Abstract
Background
Sorafenib, an orally active potent tyrosine kinase inhibitor (TKI), represented a primary treatment in patients with advanced hepatocellular carcinoma (HCC). Unfortunately, sorafenib resistance was regarded as a huge obstacle for HCC treatment.
Methods
RNA-sequencing including circRNA Sequencing (circRNA-Seq) for circular RNAs (circRNAs), miRNA Sequencing (miRNA-Seq) for microRNAs (miRNAs), as well as mRNA Sequencing (mRNA-Seq) for mRNAs in sorafenib-resistant HCC cells vs sorafenib-sensitive HCC cells, were performed. Then, interaction correlation analysis between differentially expressed circRNAs and miRNAs and their target genes in Huh7/SOR and SMMC7721/SOR cells was exhibited. The “circRNA-miRNA-mRNA” network was constructed through the Cytoscape software application, Circular RNA Interactome and Targetscan prediction, RNA binding protein immunoprecipitation (RIP), RNA pull-down, and Dual luciferase reporter assay. Furthermore, mRNA-Seq, Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for the downstream genes involved in the “circRNA-miRNA-mRNA” network was implemented. Iron detection assay, Lipid peroxidation quantification assay, ROS measurement assay, CCK-8 assay, and tumor challenge in vivo were used to determine the mechanisms promoting sorafenib resistance in HCC, where the “circRNA-miRNA-mRNA” network is clearly involved in.
Results:
circ_0001944 and circ_0078607 with upregulation and 2 downregulated expressed circRNAs (circ_0002874 and circ_0069981), as well as 11 upregulated miRNAs including miR-193a-5p, miR-197-3p, miR-27a-5p, miR-551b-5p, miR-335-3p, miR-767-3p, miR-767-5p, miR-92a-1-5p, miR-92a-3p, miR-3940-3p, and miR-664b-3p and 3 downregulated expressed miRNAs (miR-1292-5p, let-7c-5p, and miR-99a-5p) in sorafenib-resistant HCC cells were determined. Among these non-coding RNAs (ncRNAs), circ_0001944 and miR-1292-5p should not be drop out of sight; circ_0001944 has been proved to target miR-1292-5p to inhibit its expression in HCC. Subsequent findings also raise that miR-1292-5p directly targeted the 3’-noncoding region (3’-UTR) of Fibulin 2 (FBLN2) mRNA. Furthermore, circ_0001944 targets the miR-1292-5p/FBLN2 axis to inhibit cell ferroptosis in which the indicated regulators associated with iron overload and lipid peroxidation were “rearranged”. Most importantly, circ_0001944 advanced sorafenib resistance in HCC through mitigating ferroptosis, where the miR-1292-5p/FBLN2 axis cannot be left unrecognized.
Conclusion:
Circ_0001944 is a putative target for reversing sorafenib resistance in HCC. Our findings are expected to provide new targets and new directions for sorafenib sensitization in the treatment of HCC.
Keywords: Circular RNAs, Sorafenib, hepatocellular carcinoma, MicoRNAs
Introduction
Hepatocellular carcinoma (HCC) counts as the fourth leading cause of death worldwide.1 Most HCC patients are often diagnosed in the late stage with hidden clinical symptoms.2 Sorafenib, a tyrosine kinase inhibitor (TKI), has represented a first-line drug used to treat advanced HCC.3 However, many patients suffering from advanced HCC remain unsatisfied with their clinical result towards sorafenib. In particular, only 35–43% of HCC patients can respond to sorafenib, and a considerable proportion of HCC patients among them relapse within 6 months.4 Furthermore, only a 2.8-month survival advantage can be obtained after sorafenib usage in sorafenib-resistant patients.5,6 Therefore, drug resistance has taken a huge obstacle for sorafenib-based treatment for HCC.7 Thus, it’s urgent to reveal the mechanism advancing sorafenib resistance and find ways to enhance sorafenib sensitivity in HCC patients.
Ferroptosis represents a newly regulated cell death (RCD). It is charactered by the accumulated iron, which increases the production of reactive oxygen species (ROS) and lipid peroxidation.8 Ferritin heavy chain 1 (FTH1) reduction but the increase in Divalent metal-ion transporter-1 (DMT1) and Transferrin Receptor 1 (TFR1) can be regarded as armed forces to indcude Fe2+overload and mediate Fenton reaction, thus enhancing the production of lipid peroxides and ROS.9,10 Otherwise, Quinone oxidoreductase 1 (NQO1) is known as a scavenger with the ability of eliminating scavenging lipid peroxides and ROS, which is unfavourable for ferroptosis.11 New evidence has indicated that the obstruction of ferroptosis is identified to be closely associated with cisplatin resistance in gastric cancer,12 5-fluorouracil resistance in colorectal cancer,13 and gemcitabine resistance in pancreatic cancer.14
Recently, sorafenib has been recognized to promote cell death by exhibiting ferroptosis, which is caused by Fe2+overload and characterized by increased lipid peroxide generation as well as ROS accumulation.15 The obstruction of ferroptosis is also identified to be related to sorafenib resistance in HCC.16 Therefore, elucidating the underlining mechanism that inhibits ferroptosis and encourage sorafenib resistance may be a feasible strategy to confer sorafenib sensitivity in the treatment for HCC.
Non-coding RNAs (ncRNAs), especially circular RNAs (circRNAs) and microRNAs (miRNAs), started to step onto the historical stage, which have been identified to transcriptionally and posttranscriptionally regulate the progress of tumorigenesis.17 CircRNAs are charactered by a special circular structure where the 5’ and 3’ ends are covalently combined. It is generally acknowledged that circRNAs are originated from pre-mRNA through variable splicing processing. Thus, the closed structure can protect circRNAs from RNA enzyme-mediated degradation.18 Another well-known small-sized endogenous ncRNA, named miRNA, also plays a vital role in regulating the expression of target genes in tumor cells.19,20
According to the competitive endogenous RNA (CeRNA) theory, circRNA targets miRNA to regulate its transcription and expression. In particular, circRNA serves as miRNA “sponges” and targets the common miRNA response elements (MREs) to form an RNA-induced silencing complex (RISC) and inhibit miRNA expression.21 CirRNAs compete with miRNA for the same MREs and reduce miRNA inhibition, therefore increasing the mRNA expression.22 The involvement of circRNAs in cancers has been well documented. For example, Circ-E-Cad acted as oncogenic functions in glioblastoma by templating for translation of the unique E-cadherin-derived peptide (C-E-Cad).23 CircPVT1 acts as the miRNA sponge for miR-497-5p to induce cell cycle progression, thus playing an oncogenic role in neck squamous cell carcinoma (HNSCC).24 Alternatively, the anti-tumor role of Circ-Foxo3 has been identified in breast cancer where it acts as a protein scaffold to sponge for E3 ubiquitin-protein ligase MDM2 and p53 and promote cell apoptosis.25 CircNDUFB2 also functions as protein scaffold for E3 ubiquitin/ISG15 ligase TRIM25 and IGF2BPs to trigger antitumor activity in non-small cell lung cancer (NSCLC).26 More interesting, ciRS-7 located in the cytoplasm and acted as a miRNA sponge for miR-7 in multiple cancers but played oncogenic or tumour.
Suppressor function depending on the context.27,28 Recently, the understanding as to a ceRNA-based regulatory mechanism in metabolic reprogramming,29 immune microenvironment remodeling,30 tumor progression,31 and metastasis32 in HCC has been gradually improved. However, there is no clear consensus on the detailed mechanism by which circRNAs promote sorafenib resistance by monitoring ferroptosis in HCC.
Here, 70 upregulated but 45 downregulated circRNAs in SMMC7721/SOR cells; 20 upregulated but 13 downregulated circRNAs in Huh7/SOR cells compared to their respective parental cells, were identified using circRNA Sequencing (circRNA-Seq). Alternatively, 91 upregulated and 90 downregulated miRNAs in SMMC7721/SOR cells; 50 upregulated and 38 downregulated clusters in Huh7/SOR cells have been determined using miRNA Sequencing (miRNA-Seq). In particular, circ_0001944 and circ_0078607 with upregulated expression and 2 downregulated expressed circRNAs (circ_0002874 and circ_0069981) in sorafenib-resistant HCC cells vs sorafenib-sensitive HCC cells were determined. Eleven upregulated miRNAs (miR-193a-5p, miR-197-3p, miR-27a-5p, miR-551b-5p, miR-335-3p, miR-767-3p, miR-767-5p, miR-92a-1-5p, miR-92a-3p, miR-3940-3p, and miR-664b-3p) and three downregulated miRNAs (miR-1292-5p, let-7c-5p, and miR-99a-5p) were also identified.
