Abstract
Histone H2B monoubiquitination (H2Bub1) is a dynamic posttranslational modification which are linked to DNA damage and plays a key role in a wide variety of regulatory transcriptional programs. Cancer cells exhibit a variety of epigenetic changes, particularly any aberrant H2Bub1 has frequently been associated with the development of tumors. Nevertheless, our understanding of the mechanisms governing the histone H2B deubiquitination and their associated functions during stem cell differentiation remain only partially understood. In this study, we wished to investigate the role of deubiquitinating enzymes (DUBs) on H2Bub1 regulation during stem cell differentiation. In a search for potential DUBs for H2B monoubiquitination, we identified Usp7, a ubiquitin-specific protease that acts as a negative regulator of H2B ubiquitination during the neuronal differentiation of mouse embryonic carcinoma cells. Loss of function of the Usp7 gene by a CRISPR/Cas9 system during retinoic acid-mediated cell differentiation contributes to the increase in H2Bub1. Furthermore, knockout of the Usp7 gene particularly elevated the expression of neuronal differentiation related genes including astryocyte-specific markers and oligodendrocyte-specific markers. In particular, glial lineage cell-specific transcription factors including oligodendrocyte transcription factor 2, glial fibrillary acidic protein, and SRY-box transcription factor 10 was significantly upregulated during neuronal differentiation. Thus, our findings suggest a novel role of Usp7 in gliogenesis in mouse embryonic carcinoma cells.
Keywords: Deubiquitinase, Epigenetics, Gliogenesis, Histone H2A, Histone H2B, Transcriptional regulation
Introduction
The genomic DNA in a eukaryotic nucleus is wrapped around chromatin to form nucleosomes consisting of histone octamers. The five types of histone, i.e., H1, H2A, H2B, H3, and H4, generally undergo several posttranslational modifications such as phosphorylation, methylation, acetylation, sumoylation and ubiquitination (1). The developmental process is also regulated by histone post-translational modifications, where any dysregulation can lead to a wide range of pathologies (2). Among these posttranslational modifications, ubiquitination has a critical role in regulating gene transcription and DNA repair. Generally, a protein substrate tagged by polyubiquitination is guided for degradation through the 26S proteasomal pathway, while a single ubiquitin molecule can bind to a protein substrate, resulting in monoubiquitination, which primarily modifies the molecular properties and subsequently the function of the targeted proteins. Among the histone proteins, H2A and H2B are heavily ubiquitinated and especially likely to undergo monoubiquitination (3).
Monoubiquitination of H2B has a key role in the regulation of gene transcription, replication, and DNA repair processes (4). The monoubiquitination of H2B is usually caused by several E3-ligases, including RNF20 and RNF40 (5, 6). The knockdown of RNF40 impairs human mesenchymal stem cell differentiation towards osteoblasts and adipocytes (6). The H2Bub1 level was reported to be increased during the differentiation of human and mouse embryonic stem cells (ESCs). Moreover, the H2Bub1 increase plays a crucial role during differentiation and is especially important for the effective transcriptional induction of relatively long genes, which is selectively required for optimal differentiation (5). On the other hand, these monoubiquitinated H2Bs can also be reversed by deubiquitinating enzymes (DUBs). Currently, there are a few ubiquitin-specific proteases (USPs) belonging to the DUB protein family that are reported to regulate H2B protein (7-17). Importantly, USP44 was identified as a negative regulator or H2B ubiquitination, where depletion of USP44 during ESCs differentiation leads to an increase in H2B ubiquitination, suggesting its importance during differentiation (5). There are several DUBs that modulate H2B protein and play an important role in pathways associated with normal and cancerous tissue development (18), including oncogenic gene expression and malignancy (19, 20), autophagy, DNA damage response (21, 22), and multiple differentiation pathways (5, 6, 23, 24).
Screening for potential DUBs that regulate stem cell maintenance and differentiation promises to shed light on the molecular mechanism that determines the cell fate of ESCs. Importantly, several DUBs such as USP22, USP7, and USP3 are reported to regulate neuronal differentiation (25-27). However, DUBs that regulate histone H2B ubiquitination and their molecular functions during stem cell differentiation are not well studied. Thus, to find potential DUBs regulating H2B monoubiquitination, we considered H2B-binding DUBs based on previous reports (7-13). A handful of DUBs, including USP22, USP44, USP42, USP49, USP3, and USP15 have already been reported with regard to their role in regulating H2B monoubiquitination linked with chromatin modification (7-13). Although a few DUBs such as USP7, USP12, USP21, and USP37 regulate cell proliferation (28-31) their role in cell differentiation via H2B ubiquitination regulation has not yet been studied.
In this study, we showed that H2B monoubiquitination patterns during mouse neuronal differentiation were regulated by Usp7, resulting in altered expression of a glial lineage cell-specific transcription factor and suggesting a novel role of Usp7 in glial lineage cell differentiation from mouse embryonic carcinoma cells.
Materials and Methods
Nuclease construct and single guide RNA constructs
An hSpCas9 nuclease tagged with a red fluorescent protein (RFP)-expressing plasmid along with a single guide RNA (sgRNA)-expressing plasmid was used for CRISPR/Cas9-mediated gene disruption. The sgRNA target seque-nces of each USP were cloned into the cloning vectors encoding a U6 promoter. Briefly, oligonucleotides containing each target sequence were synthesized (Bioneer) and annealed in vitro using a thermocycler. The vector was digested with BsaI restriction enzyme and ligated with annealed oligonucleotides. The oligonucleotide sequences are listed in Table 1.
