Abstract
Pariacoto virus (PaV) is a nodavirus that was recently isolated in Peru from the Southern armyworm, Spodoptera eridania. Virus particles are non enveloped and about 30 nm in diameter and have T=3 icosahedral symmetry. The 3.0-Å crystal structure shows that about 35% of the genomic RNA is icosahedrally ordered, with the RNA forming a dodecahedral cage of 25-nucleotide (nt) duplexes that underlie the inner surface of the capsid. The PaV genome comprises two single-stranded, positive-sense RNAs: RNA1 (3,011 nt), which encodes the 108-kDa catalytic subunit of the RNA-dependent RNA polymerase, and RNA2 (1,311 nt), which encodes the 43-kDa capsid protein precursor α. In order to apply molecular genetics to the structure and assembly of PaV, we identified susceptible cell lines and developed a reverse genetic system for this virus. Cell lines that were susceptible to infection by PaV included those from Spodoptera exigua, Helicoverpa zea and Aedes albopictus, whereas cells from Drosophila melanogaster and Spodoptera frugiperda were refractory to infection. To recover virus from molecular clones, full-length cDNAs of PaV RNAs 1 and 2 were cotranscribed by T7 RNA polymerase in baby hamster kidney cells that expressed T7 RNA polymerase. Lysates of these cells were infectious both for cultured cells from Helicoverpa zea (corn earworm) and for larvae of Galleria mellonella (greater wax moth). The combination of infectious cDNA clones, cell culture infectivity, and the ability to produce milligram amounts of virus allows the application of DNA-based genetic methods to the study of PaV structure and assembly.
Members of the Nodaviridae family are small positive-sense RNA viruses with T=3 icosahedral symmetry (for a review, see references 4, 5, and 37). Each 30-nm particle is assembled from 180 copies of the capsid protein precursor α and one copy of each of the two unique segments of the viral RNA genome (24, 30, 41). Viruses from the alphanodavirus genus infect primarily insects. In alphanodaviruses, the 44-kDa capsid protein precursor α is autocatalytically cleaved following virion assembly to yield the two mature capsid proteins β (40 kDa) and γ (4 kDa) (20). The maturation cleavage is required for infectivity (38). The larger genome segment (RNA1) contains 3.0 to 3.2 kb and encodes protein A, the catalytic subunit of the RNA-dependent RNA polymerase (26). The smaller genome segment (RNA2) contains 1.3 to 1.4 kb and encodes the capsid protein precursor, α. Both genomic RNAs are capped but not polyadenylated (9, 10, 22, 28). During replication, a subgenomic RNA (RNA3) which is 3′ coterminal with RNA1 is transcribed. RNA3 contains about 400 nucleotides (nt) and encodes one or two small proteins (B1 and B2) of unknown function (5, 16, 22).
The best studied alphanodavirus is Flock House virus (FHV), and reverse genetic systems that allow the infectious cycle of this virus to be reconstructed from cDNA clones have been developed. FHV replication can be initiated and infectious virus can be recovered from in vitro transcripts of RNA1 and RNA2 transfected into Drosophila melanogaster cells (11) or from specialized cDNA transcription plasmids transfected into mammalian cells (2). These approaches have advanced our understanding of both RNA replication and capsid assembly of this model virus (5, 6, 25, 37). For example, two regions in the FHV capsid protein precursor that are involved in RNA encapsidation have been identified: (i) protein α that lacks amino acid residues 1 to 31 assembles into virus-like particles (VLPs) that encapsidate RNA1 but fail to encapsidate RNA2 (13); (ii) deleting the C-terminal 26 amino acids (aa) results in the assembly of VLPs that encapsidate cellular RNAs in place of the viral genome segments (36). However, most of the capsid protein regions involved in the specificity of FHV RNA encapsidation are not visible in the crystal structure (8, 14).
The structure of the alphanodavirus Pariacoto virus (PaV) was recently determined, and it significantly extended our understanding of RNA-protein interactions in nodavirus virions (43). PaV was isolated in Peru in 1996 from moribund larvae of the Southern armyworm, Spodoptera eridania (45), and is the most recent member of the alphanodavirus genus to be characterized. PaV is the most distantly related of the insect nodaviruses, with its RNA-dependent RNA polymerase and capsid protein sharing less than 29 and 41% sequence identity, respectively, with those of the other alphanodaviruses (26, 27).
The three-dimensional structure of PaV is generally similar to that of the other nodaviruses, but with several novel features (43). First, along each twofold axis of the capsid lies a 25-nt A-form RNA duplex that is visible at high resolution in the crystal structure and accounts for 1,500 nt, or approximately 35% of the single-stranded genomic RNA. Thirty such RNA duplexes are arranged as a dodecahedral cage, with discontinuous vertices where the RNA is less ordered and presumably loops into the interior of the virion. Second, for the first time in any nodavirus structure, the basic N terminus of one of the three quasiequivalent capsid protein monomers in the asymmetric unit is clearly visible from aa 7 to 51, making numerous contacts with the RNA duplex. Third, the C-terminal 8 aa of the same subunit are also visible for the first time, lying in a protein channel at the quasi-threefold axis. Thus, regions of the capsid protein that likely influence the specificity of RNA encapsidation and particle assembly are uniquely visible in the PaV structure, making it an attractive system for analysis by molecular genetics.
Such studies require a method by which infectious PaV particles and VLPs could be recovered from cDNA clones in quantities large enough for structural analysis. In our previous work we showed that transfection of PaV virion RNAs into BHK-21 cells resulted in RNA replication and the synthesis of both protein A and the capsid protein precursor α. To develop a reverse genetic system, cDNA transcription plasmids encoding PaV RNA1 and RNA2 were transfected into BHK-21 cells infected with a recombinant vaccinia virus that expressed T7 RNA polymerase to drive primary transcription (27). The cDNA clones initiated RNA replication and viral protein synthesis, but since BHK-21 cells are not susceptible to infection with PaV no evidence of infectivity was obtained. In the present study, we surveyed several insect cell lines for their susceptibility to infection with PaV and then used the infectious cell culture system to validate the recovery of clonally derived virus.
MATERIALS AND METHODS
Cells and virus.
