Abstract
The emergence and spread of antibiotic resistance in microorganisms pose significant challenges to public health, especially in hospitals. This study investigated the existence or occurrence of bacterial bioaerosol and their antibiotic resistance patterns in particulate matter (PM) collected from hospitals in the greater Dhaka region, Bangladesh. The real-time particulate matter concentrations (PM1.0, PM2.5, and PM10) were measured in four hospitals and two ambient locations. Air sampling was conducted using a filter-based method with a low-volume air sampler, while AEROCET-531 S (USA) was employed to monitor particulate matter concentrations. Bacterial bioaerosol concentration was determined using a culture-based method, and eleven bacterial species, including nine individual species, i.e., Staphylococcus aureus, Pseudomonas aeruginosa, P. stutzeri, Bacillus cereus, Acinetobacter schindleri, Proteus vulgaris, B. subtilis, Escherichia coli, and B. aerius, were isolated. Antibiotic susceptibility testing was conducted using the Kirby-Bauer disk diffusion method with 21 antibiotics. Bacterial isolates were detected using partial sequencing of the 16 S rRNA gene. Bioaerosol concentration ranged from 194.65 ± 22.48 CFU/m3 to 948.39 ± 84.14 CFU/m3, showing significant correlations with PM1.0 and PM2.5 concentrations (R2 = 0.80 and 0.85, respectively). All bacterial isolates collected from the hospitals exhibited resistance against four or more antibiotics, indicating multidrug resistance (MDR). Notably, the bacterial isolates displayed the highest resistance rate against ampicillin (90.90%), azithromycin (81.81%), erythromycin (81.81%), cefixime (81.81%), and cotrimoxazole (54.54%), among the tested antibiotics. Except B. aerius, all other bacterial isolates were associated with hospital-acquired infections (HAIs). These findings highlight the high rates of antibiotic resistance, underscoring the pressing requirement for infection control measures and continuous surveillance strategies in hospital settings. These findings emphasize the necessity for global hospital infection control strategies focusing airborne multidrug-resistant microorganisms.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81376-0.
Keywords: Bioaerosols, Particulate matter, Antibiotic resistance, Hospital-acquired infections, Nosocomial infections, Air Quality
Subject terms: Environmental chemistry, Environmental impact, Environmental sciences, Chemistry
Introduction
Quantitative studies of airborne particulate matter (PM) have focused a great deal of attention on bioaerosols in recent years, with an increasing recognition of their massive significance and impact on health, climate, and environmental pollution concerns1. Bioaerosols contain living as well as non-living elements, such as pollens, bacteria, fungi, and viruses2. Therefore, they can consist of both pathogenic and non-pathogenic microbes, whether dead or alive3. According to Humbal et al. (2019)4, these bioaerosols comprise solids and semi-solids containing biotic and abiotic components whose size ranges from 0.001 nm to 100 μm. Sneezing, coughing, talking, washing, flushing the toilet, etc., can all cause biological particulate matter to become airborne5.
In hospitals, the scenario regarding bioaerosols is more complex compared to other commercial and residential buildings. The airborne form of bacteria can cause infections in patients and hospital staff, with heightened vulnerability observed in operating theaters, intensive care units (ICUs), and delivery rooms6. Hospitals commonly use and expose pathogens to antibiotics, resulting in high levels of antibiotic resistance7. Antibiotic-resistant bacteria (ARB) create significant challenges in treating infectious diseases8. A variety of chemicals produced by various items, such as antibacterial agents, sterilizers, laboratory materials, various medical operations, and therapeutic management, as well as medical wastes, might be significant indoor sources of pollution in hospitals9. The air pollutants dictated by zonal origins and long-range transmission (prevailing wind direction) may pertain to indoor air quality (IAQ), but localized air pollution sources, such as hospital parking lots’ emissions, road traffic etc. may also contribute10. Furthermore, outdoor and interior construction, humidifiers, contaminated carpets, and cooling towers can all contribute to hospital pollution11.
In particular, PM2.5 and PM10, which are significant sources of airborne microorganisms, have been demonstrated to correlate with the concentration of bacterial colonies12. Air movements can carry these airborne bacteria upward, as they are tiny enough to survive for a long time in the environment13. Bacteria may be eliminated from the air by procedures like dry deposition and/or wet deposition (being removed by precipitation, i.e. rain or snow)14. Depending on the amount of haze and the severity of the pollution, the influence of temperature on bioaerosols might differ. For instance, in the winter, hub bacteria were more prevalent in PM2.5 on days with high pollution levels than on days with lower levels15. However, during foggy days in the fall and early winter in Beijing, the airborne bacterial load and community structure were mostly influenced by relative humidity and particulate matter concentrations16. These findings emphasize the complex connections among bioaerosols, temperature, humidity, and particulate matter in different environmental conditions. Bioaerosols have been linked to chronic health difficulties, respiratory disorders, and infectious infections17. Furthermore, there is a remarkable correlation between the Air Quality Index (AQI) and the quantity of airborne bacteria and fungi. The quantity of airborne fungus and the number of airborne bacteria steadily rise when the AQI hits 20018. The interactions between PM and AQI, and subsequently, the types of bacteria present, depend significantly on the chemical and microbiological compositions19. The composition and variety of bioaerosols have been reported to significantly change at high or extreme pollution levels15.
