Abstract
Prime editing (PE) is a recently developed genome-editing technique that enables versatile editing. Despite its flexibility and potential, applying PE in human induced pluripotent stem cells (hiPSCs) has not been extensively addressed. Genetic disease models using patient-derived hiPSCs have been used to study mechanisms and drug efficacy. However, genetic differences between patient and control cells have been attributed to the inaccuracy of the disease model, highlighting the significance of isogenic hiPSC models. Hereditary hemorrhagic telangiectasia 1 (HHT1) is a genetic disorder caused by an autosomal dominant mutation in endoglin (ENG). Although previous HHT models using mice and HUVEC have been used, these models did not sufficiently elucidate the relationship between the genotype and disease phenotype in HHT, demanding more clinically relevant models that reflect human genetics. Therefore, in this study, we used PE to propose a method for establishing an isogenic hiPSC line. Clinically reported target mutation in ENG was selected, and a strategy for PE was designed. After cloning the engineered PE guide RNA, hiPSCs were nucleofected along with PEmax and hMLH1dn plasmids. As a result, hiPSC clones with the intended mutation were obtained, which showed no changes in pluripotency or genetic integrity. Furthermore, introducing the ENG mutation increased the expression of proangiogenic markers during endothelial organoid differentiation. Consequently, our results suggest the potential of PE as a toolkit for establishing isogenic lines, enabling disease modeling based on hiPSC-derived disease-related cells or organoids. This approach is expected to stimulate mechanistic and therapeutic studies on genetic diseases.
Keywords: Induced pluripotent stem cells, Gene editing, Genetic diseases, Endoglin, Hereditary hemorrhagic telangiectasia
Introduction
Disease models are used to investigate disease mechanisms and develop treatment strategies. While numerous in vitro and in vivo models have been utilized, several limitations remain due to the insufficient replication of human diseases (1). In genetic disease modeling, knock-out/knock-in techniques or transient transfection are usually applied for loss-of-function or gain-of-function studies (2, 3). These approaches have provided basic knowledge on human genetic diseases. Nevertheless, the complexity of genetics and the various disease phenotypes caused by numerous mutations, even in a single gene, cannot be sufficiently addressed by knock-out/knock-in or transfection studies. Moreover, many in vitro disease models use established cell lines, such as HEK293T; however, these models often do not utilize disease-relevant cell types (4, 5). Furthermore, although in vivo mouse models allow for the study of disease-related organs, they cannot fully replicate human genetics and pathophysiology (6).
To overcome the limitations of previous disease models, patient-derived human induced pluripotent stem cell (hiPSC) disease models have been suggested as alternatives (7). hiPSC-based disease modeling has strengths in that it utilizes disease-related cells with identical patient genomes, reflecting the functionality of the affected cell types and disease phenotypes caused by specific mutations (8). Nevertheless, acquiring samples from patients with rare genetic diseases remains a challenge (9), which limits the use of patient-derived hiPSCs. In addition, selecting a healthy control hiPSC line for comparison with a patient-derived hiPSC line remains challenging, because differences in genetic background can disturb the disease phenotype (10). Therefore, isogenic hiPSC models have emerged as alternatives to patient-derived hiPSC models, utilizing a hiPSC line with an identical genetic background to the control line except for the pathogenic mutation (11). This approach reflects only changes caused by interested mutation and has the advantage of not requiring patient-derived samples.
The CRISPR/Cas9 system has been used for genome-editing in hiPSCs by double-strand breaks (DSBs)-mediated homology-directed repair (HDR) (12). However, the efficiency of HDR is low and DSBs can induce uninten-ded indels and genetic instability (13-15), making it difficult to establish isogenic hiPSCs. Recently, base editing and prime editing techniques have been developed to overcome the limitations of conventional CRISPR/Cas9 technique. In the base editing system, editing is performed by a deaminase attached to a mutated Cas9 nickase (16). In prime editing, prime editing guide RNA (pegRNA) and a reverse transcriptase linked to a mutated Cas9 nickase perform genome-editing (17). Compared to base editing, prime editing has a relatively lower editing efficiency, but offers the advantages of enabling various types of edits, such as insertion, deletion, and substitution (18). These features make it a promising genome-editing tool. However, the application of prime editing in hiPSCs has not been widely explored.
Hereditary hemorrhagic telangiectasia (HHT) is a genetic disease characterized by arteriovenous malformation, telangiectasia, and epistaxis (19). Among various types, HHT1 is caused by autosomal dominant mutation in the ENG gene, which leads to haploinsufficiency and disturbance of BMP9/10-ALK1-Smad1/5/9 signaling pathway (20). Although HUVEC and mouse models were applied to study HHT1 (21, 22), the relationship between the mutation type and phenotype remains inadequately clarified, which demands an improved disease model relevant with human genetics and pathophysiology. In this study, to suggest an improved option for studying HHT, we developed a strategy for applying prime editing in hiPSCs, thereby generating an isogenic HHT hiPSC line carrying ENG c.360+1G>A mutation.
Materials and Methods
Plasmids for prime editing
The pCMV-PEmax-P2A-GFP plasmid (#180020; Addgene) and pEF1a-hMLH1dn plasmid (#174824; Addgene) were used to express the prime editor and inhibit MLH1, respectively. Engineered pegRNA (epegRNA) targeting the human ENG gene was designed using the PrimeDesign tool (https://drugthatgene.pinellolab.partners.org). The designed epegRNA was cloned into the pU6-tevopreq1-GG-acceptor plasmid (#174038; Addgene) using Golden Gate assembly, as previously described (17). After mini-prepping the colonies, the cloning results were analyzed by Sanger sequencing. The oligonucleotide sequences used for cloning are listed in Table 1.
