Abstract
Tacaribe virus (TV), the prototype of the New World group of arenaviruses, comprises a single phylogenetic lineage together with four South American pathogenic producers of hemorrhagic disease. The TV genome consists of two single-stranded RNA segments called S and L. A reconstituted transcription-replication system based on plasmid-supplied TV-like RNAs and TV proteins was established. Plasmid expression was driven by T7 RNA polymerase supplied by a recombinant vaccinia virus. Plasmids were constructed to produce TV S segment analogs containing the negative-sense copy of chloramphenicol acetyltransferase (CAT) flanked at the 5′ and 3′ termini by sequences corresponding to those of the 5′ and 3′ noncoding regions of the S genome (minigenome) or the S antigenome (miniantigenome). In cells expressing N and L proteins, input minigenome or miniantigenome produced, respectively, encapsidated miniantigenome or minigenome which in turn produced progeny minigenome or progeny miniantigenome. Both minigenome and miniantigenome in the presence of N and L mediated transcription, which was analyzed as CAT expression. Coexpression of the small RING finger Z (p11) protein was highly inhibitory to both transcription and replication mediated by the minigenome or the miniantigenome. The effect depended on synthesis of Z protein rather than on plasmid or the RNA and was not ascribed to decreased amounts of plasmid-supplied template or proteins (N or L). N and L proteins were sufficient to support full-cycle RNA replication of a plasmid-supplied S genome analog in which CAT replaced the N gene. Replication of this RNA was also inhibited by Z expression.
Tacaribe virus (TV) is the prototype of the New World group of arenaviruses. Within this group the viruses form three phylogenetically distinct clades, one of which includes TV together with the four known South American pathogens that produce severe hemorrhagic disease (Junin, Machupo, Guanarito, and Sabia viruses) (5). TV, however, seems not to be a human pathogen.
TV, like all arenaviruses, is an enveloped virus with its genetic information contained in two single-stranded RNA segments called S (ca. 3.4 kb) and L (ca. 7.1 kb). The S RNA contains two genes encoding, respectively, the nucleoprotein (N, 64 kDa) and the glycoprotein precursor (GPC) (55 kDa) (10). The L RNA also posseses two genes which encode the presumptive RNA-dependent RNA polymerase (L protein, 240 kDa) (16) and a small protein with a RING finger motif (Z, 11 kDa; this protein was originally named p11 [17]). The genes in both the S and L RNAs are arranged in opposite orientations and are separated by a noncoding sequence that has the potential to form stable secondary structures in the form of hairpins (11) (the organization of the S RNA is shown in Fig. 1A). Although the 5′ region of arenavirus genomes and antigenomes are positive stranded, they are not in fact translated directly into proteins. Rather, genomes and antigenomes are found only as nucleocapsids tightly bound to N protein (11, 20), and the coding sequences are expressed from mRNAs transcribed from the 3′ region of the genomes or antigenomes (2, 11, 26). These mRNAs contain short stretches of nontemplated nucleotides at their 5′ end and appear to be capped, as they react with anticap antibodies (20). Transcription termination in TV occurs at the base of the hairpin on the distal side. This determines a hairpin structure at the 3′ end of the transcripts that has been suggested to operate as a signal for the termination of transcription (18). By analogy with other negative-strand RNA viruses, it is assumed that L constitutes the viral polymerase together with N when this protein is associated with RNA in nucleocapsid structures. Studies in TV-infected cell extracts immunodepleted with antiserum to Z led to the proposal that this protein is required for TV transcription and RNA replication (13). However, it was recently reported that in cells transfected with a lymphocytic choriomeningitis virus (LCMV)-like RNA containing the chloramphenicol acetyltransferase (CAT) reporter gene, CAT activity was generated in cells expressing only N and L (21). Z might also play other roles in arenavirus biology, as suggested in recent reports on interactions between Z and several cellular proteins (3, 4, 7).
FIG. 1.
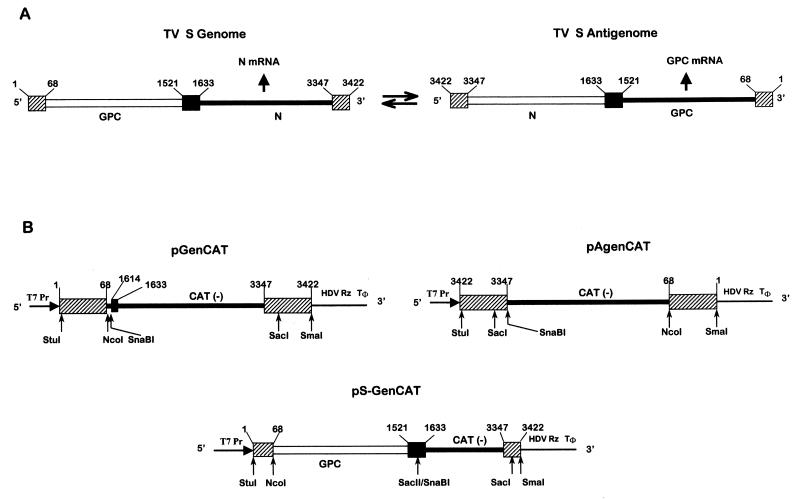
Schematic diagram (not to scale) showing the TV S RNA organization and expression (arrows show the direction of RNA synthesis) (A) and plasmids used to generate TV RNA analogs (B). The position of T7 polymerase promoter (T7Pr), the HDV Rz, and the T7 RNA polymerase terminator (Tφ) are indicated. Constructions were performed as indicated in Materials and Methods. pGenCAT transcript (842 nt) contains (5′ to 3′) the entire TV S genome 5′ NCR sequence (68 nt), a short linker (16 nt) including a SnaBI site, a short sequence (nt 1614 to 1633) of the S genome intergenic region (IGR) which is not involved in the hairpin structure (18), CAT ORF in an antisense orientation (660 nt), and the complete S genome 3′ NCR sequence (76 nt). pAgenCAT transcript (810 nt) contains (5′ to 3′) the entire TV S antigenome 5′ NCR sequence, a SnaBI site, CAT ORF in an antisense orientation, and the complete S antigenome 3′ NCR. pS-GenCAT (2,343 nt), contains (5′ to 3′) nt 1 to 1633 of the TV S genome followed by the negative-sense copy of CAT and the entire S genome 3′ NCR sequence. Sizes of transcripts refer to the processed RNA, which includes two nonviral Gs at the 5′ end (23). Nucleotides were numbered considering as 1 the 5′ end of the S genome. Open lines represent positive-sense coding regions; negative-sense coding regions are indicated in blackened thicker lines. S RNA IGR is represented as a black box, and the 3′ and 5′ NCR termini are indicated as shaded boxes. Key restriction endonuclease sites used in the assembly of the DNA constructs are shown.