Exposure to biological agents, for example, fungi, bacteria, parasites, and viruses, causes infectious diseases, which can spread through indirect contact, such as coughing or sneezing, or direct contact, such as biting, touching, or licking20. Mycobacterium tuberculosis, which infected individuals expel into the environment when they cough, sneeze, or talk, causes tuberculosis (TB) through air contamination21. Bacillus anthracis causes anthrax, which individuals can contract through ingestion, inhalation, or skin contact with infected animals22. Recognizing the potential health risks posed by bioaerosol in occupational settings is crucial, and industries where bioaerosol exposure is a concern should take appropriate measures to mitigate exposure and protect worker health. Bioaerosol components may threaten hospital facilities’ indoor air quality (IAQ). The microorganisms found in bioaerosols have the potential to affect patients and healthcare workers by increasing the prevalence of occupational illnesses and hospital-associated infections. The objectives of this research work are to estimate the concentrations of bacterial bioaerosols and particulate matter from different hospitals and two ambient locations in the greater Dhaka region. In addition, the identification of the bacterial isolates obtained from the sampling sites was identified to get information about nosocomial infection and other health impacts in the hospital environments and to evaluate the antibiotic resistance of the identified bacterial isolates.
Results
Particulate matter (PM) concentrations
The average and standard deviation of particulate matter concentrations (PM1.0, PM2.5, and PM10) for all sampling locations are given in Supplementary Table S1 and Fig. 1. Differences in average PM1.0, PM2.5 and PM10 concentrations were found to be statistically significant (p < 0.05) across all study locations.
Fig. 1.
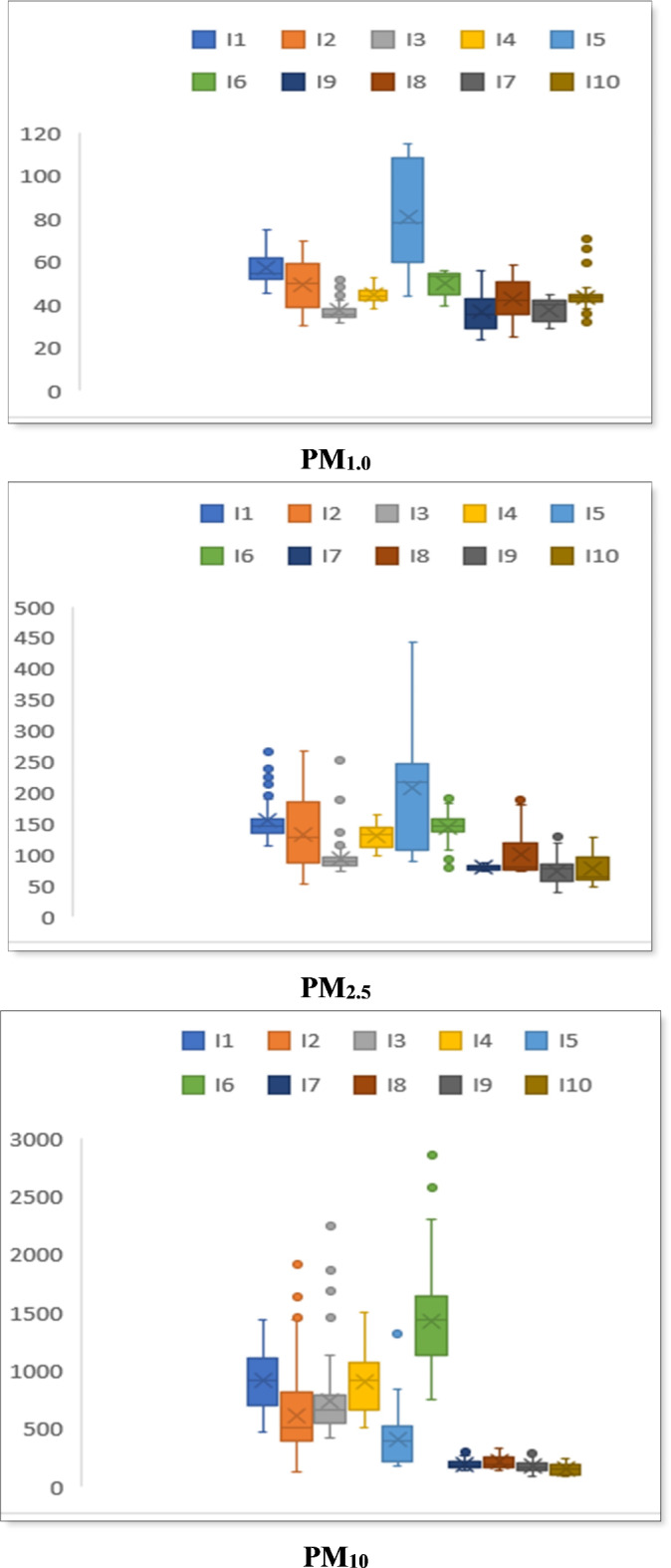
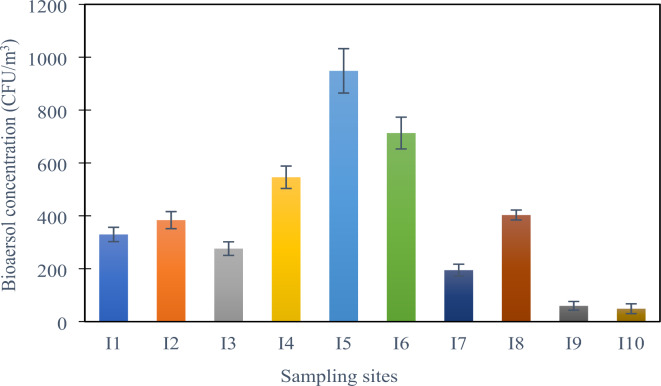
Particulate matter (PM) and bioaerosol concentrations at different sampling sites (all units are in µg/m3 except bioaerosol concentration which is in CFU/m3). Here, I1 & I2 = Bangabandhu Sheikh Mujib Medical University Hospital (BSMMUH), I3 & I4 = Khwaja Badrudduja Modern Hospital (KBMH), I5 & I6 = Dhaka Medical College Hospital (DMCH), I7 & I8 = Monno Medical College Hospital (MMCH), I9 = Green Model Town (GMT), I10 = Mukarram Hussain Khundker Bhaban (MHKB).