Table 1.
List of oligos used for cloning
| Name | Sequence (5’ to 3’) | Purpose |
|---|---|---|
| ENG c.360+1G>A_spacer_F | caccGTGGAGGGAACACACTCACGTgtttt | Golden Gate assembly cloning into pU6-tevopreq1-GG-acceptor plasmid |
| ENG c.360+1G>A_spacer_R | ctctaaaacACGTGAGTGTGTTCCCTCCAC | |
| epegRNA scaffold_F | AGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCG | |
| epegRNA scaffold_R | GCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAG | |
| ENG c.360+1G>A_extension_F | gtgcACTTGGCCTACaTGAGTGTGTTCCC | |
| ENG c.360+1G>A_extension_R | cgcgGGGAACACACTCAtGTAGGCCAAGT | |
| hU6 promoter_F | GAGGGCCTATTTCCCATGATT | Sanger sequencing |
Cell culture
hiPSCs generated from BJ fibroblasts were seeded on culture dishes coated with hESC-qualified Matrigel (#354277; Corning) containing mTeSRTM1 medium (#85850; STEMCELL technologies) with 10 μM Y-27632 (#1254; Tocris Bioscience) and maintained in humidified incubator (5% CO2 and 37℃). Each day, the medium was refreshed until the cells reached approximately 90% confluence, which generally required approximately 3 days.
Nucleofection
hiPSCs were received fresh mTeSRTM1 medium containing 10 μM of Y-27632. After 1 hour of incubation, the cells were dissociated using Accutase solution (#A6964; Sigma-Aldrich) before nucleofection. For prime editing, 1.0×106 cells were resuspended in P3 Primary Cell 4D-NucleofectorTM X kit solution (#V4XP-3024; Lonza) with pCMV-PEmax-P2A-GFP plasmid (4.5 μg), pEF1a-hMLH1dn plasmid (1.5 μg), and pU6-tevopreq1-GG-acceptor plasmid (2.2 μg). The cells were immediately nucleofected with AmaxaTM 4D-NucleofectorTM (Lonza) using CA-137 program. Transfected cells were treated with fresh medium after 24 hours, and GFP-expressing cells were sorted with an SH800S Cell Sorter (SONY) after 24 hours of nucleofection. The sorted cells were seeded on 6-well plate at a low density and cultured to form single-cell colonies. After 12 days, the colonies were picked and transferred to a 96-well plate for expansion. Finally, each clone was analyzed by Sanger sequencing to determine the genotype of ENG.
Genomic DNA extraction and Sanger sequencing
Genomic DNA (gDNA) was extracted from each clone using a lysis buffer containing proteinase K (KB-0111; Bioneer). The cell lysis buffer consists of 10% (w/v) sodium dodecyl sulfate solution (250 μL), 1 M pH 8.0 Tris-HCl (500 μL), and sterile distilled water to a final volume of 50 mL. Diluted proteinase K (1:300, v/v) was added immediately prior to lysis. Polymerase chain reaction (PCR) was conducted with the AccuPowerⓇ PCR PreMix kit (K-2012; Bioneer) and specific primers. The PCR protocol involved an initial denaturation at 95℃ for 5 minutes, followed by 30 cycles comprising denaturation at 95℃ for 20 seconds, annealing at 65℃ for 20 seconds, and extension at 72℃ for 30 seconds. The final extension step occurred at 72℃ for 5 minutes. PCR products were purified by gel extraction and subjected to Sanger sequencing. The sequencing results were analyzed using the EditR tool (http://baseeditr.com/). The oligonucleotides used for PCR and Sanger sequencing are listed in Table 2.
Table 2.
List of PCR primers for Sanger sequencing
| Name | Sequence (5’ to 3’) | Size (bp) | Purpose |
|---|---|---|---|
| ENG_e3_F | CTGCCTGTCTGGGTGGCACAACCT | 269 | gDNA PCR, Sanger sequencing |
| ENG_e3_R | CAGTAGGGACCTCCCATGGCCAGA | ||
| ENG OT(1)_F | GCAAACGCTGTCCCTATCCT | 262 | Off-target (1) Sanger sequencing |
| ENG OT(1)_R | CTCTCCCACCAACCTGGAAC | ||
| ENG OT(2)_F | CCCCAGAGAGGTGATCGAGA | 333 | Off-target (2) Sanger sequencing |
| ENG OT(2)_R | CATGGCAGGGTTTAGCCTCA |
gDNA PCR: genomic DNA polymerase chain reaction.
Immunocytochemistry
For Immunocytochemistry, cells were fixed for 10 minutes with 4% paraformaldehyde. Blocking and permeabilization were performed using a solution containing 0.3% Triton X-100 and 3% normal goat serum in phosphate-buffered saline. After overnight incubation with the primary antibody in the blocking solution, the cells were washed three times. Primary antibodies used were as follow: anti-octamer-binding transcription factor 4 (OCT4) (sc-5279, 1:200; Santa Cruz Biotechnology), anti-TRA-1- 60 (ab16288, 1:200; Abcam), anti-SOX2 (ab97959, 1:200; Abcam). Subsequently, after a 2 hours incubation with secondary antibody, the cells were washed again thrice, and nuclear staining was performed using 4’,6-diamidino-2-phenylindole (DAPI). The following secondary antibodies were used: AlexaFluor 488 goat anti-mouse IgG (A-11001, 1:1,000; Invitrogen) and AlexaFluor 488 goat anti-rabbit IgG (A-11008, 1:1,000; Invitrogen). Fluorescence images were acquired using a fluorescence microscope.