Except where indicated, all insect cell lines were maintained at 28°C in medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. Drosophila line 1 and line 2 (DL-1 and DL-2) (39) cells were maintained in Schneider's medium (Gibco/BRL), and Spodoptera frugiperda (Sf9) cells (44) were maintained in Grace's medium (Gibco/BRL). Cell lines derived from Aedes aegypti whole larvae (Ae-59) (34), Aedes albopictus whole larvae (ATC-15) (42), Spodoptera exigua larvae (Se-1) (21), and Helicoverpa zea fat body cell lines (BCIRL-HZ-FB27 and BCIRL-HZ-FB33, referred to here as FB27 and FB33, respectively) and a midgut cell line (BCIRL-HZ-MG8, referred to here as MG8) (29; A. H. McIntosh, personal communication) were maintained in ExCell-401 medium (JRH Biologicals) with 10% heat-inactivated fetal bovine serum, except for the MG8 line, which was maintained in serum-free medium.
Baby kidney hamster-derived cell line BSR-T7/5 cells (7) were grown at 37°C as monolayer cultures in modified Eagle's medium (Gibco/BRL catalogue number 41200-015) supplemented with 5% fetal calf serum and 5% newborn calf serum in an atmosphere containing 5% CO2. BSR-T7/5 cells were subcultured in the continuous presence of the antibiotics penicillin and streptomycin, and in alternate passages G418 was added to 1 mg/ml to ensure maintenance of the T7 polymerase gene. Wild-type PaV was purified as described previously (27) from Galleria mellonella larvae inoculated with the original field material collected in 1996 (45).
cDNA clones.
The PaV cDNA clones PaV1(0,0) and PaV2(0,0) were described previously (27). Briefly, reverse transcription (RT)-PCR copies of the two PaV genome segments were ligated into the transcription vector TVT7R(0,0) between the T7 promoter and cDNA sequences that encode the hepatitis delta virus antigenomic ribozyme, followed by the T7 terminator. Following autocatalytic cleavage by the hepatitis delta virus ribozyme, the resulting T7 transcripts had precise 5′ and 3′ termini with no additional nucleotides at either end, hence the designation (0,0). When these plasmids were transfected into BHK-21 cells previously infected with a recombinant vaccinia virus that expressed T7 RNA polymerase, we observed a low level of replication of PaV RNA1 and RNA2. However, for use in BSR-T7/5 cells which express T7 RNA polymerase constitutively (7) and thus eliminate the need for vaccinia virus infection, it proved necessary to optimize the PaV RNA1 clone by adding a single guanylate residue at the transcription initiation site (see Results and Discussion). The additional nucleotide was introduced into plasmid PaV1(0,0) using Quik Change as described by the supplier (Stratagene). A small DNA fragment spanning the mutation was substituted into PaV1(0,0) to give PaV1(1,0), and the nucleotide sequence of the substituted fragment was verified.
Screening for cells susceptible to infection with PaV.
To screen cells for susceptibility to infection with PaV, insect cell lines were plated in 35-mm-diameter wells of six-well plates at an appropriate cell density to achieve 50 to 90% confluence. Cells were allowed to attach for at least 1 h at 28°C, washed once with serum-free medium, and overlayed with 1 ml of serum-free medium containing 5 × 1010 PaV or FHV particles. After 2 h of adsorption, 1 ml of serum-containing medium was added and incubation was continued at 28°C. RNA replication was assayed 24 h postinfection by metabolic labeling as described below.
Transfection of BSR-T7/5 cells.
For transfection with virion RNA (vRNA) or with plasmids, BSR-T7/5 cells were plated in 35-mm-diameter wells of six-well tissue culture plates and grown overnight at 37°C to reach 80 to 90% confluency. The cells were washed twice with Dulbecco's minimal essential medium (DMEM) (catalogue number 12100-103; Gibco/BRL) and then overlayed with 1 ml of DMEM containing 20 μl of Lipofectamine 2000 (Gibco/BRL) and 0.1 to 0.5 μg of PaV vRNA or 2.5 μg of PaV1(1,0) with or without 2.5 μg of PaV2(0,0). After incubation for 24 h at 28°C, the transfection mix was removed and replaced by MEM containing serum. Incubation was continued at 28°C until 48 h posttransfection, at which time the cells were either harvested or radiolabeled as described below.
RNA and protein labeling, extraction, and analysis.
The products of RNA replication were radiolabeled by metabolic incorporation of [3H]uridine for 2 or 4 h in the presence of actinomycin D as previously described (2). For BSR-T7/5 cells actinomycin D was used at a working concentration of 5 μg/ml, whereas for insect cell lines it was necessary to increase the concentration of actinomycin D to 20 μg/ml to satisfactorily inhibit DNA-dependent RNA synthesis. Total RNA was extracted from cells or virions using an RNAgents kit (Promega) as suggested by the manufacturer. RNAs were resolved by electrophoresis on 1% agarose-formaldehyde gels and visualized by fluorography (32).
The products of protein synthesis were metabolically labeled by incorporation of either [35S]methionine-cysteine or [3H]leucine for 1 h in DMEM lacking either methionine and cysteine or leucine, respectively. Total cell proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using standard techniques (31). For fluorography, SDS-PAGE gels were treated with 1 M sodium salicylate for 10 min before drying and exposure to X-ray film.
Western blot analysis.
Proteins were transferred from SDS-polyacrylamide gels to polyvinylidene difluoride membranes in Towbin buffer (10 mM Tris base, 96 mM glycine in 10% methanol) by using a semidry graphite electroblotter (Millipore) for 2 h at a constant current of 80 mA. PaV antiserum raised in rabbits against the original isolate of PaV (45) was a gift from Jean-Louis Zeddam. PaV antiserum was diluted 3,000-fold for use as the primary antibody in the Immun-Star chemiluminescent protein detection system (Bio-Rad) according to the manufacturer's instructions.
Virus neutralization.
For neutralization assays, PaV antiserum was diluted 20-fold in phosphate-buffered saline before addition of 1 μl to each 20 μl of virus or BSR-T7/5 lysate sample. The samples were then incubated at room temperature for 30 min.
Propagation and purification of clonally derived PaV.
