Abstract
Accurate knowledge of species distributions is foundational for effective conservation efforts. Bats are a diverse group of mammals, with important roles in ecosystem functioning. However, our understanding of bats and their ecological importance is hindered by poorly defined ranges, mostly as a result of under-recording. This issue is exacerbated in Africa by the ongoing rapid discovery of new species, both de novo and splits of existing species, and by inaccessibility to museum specimens that are mostly hosted outside of the continent. Here we present the African bat database – a curated set of 17,285 unique locality records of all 266 species of bats from sub-Saharan Africa, vouched for by specimens and/or genetic sequencing, and aligned with current taxonomy. Based on these records, we also present Maxent-based distribution models and calculate the IUCN Red List metrics for Extent of Occurrence and Area of Occupancy. This database and online visualization tool provide an important open-source resource and is expected to significantly advance studies in ecology, and aid in bat conservation.
Subject terms: Biodiversity, Macroecology, Taxonomy, Conservation biology
Background & Summary
Accurate taxonomic assessments and a thorough understanding of species distributions provide the foundation for studying our environment across time and space1. Species occurrence data are often used to test hypotheses in evolution and ecology, with important applications in assessing the vulnerability of species and ecosystems in changing environments, e.g., through species distribution modelling and IUCN Red List assessments2,3. However, these efforts are hindered by the fact that many mammal species remain unknown, under-sampled and under-reported, especially in tropical regions such as Africa4,5. Furthermore, many researchers have highlighted the underrepresentation of sub-Saharan Africa, in particular in global biodiversity research6,7.
Approximately a quarter of mammalian biodiversity is comprised of bats, with 1,482 species currently recognized globally, of which over a fifth occur in Africa8,9. Bats provide essential ecosystem functions such as pollination, seed dispersal and insect pest suppression10–12. Despite their importance, bats remain poorly documented, especially in Africa13,14. This is partly because voucher specimens are not easily accessible for evaluation by African biologists, as most specimens of African bats are housed in European and North American museums15. An additional challenge is the need for continual updating of species names in museum collections or databases, arising from the rapid description of new bat species and revision of systematic relationships, driven by advances in genetic and acoustic technologies, and increased sampling16–21 (Fig. 1). This complicates our understanding of which species are represented in past records, and highlights the shortcomings of existing global datasets of mammalian distributions22 for local and regional studies.
Fig. 1.
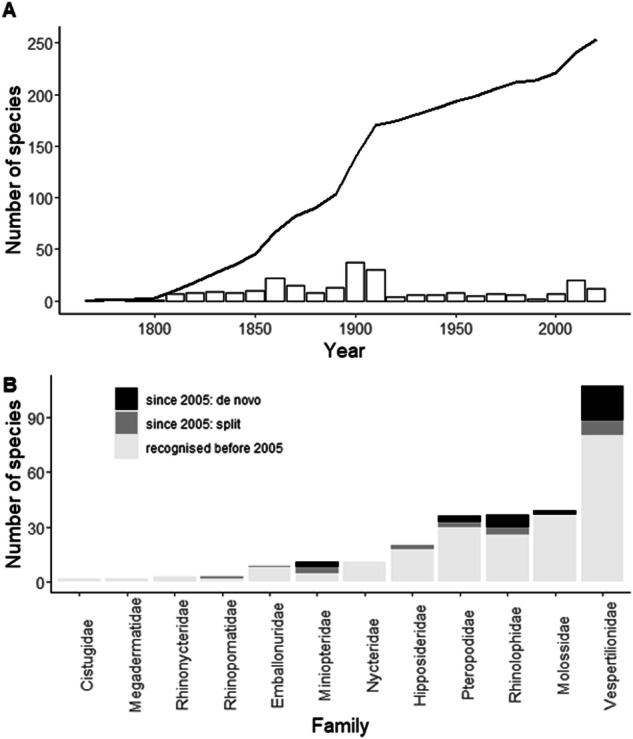
Rapid increase of described bat species in sub-Saharan Africa: (A) cumulative increase in the number of new species over time (solid line), and the number of new species per decade (bar plot); (B) the number of species recognized per family, categorized into those recognized before 2005, de novo species recognized since 2005, and those split from existing species since 2005, based on the Mammal Diversity Database9.