Teratoma assay
The teratoma assay for in vivo pluripotency assessment was approved by the Institutional Animal Care and Use Committee of Konkuk University (IACUC No. KU23225). All animal care and experimental procedures were conducted according to the institutional guidelines. Generated isogenic hiPSCs were harvested and resuspended in a 1:1 mixture of the medium and Matrigel (1×106 cells/100 μL), then immediately injected into the dorsal flank of a 5-week-old male immunocompromised BALB/c nude mouse. After 5 weeks, euthanasia was conducted in a CO2 chamber, and the tumor was resected, followed by fixation and tissue processing. Processed tumor tissues were embedded in paraffin, and sectioned samples (4 μm) were stained with hematoxylin and eosin. The morphologies of the endoderm, mesoderm, and ectoderm were observed under a microscope.
Alkaline phosphatase staining
hiPSCs fixed with 4% paraformaldehyde were stained using the StemAbTM Alkaline Phosphatase Staining Kit II (00-0055; Reprocell) according to the manufacturer’s guide.
Karyotyping
Chromosomal analysis of the established isogenic hiPSC line was performed using a standard method with slight modifications. Briefly, cells were incubated with colcemid (9311; FUJIFILM) for 3 hours at 37℃ and detached using 0.25% trypsin-ethylenediamine tetraacetic acid. Cells were treated with a hypotonic solution containing 1% sodium citrate, and the lysed cells were fixed with a fixation solution (methanol:acetic acid=3:1). G-banding analysis was performed to identify chromosomes, followed by microscopic observation.
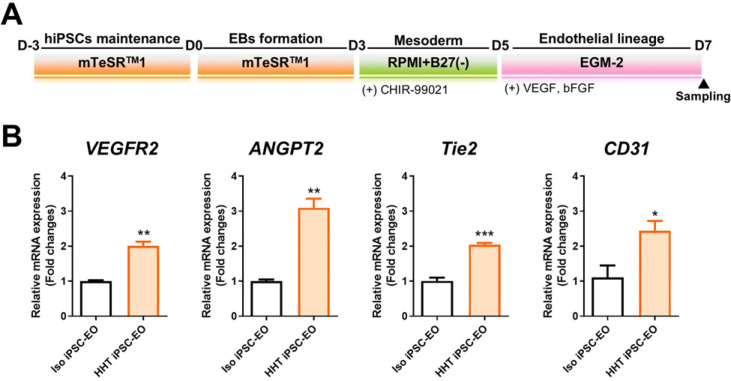
Endothelial organoid formation and quantitative RCR
Endothelial organoids (EOs) were generated from isogenic control and established HHT hiPSCs. The Cells were cultured in Matrigel-coated dishes for 3 days, then detached for embryoid body (EB) generation in petri dishes under shaking condition (60 RPM) for 3 days. Generated EBs were exposed to RPMI 1640 (#11875093; Gibco) containing B-27TM, without insulin (#A1895601; Gibco) supplemented with 6 μM of CHIR-99021 (#4423; Tocris Bioscience). After 2 days, endothelial lineage differentiation was induced by EGMTM-2 (CC-3202; Lonza) supplemented with 100 ng/mL VEGF (100-20; PeproTech) and 20 ng/mL bFGF (100-18B; PeproTech) for 2 days. RNA was extracted from hiPSC-derived EOs using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol, and the concentration of the extracted RNA was measured using a NanoDrop One/OneC spectrophotometer (Thermo Fisher Scientific). Subsequently, cDNA synthesis was performed using AccuPowerⓇ RT PreMix and Oligo dT 20 mer (2 nmol). The synthesized cDNA was mixed with AccuPowerⓇ 2X GreenStarTM quantitative RCR (qPCR) Master Mix and primers, and qPCR analysis was performed using the LightCyclerⓇ 480 instrument (Roche). Thermal cycling conditions consisted of initial denaturation at 95℃ for 5 minutes, followed by 45 cycles of 95℃ for 10 seconds, 58℃ for 10 seconds, and 72℃ for 10 seconds. Each mRNA expression level was normalized to that of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated using the 2−ΔΔCt method. The primer sequences are detailed in Table 3.
Table 3.
List of primers for qPCR
| Name | Sequence (5’ to 3’) | Purpose |
|---|---|---|
| VEGFR2_F | GGAACCTCACTATCCGCAGAGT | qPCR |
| VEGFR2_R | CCAAGTTCGTCTTTTCCTGGGC | |
| ANGPT2_F | ACCCCACTGTTGCTAAAGAAGA | qPCR |
| ANGPT2_R | CCATCCTCACGTCGCTGAATA | |
| Tie2_F | TCCCGAGGTCAAGAGGTGTA | qPCR |
| Tie2_R | AGGGTGTGCCTCCTAAGCTA | |
| CD31_F | AAGTGGAGTCCAGCCGCATATC | qPCR |
| CD31_R | ATGGAGCAGGACAGGTTCAGTC | |
| GAPDH_F | CATCAATGGAAATCCCATCAC | qPCR |
| GAPDH_R | GCAGAGATGATGACCCTTTTG |
qPCR: quantitative polymerase chain reaction.
Statistical analysis
qPCR analysis data are presented as the mean±SEM. Statistical significance was evaluated using unpaired t-test in GraphPad Prism 9.2.0. p-values<0.05 were considered statistically significant (*p<0.05, **p<0.01, ***p<0.001).
