Abstract
The difference in membrane (M) protein compositions between the transmissible gastroenteritis coronavirus (TGEV) virion and the core has been studied. The TGEV M protein adopts two topologies in the virus envelope, a Nexo-Cendo topology (with the amino terminus exposed to the virus surface and the carboxy terminus inside the virus particle) and a Nexo-Cexo topology (with both the amino and carboxy termini exposed to the virion surface). The existence of a population of M molecules adopting a Nexo-Cexo topology in the virion envelope was demonstrated by (i) immunopurification of 35S-labeled TGEV virions using monoclonal antibodies (MAbs) specific for the M protein carboxy terminus (this immunopurification was inhibited only by deletion mutant M proteins that maintained an intact carboxy terminus), (ii) direct binding of M-specific MAbs to the virus surface, and (iii) mass spectrometry analysis of peptides released from trypsin-treated virions. Two-thirds of the total number of M protein molecules found in the virion were associated with the cores, and one-third was lost during core purification. MAbs specific for the M protein carboxy terminus were bound to native virions through the M protein in a Nexo-Cexo conformation, and these molecules were removed when the virus envelope was disrupted with NP-40 during virus core purification. All of the M protein was susceptible to N-glycosidase F treatment of the native virions, which indicates that all the M protein molecules are exposed to the virus surface. Cores purified from glycosidase-treated virions included M protein molecules that completely or partially lost the carbohydrate moiety, which strongly suggests that the M protein found in the cores was also exposed in the virus envelope and was not present exclusively in the virus interior. A TGEV virion structure integrating all the data is proposed. According to this working model, the TGEV virion consists of an internal core, made of the nucleocapsid and the carboxy terminus of the M protein, and the envelope, containing the spike (S) protein, the envelope (E) protein, and the M protein in two conformations. The two-thirds of the molecules that are in a Nexo-Cendo conformation (with their carboxy termini embedded within the virus core) interact with the internal core, and the remaining third of the molecules, whose carboxy termini are in a Nexo-Cexo conformation, are lost during virus core purification.
Transmissible gastroenteritis coronavirus (TGEV) is a member of Coronaviridae, a family of viruses that infects birds and mammals and causes a variety of diseases (8, 18). The TGEV is an enveloped virus with a positive-sense RNA genome of 28.5 kb (24) for which an infectious cDNA clone has been engineered (1). The TGEV virion structure presents three structural levels (9): (i) the envelope, in which the spike (S), envelope (E), and membrane (M) proteins are embedded (6, 12, 15, 16); (ii) the internal core, made of the nucleocapsid and the C terminus of the M protein (9); and (iii) the nucleocapsid, consisting of the RNA genome and the nucleoprotein (N) (34). The TGEV core was purified and characterized using ultrastructural and biochemical techniques (9, 28), and a significant proportion of M molecules were found associated with the purified cores.
It has been proposed that the M protein molecules of the TGEV adopt two different topologies in the viral envelope, a Nexo-Cendo and a Nexo-Cexo topology (Fig. 1) (29). The Nexo-Cendo topology is adopted by the M molecules of all coronaviruses (3, 27). The Nexo-Cexo topology has been detected in TGEV virions by immunogold labeling and virus neutralization using monoclonal antibodies (MAbs) (29). The presence of these two topologies raises the question of their localization in the virion, i.e., whether both populations of M protein copurify with the TGEV core.
FIG. 1.
TGEV M protein topologies in the virus envelope. Scheme of the topologies adopted by the M protein, the Nexo-Cendo topology (left) and the Nexo-Cexo topology (right), showing the MAbs used in this report and the approximate locations of the M protein epitopes recognized by them. The domain of the M protein that interacts with the nucleocapsid is indicated as a wide bar between residues 237 and 252. Ext, external surface of the virion; Int, virion interior. Arrows indicate approximate amino acid positions in the M protein.
In this article, the biochemical characterization of the M protein topologies and the difference in M protein compositions between the TGEV virions and purified cores were studied. Evidence for the presence within TGEV virions of the M protein in two conformations has been provided. The existence of an M protein topology with the carboxy terminus facing the external surface of the virion has been shown by using a combination of approaches, including the detection of the M protein carboxy terminus by immunopurification with MAbs and by mass spectrometry to identify peptides liberated from the virion surface by controlled proteolysis. A working model for the TGEV virion structure is proposed.
MATERIALS AND METHODS
Cells and viruses.
Swine testicle (ST) cells (20) were grown in Dulbecco modified Eagle's medium (DMEM) supplemented with fetal calf serum. Baby hamster kidney cells (BHK-21) stably transformed with the gene coding for the porcine aminopeptidase N (BHK-pAPN) were grown in DMEM supplemented with 2% fetal calf serum and with Geneticin G418 (1.5 mg/ml) as a selection agent (7).
TGEV strain PUR46-MAD was grown, purified, and titrated in ST cells as described previously (15). Mouse hepatitis virus (MHV) strain A59 (ATCC VR-764) was grown in 3T3 cells (33) and concentrated by centrifugation at 27,000 rpm for 50 min in an SW60.Ti Beckmann rotor.
Recombinant vaccinia virus vT7 (ATCC VR-2153) was used to express the T7 bacteriophage DNA-dependent RNA polymerase (10).
Antibodies.
The murine MAbs 9D.B4, 3D.E3, 3D.C10, and 25.22 have been described previously (9, 11, 15, 28, 30). MAb 9D.B4 is directed to an epitope located around leucine 216 of the TGEV M protein. MAb 3D.E3 binds an epitope located within the last 10 residues of the M protein. MAb 25.22 is specific for the M protein amino terminus (5, 17). MAb 3D.C10 is specific for the N protein. Peroxidase-conjugated rabbit anti-mouse MAb and rhodamine-conjugated goat anti-mouse F(ab)2 were purchased from Cappel.
TGEV core purification.
Viral membranes were disrupted with 1% NP-40, and cores were purified by ultracentrifugation through a sucrose gradient as described previously (9).
Estimation of the M-to-N molar ratio.
Purified virions and cores (2 μg) were analyzed by gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (5 to 20% polyacrylamide), and the structural proteins were stained with silver (2). The protein bands were quantified by band densitometry using a Gel Doc 2000 system (Bio-Rad). Seven independent virus purifications were used to estimate the M-to-N molar ratios in virions and cores. The normal distribution of the M-to-N molar ratio was tested by the chi-square test (19). M-to-N stoichiometries of virions and cores were compared by the nonparametric t test of Wilcoxon (21).
Endoglycosidase treatment of TGEV virions.
Purified virions (1 μg) were incubated with endoglycosidase H or N-glycosidase F (Roche) to a final concentration of 0.3 U/μl for the times indicated in the figure legends in phosphate-buffered saline (PBS) at 37°C in a final volume of 10 μl. The proteins were then resolved by SDS-PAGE and analyzed by Western blotting. For Western blot analysis, the proteins were transferred to a nitrocellulose membrane with a Bio-Rad Mini Protean II electroblotting apparatus at 150 mA for 1 h in 25 mM Tris–192 mM glycine buffer (pH 8.3) containing 20% methanol. Membranes were saturated with TBS (20 mM Tris-HCl [pH 7.5], 150 mM NaCl) containing 5% dried milk and incubated with MAbs directed to the M protein in the appropriate dilutions. Bound antibody was detected with horseradish peroxidase-conjugated rabbit anti-mouse MAb by the enhanced chemiluminescence detection system (ECL Western blotting detection reagents; Amersham Pharmacia Biotech).