Table 1.
List of oligonucleotides used for sgRNA cloning
| sgRNA | 5’-sequence-3’ | Usage |
|---|---|---|
| mUsp7-F | CACCGGAGTGATGGGCACAGCAACG | CRISPR/Cas9-based KO |
| mUsp7-R | AAACCGTTGCTGTGCCCATCACTCC | CRISPR/Cas9-based KO |
| mUsp12-F | CACCGTCGGCATTAGAGAAAGAGAT | CRISPR/Cas9-based KO |
| mUsp12-R | AAACATCTCTTTCTCTAATGCCGAC | CRISPR/Cas9-based KO |
| mUsp21-F | CACCGCTCAAGAAACTGGAGCTGGG | CRISPR/Cas9-based KO |
| mUsp21-R | AAACCCCAGCTCCAGTTTCTTGAGC | CRISPR/Cas9-based KO |
| mUsp37-F | CACCGGTAAGGATGCAGAGGAAATG | CRISPR/Cas9-based KO |
| mUsp37-R | AAACCATTTCCTCTGCATCCTTACC | CRISPR/Cas9-based KO |
sgRNA: single guide RNA, KO: knockout.
Cells and CRISPR/Cas9-mediated knockout
Mouse embryonic carcinoma cell line P19 cells were purchased from the American Type Culture Collection. P19 cells were cultured in α-minimum essential media (α-MEM) supplemented with 7.5% newborn calf serum (NBCS; Hyclone), 2.5% fetal bovine serum (FBS; Young-In Frontier) and incubated in a humid 37℃, 5% CO2 incubator. Mouse USP gene knockout (KO) using a CRISPR/Cas9 system was performed through the transfection of two kinds of vectors: CRISPR/Cas9-RFP-Puro and USP gene-specific sgRNA-expressing vectors via lipofectamine (Invi-trogen). Gene disruption was confirmed by T7E1 assay according to the previously reported protocol (32).
Cell culture and differentiation
P19 cells cultured in α-MEM with NBCS and FBS (P19 growth medium) were sub-cultured at 1:8 ratios every 48 hours to maintain exponential growth. For neuronal differentiation of P19 cells, we used an optimized protocol previously described (33).
The cells were transfected with 1 μg of CRISPR/Cas9 vector and 2 μg of sgRNA vector with the appropriate amount of lipofectamine reagent per well for gene specific KO. We observed 40% to 50% transfection efficiency by monitoring RFP fluorescence tagged with a CRISPR/Cas9 vector. Transfected cells were incubated for 96 hours with retinoic acid (RA) to induce neuronal differentiation. Cells were then seeded to reach 80% to 90% confluence in 6-well plates. Differentiating cells were harvested at appropriate time points and media were changed every 24 hours during the differentiation period.
Antibodies
Anti-Oct3/4 antibody, anti-βIII-tubulin antibody, anti-Usp7 antibody, anti-oligodendrocyte transcription factor 2 (Olig2) antibody, and anti-glial fibrillary acidic protein (GFAP) antibody (Santa Cruz Biotechnology), anti-histone H2B monoubiquitination (H2Bub1; Millipore) antibody. Anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (Santa Cruz Biotechnology) or anti-H2B antibody (Abcam) was used for Western blotting.
Real-time polymerase chain reaction
Cells washed with phosphate buffered saline were harvested and lysed with Trizol reagent (Favorgen). Then, 500 μL of Trizol reagent, 100 μL of chloroform solution isopropanol and 70% ethanol were used to isolate and purify the RNA. Next, complementary DNA (cDNA) synthesis was performed following the manufacturer’s instructions for the SuperScript III First-Strand Synthesis System (Invitrogen) with the purified RNA, and each gene’s messenger RNA (mRNA) was quantified by real-time polymerase chain reaction (PCR). Synthesized cDNA and SYBR Green (Kapa Biosystems) were applied to real-time PCR with specific primers for the genes of interest. The mRNA was quantified by the relative standard curve method, and fold change differences were compared between two or more groups. The relative amounts of the respective mRNAs were normalized against that of GAPDH. The primer details are provided in Table 2. Results represent the average of at least three independent replicates. Standard deviations were calculated by the Mann–Whitney test.
Table 2.