Results
Selection of ENG target mutation and genome-editing method
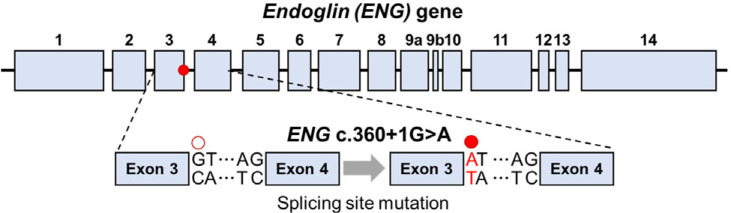
We explored previous publications to select a mutation among the clinically reported ENG mutations (23, 24). The severity of the phenotype caused by each mutation was determined by comparing clinical manifestations, such as telangiectasia, epistaxis, pulmonary arteriovenous malformation, central nervous system arteriovenous shunt, and hepatic arteriovenous shunt. Among the various ENG mutations, the c.360+1G>A mutation has been reported to exhibit the most manifestations of HHT and has been observed in a relatively large number of patients in Korea (23). The ENG c.360+1G>A mutation is located at the splicing site of intron 3 (Fig. 1), leading to a splicing error that causes haploinsufficiency. The CRISPR/Cas9-based HDR method has low efficiency in hiPSCs (25), and base editing techniques like adenine base editing (ABE) and cytosine base editing (CBE) are limited to only A·T to G·C and C·G to T·A substitutions within editing window (26). Therefore, in this study, we aimed to explore the potential of prime editing as an improved genome-editing tool for hiPSCs and applied it to introduce the ENG c.360+1 G>A mutation. Prime editing is a CRISPR/Cas9-based genome editing technology that utilizes a prime editor composed of Cas9 nickase, which does not induce DSBs, and fused reverse transcriptase (27). Prime editing is performed by prime editor and pegRNA, which consisted of spacer, reverse transcriptase template (RTT), and primer binding site (PBS) regions. The spacer in pegRNA guides Cas9 to the target site, where Cas9 induces DNA nicking. PBS then binds to the cleaved DNA, after which the reverse transcriptase uses the RTT region as a template to synthesize DNA, thereby introducing the desired editing (28). Prime editing, facilitated by the design of the RTT region, enables a variety of editing types such as insertion, deletion, and substitution. This makes it suitable for inducing numerous pathogenic mutations in human including the ENG c.360+1G>A mutation targeted in this study.
Fig. 1.
Structure of ENG gene and position of target mutation. ENG c.360+1G>A mutation. The ENG c.360+1G>A mutation (red mark) is located at the splicing site in intron 3 and disrupts normal splicing.
Establishing a strategy of prime editing in hiPSCs to introduce ENG mutation
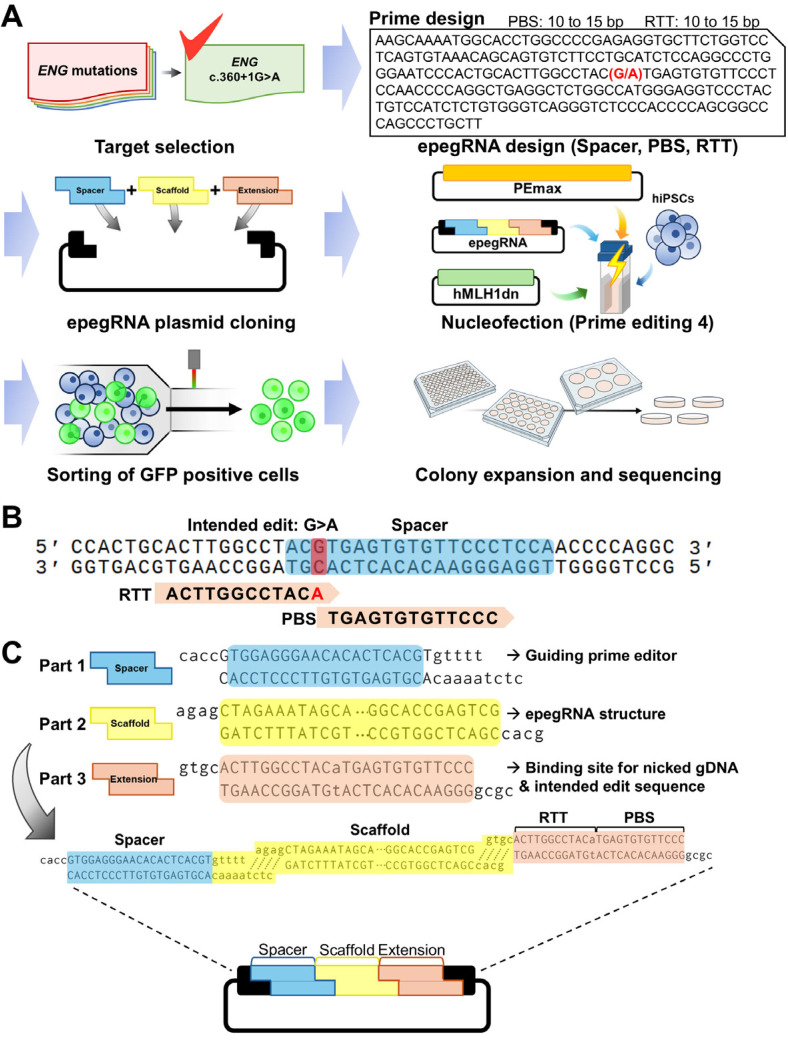
After selecting the target mutation of ENG and the genome-editing method, we established a strategy for efficient prime editing of hiPSCs (Fig. 2A). The epegRNA sequence was designed using the PrimeDesign software (Fig. 2B). Following the recommendations of previous studies (17, 29), the spacer was set to the sequence closest to the intended edit (c.360+1G>A), and PBS and RTT were set to 13 bp and 12 bp, respectively, within the recommended length range. Subsequently, spacer, scaffold, and extension oligos (RTT and PBS) were annealed according to the designed epegRNA sequence, and cloning was performed on the epegRNA plasmid through Golden Gate assembly (Fig. 2C). The cloned epegRNA plasmid was then introduced into wild-type hiPSCs via nucleofection along with the PEmax plasmid expressing the GFP marker and the hMLH1dn plasmid to enhance editing efficiency. To further improve the editing efficiency, GFP-expressing cells were isolated, expanded into colonies, and single-cell colonies were sequenced.