Reconstitution of transcription and RNA replication from plasmid-supplied RNAs and proteins would open TV to the analysis of cis-acting signals on the RNA genome, such as those required for encapsidation, initiation, and termination of RNA synthesis, and would render each of the TV proteins accessible to reverse genetic analysis.
In this study we describe the establishment of a reconstituted system based on plasmid-supplied TV RNAs and proteins. Using this system we demonstrate that (i) the noncoding sequences at the 5′ and 3′ termini of the S genome and S antigenome contain all of the cis-acting signals required for transcription, RNA replication, and encapsidation; (ii) both N and L proteins are sufficient to drive transcription and full-cycle replication mediated by the S genomic and antigenomic-like RNAs; and (iii) Z protein is a potent inhibitor of transcription and replication.
MATERIALS AND METHODS
Cells and viruses.
CV1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (GIBCO-BRL, Gaithersburg, Md.). Recombinant vaccinia virus vTF7-3, which expresses the T7 RNA polymerase, was kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.) (12). Cells were routinely infected with 3 to 5 PFU of vTF7-3 per cell so that more than 90% of the cells were infected. A TV working stock, prepared as indicated previously (19), was used to infect CV1 cells at a multiplicity of infection of 1 PFU per cell, and cell extracts were prepared at 2 days postinfection, when the virus is actively engaged in transcription and replication (19).
Plasmids expressing TV-like RNAs.
Plasmids were constructed in transcription vector pTV2.0, a generous gift of Andrew Ball, University of Alabama at Birmingham. This vector provides a T7 RNA polymerase promoter sequence immediately upstream of a unique StuI restriction site and a unique SmaI site 5′ of the hepatitis delta virus ribozyme sequence (HDV Rz) preceding a T7 polymerase terminator sequence (Tφ) (23). To eliminate the SacI site from pTV2.0, the plasmid was digested with SacI, treated with Klenow, and religated. The modified vector, named pTV2.0(SacI−), was used for the construction of two plasmids transcribing TV S genome analogs (pGenCAT and pS-GenCAT) and one plasmid expressing a TV S antigenome analog (pAgenCAT) (Fig. 1B). Construction of pGenCAT was performed as follows: the 5′ and 3′ regions of TV S RNA, comprising, respectively, nucleotides (nt) 1 to 334 and 3031 to 3422 (numbers indicate positions relative to the 5′ end of the genome-sense sequence) (10, 18), were amplified by reverse transcription-PCR (RT-PCR) using total RNA from TV-infected cells. The PCR product corresponding to the 5′ region of the S genome was inserted into the StuI site of pTV2.0(SacI−), downstream of the T7 RNA polymerase promoter, and that corresponding to the 3′ region was inserted into the SmaI site, 5′ of the HDV Rz sequence. This construction, designated p13-6, was cleaved with NcoI, treated with Klenow to fill in the end, and digested with SacI. Cleaved sites (which corresponded, respectively, to nt 67 and 3381 of the TV S genome) were used to position the CAT open reading frame (ORF) in an antisense polarity with respect to the T7 RNA polymerase promoter as follows: CAT ORF was obtained by PCR using plasmid pCAT3-Basic Vector (Promega, Madison, Wis.) as template and a forward primer containing (5′ to 3′) a short linker sequence with a SnaBI site (inserted for cloning purposes), a sequence corresponding to nt 1614 to 1633 of TV S genome, and a sequence complementary to positions 660 to 636 of the CAT ORF. The reverse primer, designed so that CAT ORF would replace the TV N ORF, encompasses (5′ to 3′) a sequence complementary to nt 3390 to 3347 of the TV S genome plus nt 1 to 27 of the CAT ORF. The PCR product was digested with SacI, purified, and inserted into p13-6 excised as indicated above. To generate plasmid pS-GenCAT, the complete TV S genomic sequence was first cloned into pTV2.0(SacI−) by insertion of a NcoI-BstEII DNA fragment from plasmid p2b2 (10) into p13-6 that had been digested with the same enzymes. The resultant plasmid (pS-Gen) was then used to introduce the CAT ORF instead of the N ORF. To this end, a SnaBI-SacI fragment from pGenCAT was cloned into pS-Gen that had been previously cleaved with SacII, treated with Klenow, and digested with SacI. Construction of pAgenCAT was performed as follows: the 5′ and 3′ regions of TV S RNA were amplified by RT-PCR as described above and cloned into pTV2.0(SacI−) so that the PCR product corresponding to the 5′ region of the S antigenome was inserted immediately downstream of the T7 RNA polymerase promoter and the fragment corresponding to the 3′ region of the S antigenome was cloned immediately upstream of the HDV Rz sequence. The plasmid was designated p15-2 and was used to insert the CAT ORF in an antisense orientation with respect to the T7 RNA polymerase promoter. The CAT ORF was amplified by PCR from pCAT3-Basic Vector using a forward primer which included (5′ to 3′) a sequence complementary to positions 3388 to 3347 of the TV S genome and 4 nt which complete a SnaBI site (added for cloning purposes), all followed by a sequence complementary to nt 660 to 638 of the CAT ORF. The reverse primer consisted (5′ to 3′) of nt 60 to 68 of the TV S genome plus nt 1 to 30 of the CAT ORF. The PCR product was digested with NcoI and SacI and inserted into p15-2 that had been cleaved with the same enzymes.
Plasmids expressing TV proteins.
Plasmids expressing TV L, N, and Z proteins (designated, respectively, pL, pN, and pZ) were constructed as follows: (i) for construction of pL, the L ORF was engineered from plasmids p96 and p14. These plasmids harbor inserts covering, respectively, positions 161 to 2240 and 2025 to 7102 relative to the 5′ end of the L genome (16). To serve as a bridge between p96 and p14 inserts in the cloning procedure, a region covering nt 1586 to 2806 of the L genome was amplified by RT-PCR using total RNA from TV-infected cells. The reverse primer included a BamHI site at its 5′ end. The RT-PCR product was digested with SnaBI and BamHI and cloned into p96 cleaved with SnaBI and BamHI, the latter site corresponding to the polylinker of p96. The resultant plasmid was digested with PvuII and BamHI and used to insert a PvuII-BamHI fragment excised from p14. This last construction was finally digested with BamHI and SmaI, and the 6,669-bp fragment containing the complete L ORF plus 22 nt of the noncoding region (NCR) 5′ of the L AUG was inserted into the corresponding sites of pGEM-4 (Promega). (ii) For construction of pN, a DNA fragment containing the complete N ORF plus 31 nt of the NCR 5′ of the N start codon was obtained by cleavage of p2b2 (10) with SacI, treatment with Klenow, and digestion with SmaI. The fragment was then cloned into pGEM-3 (Promega) previously digested with SmaI. (iii) For pZ construction, a cDNA covering the entire Z ORF and 43 nt of the NCR 5′ of the Z start codon (kindly provided by D. Kolakofsky, University of Geneva, Geneva, Switzerland) was cloned into the SmaI site of pGEM-3. (iv) Plasmid pZmut is a mutant of pZ plasmid in which the Z start codon has been replaced by a stop codon. This mutation was inserted by PCR with plasmid pZ as the template and with a forward primer containing (5′ to 3′) an EcoRI restriction site followed by nt 27 to 93 of the L genome, which included changes at positions 70 and 71. This resulted in the replacement of the Z start site by a stop codon (AUG for UAG). The reverse primer contained (5′ to 3′) a BamHI site followed by a sequence complementary to nt 372 to 354 of the L genome. The PCR product, containing the complete copy of the Z ORF with the mutated Z start site, was inserted into the EcoRI-BamHI sites of pGEM-3 downstream of the T7 RNA polymerase promoter. Plasmid constructions were verified by DNA sequencing of the junctions. DNA fragments obtained by PCR were totally sequenced. All plasmids were purified by using Qiagen Tip-100 (Qiagen Inc., Valencia, Calif.).
