Abstract
Stem cells and the cells they produce are unique because they vary from one cell to another. Traditional methods of studying cells often overlook these differences. However, the development of new technologies for studying individual cells has greatly changed biological research in recent years. Among these innovations, single-cell RNA sequencing (scRNA-seq) stands out. This technique allows scientists to examine the activity of genes in each cell, across thousands or even millions of cells. This makes it possible to understand the diversity of cells, identify new types of cells, and see how cells differ across different tissues, individuals, species, times, and conditions. This paper discusses the importance of scRNA-seq and the computational tools and software that are essential for analyzing the vast amounts of data generated by scRNA-seq studies. Our goal is to provide practical advice for bioinformaticians and biologists who are using scRNA-seq to study stem cells. We offer an overview of the scRNA-seq field, including the tools available, how they can be used, and how to present the results of these studies effectively. Our findings include a detailed overview and classification of tools used in scRNA-seq analysis, based on a review of 2,733 scientific publications. This review is complemented by information from the scRNA-tools database, which lists over 1,400 tools for analyzing scRNA-seq data. This database is an invaluable resource for researchers, offering a wide range of options for analyzing their scRNA-seq data.
Keywords: Single-cell gene expression analysis, Stem cells, Computational biology
Introduction
With the growing use of technologies that allow us to study individual cells, the quality of computational and statistical analysis plays a crucial role in extracting meaningful insights from sequencing datasets (1). As these technologies advance, researchers now have many methods to choose from when analyzing single-cell RNA sequencing (scRNA-seq) data. This variety can be overwhelming, especially for those new to scRNA-seq. To help researchers navigate these options, benchmarking efforts have been undertaken to assess the performance of common tasks such as cell clustering, differential expression analysis, and sample integration (2). These evaluations aim to find the most reliable methods and identify any that might not work well in certain situations, especially in stem cell research. As a result, the scientific community has developed tutorials, workshops, and recommended best practices (3). These resources provide valuable guidance for researchers navigating the complex landscape of scRNA-seq data analysis and help ensure robust and reproducible results in this rapidly evolving field.
In addition to computational methods and software tools, bibliometric analysis has been employed to evaluate the productivity and impact of scRNA-seq research. For example, Patra and Mishra (4), as well as Glänzel et al. (5), utilized bibliometric methods to analyze the growth of scientific literature in bioinformatics, including scRNA-seq research. They identified core journals, author productivity patterns, and research impact. Similarly, Song and Kim (6) evaluated productivity and influence based on measures such as the most productive authors, countries, organiza-tions, and popular subject terms, as well as the most cited papers, authors, emerging stars, and leading organizations. This roadmap provides a comprehensive overview of scRNA-seq research, highlighting expanding areas and potential gaps in knowledge in fields such as stem cell studies, thereby helping researchers navigate the complex landscape of scRNA-seq analysis.
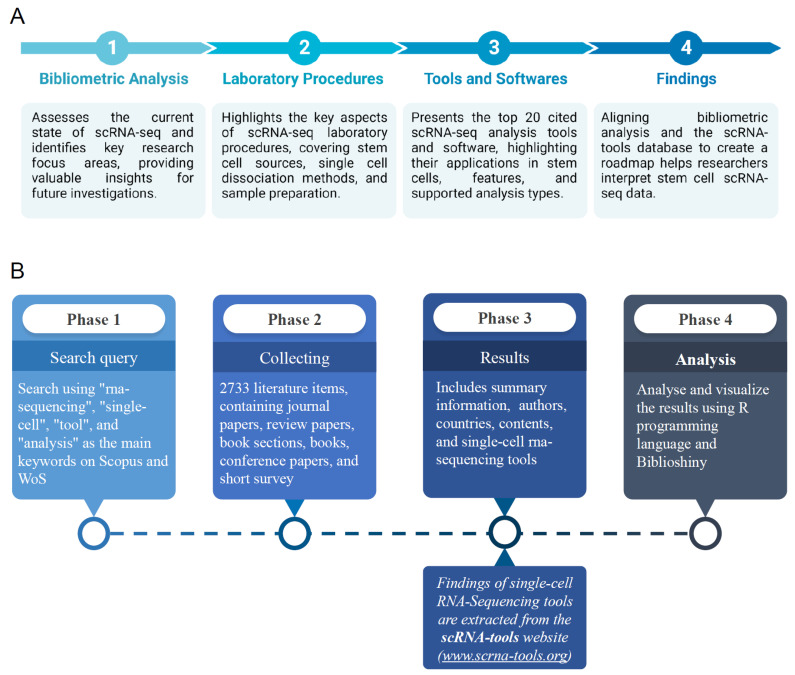
This paper covers several important areas: it starts with a bibliometric analysis to show where scRNA-seq research stands today and where it might go next. It then explains key laboratory techniques, including how to prepare samples and isolate single cells from stem cells. Lastly, it reviews the most frequently cited tools and software for scRNA-seq analysis, highlighting their features and what types of analysis they support. Together, this information creates a roadmap for interpreting scRNA-seq data, offering a clear path forward for researchers, especially those working with stem cells (Fig. 1A).
Fig. 1.
Schematic diagram of the overview of the study. (A) Study overview; comprising: (1) A bibliometric assessment of single-cell RNA sequencing (scRNA-seq); (2) Laboratory procedures with an emphasis on stem cell sourcing and sample preparation; (3) A synopsis of prevalent scRNA-seq tools and software; and (4) Inte-grated findings to aid data interpretation. (B) Process diagram; illustrating the four-phases procedure of the study. WoS: Web of Science.
Methodology of Bibliometric Analysis
Reviewing literature is crucial for understanding the current state of research on a specific topic. It helps identify what has already been explored, points out missing pieces in existing studies, and lays the groundwork for future investigations (7). Bibliometric analysis is a method that quantitatively examines scientific literature, providing an unbiased look at the impact and productivity of research. This approach involves creating a research question, gathering relevant literature, applying specific metrics, and analyzing the findings. It highlights key areas of research, sheds light on recent studies, and emphasizes significant contributions that guide future work. Our study employs bibliometric analysis as illustrated in Fig. 1B.
The first step in gathering literature is accessing the right databases. Scopus and Web of Science (WoS) are notable for allowing the export of bibliometric data (8). WoS is renowned for its extensive collection of high-impact publications (9), while Scopus is the largest database of peer-reviewed literature across many research fields, featuring over 20,000 journals from a variety of publishers (10). It includes papers indexed by both Clarivate’s WoS and Scopus. Considering these points, our search covered keywords within both the WoS and Scopus databases.
The search strategy from databases involves using keywords combined with Boolean operators. For this study, we focused on scRNA-seq analysis tools, using “RNA-sequencing,” “single-cell,” “tool,” and “analysis” as primary key-words. The “AND” operator links these keywords, while “OR” separates synonyms keywords within each category, such as (“RNA-sequencing,” “RNA-sequence,” “RNA-seq”) for RNA-sequencing, and (“tool,” “software,” “library”) for tools. This precise use of Boolean logic narrows the search to align with our study’s objectives. We collected a variety of scientific publications including journal papers, review papers, conference papers, book sections, and books published from the last decade (2013∼2022) from WoS and Scopus, totaling 2,733 unique literature items.
Various software tools like VOSviewer, SCImago, the WoS analysis tool, HistCite, Pajek, Gephi, BibExcel, and the bibliometrix package in R, facilitate bibliometric studies. HistCite and the WoS tool are limited to WoS data (10). Gephi stands out for its flexibility and efficiency but lacks necessary data preparation features, requiring additional tools like BibExcel for this task (10). Although BibExcel is powerful, it demands significant expertise for straightforward analyses. Our bibliometric analysis was performed using R, a statistical computing software, with the bibliometrix package (11). This package offers an intuitive interface, combines results from WoS and Scopus into a unified dataset for analysis, and utilizes the “mergeDbSources” function from the bibliometrix library to merge these datasets, following a method proposed by the authors of another study (12).