NP-40 was added where indicated in the figure and legend to a final concentration of 1% to dissolve the virus envelope. For treatments under denaturing conditions, SDS was added to a final concentration of 1% and the virus was dissociated by boiling for 5 min before glycosidase treatment.
Cloning and expression of mutant M genes.
The wild-type M gene (Mwt) and the carboxy-terminal deletion mutant proteins MΔ253–262 and MΔ146–262, lacking the last 10 and 117 amino acids, respectively, were cloned in the pcDNA3 plasmid under the control of both T7 and cytomegalovirus promoters, generating plasmids pcDNA3-M, pcDNA3-MΔ253–262, and pcDNA3-MΔ146–262 (9). The amino-terminal deletion mutant protein MΔ1–50 was obtained by PCR using the synthetic oligonucleotide 5′-GCCGGATCCAAAATGCTTGCAAACTGGAACTTCAGCTGGTC-3′, which introduces a BamHI restriction site at the 5′ end of the M gene followed by an ATG codon fused to the sequence coding for the first transmembrane domain, together with the SP6 primer and the pcDNA3-M plasmid as a template. The PCR product was restricted with BamHI and cloned into the pcDNA3 plasmid digested with BamHI, generating the pcDNA3-MΔ1–50 plasmid. This mutant construct lacks the first 50 amino acids of the M protein, including the signal peptide.
BHK-pAPN cells were grown to a 60% confluence in 60-mm-diameter-culture dishes. Cells were transfected in OPTIMEM medium (GIBCO BRL) with 15 μl of Lipofectin reagent (GIBCO BRL) and 5 μg of pcDNA3 plasmids coding for the Mwt protein and M gene mutant proteins for 5 h. Where indicated, cells were previously infected with the vaccinia vT7 virus at a multiplicity of infection of 10. Transfected cells were washed and complete DMEM was added for 24 h. The expression of Mwt protein and M gene mutant proteins was evaluated by immunofluorescence microscopy using M-specific MAbs, with the 3D.C10 MAb as a control. Briefly, untransfected or transfected BHK-pAPN cells were seeded on glass coverslips. Cells were washed twice with PBS and fixed with methanol for 8 min at −20°C. The methanol was washed twice with PBS, and ascitic fluids containing specific MAbs were added to the cells in a 1:100 dilution in PBS–1% bovine serum albumin (BSA) and incubated for 1 h at room temperature. Cells were washed four times in PBS, and rhodamine-conjugated goat anti-mouse F(ab′)2 was added to a 1:200 dilution in PBS–1% BSA and incubated for 30 min at room temperature. Cells were washed five times, and the preparations were mounted in Mowiol (Aldrich-chemie). Cells were observed under a Zeiss Axiovert fluorescence microscope using the appropriate UV light filters for rhodamine. TGEV-infected ST cells were used as an expression control.
Immunopurification of 35S-labeled TGEV virions.
ST cells grown in 60-mm-diameter culture dishes were infected with TGEV at a multiplicity of infection of 1 for 1 h. Monolayers were washed twice in methionine- and cysteine-depleted DMEM. One hundred microcuries of [35S]methionine-cysteine (Pro-mix l-35S in vitro cell labeling mix; Amersham Pharmacia Biotech) was added, and the infected cells were incubated for 24 h. Supernatants were recovered and cleared by low-speed centrifugation. Labeled virions were concentrated 100 times by ultracentrifugation at 27,000 rpm in an SW60.Ti Beckmann rotor for 50 min at 4°C through a 30% sucrose cushion in TNE buffer (10 mM Tris-HCl [pH 7.4] 1 mM EDTA, 100 mM NaCl). Concentrated virions were recovered in 50 μl of TNE buffer.
Protein A (5 μg per well) was bound to a 96-well vinyl assay plate (Data Packaging Corporation) for 12 h at 37°C, and unbound sites were blocked with 5% BSA in PBS buffer for 2 h at 37°C. Wells were washed with 120 μl of washing buffer (0.1% BSA in PBS buffer), and 10 μg of rabbit anti-mouse MAbs (Cappel) was bound by incubation for 1 h at 37°C (60-μl final volume). M- and N-specific MAbs (6 μg) were complexed to the rabbit anti-mouse MAbs by incubation at 37°C for 1 h. Wells were washed, 30 μl of concentrated labeled virions (about 5 μg) was added, and the plates were incubated overnight at 4°C. The wells were washed seven times in washing buffer. Bound virion proteins were recovered in 30 μl of SDS-PAGE loading buffer and resolved by SDS-PAGE. Proteins were detected by fluorography as described previously (9).
Inhibition of labeled TGEV immunopurification by unlabeled mutant M proteins.
Unlabeled mutant M proteins were expressed in BHK-pAPN cells infected with vaccinia virus strain vT7 and transfected with expression plasmids as described above. After 24 h, the monolayers were scraped off and the cells were lysed with 100 μl of lysis buffer containing 1% NP-40, 150 mM NaCl, and protease inhibitors (Complete Protease Inhibitor Cocktail tablets; Boehringer Mannheim). Nuclei were removed by low-speed centrifugation, and the cytoplasmic fraction was recovered and stored at −20°C. The protein expression levels were estimated by Western blotting using a specific swine antiserum and purified TGEV as a standard.
M-specific MAbs were bound to vinyl assay plates as described above. Increasing amounts of BHK-pAPN cell lysates containing Mwt protein and mutant M proteins were added and incubated overnight at 4°C. Wells were washed once with PBS–0.1% BSA–NP-40 (0.05%) and twice with PBS–0.1% BSA to remove cellular contaminating proteins. Labeled TGEV virions (7 μl) were added and incubated for 5 h at 4°C. Wells were washed 20 times in PBS–0.1% BSA, and bound virion proteins were recovered in 30 μl of SDS-PAGE loading buffer. Virion proteins were resolved by gradient SDS-PAGE and revealed by fluorography. Lysates of untransformed vT7-infected BHK-pAPN cells were used as a control.
Binding of TGEV-specific MAbs to the virion surface.
Purified TGEV virions (15 μg) were incubated overnight at 4°C with purified M- and N-specific MAbs in 1 ml of TNE (16 μg/ml) containing 0.05% Tween 20. Virions were centrifuged in an SW60. Ti Beckmann rotor at 27,000 rpm for 50 min at 4°C through a 30% sucrose cushion in TNE buffer. Virus sediment was resuspended in 50 μl of TNE. Samples were resolved by SDS-PAGE, and bound immunoglobulin chains were detected by Western blot analysis using horseradish peroxidase-conjugated rabbit anti-mouse MAb and ECL detection. NP-40 was added to the indicated samples in Fig. 7 at a final concentration of 1% to disrupt the viral envelope. Cores were purified by ultracentrifugation through a 30% sucrose cushion in TNE containing 1% NP-40 as described above. Core proteins were recovered in 30 μl of SDS-PAGE loading buffer. Immunoglobulins were detected by Western blotting as described above.
