Abstract
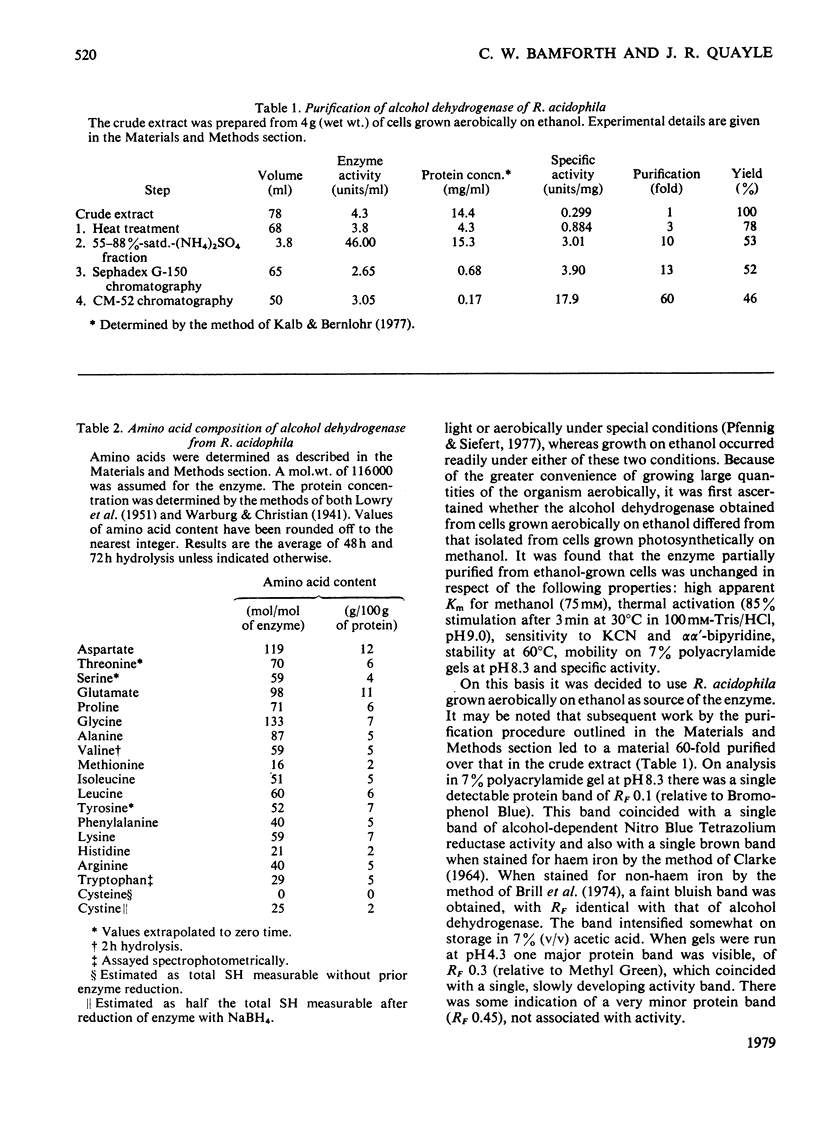
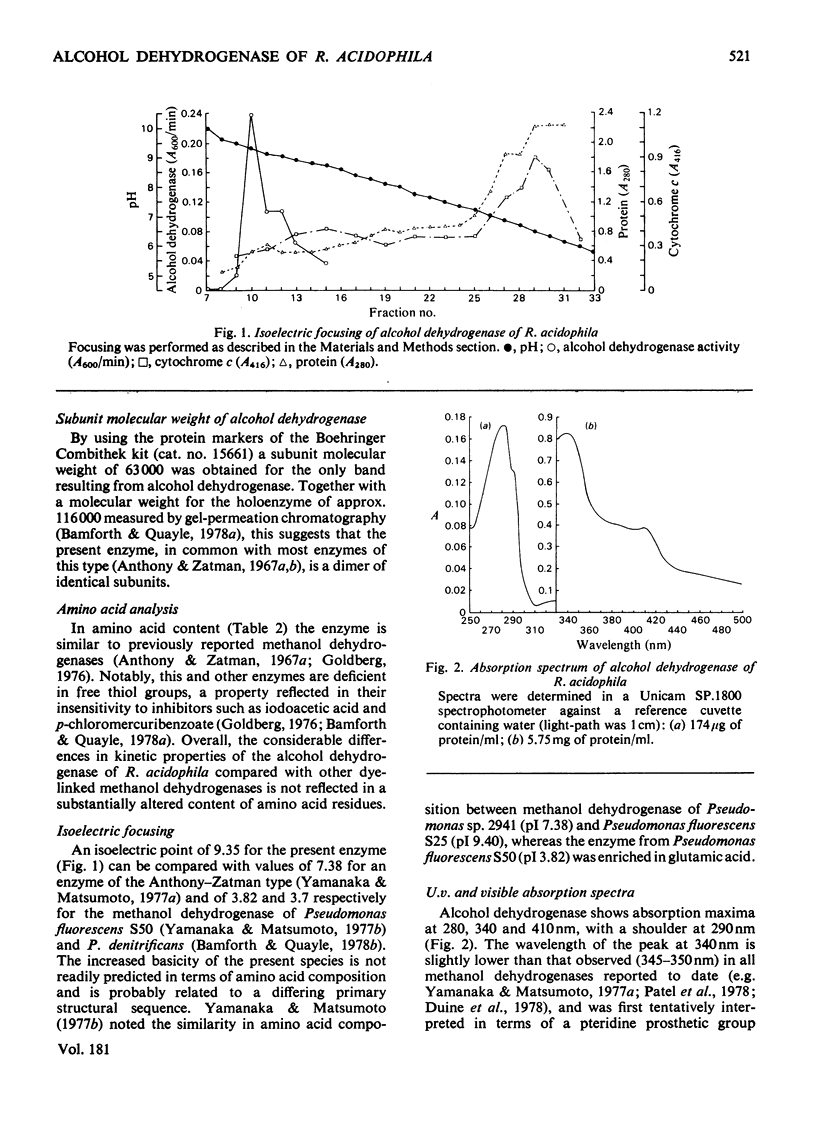
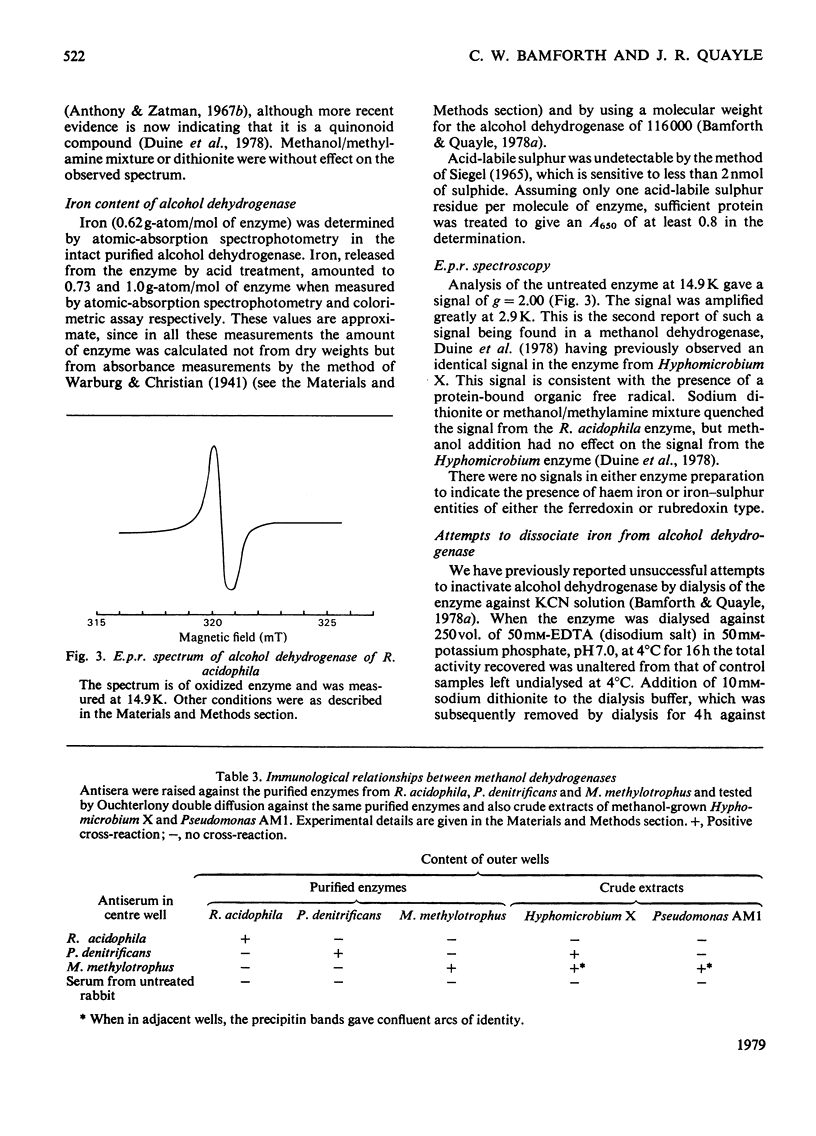
1. A dye-linked alcohol dehydrogenase was purified 60-fold from extracts of Rhodopseudomonas acidophila 10050 grown aerobically on ethanol. 2. The properties of this enzyme were identical with those of the alcohol dehydrogenase synthesized by this organism during growth on methanol anaerobically in the light, and they are judged to be the same enzyme. 3. The enzyme gave a single protein band, coincident with alcohol dehydrogenase activity, during electrophoresis on polyacrylamide gel. 4. The amino acid composition, ioselectric point, u.v. and visible absorption spectra of the enzyme were determined and compared with those of other similar enzymes. 5. The presence of 0.7--1.0 g-atom of non-haem, acidlabile iron/mol of enzyme was shown by atomic absorption spectrophotometry and colorimetric assay. The iron could not be dissociated from the enzyme by dialysis against chelating agents. 6. E.p.r. spectroscopy of the enzyme did not indicate any redox function for the iron during alcohol dehydrogenation, but showed a signal at g = 2.00 consistent with the presence of a protein-bound organic free radical. 8. Antisera were raised against alcohol (methanol) dehydrogenases purified from Rhodopseudomonas acidophila, Paracoccus denitrificans and Methylophilus methylotrophus. 9. The antiserum to the Rhodopseudomonas acidophila enzyme cross-reacted with neither of the two other antisera, nor with crude extracts of methanol-grown Hyphomicrobium X and Pseudomonas AM1, thus emphasizing its singular biochemical properties.