The concentration of PM1.0 was found to be much higher in all the hospital sites. The greatest values of PM1.0 and PM2.5 were found at sampling point I5 of Dhaka Medical College Hospital (DMCH) (80.46 ± 11.32 µg/m3, 220.60 ± 16.52 µg/m3), while the highest PM10 concentration was 1452.21 ± 189.78 µg/m3 for sampling point I6 of DMCH. The PM10 concentrations were highest near 1:00 p.m. at BSMMUH and KBMH (Supplementary Fig. S1) and it might be due to the increasing activities of the people coming to the hospitals and also due to the temperature elevation28.
Bioaerosol concentration at the sampling locations
The concentration of bacterial bioaerosols at various sample locations of the hospitals was considerably varied based on the data acquired (Supplementary Table S1). The hospitals considered include a mix of public and private institutions with different patient volumes, infrastructure, and air handling systems, providing a broad view of hospital indoor air conditions in greater Dhaka. We explored various environmental parameters affecting bioaerosol load and resistance. Following an 8-hour sampling period, the location I5 of DMCH exhibited the highest concentration of culturable total aerobic bacterial colonies in PM (948.39 ± 84.14 CFU/m3). The total concentration range of bacterial colonies across the hospital sites varied from 194.65 ± 22.48 CFU/m3 to 948.39 ± 84.14 CFU/m3.
The lowest mean concentrations of culturable bacterial bioaerosol were 59.37 ± 16.51 and 48.65 ± 18.47 CFU/m3 found at I9 and I10 or the control sites, respectively. Figure 2 shows the bioaerosol concentration with standard deviation at different sampling sites. Bacterial bioaerosol concentration was significantly lower at sampling location I3 of KBMH, which has an air conditioning system.
Fig. 2.
Bioaerosol concentration (CFU/m3) at different sampling sites. Here, I1 & I2 = Bangabandhu Sheikh Mujib Medical University Hospital (BSMMUH), I3 & I4 = Khwaja Badrudduja Modern Hospital (KBMH), I5 & I6 = Dhaka Medical College Hospital (DMCH), I7 & I8 = Monno Medical College Hospital (MMCH), I9 = Green Model Town (GMT), I10 = Mukarram Hussain Khundker Bhaban (MHKB).
Correlation between bioaerosol concentration and particulate matter (PM)
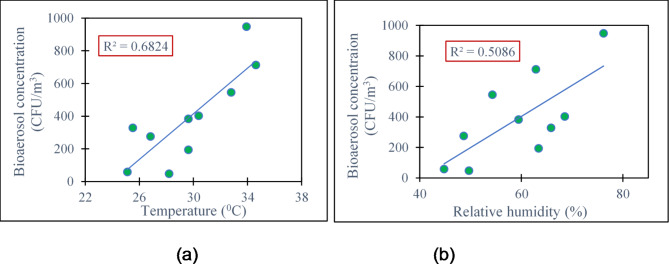
A significant relationship (p < 0.05) between bacterial concentration and particulate matter has been shown in Fig. 3, in which an increase in bacterial concentration was observed with the increasing concentration of particulate matter. The regression parameters in Fig-3(c) show the modest connection (R2 = 0.27) between the PM10 concentration and the concentration of bacterial bioaerosol, though the value of the concentration of microbes and PM10 is significant (p < 0.05).