Results of Bibliometric Analysis
In this study, we explored the range and classification of tools used in scRNA-seq analysis through an inductive approach (13), starting from the data itself. Our findings provide important insights that will help researchers choose the most appropriate tools and features for their specific goals, including projects focused on stem cell research. When we narrowed our search to include “stem cells” as a keyword specifically for stem cell studies, we discovered approximately 450 articles out of the 2,733 included in our broader analysis. This difference suggests that stem cell researchers may use a variety of terms, such as “organoids” or “iPSCs,” rather than just “stem cells.” The collection of 2,733 unique pieces of literature serves as the basis for our bibliometric analysis. We divided our findings into four main categories: an overview of the data, the yearly growth of publications, the most influential journals and articles, and the most frequently cited works in the field.
Descriptive analysis
This section offers a summary of the literature we gathered to gain a broad understanding of the field of scRNA-seq analysis. Table 1A presents the main features of the 2,733 literature items published over the last decade. These works come from 615 different sources, including books, conference proceedings, and journals. Despite spanning the past 10 years, the average “age” of these publications is under 3 years, highlighting the rapid growth and current relevance of this research area. The collection includes a large number of references, indicating the extensive research activity and interest in this field.
Table 1.
List of main features and types of literature items published over the last decade
| A | B | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Description of the dataset | Value | Description of the dataset | Value | Dataset (%) | |
| Time span (yr) | 2013∼2022 | Article | 2,168 | 79.33 | |
| Sources (journals, books, etc.) | 615 | Article; proceedings paper | 10 | 0.37 | |
| Documents | 2,733 | Book chapter | 109 | 3.99 | |
| Annual growth rate (%) | 50.37 | Conference paper | 48 | 1.76 | |
| Document average age (yr) | 2.98 | Conference review | 1 | 0.04 | |
| Average citations per document | 39.16 | Correction | 1 | 0.04 | |
| References | 103,931 | Data paper | 2 | 0.07 | |
| Keywords Plus (ID) | 13,233 | Editorial | 8 | 0.29 | |
| Author’s keywords (DE) | 4,160 | Erratum | 4 | 0.15 | |
| Authors | 13,409 | Letter | 9 | 0.33 | |
| Authors of single-authored documents | 53 | Meeting abstract | 1 | 0.04 | |
| Single-authored documents | 67 | Note | 9 | 0.33 | |
| Co-authors per document | 7.84 | Review | 350 | 12.81 | |
| International co-authorships (%) | 23.02 | Short survey | 13 | 0.48 | |
| Average citation per document | 39.15 | ||||
(A) This legend summarizes the key characteristics of 2,733 publications from the past decade in the field, showcasing it as a recent and trending area with an average publication age of under three years. The dataset encompasses over 100,000 unique references. Through content analysis, 13,233 Keyword Plus terms and 4,160 authors’ keywords were identified, offering deep insights into the literature’s traits. Values are presented as number.
ID: index term, DE: descriptive term.
(B) This part provides a breakdown of the types of documents included in the analysis. Articles make up the majority, indicating a strong academic interest in single-cell RNA sequencing analysis. Reviews form over 12% of the collection,highlighting their importance for synthesizing knowledge in this field. Book chapters and conference papers represent 4% and 1.76%, respectively, showing diverse formats of scholarly communication. Other document types such as proceedings papers, conference reviews, corrections, data papers, editorials, errata, letters, meeting abstracts, and notes each account for less than 1% of the total, illustrating a wide array of contributions to the literature. Values are presented as number.
Regarding keyword analysis, there were 13,233 “Key-words Plus” and 4,160 authors’ keywords identified. “Key-words Plus” are derived from commonly occurring terms in the titles of the references of a given literature item, while authors’ keywords are the terms most frequently used by the authors in the literature items themselves.
In terms of authorship, the 2,733 literature items were authored by 13,409 individuals. The average number of citations per document is quite high at 39.15, suggesting that the documents have significant impact. The total number of cited references across all documents reached 107,022. Table 1B provides additional details on the types of documents in the collection. Articles formed the majority of the literature items, indicating their significant scholarly contribution to scRNA-seq analysis. Review papers were the second most common document type, making up over 12% of the dataset. Book chapters accounted for about 4% of the items, and conference papers 1.76%. Other document types, such as proceedings papers, conference reviews, corrections, data papers, editorials, errata, letters, and meeting abstracts, comprised less than 1% of the dataset, highlighting the diversity of publication types in the field.
The annual growth of publications
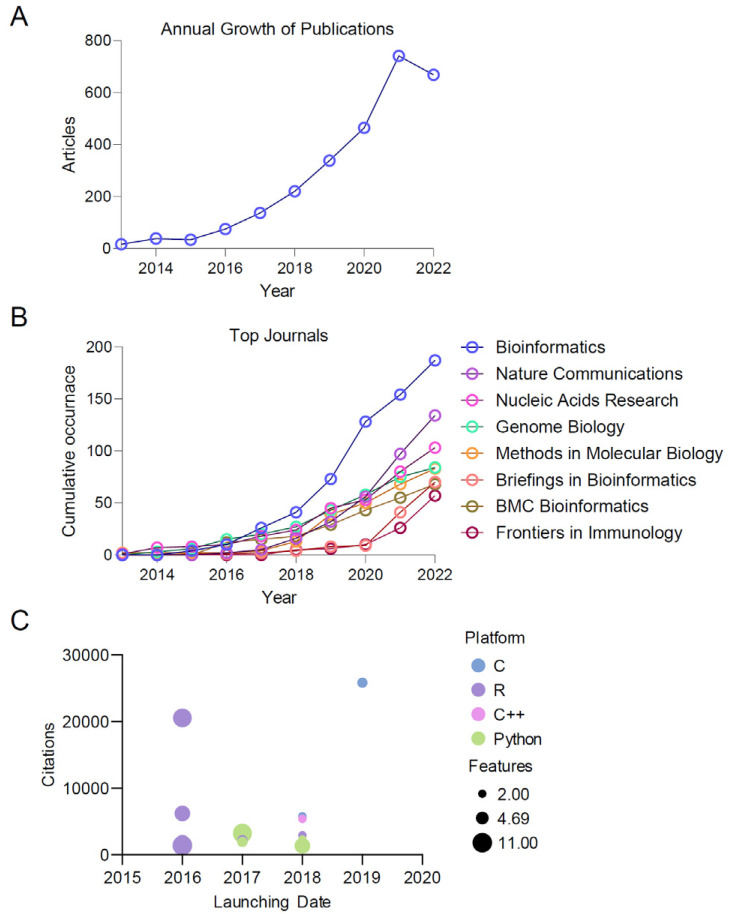
The metric for annual growth rate is determined by calculating the average number of literature items published each year over a specified period (2013∼2022). The growth in the number of publications (NP) related to scRNA-seq research within our dataset is notably high, with an average annual increase of 50.37%. Fig. 2A illustrates a significant upward trend in scRNA-seq research. Initially, up until 2015, the growth was modest, with fewer than 50 publications annually. However, starting in 2016, there was a sharp rise, reaching a peak in 2021 with over 700 publications in just one year. This surge reflects growing interest from both the academic and industrial sectors in the unique challenges and opportunities presented by scRNA-seq.
Fig. 2.
Exploring the rapid evolution of single-cell RNA sequencing (scRNA-seq) analysis. (A) The annual growth of publications in the field of scRNA-seq analysis. (B) Shows the top 8 sources’ cumulative growth in the field of scRNA-seq analysis. (C) Top 20 cited tools for analysing scRNA-seq data.