Among these differentially expressed ncRNAs (DENs), circ_0001944 has been identified to target miR-1292-5p directly using Circular RNA Interactome prediction, RNA binding protein immunoprecipitation (RIP) and Dual luciferase reporter assay. CircRNA and miRNA regulated mRNA expression via the “co-expression” mechanism based on ceRNA theory. The differentially expressed mRNAs obtained from mRNA Sequencing (mRNA-Seq) and the target mRNAs of circ_0001944 and miR-1292-5p, were then prepared for intersection analysis using the Cytoscape software to determine the target mRNAs regulated by circ_0001944 and miR-1292-5p. Accordingly, the network “circ_0001944-miR-1292-5p-mRNA” firstly stood head and shoulders, which was constructed based on the dependence of target prediction results on the strength of co-expression correlation between miR-1292-5p and mRNAs. FBLN2, HRH3, GSTP1, GNB1L, TMEM45B, STK25, SLC24A1, ADAMTS10, PRKCSH, MYADM, ARPC1B, NSMF, PMPCA which tightly surround the “circ_0001944-miR-1292-5p-mRNA” network, attracted a lot of attentions. Most interestingly, miR-1292-5p was identified to target 3’-noncoding region (3’-UTR) of Fibulin 2 (FBLN2) directly via Targetscan prediction, RNA pull-down assay, and Dual luciferase reporter assay. Furthermore, mRNA-Seq, Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis indicated that FBLN2 downregulates TFR1 and DMT1 but upregulates FTH1 and NQO1 expression to inhibit ferroptosis. Circ_0001944 targets the miR-1292-5p/FBLN2 axis and leads to the charge in terms of sorafenib resistance in HCC by suppressing ferroptosis. Thus, circ_0001944 should not be in the shadows, but can be valued for its role of impeding ferroptosis in HCC and promoting sorafenib resistance through targeting the miR-1292-5p/FBLN2 axis.
Materials and Methods
Cells, Chemicals, and Plasmids
HCC cell lines SMMC7721 and Huh7 were acquired from the National Collection of Authenticated cell Cultures (Shanghai, China). Sorafenib-resistant HCC cells were constructed using the continuity induction method in vitro. Sorafenib (#284461-73-0) and Ferrostatin-1 (#347174-05-4) were purchased from Sigma-Aldrich. Indicated plasmids including Len-circ_0001944-shRNA, pcDNA-hsa_circ_0001944, miR-1292-5p mimics, FBLN2-siRNA, miR-1292-5p inhibitor, and GV657-FBLN2 were obtained from GenePharma Co., Ltd (Shanghai, China). The Lipofectamine™3000 transfection reagent was used to transfect these vectors into HCC cells.
RNA Quantification
Firstly, the contamination and degradation of RNA samples were determined using 1% agarose gel electrophoresis. The NanoPhotometer ® spectrophotometer and Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer were then used to confirm RNA purity and concentration. Finally, the RNA Nano 6000 assay kit of the 2100 biological analyzer system was used to assess RNA integrity and quantity.
Library Preparation for RNA Sequencing
Ribosomal RNA (rRNA) removal was first carried out using the Epicentre Ribo Zero rRNA Removal Kit (Epicentre, USA), The NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA) was then used to construct the sequencing libraries, where a total amount of 3 µg RNA was prepared. Briefly, the first strand cDNA was synthesized via reverse-transcription with the help of fragmented RNA and dNTPs. Second strand cDNA was then synthesised using DNA polymerase I, RNase H, and dNTPs, where exonuclease/polymerase was used to complete the conversion of remaining overhangs in double-strand cDNA into blunt ends. Ligation of sequencing adaptors to the cDNA was performed after purification and adenylation of 3’ends of DNA fragments. cDNA fragments of preferentially 150–200 bp or 370–420 bp in length were obtained, followed by Uridine digestion using Uracil-N-Glycosylase and PCR amplification. Then, PCR for adaptor-ligated and size-selected cDNA was performed with USER Enzyme (NEB, USA), Universal PCR primers and Index (X) Primer, and Phusion High-Fidelity DNA polymerase. Finally, library concentration was checked and adjusted to 1ng/uL using the Qubit fluorometer. The concentration and insert size of the library were also examined using Agilent 2100 Bioanalyzer and qPCR, respectively.
Clustering and Sequencing
Cluster analysis of index encoded samples was prepared for Illumina sequencing using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) on a cBot Cluster Generation System. The sequencing for library preparations was then performed on an Illumina HiSeq platform (PE150) after cluster generation. 125 bp/150 bp paired-end reads can be generated.
Quality Control for Raw Data
Next, raw data with FASTQ format were processed through an internal perl script. Clean reads were achieved by trimming reads containing adapter, and ploy-N. Only the clean data with high quality can be prepared for downstream analyses after Q30, Q20, and GC content were calculated.
Mapping to the Reference Genome
STAR (v2.5.1b)33 and HISAT2 (V2.0.5)34 software were selected to build the index of the reference genome that was downloaded from the genome website. Then, clean reads were mapped to the annotation files based on a gene model. Reads alignment results were transferred using STAR (v2.5.1b) and HISAT2 (V2.0.5). Alternatively, the small RNA tags were also aligned to the reference genome to get the reads count of miRNA using the Bowtie-0.12.9 software.35
circRNA, miRNA and mRNA Identification
The circRNA was then identified using the find_ circ36 and CIRI2 software.37 Alternatively, the available software miRBase20.0, mirdeep2,38 and srna-tools-cli were integrated to identify known miRNA. Furthermore, a custom script was constructed to obtain miRNA counts and base deviations of identified miRNAs with a specific length. Additionally, the read alignment to the reference genome for transcript assembly of mRNAs was calculated using the StringTie program (StringTie1.3.3b), where the cuffmerge software was used for transcription mergence.
Quantification and Differential Expression Analysis
CircRNA and miRNA expressions were determined through the following criteria based on TPM (transcript per million): Normalized expression = mapped readcount/Total reads*1000000.39 Alternatively, mRNA quantification was exhibited in line with the StringTie software based on Reads Per Kilobase of transcript per Million mapped reads (RPKM).
Differential expression analysis was carried out via the DESeq2 R package (1.20.0). The threshold for significantly differential expression was set as absolute fold change of 1 or 2. The corrected P-value of 0.05 (Padj < 0.05) was adjusted using the Benjamini & Hochberg method. The overlap of DENs in both Huh7/SOR and SMMC7721/SOR cells was confirmed.
Identification for the Target Genes of DENs
Actually, the target mRNAs of differentially expressed circRNAs were identified as the source genes of circRNAs located in the reference genome map.40 Alternatively, the target mRNAs of differentially expressed miRNAs were then predicted via the RNAhybrid and miRanda software.41,42 Accordingly, the overlaps between the differentially expressed mRNAs obtained using the mRNA-Seq and predicted genes were regarded as the target genes of these DENs.
The Establishment of the “circRNA-miRNA-mRNA” Network
Downregulated circRNAs but upregulated miRNAs; upregulated circRNAs but downregulated miRNAs among these overlapping circRNAs and miRNAs were selected. The target genes of overlapped circRNAs and miRNAs in these two sorafenib-resistant HCC cell lines were then determined. Finally, the “circRNA-miRNA-mRNA” network was established using the Cytoscape software based on ceRNA theory, where the overlapped DENs and their target mRNAs in these two sorafenib-resistant HCC cells were included. The position of ncRNAs in the network was determined based on the correlation between the expression of miRNA and mRNA. The screening criteria require a Pearson correlation coefficient greater than 0.95.
GO Function and KEGG Enrichment Analysis
The GO can be regarded as a major bioinformatics initiative that aimed to unify the representation of gene attributes across all species that covered biological process (BP), cellular component (CC), and molecular function (MF). The KEGG PATHWAY database can be seen as a collection of pathway maps that handle biological pathways and integrate many entities such as genes, diseases, chemical and drug substances. They are stored as separate entries in other KEGG databases (http://www.genome.jp/kegg/).43 GO function and KEGG enrichment analysis of target genes were performed by the clusterProfiler R package. The enrichment was considered to be significant when the corrected p-values were less than 0.05.
The most 10 significant GO terms, as well as most 20 KEGG enrichment pathways are included in a bar chart. The red box was defined in the highest bar column to highlight the GO terms and KEGG pathways clearly related to these genes.
More detailed descriptions of RNA sequencing, network construction, and functional validation steps, particularly in data analysis and interpretation, have been provided in Supplementary Materials and methods.