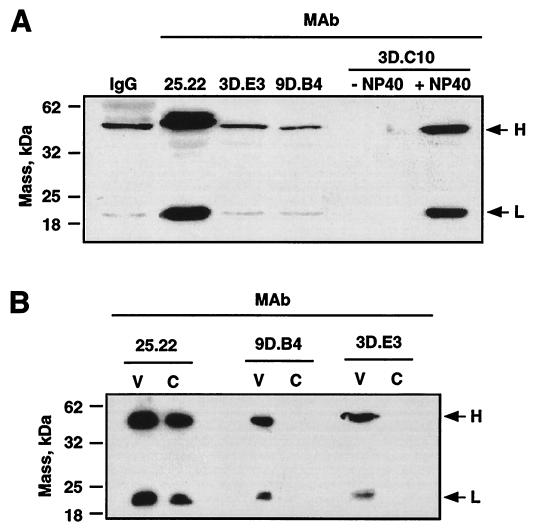
FIG. 7.
Binding of M-specific MAbs to the virus surface. (A) Western blot analysis of the heavy and light antibody chains from MAbs specifically bound to the virion surface. IgG, purified immunoglobulin G. + NP-40 or − NP-40, presence or absence of NP-40 in virus preparations. (B) Western blot analysis of the heavy and light chains of the M-specific MAbs bound to the surfaces of purified virions (V) and their cores (C). Heavy (H) and light (L) immunoglobulin chains are indicated by arrows.
Analysis of tryptic peptides released from the surfaces of trypsin-treated TGEV virions by mass spectrometry.
Purified TGEV (10 μg) was subjected to proteolysis by trypsin (0.33 ng/μl) in 25 mM ammonium bicarbonate in a final volume of 10 μl. Digestions were performed for 15 and 45 min at both 25 and 37°C. These conditions of low trypsin concentration and less than 1 h of digestion were selected for limited surface proteolysis to avoid virion damage. The virion envelope was disrupted with NP-40 where indicated in Table 1, and proteolysis was performed as described above. Released tryptic peptides were analyzed by mass spectrometry by using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. Briefly, 0.5-μl aliquots of the digestion solution were manually deposited onto a stainless steel MALDI probe and allowed to dry at room temperature. Then 0.5 μl of matrix solution (saturated α-cyano-4-hydroxycinnamic acid in 33% aqueous acetonitrile and 0.1% trifluoroacetic acid) was added and again the samples were allowed to dry at room temperature. Samples were measured automatically on a Bruker Reflex III MALDI-TOF mass spectrometer (Bruker-Franzen Analytic GmbH, Bremen, Germany) equipped with the SCOUT source in the positive ion reflector mode by using delayed extraction and AutoXecute acquisition software. The ion acceleration voltage was 25 kV. The equipment was first externally calibrated by employing protonated mass signals from a peptide mixture covering the 1,000- to 4,000-m/z (mass-to-charge ratio) range, and thereafter, every spectrum was internally calibrated using selected signals arising from trypsin autoproteolysis to reach a typical mass measurement accuracy of ±30 ppm. The measured tryptic peptide masses were transferred automatically through the mass spectrometry BioTools program as inputs to search automatically the NCBInr database using Mascot software (Matrix Science). No restrictions were placed on the species of origin of the protein, and the allowed protein molecular mass was 1 to 200 kDa. Up to one missed tryptic cleavage was considered, and a mass accuracy of 100 ppm was used for all tryptic-mass searches. N and M tryptic peptides were identified by using the TGEV PUR46-MAD genome sequence available at the GenBank nucleotide sequence database (accession number AJ271965). Trypsin or TGEV virions alone were also analyzed by MALDI-TOF as reaction controls.
TABLE 1.
Peptides identified by mass spectrometry matching proteolytic peptides predicted by the database
| Protein | Sequence of peptide(s) released from virions treated with:
|
m/z (mean ± SD)a | |
|---|---|---|---|
| Trypsin | Trypsin + NP-40 | ||
| M | TDNLSEQEKLLMHMVb | TDNLSEQEKLLMHMV | 1,656.8 ± 0.0261 |
| SDTDLSCR | 952.88 ± 0.693 | ||
| SWWSFNPETK | 1,28.1 ± 0.672 | ||
| ASSATGWAYYVK | 1,303.12 ± 0.707 | ||
| N | None | GIGNRDQQIGYWNR | 1,676.85 ± 0.029 |
| VPGEFQLEVNQSR | 1,502.72 ± 0.059 | ||
Expressed as atomic mass units per proton charge. Only the peptides with an m/z around 1,656.8 are shown.
The mean peptide mass ± SD is 1,656.8 ± 0.091.
For further identification of the released tryptic peptides, peptide mass fingerprint maps from three independent virus samples were obtained from the gel-purified structural proteins. Briefly, M, N, S, and E proteins were resolved by SDS-PAGE and stained using Gelcode blue staining reagent (Pierce). Protein bands were excised manually and then processed automatically using an Investigator ProGest protein digestion station (Genomic Solutions, Huntingdon, Cambridgeshire, United Kingdom) (14). The digestion protocol previously reported (31) was used with minor modifications: gel plugs were washed with 25 mM ammonium bicarbonate and acetonitrile prior to reduction with 10 mM dithiothreitol in 25 mM ammonium bicarbonate and alkylation with 100 mM iodoacetamide in 50 mM ammonium bicarbonate. The gel pieces were then rinsed with 50 mM ammonium bicarbonate and acetonitrile and dried under a stream of nitrogen. Modified porcine trypsin (sequencing grade; Promega, Madison, Wis.) at a final concentration of 16 ng/μl in 25 mM ammonium bicarbonate was added to the dry gel pieces, and the digestion proceeded at 37°C for 12 h. Peptides were eluted with acetonitrile, 25 mM ammonium bicarbonate, and 10% (vol/vol) formic acid for a final extraction volume of 100 μl. The tryptic peptides were identified by MALDI-TOF as described above and matched with the peptides obtained from the solubilized TGEV virion surface tryptic peptides.
To determine the peptide sequence, the fraction containing the putative carboxy terminus peptide was identified by MALDI-TOF and this peptide was sequenced by using the post-source decay MALDI spectrum (36). This spectrum was measured on a Bruker Reflex III MALDI-TOF mass spectrometer (Bruker-Franzen Analytic GmbH) equipped with the SCOUT source in the positive ion reflector mode using delayed extraction. The spectrum was recorded in 14 segments, with each successive segment representing a 20% reduction in reflector voltage. The precursor ion was selected by deflecting pulses. About 200 shots were averaged per segment, and the segments were pasted, calibrated, and smoothed under the control of Bruker XTOF 5.0.3 software. Data analysis was performed using Bruker BioTools 2.0 software.
RESULTS
Protein composition of sucrose gradient-purified virions and cores.