Fig. 3.
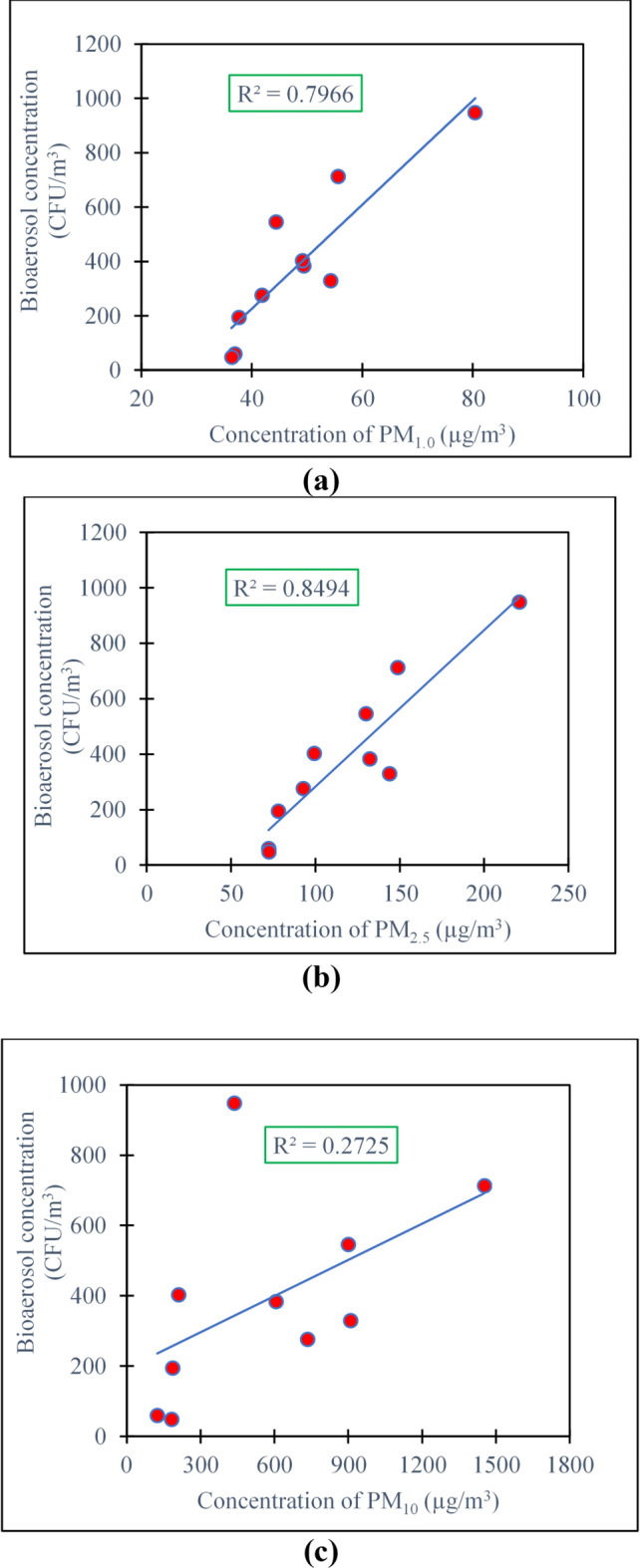
Correlation between concentration of particulate matter (a) PM1.0, (b) PM2.5, (c) PM10 with concentration of bioaerosol.
Based on the information provided in Fig. 3 (a) and (b), it can be deduced that there is a strong connection between bacterial bioaerosols and fine particles, especially PM1.0 and PM2.5. The high R2 values for PM1.0 and PM2.5 (0.80 and 0.85) suggest a significant correlation between bacterial bioaerosols and these fine particulate matter fractions.
Correlation between bioaerosol concentration and meteorological parameters
The meteorological parameters (i.e. temperature, relative humidity) were measured simultaneously to study the influence of these on the number and growth of the bacterial bioaerosol (Supplementary Table S2).
A substantial (p < 0.05) correlation was found by the ANOVA-single component test between temperature and the number and growth of bacteria. Temperature change and R2 = 0.68 showed a positive correlation for indoor microorganism variation (Fig. 4a). Relative humidity (RH) functions similarly to temperature and has a significant impact on airborne microbial concentration, diversity, and composition, among other factors. The R2 value was 0.51 (Fig. 4b), demonstrating a positive association between the concentration of bacterial bioaerosol and relative humidity.
Fig. 4.
Correlation between Temperature (a) and Relative Humidity (b) with bioaerosol concentration at different sampling sites.