Tumor Challenge and Treatment in vivo
Huh7/SOR cells were being injected into the left armpit of female nude mice (5–6 weeks old). HCC-bearing mice were grouped (four or five nude mice per group) to receive different therapies: (1) the ctrl group with PBS treatment; (2) the sorafenib mono-treatment group that was given sorafenib (30mg/kg) by intraperitoneal injection every three days for a total of three weeks; (3) the combination group that was treated with sorafenib (30mg/kg), Ferrostatin-1, and Len-circ_0001944-shRNA plasmid. The mice were sacrificed, of which tumor weight and volume were summarized. The Qingdao University Animal Care and Use Committee approved the experimental studies in vivo. This study was conducted in line with the International Council for Laboratory Animal Science (ICLAS) and NC3Rs ARRIVE guidelines.
Real-Time PCR Assay
Total RNA was prepared using a TRizol regent (Invitrogen). Indicated RNA was amplified using SYBR Green qPCR Master Mix (Bio-Rad). Relative expression of miR-1292-5p, circ_0001944, and FBLN2 was ascertained in comparison with that of U6 or GAPDH.
Western Blot Assay
Protein samples from the indicated cells were collected using RIPA Lysis Buffer (Solarbio) and were separated by 12% SDS-PAGE agarose gel electrophoresis. These protein samples were transferred to the nitrocellulose membranes and sealed in 5% (w/v) non-fat dry milk with Tris-buffered saline. These membranes were incubated with primary antibodies and Goat Anti-Rabbit IgG (Abcam) at room temperature for 1 hour. Finally, immunoreactive proteins were visualized using the Molecular Imager ChemiDocTM XRS+ System (Bio-Rad).
Dual Luciferase Reporter Assay
The binding sequences of circ_0001944 in miR-1292-5p and the mutant sequences were cloned into the psiCHECK-1 vector (Promega) to establish the dual luciferase reporter vectors. miR-NC and miR-1292-5p mimics were then transfected with the dual luciferase reporter vectors. Alternatively, the 3′UTR of FBLN2 mRNA was PCR-amplified and inserted into the downstream region of SV40 promoter-driven Renilla luciferase cassette in the psiCHECK-1 plasmid. The mutant (Δ) 3′UTR of FBLN2 containing a mutated sequence of the miR-1292-5p seed region was also constructed from the wild-type FBLN2 3′UTR plasmid. Next, Huh7/SOR and SMMC7721/SOR cells were transfected with miR-1292-5p mimics and psiCHECK-1 vector. The luciferase activity in these cells was determined using the Dual Luciferase Assay System (Promega).
RIP Assay
Indicated cells were transfected with pcDNA-hsa_circ_0001944 and miR-1292-5p mimics. AGO2-RIP expression was detected using Real-time PCR following the manufacturer’s specifications provided by the EZ-Magna RIP kit (Millipore, USA).
RNA Pull-Down Assay
Indicated cells were treated with GV657-FBLN2 vector, followed by the transfection with biotinylated miR-1292-5p mimics or miR-NC using Lipofectamine 2000. Next, these cells were collected and sonicated, followed by the incubation with M-280 streptavidin magnetic beads (Invitrogen). The binding RNAs were checked using real-time PCR.
Iron Detection Assay
Intracellular levels of Fe2+ were evaluated according to the instructions provided by the Iron Assay Kit (#ab83366, Abcam).
Lipid Peroxidation Quantification Assay
The Lipid Peroxidation Assay Kit (#ab118970, Abcam) was used to detect the Malondialdehyde (MDA) concentration.
ROS Measurement Assay
Indicated cells were collected and incubated with DCFH-DA probe for 20 min at 37°C. The fluorescence intensity of DCF was detected at 530 nm for emission and 485 nm for excitation.
Statistical Analysis
SPSS 22.0 software was used to perform statistical analysis. Group comparisons were performed using the paired Student t test. P* < 0.05 was considered to be statistically significant.
CCK-8 Assay
HCC cells were treated with plasmids for 24 h, and stimulated with sorafenib (0, 5, 10, 15 20, and 25μM) for another 48 h. Drug-free medium and CCK-8 (10 µL) were added to each well. The growth inhibition (%) of the indicated cells was calculated.
Results
The Determination for DENs in Sorafenib-Resistant HCC Cells
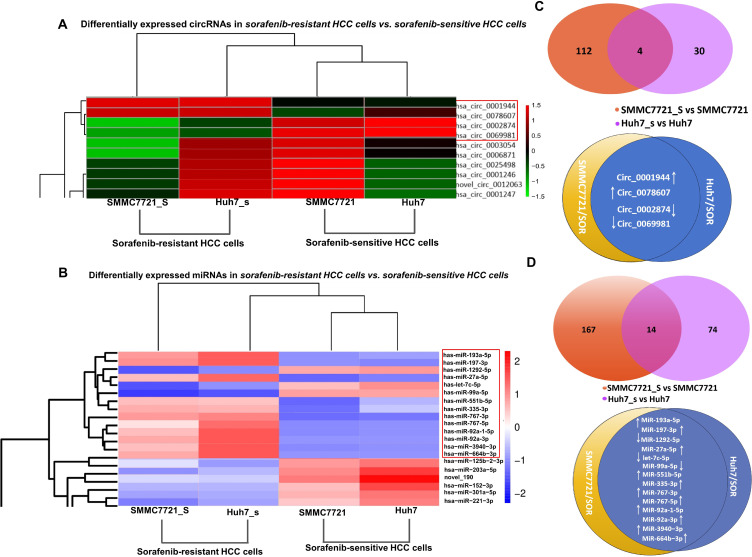
Surely, RNA sequencing in sorafenib-sensitive HCC cells and sorafenib-resistant HCC cells may after all be accepted to sense how ncRNAs promote sorafenib resistance. Herein,70 and 20 upregulated circRNAs; but 45 and 13 downregulated circRNAs were identified in SMMC7721/SOR and Huh7/SOR cells, compared to SMMC7721 and Huh7 cells, respectively (Figure 1A and B). Ninety downregulated but 91 upregulated miRNAs in SMMC7721/SOR cells; 38 downregulated but 50 upregulated clusters in Huh7/SOR cells were determined (Figure 1C and D). These results indicate a direction for exploring the role of DENs involved in sorafenib resistance in HCC.
Figure 1.
The determination for DENs in sorafenib-resistant cells, compared to sorafenib-sensitive HCC cells. (A) The gene counts of differentially expressed circRNAs in SMMC7721/SOR vs SMMC7721 and in Huh7/SOR vs Huh7 cells. (B) Heat map depicting differentially expressed circRNAs was generated using R package. (C) The gene counts of differentially expressed miRNAs in SMMC7721/SOR vs SMMC7721 and in Huh7/SOR vs Huh7 cells. (D) Heat map depicting differentially expressed miRNAs was generated using R package. For the heat map in Figure 1, the horizontal axis represents the samples, and the vertical axis represents differential genes. The left side clusters genes based on their expression similarity, while the upper side clusters each sample based on their expression profile similarity. The expression level gradually increases from blue or green to red, and the number represents the relative expression level after homogenization.Fold Change≥1.0 and Padj<0.05. For Figure 1, the transcriptome sequencing was performed with three samples per group.
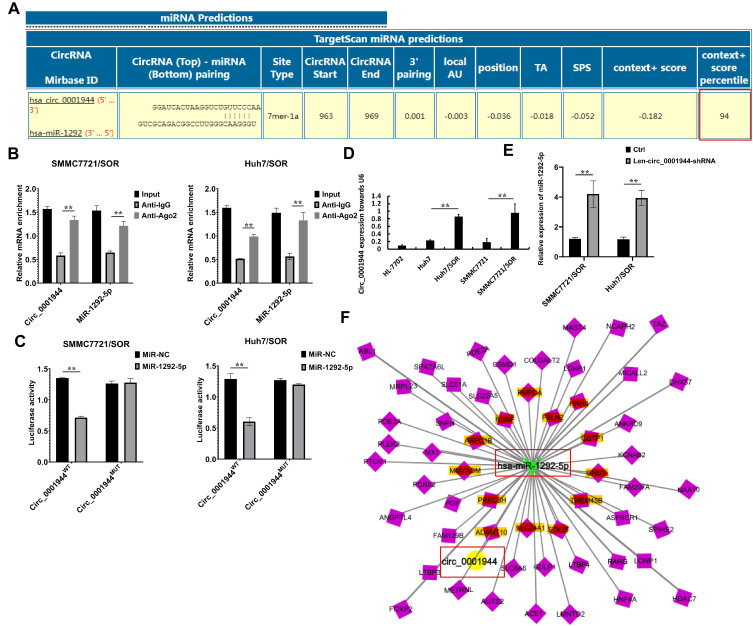
Screening for DENs Putatively Involved in Sorafenib Resistance in HCC
More concentrated DENs in both SMMC7721/SOR and Huh7/SOR cells can be focused because these DENs are endowed with greater “energy” to advance sorafenib resistance in HCC. Thus, overlapped DENs from SMMC7721/SOR and Huh7/SOR have attracted more attention. Herein, the top 10 circRNAs (Figure 2A) and the top 20 miRNAs (Figure 2B) were selected based on the average value of fold change in terms of their expressions in SMMC7721/SOR and Huh7/SOR cells compared to their parental cells. Cluster analysis was then performed to determine these DENs with similar expressed patterns in sorafenib-resistant HCC cells vs sorafenib-sensitive HCC cells. As demonstrated in Figure 2A and C, circ_0001944 and circ_0078607 with upregulated expression and 2 downregulated expressed circRNAs (circ_0002874 and circ_0069981) with the most significant change and similar expressed patterns in sorafenib-resistant HCC cells fellow with interest (as shown in red box in Figure 2A and C). Similarly, 11 upregulated miRNAs (miR-193a-5p, miR-197-3p, miR-27a-5p, miR-551b-5p, miR-335-3p, miR-767-3p, miR-767-5p, miR-92a-1-5p, miR-92a-3p, miR-3940-3p, and miR-664b-3p) and 3 downregulated miRNAs (miR-1292-5p, let-7c-5p, and miR-99a-5p) in sorafenib-resistant HCC cells were also confirmed (as shown in red box in Figure 2B and D).