Fig. 2.
Strategy for prime editing to generate isogenic human induced pluripotent stem cells (hiPSCs) carrying clinically reported ENG mutation. (A) Scheme of the process for prime editing using hiPSCs. After choosing target mutation, engineered prime editing guide RNA (epegRNA) is designed using the PrimeDesign tool. The spacer sequence is preferably selected to be closest to the editing site, while reverse transcriptase template (RTT) and primer binding site (PBS) are typically chosen to be between 10 and 15 bp in length. To introduce designed sequences into epegRNA plasmid, annealed oligos for spacer, scaffold, extension including RTT and PBS are assembled into plasmid by golden gate cloning. Nucleofection is performed to introduce cloned epegRNA plasmid combined with PEmax plasmid and hMLH1dn plasmid, followed by isolating transfected cells via flow cytometry. Then, single-cell colonies are expended and sequenced to examine the genotype. (B) epegRNA design for targeting ENG c.360+1G>A mutation. The spacer sequence was selected as the closest to the target, and an A was added to the RTT sequence to change the target base to the desired sequence (G>A). (C) Scheme of Golden gate cloning for epegRNA plasmid construction. Oligo sets corresponding to spacer, scaffold, extension (RTT and PBS) sequence are shown.
Generation of an isogenic HHT hiPSC line by prime editing
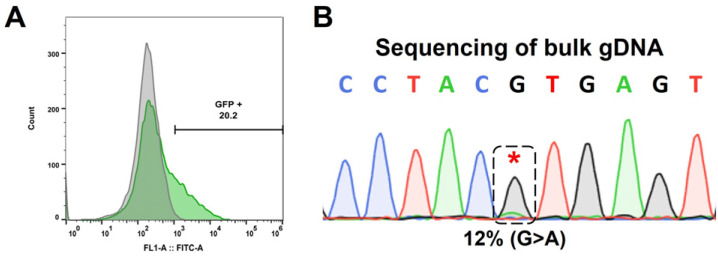
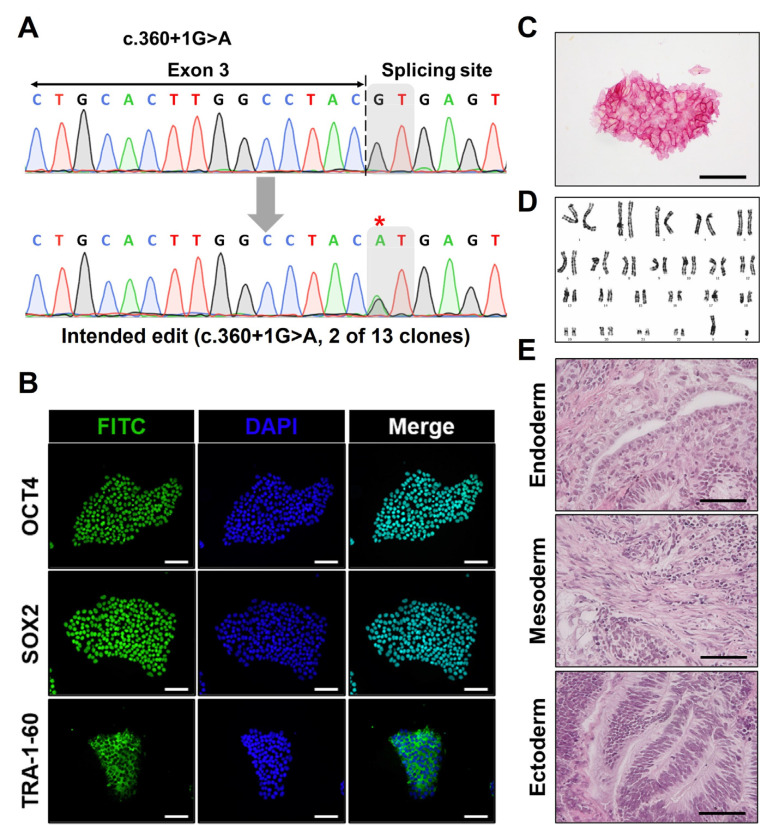
Based on the established strategy, we nucleofected hiPSCs and isolated GFP-expressing cells to selectively identify cells with successful nucleofection (Fig. 3A). Before the formation of single-cell colonies, bulk gDNA sequencing was conducted to assess the overall editing efficiency, which was determined to be 12% by EditR analysis (Fig. 3B). Following the confirmation of editing via bulk gDNA sequencing, we proceeded with the expansion and sequencing of single-cell colonies to secure an hiPSC line containing the intended edit. As a result, we verified the heterozygous introduction of the intended ENG c.360+1G>A mutation in two out of the 13 clones obtained (Fig. 4A). We conducted pluripotency marker staining and alkaline phosphatase staining to assess the impact of prime editing on the pluripotency of hiPSCs and confirmed that there was no effect (Fig. 4B, 4C). Also, karyotyping and teratoma formation assays were performed to evaluate the chromosomal integrity and in vivo pluripotency, respectively (Fig. 4D, 4E). Additionally, it was confirmed that no unintended editing occurred at potential off-target sites with sequences similar to the spacer sequence (Supplementary Fig. S1). Through these analyses, we verified that the isogenic hiPSC line generated via prime editing maintained stable pluripotency and integrity. Finally, isogenic control and HHT hiPSCs were differentiated into EOs to assess the functional changes (Fig. 5A). Subsequently, expression of genes associated with angiogenesis in HHT, such as VEGFR2, ANGPT2, Tie2, was analyzed in EOs (20). As a results, mRNA expression levels of VEGFR2, ANGPT2, Tie2, and CD31, were significantly higher in HHT hiPSC-derived EOs than in EOs from isogenic control hiPSCs (Fig. 5B). These results mean that our isogenic model reflects the proangiogenic molecular features of HHT.