Gram positive and negative bacteria in bioaerosol samples
The colony features and structural morphology of these isolates were recorded (Supplementary Table S3). Eleven different bacterial colonies were isolated based on their apparent colony characteristics. Eight (isolates 2, 3, 5, 6, 7, 8, 9) were gram-negative (73% of the total isolated samples), and the other four (isolates 1, 4, 10, 11) were gram-positive (27% of the total isolated samples).
Antibiotic susceptibility profile of isolates
Twenty-one different antibiotics were employed in the antibiotic susceptibility test. With the exception of isolate-11, every examined bacterial isolate shown resistance to the majority of drugs. Isolate-11 was sensitive to all the antibiotics, and it was obtained from the bioaerosol sample of one of the controlled sites (I10) (Table 1; Fig. 5). All bacterial isolates were sensitive to only 3 antibiotics (gentamicin, tigecycline, and vancomycin). Only one isolate was found to be resistant against imipenem, meropenem, and colistin (isolate-9 against imipenem, meropenem and isolate-6 against colistin). The isolates’ multi-drug resistance (MDR) was demonstrated by these findings.
Table 1.
Antibiogram of the isolates obtained from bioaerosol samples of different hospital environments.
| Name of the Antibiotics | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | Azithromycin | Erythromycin | Gentamicin | Chloramphenicol | Ampicillin | Ciprofloxacin | Cotrimoxazole | Cefixime | Ceftazidime | Tigecycline | Aztreonam | Imipenem | Cloxacillin | Cefepime | Vancomycin | Cefalexin | Ceftriaxone | Cefotaxime | Cefuroxime | Meropenem | Colistin |
| 1 | R | R | S | R | R | R | S | R | R | S | R | S | R | S | S | S | S | S | S | S | S |
| 2 | R | R | S | R | R | S | S | R | R | S | R | S | S | R | S | R | R | S | R | S | S |
| 3 | R | R | S | R | R | S | S | R | S | S | S | S | R | S | S | R | R | R | R | S | S |
| 4 | R | R | S | R | R | R | R | R | R | S | S | S | S | R | S | S | S | S | S | S | S |
| 5 | R | R | S | S | R | S | R | R | S | S | R | S | R | S | S | S | S | S | S | S | S |
| 6 | S | S | S | R | R | S | R | S | S | S | S | S | R | S | S | S | S | S | S | S | R |
| 7 | R | R | S | S | R | R | R | R | S | S | S | S | R | S | S | S | S | S | S | S | S |
| 8 | R | R | S | S | R | R | R | R | S | S | S | S | R | S | S | S | S | S | S | S | S |
| 9 | R | R | S | S | R | R | R | R | R | S | S | R | R | R | S | R | R | R | R | R | S |
| 10 | R | R | S | S | R | S | S | R | S | S | S | S | S | S | S | S | S | S | S | S | S |
| 11 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
S= Sensitive, R= Resistant .
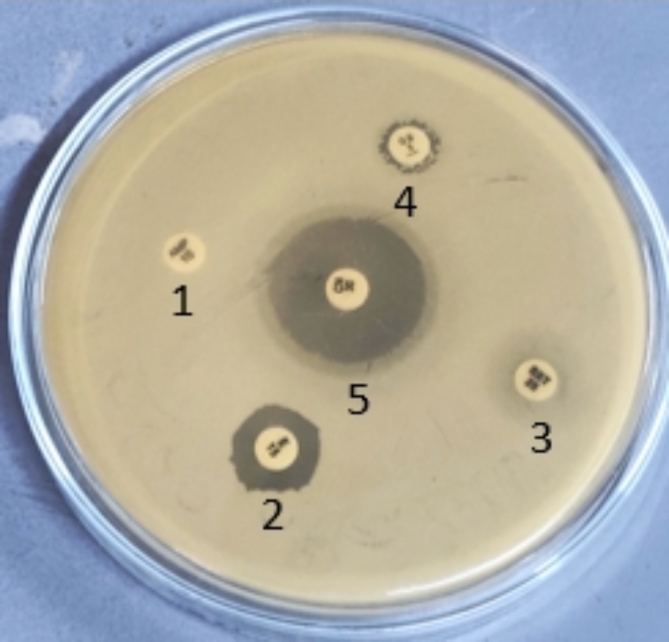
Fig. 5.
A representative antibiogram of the isolate-5. 1- Ampicillin; 2-Erithromycin; 3- Cotrimoxazole; 4-Cloxacillin; 5- Gentamicin.
The highest antibiotic resistance was observed in the case of isolate-9, which was about 71.43% of the total antibiotics used, and the lowest was observed in isolate-10 at 19.05% (Table 2). Isolate-11 showed no resistance against the antibiotics used. Isolates of bioaerosol samples collected from BSMMUH (I1, I2) and DMCH (I5, I6) showed higher resistance than other hospitals.
Table 2.