The lack of accurate distribution maps for many bat species complicates their conservation23, as knowing where a species occurs is essential for informing where it may be best protected. Robust occurrence records are necessary for calculating crucial conservation metrics, such as the extent of occurrence (EOO) and the area of occupancy (AOO) of a species24. These are key parameters in assessments undertaken by the International Union for Conservation of Nature (IUCN) Red List of Threatened Species to evaluate the conservation status and risk of extinction of a species. Distribution records are also needed for accurate species distribution modelling, increasingly used to present the ranges of animals, and applied to their conservation25. Importantly, these applications depend on accurate records, as poor-quality input data can result in misleading metrics and distribution models, which in turn can significantly impact conservation planning, environmental impact assessments, and subsequent mitigation strategies26.
The proportion of species facing extinction continues to grow, with over a quarter of mammals currently threatened and a third of bats defined as threatened or data deficient by the IUCN27. As the number of species facing elevated extinction risk rises each year28, it becomes increasingly important to accurately assess each species’ conservation status. This underscores the need for comprehensive distributional knowledge of each species. Without this, we cannot compare current distributions with those of the past and/or predicted future, to detect declines and changes that provide warnings of how different drivers are altering the structure and functioning of biodiversity over time29–31.
Here, we present a comprehensive database of bat occurrence records for species in sub-Saharan Africa, with each record verified by voucher specimens and/or genetic data32. This database builds on the 6,042 records of southern and central African bats in Monadjem et al.13, to which we have added over 11,200 additional records. We present unique locality records and models of predicted suitable habitat for each species of bat known from this region. We then calculate two distributional metrics that can be used in future Red List assessments for African bats as they become outdated and for species still requiring initial assessments. Our aim is to facilitate and expedite this process not only with the newly calculated metrics but also by presenting a standardised, replicable method for calculating these metrics in the future. Furthermore, this database complements the recently published African bat traits (AfroBaT) dataset33. The data are released as open source and presented via an R Shiny app tool that allows easy visualization of the distribution records of any selected species, all made available for use by the wider research community.
Methods
Taxonomic and geographic coverage
Our dataset includes occurrence records of 266 bat species from 53 genera and 12 families (order Chiroptera, class Mammalia) from sub-Saharan Africa. We define the region as all the countries of Africa partially or completely south of the Sahara, excluding oceanic islands such as Madagascar, the Comoros, and Seychelles34, but including Bioko, Zanzibar and Mafia islands, as these lie close to the main continent and have shared bat communities with the mainland35,36. Although our database only includes species that occur in sub-Saharan Africa, we include records for these species from across the continent, including North Africa (north of the Sahara). We include the year of collection for 10,960 records (63%), with most records collected in the decades since 1970 (Figure S1). We included records up until 28 April 2024. We follow the taxonomy of Simmons & Cirranello8, and use it along with the Mammal Diversity database9, to quantify the number of species in sub-Saharan Africa over time, and the number of species now recognized by family (Fig. 1). The African bat database includes 16 taxa for which a specific species identity cannot be claimed. These species are included in the database with species epithets beginning with “cf.” followed by the name of the taxon closest in morphology to it, but from which it is distinct. The inclusion of these unnamed species allows for the recognition of morphologically similar taxa that require further investigation for definitive identification and aims to enhance the comprehensiveness of the dataset while acknowledging taxonomic uncertainties.
Data categories
All records are provided as individual-level data, with taxonomic information including family, genus and species. Each record is accompanied by a location (with latitude and longitude coordinates in decimal degrees), name of the country and location, and data source or reference. Other columns in the database include: i) date of collection (41% of records); and ii) museum or collection number (66% of the records have a museum assigned to them). We indicate the specimens that we have inspected ourselves (30%), and we also indicate whether the specimen is a holotype (247 specimens) for currently recognized species only. We attempted to apply the most recent nomenclature to each record, which we were able to do for most specimens. However, in a few cases this was not possible, and we referred to them as cf. species (16 taxa). These fell into two groups. On the one hand, these include taxa with deeply divergent lineages whose taxonomy has not yet been resolved, such as Hipposideros cf. ruber37,38 and Miniopterus cf. inflatus39. On the other hand, these include specimens that we were unable to inspect and for which no genetics existed. Furthermore, these latter specimens were far beyond the known range of the species. We could have removed these records, but we felt that keeping this data might be useful for certain future analyses.
Data sources
The African bat database was meticulously compiled by considering only records with confirmed species identification that could be interrogated against current taxonomic knowledge. All specimens included in this database are vouched for by one of the following: a museum specimen, a genetically sequenced tissue sample, or a photograph. A huge effort was undertaken to catalogue museum specimens, which were examined by the authors (over 5,100 specimens), alongside other published literature records of museum specimens, as well as unpublished data of the authors (300 records). Records of bats that were captured and released were included only when tissue samples were taken and genetically sequenced to confirm species. Photographs were used very scarcely to confirm ID and only in cases where the identification was unquestionable (e.g., for unmistakable species like Myotis welwitschii, Taphozous mauritianus or Glauconycteris variegata).