List of primers used for qPCR and ChIP assays in this study
| Primer | 5’-sequence-3’ | Usage |
|---|---|---|
| mUsp7-F | CCTTAGCCCTCCGTGTTTTGT | qPCR |
| mUsp7-R | CCAGTCGTTTTCCTTGTGGAAG | qPCR |
| mAscl1-F | AGATGAGCAAGGTGGAGACG | qPCR |
| mAscl1-R | TGGAGTAGTTGGGGGAGATG | qPCR |
| mNgn1-F | ATGCCTGCCCCTTTGGAGAC | qPCR |
| mNgn1-R | TGCATGCGGTTGCGCTCGC | qPCR |
| mNeuroD1-F | CATGCCCCCGCATCTGCCAA | qPCR |
| mNeuroD1-R | GCCATTGATGCTGAGCGGCG | qPCR |
| mMap2-F | AAACAGGCGAAGGATAAAGT | qPCR |
| mMap2-R | TGTTGTCAGTTGATCCGATT | qPCR |
| mβIII-tubulin-F | AAGGTAGCCGTGTGTGACATC | qPCR |
| mβIII-tubulin-R | ACCAGGTCATTCATGTTGCTC | qPCR |
| mOlig-F | CACAGGAGGGACTGTGTCCT | qPCR |
| mOlig-R | GGTGCTGGAGGAAGATGACT | qPCR |
| mSox10-F | GTCAACGGTGCCAGCAAGAG | qPCR |
| mSox10-R | TCAATGAAGGGGCGCTTGTC | qPCR |
| mO4-F | CATCCGTCACAACCTGTCCT | qPCR |
| mO4-R | GGCTTCTTCTTGGGGCCTTT | qPCR |
| mGFAP-F | AGCGGCAAATGCGCGAACAG | qPCR |
| mGFAP-R | GTGCTTTTGCCCCCTCGGAT | qPCR |
| mAldh1L1-F | GAGCCACCTATGAGGGCATT | qPCR |
| mAldh1L1-R | AAGAGGATGAGTCCCGCTTT | qPCR |
| mS100β-F | CTGATCGCCTACACCCTTCC | qPCR |
| mS100β-R | AAAGGAGAAGTCTGCCGAGC | qPCR |
| mActin-β-F | GGACCTGACAGACTACCTCA | qPCR |
| mActin-β-R | GTTGCCAATAGTGATGACCT | qPCR |
| mOlig2-F | TGCTTATTACAGACCGAGCCAAC | ChIP |
| mOlig2-R | CTAAATCCTAGCCACTTTGGAGAAGT | ChIP |
| mGFAP-F | CGAGTGACTCACCTTGGCATAG | ChIP |
| mGFAP-R | CCAGGATGCCAGGATGTCAG | ChIP |
| mPax6-F | CACCAGACTCACCTGACACC | ChIP |
| mPax6-R | GAACACACAGGTTGCACGTC | ChIP |
| mActin-β-F | CCGTAAAGACCTCTATGCCAACAC | ChIP |
| mActin-β-R | GCTAGGAGCCAGAGCAGTAATCTC | ChIP |
qPCR: quantitative polymerase chain reaction, ChIP: chromatin immunoprecipitation.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were conducted as previously described (34) using a Pierce Agarose ChIP kit (#26156; Thermo Scientific). Anti-H2Bub1 and anti-H2B were added as primary antibodies and normal rabbit immunoglobin G was used as a negative control. Immunoprecipitated DNA fragments were analyzed by quantitative PCR (qPCR). Gene-specific primers were added to each sample, and the H2B gene primer was used as a control. The primer details are provided in Table 2. All qPCR reactions were performed in triplicate.
Results
Identification of potential DUB candidates regulating H2B monoubiquitination in P19 cells
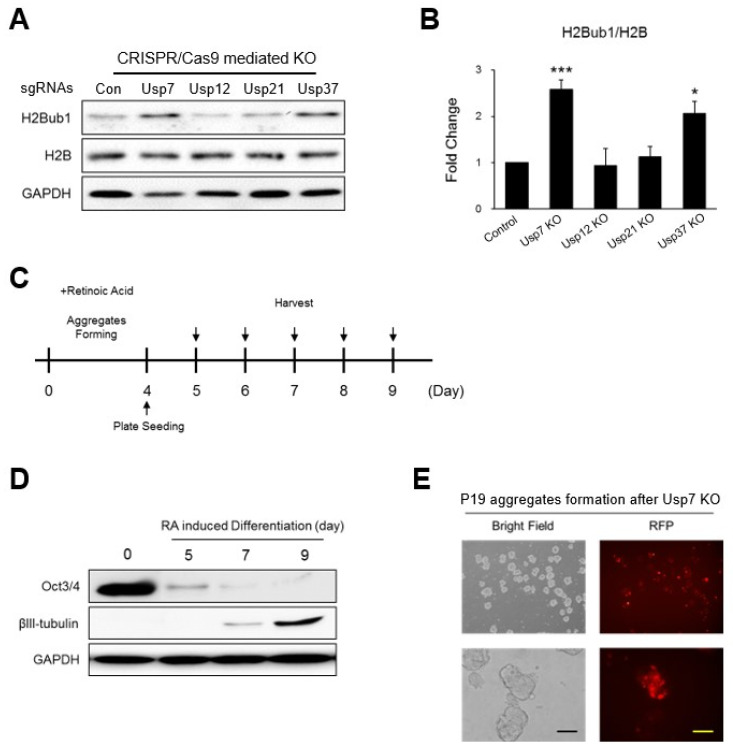
To find potential DUBs regulating H2B monoubiquitination we used CRISPR/Cas9 system to knock down individual DUBs in P19 cells. For this purpose, sgRNAs specifically targeting the mouse Usp7, Usp12, Usp21, and Usp37 genes were designed and transfected along with a CRISPR/Cas9 vector to induce gene disruption. Global H2Bub1 levels after the KO of each USP were detected by Western blot. The KO of Usp7 and Usp37 resulted in increased levels of global H2Bub1 in the P19 cells, but less or no significant changes were observed in H2Bub1 expression when USP12 and USP21 were knocked out (Fig. 1A, 1B). Thus, we hypothesized that Usp7 and Usp37 might critically regulate the H2B monoubiquitination level of undifferentiated P19 cells and thus might have a role in neuronal differentiation.
Fig. 1.