Larvae of the greater wax moth (G. mellonella) were reared at 31°C as previously described (27). Late-instar larvae (average weight, 150 mg) were injected with lysates derived from either mock-transfected BSR-T7/5 cells (12 larvae) or cells transfected with plasmids PaV1(1,0) + PaV2(0,0) (58 larvae). Larvae were incubated at 31°C for 8 days before being frozen at −20°C. Larval homogenates were clarified by low-speed centrifugation, and the virus was pelleted through a 30% sucrose cushion. The redissolved virus pellet was injected into 92 larvae (average weight, 220 mg), with 12 larvae left uninfected. The larvae were collected after 8 days of incubation and frozen at −20°C until virus purification. Clonally derived PaV was purified from frozen infected larvae as previously described (27) except that 50 mM Tris (pH 7.4) buffer was used throughout purification. Virus was resuspended in 50 mM Tris (pH 7.4) buffer and stored in aliquots at −80°C. Virus concentration was determined using an extinction coefficient at 260 nm of 4.15/mg and a particle mass of 9.2 × 106 g/mol, which was calculated on the basis of the protein and RNA content of a PaV particle (27, 43).
RESULTS AND DISCUSSION
Cell lines susceptible to infection with PaV.
Nine insect cell lines were assayed for susceptibility to infection with PaV (Table 1). We determined infectivity based on replication of PaV RNA in cells following exposure to 5 × 1010 PaV particles, which corresponds to a multiplicity of infection (MOI) of about 104 particles per cell. RNA replication was assayed by metabolic labeling of RNAs with [3H]uridine in the presence of actinomycin D, followed by separation of RNAs on denaturing agarose-formaldehyde gels and detection of the RNAs by fluorography. Input RNA was not detected by this method because only the products of RNA replication are actinomycin D resistant and became labeled. PaV infection led to detection of RNA replication in five of the cell lines: those from A. albopictus (mosquito; ATC-15), S. exigua (beet armyworm; Se-1) and H. zea (corn earworm; FB33, FB27 and MG8). The level of RNA labeled varied among these cell lines but was highest in FB33 cells. However, even in FB33 cells the level of RNA labeled was at least 100-fold less than during FHV infection of Drosophila DL-1 or DL-2 cells. FHV infection of FB33 cells led to a similarly low level of RNA labeling. Under the conditions used for the infectivity assay, there was no evidence of PaV infection in the mosquito cell line Ae-59, a fall armyworm line (Sf9), or the two Drosophila cell lines (DL-1 and DL-2) that are permissive for other alphanodaviruses such as FHV, Black beetle virus (BBV), and Boolarra virus (12, 15, 35). The lack of detection of RNA replication in Sf9 cells following exposure to PaV is consistent with the previous finding by Zeddam et al. (45) that S. frugiperda larvae were not susceptible to infection by PaV. Based on the results of the infectivity assay, the H. zea fat body cell line FB33 was chosen for use in further experiments with PaV.
TABLE 1.
Susceptibility of cell lines to infection with Pariacoto virusa
| Cell line | Insect | PaV | FHV |
|---|---|---|---|
| Ae-59 | Aedes aegypti (mosquito) | −b | + |
| ATC-15 | Aedes albopictus (mosquito) | + | − |
| DL-1 | Drosophila melanogaster (vinegar fly) | − | + |
| DL-2 | Drosophila melanogaster | − | + |
| FB27 | Helicoverpa zea (corn earworm) | + | − |
| FB33 | Helicoverpa zea | + | + |
| MG8 | Helicoverpa zea | + | − |
| Se-1 | Spodoptera exigua (beet armyworm) | + | − |
| Sf9 | Spodoptera frugiperda (fall armyworm) | − | − |
Susceptibility of cell lines to infection by either PaV or FHV as defined by the infectivity assay described in Materials and Methods; +, infectivity was detected; −, no infectivity was detected.
The results of the infectivity assays suggested that PaV and FHV may have cell specificities that only partially overlap. Of the nine cell lines tested, only FB33 cells were susceptible to both viruses. This is understandable in view of the different isolation sites of the two viruses, their highly diverged genome sequences (26, 27), and the distinct virion surface structures in the putative receptor-binding region (43).
Titration of infectivity of PaV in FB33 cells.
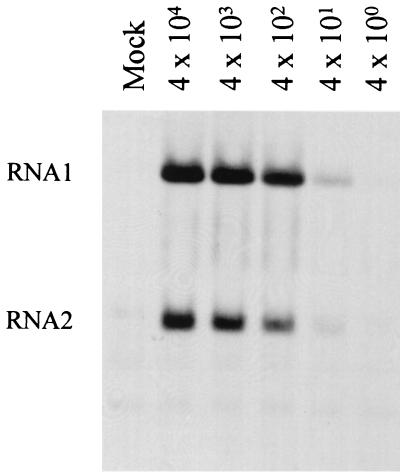
The initial infectivity assays used arbitrary cell densities and a high MOI. To optimize the infectivity assay for FB33 cells, we examined the effect of cell density on RNA replication. Cells were plated at a range of densities (5 × 105 to 4 × 106 cells per 35-mm-diameter well), and replicating RNAs were labeled 24 h postinfection. The optimal cell density for infection with PaV was found to be 2 × 106 cells in a 35-mm-diameter well, equivalent to 70 to 80% confluency (data not shown). Using this optimum cell density, we tested the effects of various MOIs ranging from 4 × 104 to 4 × 10−2 PaV particles per cell (Fig. 1). RNA1 and RNA2 were clearly labeled at MOIs down to 4 × 102, and a weak signal was detected at 4 × 101 particles per cell. No signal was detected at lower MOIs. Thus, PaV infection of FB33 cells could be readily detected at or above a MOI of 400 particles per cell, a level similar to the previously reported optimal MOIs for synchronous infection of DL-1 cells with BBV (17) and consistent with the high particle-to-PFU ratio for FHV (38, 41). In addition to the intense signals seen for the PaV genomic RNAs, some weaker bands that were also present in the mock-infected cells were visible. We attribute these to residual labeling of rRNAs (Fig. 1).
FIG. 1.