Figure 2.
The identification for DENs which partially play a very important role in the development of sorafenib resistance. (A) The Heat map depicting 10 circRNAs in terms of the fold change of their expressions in sorafenib-resistant HCC cells vs sorafenib-sensitive HCC cells was obtained using the cluster analysis. (B) The Heat map depicting top 20 miRNAs in terms of the fold change of their expressions in sorafenib-resistant HCC cells vs sorafenib-sensitive HCC cells was obtained using the cluster analysis. (C) Details for these differentially expressed circRNAs in both SMMC7721/SOR and Huh7/SOR cells, compared to SMMC7721 and Huh7 cells. (D) The detail information for the differentially expressed miRNAs in both SMMC7721/SOR and Huh7/SOR cells, compared to SMMC7721 and Huh7 cells For the heat map in Figure 2, the horizontal axis represents the samples, and the vertical axis represents differential genes. The left side clusters genes based on their expression similarity, while the upper side clusters each sample based on their expression profile similarity. The expression level gradually increases from blue or green to red, and the number represents the relative expression level after homogenization. Fold Change≥1.0 and Padj<0.05.
circ_0001944 Directly Targets miR-1292-5p to Inhibit Its Expression
A preliminary selection was made for circ_0002874, circ_0001944, circ_0078607, and circ_0069981 based on the ceRNA mechanism to identify miRNAs with negative correlation in the expression of these circRNAs. Circular RNA Interactome prediction was then performed, and the Context+ score percentile was calculated as 94 (Figure 3A) (“Context+ score percentile=94” as shown in red box in Figure 3A). The closer this value is to 100, the more accurate the prediction results are. Thus, the results shown as “Context+ score percentile=94” indicted that miR-1292-5p may be the target of circ_0001944. Furthermore, the RIP assay and Dual luciferase reporter assay confirmed that circ_0001944 directly targets miR-1292-5p to inhibit its expression (Figure 3B and C). Interestingly, miR-1292-5p expression was dramatically decreased in sorafenib-resistant HCC cells compared with that in sorafenib-sensitive HCC cells (Figure 3D). Len-circ_0001944-shRNA significantly increased miR-1292-5p expression in HCC cells (Figure 3E).
Figure 3.
MiR-1292-5p is putative target downstream of circ_0001944. (A) Circular RNA Interactome predicts that miR-1292-5p may be the target of circ_0001944. The red box was meant to highlight the Context+ score percentile. (B) SMMC7721/SOR and Huh7/SOR cells were transfected with pcDNA-hsa_circ_0001944 and miR-1292-5p mimics. The RNA expression of AGO2-RIP was detected using Real-time PCR.**P<0.01. (C) SMMC7721/SOR and Huh7/SOR cells were co-transfected with reporter construct and circ_0001944 and cel-miR-936. Renilla luciferase activity of reporter constructs (psiCHECK-1) was evaluated 48 h after transfection. **P<0.01. (D) Circ_0001944 expression is determined in SMMC7721/SOR and Huh7/SOR cells, compared to SMMC7721 and Huh7 cells respectively. **P<0.01. (E) SMMC7721/SOR and Huh7/SOR cells were transfected with Len-circ_0001944-shRNA for 24h, miR-1292-5p expression was detected using Real-time PCR assay. **P<0.01. (F) The depiction of interactive network diagram “circ_0001944-miR-1292-5p-mRNA” involved in the development of sorafenib resistance in HCC. The red box was provided to highlight the location of “circ_0001944” and “miR-1292-5p” in the“circ_0001944-miR-1292-5p-mRNA”network. For the Figure B-E, data are shown as means±SD of three independent experiments.
As reportedly as in ceRNA theory, circRNA or miRNA regulates mRNA expression through “co-expression” regulation. Therefore, the target gene union of “co-expression” effects were selected, and differentially expressed mRNAs were identified. When the target gene of differentially expressed circRNA or miRNA between overlaps with differentially expressed mRNA, it indicates that the differentially expressed mRNA is highly likely to be regulated by circRNA and miRNA. Accordingly, a bold idea popped up that an interactive network based on the DENs could exist. Here, we have tried to complete the intersection association analysis for the target mRNAs of overlapped circRNAs and miRNAs in Huh7/SOR and SMMC7721/SOR cells. The “circRNA-miRNA-mRNA” regulatory relationships were then established for which circRNA, miRNA, and mRNA were regarded as the decoy, the core, and the target, respectively. Interestingly, the “circ_0001944-miR-1292-5p-mRNA” network based on circ_0001944 and miR-1292-5p was constructed using Cytoscape software (Figure 3F).
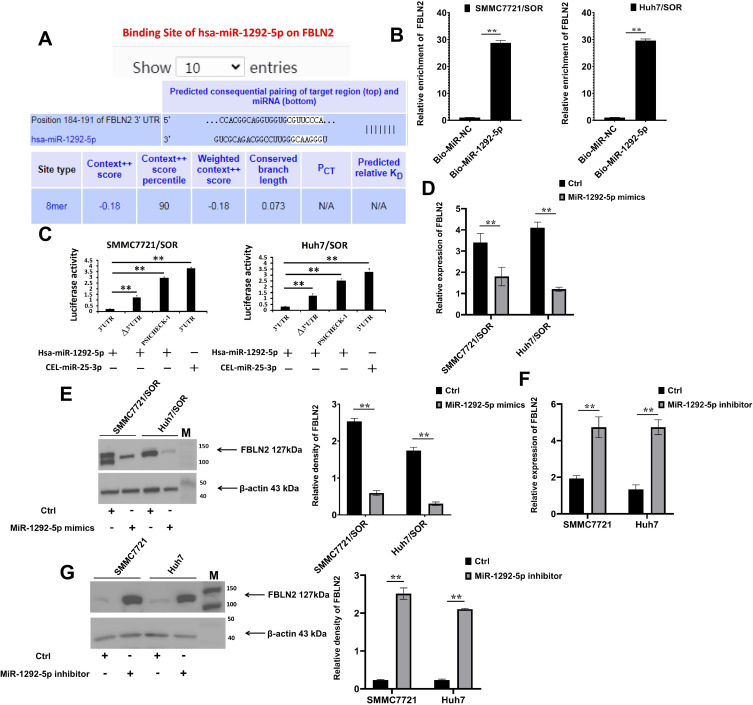
miR-1292-5p Directly Targets FBLN2 to Suppress Its Expression
Furthermore, FBLN2, HRH3, GSTP1, GNB1L, TMEM45B, STK25, SLC24A1, ADAMTS10, PRKCSH, MYADM, ARPC1B, NSMF, PMPCA (as marked that were located around the “circ_0001944-miR-1292-5p-mRNA” network were selected, to reveal the downstream molecules of miR-1292-5p. miR-1292-5p-binding sites were then confirmed via online prediction with the Targetscan chimeric algorithm (Figure 4A), followed by the RNA pull-down assay (Figure 4B). Co-transfection with miR-1292-5p mimics, but not the cel-miR-25-3p mimics, suppressed the luciferase activity of the wild-type FBLN2 3′-UTR construct (Figure 4C). Therefore, miR-1292-5p directly targeted a highly conserved sequence in the 3′-UTR of FBLN2 (Figure 4B and C) Moreover, the miR-1292-5p mimics significantly decreased the FBLN2 expression in SMMC7721/SOR and Huh7/SOR cells (Figure 4D and E); but miR-1292-5p inhibitor dramatically increased the FBLN2 expression in SMMC7721 cells and Huh7 cells (Figure 4F and G). So we can infer that miR-1292-5p is situated upstream of FBLN2 to bound to its 3′-UTR and downregulate its expression.