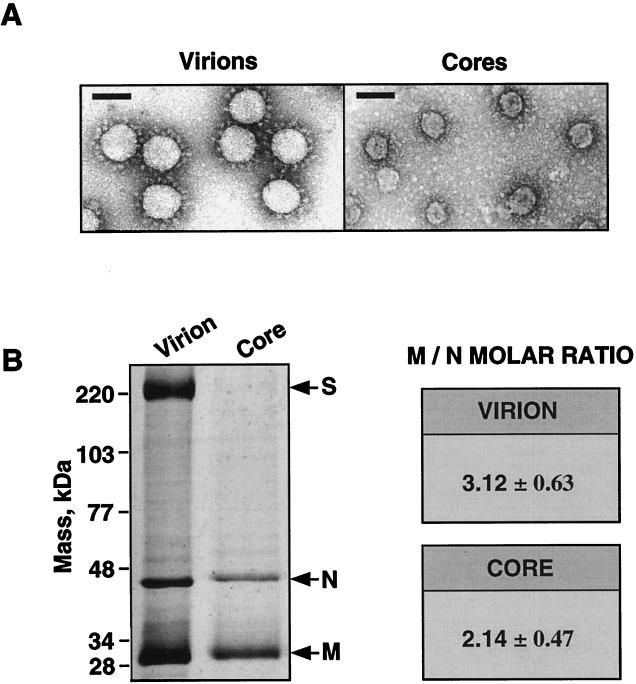
Virions and cores were purified by sucrose gradient centrifugation, and the presence of the three main structural proteins, S, M, and N, has been shown in purified virions. In contrast, purified cores (Fig. 2A) contained the M and N proteins but not the S protein (9). In purified virions, the M-to-N molar ratio was estimated to be 3, while in purified cores, this ratio was 2 (Fig. 2B). The difference in the M-to-N molar ratios was statistically significant (see Materials and Methods) and indicated that approximately one-third of the M protein molecules was lost and that two-thirds remained tightly attached to the cores during envelope removal in the core purification process.
FIG. 2.
Electron microscopy of negatively stained, purified TGEV virions and cores and estimations of the M-to-N molar ratios. (A) Electron microscopy pictures of sucrose-purified TGEV virions (left panel) and cores (right panel). Bars, 100 nm. (B) Left panel, SDS-PAGE analysis and silver staining of purified TGEV virions and cores. Arrows indicate the positions of the major structural proteins. Right panel, estimations of the M-to-N molar ratios in virions and cores presented as means ± SDs. The means of the M-to-N molar ratios were deduced from eight independent experiments.
Immunopurification of 35S-labeled TGEV virions.
To study whether a subset of the M proteins adopts a Nexo-Cexo topology in the virus envelope (Fig. 1), TGEV virions were purified using M protein carboxy-terminus-specific MAbs. To this end, 35S-labeled virions were concentrated by centrifugation through a 30% sucrose cushion from supernatants of metabolically labeled infected ST cells and were immunopurified with MAbs directed to both the M protein amino and carboxy termini (Fig. 1). The extent of virus purification was assessed by SDS-PAGE protein analysis. Viruses immunopurified with MAbs specific for either the amino or the carboxy terminus of the M protein showed the presence of the three major structural proteins (S, N, and M) and the absence of cellular contaminants (Fig. 3A). In contrast, virions were not bound either when MAb 3D.C10 specific for the internal N protein was present or when no antibodies were used to arm protein A. Immunopurification of 35S-labeled virions by M-specific MAbs was inhibited by moderate concentrations (ratio of unlabeled virus to labeled virus, 1:15) of unlabeled TGEV (Fig. 3B). Furthermore, no significant inhibition was observed when MHV was used as the competitor virus, showing the specificity of the purification procedure for TGEV virions. This result showed that both the amino and the carboxy termini of the M protein were exposed to the virion surface in a significant proportion of M protein molecules.
FIG. 3.
Immunopurification of 35S-labeled TGEV virions. (A) SDS-PAGE and fluorography of immunopurified 35S-labeled TGEV virions using the indicated MAbs. Arrows indicate the positions of the TGEV structural proteins S, N, and M. −, absence of MAb.; IC, extract from infected cells. (B) Inhibition of immunopurification of labeled virions with the indicated concentrations of unlabeled purified TGEV virions, using MAbs 25.22, 3D.E3, and 9D.B4 as indicated. Control experiments were performed using concentrated MHV virions as an unspecific competitor at the highest concentration assayed. + or −, presence or absence of unlabeled MHV, respectively. Arrows indicate the positions of the structural proteins.
To further prove that the M protein carboxy terminus was exposed to the virion surface, the specificities of M protein carboxy-terminus-specific MAbs in binding to the virus were tested by inhibiting their binding by using either full-length M proteins (Mwt) or deletion-containing M proteins from cells transfected with plasmids expressing these proteins (Fig. 4A). The Mwt and M mutant proteins were produced at high levels (between 2 and 5 μg/106 cells) using the expression system based on the recombinant poxvirus strain vT7. Proteins were detected by Western blot analysis using a TGEV-specific swine antiserum. All of the mutant proteins presented the expected sizes (Fig. 4B).
FIG. 4.
Expression of Mwt protein and deletion mutant M proteins. (A) Scheme of mutant M proteins cloned in the expression vector pcDNA3. The gray boxes indicate deletions. The numbers above the bars represent the amino acid positions immediately before and after the deletions. The approximate locations of the MAb binding sites in the M protein are indicated above the Mwt protein bar. (B) Western blot analysis of BHK-pAPN cell lysates expressing the indicated mutant M proteins using a polyvalent TGEV-specific antiserum. The M genes were cloned in plasmid pcDNA3 under the T7 polymerase promoter, and the vaccinia virus-expressing T7 polymerase was used to drive the expression. (C) Immunofluorescence microscopy patterns of BHK-pAPN cells transfected with pcDNA3 plasmids encoding Mwt and the indicated mutant M genes (left) using the M protein-specific MAbs indicated at the top. TGEV-infected ST cells were used as a positive expression control (top row).
To assess the specificities of the MAbs, the binding of these antibodies to deletion mutants M proteins was evaluated by immunofluorescence microscopy. Full-length M protein, the amino-terminal deletion mutant protein MΔ1–50, and the carboxy-terminal deletion mutant proteins MΔ253–262 and MΔ146–262 (Fig. 4A) were expressed in BHK-pAPN cells (Fig. 4C). The full-length M protein was recognized by all of the M-specific MAbs in TGEV-infected ST cells and also in BHK-pAPN cells transfected with plasmids encoding the M protein. The amino-terminal deletion mutant protein MΔ1–50 was bound by all MAbs except MAb 25.22. The MΔ253–262 mutant protein was recognized by all MAbs except 3D.E3, and the MΔ146–262 mutant protein bound only MAb 25.22. The immunofluorescence observed was specific, since the N-specific MAb 3D.C10 recognized only the N protein produced by TGEV-infected ST cells. These results confirmed the specificities of the M-specific MAbs used in the study.