We also analyzed the cumulative growth of publications in the top 8 sources identified within our dataset, as shown in Fig. 2B. Among these sources, those related to informatics showed the most substantial increase. Notably, the growth rate of these top sources was relatively steady and low until 2017 but saw a dramatic rise after 2020. Of particular interest is the journal Nature Communications, which, despite only starting to publish in this area in 2017, showed the second-highest growth rate. However, the growth trends of scRNA-seq in the International Journal of Stem Cell began in 2020. Since then, 13 articles have discussed the applications of scRNA-seq across various subtypes of cells derived from stem cells. Out of these, 3 of the 13 articles have performed scRNA-seq analysis in their research articles published in the International Journal of Stem Cell. The growth rates of publications in other sources were fairly consistent with each other.
The top 15 impacting sources
Several metrics are available to assess the impact and productivity of scientific sources in the field of scRNA-seq analysis. This section uses a variety of these metrics to give a comprehensive overview of the influence these sources have on the field. Out of 615 sources in our dataset, more than half (340) have published only one item. However, the top 15 sources account for over 40% of the publications in our dataset. This indicates a concentration of output in a small number of sources, despite the overall diversity of publication origins. These top 15 sources (Supplementary Table S1) stand out significantly among the total of 615.
Supplementary Table S1 lists these top 15 scientific sources along with their metrics: NP, local citations (LC) from the dataset, h-index, g-index, total citations (TC), and the average year of publication. Initially sorted by NP, we see that the journal Bioinformatics leads with 187 documents in scRNA-seq analysis, also ranking in the top 10 across all other metrics. Nature Communications follows, with the second-highest NP as well as the highest h-index and g-index scores, receiving a total of 5,439 citations and 5,244 LC from our dataset. This highlights the significant impact of Nature Communications in this field, especially notable since it only began publishing on this topic in 2016.
Nature Biotechnology has the highest number of TC (10,931), with Cell journal following (8,746 citations). Interestingly, Cell does not rank in the top 15 by NP (with only 19 papers) but has the highest local citation count from our dataset, underscoring its substantial influence in scRNA-seq analysis. It is important to note that more than half of the sources in our dataset received fewer than 10 TC, reflecting a wide disparity in impact among the publication venues.
Top cited literature items
This section examines the citations received by various documents in our dataset, focusing on both LC and global citations (GC). LC refer to the number of times an article is cited within the dataset we analyzed, while GC account for citations from all sources. Table 2 (14-28) lists the top 15 publications with the highest LC, including their global citation counts, the ratio of local to global citations (LC/GC), and their publication year. The ranking is based on local citation counts, which may not align with their global citation standings. Remarkably, only 15% of the documents in our dataset have not received any GC up to the time of this analysis, a relatively small fraction. About 18% of the documents have received at least the average citation count per document in our dataset, which is 39. However, around 60% of the literature items did not receive any local citation.
Table 2.
List of top 15 cited articles in scRNA-seq
| Study | Value | Citations | LC/GC ratio | Year | |
|---|---|---|---|---|---|
|
| |||||
| Local | Global | ||||
| Integrating single-cell transcriptomic data across different conditions, technologies, and species (14) | Presents a methodology for the comprehensive analysis and integration of scRNA-seq data, enabling the identification of shared populations across data sets and downstream analysis | 443 | 4,123 | 10.74 | 2018 |
| Comprehensive integration of single-cell data (15) | Develops a strategy to “anchor” various datasets simultaneously, allowing scientists to integrate single-cell across different modalities | 352 | 4,308 | 8.17 | 2019 |
| The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells (16) | Introduces Monocle, an unsupervised algorithm that enhances the resolution of transcriptome dynamics in cellular processes such as differentiation | 297 | 2,415 | 12.3 | 2014 |
| SC3: consensus clustering of single-cell RNA-seq data (17) | Proposes a consensus clustering algorithm specifically designed for scRNA-seq data, improving the accuracy of cell type identification | 188 | 683 | 27.53 | 2017 |
| Full-length RNA-seq from single cells using Smart-seq2 (18) | Describes an improved protocol for full-length RNA sequencing from single cells, enabling more detailed transcriptome analyses, especially for stem cell research | 182 | 1,942 | 9.37 | 2014 |
| Smart-seq2 for sensitive full-length transcriptome profiling in single cells (19) | Enhances the sensitivity and accuracy of single-cell transcriptome profiling with the Smart-seq2 technology | 172 | 1,216 | 14.14 | 2013 |
| Comparative analysis of single-cell RNA sequencing methods (20) | Offers a comparative study of various scRNA-seq methodologies, evaluating six prominent methods | 152 | 728 | 20.88 | 2017 |
| Computational and analytical challenges in single-cell transcriptomics (21) | Discusses the key computational and analytical challenges in single-cell transcriptomics, proposing solutions to address these issues | 144 | 691 | 20.84 | 2015 |
| Quantitative single-cell RNA-seq with unique molecular identifiers (22) | Introduces a quantitative approach to scRNA-seq that uses unique molecular identifiers, improving data accuracy | 138 | 729 | 18.93 | 2014 |
| Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris (23) | Presents a comprehensive single-cell transcriptomic atlas of mouse, providing insights into organ-specific cell types and states | 129 | 925 | 13.95 | 2018 |
| Current best practices in single-cell RNA-seq analysis: a tutorial (24) | Offers a guide on best practices for analysing scRNA-seq data, from preprocessing to downstream analysis | 119 | 610 | 19.51 | 2019 |
| Splatter: simulation of single-cell RNA sequencing data (25) | Provides a tool for simulating scRNA-seq data, aiding in the development and testing of analytical methods | 118 | 324 | 36.42 | 2017 |
| Single-cell RNA sequencing technologies and bioinformatics pipelines (26) | Reviews the latest technologies and bioinformatics pipelines for scRNA-seq, highlighting their advantages and limitations | 110 | 665 | 16.54 | 2018 |
| Recovering gene interactions from single-cell data using data diffusion (27) | Proposes a data diffusion approach called MAGIC a method that shares information across similar cells to denoise the cell count matrix and fill in missing transcripts | 109 | 590 | 18.47 | 2018 |
| Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R (28) | Introduces an R package for comprehensive preprocessing, quality control, normalization, and visualization of scRNA-seq data | 106 | 620 | 17.1 | 2017 |
Analyses thetop 15 locally cited articles and their contribution in the field of single-cell RNA sequencing (scRNA-Seq) analysis, considering both their global citations and the local/global citation (LC/GC) ratio. The “LC/GC ratio” field signifies the ratio between local and global citations, providing a measure of the extent to which these articles are cited within their immediate research community relative to their global reach. Additionally, the “Year” field indicates the year of publication foreach article. The analysis of these parameters provides valuable information on the local and global recognition of the top 15 articles in scRNA-Seq analysis, allowing for a comprehensive understanding of their significance within the field.
The publication with the highest number of LC is titled “Integrating single-cell transcriptomic data across different conditions, technologies, and species” (14), which also ranks second in GC with 443 LC and 4,123 GC. The document with the second-highest number of LC, “Comprehensive integration of single-cell data” (15), leads in GC with 352 LC and 4,308 GC. These findings highlight the significant impact of these two articles in the field of scRNA-seq analysis, both locally within our dataset and globally. Notably, “Splatter: simulation of single-cell RNA sequencing data” (25) achieved the highest citation ratio among the top 15 cited documents, and “SC3: consensus clustering of single-cell RNA-seq data” (17) had the second-highest ratio. Among these leading publications, 7 were published by Nature journals, and 3 by Cell journals, underscoring the dominant role of these publishers in advancing scRNA-seq analysis research.