Radiolabeling, immunoprecipitation, and protein gel electrophoresis.
CV1 cells transfected or infected with TV were radiolabeled as indicated in the legends for Fig. 3 and 7, below. Harvesting of the cells, radioimmunoprecipitation using monospecific serum to each protein (25), and analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as indicated previously (22).
FIG. 3.
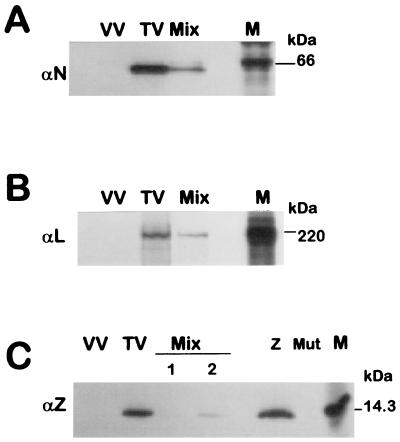
Analysis of TV proteins expressed in virus-infected and plasmid-transfected cells. CV1 cells were infected with TV (lanes TV), vTF7-3 (lanes VV), or vTF7-3 and transfected with a mixture of pGenCAT, pL, and pN as indicated in Materials and Methods (lanes Mix) without (A and B) or with the addition of 10 ng (C, lane 1) or 100 ng (C, lane 2) of pZ. Lanes Z and Mut in C, correspond to cells infected with vTF7-3 and transfected, respectively, with 1 μg of pZ or 1 μg of pZmut. Transfected cells were incubated for 18 h and TV-infected cells were incubated for 40 h and then labeled for 1.5 h with [35S]cysteine (150 μCi/ml) and cell extracts were prepared. At the time of labeling, both TV-infected and transfected cells were actively engaged in transcription and replication (see Materials and Methods). Cytoplasmic extracts were immunoprecipitated with monospecific serum against N (panel A), L (panel B), or Z (panel C) proteins. Immunoprecipitated proteins (corresponding to 2 × 105 cells per lane) were separated on an SDS-PAGE gel containing 10% (A), 7% (B), or 17% (C) polyacrylamide, followed by direct exposure to X-ray film for 2, 6, and 4 days (panels A, B, and C, respectively), using intensifying screen (Biomax, Kodak). Molecular masses of 14C-labeled markers (lanes M) are indicated in kilodaltons.
FIG. 7.
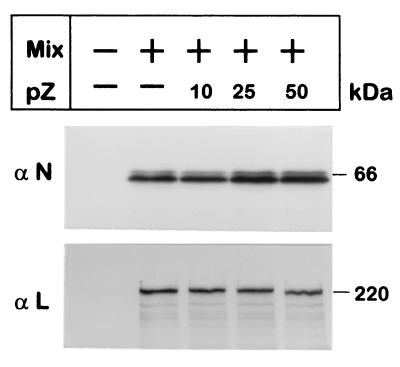
Effect of Z protein expression on the synthesis of N or L proteins. CV1 cells (4 × 105) were infected with vTF7-3 and those indicated (+) were transfected with: 2 μg of pGenCAT, 2 μg of pN, and 50 ng of pL (Mix) without or with the addition of 10, 25, or 50 ng of pZ as indicated. At 24 h posttransfection, cells were labeled for 1.5 h with a mixture of [35S]cysteine (80 μCi/ml) and [35S]methionine (80 μCi/ml), cytoplasmic extracts were prepared, and proteins were immunoprecipitated with monospecific serum directed to N (αN) or L (αL) proteins. Immunoprecipitated proteins (corresponding to 2 × 105 cells per lane) were subjected to SDS-PAGE gels containing 12% (αN) or 7% (αL) polyacrylamide. Gels were exposed to X-ray film using an intensifying screen (Biomax, Kodak). Molecular masses according to 14C-labeled markers are indicated on the right.
DNA transfections.
Subconfluent monolayers of CV1 cells were infected with vTF7-3 for 1 h at 37°C. The inoculum was removed and the cells were washed and then transfected using Lipofectamine 2000 reagent (GIBCO-BRL), according to the manufacturer's specifications. Specifically, the amount of plasmid added to approximately 4 × 105 cells grown in a 12-well dish was as follows: 2 μg of minigenome or miniantigenome plasmid, 2 μg of pN, and 50 ng of pL per well. When pS-GenCAT was transfected, the amount of plasmid was increased to 2.6 μg per well. The amount of transfected pZ and pZmut is indicated below in the respective figure legends. The total amount of transfected DNA was kept constant by addition of the appropriate amount of empty pGEM-3 DNA. Under these conditions of transfection, accumulation of CAT activity (data not shown) and amplification of plasmid-supplied template by the TV polymerase (see Fig. 5) proceeded for at least 2 days.
FIG. 5.
Replication of plasmid-supplied minigenome or miniantigenome. Subconfluent cultures of CV1 cells were infected with vTF7-3 and transfected with pGenCAT (A) or pAgenCAT (B) and pN and/or pL, as indicated. At 24 h (lanes 1 to 3, 6, and 7) and 40 h (lanes 4, 5, 8, and 9) posttransfection (h.p.t.), total RNA (lanes 1 to 5) or nucleocapsid-associated RNA (Nc RNA, lanes 6 to 9) was purified and analyzed by Northern blotting. Duplicate blots were hybridized either with an antisense CAT riboprobe [CAT(−)] (upper panels) or a sense CAT riboprobe [CAT(+)] (lower panels). Films were exposed for 30 h with the exception of lanes 1 to 5 in lower panels that were exposed for 3 h. Arrows on the left side of the panels indicate the position of the RNAs synthesized by the TV polymerase or the T7 polymerase. Sizes (in kilobases) of marker RNAs (Promega) are indicated on the right side of the panels.
CAT assay.