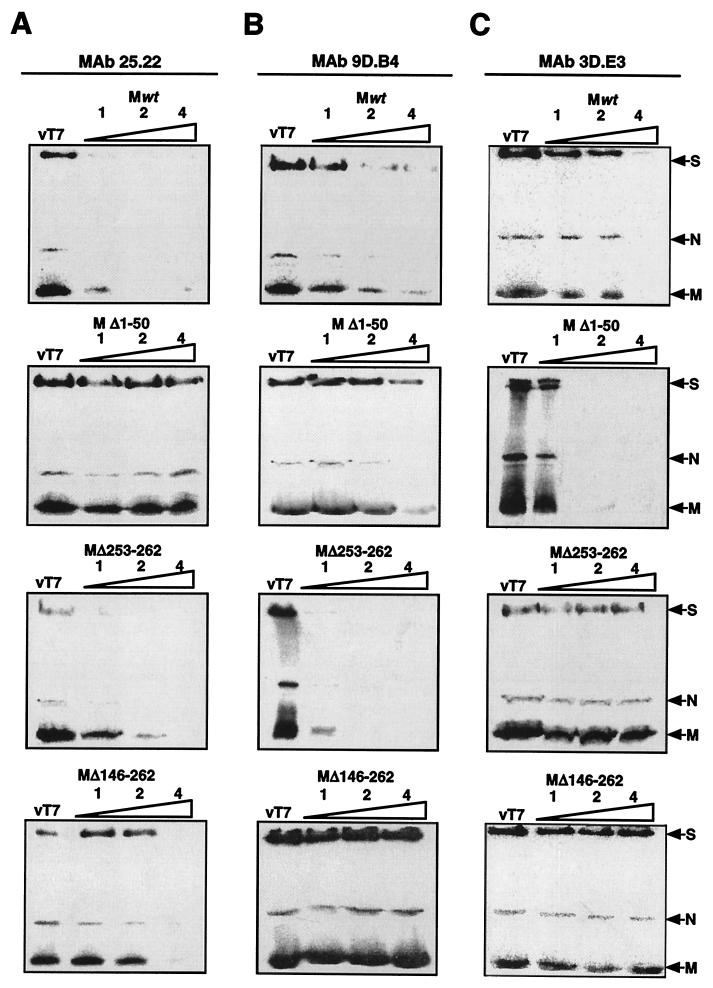
Cell extracts containing unlabeled mutant M protein expressed in transfected cells were used to inhibit the immunopurification of the TGEV virions (Fig. 5). Virus immunopurification using MAb 25.22 (Fig. 5A) was specifically inhibited by Mwt and by the deletion mutant M proteins MΔ253–262 and MΔ146–262 but not by MΔ1–50. When MAb 9D.B4 was used (Fig. 5B), virus immunopurification was specifically inhibited by Mwt and by all mutant proteins except MΔ146–262. When MAb 3D.E3 was used (Fig. 5C), immunopurification was inhibited by Mwt and by the MΔ1–50 mutant proteins but not by the MΔ253–262 and MΔ146–262 mutant proteins. These results showed that immunopurification was specifically inhibited by the deletion mutant M proteins only when these mutant proteins presented the epitope bound by the MAbs used to immunopurify the virus.
FIG. 5.
Immunounification of 35S-labeled virions by MAbs. SDS-PAGE and fluorography of labeled TGEV virions immunopurified with MAb 25.22 (A), MAb 9D.B4 (B), and MAb 3D.E3 (C) were performed in the presence of increasing amounts of cell lysates from BHK-pAPN cells expressing Mwt (top row), MΔ1–50 (second row), MΔ253–262 (third row), and MΔ146–262 (bottom row). vT7, lysate of vT7-infected BHK-pAPN cells.
Release of the M protein carboxy terminus by protease treatment.
TGEV virions were partially digested with trypsin in the absence or in the presence of the detergent NP-40, which dissolved the virus membrane, and the peptides released were compared with those cleaved from virions incubated in the absence of trypsin (Fig. 6).
FIG. 6.
Mass spectrometry (MALDI-TOF) analysis of tryptic peptides released from the TGEV virion surface. MALDI-TOF spectra of trypsin (A), untreated TGEV virions (B), and TGEV virions treated with trypsin for 15 (C) or 45 (D) min at room temperature are shown. A domain of the spectrum that includes the predicted M peptide with an m/Z of 1,656.8 is shown in the figure.
The most C-terminal M protein tryptic peptide from position 249 to the end of the protein was identified by mass spectrometry (MALDI-TOF) among the solubilized peptides in the trypsin-treated virion samples at all trypsin incubation times (Fig. 6C and D) but not in the absence of protease (Fig. 6B) or in the trypsin used for the proteolysis (Fig. 6A). The mean peptide mass and the standard deviation (SD), 1,656.8 ± 0.091 (n = 8; coefficient of variation = 0.0055%), were calculated from the results of eight independent experiments and matched the expected peptide mass of 1,656.826 m/z predicted by the NCBInr database (Table 1). As a control of virion integrity, the N protein-specific peptides were investigated. No peptides from the N protein were detected in the absence of NP-40 (Table 1). In contrast, M and N protein-derived peptides were detected when the virus envelope had been disrupted previously with NP-40 (Table 1).
To definitively assess the identity of the released peptide with an m/z of 1,656.8, the mass fingerprint maps of gel-purified TGEV structural proteins were obtained (Table 2). Only the M protein fingerprint presented a peptide with an m/z of 1,656.8 ± 0.0153 (n = 3; coefficient of variation = 0.001%), matching that of the M protein peptide released from the surfaces of trypsin-treated virions, identified above as the C terminus. This peptide was identified as the M protein carboxy terminus, since no other peptide with the same mass could be obtained either experimentally or theoretically from trypsin-treated M protein. This peptide was partially sequenced by analyzing its fragmentation spectrum. The spectrum obtained definitely confirmed the nature of this peptide, since it identified a peptide with the sequence LLHMV, which is compatible with that of the M protein carboxy-terminus peptide (TDNLSEQEKLLHMV).
TABLE 2.
Peptide mass fingerprints from SDS-PAGE-purified TGEV structural proteinsa
| Protein | Predicted peptide | Exptl mass (mean ± SD)b |
|---|---|---|
| S | FYLTPR | 795.91 ± 0.714 |
| YVCNGNPR | 978.95 ± 0.714 | |
| DVQLTLFR | 991.05 ± 0.714 | |
| NLDDKFYLTPR | 1,381.39 ± 0.583 | |
| FCLSLSPVGANCK | 1,452.23 ± 0.714 | |
| LTALNAFVSQTLTR | 1,534.37 ± 0.714 | |
| EWPNIGGSWLEGLK | 1,585.32 ± 0.714 | |
| TFGLVVKDVQLTLFR | 1,735.52 ± 0.714 | |
| ISFENQWSGTVTFGDMR | 1,974.44 ± 0.707 | |
| LNVVVNGYPYSITVTTTR | 1,996.60 ± 0.714 | |
| TLLSGLYYTSLSGDLLGFK | 2,047.63 ± 0.714 | |
| LLTQYVSACQTIEQALAMGAR | 2,323.76 ± 0.714 | |
| LPPNSDVVLGDYFPTVQPWFNCIR | 2,834.05 ± 0.714 | |
| ATTLEVAGTLVDLWWFNPVYDVSYYR | 3,078.21 ± 0.707 | |
| MTMYTASLAGGITLGALGGGAVAIPFAVAVQAR | 3,135.33 ± 0.714 | |
| N | VSWGDESTK | 1,008.46 ± 0.007 |
| YHKPKDDPK | 1,112.54 ± 0.023 | |
| DGAMNKPTTLGSR | 1,363.61 ± 0.047 | |
| KDDSVEQAVLAALK | 1,486.56 ± 0.396 | |
| DDSVEQAVLAALKK | 1,486.56 ± 0.396 | |
| VPGEFQLEVNQSR | 1,502.72 ± 0.046 | |
| EDGDQIEVTFTHK | 1,518.66 ± 0.044 | |
| WFFYLGTGPHADAK | 1,772.78 ± 0.067 | |
| FDGKVPGEFQLEVNQSR | 1,949.89 ± 0.070 | |
| TGQFLQQINAYARPSEVAK | 2,121.03 ± 0.067 | |
| NNNIPLSFFNPITLQQGSK | 2,132.10 ± 0.015 | |
| M | SIQLYRR | 935.2 ± 0.575 |
| SDTDLSCR | 953.07 ± 0.595 | |
| AGDSTEAR | 969.09 ± 0.583 | |
| TIVYTLVGK | 993.26 ± 0.577 | |
| YVMVALPSR | 1,035.23 ± 0.583 | |
| AILCVSALGR | 1,059.27 ± 0.589 | |
| TIVYTLVGKK | 1,121.35 ± 0.571 | |
| IAGGMNIDNLPK | 1,242.31 ± 0.583 | |
| SWWSFNPETK | 1,281.25 ± 0.583 | |
| ASSATGWAYYVK | 1,302.95 ± 0.577 | |
| TKSWWSFNPETK | 1,510.39 ± 0.571 | |
| TDNLSEQEKLLHMV | 1,656.81 ± 0.015 | |
| SWWSFNPETKAILCVSALGP | 2,321.83 ± 0.577 | |
| SYVLPLEGVPTGVTLTLLSGNLYAEGFK | 2,938.21 ± 0.554 | |
| E | TVIIVPAQHAYDAYK | 1,688.86 ± 0.054 |
Boldface indicates the sequence and the mass of the C terminus of the M protein.