Fig. 3.
Isolation of transfected human induced pluripotent stem cells (hiPSCs) and bulk sequencing for evaluating editing efficiency. (A) After 24 hours of nucleofection for prime editing, GFP-expressing cells were sorted by FACS to increase editing efficiency. (B) Isolated GFP-expressing cells were analyzed to examine efficiency. Bulk genomic DNA was extracted from sorted cells and the target sequence was amplified by polymerase chain reaction (PCR). Gel-extracted PCR product was sequenced and analyzed by EditR. The asterisk mark indicates the target base pair.
Fig. 4.
Generation and characterization of an isogenic human induced pluripotent stem cells (hiPSC) line with intended ENG mutation by prime editing. (A) Single-cell colonies were formed and expanded. Genomic DNA samples of each colony were amplified by polymerase chain reaction (PCR). After sequencing the PCR products, the results were analyzed to investigate the genotype of each colony. Among 13 acquired clones, two clones possessed the intended mutation (ENG c.360+1G>A) heterozygously. The asterisk mark indicates the target base pair. (B, C) Immunofluorescence staining and AP staining were performed to evaluate the pluripotency of generated isogenic mutant hiPSC line (scale bar=100 μm). (D) Chromosomal integrity was confirmed by karyotyping. (E) Teratoma assay was further conducted to verify in vivo pluripotency, resulting in the formation of all three germ layers including endoderm, mesoderm, and ectoderm (scale bar=50 μm).
Fig. 5.
Endothelial organoid (EO) formation and proangiogenic marker analysis. (A) Differentiation schedule for EOs from isogenic control and hereditary hemorrhagic telangiectasia human induced pluripotent stem cell (HHT hiPSC) line. (B) mRNA expression levels of proangiogenic markers, including VEGFR2, ANGPT2, Tie2, and CD31, were analyzed by quantitative polymerase chain reaction. Statistical significance: *p<0.05, **p<0.01, ***p<0.001.
Discussion
Conventional HDR-based genome editing using CRISPR/Cas9 has shown relatively low efficiency in hiPSCs and is associated with safety concerns due to the non-target effects caused by DSBs (25). To address these issues, base editors that combine mutated Cas9 nickase with deaminases have been developed (30). Two types of base editors, ABE and CBE, demonstrated relatively higher target correction efficiencies than CRISPR/Cas9-mediated HDR (31). However, ABE and CBE are limited to the A·T to G·C and C·G to T·A substitutions, respectively (26). They are also restricted to an editing window that is approximately 12 to 16 bp away from the PAM sequence and may cause unintended changes to nearby target base pairs (32). Despite these limitations, previous studies have reported the effectiveness of base editing for disease modeling in hiPSCs and have presented it as a genome-editing tool for hiPSCs (33, 34). However, the aforementioned limitations of base editing result in limited genome-editing capabilities, thereby reducing its versatility.
Prime editing is an advanced technology that overcomes the limitations of base editing. Similar to base editing, it uses mutated Cas9, which does not induce DSBs, in combination with reverse transcriptase (17). Prime editors use pegRNA, which includes RTT and PBS regions in addition to spacer to target specific sequences. This allows the prime editor to introduce the intended edit based on the RTT backbone, thereby enabling all types of substitutions, insertions, and deletions without unintended change of bystander (18). Additionally, the option to adjust the lengths of the RTT and PBS regions allows for the editing of targets further from the PAM sequence, providing a much broader targeting range than base editing (35). Taken together, prime editing offers significant advantages over base editing in terms of the variety of editing types, less stringent PAM sequence requirements, and absence of bystander effects.
HHT1 is caused by haploinsufficiency due to autosomal dominant mutations in ENG. Among all types of HHT, HHT1 has the highest prevalence (36). About 490 mutations in ENG associated with HHT1 were found (20), and differences in clinical severity have been reported depending on the type of ENG mutation (37). This emphasizes the significance of isogenic modeling for investigating HHT, which exhibit the relationship between disease phenotypes and mutations. In this study, we aimed to target the ENG c.360+1G>A mutation associated with severe HHT phenotype (23), generating an isogenic HHT hiPSC line. Establishing more isogenic hiPSC lines with different ENG mutations using prime editing and comparing each isogenic lines will elucidate the relationship between the genotype and phenotype in HHT. Also, clinical drug efficacy tests can be conducted using differentiated cells or vascular organoids, which can reflect differences in drug response depending on the mutation types.