Stem Cell Sources for scRNA-Seq Analysis
The collection of human samples for research can be challenging, especially when it involves foetal tissues, due to ethical concerns. Organoids serve as an excellent platform for studying human embryonic tissues. Utilizing organoids, such as brain organoids, allows for the comparative study of distinct developmental stages and the potential uncovering of pathological processes in neurodevelopmental disorders, including autism spectrum disorder and Down syndrome (29). The application of scRNA-seq to study various domains of the brain, such as the neocortex or forebrain, can provide detailed insights into the evolution of the human brain (30). Unlike bulk RNA-seq, which is more effective for analyzing single cell types, scRNA-seq is particularly suited for and often employed in the analysis of complex tissues like organoids (31), which typically display a heterogeneity of cell type composition (32). Therefore, scRNA transcriptomics offers a superior method over bulk RNA sequencing by delivering a detailed analytical approach that aids in characterizing and identifying previously unknown subpopulations of cell types. Currently, repositories such as the Single Cell Expression Atlas (https://www.ebi.ac.uk/gxa/sc/home) and the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) offer enriched datasets that biologists and bioinformaticians can use to compare transcriptomics data from organoids.
Molecular cues are provided to pluripotent stem cells (PSCs) to direct them towards the desired cell fate and organoid type, in an effort to replicate the embryonic development of a specific organ (33) or to capture an aging-like phenotype in vitro (34). As a result, organoids will resemble the target organ (35); however, general characterization methods such as immunohistochemistry, western blot, and bulk RNA sequencing will not be sufficiently informative to identify the heterogeneity of cell types generated in organoids (34). Furthermore, experiments involving organoids cannot be interpreted accurately without a clear understanding of their cellular composition. In this context, comparing scRNA-seq data of primary tissues/organs with that of organoids will demonstrate the degree of similarity and resemblance of the organoids to the target organ. For example, in cases where human ventral midbrain organoids were generated, a comparison of scRNA-seq data of the organoids with fetal dopaminergic neurons of the ventral midbrain revealed transcriptional similarities (36). Similarly, scRNA-seq data showed similarities between cerebral organoids and fetal cortex tissues (37), as well as at the single-cell level (38). While bulk RNA-seq can also be used to demonstrate the maturation similarity of organoids compared to in vivo tissue (29), scRNA-seq is particularly important for organoid research. It helps to interpret the data more precisely, which will improve the quality of organoid production for disease modelling (39).
In addition to defining the cellular composition of organoids, scRNA-seq can enhance our understanding of the genetic identity of existing stem cell types. For example, a study compared human embryonic stem cells with human epiblast cells using scRNA-seq analysis and identified approximately 1,500 genes that were differentially expressed between these cell types (40). Another study analyzing aging versus young mouse hematopoietic stem cells (HSCs) with scRNA-seq revealed that the expression of cell cycle-associated genes corresponded overall to the aged status of the HSCs (41). Transcriptomics of stem cells found in different tissues can also be compared using scRNA-seq. One study discovered that mesenchymal stem cells (MSCs) from various origins diverged due to differences in extracellular matrix protein and immunity-related genes (42). Additionally, another study identified two subpopulations within umbilical cord MSCs that had distinct differentiation capabilities based on their gene expression patterns (43). Even though these MSCs originated from the same source, it is possible that by the time of isolation, they had already activated distinct gene expression programs consistent with their distinct cellular fates.
Furthermore, scRNA-seq can also shed light onto the reprogramming of novel stem cell types. A recent study discovered a methodology for reprogramming mouse pluripotent embryonic stem cells into totipotent stem cells using a spliceosome inhibitor (44). These reprogrammed totipotent stem cells were implanted into mouse blastocysts, and lineage tracing revealed that the implanted cells could differentiate into six different cell types of extraembryonic origin, such as trophoblast cells. Transcriptomics analysis confirmed the expression of totipotency genes in this generated totipotent stem cell population. In a similar study, totipotent blastocyst-like structures were generated using human induced PSCs (hiPSCs) (45). scRNA-seq of these human blastoids showed that their composition—a mix of hypoblast-, trophoblast-, and epiblast-like cells—resembled human blastocysts. In such cases, the transcriptomic resolution provided by scRNA-seq is highly significant, as totipotent stem cells have the capacity to differentiate into extraembryonic tissues.
While single-cell transcriptome analysis methods are broadly applicable across various fields of biology, interestingly, however, Smart-seq2 is a highly sensitive method for scRNA-seq that has been further refined for stem cells to capture a wide range of gene expression levels (Table 2), particularly useful for identifying rare stem cell populations or capturing the full complexity of gene expression dynamics during differentiation (18), allowing understanding pluripotency, differentiation pathways, and cellular heterogeneity within stem cell populations. Therefore, the application of scRNA-seq in stem cell and developmental studies can significantly accelerate our understanding of developmental processes.
Single cell dissociation methods
ScRNA-seq requires samples to be prepared in a solution with individual cells, minimizing doublet formation and ensuring maximum viability (usually >90%). For scRNA-seq samples like blood or immune cells, which circulate in the blood without extracellular matrix connections to other cell types, creating single-cell suspensions is straightforward. However, tissues and organs, containing various cell types, need enzymatic dissociation. Different tissues require specific enzymes for dissociation, each with its own set of advantages and disadvantages. Typically, fresh tissue preparation involves proteases such as papain, collagenases, or trypsin, which can be performed at 37℃ or at colder temperatures (46). Cells within a tissue may respond differently to enzymatic digestion, potentially introducing bias in the preparation of single-cell solutions. For example, a study comparing various single-cell dissociation methods for mouse kidney found that podocytes were disproportionately affected by warm dissociation compared to the cold dissociation method (47). Similarly, satellite cells, a population of muscle stem cells, were shown to be impacted by dissociation methods in a manner that their transcriptome resembled an injury-induced subtype of satellite cells (48). To capture rare cell types and reduce dissociation stress-associated gene expression changes, single-nucleus RNA sequencing may be more appropriate. In another study, a combination of dispase and collagenase was used for initial tissue digestion, followed by trypsin for remaining undissociated tissue parts, improving cell dissociation and the capture of rare cell types in skin samples (49). The choice of cell dissociation method for single-cell preparation should be tailored to the target cell type and study goals to avoid biased data and the loss of rare cell types.
Quality control of sample preparation in scRNA-seq analysis
The quality of scRNA-seq data is significantly influenced by the biological material used and the method of sample preparation. To mitigate any artifacts that occur during sample dissociation and library preparation, effective quality control (QC) measures are essential during data analysis. First, the expected gene profile of the biological material intended for sequencing should be roughly estimated. For example, a high-count number for mitochondrial genes may indicate apoptotic cells. However, if the biological material is known to express mitochondrial genes at a relatively high level, this factor must be considered before discarding cells that exceed the threshold for high mitochondrial gene counts. Second, it is necessary to remove ambient RNA, which results from freely floating mRNA transcripts from dead cells during cell dissociation. Finally, genes that are less abundant or cells with a lower count of genes should be excluded before further analysis. Furthermore, noise reduction in bulk RNA sequencing is comparatively simpler than in scRNA-seq, as the amplified and sequenced transcripts are not attributed to individual cells. With the increased use of scRNA-seq technologies in this decade, several QC measures and software packages have been developed. Firstly, a threshold for identifying good quality cells is established by examining parameters such as the total number of reads per cell, the total number of gene counts, and library complexity (50). For example, sinQC (Morgridge Institute for Research), an scRNA-seq QC software tool, eliminates low-quality cells by considering the main cell population as of good quality and generates a false positive rate by calculating a minimal quantile score and a weighted combined quality score (51). Ano-ther software, named Dropkick (United Plugins), employs a more sophisticated approach to filter out ambient RNA (52). This method initially profiles a matrix based on the total gene count per cell versus barcode count to distinguish high-quality cells from empty droplets and low-quality cells. Subsequently, it identifies the most common genes found in low-quality cells to label them as ambient RNA and filters them out. However, researchers often make several common mistakes and encounter issues when analyzing scRNA-seq data. These include not adequately filtering out low-quality cells or genes, which can skew results; overlooking batch effects that arise when combining datasets from different experiments; failing to select an appropriate normalization method, potentially leading to incorrect conclusions; ignoring the complexity of cell cycle effects on gene expression; and choosing unsuitable algorithms for clustering or trajectory analysis that do not match the characteristics of the data. These oversights can significantly impact the accuracy and interpretability of scRNA-seq analyses.