Species distribution models
The theory and practice of species distribution models (SDMs) have advanced rapidly in the past two decades, with numerous approaches developed in recent years25,40. However, Maxent has remained consistently popular and robust for species distribution modelling and can be easily implemented in R software41,42. Furthermore, Maxent models have performed well even when compared with ensemble modeling43, and have been recommended as the method of choice for certain situations, particularly when the data are presence-only44. As Maxent has been used previously to develop SDMs for African bats45,46, we chose to keep the methodology consistent for this database.
We modelled the predicted suitable habitat space of each bat species that had seven or more locality records within Africa under present climatic conditions using Maxent41. Models were run at a resolution of 2.5 arc min (approximately 5 km) using BIOCLIM variables (Table S1) and elevation from the WorldClim database47. To deal with multicollinearity between variables, we used the Variance Inflation Factor (VIF) to select a set of uncorrelated environmental variables34. We calculated the VIF for the 20 variables and followed a backward model selection process to determine the minimum adequate model removing the variables with the highest VIF one at a time, until we were left with 10 variables that had VIF < 10: elevation, BIO02, BIO03, BIO08, BIO09, BIO13, BIO14, BIO15, BIO18 and BIO19 (see Table S1 for definitions).
We ran Maxent models in R version 4.1.248 using the package dismo42, following the procedure outlined in Monadjem et al.49. We used hinged and categorical variables that smooth variable responses and generally improve model performance50,51. We randomly divided bat species occurrence records from southern Africa into training (75%) and test (25%) datasets. The selection of the geographical background has important implications for the results of species distribution models52,53; a suitable background reflects the geographical space available to the species by dispersal54. Therefore, for each species, we randomly sampled 10,000 background points from 100 km circular buffers around all occurrence points for that species. We used the value of 100 km because this is the distance that Nycteris thebaica (a particularly sedentary, clutter-foraging bat species) is able to cover during dispersal55, and therefore, this buffered range represents the minimum area available to any of the species we included in our analyses51. We tested each model with the area under the receiver operating characteristic curve (AUC) statistic, which ranges from 0 to 1 with higher values signifying a better fit51. Values equal to or less than 0.5 indicate models no better than random, while values greater than 0.75 represent good model fit56. We used the same 12 environmental variables (Table S1) and Maxent parameters for all species45. We converted the predicted model outputs from Maxent (i.e., probabilities of suitability) into “presence-absence” maps using species-specific thresholds that maximized the sum of sensitivity and specificity, which is appropriate for presence-only data57.
Calculating EOO and AOO
Extent of occurrence (EOO) is defined as “the area contained within the continuous imaginary boundary which can be drawn to encompass all the known, inferred or projected sites of present occurrence of a taxon, excluding cases of vagrancy”58, and is calculated as the minimum convex polygon around all the known localities of a species24,59. We employed the same procedure to calculate EOO of African bat species, excluding any species with less than 3 locality records. For each species, we calculated a minimum convex polygon using the function mcp() in the package changeRangeR60.
Area of occupancy (AOO), in contrast to EOO, “represents the area of suitable habitat currently occupied by the taxon”58, and can be calculated by measuring the 2 × 2 km cells known or predicted to be occupied by the species59. To calculate AOO, we clipped the predicted species distribution models (i.e., outputs from the Maxent analysis described above) with the EOO so as to avoid including areas outside the known range of the species61. Finally, we also calculated minimum and maximum elevation from the predicted Maxent model (clipped to AOO), based on the BIOCLIM digital elevation model (see above). For species without SDMs (i.e., species with less than seven occurrence records, see above), we calculated AOO by multiplying the number of localities by 4 (2 × 2) km2.
Data Records
The African bat database and SDMs are available on figshare (https://figshare.com/articles/dataset/African_Bat_Database/26363308)32. The distribution of each bat species, based on our database, can be visualized through the R Shiny App (https://adam-kane.shinyapps.io/african_bat_dist/). The metadata associated with the African bat database is as follows. Family: family of bat record; Genus: genus of bat record; Species: species of bat record; Museum_number: museum accession number of bat record (see Table S2 for definition of the museum acronyms); Date: date on which the bat was collected or otherwise recorded; Year: year in which the bat was recorded; Country: country in which the bat was recorded; Location: name of the location or locality that the bat was recorded; Latitude: latitude in decimal degrees of the record; Longitude: longitude in decimal degrees of the record; Reference: source of the record (see Appendix 1 for the references); Holotype: whether the record is a type specimen; Checked: whether the bat specimen was examined by the authors.