Infectivity of PaV in FB33 cells. Helicoverpa zea FB33 cells were infected with 10-fold dilutions of purified PaV (MOIs of 4 × 104 to 4 × 100) or mock infected and incubated at 28°C. After 22 h of incubation, actinomycin D was added at 20 μg/ml, and 30 min later replicating RNAs were metabolically labeled by incorporation of [3H]uridine (20 μCi/ml) for a period of 2 h before total cellular RNAs were harvested. RNAs were resolved by electrophoresis on a 1% agarose-formaldehyde gel and visualized by fluorography. PaV genomic RNA1 and RNA2 are identified on the left. The faint bands below RNA2 are cellular RNAs that are present in all samples including that from mock-infected cells.
Despite the evidence of PaV RNA replication, infected FB33 cells showed no cytopathic effects even 5 days after infection at an MOI of 104 particles per cell, and only low levels of PaV capsid proteins were detected in cell lysates (data not shown). In view of these results, it was not surprising that attempts to develop a plaque assay for PaV in FB33 cells, as was achieved for FHV and BBV in DL-2 cells (41), were unsuccessful. Nevertheless, PaV infection could be passed repeatedly on FB33 cells, although infectivity increased by only 3 to 4 logs in a single passage. This contrasts with the infection of DL-2 cells with FHV, which is highly productive (40). Despite the low yields, FB33 cells provided a simple infectivity assay for wild-type PaV and recombinant viruses recovered from cDNA clones.
PaV RNA and protein synthesis in BSR-T7/5 cells.
We showed previously that PaV RNA replication can be initiated in BHK-21 cells from the full-length cDNA clones PaV1(0,0) and PaV2(0,0) (27). These plasmids were constructed such that the major RNA transcripts made by T7 RNA polymerase initiated on the first nucleotide of the PaV RNA (adenosine) and, after cleavage by the HDV ribozyme, had terminal nucleotides that corresponded exactly to those of the PaV genomic RNAs. In these original experiments, T7 RNA polymerase was provided by infection of BHK-21 cells with a recombinant vaccinia virus (27). However, since the presence of vaccinia virus would complicate experiments designed to produce infectious PaV, for the present study we used the BHK-derived cell line BSR-T7/5, which stably expresses T7 RNA polymerase under the control of a cytomegalovirus pol II promoter (7). The replication of FHV RNAs transcribed from cDNA clones in these cells has been characterized in detail (1).
BSR-T7/5 cells were transfected with vRNA, or with plasmid PaV1(0,0) with or without PaV2(0,0), and 48 h posttransfection RNA replication products were labeled with [3H]uridine. Despite strong replication of PaV vRNA, no RNAs were labeled in the cells transfected with PaV1(0,0) with or without PaV2(0,0) (data not shown). Therefore, while previous experiments had shown that PaV cDNA transcripts could initiate RNA replication, and BSR-T7/5 cells could support authentic vRNA replication (Fig. 2, lane 4), the original PaV transcription plasmids failed to initiate RNA replication in BSR-T7/5 cells.
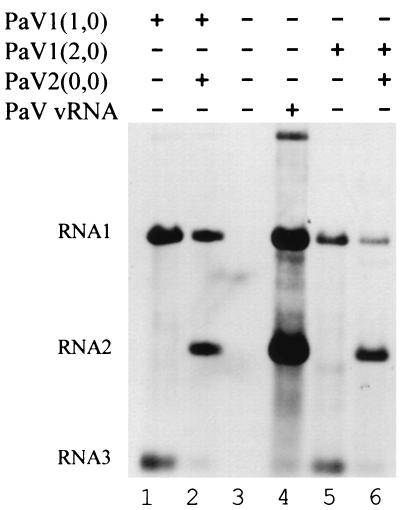
FIG. 2.
Replication of RNAs in BSR-T7/5 cells transfected with PaV cDNA clones. BSR-T7/5 cells were mock transfected (lane 3), transfected with 100 ng of vRNA (lane 4), or transfected with 2.5 μg of the following plasmids: PaV1(1,0) (lane 1); PaV1(1,0) + PaV2(0,0) (lane 2); PaV1(2,0) (lane 5); or PaV1(2,0) + PaV2(0,0) (lane 6). Following transfection, cells were incubated at 28°C for 48 h, at which time 5 μg of actinomycin D per ml was added, and incubation was continued for 30 min before the RNAs were metabolically labeled by incorporation of [3H]uridine (20 μCi/ml) for 4 h. Total cellular RNAs were harvested, resolved by electrophoresis on a 1% agarose-formaldehyde gel, and visualized by fluorography. PaV RNA1, RNA2, and RNA3 are identified on the left.
To overcome this problem, we inserted either one or two additional G residues at the transcription initiation site in the PaV1 plasmid to improve the sequence context of the T7 promoter and thereby enhance the level of DNA-templated primary transcription (33). An additional G residue at the initiation site for T7 transcription was also found necessary for replication initiated by plasmids containing FHV1 cDNA (3). BSR-T7/5 cells were transfected with PaV vRNA, PaV1(1,0), or PaV1(2,0) alone or in combination with PaV2(0,0), and 48 h posttransfection RNA was metabolically labeled with [3H]uridine as before. Total cellular RNA was extracted and resolved on denaturing agarose-formaldehyde gels, and replication products were visualized by fluorography (Fig. 2). In cells transfected with PaV1(1,0) (Fig. 2, lane 1) or PaV1(2,0) alone (Fig. 2, lane 5), RNA replication was detected by the presence of labeled RNAs corresponding to authentic PaV RNA1 and RNA3 (Fig. 2, lane 4). Additionally, a small amount of an RNA1 dimer (1, 27), seen most clearly in the vRNA sample (Fig. 2, lane 4), was noted in cells transfected with plasmids for RNA1. In cells transfected with PaV1(1,0) + PaV2(0,0) (Fig. 2, lane 2) or PaV1(2,0) + PaV2(0,0) (Fig. 2, lane 6), the RNA replication products comigrated with authentic RNA1 and RNA2. These results indicate that PaV RNA replication can be initiated in BSR-T7/5 cells from the cDNA transcription plasmids PaV1(1,0) or PaV1(2,0) in conjunction with PaV2(0,0). PaV replication initiated from these plasmids establishes an authentic pattern of RNA products, including the RNA2-mediated suppression of RNA3 synthesis (19, 46). Plasmid PaV1(1,0) was chosen for further experiments.