To ensure the quality of scRNA-seq analysis, the annotation step becomes crucial after generating a gene count matrix. Annotation leverages known biological information to assign specific identities to cells, grouping those with similar identities into clusters. This process can be conducted either by comparing with previously obtained reference scRNA-seq datasets or by utilizing publicly available biological information repositories. Recent studies have extensively reviewed these annotation methods (53). It is important to acknowledge that annotating data from primary tissues is generally more straightforward than from organoid clusters. This challenge is particularly pronou-nced in data from organoids, as the reference genes for the in vivo counterparts of organoids may exhibit slightly different transcriptomes (38). For example, a recent study investigating the relationship between brain organoid morphology and architecture discovered that unsuccessful differentiation resulted in scRNA-seq data clusters with mixed identities (54). This issue, common in hiPSC-derived organoid differentiation due to interorganoid variability, must be considered when annotating cell clusters in scRNA-seq data using reference datasets from previously obtained organoid scRNA-seq analyses.
scRNA Sequencing Tools and Software
scRNA-seq is an advanced technology that enables scientists to explore the variety of gene expression within individual cells. This capability provides a deeper insight into cellular processes, such as those occurring in stem cells and their derived tissues. As a result, a wide range of software tools and analysis pipelines has been developed to analyze scRNA-seq data efficiently (55). The main purpose of these tools is to convert raw sequencing data into detailed gene expression profiles for each cell (56). This process typically includes steps like QC, normalization, reducing data complexity, grouping similar cells (clustering), quantifying gene expression, and identifying genes that are expressed differently between cell populations (57).
Moreover, some tools offer specialized functions such as classifying cell types, analyzing gene pathways, and combining data from multiple scRNA-seq studies. These functionalities are particularly valuable in stem cell research and other specialized areas (58). While scRNA-seq excels at comparing gene expression across individual cells and uncovering cell diversity, its ultimate goal is to find transcriptional similarities and differences within groups of cells. This approach is crucial for identifying rare cell types that were often overlooked by previous methods (59). Additionally, scRNA-seq can reveal intricate gene expre-ssion details, including patterns of gene splicing, expression from single alleles, and groups of genes that are regulated together, by analyzing gene co-expression patterns at the single-cell level.
However, the accuracy of the insights gained from scRNA-seq largely depends on the experimental approaches used (60). The selection of a scRNA-seq analysis tool often hinges on the specific research questions, the nature of the data, and the complexity of the analysis required. Some tools are designed for particular data types or analytical methods, while others are more versatile, catering to a broader range of uses. For instance, ZINB-WaVE addresses the zero-inflation common in scRNA-seq data with a zero-inflated negative binomial model, improving the accuracy of further analyses by effectively managing datasets rich in zeros. Conversely, for pseudotemporal analysis, which orders cells based on their gene expression changes to infer cellular development or progression, Monocle is a leading tool. It enables the reconstru-ction of cell development pathways or progression stages from a single snapshot in time. In conclusion, scRNA-seq analysis tools and software are indispensable in advancing our comprehension of gene expression and cellular dynamics at the individual cell level. They enable researchers to sift through large scRNA-seq datasets, extract meaningful information, and contribute to scientific discoveries that have the potential to impact society positively.
Top 20 cited tools for analysing scRNA-seq data
The introduction of scRNA-seq has made it feasible to collect detailed data from a wide variety of cells at different stages of their development and maturation. This breakthrough has opened new avenues for uncovering insights into cell development, transformation, and fate, both in vivo and in vitro (61). Such extensive datasets are incredibly useful for researchers who need to choose the most appropriate tool for analyzing their scRNA-seq data. By comparing the types of data that can be processed and the functionalities offered by each tool, researchers can select one that best meets their specific needs (62).
In this section, we explore the top 20 most cited tools for analyzing scRNA-seq data, as illustrated in Fig. 2C. The primary source for this analysis is the scRNA-tools database, updated as of May 9, 2023. Table 3 (58, 63-80) includes a row for each of the top 20 scRNA-seq tools, with columns providing details about the type of input data each tool accepts and the features it offers. For instance, the first column might name the tool (like STAR), the second column describes the type of input data it works with (such as FASTQ files), and the following columns detail the features the tool provides (like QC, normalization, integration, clustering, classification, etc.).
Table 3.
Top 20 cited tools for analysing scRNA-seq data
| Tool name | System | Output | Tool overview | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| Platform | Input data | Quality control | Normalization | Integration | Clustering | Classification | Ordering | Diff. expression | Gene networks | Dim. reduction | Visuali-zation | |||
| STAR | C/C++ | FASTQ | An ultrafast universal RNA-seq aligner designed to align RNA sequencing reads to a reference genome (63) | |||||||||||
| Seurat | R | Count Matrix | ∨ | ∨ | ∨ | ∨ | ∨ | ∨ | A toolkit for quality control, analysis, and exploration of scRNA-seq data (64) | |||||
| Monocle | R | FASTQ | ∨ | ∨ | ∨ | ∨ | ∨ | A toolkit for analysing single-cell gene expression to discover, explore, and visualize cell differentiation processes (65) | ||||||
| kallisto | C/C++ | FASTQ | A program for quantifying abundances of transcripts from RNA-seq data, using pseudoalignment to speed up the process (66) | |||||||||||
| salmon | C++ | FASTQ | A tool for fast transcript-level quantification from RNA-seq data using lightweight alignments (67) | |||||||||||
| CellRanger | Python/R | FASTQ | ∨ | ∨ | ∨ | ∨ | ∨ | A set of analysis pipelines that process Chromium scRNA-seq output to align reads, generate feature-barcode matrices, and perform clustering and gene expression analysis (58) | ||||||
| Scanpy | Python | Count Matrix | ∨ | ∨ | ∨ | ∨ | ∨ | ∨ | ∨ | An open-source, scalable toolkit for analysing single-cell gene expression data using Python (68) | ||||
| inferCNV | R | FASTQ | ∨ | Uses to investigate tumor scRNA-seq data to recognise evidence for large-scale chromosomal copy number variations (69) | ||||||||||
| CellPhoneDB | Python | Count Matrix | ∨ | ∨ | A publicly available repository of curated receptors, ligands, and their interactions, intended for analysing cell-cell communication (70) | |||||||||
| BackSPIN | Python | FASTQ | ∨ | A gene clustering and ordering algorithm based on a biclustering technique, used for single-cell data analysis (71) | ||||||||||
| SCENIC | Python/R | FASTQ | ∨ | ∨ | ∨ | A computational method for finding regulators and their target genes from scRNA-seq data to reconstruct gene regulatory networks (72) | ||||||||
| AUCell | R | FASTQ | ∨ | ∨ | A tool for analysing gene sets in single-cell data, identifying cells with active gene sets (73) | |||||||||
| velocyto | Python/R | FASTQ | ∨ | ∨ | A package for estimating RNA velocity in scRNA-seq data, predicting the future state of individual cells (74) | |||||||||
| scran | R | Count Matrix | ∨ | ∨ | ∨ | Implements methods for low-level analyses of scRNA-seq data such as normalization and cell cycle phase assignment (75) | ||||||||
| Harmony | R/C++ | FASTQ | ∨ | ∨ | An algorithm for integrating scRNA-seq data across different datasets or experimental conditions (76) | |||||||||
| MAST | R | Count Matrix | ∨ | ∨ | ∨ | ∨ | A flexible statistical framework to assess differential expression in scRNA-seq data (77) | |||||||
| RaceID | R/C++ | Count Matrix | ∨ | ∨ | ∨ | ∨ | ∨ | ∨ | Identifies rare cell types from single-cell gene expression data based on clustering (78) | |||||
| scvi-tools | Python | FASTQ | ∨ | ∨ | ∨ | ∨ | ∨ | ∨ | A suite of methods for analysing single-cell genomics data, leveraging variational inference to model cell heterogeneity and dependencies (79) | |||||
| SCDE | R | Count Matrix | ∨ | ∨ | An error model and differential expression analysis for scRNA-seq data, accounting for the unique characteristics of sparse and noisy data (80) | |||||||||
Diff.: differential, Dim.: dimension, scRNA-seq: single-cell RNA sequencing.