To maximize the editing efficiency, we performed nucleofecting the epegRNA and PEmax in combination with hMLHdn plasmids (17). Additionally, to select cells transfected with the largest plasmid, PEmax, we sorted GFP-expressing cells. Bulk gDNA sequencing revealed an editing efficiency of 12%, and sequencing analysis of single-cell colonies confirmed that two of the 13 colonies carried the intended c.360+1G>A mutation in a heterozygous form. These results, obtained without additional optimization steps, were considered satisfactory. Given that the goal of genome-editing in hiPSCs is to obtain clones with intended editing, prime editing is considered a useful tool. Additionally, compared to traditional CRISPR/Cas9 HDR editing, this method is more convenient because it does not require the additional donor DNA and usually show higher efficiency than unoptimized HDR methods (38).
To date, prime editing has primarily focused on the treatment of genetic diseases, with the potential to correct approximately 89% of genetic mutation (27). This implies that prime editing can also introduce most of pathogenic mutations into normal cells, making it a valuable tool for modeling genetic diseases. This approach can address the challenges associated with the use of patient-derived iPSCs for disease modeling, such as the scarcity of patient samples and the difficulty in establishing appropriate control cells due to genomic discrepancies (9, 10). Moreover, within the same genetic disorder, disease severity and potential treatment options may vary depending on the mutation type (39). Prime editing can introduce a wide range of mutations, enabling disease modeling that reflects these differences.
Nevertheless, additional improvements are necessary in its application, especially in enhancing efficiency. In this study, the ENG gene editing efficiency, assessed through bulk gDNA sequencing, was 12%. Since HHT1 is associated with a heterozygous mutation in ENG, obtaining the cells with desired genotype was achievable. However, in the case of targeting autosomal recessive diseases, pathogenic mutations should be introduced homozygously, which means that editing efficiency of 12% might not be sufficient to acquire the desired genotype. Therefore, further research is required to enhance prime editing efficiency in hiPSCs. A recent report on improvement of prime editing system demonstrated that tagging the prime editor with the small RNA-binding protein La can significantly enhance editing efficiency, which has been suggested as prime editing 7 (40). Optimizing prime editing 7 in hiPSCs could enable high efficiency editing and further promote its application for disease modeling.
In this study, we established a strategy for the prime editing in hiPSCs and demonstrated the feasibility of introducing point mutations into hiPSCs. This approach is expected to facilitate disease modeling using prime editing. Future research will utilize the established isogenic disease hiPSCs to differentiate disease-relevant cells or organoids, enabling human-relevant disease modeling that cannot be adequately evaluated using animal models. This advancement is expected to promote studies on disease mechanisms, therapeutic development, and personalized precision medicine. Given the current surge in gene therapy development, we propose an isogenic hiPSC model generated by prime editing as a potential platform to overcome the limitations of animal models for evaluating gene therapies.
Supplementary Materials
Supplementary data including one figure can be found with this article online at https://doi.org/10.15283/ijsc24084
Footnotes
Potential Conflict of Interest
There is no potential conflict of interest to declare.
Authors’ Contribution
Conceptualization: MWK, CYK, HMC. Data curation: MWK, KSJ, JK, SGL. Formal analysis: MWK. KSJ, SGL. Funding acquisition: HMC. Investigation: MWK, KSJ. Methodology: MWK, JK, SGL. Project administration: CYK, HMC. Supervision: CYK, HMC. Visualization: KSJ. Writing – original draft: MWK, KSJ. Writing – review and editing: CYK, JK, HMC.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants from the Korea government (MSIT) (2022R1A2C2012738) and was supported by Basic Science Research Program through the NRF funded by the Ministry of Education (RS-2023-00240972). This paper was written by the support program for Research Oriented Professors of Konkuk University.