Potential ubiquitin-specific protease (USP) candidates regulating H2B monoubiquitination. (A) The effect of CRISPR/Cas9-mediated knockout (KO) of each USP candidate (Usp7, Usp12, Usp21, and Usp37) on the expression level of histone H2B monoubiquitination (H2Bub1) was estimated by Western blotting. (B) The expression of H2Bub1 was graphically represented. *p<0.05, ***p<0.001 by paired t-test. (C) Schematic representation of retinoic acid (RA)-induced P19 cell neuronal differentiation schedule. P19 cells formed aggregates for four days with 10 μM of RA. Aggregates were collected and seeded in cell culture plates on day 4 and harvested at the indicated time points. (D) RA-induced neuronal differentiation of P19 cells at different days. Samples were collected at the indicated time points and the expressions of Oct3/4 and βIII-tubulin protein were analyzed by Western blotting. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was analyzed as a loading control. (E) Fluorescence microscopy of P19 cell aggregates after transfection of Cas9 tagged with red fluorescent protein (RFP) along with single guide RNA (sgRNA) targeting Usp7. Scale bar=100 μm.
For neuronal differentiation of P19 cells, we used an optimized protocol (33), and its efficiency was confirmed by neuronal differentiation marker proteins. P19 cells were allowed to form aggregates for four days by treating them with 10 μM of RA. Aggregates were trypsinized, and single cells were plated onto a 6-well plate. Cells were collected at every day until day 9 in order to avoid a mixed population increasing after this time point (Fig. 1C). Oct4 and βIII-tubulin were validated as neuronal differentiation markers by Western blot analysis. The Oct3/4 protein showing abundant expression levels in undifferentiated P19 cells gradually decreased during differentiation and had almost disappeared by day 9. In contrast, the common neuronal differentiation marker βIII-tubulin, which was not expressed in undifferentiated P19 cells, showed a constant increase during differentiation and became prominent at day 9 (Fig. 1D). Thus, we used this standardized RA-mediated neuronal differentiation protocol to analyze the effect of knocking out Usp7 and Usp37 during neuronal differentiation.
Usp7 and Usp37 KO-mediated regulation of H2Bub1 expression during neuronal differentiation
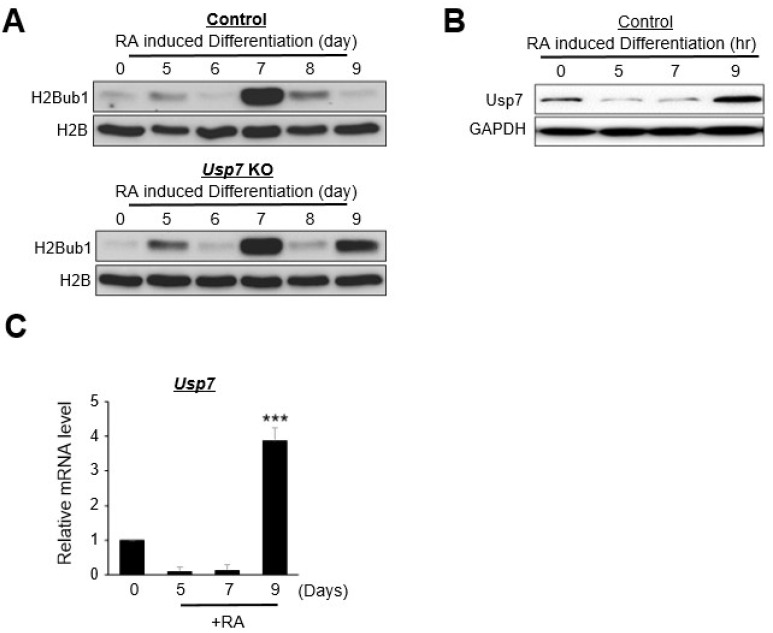
Based on the effects of Usp7 and Usp37 KO on the upregulation of H2B monoubiquitination in undifferentiated P19 cells, we were interested to analyze their roles in the RA-mediated neuronal differentiation in P19 cells. Usp7 or Usp37 was knocked out with the CRISPR/Cas9 system, as mentioned above, and the resulting cells were induced to neuronal differentiation with RA. Usp7 or Usp37 KO P19 cells were allowed to aggregate (Fig. 1E), and then the cells were differentiated for 9 days, including the aggregate formation period. We excluded the cells after nine days to reduce the effects of non-neuronal cell differentiation populations (33). Cells were collected every day, and their global H2B monoubiquitination levels were examined through Western blot. Control and Usp7 KO cells showed similar patterns of H2B monoubiquitination levels until day 6. Surprisingly, in the control group, the H2B monoubiquitination level was dramatically increased on day 7, decreased gradually on day 8 and was very low on day 9 (Fig. 2A, upper panel), while the Usp7 KO group also showed abundant expression of H2B monoubiquitination at day 7, but unlike the control group, the expression of H2B monoubiquitination was again prominent at day 9 (Fig. 2A, lower panel). We hypothesized that the regaining of H2B monoubiquitination expression on day 9 might be due to the Usp7 gene KO. To confirm this hypothesis, we analyzed the expression of Usp7 in both the control group and the Usp7 KO group at different days during RA-induced differentiation. Interestingly, the expression level of Usp7 remained moderate in the undifferentiated P19 cells, gradually decreased during differentiation and finally regained its expression level significantly by day 9 (Fig. 2B). The mRNA expression of Usp7 was also found to be significantly high on day 9 (Fig. 2C). Our results provide evidence that the significant recovery of the expression level of Usp7 on day 9 is associated with the reduced expression level of H2B monoubiquitination at day 9 during neuronal differentiation (Fig. 2A, upper panel). In contrast, the KO of Usp7 in P19 cells results in the significant increase in the expression level of H2B monoubiquitination by day 9 during neuronal differentiation (Fig. 2A, lower panel). Thus, our data signify that the restoration of H2B monoubiquitination expression by day 9 during neuronal differentiation is due to the loss of Usp7 deubiquitinating activity.