Data coverage
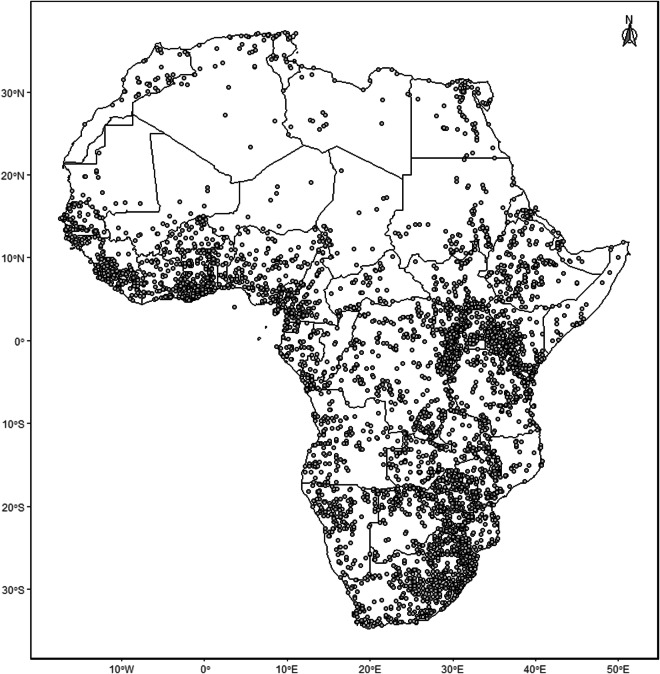
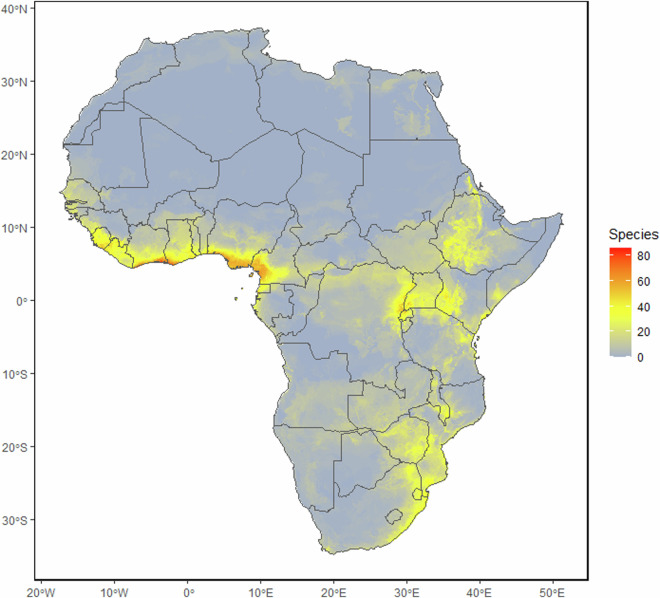
The database contains 17,285 unique locality records of all of the 266 species of bats currently recognized from sub-Saharan Africa (Fig. 2). The number of species recognized in this region has increased by 60% in the last 100 years, and 17% since 2000 (Fig. 1A). In recent years, the increase in species numbers was primarily due to new species being discovered (both de novo descriptions and epithet splits) in the families Vespertilionidae, Rhinolophidae, Pteropodidae and Miniopteridae (Fig. 1B). Species distribution models were built for the 209 species that had seven or more locality records. Species richness based on these modelled distributions is shown in Fig. 3. EOO was calculated for 242 species recorded from sub-Saharan Africa, with three or more occurrence records (Table S3), and ranged from 27 km2 in Rhinolophus hilli to 35,126,201 km2 in Nycteris thebaica. AOO was calculated for all 266 species (Table S3) and ranged from 4 km2 in several species known from just the type locality to 5,344,996 km2 in Taphozous mauritianus. Minimum and maximum elevation of all records ranged from sea level up to 4,207 m above sea level (Table S3), with 33 species having been recorded above 3,000 m above sea level. The species with the largest elevational range was Afronycteris nana, which was recorded at sea level and up to 4,207 m above sea level.
Fig. 2.
Map showing the localities at which the 17,285 occurrences of 266 species in the African bat database were obtained from.
Fig. 3.
Species richness of bat species in sub-Saharan Africa based on the 209 SDMs created in this study.