Cells (approximately 4 × 105) were washed twice with ice-cold phosphate-buffered saline and once with TNE (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA), harvested by scraping into TNE, and recovered by centrifugation. Cells were lysed in 60 μl of 250 mM Tris-HCl (pH 7.4) by three cycles of freezing and thawing. Cell lysates were clarified by centrifugation (13,000 × g for 5 min at 4°C), and endogenous CAT was inactivated by heating for 10 min at 65°C. For the assay, 5 μl of cell extract was mixed with 0.2 μCi of [14C]chloramphenicol (57 mCi/mmol; New England Nuclear, Boston, Mass.), 15 μl of 4 mM acetyl coenzyme A (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England) adjusted to 250 mM of Tris-HCl (pH 7.4) in a total volume of 130 μl, and incubated for 1 h at 37°C. Products of the CAT assay were analyzed by ascending thin-layer chromatography (TLC) developed with 19:1 chloroform-methanol followed by autoradiography. The amount of cell extract assayed was determined so that the assay was in the linear range of enzyme activity (conversion of nonacetylated to monoacetylated chloramphenicol less than 30%). CAT activity was calculated by determining the percentage of counts that were the monoacetylated chloramphenicol species relative to total counts, achieved by cutting the appropriate pieces of TLC and counting in scintillation fluid (14).
Analysis of RNA.
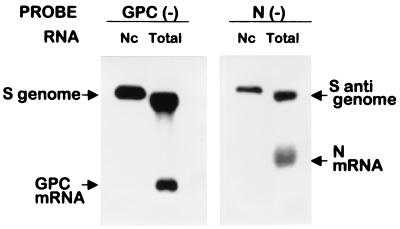
Total RNA from infected-transfected cells was purified at the indicated times by the procedure of Chomczinski and Sacchi (8). For obtaining nucleocapsid-associated RNA, cells were harvested by scraping into cold phosphate-buffered saline. Pelleted cells were lysed in TNEN (TNE containing 0.2% Nonidet P-40), and the cytoplasmic fraction was recovered after centrifugation. Encapsidated RNAs were immunoselected as follows: for each sample, 35 μl of protein A-Sepharose 4B fast flow (Sigma-Aldrich, St. Louis, Mo.) was incubated with 25 μl of antiserum to N protein for 90 min at room temperature (RT) in a total volume of 100 μl. After four washes with cold TNEN, the antibody-coupled protein A-Sepharose mixture was incubated with cell lysates (6 × 105 cells in a total volume of 300 μl) for 30 min at RT followed by 60 min at 4°C. Beads containing immunoselected nucleocapsids were washed six times as above and treated with 1% SDS, and the associated RNA was phenol extracted and ethanol precipitated with 70 μg of tRNA as carrier. When the proportion of antibody-coupled beads relative to cell extract was increased twofold, the amount of RNA recovered remained unchanged, indicating that the immunoselection was carried out under limiting conditions. As seen in Fig. 2, when this procedure was applied to cytoplasmic extracts from TV-infected cells, analysis of the immunoselected RNAs revealed only genomes and antigenomes but not subgenomic RNAs.
FIG. 2.
Northern blot analysis of immunoselected and total RNA from TV-infected cells. Cells were infected with TV as described in Materials and Methods. Immunoselection of nucleocapsids (Nc) and extraction of Nc-associated and total RNA from infected cell extracts were performed as indicated in Materials and Methods. RNAs were analyzed by Northern blotting using 32P-labeled negative-sense GPC or N riboprobes which detected, respectively, S genome and GPC mRNA or S antigenome and N mRNA. Comigration of 28S rRNA with genome and antigenome RNA in the total RNA preparation leads to a higher mobility of these RNAs than that observed for the genome and antigenome RNA extracted from immunoselected nucleocapsids.
Purified RNA (corresponding to 3 × 105 cells per lane) was separated on 1.5% agarose-morpholinepropanesulfonic acid gels containing 2.2 M formaldehyde and transferred onto a Hybond-N membrane (Amersham Pharmacia Biotech) as instructed by the manufacturer. Blots were prehybridized for 2 h at 65°C in a solution containing 50% formamide, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA) (pH 7.7), 5× Denhardt's solution, 0.5% SDS, 100 μg of boiled salmon sperm DNA/ml, and 100 μg of yeast tRNA/ml. 32P-labeled riboprobes were then added directly to the hybridization bag and allowed to hybridize for 18 h at 65°C. The filters were washed two times for 10 min each in 2× SSPE, 0.1% SDS at RT and subsequently for 15 min in 1× SSPE, 0.1% SDS at 65°C followed by two washes for 10 min each in 0.1× SSPE, 0.1% SDS at 65°C. Filters were exposed to film (BioMax Film; Kodak, Rochester, N.Y.) with an intensifying screen at −70°C.
CAT riboprobes were synthesized by in vitro transcription of a linearized pGEMCAT, using either SP6 or T7 polymerase and [32P]CTP. This plasmid was obtained by inserting the CAT ORF (without any TV sequence) between the SP6 and T7 promoters into pGEM-3 vector. Similar total incorporation was obtained with either positive- or negative-sense CAT probe. GPC and N riboprobes were synthesized similarly by using linearized pGEMGPC and pGEMN plasmids and SP6 polymerase.
RESULTS
The present model concerning arenavirus replication assumes that both genomes and antigenomes serve as a template for RNA replication and for transcription of a single mRNA (Fig. 1, S RNA). There is a high degree of sequence identity between the 3′ ends of the arenavirus genomes and antigenomes. Specifically, the TV S genome and antigenome are identical up to nt 20 with the exception of positions 6 and 8, whereas the first 20 nt of the TV L genome and antigenome differ only with regard to position 8. After nt 21, there is no sequence relatedness. As a result of these conserved sequences, both genomic and antigenomic RNAs have a high degree of complementarity between the 3′ and 5′ ends and thus are potentially able to form a panhandle structure (11). The conserved primary sequence at the 3′ end (or the conserved base pairing between the 3′ and 5′ ends) of the genomes and the antigenomes are more likely to represent a conserved promoter for transcription and replication. These sequences might also specify encapsidation signals in the nascent RNA. Assuming this hypothesis to be correct, plasmid-derived TV-like RNAs containing the 3′ and 5′ end sequences of either the genome or the antigenome should be both transcribed and replicated in cells in which the viral transcription-replication proteins are also expressed. To obtain these templates, we constructed the plasmids pGenCAT and pAgenCAT (Fig. 1). pGenCAT directs the synthesis of a TV S genome analog containing the negative-sense copy of the reporter CAT gene flanked, at its 5′ and 3′ ends, by the 5′ and 3′ NCR sequences of the TV S genome. pAgenCAT expresses a TV S antigenome analog in which the negative-sense CAT coding sequence is surrounded at its 5′ and 3′ ends by the 5′ and 3′ NCR sequences of the S antigenome. These constructions were performed in transcription vector pTV2.0 downstream of the T7 RNA polymerase promoter. As shown by Pattnaik et al. (23), pTV2.0 permits the obtention of precise termini of the transcribed RNA. The 5′ end is determined by the position of the T7 promoter relative to the insert, whereas the exact 3′ end of the transcript is generated by autocatalytic cleavage mediated by the HDV Rz placed immediately downstream of the insert (Fig. 1B). The TV-like RNAs expressed by pGenCAT and pAgenCAT are hereafter called minigenome and miniantigenome, respectively.