Fig. 2.
Usp7 knockout (KO)-mediated negative regulation of H2B ubiquitination during retinoic acid (RA)-induced neuronal differentiation. (A) The histone H2B monoubiquitination (H2Bub1) protein expression levels in the control and Usp7 KO P19 cells during RA-induced neuronal differentiation analyzed by Western blot. Differentiated samples were collected on the indicated days. The protein expression of H2Bub1 and total H2B were analyzed by Western blot. (B) RA-induced neuronal differentiation in P19 cells. Differentiated samples were collected on the indicated days and the protein expression of Usp7 were analyzed by Western blot. (C) The messenger RNA (mRNA) expression of Usp7 in control P19 cells was estimated by real-time polymerase chain reaction during RA-induced differentiation. Error bars were derived from three independent experiments. N=3. ***p<0.001.
KO of Usp7 regulates mouse neurogenesis by elevating the expression of glial progenitor lineage transcription factors
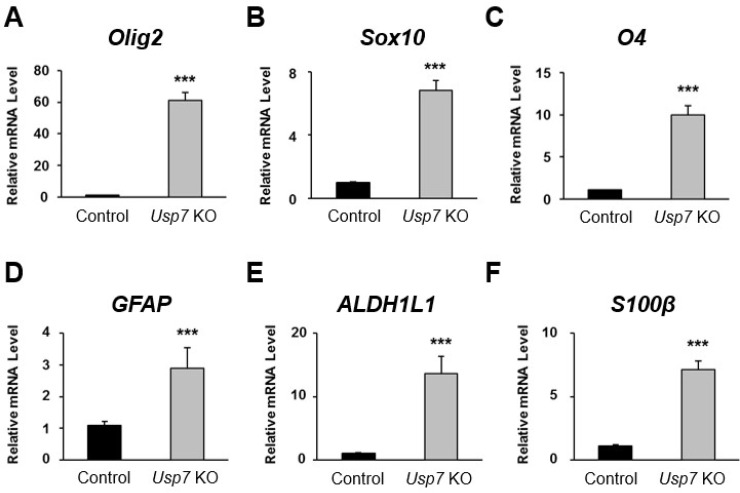
We further investigated whether the influence of Usp7 KO on the expression level of H2B monoubiquitination on day 9 might lead to any altered expression levels of genes responsible for neuronal differentiation. For this purpose, several genes involved in the neuronal differentiation pathway were selected for analysis. Ascl1, Ngn1, NeuroD1, Map2, and βIII-tubulin were selected as neuronal or neuron precursor cell markers. GFAP, ALDH1L1, and S100β were selected as astrocyte-specific markers. Olig2, Sox10, and O4 were selected as oligodendrocyte-specific markers.
The RNA was isolated at differentiation day 9 from both control and Usp7 KO P19 cells, and the mRNA expression level of each gene was validated through real-time PCR. Among the oligodendrocyte-specific marker genes, Olig2 showed the most elevated expression level of mRNA of about 60-fold higher in the Usp7 KO group than in the control group (Fig. 3A). Likewise, the mRNA levels of Sox10 and O4 also showed about 6- and 10-fold increases, respectively, in the Usp7 KO group when compared to control group (Fig. 3B, 3C). Among the astrocyte-specific marker genes, GFAP, ALDH1L1, and S100β showed about 2-, 12-, and 9-fold increases in mRNA expression levels, respec-tively, in the Usp7 KO group when compared to the control group (Fig. 3D-3F).
Fig. 3.
The expression of transcription factors related to mouse neurogenesis upon Usp7 knockout (KO). (A-C) Relative expression levels of oligodendrocyte-specific transcription factors (A) Olig2, (B) Sox10, and (C) O4 messe-nger RNA (mRNA) were analyzed by real-time polymerase chain reaction (PCR). Samples were collected on day 9 after differentiation from control and Usp7 KO P19 cells. Results were normalized with the level of GAPDH. (D-F) Relative expression of astrocyte-specific transcription factor (D) GFAP, (E) ALDH1L1, and (F) S100β were analyzed by real-time PCR. Error bars were derived from three independent experiments. N=3. ***p<0.001.