At least one additional G nucleotide following the resected T7 promoter was required to initiate PaV RNA replication in cells that expressed T7 RNA polymerase constitutively, but not in those that expressed the enzyme from a recombinant vaccinia virus (27). In the latter system, T7 transcripts are capped and methylated in the cytoplasm by the vaccinia virus guanylyl- and methyltransferases (18), which will enhance their translation efficiency. In contrast, primary transcripts made in the cytoplasm of BSR-T7/5 cells are presumably not capped and thus poorly translated. Consistent with this interpretation, infection of BSR-T7/5 cells with wild-type, nonrecombinant vaccinia virus rescued the replication of RNA1 from PaV1(0,0), confirming that vaccinia virus contributed to the recovery of RNA replication from transcripts of this plasmid (data not shown). We speculate that adding a G residue to the T7 promoter increased the level of primary transcripts made from PaV1(1,0), thus compensating for their lack of cap structures. Unlike RNA1, which encodes the RNA-dependent RNA polymerase, RNA2 need provide no translation product before it can replicate, and the transcripts made from PaV2(0,0) evidently sufficed to initiate RNA2 replication. In both expression systems, once RNA replication was established presumably all products would be capped and methylated by PaV-specific enzymes.
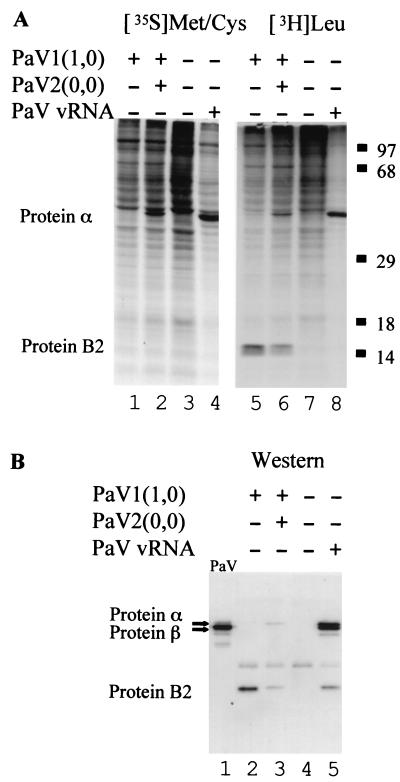
To examine PaV protein synthesis, BSR-T7/5 cells were transfected with authentic vRNA or with plasmids PaV1(1,0) + PaV2(0,0) and proteins were labeled with [35S]methionine-cysteine or [3H]leucine 48 h posttransfection. Cytoplasmic extracts were resolved by SDS-PAGE, and labeled proteins were visualized by fluorography (Fig. 3A). A protein of the size expected for protein α was labeled both in cells transfected with PaV vRNA and in cells transfected with PaV1(1,0) + PaV2(0,0) plasmids (Fig. 3A, lanes 2, 4, 6, and 8), but not in mock-transfected cells (Fig. 3A, lanes 3 and 7) or in cells that received PaV1(1,0) alone (Fig. 3A, lanes 1 and 5). As expected given the short period of labeling, little or no cleavage of protein α into the mature capsid proteins β and γ was observed. Extensive shutoff of host protein synthesis was seen in cells transfected with vRNA. In cells transfected with PaV1(1,0) alone or together with PaV2(0,0), [3H]leucine labeling revealed an additional protein with an apparent Mr of about 14 kDa (Fig. 3A, lanes 5 and 6) that was not present in the untransfected cell sample (Fig. 3A, lane 7). The identification of this as the B2 protein is described below.
FIG. 3.
Viral proteins synthesized in BSR-T7/5 cells transfected with PaV cDNA clones. (A) In vivo labeling. Cells were mock transfected (lanes 3 and 7) or transfected with 100 ng of vRNA (lanes 4 and 8), or 2.5 μg of PaV1(1,0) (lanes 1 and 5), or 2.5 μg each of PaV1(1,0) + PaV2(0,0) (lanes 2 and 6). After 48 h of incubation at 28°C, cells were preincubated for 30 min with medium lacking either methionine and cysteine (lanes 1 to 4) or leucine (lanes 5 to 8), and then proteins were metabolically labeled for 1 h with [35S]methionine-cysteine (lanes 1 to 4) or [3H]leucine (lanes 5 to 8). Cytoplasmic extracts were harvested, resolved by SDS-PAGE on 12.5% gels, and visualized by fluorography. Proteins α and B2 are indicated on the left, and the migration positions of molecular mass markers are shown on the right. (B) Western blot analysis. Samples 5 to 8 from panel A were also resolved by SDS-PAGE on a 12.5% mini-gel (lanes 2 to 5) along with 250 ng of purified wild-type PaV virus particles (lane 1). Proteins were transferred to a polyvinylidene difluoride membrane, probed with a rabbit antiserum raised against purified PaV particles, and visualized by chemiluminescence.
To confirm the identity of the major labeled protein, the samples were examined by Western blot analysis using a polyclonal rabbit antiserum raised against authentic PaV. Cytoplasmic extracts were resolved on SDS–12.5% polyacrylamide gels, transferred to membranes, and probed with antiserum against purified PaV (Fig. 3B). In purified virus particles the major protein was the mature capsid protein β, although small amounts of the capsid precursor protein α and some putative breakdown products were also present (Fig. 3B, lane 1). BSR-T7/5 cells transfected with PaV vRNA contained proteins that comigrated with both α and β (Fig. 3B, lane 5), suggesting synthesis and some cleavage of the capsid precursor protein. In comparison, cells transfected with the cDNA plasmids PaV1(1,0) + PaV2(0,0) (Fig. 3B, lane 3) contained less capsid protein precursor α and no detectable mature capsid protein β. The smaller amount of capsid proteins in this sample was consistent with both the analysis of labeled proteins (Fig. 3A) and the lower level of RNA replication compared to vRNA-transfected cells (Fig. 2).