References
- 1.Loewa A, Feng JJ, Hedtrich S. Human disease models in drug development. Nat Rev Bioeng. 2023;1:545–559. doi: 10.1038/s44222-023-00063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyle A, McGarry MP, Lee NA, Lee JJ. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 2012;21:327–349. doi: 10.1007/s11248-011-9537-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prelich G. Gene overexpression: uses, mechanisms, and interpretation. Genetics. 2012;190:841–854. doi: 10.1534/genetics.111.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dabrowska M, Ciolak A, Kozlowska E, Fiszer A, Olejniczak M. Generation of new isogenic models of huntington's disease using CRISPR-Cas9 technology. Int J Mol Sci. 2020;21:1854. doi: 10.3390/ijms21051854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Z, Xu L, Cao D, et al. Effect of orexin-A on mitochondrial biogenesis, mitophagy and structure in HEK293-APPSWE cell model of Alzheimer's disease. Clin Exp Pharmacol Physiol. 2021;48:355–360. doi: 10.1111/1440-1681.13424. [DOI] [PubMed] [Google Scholar]
- 6.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soldner F, Jaenisch R. Medicine. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Oikonomopoulos A, Sayed N, Wu JC. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development. 2018;145:dev156166. doi: 10.1242/dev.156166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escribá R, Ferrer-Lorente R, Raya Á. Inborn errors of metabolism: lessons from iPSC models. Rev Endocr Metab Disord. 2021;22:1189–1200. doi: 10.1007/s11154-021-09671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soldner F, Jaenisch R. Stem cells, genome editing, and the path to translational medicine. Cell. 2018;175:615–632. doi: 10.1016/j.cell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McTague A, Rossignoli G, Ferrini A, Barral S, Kurian MA. Genome editing in iPSC-based neural systems: from disease models to future therapeutic strategies. Front Genome Ed. 2021;3:630600. doi: 10.3389/fgeed.2021.630600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassett AR. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm Genome. 2017;28:348–364. doi: 10.1007/s00335-017-9684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira da Silva J, Oliveira GP, Arasa-Verge EA, et al. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat Commun. 2022;13:760. doi: 10.1038/s41467-022-28442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Porto EM, Komor AC, Slaymaker IM, Yeo GW. Base editing: advances and therapeutic opportunities. Nat Rev Drug Discov. 2020;19:839–859. doi: 10.1038/s41573-020-0084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doman JL, Sousa AA, Randolph PB, Chen PJ, Liu DR. Designing and executing prime editing experiments in mammalian cells. Nat Protoc. 2022;17:2431–2468. doi: 10.1038/s41596-022-00724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA base-editing and prime-editing. Int J Mol Sci. 2020;21:6240. doi: 10.3390/ijms21176240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald J, Bayrak-Toydemir P, Pyeritz RE. Hereditary hemorrhagic telangiectasia: an overview of diagnosis, management, and pathogenesis. Genet Med. 2011;13:607–616. doi: 10.1097/GIM.0b013e3182136d32. [DOI] [PubMed] [Google Scholar]
- 20.Robert F, Desroches-Castan A, Bailly S, Dupuis-Girod S, Feige JJ. Future treatments for hereditary hemorrhagic telangiectasia. Orphanet J Rare Dis. 2020;15:4. doi: 10.1186/s13023-019-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cymerman U, Vera S, Pece-Barbara N, et al. Identification of hereditary hemorrhagic telangiectasia type 1 in newborns by protein expression and mutation analysis of endo-glin. Pediatr Res. 2000;47:24–35. doi: 10.1203/00006450-200001000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Tual-Chalot S, Oh SP, Arthur HM. Mouse models of hereditary hemorrhagic telangiectasia: recent advances and future challenges. Front Genet. 2015;6:25. doi: 10.3389/fgene.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Seo EJ, Song YS, et al. Current status of clinical diagnosis and genetic analysis of hereditary hemorrhagic telangiectasia in South Korea: multicenter case series and a systematic review. Neurointervention. 2019;14:91–98. doi: 10.5469/neuroint.2019.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim BG, Jung JH, Kim MJ, et al. Genetic variants and clinical phenotypes in Korean patients with hereditary hemorrhagic telangiectasia. Clin Exp Otorhinolaryngol. 2021;14:399–406. doi: 10.21053/ceo.2020.02124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JC, Park MJ, Lee SY, et al. Gene editing with 'pencil' rather than 'scissors' in human pluripotent stem cells. Stem Cell Res Ther. 2023;14:164. doi: 10.1186/s13287-023-03394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra R, Joshi RK, Zhao K. Base editing in crops: current advances, limitations and future implications. Plant Bio-technol J. 2020;18:20–31. doi: 10.1111/pbi.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, Kuang J, Shao T, et al. Prime editing: an all-rounder for genome editing. Int J Mol Sci. 2022;23:9862. doi: 10.3390/ijms23179862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HK, Yu G, Park J, et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol. 2021;39:198–206. doi: 10.1038/s41587-020-0677-y. [DOI] [PubMed] [Google Scholar]
- 30.Jeong YK, Song B, Bae S. Current status and challenges of DNA base editing tools. Mol Ther. 2020;28:1938–1952. doi: 10.1016/j.ymthe.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molla KA, Yang Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Qi T, Wu F, Xie Y, et al. Base editing mediated generation of point mutations into human pluripotent stem cells for modeling disease. Front Cell Dev Biol. 2020;8:590581. doi: 10.3389/fcell.2020.590581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Li H, Zhu M, Han RY, Guo S, Han R. Correction of DMD in human iPSC-derived cardiomyocytes by base-editing-induced exon skipping. Mol Ther Methods Clin Dev. 2022;28:40–50. doi: 10.1016/j.omtm.2022.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 36.Kritharis A, Al-Samkari H, Kuter DJ. Hereditary hemorrhagic telangiectasia: diagnosis and management from the hematologist's perspective. Haematologica. 2018;103:1433–1443. doi: 10.3324/haematol.2018.193003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gariballa N, Badawi S, Ali BR. Endoglin mutants retained in the endoplasmic reticulum exacerbate loss of function in hereditary hemorrhagic telangiectasia type 1 (HHT1) by exerting dominant negative effects on the wild type allele. Traffic. 2024;25:e12928. doi: 10.1111/tra.12928. [DOI] [PubMed] [Google Scholar]
- 38.Ochoa-Sanchez A, Perez-Sanchez G, Torres-Ledesma AM, et al. Prime editing, a novel genome-editing tool that may surpass conventional CRISPR-Cas9. Re: GEN Open. 2021;1:75–82. doi: 10.1089/regen.2021.0016. [DOI] [Google Scholar]
- 39.Modrego A, Amaranto M, Godino A, Mendoza R, Barra JL, Corchero JL. Human α-galactosidase a mutants: priceless tools to develop novel therapies for fabry disease. Int J Mol Sci. 2021;22:6518. doi: 10.3390/ijms22126518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J, Oyler-Castrillo P, Ravisankar P, et al. Improving prime editing with an endogenous small RNA-binding pro-tein. Nature. 2024;628:639–647. doi: 10.1038/s41586-024-07259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.