Percentage of antibiotics resistant by the isolated bacterial samples cultured from bioaerosol samples of different sampling locations.
| Sampling locations | Cultured bacterial isolates | % of antibiotic resistant |
|---|---|---|
| Bangabandhu Sheikh Mujib Medical University Hospital (BSMMUH) | 1 | 42.86 |
| 2 | 52.38 | |
| 3 | 47.62 | |
| 4 | 42.86 | |
| Khwaja Badrudduja Modern Hospital | 5 | 33.33 |
| 6 | 23.81 | |
| Dhaka Medical College Hospital (DMCH) | 7 | 33.33 |
| 8 | 33.33 | |
| 9 | 71.43 | |
| Monno Medical College Hospital | 10 | 19.05 |
| Mukarram Hussain Khundker Bhaban | 11 | No resistance shown |
Identification of the bacterial isolates
The isolates obtained from the study were subjected to phylogenetic and 16 S rRNA sequence analyses, which revealed that they belong to various bacterial families, including Staphylococcaceae, Pseudomonadaceae, Bacillaceae, Acetobacteraceae, and Enterobacteriaceae.
Distance tree analysis demonstrated that isolate-3 was very closely associated with P. stutzeri (Table 3; Fig. 6), with a 100% similarity to the database’s reference sequence. Isolates-1 and 10 exhibited 99% resemblance to the standard sequence, indicating a close relationship with S. aureus. On the other hand, 98% closeness to the reference sequence indicated that isolates 7 and 9 were closely related to E. coli. Isolate-2 and 3 were P. aeruginosa and P. stutzeri found to have 99% and 100% similarities, respectively. B. cereus, B. subtilis and B. aerius were some of the species (isolate-4, 8 and 11) of bacillaceae family, also found to have 99% likeliness to the reference sequence. Isolate-6 was found to be P. vulgaris and had 99% similarity.
Table 3.
Identification of bacterial isolates by blast analysis of a partial 16 S rRNA sequence that was searched in a nucleotide database.
| Isolates | Query Recovery (%) | E Value | Identity | Species that the maximum match associated with |
|---|---|---|---|---|
| 1 | 100 | 0.00 | 99% | S. aureus |
| 2 | 100 | 0.00 | 99% | P. aeruginosa |
| 3 | 100 | 0.00 | 100% | P. stutzeri |
| 4 | 100 | 0.00 | 99% | B. cereus |
| 5 | 100 | 0.00 | 99% | A. schindleri |
| 6 | 100 | 0.00 | 99% | P. vulgaris |
| 7 | 100 | 0.00 | 98% | E. coli |
| 8 | 100 | 0.00 | 99% | B. subtilis |
| 9 | 100 | 0.00 | 98% | E. coli |
| 10 | 100 | 0.00 | 99% | S. aureus |
| 11 | 100 | 0.00 | 99% | B. aerius |
Fig. 6.
A representative figure of phylogenetic tree analysis of isolate-3, which was found closely related to P. stutzeri.
Supplementary Fig. S2 shows the DNA sequencing chromatogram of isolate-3 which was generated by using Chromas software (Version 2.6.6). The sequences generated by automated sequencers are displayed as a graph called a chromatogram, which contains a series of peaks in four different colors. The DNA sequences of the bacterial isolates were aligned with the reference sequences. According to Supplementary Fig. S3, in the isolate-3 sequence examined by Chromas software, all 643 nucleotide base pairs were matched with the reference P. stutzeri with no gaps observed.
Bacterial species identified at different sampling locations
In the hospital environments, S. aureus and E. coli were the most frequently detected bacteria (Table 4). The majority of the bacterial species, which are primarily pathogens or opportunistic pathogens, were discovered in the Bangabandhu Sheikh Mujib Medical University Hospital and the Dhaka Medical College Hospital. Bacillus spp. was mostly found in the controlled sites which rarely create any health issues. So, hospitals contained many harmful bacterial species compared to other places.
Table 4.
Identified bacterial isolates at different sample sites in Dhaka, Bangladesh.
| Sampling Sites | Bacterial isolates | Bacterial species present |
|---|---|---|
| Bangabandhu Sheikh Mujib Medical University Hospital | 1 | S. aureus |
| 2 | P. aeruginosa | |
| 3 | P. stutzeri | |
| 4 | B. cereus | |
| Khwaja Badrudduja Modern Hospital | 5 | A. schindleri |
| 6 | P. vulgaris | |
| Dhaka Medical College Hospital | 7 | E. coli |
| 8 | B. subtilis | |
| 9 | E. coli | |
| Monno Medical College Hospital | 10 | S. aureus |
| Mukarram Hussain Khundker Bhaban | 11 | B. aerius |
Discussion
The values of particulate matter concentration at the controlled sites were significantly lower than those at the hospital locations, likely due to the absence of a source of particulate matter pollution29. All the hospital sites had considerably higher PM1.0 concentrations, a significant concern given that smaller particulate matter easily deposits in the human lower respiratory tract, leading to serious public health issues26,27. There were also significant variances in the mean values of different hospitals. National Ambient Air Quality Standards (NAAQS) for Bangladesh are 65 µg/m3 for PM2.5 (24-hour average) and 150 µg/m3 for PM10 (24-hour average). So, the concentration of particulate matter at different hospitals exceeded the NAAQS value and also the WHO value, which is 15 µg/m3 for PM2.5 and 45 µg/m3 for PM10 (24-hour average)30.