In addition to the capsid proteins, two minor bands were detected in BSR-T7/5 cell extracts by Western blot analysis. One of these was present in all samples, including the mock-infected control, and therefore must represent a cellular protein. The other immunoreactive protein was present in all the lysates except the mock-transfected control (Fig. 3B, lanes 2, 3, and 5) and was similar in size to the 14-kDa protein detected by [3H]leucine labeling (Fig. 3A). This protein was RNA1 related and corresponded in size to either of the two overlapping open reading frames (ORFs) in PaV RNA3 (26) that are analogous to the ORFs that encode the B1 and B2 proteins in FHV RNA3 (16, 22, 23). Additionally, the 14-kDa protein was more abundant in the absence of PaV2(0,0), suggesting that it was translated from RNA3, which is inhibited by RNA2 (Fig. 2). To determine whether the 14-kDa protein was the product of one of these ORFs, point mutations were introduced into PaV1(1,0) to disrupt the initiation codons of either the B1 or the B2 ORF. The first AUG of the B1 ORF was mutated to GUG, which changed the initiating methionine of B1 to a valine without altering the amino acid sequence of the B2 ORF. Because B1 is in the same reading frame as protein A, the latter was also mutated at this position (M880V). The first AUG of the B2 ORF was mutated to ACG, which changed the B2-initiating methionine to threonine without altering the protein A or B1 amino acid sequences. The proteins synthesized in BSR-T7/5 cells transfected with the mutant plasmids were analyzed by [3H]leucine labeling and Western blot analysis, and the results identified the 14-kDa protein as a product of the B2 ORF (data not shown).
Reactivity of the PaV antiserum with the B2 protein was unexpected, as B2 is a nonstructural protein that is not normally present in purified virus preparations (Fig. 3B, lane 1). We prepared a second antiserum by inoculation of a rabbit with gradient-purified, clonally derived PaV (see Fig. 5, lane 5). This antiserum also detected B2 protein in the extracts of vRNA-transfected BSR-T7/5 cells shown in Fig. 3B. Since both antisera were polyclonal, we cannot exclude the possibility that both of the purified virus samples used as antigens contained undetectable levels of B2 protein. However, it seems more likely that some virus replication and concomitant B2 protein synthesis occurred in each of the rabbits in which the antisera were raised. The host-range of PaV has not been examined, and another alphanodavirus, Nodamura virus, is able to replicate in some mammals (5); thus, it is possible that PaV replicated in the immunized rabbit. In these experiments we found no evidence for expression of the B1 ORF, which in PaV RNA3, unlike the situation in FHV, begins downstream of the B2 ORF. However, the predicted amino acid sequence of the B1 ORF contains only one methionine, no cysteines, and one leucine, so it is possible that synthesis of B1 protein would not have been detected by using the methods described here.
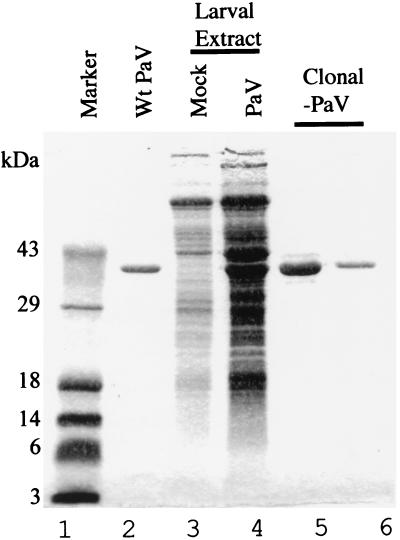
FIG. 5.
Protein composition of clonally derived virus purified from G. mellonella larvae. Larvae were injected with lysates of BSR-T7/5 cells transfected with plasmids PaV1(1,0) + PaV2(0,0) or mock transfected. After 8 days of incubation at 31°C, partially purified virus was amplified by a second passage in larvae and clonally derived PaV was purified from these larvae by sedimentation through a sucrose gradient. Samples were resolved on an SDS–12.5% polyacrylamide gel and visualized by staining with Coomassie blue. Molecular mass markers are shown on the left (lane 1). Shown are authentic PaV (lane 2; 1.5 μg); partially purified samples from larvae injected with lysates of mock-transfected BSR-T7/5 cells (lane 3) or PaV1(1,0) + PaV2(0,0)-transfected BSR-T7/5 cells (lane 4); clonally-derived, sucrose gradient-purified PaV (lanes 5 and 6; 5 and 1 μg respectively).
Recovery of infectious virus from BSR-T7/5 cells.
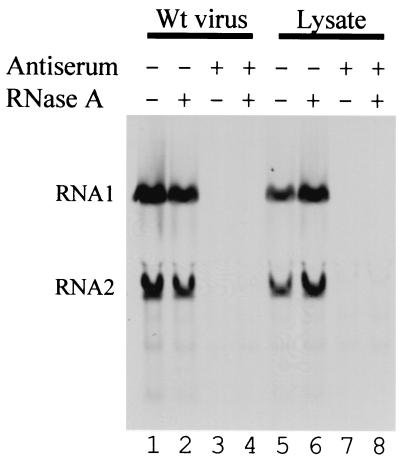
To test whether the cDNA clones yielded infectious virus, lysates were prepared from BSR-T7/5 cells that had been transfected with PaV1(1,0) + PaV2(0,0). An aliquot of the lysate and a control sample containing 109 purified PaV particles were treated with anti-PaV antiserum, with RNase A, or both. FB33 cells were infected with the treated and mock-treated samples, and 24 h postinfection RNAs were metabolically labeled with [3H]uridine, resolved on denaturing agarose gels, and visualized by fluorography (Fig. 4). RNA replication was evident in cells infected with untreated PaV particles and with the untreated lysate from BSR-T7/5 cells that had been transfected with PaV1(1,0) + PaV2(0,0) (Fig. 4, lanes 1 and 5). RNase A treatment did not affect this infectivity (Fig. 4, lanes 2 and 6). In contrast, treatment with anti-PaV antiserum decreased the infectivity of both authentic and cDNA-derived PaV to below the limits of detection (Fig. 4, lanes 3 and 7). These results demonstrate that infectious PaV was generated in BSR-T7/5 cells following transfection with plasmids that contained cDNA copies of PaV RNA1 and RNA2.
FIG. 4.