Figure 4.
MiR-1292-5p inhibits FBLN2 expression in HCC by directly targeting its 3′-UTR. (A) MiR-1292-5p and the 3′-UTR sequences of FBLN2 were complementary to each other according to the predictions using the TargetScan algorithm. (B) SMMC7721/SOR and Huh7/SOR cells were treated with GV657-FBLN2 vector, followed by the transfection with biotinylated miR-1292-5p mimics or miR-NC.The binding RNAs were checked using Real-time PCR. **P<0.01 versus miR-NC group. (C) SMMC7721/SOR and Huh7/SOR cells were co-transfected with reporter construct and miR-1292-5p mimics and cel-miR-239b. Renilla luciferase activity of reporter constructs (psiCHECK-1) containing the wild-type and mutated FBLN2 3′UTR was evaluated 48 h after transfection. **P<0.01. (D and E) FBLN2 expression in HCC cells with the transfection of miR-1292-5p mimics was detected using Real-time PCR assay (D) and Western Blot assay (E). **P<0.01 versus Ctrl group. (F) and (G) FBLN2 expression in HCC cells with the transfection of miR-1292-5p inhibitor was detected using Real-time PCR assay (F) and Western Blot assay (G). **P<0.01 versus Ctrl group. For the Figure B, C, D and F, data are shown as means±SD of three independent experiments. For the Figure E and G, Data are shown as representatives (left) or means±SD (right) from three independent experiments.
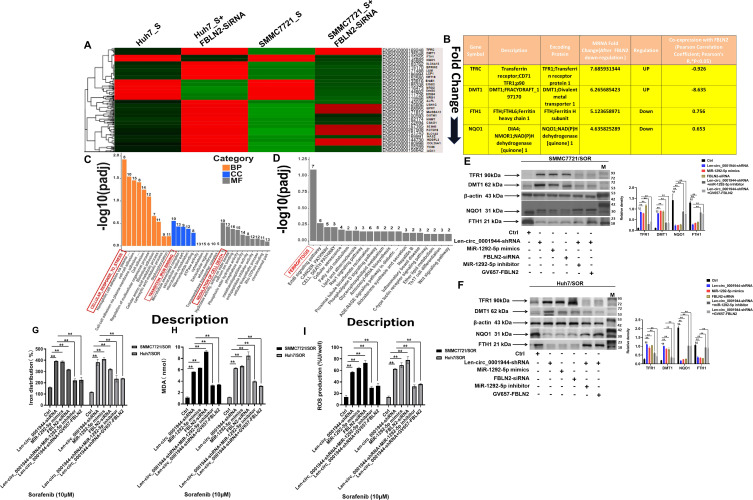
circ_0001944 Targets the miR-1292-5p/FBLN2 Axis to Inhibit Cell Ferroptosis
To confirm the role of circ_0001944 in promoting sorafenib resistance through targeting the miR-1292-5p/FBLN2 axis, the differentially expressed genes (DEGs) were then confirmed after the gene silence of FBLN2. FBLN2 expression in SMMC7721/SOR and Huh7/SOR cells after the gene silence of FBLN2 was also detected using Western Blot assay (Supplementary Figure 1). In particular, 2 upregulated DEGs including TFRC and DMT1, as well as 2 downregulated clusters including FTH1 and NQO1 demonstrated the most significant changes in mRNA levels with similar expressed patterns in both SMMC7721/SOR and Huh7/SOR cells after the gene silence of FBLN2 (Figure 5A). Moreover, these DEGs were identified to be “co-expressed” with FBLN2 using Pearson correlation analysis. They also encoded TFR1, DMT1, FTH1, and NQO1 proteins that were associated with iron homeostasis and lipid peroxidation (Figure 5B). Similarly, the TFR1 and DMT1 were expression upregulated, but the protein level of FTH1 and NQO1 was downregulated in SMMC7721/SOR and Huh7/SOR cells after the gene silence of FBLN2 (Supplementary Figure 2). Consistently, GO and KEGG functional enrichment analysis also confirmed that FBLN2 is most probably connected with CELLULAR_RESPONSE_TO_STRESS, TRANSITION_IRON BINDING, REGULATION OF CELL DEATH, and FERROPTOSIS (red box as shown in Figure 5C and D). Moreover, circ_0001944 depletion, miR-1292-5p mimics, and FBLN2 silence distinctly decreased the expression of FTH1 and NQO1 but increase the protein level of TFR1 and DMT1. However, miR-1292-5p inhibitor and FBLN2 overexpression can partially offset the effects of circ_0001944 depletion on the expression of the above-mentioned ferroptosis-related markers in HCC cells (Figure 5E and F). Most importantly, circ_0001944 silence, miR-1292-5p mimics, and FBLN2 depletion obviously increased intracellular Fe2+, lipid peroxides, and ROS levels in HCC cells. But FBLN2 overexpression reversed the effects mediated by circ_0001944 depletion on the increase in Fe2+, lipid peroxides, and ROS levels (Figure 5G–I). In conclusion, circ_0001944 targets the miR-1292-5p/FBLN2 axis to suppress ferroptosis in HCC.
Figure 5.
Circ_0001944 restrains cell ferroptosis by targeting the miR-1292-5p/FBLN2 axis. (A) A heatmap of hierarchical clustering showing the differentially expressed miRNAs in Huh7/SOR-FBLN2 silence vs Huh7/SOR and SMMC7721/SOR-FBLN2 silence vs SMMC7721 cells with three biological replicates per group (Fold Change≥2 and Padj<0.05). (B) The expression profile of DEGs in SMMC7721/SOR-FBLN2 silence cells. (C) The identification for GO function of these DEGs. (D) KEGG pathway enrichment for these differentially expressed mRNAs. The most 10 significant GO terms, as well as most 20 KEGG enrichment pathways are drawn as a bar chart; the horizontal axis in the figure represents the GO terms and KEGG pathways, and the vertical axis represents the significance level of pathway enrichment, with higher values indicating greater significance. The red box was defined in the highest bar column to highlight the GO terms and KEGG pathways associated with these genes. (E and F) Huh7/SOR (E) and SMMC7721/SOR (F) cells were pre-transfected with Len-circ_0001944-shRNA, miR-1292-5p mimics, FBLN2-siRNA, miR-1292-5p inhibitor and GV657-FBLN2 for 24h, and then were stimulated with sorafenib (10μM) for another 48h. The protein level of TFR1, DMT1, FTH1 and NQO1 were determined using Western blot assay. For Western blot assay, all of the data were manifested as the representatives (left) and the relative expression with means±SD (right) from three independent experiments. **P<0.01. (G–I) SMMC7721/SOR and Huh7/SOR cells were pre-treated with Len-circ_0001944-shRNA, miR-1292-5p mimics, FBLN2-siRNA, miR-1292-5p inhibitor and GV657-FBLN2 for 24h, and were stimulated with sorafenib (10μM) for another 48h. Intracellular levels of Fe2+(G), lipid peroxides (H) and ROS levels (I) in HCC cells were detected, respectively.**P<0.01. Data are shown as means±SD of three independent experiments.
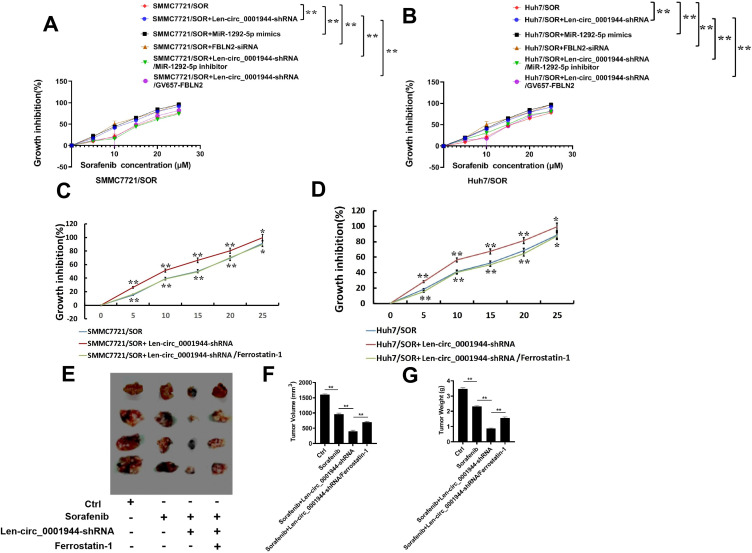
circ_0001944 Promotes Sorafenib Resistance by Discouraging Cell Ferroptosis in HCC
Interestingly, Len-circ_0001944-shRNA, miR-1292-5p mimics, and FBLN2 depletion also remarkably increased the sensitivity of SMMC7721/SOR and Huh7/SOR cells to sorafenib. But the enhancement of sensitivity induced by Len-circ_0001944-shRNA was partially abolished after the transfection of miR-1292-5p inhibitor and FBLN2 overexpression (Figure 6A and B). Thus, circ_0001944 directly targets the miR-1292-5p/FBLN2 axis to facilitate sorafenib resistance in HCC. Then we would like to confirm whether ferroptosis ablation mediated by circ_0001944 can enjoy a high exposure in the development of sorafenib resistance in HCC. As expected, circ_0001944 depletion distinctly enhanced the sensitivity of SMMC7721/SOR and Huh7/SOR cells to sorafenib. However, the sensitization effect caused by circ_0001944 knockdown can be blocked by Ferrostatin-1 which is represented as a ferroptosis inhibitor (Figure 6C and D). Furthermore, the enhancement of sorafenib-sensitivity in HCC-bearing mice that was boosted by the transfection of Len-circ_0001944-shRNA can partly be offset after the administration of Ferrostatin-1 (Figure 6E–G). Collectively, circ_0001944 targets the miR-1292-5p/FBLN2 axis to promote sorafenib resistance, in which ferroptosis inhibition is now an absolute certainty. .
