10071):847–860. doi: 10.1016/S0140-6736(16)31392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memczak S., Jens M., Elefsinioti A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature . 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 4.Conn S. J., Pillman K. A., Toubia J., et al. The RNA binding protein quaking regulates formation of circRNAs. Cell . 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America . 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J. U., Agarwal V., Guo H., Bartel D. P. Expanded identification and characterization of mammalian circular RNAs. Genome Biology . 2014;15(7):p. 409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie F., Li Y., Wang M., et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Molecular Cancer . 2018;17(1):p. 144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., Liu Y., Bian Z., et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 (Kip1) axis. Molecular Cancer . 2018;17(1):p. 151. doi: 10.1186/s12943-018-0902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One . 2012;7(2, article e30733) doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kos A., Dijkema R., Arnberg A. C., van der Meide P. H., Schellekens H. The hepatitis delta (δ) virus possesses a circular RNA. Nature . 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 11.Capel B., Swain A., Nicolis S., et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell . 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 12.Li P., Chen S., Chen H., et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica Chimica Acta . 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 13.Liang H.-F., Zhang X.-Z., Liu B.-G., Jia G.-T., Li W.-L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. American Journal of Cancer Research . 2017;7(7):1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 14.Qin M., Liu G., Huo X., et al. hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers . 2016;16(1):161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 15.Xie H., Ren X., Xin S., et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget . 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P., Wang Z., Ou X., et al. The FUS/circEZH2/KLF5/feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Molecular Cancer . 2022;21(1):p. 198. doi: 10.1186/s12943-022-01653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sang Y., Chen B., Song X., et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Molecular Therapy . 2021;29(12):3525–3527. doi: 10.1016/j.ymthe.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T., Wilhite S. E., Ledoux P., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research . 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M., Wang Q., Shen J., Yang B. B., Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biology . 2019;16(7):899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Research . 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeary S. E., Lin K. S., Shi C. Y., et al. The biochemical basis of microRNA targeting efficacy. Science . 2019;366(6472) doi: 10.1126/science.aav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman B. T., Hao M., Qiu J., et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Research . 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warde-Farley D., Donaldson S. L., Comes O., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research . 2010;38(Supplement_2):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research . 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research . 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA . 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong S., Feng J. CircPrimer 2.0: a software for annotating circRNAs and predicting translation potential of circRNAs. BMC Bioinformatics . 2022;23(1):p. 215. doi: 10.1186/s12859-022-04705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greene J., Baird A. M., Brady L., et al. Circular RNAs: biogenesis, function and role in human diseases. Frontiers in Molecular Biosciences . 2017;4:p. 38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z.-J., Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biology . 2017;14(5):514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enuka Y., Lauriola M., Feldman M. E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Research . 2016;44(3):1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv W., Tan Y., Xiong M., et al. Analysis and validation of m6A regulatory network: a novel circBACH2/has-miR-944/HNRNPC axis in breast cancer progression. Journal of Translational Medicine . 2021;19(1):p. 527. doi: 10.1186/s12967-021-03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dou D., Ren X., Han M., et al. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Molecular Oncology . 2021;15(2):697–709. doi: 10.1002/1878-0261.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Gao X., Zhang Z., et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Molecular Cancer . 2021;20(1):p. 138. doi: 10.1186/s12943-021-01444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Our study used a public online database. The data can be accessed by the following websites: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182471, and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143564.