Neutralization of authentic and clonally derived PaV. BSR-T7/5 cells were transfected with 2.5 μg of PaV1(1,0) + PaV2(0,0) and incubated at 28°C for 48 h before the cells were washed and lysed by freezing and thawing. These lysates (lanes 5 to 8) and purified wild-type PaV particles (lanes 1 to 4) were incubated with RNase A (lanes 2 and 6), PaV antiserum (lanes 3 and 7), or both RNase A and PaV antiserum (lanes 4 and 8), or mock-treated with PBSM (lanes 1 and 5). The treated samples were used to infect FB33 cells. After 22 h of incubation at 28°C, actinomycin D was added at 20 μg/ml, and 30 min later replicating RNAs were metabolically labeled by incorporation of [3H]uridine (20 μCi/ml) for a period of 2 h before total cellular RNAs were harvested. RNAs were resolved by electrophoresis on a 1% agarose-formaldehyde gel and visualized by fluorography. PaV RNA1 and RNA2 are indicated on the left.
Growth of PaV in G. mellonella larvae.
In order to prepare a large stock of clonally derived virus, a lysate of BSR-T7/5 cells transfected with PaV1(1,0) + PaV2(0,0) was injected into G. mellonella larvae. Following 8 days of incubation at 31°C, the nonpupated larvae (approximately 70%) were collected and frozen at −20°C. Cadavers were homogenized in 50 mM Tris (pH 7.4), and the clarified homogenates were pelleted through a 30% sucrose cushion and resuspended in 50 mM Tris (pH 7.4). The resuspended pellets were subjected to SDS-PAGE, and the proteins were stained with Coomassie brilliant blue (Fig. 5). Two major proteins were visible in extracts from infected larvae (Fig. 5, lane 4) that were not present in uninfected samples (Fig. 5, lane 3). One of these bands comigrated with the major capsid protein from purified wild-type PaV (Fig. 5, lane 2), and the other was about 4 kDa larger, which is the size expected for the PaV capsid protein precursor α. As described previously (27), the small capsid protein γ was not visualized by Coomassie blue staining. These results indicated that BSR-T7/5 cells transfected with PaV1(1,0) + PaV2(0,0) generated PaV particles that were infectious for G. mellonella larvae.
To amplify the clonally derived virus, the larval extracts described above were repassaged in G. mellonella larvae, and after 8 days of incubation at 31°C the virus was purified as described in Materials and Methods. Seven milligrams of recombinant virus was purified from 92 G. mellonella larvae. The purified clonally derived virus was examined by SDS-PAGE and found to contain a single major protein that comigrated with the major capsid protein of authentic wild-type PaV (Fig. 5, lanes 5 and 6). Furthermore, at equivalent MOIs (similar to those shown in Fig. 1), the recombinant and wild-type viruses produced similar patterns of labeled RNAs in infected FB33 cells (data not shown).
In summary, we have described a cell-based infectivity assay for PaV and used it to recover infectious virus from cDNA clones. Although infected FB33 cells produced only modest amounts of virus, amplification of the clonally derived virus in G. mellonella larvae provided quantities of virus which have previously been sufficient for structural examination. Previous studies of nodavirus assembly have focused on FHV partly because of the availability of susceptible cell lines and infectious cDNA clones (25, 37). The work described above makes PaV only the second nodavirus for which these experimental tools are available and provides methods to investigate the mechanisms of RNA encapsidation and virion assembly. These studies will be guided by the high-resolution structure of PaV, which revealed many contacts between the capsid proteins and the genomic RNAs of this virus.
ACKNOWLEDGMENTS
We thank Jean-Louis Zeddam (Station de recherches de pathologie comparée, INRA-CNRS, 30380 Saint-Christol-les-Alès) for the gift of PaV antiserum, K. Conzelmann and M. Schnell (Max von Pettenkofer Institute & Gene Center, Ludwig-Maximilians-University Munich, D-81377 Munich, Germany) for BSR-T7/5 cells, and Art McIntosh and Cindy Goodman (USDA, Agricultural Research Service, Biological Control of Insects Research Laboratory, Columbia, Mo.) for providing the following cell lines: Se-1, Ae-59, ATC-15, FB33, FB27 and MG8. We thank members of the A. Ball and Gail Wertz laboratories for discussions and critical reading of the manuscript.
This work was supported by NIH grant R01AI18270.
REFERENCES
- 1.Albariño, C. G., B. D. Price, L. D. Eckerle, and L. A. Ball. Characterization and template properties of RNA dimers generated during Flock House Virus RNA replication. Virology, in press. [DOI] [PubMed]
- 2.Ball L A. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J Virol. 1992;66:2335–2345. doi: 10.1128/jvi.66.4.2335-2345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball L A. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball L A, Hendry D A, Johnson J E, Rueckert R R, Scotti P D. Nodaviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, McGeoch D J, Maniloff J, Mayo M A, Pringle C R, Wickner R B, editors. Virus taxonomy, 7th Report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 2000. pp. 747–755. [Google Scholar]
- 5.Ball L A, Johnson K L. Nodaviruses of insects. In: Miller L K, Ball L A, editors. The insect viruses. New York, N.Y: Plenum Publishing Corporation; 1998. pp. 225–267. [Google Scholar]
- 6.Ball L A, Johnson K L. Reverse genetics of nodaviruses. Adv Virus Res. 1999;53:229–244. doi: 10.1016/s0065-3527(08)60350-4. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz U J, Finke S, Conzelmann K K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng R H, Reddy V S, Olson N H, Fisher A J, Baker T S, Johnson J E. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure. 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasgupta R, Ghosh A, Dasmahapatra B, Guarino L A, Kaesberg P. Primary and secondary structure of black beetle virus RNA2, the genomic messenger for BBV coat protein precursor. Nucleic Acids Res. 1984;12:7215–7223. doi: 10.1093/nar/12.18.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasmahapatra B, Dasgupta R, Ghosh A, Kaesberg P. Structure of the black beetle virus genome and its functional implications. J Mol Biol. 1985;182:183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasmahapatra B, Dasgupta R, Saunders K, Selling B, Gallagher T, Kaesberg P. Infectious RNA derived from transcription from cloned cDNA copies of the genomic RNA of an insect virus. Proc Natl Acad Sci USA. 1986;83:63–66. doi: 10.1073/pnas.83.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dearing S C, Scotti P D, Wigley P J, Dhana S D. A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae) N Z J Zool. 1980;7:267–269. [Google Scholar]