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony C., Zatman L. J. The microbial oxidation of methanol. Purification and properties of the alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J. 1967 Sep;104(3):953–959. doi: 10.1042/bj1040953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J. 1967 Sep;104(3):960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin G. E., Ferdinand W. Accelerated amino acid analysis with lithium citrate buffers. The effect of resin cross-linkage on elution patterns and the avoidance of microbial contamination of columns. J Chromatogr. 1971 Nov 26;62(3):373–381. doi: 10.1016/s0021-9673(00)91388-8. [DOI] [PubMed] [Google Scholar]
- Atkin G. E., Ferdinand W. Accelerated amino acid analysis: studies on the use of lithium citrate buffers and the effect of n-propanol, in the analysis of physiological fluids and protein hydrolyzates. Anal Biochem. 1970 Dec;38(2):313–329. doi: 10.1016/0003-2697(70)90456-2. [DOI] [PubMed] [Google Scholar]
- BARD R. C., GUNSALUS I. C. Glucose metabolism of Clostridium perfringens: existence of metallo-aldolase. J Bacteriol. 1950 Mar;59(3):387–400. doi: 10.1128/jb.59.3.387-400.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEAVEN G. H., HOLIDAY E. R. Ultraviolet absorption spectra of proteins and amino acids. Adv Protein Chem. 1952;7:319–386. doi: 10.1016/s0065-3233(08)60022-4. [DOI] [PubMed] [Google Scholar]
- Bamforth C. W., Large P. J. The molecular size of N-methylglutamate dehydrogenase of Pseudomonas aminovorans. Biochem J. 1977 Nov 1;167(2):509–512. doi: 10.1042/bj1670509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth C. W., Quayle J. R. Aerobic and anaerobic growth of Paracoccus denitrificans on methanol. Arch Microbiol. 1978 Oct 4;119(1):91–97. doi: 10.1007/BF00407934. [DOI] [PubMed] [Google Scholar]