Genes encoding TV proteins that are likely involved in transcription and replication —those of the putative RNA polymerase (L), the nucleocapsid protein (N), and the Z protein—were cloned under the control of the promoter for T7 RNA polymerase. To analyze the proteins expressed by these plasmids, each plasmid was transfected in CV1 cells that had been infected with vaccinia virus vTF7-3 expressing T7 RNA polymerase to drive plasmid expression. At 18 h posttranfection, plasmid-expressed proteins were examined by metabolic labeling and SDS-PAGE following radioimmunoprecipitation with monospecific serum to each protein. TV-expressed proteins were simultaneously analyzed. As shown in Fig. 3, plasmid-expressed proteins showed electrophoretic mobility identical to those of authentic TV proteins.
Transcription of minigenome and miniantigenome by plasmid-expressed TV proteins.
Transcription mediated by the minigenome or the miniantigenome was analyzed as CAT expression. After transfection of either the minigenome or miniantigenome together with support plasmids, CAT activity would only be detected if the RNAs were recognized and transcribed into message-sense RNA by the TV polymerase. CAT mRNA copied off the minigenome or the miniantigenome contained, 5′ of the CAT start codon, the 5′ NCR of the N mRNA or the GPC mRNA, respectively (Fig. 1). These sequences share 70% overall identity, and each start codon is surrounded by a sequence very favorable for initiation by eukaryotic ribosomes (10). This implies that the translational efficiency of CAT mRNA transcribed from the minigenome or the miniantigenome would be very similar. In preliminary experiments, we detected CAT activity when the minigenome or the miniantigenome was transfected in cells expressing L and N. Then, we studied the amounts and ratios of transfected plasmids to obtain the highest level of CAT activity. The best conditions were obtained when the plasmid-supplied template (either the minigenome or the miniantigenome) and pN were transfected in equal amounts at a pN/pL ratio of about 40 (see Materials and Methods). Under these conditions, the expression levels of N and L proteins in plasmid-transfected cells were lower than those observed in TV-infected cells and the proportion of N to L in transfected cells and in TV-infected cells was quite similar (Fig. 3A and B).
The experiment depicted in Fig. 4 was performed under the optimum conditions for obtaining maximal reporter gene expression. Cells were infected with vTF7-3 and subsequently transfected with pGenCAT, expressing the minigenome (Fig. 4A), or with pAgenCAT, expressing the miniantigenome (Fig. 4B), together with various combinations of plasmids encoding the TV L, N, and Z proteins. At 24 h posttransfection, cell extracts were prepared and CAT activity was assayed. In cells expressing L, N, or Z proteins individually (lanes 1, 2, and 3) or expressing Z in combination with L or N (lanes 4 and 5), CAT activity was undetectable. In cells expressing N and L together, CAT expression was readily detected (lane 6), indicating that L and N are the minimal trans-acting viral factors required for CAT expression mediated by the minigenome or by the miniantigenome. Estimations from three independent experiments indicated that CAT activity produced by transfected miniantigenome was 4 to 7% of that yielded by input minigenome (not shown). Unexpectedly, inclusion of 10 ng of pZ in cells expressing L and N caused a significant reduction of CAT activity mediated by either the minigenome or the miniantigenome, and cotransfection with 100 ng of pZ almost abolished CAT expression (lanes 7 and 8). CAT activity expressed from the minigenome or the miniantigenome appeared almost equally reduced (Fig. 4C).
FIG. 4.
Detection of CAT activity mediated by the minigenome or the miniantigenome. Duplicate dishes were transfected with pGenCAT (A) or pAgenCAT (B) with (+) or without (−) the addition of pN and pL as indicated. When pZ or pZmut was included, its amount is indicated in nanograms. At 24 h posttransfection, cell extracts were prepared, CAT activity was assayed, and the reaction products were analyzed as described in Materials and Methods. Films were exposed for 18 h (A) or 48 h (B). (C) CAT activity expressed from the minigenome (▴) or the miniantigenome (●) was quantified in three independent experiments in which cells were transfected as in lanes 6 to 8. The mean values obtained are shown as the percentage of the corresponding value without added pZ.
We then investigated whether the inhibition associated with cotransfection with Z plasmid depended upon its ability to express Z protein or whether the inhibitory effect might be an artifact of transfection of the Z plasmid or synthesis of Z mRNA. To distinguish between these possibilities, we first mutated pZ in order to preclude synthesis of Z protein. This was done by replacing the Z start site by a stop codon. As expected, Z protein was not expressed in cells transfected with the mutant plasmid (designated pZmut), while the original plasmid produced Z protein (Fig. 3C). When pZmut was evaluated for its effect on CAT activity, it was found that CAT expression was undiminished in cells cotransfected with 10 ng and 100 ng of pZmut, whereas inclusion of these amounts of pZ strongly inhibited CAT expression (Fig. 4, compare lanes 7 and 8 with lanes 9 and 10). Note that inhibition by pZ was associated with very small amounts of transfected plasmid and was observed at levels of Z protein synthesis barely detected by radioimmunoassay (Fig. 3C) and below the limit of detection by Western blotting (results not shown). Intriguingly, the proportion of expressed Z protein relative to N or L in the transfected cells when the inhibitory effect occurred was lower than that observed in TV-infected cells (Fig. 3, A to C).
RNA encapsidation and replication.
Transcription of CAT mRNA implied that the added templates (minigenome or miniantigenome) had associated with the N protein in the form of nucleocapsid structures that, together with L protein, formed functional nucleocapsids. We speculated then whether these putative nucleocapsids would also support RNA replication. Thus, the plasmid-supplied minigenome would produce, in the first step of replication, an encapsidated CAT(+) miniantigenome that would serve in turn as the template for the second step in RNA replication, namely, the production of progeny encapsidated minigenome. Conversely, the plasmid-supplied miniantigenome would first produce an encapsidated CAT(+) minigenome that would be the template for synthesis of progeny encapsidated miniantigenome. To analyze RNA replication, we performed the experiment depicted in Fig. 5. Plasmids encoding either the minigenome (Fig. 5A) or the miniantigenome (Fig. 5B) were transfected, together with support plasmids, into CV1 cells that had been infected with vTF7-3 to drive plasmid expression. The amounts of transfected plasmids used were those previously found to be optimum for CAT expression. At 24 and 40 h posttransfection, total intracellular RNA was extracted directly from a set of transfected wells, whereas cell extracts for nucleocapsid analysis were prepared from duplicate transfections. Nucleocapsids were selectively immunoprecipitated with antibody to TV N protein, followed by purification of the RNA present in the immunoprecipitate. This procedure was shown to be reliable for the analysis of TV nucleocapsids, since only TV genomes and antigenomes—which are always encapsidated (18)—were present in the immunoprecipitate, whereas mRNAs were excluded (Fig. 2). Total intracellular RNA and nucleocapsid-associated RNA were then analyzed by Northern blotting using strand-specific probes.