Figure 6.
Circ_0001944 advances sorafenib resistance in HCC by inhibiting ferroptosis. (A and B) SMMC7721/SOR and Huh7/SOR cells were transfected with Len-circ_0001944-shRNA, miR-1292-5p mimics, FBLN2-siRNA, miR-1292-5p inhibitor and GV657-FBLN2 for 24h, Then they were stimulated with sorafenib for another 48h. The sensitivity of these cells as shown as growth inhibition (%), was confirmed using CCK-8 assay.**P<0.01. For Figure A and B, **P<0.01 was considered to be statistically significant in Len-circ_0001944-shRNA group versus Ctrl group, in miR-1292-5p mimics group versus Crl group, in FBLN2-siRNA group versus Ctrl group, as well as in Len-circ_0001944-shRNA/miR-1292-5p inhibitor group versus Len-circ_0001944-shRNA group and in Len-circ_0001944-shRNA/GV657-FBLN2 group versus Len-circ_0001944-shRNA group. Data are shown as means±SD of three independent experiments. (C and D) SMMC7721/SOR and Huh7/SOR cells were transfected with Len-circ_0001944-shRNA.The sensitivity of these cells as shown as growth inhibition (%), was confirmed using CCK-8 assay. For Figure A and B, **P<0.01 and *P<0.05 were considered to be statistically significant in Len-circ_0001944-shRNA group versus Ctrl group, as well as in Len-circ_0001944-shRNA/Ferrostatin-1 group versus Len-circ_0001944-shRNA group. Data are shown as means±SD of three independent experiments. (E–G) Tumor-bearing mice were administrated with Len-circ_0001944-shRNA plasmid or Ferrostatin-1, and then given sorafenib (30mg/kg). Tumor presentations (E), tumor volume (F) and tumor weight (G) of each group were demonstrated. Data are representative of three independent experiments with four mice per group. **P<0.01 was considered to be statistically significant in sorafenib-treated group versus Ctrl group, and in Len-circ_0001944-shRNA/sorafenib group versus sorafenib-treated group, as well as in Len-circ_0001944-shRNA/sorafenib/Ferrostatin-1 group versus Len-circ_0001944-shRNA/sorafenib group.
Discussion
Sorafenib represents a standard treatment for advanced HCC, but offers unfavorable outcomes depressingly with resistance conditions. At present, the mechanisms underpinning sorafenib resistance in HCC have not been fully elucidated.
It is recognized that enhanced DNA repair and off-target effects, increased drug efflux but reduced drug absorption contribute to drug resistance development in tumor cells.44 But it is newly identified that accumulated genetic alterations in tumor cells help them adapt to the complicated TME.45 The “successful” tumor cells optimized themselves progressively to exhibit intratumor heterogeneity, where lncRNAs, circRNAs, and miRNAs cooperated with each other. Some of them trigger drug resistance; others may be considered as a sentry to combat drug resistance.46,47
Herein, SMMC7721/SOR and Huh7/SOR cells were both prepared in our study to find common issues involved in sorafenib resistance. CircRNA-Seq and miRNA-Seq were carried out, by which some circRNAs and miRNAs were expounded. Accordingly, the overlap of differentially expressed circRNAs and miRNAs from SMMC7721/SOR and Huh7/SOR cells compared to their parental cells was determined. For the first time, circ_0078607, circ_0001944, circ_0002874, and circ_0069981; miR-193a-5p, miR-197-3p, miR-27a-5p, miR-551b-5p, miR-335-3p, miR-767-3p, miR-767-5p, miR-92a-1-5p, miR-92a-3p, miR-3940-3p, miR-664b-3p, miR-1292-5p, miR-99a-5p, and let-7c-5p taken the lead and caught our attentions in sorafenib-resistant HCC cells.
CircRNAs competitively bind to MREs and initiate Targeted MicroRNA degradation (TDMD) to inhibit miRNA splicing and transcription, thus serving their purpose to affect the expression of target genes through the “circRNA-miRNA-mRNA” axis.48 Furthermore, we can boldly guess that circRNA and miRNA targeted mRNAs, between which there exist complementary and intercrossing relationships based on ceRNA network involved in sorafenib resistance. Accordingly, “circ_0001944-miR-1292-5p-mRNA” regulatory network-based circ_0001944 and miR-1292-5p has been depicted successfully using transcriptome association analysis in the present study, thus providing more integrated evidence to investigate the mechanisms promoting sorafenib resistance. As far as we know, this is at the forefront in terms of interaction relationship with circ_0001944, miR-1292-5p, and indicated mRNA.
Actually, circ_0001944 is first demonstrated in breast cancer, where it acted as a proto-oncogene to encourage tumor metastasis by targeting the miR-125a/BRD4 axis.49 Circ_0001944 also promote tumor progression in colorectal cancer by targeting miR-548.50 Alternatively, circ_0001944 facilitates glycolysis by targeting miR-142-5p, and thus encouraging tumor proliferation and invasion in non-small cell NSCLC.51 However, there is no understanding about the role of circ_0001944 in HCC. Recent evidence indicated that miR-1292-5p inhibits tumor growth and migration only in gastric cancer.52 But there are no reports indicating its role in tumors other than gastric cancer, let alone in HCC. FBLN2, also known as Fibulin-2, is a newly extracellular matrix glycoprotein that is associated with tissue development and remodelling. It has been revealed that FBLN2 can be a candidate tumor-suppressor gene in urothelial carcinoma,53 NSCLC,54 and gastric cancer.55 However, the role of FBLN2 in HCC has not yet been clarified. Herein, circ_0001944, miR-1292-5p, and FBLN2 were first discovered in HCC. It is expected to write a rich and colorful stroke in the long history of studying sorafenib resistance in HCC. Most importantly, FBLN2 as the putative downstream regulator of miR-1292-5p, having emerged as the times required and stepping onto the historical stage in sorafenib resistance. Therefore, this has taken a solid step towards exploring the role of circ_0001944 involved in the sorafenib resistance in HCC.
CircRNAs with cancer-specific expression patterns have newly emerged as to play an important role in drug resistance in HCC. For example, circ_0003998 targets the miR-218-5p/EIF5A256 and miR-513a-5p/ARK557 axis to elevate the doxorubicin and 5-Fluorouracil resistance, respectively. Alternatively, circMRPS35 and circ_0031242 have been identified to be widely distributed in cisplatin resistance in HCC by targeting the miR-148a/STX358 and miR-924/POU3F259 axis, respectively. In particular, circ_0006988, circRNA-001241, and circFOXM1 markedly enhanced the sorafenib resistance in HCC by modulating the miR-15a-5p/IGF1,60 miR-21-5p/TIMP361 and miR-1324/MECP262 axis, respectively. Additionally, circ_0035944 and circ_0006294 expression were determined to be consistently downregulated in sorafenib-resistant HCC cells.63 Conversely, circ_0000615 was shown to be distinctly upregulated in sorafenib-resistant HCC patients.64 Thus, the expression profile of circ_0035944, circ_0006294, and circ_0000615 highlighted them as promising targets and prognostic biomarkers for the treatment of sorafenib in HCC patients.
Recently, the understanding about circRNA-based mechanism that was associated with suppression of ferroptosis and drug resistance in cancers has been continually improved. A novel circular RNA Circ-BGN was evidently confered trastuzumab resistance by inhibiting ferroptosis in human epidermal growth factor receptor 2-positive breast cancer.65 CircSnx12 also targets the miR-194-5p/SLC7A11 axis to suppress ferroptosis, thus promoting cisplatin resistance in ovarian cancer.66 However, it is rarely reported for ferroptosis-regulating circRNA in drug resistant HCC. Moreover, the implications of circRNAs involved in the inhibition of ferroptosis and sorafenib resistance in HCC remain to be clearly established.