Limitations
We acknowledge that there are still significant data gaps, especially for under-recorded species and regions that require more sampling efforts; which applies to most of Africa23. This is clearly seen in the 24 species for which there were insufficient occurrence records (<3 records) to calculate EOO (Table S3). These included species only known from the type locality, e.g., Mops gallagheri, Glauconycteris kenyacola, G. machadoi, Myotis nimbaensis, Pipistrellus permixtus, and Pseudoromicia kityoi. African regions that are clearly under-sampled for bats include the Congo Basin (mostly within the Democratic Republic of Congo), the Lower Guinea Forest (especially Equatorial Guinea, Gabon and the Republic of Congo), the Guinean and Sudanian savannas of Central African Republic, Chad and South Sudan, and the Horn of Africa (Ethiopia, Eritrea and Somalia). However, even within relatively well surveyed regions such as South Africa and Eswatini13, new species are still being discovered and described19, highlighting the urgent need for more sampling across the continent.
Additionally, new species continue to be described (Fig. 1), and some taxa remain unresolved. Perhaps the best example here is the Hipposideros cf. ruber complex, which has at least five distinct lineages38 that may represent separate species, but it still awaits taxonomic revision. The lack of specimen collection dates for a large portion of the database is also undesirable, however, this is somewhat ameliorated by the fact that almost two-thirds of the specimens have the year of collection. Where needed, missing dates will have to be obtained from the museums where the specimens have been deposited.
Accurate species identification is increasingly reliant on genetic data, especially to distinguish cryptic species19, and these data are generally available online (e.g., on Genbank). We acknowledge that incorrect identifications or outdated taxonomy in the specimens represented in these databases can result in inaccuracies. While we do not provide genetic data in the current version of the African bat database, this would strengthen future versions by improving traceability across records and genetic reference material.
Technical Validation
All data were entered manually and were cross-checked by multiple authors. A series of checks were undertaken to examine the validity of the data, and the accuracy of the location metadata. We examined the distributions of each species by plotting all the occurrence points and then carefully identifying possible outliers, or incorrect records plotting over water. For records that were marginally plotted over water (within 2 km of the shoreline), we slightly adjusted the latitude or longitude if these were not based on Global Positioning Systems (i.e., records based on quarter-degree squares). Records plotting obviously outside the core of a species range were also checked to ensure that they were not mapping errors. We encourage users to notify the corresponding author of any errors or additional records to ensure the dataset remains accurate and up to date. All updates will be subject to the same stringent technical validation process outlined previously.
Usage Notes
The African bat database32 is the first accessible dataset of occurrence records, distribution models and conservation metrics for all bat species in sub-Saharan Africa where all the records have been carefully examined and corrected for current taxonomy. This database has been a long time in the making and will be crucial in resolving the distributions of poorly recorded bat taxa in sub-Saharan Africa.
Existing databases, such as iNaturalist, GBIF, and other citizen science platforms, have severely limited accuracy in bat records because many bats cannot be identified based solely on appearance. Most published resources on African bat distributions present range maps62,63 rather than point localities, and have out-of-date records and taxonomy.
To the best of our knowledge, the African Chiroptera Report15 is the only published resource that contains point localities for all African bats. However, because this report is a document rather than a database, the underlying data is not readily available for analyses, and inaccuracies frequently persist over time. Distributional mistakes and outliers (for example, in the distribution of Stenonycteris lanosus, which is restricted to the highlands of East Africa63, but yet has records in the report in central and western Democratic Republic of Congo) have an impact on any future analysis and may have a negative effect on conservation planning. Our versioned digital dataset aims to overcome these issues by offering regularly updated, curated data with accurate point localities, and providing a reliable, accessible resource for research and conservation planning.
We hope this comprehensive digital dataset will integrate previously disconnected data resources, and bring African bats into focus for novel and integrative applications that have historically been limited to other regions and taxonomic groups64. It may be useful in ecological and evolutionary research, for species distribution modeling, macroecology, biogeography, and community ecology studies65–67. Ideally, it will help inform biodiversity monitoring and climate change adaptation policy, by predicting the responses of species and ecosystems to increasing threats and changing environments, including habitat destruction and climate change. For taxonomy and systematics, our dataset supports species identification and phylogenetic studies, highlights unresolved taxa, and provides a tool for continuously updating records. In conservation planning and management, we expect that our database will aid both regional and global IUCN Red List assessments by providing systematic and replicable calculations of distribution metrics based on records in the database, which in turn may help inform and facilitate protected area designation as well as environmental impact assessments. Moreover, the online R Shiny App visualization tool allows for interactive exploration of bat distributions, which we hope will be used for educational and outreach efforts, and to support citizen science initiatives.
The database is intended to be a dynamic, living dataset that will be updated regularly, at least once a year, or as new data becomes available, and errors are identified and corrected. Future updates will welcome new or previously unavailable data, will incorporate taxonomic classification changes, and will include the distribution of newly resolved and described species. Updates of the dataset will be made directly on Github (https://github.com/kanead/Bat_database) and will be reflected in the associated R Shiny App. All updates will follow the same technical validation described in this manuscript.