To visualize RNA products copied from the supplied templates, blots shown in the upper panels of Fig. 5 were hybridized with a 32P-labeled negative-sense CAT riboprobe. Transfection of both minigenome and miniantigenome would produce CAT(+) miniantigenome and CAT(+) minigenome, respectively. Each input template would transcribe, in addition, CAT mRNA. Since plasmid-supplied RNAs did not include the intergenic region (IGR) hairpin structure thought to be responsible for transcription termination (18), CAT mRNA and the corresponding replication product would be of the same size (ca. 800 nt) and, therefore, indistinguishable when total RNA is analyzed (lanes 1 to 5). As shown in lanes 3 and 5, a band of the size and polarity predicted for TV polymerase products appeared in cells expressing both N and L proteins but not when N or L were expressed individually (lanes 1, 2, and 4). The fact that a fraction of CAT(+) RNA products was associated with N protein to form nucleocapsids (lanes 7 and 9) demonstrated that the reconstituted system derived from either the minigenome or the miniantigenome supported the first step of RNA replication, namely, the production of encapsidated miniantigenome or encapsidated minigenome, respectively.
In order to recognize progeny minigenome and progeny miniantigenome as well as any plasmid-expressed RNAs, Northern blots were hybridized with a positive-sense CAT riboprobe (Fig. 5, lower panels). Analysis of total cellular RNA (lanes 1 to 5) showed a band that migrated slightly more slowly than expected for correctly sized minigenome or miniantigenome. This band appeared independently of whether L or N were expressed individually or coexpressed. The size estimated for these larger-than-expected RNAs corresponded to that predicted for the uncleaved minigenome or the uncleaved miniantigenome T7 transcripts (ca. 1,000 nt). Levels of plasmid-supplied minigenome or miniantigenome were quite similar, with each level decreasing at 40 h posttransfection. In cells expressing both N and L (lanes 3 and 5), a band of the size predicted for the minigenome or the miniantigenome was clearly detected at 40 h posttransfection, when the background of T7 transcript had decreased. Analysis of nucleocapsid-associated RNA (lanes 6 to 9) confirmed that minigenome or miniantigenome had accumulated in cells expressing the TV polymerase complex (N and L) (lanes 7 and 9) but not in cells expressing N individually (lanes 6 and 8). In the latter cells, correctly sized RNA was not detected even after overexposure of the film. At the earliest posttransfection time, the nucleocapsid-enriched fraction revealed barely detectable levels of uncleaved T7 transcript (lanes 6 and 7) (note that films for lanes 6 to 9 were exposed for 10-fold longer than were lanes 1 to 5). These results showed that plasmid-supplied templates accumulated intracellularly in the larger uncleaved form and that these RNAs were not encapsidated. The results also showed that the amount of plasmid-expressed template that was cleaved and encapsidated was too low for detection, as the only encapsidated minigenome or miniantigenome detected was that amplified by TV polymerase (Fig. 5, lower panels, compare lanes 6 and 8 with lanes 7 and 9).
Effect of pZ cotransfection on RNA replication.
We also examined the effect of pZ cotransfection on RNA replication. In the experiment depicted in Fig. 6A and B, the plasmid-supplied templates (minigenome or miniantigenome) were complemented with support plasmids pN and pL as indicated, with or without the plasmid expressing Z (pZ). pZ was included at the amounts shown to inhibit transcription, measured as CAT activity. Hybridization with a negative-sense CAT riboprobe (Fig. 6A, upper panel, lanes 1 to 4) showed that cotransfection with 10 ng and 100 ng of pZ resulted in a growing decrease of the accumulation of nucleocapsid-associated CAT(+) miniantigenome from transfected minigenome. The inhibitory effect of Z was also detected when total cellular RNA was analyzed (lanes 5 to 7). As shown in Fig. 6B, pZ inclusion similarly reduced accumulation of nucleocapsid-associated CAT(+) minigenome from transfected miniantigenome.
FIG. 6.
Effect of pZ cotransfection on RNA replication. Subconfluent cultures of CV1 cells previously infected with vTF7-3 were transfected with minigenome (A), miniantigenome (B), or pS-GenCAT (C) together with pN with or without pL. When pZ was included, its amount is indicated in nanograms. Total RNA (A, lanes 5 to 7) was obtained at 24 h posttransfection, whereas nucleocapsid-associated RNA (Nc RNA; A and B, lanes 1 to 4; and C, lanes 1 to 5) was immunoselected at 40 h posttransfection. Purified RNAs were then analyzed by Northern blotting. Duplicate blots were hybridized either with an antisense CAT riboprobe [CAT(−)] (upper panels) or a sense CAT riboprobe [CAT(+)] (lower panels). Arrows show the positions of the RNAs synthesized by the TV polymerase or the T7 polymerase (T7 transcript). “kb” indicates the sizes of RNA markers resolved in parallel. In panels A and B, films were exposed for 20 h, with the exception of lanes 5 to 7 (A), which were exposed for 2 h. In panel C, films were exposed for 4 days.
Hybridization with a positive-sense CAT riboprobe (lower panels) confirmed the above results, indicating that both encapsidated minigenome (Fig. 6A) and encapsidated miniantigenome (Fig. 6B) represent RNAs amplified by TV polymerase, since they appeared only when L was included (lanes 1 and 2), and demonstrated that cotransfection with pZ reduced both progeny minigenome and progeny miniantigenome accumulation (lanes 2 to 4). Analysis of total cellular RNA (Fig. 6A, lanes 5 to 7) showed that while accumulation of minigenome had decreased as result of pZ inclusion, the amount of uncleaved T7 transcript remained unchanged. This indicated that the lower levels of RNA replication as result of pZ cotransfection cannot be ascribed to decreased amounts of plasmid-supplied template.
Considering that the minigenome and the miniantigenome did not include the IGR sequence and that their length is approximately 25% that of TV S RNA, we sought to examine replication of a TV RNA analog more closely resembling the authentic TV S genome and to study the effect of pZ inclusion on this process. To this end, we constructed plasmid pS-GenCAT (Fig. 1), which expressed a TV S genome analog in which the negative-sense N ORF was replaced by the negative-sense copy of CAT. Cotransfection of pS-GenCAT together with support plasmids pN and pL (Fig. 6C) produced a positive-sense CAT RNA of the predicted size, S-AgenCAT(+), which was encapsidated (upper panel, lane 2). Synthesis of this RNA was not detected when pL was omitted (lanes 1 and 5). In blots hybridized with a positive-sense CAT riboprobe (lower panel), an encapsidated RNA of the predicted size for S-GenCAT appeared only in cells expressing the TV polymerase complex (N plus L) (compare lanes 1, 2, and 5). Cotransfection with pZ inhibited production of both S-AgenCAT(+) and progeny S-GenCAT (lanes 2 to 4).