- Bamforth C. W., Quayle J. R. The dye-linked alcohol dehydrogenase of Rhodopseudomonas acidophila. Comparison with dye-linked methanol dehydrogenases. Biochem J. 1978 Mar 1;169(3):677–686. doi: 10.1042/bj1690677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakesley R. W., Boezi J. A. A new staining technique for proteins in polyacrylamide gels using coomassie brilliant blue G250. Anal Biochem. 1977 Oct;82(2):580–582. doi: 10.1016/0003-2697(77)90197-x. [DOI] [PubMed] [Google Scholar]
- Brill W. J., Westphal J., Stieghorst M., Davis L. C., Shah V. K. Detection of nitrogenase components and other nonheme iron proteins in polyacrylamide gels. Anal Biochem. 1974 Jul;60(1):237–241. doi: 10.1016/0003-2697(74)90149-3. [DOI] [PubMed] [Google Scholar]
- CLARKE J. T. SIMPLIFIED "DISC" (POLYACRYLAMIDE GEL) ELECTROPHORESIS. Ann N Y Acad Sci. 1964 Dec 28;121:428–436. doi: 10.1111/j.1749-6632.1964.tb14214.x. [DOI] [PubMed] [Google Scholar]
- Cavallini D., Graziani M. T., Dupré S. Determination of disulphide groups in proteins. Nature. 1966 Oct 15;212(5059):294–295. doi: 10.1038/212294a0. [DOI] [PubMed] [Google Scholar]
- Cox R. B., Quayle J. R. Synthesis and hydrolysis of malyl-coenzyme A by Pseudomonas AM1: an apparent malate synthase activity. J Gen Microbiol. 1976 Jul;95(1):121–133. doi: 10.1099/00221287-95-1-121. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Westerling J. Purification and properties of methanol dehydrogenase from Hyphomicrobium x. Biochim Biophys Acta. 1978 Jun 9;524(2):277–287. doi: 10.1016/0005-2744(78)90164-x. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Jarman T. R., Large P. J. Microbial oxidation of amines. Partial purification of a mixed-function secondary-amine oxidase system from Pseudomonas aminovorans that contains an enzymically active cytochrome-P-420-type haemoprotein. Biochem J. 1971 Nov;125(2):449–459. doi: 10.1042/bj1250449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg I. Purification and properties of a methanol-oxidizing enzyme in Pseudomonas C. Eur J Biochem. 