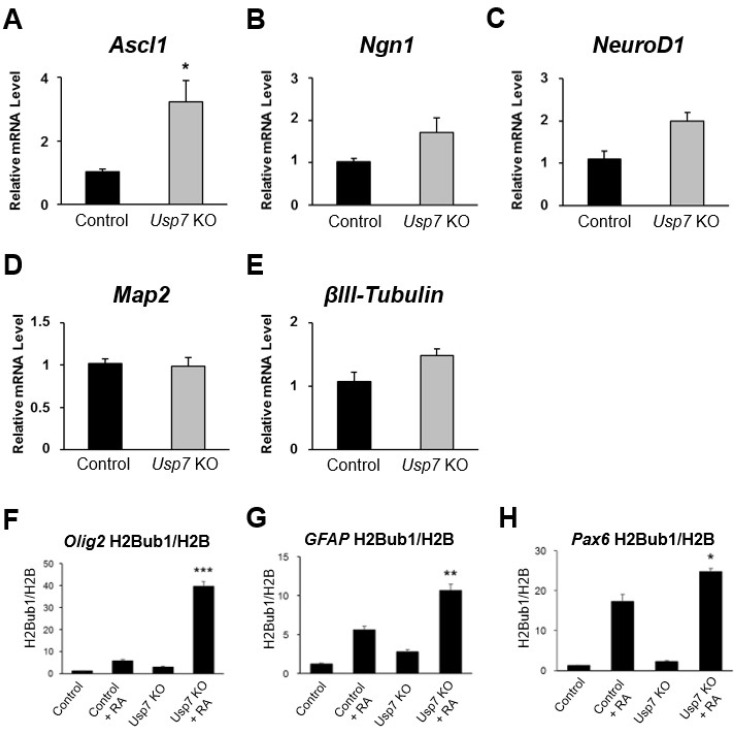
We next checked the mRNA expression levels of genes such as Ascl1, Ngn1, NeuroD1, Map2, and βIII-tubulin, which are involved in neuron-like cell differentiation. However, except for Ascl1 no other genes showed significant changes at the mRNA level of genes regulating neuron-like cell differentiation between the Usp7 KO cells and the control cells (Fig. 4A-4E).
Fig. 4.
Knockout (KO) of Usp7 alters histone H2B monoubiquitination (H2Bub1) of genes involved in neuronal differentiation. (A-C) Relative expression levels of neuronal lineage cell-specific transcription factors (A) Ascl1, (B) Ngn1, (C) NeuroD1, and (D, E) marker genes (D) Map2 and (E) βIII-tubulin messenger RNA (mRNA) were analyzed with real-time polymerase chain reaction (PCR). Samples were collected on day 9 after differentiation from control and Usp7 KO P19 cells. Results were normalized with the level of GAPDH. (F-H) Samples were collected on day 9 after differentiation from control and Usp7 KO P19 cells and subjected to chromatin immunoprecipitation analysis with antibodies specific for H2Bub1 and H2B. Immunoprecipitated DNA was quantified by real-time PCR with primers specific for the promoter regions of (F) Olig2, (G) GFAP, and (H) Pax6. Error bars were derived from three independent experiments. N=3. RA: retinoic acid. *p<0.05, **p<0.01, ***p<0.001.
KO of Usp7 alters H2Bub1 of genes involved in neuronal differentiation
To demonstrate the role of Usp7 in neuronal differentiation, we conducted a ChIP assay to investigate whether it showed an altered pattern of monoubiquitination of genes involved in neuronal differentiation between the control group and the Usp7 KO cells. Olig2, GFAP, and Pax6 genes were tested as specific markers of oligodendrocytes, astrocytes and neuronal precursor cells, respectively. Each differentiated cell at day 9 was fixed, and its chromatin fragments were collected and processed for ChIP assays. DNA fragments were analyzed with gene-specific primers through real-time PCR. All of the results were normalized with H2B expression level. Our results showed that the monoubiquitination levels of Olig2 increased up to 40-fold in differentiated Usp7 KO cells, while only a 5-fold increase was observed in the control cells (Fig. 4F). Similarly, GFAP showed monoubiquitination levels that increased up to 10-fold in differentiated Usp7 KO cells, while only a 5-fold increase was observed in the control cells (Fig. 4G). Even Pax6 also showed increased monoubiquitination levels in differentiated Usp7 KO cells when compared to control cells (Fig. 4H). However, the fold change of the Pax6 gene in the Usp7 KO cells when compared with the control cells was lower than the fold changes observed in the Olig2 and GFAP genes. Thus, it is evident that Usp7 alters the H2Bub1 of genes involved in glial lineage cell differentiation.
Discussion
Histone ubiquitination is a reversible posttranslational modification that is responsible for the regulation of genomic DNA stability and transcription in eukaryotic cells. The enzymes that are involved in the regulation of both histone ubiquitination and deubiquitination play an important role in gene transcription and DNA repair. Histone ubiquitination is also a key regulator of stem cell differentiation, where global upregulation of H2Ub1 levels is linked with the onset of differentiation in ESCs and ESC-like cells (5). In this study, we wished to identify DUBs that modulate histone ubiquitination and their role during stem cell differentiation.
We showed that the Usp7 expression level was moderate in the early stage of cell differentiation and that it accumulated further by differentiation day 9 (Fig. 2C). Thus, we predicted that the increased expression of Usp7 on differentiation day 9 was associated with lower expression of H2Bub1 on differentiation day 9 in normal P19 cells. We confirmed our hypothesis by knocking out the Usp7 gene in P19 cells and allowed for differentiation. As expected, the results showed exactly the opposite behavior in the expression of H2Bub1 on differentiation day 9 (Fig. 2B). One of the reasons for the fluctuation in the expression of H2Bub1 protein during cell differentiation could be that different DUBs might be targeting the H2Bub1 protein at different stages of cellular differentiation. In our study, we confirmed that the significant increase in the expression level of H2Bub1 on differentiation day 9 was due to the loss of Usp7’s deubiquitinating function. Different USPs might switch on their deubiquitinating functions at different stages of cellular differentiation by regulating their protein turnover. The sudden and dramatic increase in the expression level of H2Bub1 protein on differentiation day 7 were particularly interesting. This observation allows us to predict that there could be several other DUBs that might be regulating H2Bub1 protein expression on differentiation day 7. Thus, our future studies focusing on screening for potential DUBs that can regulate stem cell differentiation at different days might provide some clue towards the molecular mechanism that determines the fate of ESCs during neuronal differentiation.