Finally, the possibility that pZ cotransfection inhibited synthesis of N and L proteins from T7 polymerase-based expression plasmids was considered. As shown in Fig. 7, inclusion of pZ in amounts producing substantial inhibition of transcription and replication did not affect expression of the polymerase complex proteins.
DISCUSSION
In this article, we report the establishment of a transcription and replication system based on plasmid-supplied TV RNAs and proteins. Using this system we demonstrated that in cells expressing both N and L proteins, the 3′ and 5′ NCR sequences of each the genome or the antigenome were sufficient to promote transcription, measured as CAT activity, and replication, analyzed as nucleocapsid formation. In arenavirus, as in other negative-strand RNA viruses, the nucleocapsids should serve as the functional templates for transcription and replication. This implies that in the reconstituted system, plasmid-supplied RNAs should become encapsidated to form an active template. When Northern blots were hybridized in order to examine plasmid-supplied RNAs (Fig. 5), we detected an abundant accumulation of an RNA of the size expected for T7 transcripts linked to uncleaved ribozyme. These RNAs were not encapsidated, indicating that a correct 3′ end sequence is critical for encapsidation. However, plasmid-transcribed minigenome or miniantigenome of the correct size were undetectable despite the concurrent transcription and N protein synthesis (lanes 2, 4, 6, and 8). These results might be explained by the low self-cleavage efficiency of the transcripts and perhaps by the rapid degradation of the cleaved products. We found that less than 1% of the RNA transcribed in vitro by pGenCAT or pAgenCAT was autolytically processed (not shown). Other authors have reported that RNAs transcribed by the T7 RNA polymerase could be encapsidated very inefficiently (9, 23, 24). Nevertheless, a fraction of plasmid-supplied RNAs were encapsidated in amounts sufficient to initiate replication (and possibly transcription), as demonstrated by the abundant accumulation of RNA in the presence of L (Fig. 5, lanes 3, 5, 7, and 9). The products of replication by the TV polymerase were readily encapsidated. In summary, in cells expressing L and N, the input minigenome or the input miniantigenome were amplified producing, respectively, miniantigenome or minigenome, which were in turn encapsidated and amplified, producing progeny minigenome or progeny miniantigenome. Similarly the minigenome, L and N proteins but not Z, supported full-cycle replication of an S genome analog in which the N gene had been replaced by CAT (Fig. 6C). This RNA replicated somewhat less efficiently than the minigenome. A possible reason for this behavior might be a very low self-cleavage efficiency of the T7 transcript. Another feature of RNA replication by the reconstituted system deserves further comment, namely, that whatever the input template, genomes appear to accumulate to a greater extent than antigenomes. This has been repeatedly observed in all experiments to date, mainly at 40 h posttransfection when multiple rounds of full-cycle replication by the TV polymerase have occurred (i.e., Fig. 5A and B, lane 9, and Fig. 6, lanes 2 to 4, compare lower and upper panels). In this particular aspect, RNA replication by the reconstituted system appears to reflect TV RNA replication, since in the infected cells S genome is always more abundant than S antigenome (unpublished observations).
Both genome and antigenome specify transcription signals, as evidenced by CAT activity, mediated by the minigenome or the miniantigenome. However, in cells transfected with the miniantigenome, CAT activity comprises less than 10% of that in cells expressing the minigenome. Since CAT mRNAs would be translated with similar efficiency, lower CAT expression implies a lower level of transcription mediated by the miniantigenome relative to that with the minigenome. This result most likely reflects the different intracellular accumulation of template to be transcribed by the polymerase, as miniantigenome accumulation comprises 5 to 20% of that of the minigenome (Fig. 5, lower panels, compare lanes 7 and 9 in panels A and B; also, Fig. 6, lower panel, compare lane 2 in panels A and B). This could be a feasible mechanism for the regulation of gene expression.
Our results with TV are in agreement with a recent finding showing that LCMV L and N proteins are sufficient to support CAT expression and synthesis of positive-sense CAT RNA from an LCMV genome analog containing the 3′ and 5′ termini together with the IGR of the S genome (21). We went further by demonstrating that all cis-acting signals required for transcription, replication, and encapsidation are contained in the 3′ and 5′ NCR sequences of the S genome or the S antigenome. In addition, the recreation of full-cycle RNA replication in our system made possible the analysis of certain features of transcription and replication not contemplated in the study with the LCMV-reconstituted system.
Using the reconstituted system, we demonstrated that TV Z protein is a potent inhibitor of RNA replication mediated by the minigenome and the miniantigenome, with an input pZ plasmid concentration of 100 ng per dish almost suppressing replication (Fig. 6). The Z inhibitory effect was not restricted to the minireplicons, as replication of a larger TV-like RNA containing all of the noncoding sequences of the S genome was similarly affected (Fig. 6). Our results also showed that Z affected transcription as analyzed as CAT expression (Fig. 4). However, since RNA replication involved extensive amplification of the plasmid-supplied template by the TV polymerase (Fig. 5 and 6), it is not possible to determine whether the reduced CAT activity was due to a direct effect on mRNA synthesis or was an indirect effect of reduced template availability for transcription because of reduced RNA replication (Fig. 6). Our results did rule out certain trivial possibilities as causing the effect of pZ cotransfection. It was demonstrated that inhibition by pZ depended on Z protein synthesis rather than on perturbation by pZ transfection or synthesis of Z RNA, since replacement of the Z start codon by a stop codon abolished the Z effect (Fig. 4). In addition, cotransfection with pZ did not affect the synthesis of plasmid-supplied template or proteins required for replication and transcription (Fig. 6 and 7). It is noteworthy that the inhibitory effect of Z was associated with levels of Z expression barely detectable by radioimmunoprecipitation and with ratios of Z to L or N lower than those observed in TV-infected cells (Fig. 3). Taken together, our results support the notion that the effect of Z could be an actual effect which could operate during TV infection. The inhibitory action of Z is not restricted to TV, as it was recently reported that LCMV Z protein strongly inhibits transcription and replication in an LCMV minigenome model system (T. Cornu and J. C. de la Torre, Abstr. 20th Annu. Meet. Am. Soc. Virol., abstr. W48-8, 2001). Precedents for other negative-stranded viruses encoding proteins with roles as negative regulators can be found, including Sendai virus (6), respiratory syncytial virus (1), and Bunyamwera virus (27).
Coding for a negative regulatory protein could be one of the reasons why arenavirus shows slow replication and gene expression and readily establishes persistent infections in both its natural hosts and cells in culture. In support of this idea, we would like to mention previous results indicating that TV stocks producing noncytopathic infections characterized by low levels of synthesis of viral RNAs and proteins contained a higher proportion of Z to L RNAs than virus stocks producing cytopathic infections associated with high levels of viral gene expression (19).