The features outlined in Supplementary Table S2 and S3 cover various steps in the scRNA-seq analysis workflow. QC is the preliminary step, ensuring the raw sequencing data is of high quality. Normalization adjusts gene expression levels to minimize technical differences. Integration combines datasets from various scRNA-seq experiments into a cohesive dataset. Clustering groups cells with similar gene expression patterns together. Classification assigns cell types to cells based on their gene expression profiles, facilitating deeper insights into their functions and identities.
Findings
This study provides a comparative analysis of the research published in the area of scRNA-seq, focusing on the software and tools developed for this purpose. It distinguishes between the most commonly provided features of these tools and those that are essential for effective research, especially in the context of stem cell studies. The insights offered here are intended to assist researchers in choosing the most appropriate tools and features for their specific research goals. The paper highlights the crucial factors that should be taken into account when selecting software for scRNA-seq data analysis and suggests directions for future research.
The roadmap provided in this paper serves as an invaluable resource for professionals working in bioinformatics, data science, biology, and especially those involved in stem cell research. It aims to simplify the process of navigating the complex field of scRNA-seq analysis, enabling researchers to make well-informed choices about the studies, methodologies, and tools that are best suited for their work. By comparing the features of various tools and software with the needs highlighted in published studies, this paper facilitates a more straightforward selection process for researchers working with scRNA-seq data, ensuring that their chosen tools align well with both common and critical research requirements.
Essential and advanced analysis features in scRNA-seq
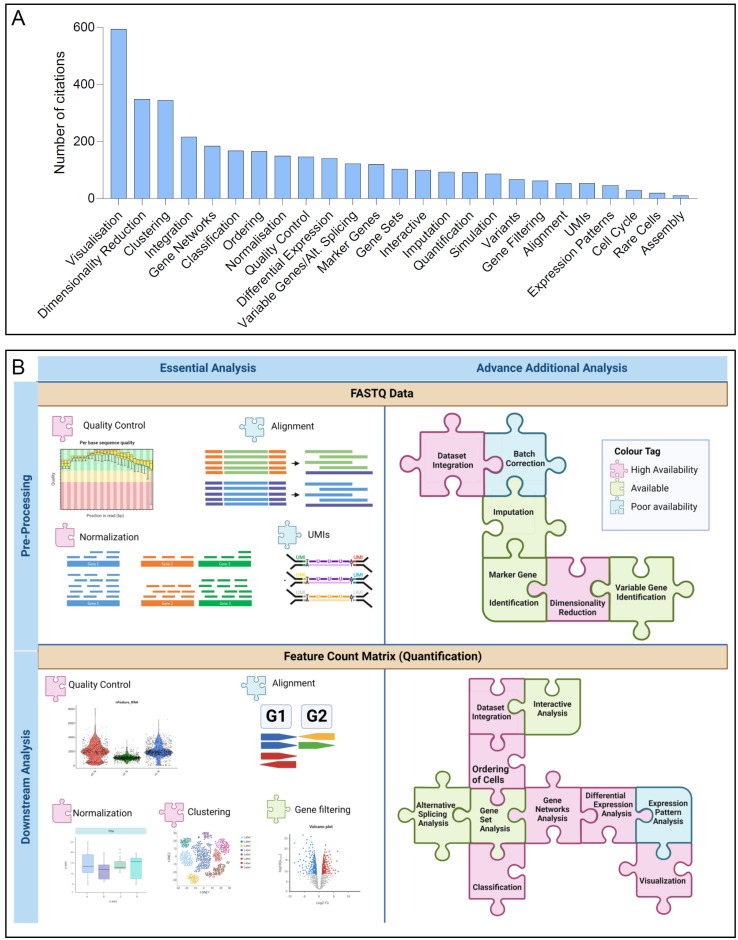
Fig. 3A illustrates the prevalence of certain features within current analysis tools as derived from the litera-ture. This review scRNA-seq analysis and the popularity of tool features has led to the classification of scRNA-seq analysis features into two primary phases: pre-processing and downstream analysis, as illustrated in Fig. 3B. Pre-processing includes tasks like alignment, normalization, and QC, whereas downstream analysis encompasses clustering, gene filtering, and visualization. Additionally, we have identified tasks as either essential or advanced based on their significance and applicability to most scRNA-seq studies (Fig. 3B). Essential tasks, such as QC, alignment, and normalization, are fundamental across both processing stages and are crucial for the majority of scRNA-seq experiments. In contrast, advanced tasks, like allele-specific expression analysis or immune receptor analysis, may be pertinent to specific research inquiries.
Fig. 3.
Features of current single-cell RNA sequencing (scRNA-seq) analysis tools and availability of analysis options. (A) Number of features included in the current developed tools. (B) Essential and advanced analysis features in scRNA-Seq availability. UMIs: unique molecular identifiers.
We have provided a comprehensive overview of the tools and methods available for scRNA-seq data analysis, categorizing features to assist researchers in selecting the most suitable tools for their analysis requirements. Fig. 3B synthesizes our in-depth review of scRNA-seq analysis, employing a systematic examination of the diverse features and tasks invol-ved. It presents these components as parts of a broader framework that researchers can tailor to their specific needs. Additionally, Fig. 3B employs a color-coding system to denote the availability of each feature across the current scRNA-seq analysis tools and software, thereby enabling researchers to swiftly identify tools that support their required features, facilitating the selection process. Consequently, Fig. 3B not only provides a detailed overview of scRNA-seq analysis but also serves as a practical aid for researchers to match their desired features with available tools and software, thereby enhancing informed decision-making and advancing cellular biology and disease research through precise and efficient scRNA-seq data analysis.
Supplementary Table S2 and S3 list the most commonly used analysis features in scRNA-seq, selected for their popularity and utility within the research community. The table also details the programming languages used to develop these tools, offering insights into the technical execution of the analysis. This table is intended as a resource for scientists and researchers involved in scRNA-seq analysis, listing tools alongside their programming languages to help researchers find the most appropriate tool that matches their analysis needs and programming proficiency. It allows researchers to navigate through various options and make knowledgeable choices in tool selection for their projects. Additionally, it helps in assessing tool compatibility with preferred programming languages, ensuring smooth integration and effective use of the chosen tool within existing computational workflows, thus enabling researchers to utilize their programming skills effectively with compatible tools.
Future recommendations
This study presents several important recommendations for future work in the field of scRNA-seq analysis. First, there is a clear need for the research community to focus on improving and expanding the range of tools that include essential features for thorough scRNA-seq analysis. Our review identified a limited number of tools, such as STAR and CellRanger, that provide critical functionalities like unique molecular identifiers (UMIs) and alignment analysis. Developing a wider array of tools that offer these and other key capabilities is essential. Additionally, there’s a priority to integrate these essential features into comprehensive analysis frameworks, which would provide holistic solutions that meet current needs and anticipate future advancements in scRNA-seq technologies. Tools should ideally incorporate features like UMIs, QC, alignments, and normalization to offer all-encompassing solutions for scRNA-seq data analysis.
Moreover, there is a significant emphasis on the need for user-friendly interfaces and intuitive workflows to make scRNA-seq analysis tools more accessible. Making these tools easy to use will allow researchers with varying levels of computational expertise to fully utilize scRNA-seq tech-nology. As scRNA-seq methodologies and research fields continue to evolve rapidly, it is crucial for resources like the scRNA-tools database to be regularly updated and expanded, adding new categories and tools designed for specific research tasks to maintain its value as a comprehensive resource.