There are reports on USP7 that regulate proteins implicated in nervous system disorders, neurodegenerative illnesses, and brain tumors (35). For an instance, USP7 interacts with ATXN1, which is linked to spinocerebellar ataxia type 1 (SCA1), an autosomal dominant neurodege-nerative disease with multiple neurological abnormalities (36). The stability of p53 mediated by USP7 is essential for brain development, where KO of USP7 causes neonatal lethality, hypoplasia, and deficiencies in neuronal development (37). Additionally, USP7 and LSD1 expression is elevated in brain tumors such as gliomas. USP7 deubiquitinates LSD1 and promotes cell proliferation and invasion of glioblastoma cells, suggesting that USP7 is a prognostic marker and therapeutic target for gliomas and other neuronal disorders (38).
A number of DUBs, such as USP3, USP8, USP7, USP22, and USP51 are associated with the neuronal differentiation. Neuronal cell morphology, survival, functions, and its synaptic plasticity are all regulated by the tropomyosin-related kinase (Trk) family of receptor tyrosine kinases. USP8 inhibits neuronal differentiation in PC12 cells through its nerve growth factor-dependent manner by interacting with and deubiquitinating the TrkA receptor (39). Another study demonstrated that USP27x, Usp22, and Usp51 deubiquitinate and stabilize Hes1 protein, a transcriptional repressor necessary for the maintenance of neural stem/progenitor cells. Moreover, depletion of Usp22 significantly reduced the half-life of the Hes1 protein and affected Hes1 oscillation, which in turn enhanced neuronal differentiation in the developing mouse brain (25). However, Usp27x and Usp51 did not show a significant effect on Hes1 half-life and also showed lower expression in mouse developing brains when compared with Usp22 expression, suggesting Usp22 is the main potential DUB for Hes1 during mouse brain development (25).
REST is a transcriptional factor that regulates and maintains neural stem and progenitor cells. Recently, we performed genome-wide screening for DUBs regulating REST protein and identified USP3 as a bonafide DUB interacting, stabilizing, and deubiquitinating REST protein (27). Depletion of USP3 resulted in a significant reduction in REST protein levels, which impacted RA-induced neuronal differentiation in neuroblastoma cells. USP3 depleted cells showed decreased neural stem cell marker NESTIN-positive cells, suggesting that USP3 plays a key role in maintaining REST protein abundance to determine neurobla-stoma differentiation (27). USP7 interacts with and extends the half-life of the REST protein through its deubiquitinating activity (26). Silencing of USP7 induces neuronal differentiation, while overexpression stabilizes REST protein and promotes neuronal progenitor stem cell maintenance (26). This study suggested that Usp7 modulates histone ubiquitination during stem cell differentiation. Knockdown of USP7 resulted in the elevated expression of oligodendrocyte-specific marker genes such as Olig2, Sox10, and O4 (Fig. 3, 4). Additionally, depletion of Usp7 upregulated astrocyte-specific markers such as GFAP, ALDH1L1, and S100β (Fig. 3, 4), suggesting that Usp7 plays an important role in the modification of histone ubiquitination and thereby regulate gliogenesis in mouse ESCs.
Footnotes
Potential Conflict of Interest
There is no potential conflict of interest to declare.
Authors’ Contribution
Conceptualization: DHK, SR. Data curation: DHK, SLK, VS. Formal analysis: DHK, VS, SR. Funding acquisition: SR. Investigation: DHK, SLK. Methodology: DHK. Project administration: SR. Resources: SR. Software: SR. Supervision: VS, SR. Validation: DHK, SLK, VS, SR. Visualization: DHK. Writing – original draft: DHK. Writing – review and editing: VS, SR.
Funding
This research was supported by the National Research Foundation of Korea grant (2021M3A9H3015390, RS-2023-00279214 and RS-2024-00341469) and the Korean Fund for Regenerative Medicine grant funded by the Korean government (the Ministry of Science and ICT, the Ministry of Health & Welfare) (22A0304L1-01).