- 13.Dong X F, Natarajan P, Tihova M, Johnson J E, Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasi-equivalent icosahedral virus. J Virol. 1998;72:6024–6033. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher A J, Johnson J E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature (London) 1993;361:176–182. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 15.Friesen P, Scotti P, Longworth J, Rueckert R. Black beetle virus: propagation in Drosophila line 1 cells and an infection-resistant subline carrying endogenous black beetle virus-related particles. J Virol. 1980;35:741–747. doi: 10.1128/jvi.35.3.741-747.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen P D, Rueckert R R. Black beetle virus: messenger RNA for protein B is a subgenomic viral RNA. J Virol. 1982;42:986–995. doi: 10.1128/jvi.42.3.986-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesen P D, Rueckert R R. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher T M, Friesen P D, Rueckert R R. Autonomous replication and expression of RNA1 from black beetle virus. J Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher T M, Rueckert R R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelernter W D, Federici B A. Continuous cell line from Spodoptera exigua (Lepidoptera: Noctuidae) that supports replication of nuclear polyhedrosis viruses from Spodoptera exigua and Autographa californica. J Invertebr Pathol. 1986;48:199–207. [Google Scholar]
- 22.Guarino L A, Ghosh A, Dasmahapatra B, Dasgupta R, Kaesberg P. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology. 1984;139:199–203. doi: 10.1016/0042-6822(84)90342-8. [DOI] [PubMed] [Google Scholar]
- 23.Harper T A. Characterization of the proteins encoded from the nodaviral subgenomic RNA. Ph.D. Thesis. University of Wisconsin—Madison; 1994. [Google Scholar]
- 24.Hosur M V, Schmidt T, Tucker R C, Johnson J E, Gallagher T M, Selling B H, Rueckert R R. Structure of an insect virus at 3.0 angstrom resolution. Proteins: Struct. Funct Genet. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- 25.Johnson J E, Reddy V. Structural studies of nodaviruses and tetraviruses. In: Miller L K, Ball L A, editors. The insect viruses. New York, N.Y: Plenum Publishing Corporation; 1998. pp. 171–223. [Google Scholar]
- 26.Johnson K N, Johnson K L, Dasgupta R, Gratsch T, Ball L A. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J Gen Virol. 2001;82:1855–1866. doi: 10.1099/0022-1317-82-8-1855. [DOI] [PubMed] [Google Scholar]
- 27.Johnson K N, Zeddam J, Ball L A. Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia. J Virol. 2000;74:5123–5132. doi: 10.1128/jvi.74.11.5123-5132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaesberg P, Dasgupta R, Sgro J-Y, Wery J-P, Selling B H, Hosur M V, Johnson J E. Structural homology among four nodaviruses as deduced by sequencing and X-ray crystallography. J Mol Biol. 1990;214:423–435. doi: 10.1016/0022-2836(90)90191-N. [DOI] [PubMed] [Google Scholar]
- 29.Kariuki C W, McIntosh A H, Goodman C L. In vitro host range studies with a new baculovirus isolate from the diamondback moth Plutella xylostella (L.) (Plutellidae: Lepidoptera) In Vitro Cell Dev Biol. 2000;36:271–276. doi: 10.1290/1071-2690(2000)036<0271:IVHRSW>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 30.Krishna N K, Schneemann A. Formation of an RNA heterodimer upon heating of nodavirus particles. J Virol. 1999;73:1699–1703. doi: 10.1128/jvi.73.2.1699-1703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Laskey R A, Mills A D. Quantitative film detection of 3H and 14C polyacrylamide gels by fluorography. Eur J Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 33.Ling M L, Risman S S, Klement J F, McGraw N, McAllister W T. Abortive initiation by bacteriophage T3 and T7 RNA polymerases under conditions of limiting substrate. Nucleic Acids Res. 1989;17:1605–1618. doi: 10.1093/nar/17.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peleg J, Shahar A. Morphology and behaviour of cultured Aedes aegypti mosquito cells. Tissue Cell. 1972;4:55–62. doi: 10.1016/s0040-8166(72)80006-5. [DOI] [PubMed] [Google Scholar]
- 35.Reinganum C, Bashiruddin J B, Cross G F. Boolarra virus: a member of the Nodaviridae isolated from Oncopera intricoides (Lepidoptera: Hepialidae) Intervirology. 1985;24:10–17. doi: 10.1159/000149613. [DOI] [PubMed] [Google Scholar]
- 36.Schneemann A, Marshall D. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J Virol. 1998;72:8738–8746. doi: 10.1128/jvi.72.11.8738-8746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneemann A, Reddy V, Johnson J E. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 38.Schneemann A, Zhong W, Gallagher T M, Rueckert R R. Maturation cleavage required for infectivity of a nodavirus. J Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morph. 1972;27:353–365. [PubMed] [Google Scholar]
- 40.Scotti P D, Dearing S, Mossop D W. Flock house virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae) Arch Virol. 1983;75:181–189. doi: 10.1007/BF01315272. [DOI] [PubMed] [Google Scholar]
- 41.Selling B H, Rueckert R R. Plaque assay for black beetle virus. J Virol. 1984;51:251–253. doi: 10.1128/jvi.51.1.251-253.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh K R. Propagation of arboviruses in Singh's Aedes cell lines. I. Growth of arboviruses in Aedes albopictus and A. aegypti cell lines. Curr Top Microbiol Immunol. 1971;55:127–133. doi: 10.1007/978-3-642-65224-0_20. [DOI] [PubMed] [Google Scholar]
- 43.Tang L, Johnson K N, Ball L A, Lin T, Yeager M, Johnson J E. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat Struct Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 44.Vaughn J L, Goodwin R H, Tompkins G J, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 45.Zeddam J L, Rodriguez J L, Ravallec M, Lagnaoui A. A noda-like virus isolated from the sweetpotato pest Spodoptera eridania (Cramer) (Lep.; Noctuidae) J Invertebr Pathol. 1999;74:267–274. doi: 10.1006/jipa.1999.4881. [DOI] [PubMed] [Google Scholar]
- 46.Zhong W D, Rueckert R R. Flock House Virus-Down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J Virol. 1993;67:2716–2722. doi: 10.1128/jvi.67.5.2716-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]