Supplementary information
Author contributions
A.M. conceived the idea. A.M. created and populated the database with contributions from all coauthors. A.M., C.M., P.J.T. and A.K. performed the analysis and wrote the first draft. All coauthors read and revised drafts of the manuscript.
Code availability
Two R scripts are available on Figshare (African Bat Database)32. One generates the species distribution models based on bioclim data (Historical climate data — WorldClim 1 documentation). The other generates area of occurrence (AOO) and extent of occurrence (EOO) information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04170-7.
References
- 1.Cotterill, F. P. D. Systematics, biological knowledge and environmental conservation. Biodivers. Conserv.4, 183–205 (1995). [Google Scholar]
- 2.Razgour, O., Rebelo, H., Di Febbraro, M. & Russo, D. Painting maps with bats: Species distribution modelling in bat research and conservation. Hystrix27, 11753 (2016). [Google Scholar]
- 3.Pio, D. V. et al. Climate change effects on animal and plant phylogenetic diversity in southern Africa. Glob. Chang. Biol.20, 1538–1549 (2014). [Google Scholar]
- 4.Fisher, M. A., Vinson, J. E., Gittleman, J. L. & Drake, J. M. The description and number of undiscovered mammal species. Ecol. Evol.8, 3628–3635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanshi, I., Obitte, B. C., Monadjem, A. & Kingston, T. Hidden Afrotropical bat diversity in Nigeria: ten new country records from a biodiversity hotspot. Acta Chiropterologica23, 313–343 (2021). [Google Scholar]
- 6.Tydecks, L., Jeschke, J. M., Wolf, M., Singer, G. & Tockner, K. Spatial and topical imbalances in biodiversity research. PLoS One13(7), e0199327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clements, H. S. et al. The bii4africa dataset of faunal and floral population intactness estimates across Africa’s major land uses. Sci. Data11, 191 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons, N. B. & Cirranello, A. L. Bat species of the world: a taxonomic and geographic database. Version 1.5. Accessed on 5 August 2024 (2024).
- 9.Burgin, C. J., Colella, J. P., Kahn, P. L. & Upham, N. S. How many species of mammals are there? J. Mammal.99, 1–14 (2018). [Google Scholar]
- 10.Jones, G., Jacobs, D. S., Kunz, T. H., Wilig, M. R. & Racey, P. A. Carpe noctem: The importance of bats as bioindicators. Endanger. Species Res.8, 93–115 (2009). [Google Scholar]
- 11.Kunz, T. H., de Torrez, E. B., Bauer, D., Lobova, T. & Fleming, T. H. Ecosystem services provided by bats. Ann. N. Y. Acad. Sci.1223, 1–38 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Tuneu-Corral, C. et al. Pest suppression by bats and management strategies to favour it: a global review. Biol. Rev.98, 1564–1582 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Monadjem, A., Taylor, P. J., Cotterill, F. P. D. & Schoeman, M. C. Bats of southern and central Africa: a biogeographic and taxonomic synthesis. (Wits University Press, 2020).
- 14.Patterson, B. D. & Webala, P. W. Keys to the Bats (Mammalia: Chiroptera) of East Africa. Fieldiana Life Earth Sci.1563, 1–60 (2012). [Google Scholar]
- 15.Van Cakenberghe, V. & Seamark, E. C. J. Africa Chiroptera Report 2022. (AfricanBats NPC, 2022).
- 16.Monadjem, A., Richards, L., Taylor, P. J. & Stoffberg, S. High diversity of pipistrelloid bats (Vespertilionidae: Hypsugo, Neoromicia, and Pipistrellus) in a West African rainforest with the description of a new species. Zool. J. Linn. Soc.167, 191–207 (2013). [Google Scholar]
- 17.Monadjem, A. et al. A revision of pipistrelle-like bats (Mammalia: Chiroptera: Vespertilionidae) in East Africa with the description of new genera and species. Zool. J. Linn. Soc.191, 1114–1146 (2021). [Google Scholar]
- 18.Grunwald, A. L. et al. A review of bats of the genus Pseudoromicia (Chiroptera: Vespertilionidae) with the description of a new species. Syst. Biodivers. 21 (2023).
- 19.Taylor, P. J. et al. Integrative taxonomic analysis of new collections from the central Angolan highlands resolves the taxonomy of African pipistrelloid bats on a continental scale. Zool. J. Linn. Soc.196, 1570–1590 (2022). [Google Scholar]
- 20.Taylor, P. J., Denys, C. & Cotterill, F. P. D. Taxonomic anarchy or an inconvenient truth for conservation? Accelerated species discovery reveals evolutionary patterns and heightened extinction threat in Afro-Malagasy small mammals. Mammalia (2019).