In our present study, circ_0001944 was first proven to target the miR-1292-5p/FBLN2 axis and inhibit cell ferroptosis by “rearranging” iron homeostasis and lipid peroxidation. Ferroptosis frustration mediated by circ_0001944 is indispensable in the development of sorafenib resistance in HCC. Taken together, our results can replenish previous achievements, which provide a new perspective for enunciating the role of circRNAs in encouraging sorafenib resistance in HCC.
Although these findings are somewhat innovative, there are still many shortcomings to the present study. Firstly, the clinical implications of these ncRNAs in sorafenib resistance in HCC should be further excavated. In particular, the validation of the clinical potential of circ_0001944 as well as miR-1292-5p and FBLN2, is a long way to achieve “clinical reversal” through “theoretical exploration”. Clinical validation is crucial for exploring new strategies to enhance the sorafenib sensitivity towards HCC patients based on circ_0001944, miR-1292-5p, and FBLN2 will provide new ideas for exploring the malignant behavior especially sorafenib resistance in HCC. Similarly, there is still a great need to delve deeper into the role of “circ_0001944-miR-1292-5p-FBLN2” axis in suppressing ferroptosis and facilitating sorafenib resistance in HCC. Furthermore, the signaling pathways that inhibit ferroptosis to encourage sorafenib resistance based on “circ_0001944-miR-1292-5p-FBLN2” axis should be determined. We intended to focus on the erythroblastic leukaemia viral oncogene homologues (ErbB) signaling pathway in the follow-up study, which ranked second in the bar chart obtained from KEGG pathway enrichment analysis (Figure 5D). Consistently, ErbB1, ErbB2, ErbB3, ErbB4, neuregulin 1 (NRG1), and neuregulin 2 (NRG2) were confirmed to be differentially expressed and also rank among the top in terms of the fold change of their expressions in Huh7/SOR-FBLN2 silence vs Huh7/SOR cells and SMMC7721/SOR-FBLN2 silence vs SMMC7721 cells (Figure 5A). In fact, the dimerization of ErbB2 and ErbB3 can produce a high-affinity receptor for NRG-1. The activation of the NRG1/ErbB2 signaling pathway had been implicated in promoting neurodevelopment.67 Most of the patients who are sensitive to ErbB receptor-targeted therapies become resistant. Abnormalities in the ErbB family can be found in a variety of human cancers. Alternatively, activated EGFR receptor can recruit TKI to interact with intracellular signaling pathways by transmitting signals.68 Therefore, we will integrate the new findings with these existing concepts and discuss how the identified DENs and targeted genes contribute to or interact with known resistance pathways. Especially, the mechanisms that restrain ferroptosis and promote sorafenib resistance should be explored. How the activation of the NRG/ErbB signaling pathway was regulated by “circ_0001944-miR-1292-5p-FBLN2” axis should be illuminated.
Conclusions
Our data provided new ideas for exploring the malignant behavior especially sorafenib resistance in HCC. Especially, circ_0001944 targets the miR-1292-5p/FBLN2 axis to impede ferroptosis, thus providing fuel for encouraging sorafenib resistance in HCC. However, more clinical verification about the role of circ_0001944, miR-1292-5p even FBLN2 implicated in the reversal of drug resistance and the treatment for HCC, should be conducted. New evidence should be certificated that the suppression of ferroptosis associated with the activation of the NRG/ErbB signaling pathway regulated by the “circ_0001944-miR-1292-5p-FBLN2” axis can lead to sorafenib resistance in HCC.
Acknowledgments
The authors would like to thank LetPub for editing our manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No.82204433), and the Natural Science Foundation of Shandong Province, China (No.ZR2020QH362) and the clinical medicine X+ project of the affiliated hospital of Qingdao University.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Alawyia B, Constantinou C. Hepatocellular carcinoma: a narrative review on current knowledge and future prospects. Curr Treat Options Oncol. 2023;24(7):711–724. doi: 10.1007/s11864-023-01098-9 [DOI] [PubMed] [Google Scholar]
- 2.Fan Z, Zhou P, Jin B, et al. Recent therapeutics in hepatocellular carcinoma. Am J Cancer Res. 2023;13(1):261–275. [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelgalil AA, Alkahtani HM, Al-Jenoobi FI. Sorafenib. Profiles Drug Subst Excip Relat Methodol. 2019;44:239–266. doi: 10.1016/bs.podrm.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 4.Jiang Z, Dai C. Potential treatment strategies for hepatocellular carcinoma cell sensitization to sorafenib. J Hepatocell Carcinoma. 2023;10:257–266. doi: 10.2147/jhc.S396231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan Y, Xue H, Zheng H. Systemic therapy for hepatocellular carcinoma: current updates and outlook. J Hepatocell Carcinoma. 2022;9:233–263. doi: 10.2147/jhc.S358082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C, Zhang H, Zhang L, et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20(4):203–222. doi: 10.1038/s41575-022-00704-9 [DOI] [PubMed] [Google Scholar]
- 7.Wei S, Wei F, Li M, et al. Target immune components to circumvent sorafenib resistance in hepatocellular carcinoma. Biomed Pharmacother. 2023;163:114798. doi: 10.1016/j.biopha.2023.114798 [DOI] [PubMed] [Google Scholar]
- 8.Xu L, Liu Y, Chen X, Zhong H, Wang Y. Ferroptosis in life: to be or not to be. Biomed Pharmacother. 2023;159:114241. doi: 10.1016/j.biopha.2023.114241 [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Wang X, Xiao A, Han J, Wang Z, Wen M. Ketogenic diet prevents chronic sleep deprivation-induced Alzheimer’s disease by inhibiting iron dyshomeostasis and promoting repair via Sirt1/Nrf2 pathway. Front Aging Neurosci. 2022;14:998292. doi: 10.3389/fnagi.2022.998292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng X, An H, Yu F, et al. Benefits of iron chelators in the treatment of Parkinson’s disease. Neurochem Res. 2021;46(5):1239–1251. doi: 10.1007/s11064-021-03262-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajoolabady A, Tang D, Kroemer G, Ren J. Ferroptosis in hepatocellular carcinoma: mechanisms and targeted therapy. Br J Cancer. 2023;128(2):190–205. doi: 10.1038/s41416-022-01998-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zheng L, Shang W, et al. Wnt/beta-catenin signaling confers ferroptosis resistance by targeting GPX4 in gastric cancer. Cell Death Differ. 2022;29(11):2190–2202. doi: 10.1038/s41418-022-01008-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Zhao F, Ke B, Chen D, Liu F. Harnessing ferroptosis to overcome drug resistance in colorectal cancer: promising therapeutic approaches. Cancers. 2023;15(21). doi: 10.3390/cancers15215209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi R, Bai Y, Li K, et al. Cancer-associated fibroblasts suppress ferroptosis and induce gemcitabine resistance in pancreatic cancer cells by secreting exosome-derived ACSL4-targeting miRNAs. Drug Resist Updat. 2023;68:100960. doi: 10.1016/j.drup.2023.10096015 [DOI] [PubMed] [Google Scholar]
- 15.Bai T, Wang S, Zhao Y, Zhu R, Wang W, Sun Y. Haloperidol, a sigma receptor 1 antagonist, promotes ferroptosis in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2017;491(4):919–925. doi: 10.1016/j.bbrc.2017.07.136 [DOI] [PubMed] [Google Scholar]
- 16.Guo L, Hu C, Yao M, Han G. Mechanism of sorafenib resistance associated with ferroptosis in HCC. Front Pharmacol. 2023;14:1207496. doi: 10.3389/fphar.2023.1207496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good DJ. Non-Coding RNAs in Human Health and Diseases. Genes. 2023;14(7):1429. doi: 10.3390/genes14071429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Zhong Y, Wang X, Shen J, An W. Advances in circular RNA and its applications. Int J Med Sci. 2022;19(6):975–985. doi: 10.7150/ijms.71840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson BS, Pye H, Whitaker HC. The oncological relevance of fragile sites in cancer. Commun Biol. 2021;4(1):567. doi: 10.1038/s42003-021-02020-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: a promising therapeutic target in cancer. Int J Mol Sci. 2022;23(19):11502. doi: 10.3390/ijms231911502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S, Sinha T, Panda AC. Regulation of microRNA by circular RNA. Wiley Interdiscip Rev RNA. 2023;e1820. doi: 10.1002/wrna.1820 [DOI] [PubMed] [Google Scholar]