1976 Mar 16;63(1):233–240. doi: 10.1111/j.1432-1033.1976.tb10225.x. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Patel R. N., Hou C. T., Felix A. Microbial oxidation of methane and methanol: crystallization of methanol dehydrogenase and properties of holo- and apomethanol dehydrogenase from Methylomonas methanica. J Bacteriol. 1978 Feb;133(2):641–649. doi: 10.1128/jb.133.2.641-649.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle J. R., Pfennig N. Utilization of methanol by rhodospirillaceae. Arch Microbiol. 1975 Mar 10;102(3):193–198. doi: 10.1007/BF00428368. [DOI] [PubMed] [Google Scholar]
- REISFELD R. A., LEWIS U. J., WILLIAMS D. E. Disk electrophoresis of basic proteins and peptides on polyacrylamide gels. Nature. 1962 Jul 21;195:281–283. doi: 10.1038/195281a0. [DOI] [PubMed] [Google Scholar]
- Ruettinger R. T., Griffith G. R., Coon M. J. Characterization of the omega-hydroxylase of Pseudomonas oleovorans as a nonheme iron protein. Arch Biochem Biophys. 1977 Oct;183(2):528–537. doi: 10.1016/0003-9861(77)90388-5. [DOI] [PubMed] [Google Scholar]
- SIEGEL L. M. A DIRECT MICRODETERMINATION FOR SULFIDE. Anal Biochem. 1965 Apr;11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- Sahm H., Cox R. B., Quayle J. R. Metabolism of methanol by Rhodopseudomonas acidophila. J Gen Microbiol. 1976 Jun;94(2):313–322. doi: 10.1099/00221287-94-2-313. [DOI] [PubMed] [Google Scholar]
- Wolf H. J., Hanson R. S. Alcohol dehydrogenase from Methylobacterium organophilum. Appl Environ Microbiol. 1978 Jul;36(1):105–114. doi: 10.1128/aem.36.1.105-114.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