- 21.Patterson, B. D. et al. Systematics of the Rhinolophus landeri complex, with evidence for 3 additional Afrotropical bat species. J. Mammal. (2024).
- 22.Marsh, C. J. et al. Expert range maps of global mammal distributions harmonised to three taxonomic authorities. J. Biogeogr.49, 979–992 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farooq, H., Azevedo, J. A. R., Soares, A., Antonelli, A. & Faurby, S. Mapping Africa’s biodiversity: more of the same is just not good enough. Syst. Biol.70, 623–633 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fivaz, F. P. & Gonseth, Y. Using species distribution models for IUCN Red Lists of threatened species. J. Insect Conserv.18, 427–436 (2014). [Google Scholar]
- 25.Zurell, D. et al. A standard protocol for reporting species distribution models. Ecography (Cop.).43, 1–17 (2020). [Google Scholar]
- 26.Herkt, K. M. B., Skidmore, A. K. & Fahr, J. Macroecological conclusions based on IUCN expert maps: A call for caution. Glob. Ecol. Biogeogr.26, 930–941 (2017). [Google Scholar]
- 27.Frick, W. F., Kingston, T. & Flanders, J. A review of the major threats and challenges to global bat conservation. Ann. N. Y. Acad. Sci.1469, 5–25 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Andermann, T., Faurby, S., Turvey, S. T., Antonelli, A. & Silvestro, D. The past and future human impact on mammalian diversity. Sci. Adv.6, eabb2313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, P. J., Nengovhela, A., Linden, J. & Baxter, R. M. Past, present, and future distribution of Afromontane rodents (Muridae: Otomys) reflect climate-change predicted biome changes. Mammalia80, 359–375 (2016). [Google Scholar]
- 30.Monadjem, A., Virani, M. Z., Jackson, C. & Reside, A. Rapid decline and shift in the future distribution predicted for the endangered Sokoke Scops Owl Otus ireneae due to climate change. Bird Conserv. Int.23, 247–258 (2013). [Google Scholar]
- 31.Taylor, P. J. et al. Southern Africa’s Great Escarpment as an amphitheater of climate-driven diversification and a buffer against future climate change in bats. Glob. Chang. Biol.30, e17344 (2024). [DOI] [PubMed] [Google Scholar]
- 32.Monadjem, A. et al. African Bat Database, Figshare Dataset, 10.6084/m9.figshare.26363308.v5 (2024).
- 33.Cosentino, F., Castiello, G. & Maiorano, L. A dataset on African bats’ functional traits. Sci. Data10, 623 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monadjem, A., Healy, K., Guillerme, T. & Kane, A. Dispersal ability is associated with contrasting patterns of beta diversity in African small mammal communities. J. Biogeogr.50, 539–550 (2023). [Google Scholar]
- 35.Kock, D. & Stanley, W. T. Mammals of Mafia Island, Tanzania. Mammalia73, 339–352 (2009). [Google Scholar]
- 36.Juste, J. B. & Ibanez, C. Contribution to the knowledge of the bat fauna of Bioko island, Equatorial Guinea (Central Africa). Zeitschrift fur Saugetierkd.59, 274–281 (1994). [Google Scholar]
- 37.Vallo, P. et al. Morphologically uniform bats Hipposideros aff. ruber (Hipposideridae) exhibit high mitochondrial genetic diversity in southeastern Senegal. Acta Chiropterologica13, 79–88 (2011). [Google Scholar]
- 38.Monadjem, A. et al. Diversity of Hipposideridae in the mount Nimba massif, west Africa, and the taxonomic status of Hipposideros lamottei. Acta Chiropterologica15, 341–352 (2013). [Google Scholar]
- 39.Monadjem, A. et al. Systematics of West African Miniopterus with the description of a new species. Acta Chiropterologica21, 237–256 (2019). [Google Scholar]
- 40.Gandaho, S. M., Sogbohossou, E. A. & Thompson, L. J. NIMO: A graphical user interface-based R package for species distribution modelling. Ecol. Solut. Evid.5, e12385 (2024). [Google Scholar]
- 41.Phillips, S. J., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Modell.190, 231–259 (2006). [Google Scholar]
- 42.Hijmans, R. J., Phillips, S. J., Leathwick, J. & Elith, J. dismo: species distribution modeling. R package version 1.1-4. https://cran.r-project.org/web/packages/dismo/index.html. Cran 1–68 (2017).