Encouraging collaboration between software engineers, bioinformaticians, data scientists, and biologists is critical for fostering interdisciplinary innovation in scRNA-seq analysis. Such collaborative efforts can address existing challenges more effectively, leading to the development of higher quality and more versatile tools for scRNA-seq data analysis. It’s also vital to continually refine existing tools and develop new ones with a focus on user-centric features, particularly in areas such as alignment analysis. Keeping the scRNA-tools database responsive to the diverse needs of the research community is another key recommendation, ensuring it remains a vital tool for advancing our understanding of stem cell biology, disease mechanisms, and opening up new avenues for therapeutic developments. By focusing on these strategic areas and promoting strong interdisciplinary partnerships, we can expect to achieve deeper insights and innovations in the field of scRNA-seq.
Spatial Single-Cell mRNA Sequencing in Stem Cell Research
While this study primarily focuses on the analysis and application of scRNA-seq in stem cell research, it is important to acknowledge the emerging relevance of spatial single-cell mRNA sequencing technology within this field. Its potential to provide spatial context to gene expression in stem cells represents a significant advancement in unravelling the complex spatial heterogeneity of stem cell populations (81). Spatial single-cell mRNA sequencing enables researchers to map the transcriptomic profiles of individual cells within their native tissue environments, offering insights into the spatial dynamics of stem cell differentiation, tissue development, and disease progression. This method complements scRNA-seq by adding a layer of spatial information, thus enhancing our understanding of the complex cellular landscapes in stem cell biology. However, the current main limitation of spatial transcriptomics is achieving sequencing and visualizing the transcrip-tomic maps at the single-cell level. As this technology continues to evolve, it promises to shed light into the spatial aspects of gene expression at the single-cell level, which is crucial for comprehending the full spectrum of stem cell function and regulation before and after differentiation into specific cell types during development in vivo as well as in vitro using human cellular models.
Conclusions
Since its discovery a decade ago, scRNA-seq technology has made extraordinary strides. It has made significant contributions across several areas, including the development of comprehensive cellular maps for tissues, organs, and whole organisms, the redefinition of cell types, the discovery of new marker genes, and the identification of unique cell subpopulations. Furthermore, scRNA-seq has enabled the tracing of cell differentiation and developmental pathways, the identification of tumor-specific molecular markers, and the exploration of tumor heterogeneity and the tumor microenvironment. Additionally, this technology has been instrumental in advancing our understanding of disease mechanisms and the impact of therapeutic interventions.
The roadmap outlined in this paper offers valuable insights for researchers looking to select and utilize the most effective features and tools for scRNA-seq data analysis. Emphasizing the importance of essential features, regularly updating the scRNA-tools database, and promoting collaboration across disciplines are key steps for further progress in scRNA-seq analysis. These efforts will not only facilitate a deeper understanding of stem cells and disease mechanisms but also open up new avenues for discovery and therapeutic development. Researchers are encouraged to consider these recommendations into account to continue advancing the field of stem cells and contribute to the broader progress in scRNA-seq analysis.
Supplementary Materials
Supplementary data including three tables can be found with this article online at https://doi.org/10.15283/ijsc23170
Footnotes
Potential Conflict of Interest
There is no potential conflict of interest to declare.
Authors’ Contribution
Conceptualization: MA. Data curation: MA, HA, SP, BA. Formal analysis: MA, HA, BA. Funding acquisition: BA, EJW, MRS. Investigation: MA, EJW, MRS. Methodology: MA, HA, SP, BA. Project administration: MA, EJW, MRS. Resources: EJW, MRS. Software: MA, EJW, MRS. Super-vision: EJW, MRS. Validation: MA, MRS. Visualization: MA, HA, BA, MRS. Writing – original draft: MA, EJW, MRS. Writing – review and editing: EJW, MRS.
Funding
BA is supported by the University of Queensland (UQ) Research Training Scholarship, and by the UQ Entrepre-neurial PhD Top-up Scholarship. MRS is supported by the Children Hospital Foundation (PCC0252021). EJW and MRS are supported by the Medical Research Future Fund-Stem Cell Mission (APP2007653).
References
- 1.Pullin JM, McCarthy DJ. A comparison of marker gene selection methods for single-cell RNA sequencing data. Genome Biol. 2024;25:56. doi: 10.1186/s13059-024-03183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das S, Rai A, Rai SN. Differential expression analysis of single-cell RNA-Seq data: current statistical approaches and outstanding challenges. Entropy (Basel) 2022;24:995. doi: 10.3390/e24070995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malhotra A, Das S, Rai SN. Analysis of single-cell RNA-sequencing data: a step-by-step guide. BioMedInformatics. 2022;2:43–61. doi: 10.3390/biomedinformatics2010003. [DOI] [Google Scholar]
- 4.Patra SK, Mishra S. Bibliometric study of bioinformatics literature. Scientometrics. 2006;67:477–489. doi: 10.1556/Scient.67.2006.3.9. [DOI] [Google Scholar]
- 5.Glänzel W, Janssens F, Thijs B. A comparative analysis of publication activity and citation impact based on the core literature in bioinformatics. Scientometrics. 2009;79:109–129. doi: 10.1007/s11192-009-0407-1. [DOI] [Google Scholar]
- 6.Song M, Kim SY. Detecting the knowledge structure of bioinformatics by mining full-text collections. Scientometrics. 2013;96:183–201. doi: 10.1007/s11192-012-0900-9. [DOI] [Google Scholar]
- 7.Tranfield D, Denyer D, Smart P. Towards a methodology for developing evidence-informed management knowledge by means of systematic review. Br J Manag. 2003;14:207–222. doi: 10.1111/1467-8551.00375. [DOI] [Google Scholar]
- 8.Mongeon P, Paul-Hus A. The journal coverage of Web of Science and Scopus: a comparative analysis. Scientometrics. 2016;106:213–228. doi: 10.1007/s11192-015-1765-5. [DOI] [Google Scholar]
- 9.Chadegani AA, Salehi H, Yunus MM, et al. A comparison between two main academic literature collections: Web of Science and Scopus databases. Asian Soc Sci. 2013;9:18–26. doi: 10.5539/ass.v9n5p18. [DOI] [Google Scholar]
- 10.Altarturi HHM, Saadoon M, Anuar NB. Cyber parental control: a bibliometric study. Child Youth Serv Rev. 2020;116:105134. doi: 10.1016/j.childyouth.2020.105134. [DOI] [Google Scholar]
- 11.Aria M, Cuccurullo C. bibliometrix: an R-tool for comprehensive science mapping analysis. J Informetr. 2017;11:959–975. doi: 10.1016/j.joi.2017.08.007. [DOI] [Google Scholar]
- 12.Echchakoui S. Why and how to merge Scopus and Web of Science during bibliometric analysis: the case of sales force literature from 1912 to 2019. J Market Anal. 2020;8:165–184. doi: 10.1057/s41270-020-00081-9. [DOI] [Google Scholar]
- 13.Fahimnia B, Sarkis J, Davarzani H. Green supply chain management: a review and bibliometric analysis. Int J Prod Econom. 2015;162:101–114. doi: 10.1016/j.ijpe.2015.01.003. [DOI] [Google Scholar]
- 14.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiselev VY, Kirschner K, Schaub MT, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 19.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 20.Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65:631–643.e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 22.Islam S, Zeisel A, Joost S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 23.Tabula Muris Consortium, author; Overall coordination, author; Logistical coordination, author. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zappia L, Phipson B, Oshlack A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 2017;18:174. doi: 10.1186/s13059-017-1305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dijk D, Sharma R, Nainys J, et al. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaker MR, Slonchak A, Al-mhanawi B, et al. Choroid plexus defects in Down syndrome brain organoids enhance neurotropism of SARS-CoV-2. bioRxiv 544552 [Preprint] 2023. [cited 2024 Feb 7]. Available from: https://doi.org/10.1101/2023.06.12.544552 . [DOI] [PMC free article] [PubMed]