The mean was determined from the results of three independent experiments.
Binding of M-specific MAbs to the virion surface to identify the M protein domains externally exposed.
M-specific MAbs were directly bound to exposed epitopes of the M protein (Fig. 7A), since bound immunoglobulin chains were specifically detected in purified virions by Western blotting when the virions were incubated with MAb 25.22, which is specific for the amino terminus of the M protein, or MAbs 9D.B4 and 3D.E3, both of which are specific for different peptides of the M protein carboxy terminus. No antibody chains were detected with MAb 3D.C10 which is specific for the N protein, an internal structural protein. In contrast, strong binding of MAb 3D.C10 was detected when the viral membrane was previously disrupted with NP-40 due to the exposure of the N protein epitopes in the virions without the envelope.
Virus cores were purified from intact virions with MAbs 9D.B4, 3D.E3, and 25.22 bound to virion surfaces. Interestingly, during virus core purification, MAbs 3D.E3 and 9D.B4, directed to the carboxy terminus of the M protein, were lost (Fig. 7B), while no significant loss of MAb 25.22, directed to the M protein amino terminus, was detected.
M protein composition of glycosidase-treated virions.
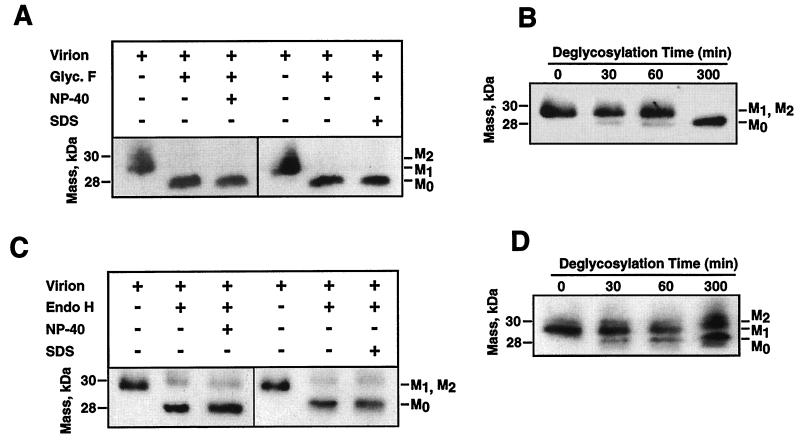
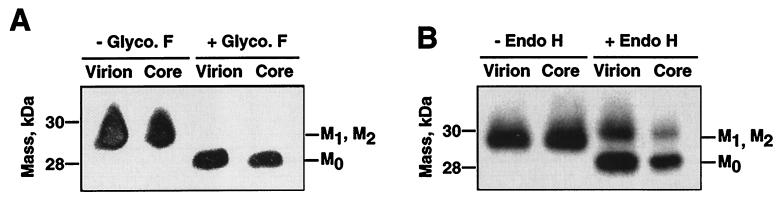
The M protein present in native virions was different in size from the M protein derived from the glycosidase-treated virions after these proteins were resolved by SDS-PAGE and Western blot analysis using the M-specific MAb 9D.B4. We observed bands for three M proteins that were unglycosylated (M0) or had low (M1) and high (M2) glycosylation levels. Nevertheless, the two glycosylation bands (M1 and M2) often were not resolved by standard SDS-PAGE. N-Glycosidase F treatment reduced all M protein bands regularly observed in native virions to 28 kDa, indicating that the different M proteins corresponded to different degrees of N glycosylation (Fig. 8A and B). In contrast, endoglycosidase H treatment led to the observation of two protein bands, a major one corresponding to unglycosylated M protein and another corresponding to the M protein with high glycosylation levels (Fig. 8C and D). The highly glycosylated M protein was resistant to endoglycosidase H treatment under both denaturing (in the presence of 1% SDS) and nondenaturing conditions. Prolonged incubation or the addition of higher concentrations of endoglycosidase H did not reduce the endoglycosidase H-resistant M protein species (Fig. 8D). Virion integrity was shown by MALDI-TOF analysis of trypsin-treated TGEV virions that showed the absence of N protein-derived peptides (Table 1). Cores purified by incubation in the presence of 1% NP-40 from glycosidase-treated virions presented an M protein with the same deglycosylation pattern as that from the native virions (Fig. 9). Untreated virions and cores presented the same M protein species (Fig. 10A and B). N-Glycosidase F-treated virions and cores derived from these virions presented deglycosylated M protein (Fig. 10A). When endoglycosidase H was used, both virions and cores presented two M protein bands, a highly glycosylated M protein and a deglycosylated M protein (Fig. 10B). These data strongly suggested that the M protein found in cores is the same as that present within the virus envelope (Fig. 9).
FIG. 8.
Susceptibility of TGEV M protein to deglycosylation. (A) Western blot of N-glycosidase F (Glyc. F)-treated virions in the presence (+) or absence (−) of the detergents NP-40 and SDS. (B) Deglycosylation kinetics of TGEV virion M protein by N-glycosidase F at the indicated times. (C) Western blot of endoglycosidase H (Endo H)-treated virions in the presence (+) or absence (−) of the detergents NP-40 and SDS. (D) Deglycosylation kinetics of TGEV virion M protein by glycosidase H at the indicated times.
FIG. 9.
Working model for the TGEV virion structure and structural dissociation. A working scheme of the chemical dissociation of TGEV virions compatible with all the experimental observations obtained is shown. According to this model, cores from purified virions were purified by removal of the lipid bilayer with the M protein in a Nexo-Cexo topology. Cores treated with high salt concentrations were disrupted, lost their M proteins, and became unstable, which led to release of the helical nucleocapsids that form the cores. M and M′, membrane protein molecules adopting Nexo-Cendo and Nexo-Cexo topologies, respectively.
FIG. 10.