- 22.Li R, Xu H, Gao X. The ceRNA network regulates epithelial-mesenchymal transition in colorectal cancer. Heliyon. 2023;9(3):e14143. doi: 10.1016/j.heliyon.2023.e14143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X, Xia X, Li F, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23(3):278–291. doi: 10.1038/s41556-021-00639-4 [DOI] [PubMed] [Google Scholar]
- 24.Verduci L, Ferraiuolo M, Sacconi A, et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18(1):237. doi: 10.1186/s13059-017-1368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1):295. doi: 10.1038/s41467-020-20527-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanniford D, Ulloa-Morales A, Karz A, et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37(1):55–70.e15. doi: 10.1016/j.ccell.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi: 10.1038/s41571-021-00585-y [DOI] [PubMed] [Google Scholar]
- 29.Liao W, Du J, Wang Z, et al. The role and mechanism of noncoding RNAs in regulation of metabolic reprogramming in hepatocellular carcinoma. Int, J, Cancer. 2022;151(3):337–347. doi: 10.1002/ijc.34040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Q, Tian H, Guo Z, Ma Z, Wang G. The role of noncoding RNAs in the tumor microenvironment of hepatocellular carcinoma. Acta Biochim Biophys Sin. 2023;55(11):1697–1706. doi: 10.3724/abbs.2023231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng H, Niu R, Huang C, Li J. Circular RNA as a novel biomarker and therapeutic target for HCC. Cells. 2022;11(12):1948. doi: 10.3390/cells11121948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu ZS, Wang WH. Circular RNAs in hepatocellular carcinoma: recent advances. World J Gastrointest Oncol. 2022;14(6):1067–1085. doi: 10.4251/wjgo.v14.i6.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, Gingeras TR. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics. 2015;51:11.14.1–11.14.19. doi: 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):7441):333–8. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2018;19(5):803–810. doi: 10.1093/bib/bbx014 [DOI] [PubMed] [Google Scholar]
- 38.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, Chen J, Li Z, et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One. 2010;5(12):e15224. doi: 10.1371/journal.pone.0015224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- 41.Bo X, Wang S. TargetFinder: a software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics. 2005;21(8):1401–1402. doi: 10.1093/bioinformatics/bti211 [DOI] [PubMed] [Google Scholar]
- 42.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–4. doi: 10.1093/nar/gkl243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–3793. doi: 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 44.Duan C, Yu M, Xu J, Li BY, Zhao Y, Kankala RK. Overcoming cancer multi-drug resistance (MDR): reasons, mechanisms, nanotherapeutic solutions, and challenges. Biomed Pharmacother. 2023;162:114643. doi: 10.1016/j.biopha.2023.114643 [DOI] [PubMed] [Google Scholar]
- 45.Li J, Yu N, Li X, Cui M, Guo Q. The single-cell sequencing: a dazzling light shining on the dark corner of cancer. Front Oncol. 2021;11:759894. doi: 10.3389/fonc.2021.759894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro-Muñoz LJ, Ulloa EV, Sahlgren C, Lizano M, De La Cruz-Hernández E, Contreras-Paredes A. Modulating epigenetic modifications for cancer therapy (Review). Oncol Rep. 2023;49(3). doi: 10.3892/or.2023.8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. 2022;7(1):121. doi: 10.1038/s41392-022-00975-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheu-Gruttadauria J, Pawlica P, Klum SM, et al. Structural basis for target-directed microRNA degradation. Mol Cell. 2019;75(6):1243–1255.e7. doi: 10.1016/j.molcel.2019.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu B, Liu W, Zhu C, et al. Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. Int J Biol Sci. 2021;17(12):3104–3117. doi: 10.7150/ijbs.58916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan H, Qiu J. Association of has_circ_0001944 upregulations with prognosis and cancer progression in patients with colorectal cancer. Discov Oncol. 2022;13(1):23. doi: 10.1007/s12672-022-00485-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dou Y, Tian W, Wang H, Lv S. Circ_0001944 contributes to glycolysis and tumor growth by upregulating NFAT5 through acting as a decoy for miR-142-5p in non-small cell lung cancer. Cancer Manag Res. 2021;13:3775–3787. doi: 10.2147/cmar.S302814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hui W, Ma X, Zan Y, Song L, Zhang S, Dong L. MicroRNA-1292-5p inhibits cell growth, migration and invasion of gastric carcinoma by targeting DEK. Am J Cancer Res. 2018;8(7):1228–1238. [PMC free article] [PubMed] [Google Scholar]
- 53.Li WM, Chan TC, Huang SK, et al. Prognostic utility of FBLN2 expression in patients with urothelial carcinoma. Front Oncol. 2020;10:570340. doi: 10.3389/fonc.2020.570340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma Y, Nenkov M, Schröder DC, Abubrig M, Gassler N, Chen Y. Fibulin 2 is hypermethylated and suppresses tumor cell proliferation through inhibition of cell adhesion and extracellular matrix genes in non-small cell lung cancer. Int J Mol Sci. 2021;22(21):11834. doi: 10.3390/ijms222111834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen K, Xia W, Wang K, et al. ITGBL1 promotes anoikis resistance and metastasis in human gastric cancer via the AKT/FBLN2 axis. J Cell Mol Med. 2024;28(4):e18113. doi: 10.1111/jcmm.18113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, He J, Ren X, Zhao H, Zhao H. Circ_0003998 enhances doxorubicin resistance in hepatocellular carcinoma by regulating miR-218-5p/EIF5A2 pathway. Diagn Pathol. 2020;15(1):141. doi: 10.1186/s13000-020-01056-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue L, He J, Chen H, Ren C, Fu X. Circ_0003998 upregulates ARK5 expression to elevate 5-Fluorouracil resistance in hepatocellular carcinoma through binding to miR-513a-5p. Anticancer Drugs. 2022;33(10):1103–1113. doi: 10.1097/cad.0000000000001359 [DOI] [PubMed] [Google Scholar]
- 58.Li P, Song R, Yin F, et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther. 2022;30(1):431–447. doi: 10.1016/j.ymthe.2021.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fan W, Chen L, Wu X, Zhang T. Circ_0031242 silencing mitigates the progression and drug resistance in DDP-resistant hepatoma cells by the miR-924/POU3F2 axis. Cancer Manag Res. 2021;13:743–755. doi: 10.2147/cmar.S272851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu R, Zeng Z. Hsa_circ_0006988 promotes sorafenib resistance of hepatocellular carcinoma by modulating IGF1 using miR-15a-5p. Can J Gastroenterol Hepatol. 2022;2022:1206134. doi: 10.1155/2022/1206134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Q, Wu G. CircRNA-001241 mediates sorafenib resistance of hepatocellular carcinoma cells by sponging miR-21-5p and regulating TIMP3 expression. Gastroenterol Hepatol. 2022;45(10):742–752. doi: 10.1016/j.gastrohep.2021.11.007 [DOI] [PubMed] [Google Scholar]
- 62.Weng H, Zeng L, Cao L, et al. circFOXM1 contributes to sorafenib resistance of hepatocellular carcinoma cells by regulating MECP2 via miR-1324. Mol Ther Nucleic Acids. 2021;23:811–820. doi: 10.1016/j.omtn.2020.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu MY, Tang YP, Liu JJ, Liang R, Luo XL. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer. 2020;11(10):2993–3001. doi: 10.7150/jca.39854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, Xu T, Li Y, Pang Q, Ding X. Serum hsa_circ_0000615 is a prognostic biomarker of sorafenib resistance in hepatocellular carcinoma. J Clin Lab Anal. 2022;36(11):e24741. doi: 10.1002/jcla.24741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S, Wang Y, Li Q, Li X, Feng X. A novel circular RNA confers trastuzumab resistance in human epidermal growth factor receptor 2-positive breast cancer through regulating ferroptosis. Environ Toxicol. 2022;37(7):1597–1607. doi: 10.1002/tox.23509 [DOI] [PubMed] [Google Scholar]
- 66.Qin K, Zhang F, Wang H, et al. circRNA circSnx12 confers Cisplatin chemoresistance to ovarian cancer by inhibiting ferroptosis through a miR-194-5p/SLC7A11 axis. BMB Rep. 2023;56(2):184–189. doi: 10.5483/BMBRep.2022-0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kataria H, Alizadeh A, Karimi-Abdolrezaee S. Neuregulin-1/ErbB network: an emerging modulator of nervous system injury and repair. Prog Neurobiol. 2019;180:101643. doi: 10.1016/j.pneurobio.2019.101643 [DOI] [PubMed] [Google Scholar]
- 68.Sahu A, Verma S, Varma M, Yadav MK. Impact of ErbB receptors and anticancer drugs against breast cancer: a review. Curr Pharm Biotechnol. 2022;23(6):787–802. doi: 10.2174/1389201022666210719161453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.