- 30.Kanton S, Boyle MJ, He Z, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain deve-lopment. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Shin H, Shaker MR, et al. Production of human spinal-cord organoids recapitulating neural-tube morphoge-nesis. Nat Biomed Eng. 2022;6:435–448. doi: 10.1038/s41551-022-00868-4. [DOI] [PubMed] [Google Scholar]
- 32.Shaker MR, Pietrogrande G, Martin S, Lee JH, Sun W, Wolvetang EJ. Rapid and efficient generation of myelinating human oligodendrocytes in organoids. Front Cell Neurosci. 2021;15:631548. doi: 10.3389/fncel.2021.631548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaker MR, Hunter ZL, Wolvetang EJ. Robust and highly reproducible generation of cortical brain organoids for mo-delling brain neuronal senescence in vitro. J Vis Exp. 2022;183:e63714. doi: 10.3791/63714. [DOI] [PubMed] [Google Scholar]
- 34.Shaker MR, Aguado J, Chaggar HK, Wolvetang EJ. Klotho inhibits neuronal senescence in human brain organoids. npj Aging Mech Dis. 2021;7:18. doi: 10.1038/s41514-021-00070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaker MR, Kahtan A, Prasad R, et al. Neural epidermal growth factor-like like protein 2 is expressed in human oligodendroglial cell types. Front Cell Dev Biol. 2022;10:803061. doi: 10.3389/fcell.2022.803061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiorenzano A, Sozzi E, Birtele M, et al. Single-cell transcrip-tomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nat Commun. 2021;12:7302. doi: 10.1038/s41467-021-27464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camp JG, Badsha F, Florio M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Mhanawi B, Marti MB, Morrison SD, et al. Protocol for generating embedding-free brain organoids enriched with oligodendrocytes. STAR Protoc. 2023;4:102725. doi: 10.1016/j.xpro.2023.102725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Shaker MR, Park SH, Sun W. Transcriptional signature of valproic acid-induced neural tube defects in human spinal cord organoids. Int J Stem Cells. 2023;16:385–393. doi: 10.15283/ijsc23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan L, Yang M, Guo H, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczyk MS, Tirosh I, Heckl D, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Han X, Liu J, et al. Single-cell transcriptomic analysis reveals the cellular heterogeneity of mesenchymal stem cells. Genom Proteom Bioinform. 2022;20:70–86. doi: 10.1016/j.gpb.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Chai C, Wang R, et al. Single-cell transcriptome atlas of human mesenchymal stem cells exploring cellular heterogeneity. Clin Transl Med. 2021;11:e650. doi: 10.1002/ctm2.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen H, Yang M, Li S, et al. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell. 2021;184:2843–2859.e20. doi: 10.1016/j.cell.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 45.Yu L, Wei Y, Duan J, et al. Blastocyst-like structures generated from human pluripotent stem cells. Nature. 2021;591:620–626. doi: 10.1038/s41586-021-03356-y. [DOI] [PubMed] [Google Scholar]
- 46.Vieira Braga FA, Miragaia RJ. Tissue handling and disso-ciation for single-cell RNA-Seq. Methods Mol Biol. 2019;1979:9–21. doi: 10.1007/978-1-4939-9240-9_2. [DOI] [PubMed] [Google Scholar]
- 47.Denisenko E, Guo BB, Jones M, et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21:130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Brink SC, Sage F, Vértesy Á, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 49.Burja B, Paul D, Tastanova A, et al. An optimized tissue dissociation protocol for single-cell RNA sequencing analysis of fresh and cultured human skin biopsies. Front Cell Dev Biol. 2022;10:872688. doi: 10.3389/fcell.2022.872688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang P, Thomson JA, Stewart R. Quality control of single-cell RNA-seq by SinQC. Bioinformatics. 2016;32:2514–2516. doi: 10.1093/bioinformatics/btw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heiser CN, Wang VM, Chen B, Hughey JJ, Lau KS. Automated quality control and cell identification of droplet-based single-cell data using dropkick. Genome Res. 2021;31:1742–1752. doi: 10.1101/gr.271908.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarke ZA, Andrews TS, Atif J, et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat Protoc. 2021;16:2749–2764. doi: 10.1038/s41596-021-00534-0. [DOI] [PubMed] [Google Scholar]
- 54.Chiaradia I, Imaz-Rosshandler I, Nilges BS, et al. Tissue morphology influences the temporal program of human brain organoid development. Cell Stem Cell. 2023;30:1351–1367.e10. doi: 10.1016/j.stem.2023.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zappia L, Theis FJ. Over 1000 tools reveal trends in the single-cell RNA-seq analysis landscape. Genome Biol. 2021;22:301. doi: 10.1186/s13059-021-02519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macosko E. Novel technologies for single-cell resolution whole-transcriptome analysis in CNS tissue [Internet] Society for Neuroscience; Washington, D.C.: 2016. [cited 2023 Oct 18]. Available from: https://www.sfn.org/-/media/Project/Neuronline/PDFs/2017/Novel-Technologies-for-Single-Cell-Resolution-Whole-Transcriptome-Analysis-in-CNS-Tissue.pdf . [Google Scholar]
- 57.Gierahn TM, Wadsworth MH, 2nd, Hughes TK, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 60.Spiro A, Shapiro E. Accuracy of answers to cell lineage questions depends on single-cell genomics data quality and quantity. PLoS Comput Biol. 2016;12:e1004983. doi: 10.1371/journal.pcbi.1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conrad S, Azizi H, Skutella T. Single-cell expression profiling and proteomics of primordial germ cells, spermatogonial stem cells, adult germ stem cells, and oocytes. Adv Exp Med Biol. 2018;1083:77–87. doi: 10.1007/5584_2017_117. [DOI] [PubMed] [Google Scholar]
- 62.Chen T, Li J, Jia Y, et al. Single-cell sequencing in the field of stem cells. Curr Genomics. 2020;21:576–584. doi: 10.2174/1389202921999200624154445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaminow B, Yunusov D, Dobin A. Accurate quantification of single-cell and single-nucleus RNA-seq transcripts using distinguishing flanking k-mers. bioRxiv 442755 [Preprint] 2021. [cited 2023 Oct 18]. Available from: https://doi.org/10.1101/2021.05.05.442755 . [DOI]
- 64.Hao Y, Stuart T, Kowalski MH, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. 2024;42:293–304. doi: 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu X, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kristján Eldjárn H, Delaney KS, Nikhila PS, Guillaume H, Páll M, Lior P. Accurate quantification of single-cell and single-nucleus RNA-seq transcripts using distinguishing flanking k-mers. bioRxiv 518832 [Preprint] 2024. [cited 2023 Oct 18]. Available from: https://doi.org/10.1101/2022.12.02.518832 . [DOI]
- 67.Srivastava A, Malik L, Smith T, Sudbery I, Patro R. Alevin efficiently estimates accurate gene abundances from dscRNA-seq data. Genome Biol. 2019;20:65. doi: 10.1186/s13059-019-1670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary gliobla-stoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 71.Zeisel A, Muñoz-Manchado AB, Codeluppi S, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 72.Van de Sande B, Flerin C, Davie K, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 73.Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA seque-ncing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grün D. Revealing dynamics of gene expression variability in cell state space. Nat Methods. 2020;17:45–49. doi: 10.1038/s41592-019-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boyeau P, Regier J, Gayoso A, Jordan MI, Lopez R, Yosef N. An empirical Bayes method for differential expression analysis of single cells with deep generative models. Proc Natl Acad Sci U S A. 2023;120:e2209124120. doi: 10.1073/pnas.2209124120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan J, Salathia N, Liu R, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods. 2016;13:241–244. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedi-cal research. Genome Med. 2022;14:68. doi: 10.1186/s13073-022-01075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.