M protein compositions of cores purified from glycosidase-treated virions. Shown are Western blots of virions and cores purified from virions treated with N-glycosidase glycosidase F (Glyco. F) (A) or endoglycosidase H (Endo H) (B).
DISCUSSION
We have presented evidence for the existence of two topologies for the M protein in the envelope of the TGEV virion, one Nexo-Cendo and the other Nexo-Cexo. One-third of the total amount of M protein of the virions was found in the Nexo-Cexo conformation and associated exclusively with the envelope, while the remaining two-thirds was in the Nexo-Cendo conformation, with the carboxy terminus associated with the virion core.
Both purified virions and cores included M protein N glycosylated to different extents in the virion membrane. The existence of a population of M protein molecules in a Nexo-Cexo topology in the viral envelope was demonstrated by a variety of experimental approaches, including (i) binding of M-specific MAbs to the virion surface; (ii) immunopurification of labeled TGEV virions, including inhibition of this binding by M protein mutants; and (iii) mass spectrometry analysis of the trypsin-treated virions.
The presence of the M protein in association with purified cores as previously reported (9, 28) was confirmed by showing that the M protein specifically interacts with the TGEV internal core by means of a domain of 16 residues in the carboxy terminus (9). These results strongly suggested that the M protein carboxy terminus is embedded in the virus core. Furthermore, it is an essential component of the core, since its removal led to core disruption.
Previous data revealed the existence of a significant proportion of M protein molecules in a Nexo-Cexo topology based mostly on immune electron microscopy evidence (29). In fact, TGEV virions were weakly but significantly neutralized by M-specific MAbs directed to the C domain, which reinforces the concept that C-terminal epitopes were exposed on the surfaces of infective virions. The presence of this topology of the M protein in TGEV particles has been confirmed by alternative methods.
It has been demonstrated that the M protein carboxy-terminal domain was exposed to the virion surface, at least in a significant proportion of the M protein molecules, by immunopurification of 35S-labeled virions with MAbs directed to the amino- and carboxy-terminal domains of the M protein. Similar studies of MHV virions demonstrated that the M protein was found only in a single Nexo-Cendo topology (27). Therefore, exposure of the carboxy terminus seems a characteristic feature of TGEV virions. This is possibly a consequence of the differences in the hydrophilicity patterns of the M proteins from TGEV and MHV (data not shown). Although the two patterns are very similar, the TGEV M protein carboxy terminus is more hydrophobic than the homologous domain in the M protein of MHV, which may have contributed to the M protein topology with a fourth transmembrane domain. Alternatively, since the primary sequences of the two M proteins, as with those of the other structural proteins of TGEV and MHV, are different, the surface exposition of a domain may be conditioned by the interactions between these proteins.
The demonstration of the presence of the M protein carboxy terminus in the external surface of the TGEV virion was in part based on the binding of MAbs specific for different domains of the M protein. It was therefore essential to ensure the specificities of these MAbs before proceeding. Their specificities were determined previously in a bacterial (28) and in a rabbit reticulocyte lysate (9) expression system. To further confirm the domain specificities of these MAbs, we expressed Mwt and three deletion mutant M proteins under the control of the cytomegalovirus promoter in BHK-pAPN cells. The full-length M protein and the deletion mutant M proteins were recognized by the M-specific MAbs, as expected, according to their specificities and displayed the expected pattern by immunofluorescence microscopy. The immunopurification of labeled TGEV virions with M-specific MAbs was inhibited by unlabeled deletion mutant M proteins according to the pattern expected for the existence of an M protein population with a Nexo-Cexo topology in the virion surface (Fig. 5).
Exposure of the C terminus was also confirmed by identifying the virus surface-exposed peptides cleaved with trypsin. These peptides were analyzed by mass spectrometry (MALDI-TOF). The M protein carboxy terminus was readily and clearly detected. The rest of the C-terminal peptides further trimmed by the protease could also be detected after longer incubation times in the presence of trypsin (results not shown). Interestingly, release of the C terminus from the M protein in native purified virions was detected after a short incubation time (15 min) at room temperature with the protease, which suggests that this M protein domain is easily accessible to trypsin in the virion surface. M protein C terminus cleavage by the protease was a consequence of its exposure to the external surface of the virus and not to the presence of open virions, since no peptides from the internal N protein were detected unless the virus envelope was disrupted with NP-40.
The nature of the trypsin-released peptide was confirmed by comparison with the peptide mass fingerprint of the gel-purified TGEV structural proteins. The equivalent peptide was identified only in the M protein fingerprint. Furthermore, the five amino acids identified in the sequence of this peptide matched those of the M protein carboxy terminus. This specific amino acid sequence is present only in the M protein of TGEV, which strongly suggests that it corresponded to a domain of the M protein carboxy terminus.
The M-to-N molar ratio was accurately estimated, and it was found that a significant proportion (two-thirds) of the M protein was present in TGEV cores and that one-third was lost after the membrane disruption. To investigate whether the M protein associated with TGEV cores (two-thirds) corresponded to M protein molecules embedded in the virus envelope, the susceptibility of virion M protein to glycosidase digestion was studied. If the M protein found in the cores had been a subset of the M proteins present only inside the virion particle, then this subset of M protein molecules would have been resistant to glycosidase treatments in purified native virions. However, all the virion M protein was susceptible to deglycosylation by N-glycosidase F, showing that it was embedded in the virus envelope, which exposed glycosylated domains to the virus surface. Furthermore, when virion M protein was deglycosylated and cores were purified from it, the cores included deglycosylated M protein as well, which shows that these M protein species actually came from the virus envelope.
Three M protein bands were detected in virions and cores (M0, M1, and M2). A fraction of the slower-migrating species was resistant to endoglycosidase H treatment under both denaturing and nondenaturing conditions. The resistance to endoglycosidase H treatment was not due to protection by the lipid bilayer of the envelope, since disruption of the envelope with NP-40 and SDS did not modify the deglycosylation pattern. Furthermore, these M protein species were not affected by endoglycosidase H under denaturing conditions even in the presence of SDS, which strongly suggests that they were intrinsically resistant to the glycosidase. This suggestion is in agreement with previously reported results indicating that the M protein molecules have incorporated complex N-linked glycan chains, which are frequently resistant to endoglycosidase H removal (4, 32, 35).
To test whether the difference in M protein composition between virions and cores was due to the different M protein topologies adopted in the viral envelope, cores were purified from virions with MAbs specific for the M protein amino and carboxy termini bound to their surfaces. The 3D.E3 and 9D.B4 MAbs, bound to the M protein in a Nexo-Cexo topology in virions, were lost when the cores were purified. This was not the case with MAb 25.22, which is specific for the M protein amino terminus. These results showed that, although a majority (two-thirds) of the M protein molecules remained associated with the core, the M protein in a Nexo-Cexo topology was completely lost when the viral envelope was disrupted. Consequently, the M protein in a Nexo-Cendo topology copurified with TGEV cores due to embedding of the M protein carboxy terminus within the core. The M protein in a Nexo-Cexo topology would not interact with the internal core due to the exposure of the carboxy terminus to the virion surface. The copurification of MAb 25.22 with the TGEV core when this antibody was bound to the M protein in the virion surface confirmed that the M protein found in purified cores was also associated with the viral envelope. Since the virus cores have associated M protein molecules, these data strongly suggest that the epitopes recognized by MAbs 3D.E3 and 9D.B4 were not exposed in the core surface because the M protein carboxy terminus is embedded within the core structure.