- 43.Ahmadi, M., Hemami, M. R., Kaboli, M. & Shabani, F. MaxEnt brings comparable results when the input data are being completed; model parameterization of four species distribution models. Ecol. Evol.13, e9827 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaky, E., Nolan, V., Alatawi, A. & Gilbert, F. A comparison between Ensemble and MaxEnt species distribution modelling approaches for conservation: a case study with Egyptian medicinal plants. Ecol. Inform.60, 101150 (2020). [Google Scholar]
- 45.Cooper-Bohannon, R. et al. Predicting bat distributions and diversity hotspots in Southern Africa. Hystrix, Ital. J. Mammal.27, 38–48 (2016). [Google Scholar]
- 46.Herkt, K. M. B., Barnikel, G., Skidmore, A. K. & Fahr, J. A high-resolution model of bat diversity and endemism for continental Africa. Ecol. Modell.320, 9–28 (2016). [Google Scholar]
- 47.Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol.25, 1965–1978 (2005). [Google Scholar]
- 48.R Core Team. R: a language and environment for statistical computing. (R Foundation for Statistical Computing, 2021).
- 49.Monadjem, A. et al. Using species distribution models to gauge the completeness of the bat checklist of Eswatini. Eur. J. Wildl. Res.67, 21 (2021). [Google Scholar]
- 50.Phillips, S. J. & Dudík, M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography (Cop.).31, 161–175 (2008). [Google Scholar]
- 51.Merow, C., Smith, M. J. & Silander, J. A. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography (Cop.).36, 1058–1069 (2013). [Google Scholar]
- 52.Acevedo, P., Jiménez-Valverde, A., Lobo, J. M. & Real, R. Delimiting the geographical background in species distribution modelling. J. Biogeogr.39, 1383–1390 (2012). [Google Scholar]
- 53.Phillips, S. J. et al. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl.19, 181–197 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Zhu, G. P., Rédei, D., Kment, P. & Bu, W. J. Effect of geographic background and equilibrium state on niche model transferability: Predicting areas of invasion of Leptoglossus occidentalis. Biol. Invasions16, 1069–1081 (2014). [Google Scholar]
- 55.Monadjem, A. Longevity and movement of the common slit-faced bat Nycteris thebaica. African Bat Conserv. News9, 7 (2006). [Google Scholar]
- 56.Elith, J. et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography (Cop.).29, 129–151 (2006). [Google Scholar]
- 57.Liu, C., White, M. & Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr.40, 778–789 (2013). [Google Scholar]
- 58.IUCN. Mapping standards and data quality for the IUCN Red List spatial data, version 1.19 (2021).
- 59.Cardoso, P. red - An R package to facilitate species red list assessments according to the IUCN criteria. Biodivers. Data J.5, e20530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galante, P. J. et al. changeRangeR: An R package for reproducible biodiversity change metrics from species distribution estimates. Conserv. Sci. Pract.5, e12863 (2023). [Google Scholar]
- 61.Palacio, R. D., Negret, P. J., Velásquez-Tibatá, J. & Jacobson, A. P. A data-driven geospatial workflow to map species distributions for conservation assessments. Divers. Distrib.27, 2559–2570 (2021). [Google Scholar]
- 62.IUCN. The IUCN Red List of Threatened Species. Version 2024-1 (2024).
- 63.Mammals of Africa Vol IV: hedgehogs, shrews and bats. (Bloomsbury Publishing, 2013).
- 64.Ball-Damerow, J. E. et al. Research applications of primary biodiversity databases in the digital age. PLoS One14(9), e0215794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zurell, D. et al. Testing species assemblage predictions from stacked and joint species distribution models. J. Biogeogr.47, 101–113 (2020). [Google Scholar]
- 66.Delgado-Jaramillo, M., Aguiar, L. M. S., Machado, R. B. & Bernard, E. Assessing the distribution of a species-rich group in a continental-sized megadiverse country: bats in Brazil. Divers. Distrib.26, 632–643 (2020). [Google Scholar]
- 67.Monadjem, A., Farooq, H. & Kane, A. Elevation filters bat, rodent and shrew communities differently by morphological traits. Divers. Distrib.30, e13801 (2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Monadjem, A. et al. African Bat Database, Figshare Dataset, 10.6084/m9.figshare.26363308.v5 (2024).
Supplementary Materials
Data Availability Statement
Two R scripts are available on Figshare (African Bat Database)32. One generates the species distribution models based on bioclim data (Historical climate data — WorldClim 1 documentation). The other generates area of occurrence (AOO) and extent of occurrence (EOO) information.