At variance with our results indicating that N and L were sufficient to support transcription and replication in the reconstituted system, Garcin et al. (13) proposed that TV Z is required for both processes. This suggestion was supported by the finding that immunodepletion of TV-infected cell extracts with antiserum to Z strongly reduced mRNA and S genome synthesis and that extracts from Z-expressing cells restored these activities. Considering the probable implication of host cell factors in arenavirus replication (15) and recent findings indicating that LCMV Z interacts with several cellular proteins (3, 4, 7), a possible explanation of these results is that treatment of the TV-infected cell extracts with antibody to TV Z could have been depleted through its interaction with Z, a cellular activator required for RNA synthesis which might have been restored by the Z-expressing cell extracts. However, it is difficult to explain the lack of inhibition by the Z protein included in the extracts. On this regard it should be considered that in the authentic infection, the effect of Z was studied in extracts prepared from cells that were infected for 2 days prior to the experiment and that the measurement of RNA synthesis might have reflected RNA chains already initiated in vivo, whereas in the reconstituted system we added Z from the onset of the replication process and evaluated its effect on multiple rounds of RNA replication. Under these two conditions, the interplay between Z and viral and cellular proteins could differ substantially. It is likely that the apparent discrepancy between the results of Garcin et al. (13) and those reported in this study simply reflects the poor understanting of the role of viral and cellular proteins on the arenavirus replication cycle.
The system described in this report opens the possibility for studying the molecular bases of TV transcription and replication and their regulation, and it provides the foundation for the establishment of rescue systems for TV and its closely related pathogens. This will be useful in studying the attenuating effects of defined mutations and might eventually facilitate the generation of new vaccines.
ACKNOWLEDGMENTS
We are very grateful to Andrew Ball (University of Alabama at Birmingham), who kindly provided plasmid pTV2.0. We also thank D. Kolakofsky (University of Geneva, Geneva, Switzerland) for supplying the cDNA encoding Z protein.
This work was supported by grants from ANPCyT and CONICET. We also thank Laboratorios Bago (Argentina) for financial support. N.L. and M.T.F.-F. are research investigators of CONICET. R.J. is a recipient of a fellowship from this institution.
REFERENCES
- 1.Atreya P L, Peeples M E, Collins P L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop D H L, Auperin D D. Arenavirus gene structure and organization. Curr Top Microbiol Immunol. 1987;133:5–17. doi: 10.1007/978-3-642-71683-6_2. [DOI] [PubMed] [Google Scholar]
- 3.Borden K L B, Campbell Dwyer E J, Salvato M S. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758–766. doi: 10.1128/jvi.72.1.758-766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden K L B, Campbell Dwyer E J, Carlile G W, Djavani M, Salvato M S. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J Virol. 1998;72:3819–3826. doi: 10.1128/jvi.72.5.3819-3826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen M D, Peters C J, Nichol S T. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology. 1996;219:285–290. doi: 10.1006/viro.1996.0248. [DOI] [PubMed] [Google Scholar]
- 6.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell Dwyer E J, Lai H, MacDonald R C, Salvato M S, Borden K L B. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol. 2000;74:3293–3300. doi: 10.1128/jvi.74.7.3293-3300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:150–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 10.Franze-Fernández M T, Zetina C, Iapalucci S, Lucero M A, Bouissou C, López R, Rey O, Daheli M, Cohen G N, Zakin M M. Molecular structure and early events in the replication of Tacaribe arenavirus S RNA. Virus Res. 1987;7:309–324. doi: 10.1016/0168-1702(87)90045-1. [DOI] [PubMed] [Google Scholar]
- 11.Franze-Fernández M T, Iapalucci S, López N, Rossi C. Subgenomic RNAs of Tacaribe virus. In: Salvato M S, editor. The Arenaviridae. New York, N.Y: Plenum Press; 1993. pp. 113–132. [Google Scholar]
- 12.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin D, Rochat S, Kolakofsky D. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J Virol. 1993;67:807–812. doi: 10.1128/jvi.67.2.807-812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harnish D G, Polyak S J, Rawls W E. Arenavirus replication: molecular dissection of the role of viral protein and RNA. In: Salvato M S, editor. The Arenaviridae. New York, N.Y: Plenum Press; 1983. pp. 157–174. [Google Scholar]
- 16.Iapalucci S, López R, Rey O, López N, Franze-Fernández M T, Cohen G N, Lucero M, Ochoa A, Zakin M M. Tacaribe virus L gene encodes a protein of 2210 amino acid residues. Virology. 1989;170:40–47. doi: 10.1016/0042-6822(89)90349-8. [DOI] [PubMed] [Google Scholar]
- 17.Iapalucci S, López N, Rey O, Zakin M M, Cohen G N, Franze-Fernández M T. The 5′ region of Tacaribe virus L RNA encodes a protein with a potential metal binding domain. Virology. 1989;173:357–361. doi: 10.1016/0042-6822(89)90257-2. [DOI] [PubMed] [Google Scholar]
- 18.Iapalucci S, López N, Franze-Fernández M T. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology. 1991;182:269–278. doi: 10.1016/0042-6822(91)90670-7. [DOI] [PubMed] [Google Scholar]
- 19.Iapalucci S, Cherñavsky A, Rossi C, Burgin M J, Franze-Fernández M T. Tacaribe virus gene expression in cytopathic and non-cytopathic infections. Virology. 1994;200:613–622. doi: 10.1006/viro.1994.1224. [DOI] [PubMed] [Google Scholar]
- 20.Kolakofsky D, Garcin D. The unusual mechanism of arenavirus RNA synthesis. In: Salvato M S, editor. The Arenaviridae. New York, N.Y: Plenum Press; 1993. pp. 103–112. [Google Scholar]
- 21.Lee K J, Novella I S, Teng M N, Oldstone M B A, de la Torre J C. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López N, Scolaro L, Rossi C, Jácamo R, Candurra N, Pujol C, Damonte E B, Franze-Fernández M T. Homologous and heterologous glycoproteins induce protection against Junin virus challenge in guinea pigs. J Gen Virol. 2000;81:1273–1281. doi: 10.1099/0022-1317-81-5-1273. [DOI] [PubMed] [Google Scholar]
- 23.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 24.Peeples M E, Collins P L. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi C, Rey O, Jenik P, Franze-Fernández M T. Immunological identification of Tacaribe virus proteins. Res Virol. 1996;147:203–211. doi: 10.1016/0923-2516(96)89650-6. [DOI] [PubMed] [Google Scholar]
- 26.Salvato M S. Molecular biology of the prototype arenavirus, lymphocytic choriomeningitis virus. In: Salvato M S, editor. The Arenaviridae. New York, N.Y: Plenum Press; 1993. pp. 133–156. [Google Scholar]
- 27.Weber F, Dunn E F, Bridgen A, Elliot R M. The Bunyamwera virus non-structural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281:67–74. doi: 10.1006/viro.2000.0774. [DOI] [PubMed] [Google Scholar]