The higher concentrations of bioaerosol found in this study might have been attributed to a number of factors, including a high patient and visitor volume, inadequate air conditioning, and prolonged window closures. Artificial air cooling has been proven in several studies to have the potential to lower indoor bacterial counts31. The highest values in I5 and I6 may also be associated with the building’s age, non-standard flooring, consumable materials, wall seams, a high percentage of outdated beds in the wards, natural ventilation, and a high patient density in the wards32. The number of patient beds is one of the primary factors promoting the generation and release of airborne bioaerosols, according to several studies33.
Bacteria, particularly pathogenic bacteria, positively correlate with PM’s physical and chemical makeup34. A moderate correlation (R² = 0.27) was found between PM10 levels and bacterial bioaerosol concentrations, with both showing statistically significant values (p < 0.05). This might be because of the high concentration of fungi that greatly contributes to the PM10 concentration28. Bacterial bioaerosols and fine particulate matter fractions appear to be significantly correlated, as indicated by the high R2 values for PM1.0 and PM2.5 (0.80 and 0.85). It appears that the elements in the particulate matter provide ideal circumstances for the development of microorganisms.
A study by Hoeksma et al. (2015)35 investigated the effects of temperature on E. coli, M. synoviae, and E. mundtii by observing bacterial decay. The results demonstrated that different microbial species exhibited varying abundances at different temperature ranges, with some thriving in high temperatures, others in low temperatures, and a majority occurring at moderate air temperature values. In an indoor environment at the University of Dhaka, RS et al. (2022)36 conducted a study that showed a positive correlation between bacterial concentration and temperature (R2 value 0.73) and a R2 value of 0.68 was found between the relation of bioaerosol concentration and temperature in this study which is also supported by the previous one. The growth of culturable microorganisms was more pronounced during the spring or winter compared to the summer season, primarily due to temperature fluctuations4. Relative humidity (RH) functions similarly to temperature and has a significant impact on airborne microbial concentration, diversity, and composition, among other factors. The combined effects of relative humidity and temperature are likely to play a crucial role in shaping the behavior of airborne microorganisms16. High relative humidity was advantageous for bacterial release and proliferation, but it may potentially lower bacterial viability37. Another study, which was performed at the University of Dhaka, found a positive correlation between indoor bacterial concentration and relative humidity, and the R2 value was 0.6837, which supports the result obtained here (R2 = 0.51). The relative humidity and bacterial concentration were discovered to be negatively correlated38. According to Knudsen et al. (2017)39, various microorganisms react to relative humidity differently, and occasionally they don’t react at all.
In a study at a hospital in Iran, it was found that the highest antibiotic resistance was in cefixime (45.8%), ceftazidime (30.2%), gentamicin (12%) and ciprofloxacin (12%)8. One contributing factor to the rise of antibiotic resistance is the overuse of antibiotics, which can promote the development of antibiotic-resistant bacteria and antibiotic-resistant genes40. This overuse creates a selection pressure that favors the survival and proliferation of resistant strains, reducing the effectiveness of antibiotics and making it more challenging to treat bacterial infections effectively.
P. aeruginosa, P. stutzeri, (A) schindleri, and P. vulgaris were also identified as potential sources of nosocomial infections in immunocompromised patients within hospital settings44. (B) cereus and B. subtilis were also found in the hospital environment, according to the research. According to reports, E. coli and K. pneumoniae are the two most prevalent nosocomial pathogens in hospitals that cause urinary tract infections (UTIs) in Europe45. Others have found similar patterns, with high incidence of Staphylococcus, Micrococcus, and Bacillus in various hospital settings46,47. Among the Gram-negative bacteria, Acinetobacter spp., P. aeruginosa, and E. aerogenes were detected on the plate surface, highlighting their potential association with healthcare-related infections through hospital indoor air48. These findings underscore the importance of monitoring and understanding the presence of different bacterial species in hospital environments to implement effective infection control measures and protect the health of patients and healthcare workers.