References
- 1.Van HT, Santos MA. Histone modifications and the DNA double-strand break response. Cell Cycle. 2018;17:2399–2410. doi: 10.1080/15384101.2018.1542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik SR, Smith E, Shilatifard A. Covalent modifica-tions of histones during development and disease pathoge-nesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava A, McGrath B, Bielas SL. Histone H2A monoubiquitination in neurodevelopmental disorders. Trends Genet. 2017;33:566–578. doi: 10.1016/j.tig.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/S1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs G, Shema E, Vesterman R, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B mono-ubiquitylation. Mol Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpiuk O, Najafova Z, Kramer F, et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Joo HY, Jones A, Yang C, et al. Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J Biol Chem. 2011;286:7190–7201. doi: 10.1074/jbc.M110.158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emre NC, Ingvarsdottir K, Wyce A, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XY, Varthi M, Sykes SM, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicassio F, Corrado N, Vissers JH, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum Pathol. 2015;46:1006–1014. doi: 10.1016/j.humpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Lancini C, van den Berk PC, Vissers JH, et al. Tight regulation of ubiquitin-mediated DNA damage response by USP3 preserves the functional integrity of hematopoietic stem cells. J Exp Med. 2014;211:1759–1777. doi: 10.1084/jem.20131436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hock AK, Vigneron AM, Vousden KH. Ubiquitin-specific peptidase 42 (USP42) functions to deubiquitylate histones and regulate transcriptional activity. J Biol Chem. 2014;289:34862–34870. doi: 10.1074/jbc.M114.589267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long L, Thelen JP, Furgason M, et al. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J Biol Chem. 2014;289:8916–8930. doi: 10.1074/jbc.M114.551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Jones A, Joo HY, et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA spli-cing. Genes Dev. 2013;27:1581–1595. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeVine T, Sears RC, Dai MS. The ubiquitin-specific protease USP36 is a conserved histone H2B deubiquitinase. Biochem Biophys Res Commun. 2018;495:2363–2368. doi: 10.1016/j.bbrc.2017.12.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole AJ, Clifton-Bligh R, Marsh DJ. Histone H2B monoubiquitination: roles to play in human malignancy. Endocr Relat Cancer. 2015;22:T19–T33. doi: 10.1530/ERC-14-0185. [DOI] [PubMed] [Google Scholar]
- 19.Prenzel T, Begus-Nahrmann Y, Kramer F, et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 2011;71:5739–5753. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- 20.Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet. 2012;21:559–568. doi: 10.1093/hmg/ddr490. [DOI] [PubMed] [Google Scholar]
- 21.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcri-bed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 22.Kari V, Shchebet A, Neumann H, Johnsen SA. The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle. 2011;10:3495–3504. doi: 10.4161/cc.10.20.17769. [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Jones AE, Yang W, et al. The histone H2A deubiquitinase Usp16 regulates hematopoiesis and hematopoietic stem cell function. Proc Natl Acad Sci U S A. 2016;113:E51–E60. doi: 10.1073/pnas.1517041113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Q, Xia W, Li W, Jiao J. RNF20 controls astrocytic differentiation through epigenetic regulation of STAT3 in the developing brain. Cell Death Differ. 2018;25:294–306. doi: 10.1038/cdd.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi T, Iwamoto Y, Takashima K, et al. Deubiquiti-nating enzymes regulate Hes1 stability and neuronal differ-entiation. FEBS J. 2015;282:2411–2423. doi: 10.1111/febs.13290. [DOI] [PubMed] [Google Scholar]
- 26.Huang Z, Wu Q, Guryanova OA, et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karapurkar JK, Kim MS, Colaco JC, et al. CRISPR/Cas9-based genome-wide screening of the deubiquitinase subfamily identifies USP3 as a protein stabilizer of REST blocking neuronal differentiation and promotes neuroblas-toma tumorigenesis. J Exp Clin Cancer Res. 2023;42:121. doi: 10.1186/s13046-023-02694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya S, Chakraborty D, Basu M, Ghosh MK. Emer-ging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct Target Ther. 2018;3:17. doi: 10.1038/s41392-018-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang LJ, Li Y, Liu YL, Wang JM, Liu DW, Tian QB. USP12 regulates cell cycle progression by involving c-Myc, cyclin D2 and BMI-1. Gene. 2016;578:92–99. doi: 10.1016/j.gene.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Zhou B, Chen D. USP21 promotes cell prolifera-tion and metastasis through suppressing EZH2 ubiquitination in bladder carcinoma. Onco Targets Ther. 2017;10:681–689. doi: 10.2147/OTT.S124795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das CM, Taylor P, Gireud M, et al. The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation. Oncogene. 2016;35:6153–6154. doi: 10.1038/onc.2016.141. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishna S, Cho SW, Kim S, et al. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun. 2014;5:3378. doi: 10.1038/ncomms4378. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama Y, Wada A, Inoue R, et al. A rapid and efficient method for neuronal induction of the P19 embryonic carcinoma cell line. J Neurosci Methods. 2014;227:100–106. doi: 10.1016/j.jneumeth.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/S0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 35.Das S, Ramakrishna S, Kim KS. Critical roles of deubiquitinating enzymes in the nervous system and neurodegene-rative disorders. Mol Cells. 2020;43:203–214. doi: 10.14348/molcells.2020.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong S, Kim SJ, Ka S, Choi I, Kang S. USP7, a ubiquitin-specific protease, interacts with ataxin-1, the SCA1 gene product. Mol Cell Neurosci. 2002;20:298–306. doi: 10.1006/mcne.2002.1103. [DOI] [PubMed] [Google Scholar]
- 37.Kon N, Zhong J, Kobayashi Y, et al. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. 2011;18:1366–1375. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi L, Cui Y, Xu Q, Jiang Y. Stabilization of LSD1 by deubiquitinating enzyme USP7 promotes glioblastoma cell tumorigenesis and metastasis through suppression of the p53 signaling pathway. Oncol Rep. 2016;36:2935–2945. doi: 10.3892/or.2016.5099. [DOI] [PubMed] [Google Scholar]
- 39.Ceriani M, Amigoni L, D'Aloia A, Berruti G, Martegani E. The deubiquitinating enzyme UBPy/USP8 interacts with TrkA and inhibits neuronal differentiation in PC12 cells. Exp Cell Res. 2015;333:49–59. doi: 10.1016/j.yexcr.2015.01.019. [DOI] [PubMed] [Google Scholar]