The existence of different topologies is not uncommon in viral membrane proteins. This is the case for the L protein of the hepatitis B virus, which adopts two topologies in the viral envelope, each of them playing a different role (13, 25, 26). The L protein that interacts with the cellular receptor adopts a topology in which the amino-terminal part is exposed to the virion surface. The L protein that interacts with the core, and possibly drives the encapsidation of the nucleocapsid, presents the amino-terminal domain inside the viral particle. Also, nonviral proteins can adopt more than one topology when they are embedded in membranes (23). It is likely that the TGEV M protein in a Nexo-Cendo topology plays an important role in the assembly of the nucleocapsid (9, 22). However, the role of the M protein in a Nexo-Cexo topology is still unknown.
In consideration of all these data together, a model for the TGEV structural dissociation is proposed (Fig. 9). All of the M protein remained embedded in the viral envelope, with two-thirds of the molecules being in a Nexo-Cendo topology and with their carboxy termini being embedded within the core and possibly directly interacting with the N protein or the genomic RNA (9, 22; K. Narayanan, J. Maeda, A. Maeda, and S. Makino, Abstr. 19th Annu. Meet. Am. Soc. Virol., p. 84, 2000). It was estimated that one-third of the M protein molecules do not interact with the internal core due to a Nexo-Cexo topology in the viral membrane and that they are lost during the disruption of the virus envelope with NP-40. It was also previously shown (9) that the remaining M protein was tightly attached to the core by ionic interactions and that its removal by high concentrations of an ionic agent led to complete core disruption and release of the helical nucleocapsid, which suggests that the M protein is essential for core stability.
ACKNOWLEDGMENTS
This work has been supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Communities (Frame V, Key Action 2, Control of Infectious Disease Projects QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874). D.E. and J.O. received a fellowship and contract from the Spanish Department of Education and Culture. E.C. received a contract from the Consejo Superior de Investigaciones Científicas (CSIC).
REFERENCES
- 1.Almazán F, González J M, Pénzes Z, Izeta A, Calvo E, Plana-Durán J, Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansorge W. Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J Biochem Biophys Methods. 1985;11:13–20. doi: 10.1016/0165-022x(85)90037-5. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D. Coronavirus IBV glycopolypeptides: size of their polypeptide moieties and nature of their oligosaccharides. J Gen Virol. 1983;64:1187–1191. doi: 10.1099/0022-1317-64-5-1187. [DOI] [PubMed] [Google Scholar]
- 5.Charley B, McCullough K, Martinod S. Antiviral and antigenic properties of recombinant porcine interferon gamma. Vet Immunol Immunopathol. 1988;19:95–103. doi: 10.1016/0165-2427(88)90001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correa I, Gebauer F, Bullido M J, Suñé C, Baay M F D, Zwaagstra K A, Posthumus W P A, Lenstra J A, Enjuanes L. Localization of antigenic sites of the E2 glycoprotein of transmissible gastroenteritis coronavirus. J Gen Virol. 1990;71:271–279. doi: 10.1099/0022-1317-71-2-271. [DOI] [PubMed] [Google Scholar]
- 7.Delmas B, Gelfi J, L'Haridon R, Vogel L K, Norén O, Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enjuanes L, Brian D, Cavanagh D, Holmes K, Lai M M C, Laude H, Masters P, Rottier P, Siddell S G, Spaan W J M, Taguchi F, Talbot P. Coronaviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carsten E B, Estes M K, Lemon S M, McGeoch D J, Maniloff J, Mayo M A, Pringle C R, Wickner R B, editors. Virus taxonomy. Classification and nomenclature of viruses. New York, N.Y: Academic Press; 2000. pp. 835–849. [Google Scholar]
- 9.Escors D, Ortego J, Laude H, Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebauer F, Posthumus W A P, Correa I, Suñé C, Sánchez C M, Smerdou C, Lenstra J A, Meloen R, Enjuanes L. Residues involved in the formation of the antigenic sites of the S protein of transmissible gastroenteritis coronavirus. Virology. 1991;183:225–238. doi: 10.1016/0042-6822(91)90135-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godet M, L'Haridon R, Vautherot J F, Laude H. TGEV coronavirus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo J-T, Pugh J C. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houthaeve T, Gausepohl H, Mann M, Ashman K. Automation of micro-preparation and enzymatic cleavage of gel electrophoretically separated proteins. FEBS Lett. 1995;376:91–94. doi: 10.1016/0014-5793(95)01242-7. [DOI] [PubMed] [Google Scholar]
- 15.Jiménez G, Correa I, Melgosa M P, Bullido M J, Enjuanes L. Critical epitopes in transmissible gastroenteritis virus neutralization. J Virol. 1986;60:131–139. doi: 10.1128/jvi.60.1.131-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laude H, Chapsal J M, Gelfi J, Labiau S, Grosclaude J. Antigenic structure of transmissible gastroenteritis virus. I. Properties of monoclonal antibodies directed against virion proteins. J Gen Virol. 1986;67:119–130. doi: 10.1099/0022-1317-67-1-119. [DOI] [PubMed] [Google Scholar]
- 17.Laude H, Gelfi J, Lavenant L, Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laude H, Vanreeth K, Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res. 1993;24:125–150. [PubMed] [Google Scholar]
- 19.Martín-Andrés A, Luna-Castillo J D. Bioestadistica para las ciencias de la salud. 4th ed. Madrid, Spain: Ediciones Norma, S.A.; 1994. [Google Scholar]
- 20.McClurkin A W, Norman J O. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can J Comp Med Vet Sci. 1966;30:190–198. [PMC free article] [PubMed] [Google Scholar]
- 21.Motulsky H. Intuitive biostatistics. New York, N.Y: Oxford University Press; 1995. pp. 225–229. [Google Scholar]
- 22.Narayanan K, Maeda A, Maeda J, Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishiyama K, Suzuki T, Tokuda H. Inversion of the membrane topology of secG coupled with secA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 24.Penzes Z, González J M, Calvo E, Izeta A, Smerdou C, Mendez A, Sánchez C M, Sola I, Almazán F, Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 26.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raamsman M J B, Locker J K, de Hooge A, de Vries A A F, Griffiths G, Vennema H, Rottier P J M. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risco C, Antón I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risco C, Antón I M, Suñé C, Pedregosa A M, Martín-Alonso J M, Parra F, Carrascosa J L, Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez C M, Jiménez G, Laviada M D, Correa I, Suñé C, Bullido M J, Gebauer F, Smerdou C, Callebaut P, Escribano J M, Enjuanes L. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 32.Stern D F, Sefton B M. Coronavirus proteins: structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J Virol. 1982;44:804–812. doi: 10.1128/jvi.44.3.804-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturman L S, Holmes K V. The molecular biology of coronaviruses. Adv Virus Res. 1983;28:36–112. doi: 10.1016/S0065-3527(08)60721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vennema H, Heijnen L, Zijderveld A, Horzinek M C, Spaan W J M. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates J R., III . Peptide sequencing by tandem mass spectrometry. In: Celis J E, editor. Cell biology: a laboratory handbook, 2 ed. Vol. 4. San Diego, Calif: Academic Press; 1998. pp. 529–538. [Google Scholar]