Coagulase-negative staphylococcus (CONS) is a common cause of nosocomial infections, particularly in neonatal and pediatric intensive care units, and is associated with significant patient mortality and morbidity42. Similar findings were reported by Memon et al. (2016)43, who observed a high prevalence of S. aureus in various hospital wards, highlighting its role as a notorious pathogen responsible for nosocomial infections in immunocompromised patients. S. aureus, P. aeruginosa, P. stutzeri, B. cereus, (A) schindleri, P. vulgaris, (B) subtilis, E. coli, and B. aerius were found in this study, all of which are associated with creating nosocomial infections or hospital-acquired infections (HAI) in patients and healthcare workers, except Bacillus aerius44,51. Most of the bacterial species were found to be opportunistic pathogens. B. cereus and E. coli were identified as pathogens based on other studies. B. aerius was the one with no pathogenic report so far. Among the bacteria responsible for causing a wide range of clinical infections, S. aureus is a major human pathogen. It is known to be a leading cause of various infections, including bacteremia, infective endocarditis, osteoarticular infections, skin and soft tissue infections, pleuropulmonary infections, and device-related infections52. P. aeruginosa and P. stutzeri are also implicated in causing infections such as bacteremia, urinary tract infections (UTIs), and respiratory infections53,54. Additionally, A. schindleri can lead to nosocomial infections, with a predilection for aspiration pneumonia and catheter-associated bacteremia. These bacteria can pose significant health risks in hospital settings, particularly to immunocompromised patients and those with underlying medical conditions. Recent works have proposed hospitals as emission hotspots of antibiotic-resistant bacteria in urban environments49,50 which is in accordance with this study.
The World Health Organization (WHO) has provided 100 CFU/m3 as the maximum number for hospital guidelines for bacteria55. Given that each patient and staff member have a different level of immunosuppression and susceptibility to infection, the study of bioaerosol concentration and the evaluation of bacterial resistance to antibiotics is crucial for the prevention of hospital-acquired infections (HAIs) or nosocomial infections and may be impacted by ineffective management of these factors (11). Healthcare facilities and hospitals stand out among all building types for their link with pathogenic bacteria. Nosocomial infections, which impact 15% of inpatients, are particularly prone to hospitalized patients56. Antibiotic resistance will cause at least 700,000 deaths annually, and the rise in ARGs will cause 10 million fatalities annually by 205050. A study estimated a global antibiotic consumption rate of 14.3 defined daily doses (DDD) per 1000 population per day in 2018, with a 95% uncertainty interval of 13.2 to 15.6 DDD57. This amounted to a total of 40.2 billion DDD consumed worldwide in 2018. This represented a significant increase of 46% from the antibiotic consumption rate of 9.8 DDD per 1000 per day in 2000, with a 95% uncertainty interval of 9.2 to 10.5 DDD. The rise in antibiotic consumption over this period raises concerns about the potential impact on antimicrobial resistance and the need for appropriate stewardship and control measures to ensure responsible and effective use of antibiotics globally. These findings emphasize the need for national and international hospital infection control guidelines to address airborne antibiotic-resistant bioaerosol threats, especially in locations with limited resources.
Methods
Characteristics of the sampling sites and hospital building
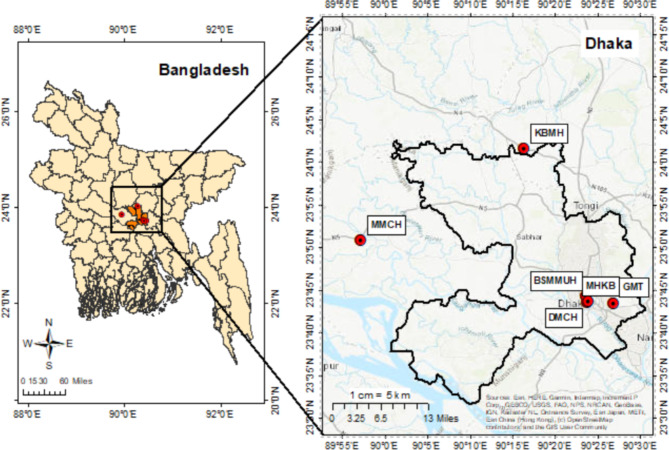
In the greater Dhaka region, the samples were collected from two public and two private hospitals (Fig. 7). The Bangabandhu Sheikh Mujib Medical University Hospital (BSMMUH), with a capacity of 1900 beds, is Bangladesh’s first public and second largest hospital with numerous departments. The second hospital was Khwaja Badrudduja Modern Hospital is a compact healthcare facility comprising 20 beds primarily catering to primary care needs. The third hospital was the Dhaka Medical College Hospital (DMCH).With 2600 beds and multiple departments, it’s one of the largest and most established hospitals in Bangladesh. The fourth hospital was Monno Medical College Hospital, which is located in a rural region. The hospital is a significant medical facility equipped with 500 beds and multiple departments.
Fig. 7.
Map of Bangladesh (Left); four hospitals and two sample-control sites in the greater Dhaka area, Bangladesh (Right).
As the ambient sites, Green Model Town Residential Area and Mukarram Hussain Khundker Bhaban were chosen. In the Green Model Town area, the living room space of a six-story building was chosen for the sampling point. The area is full of trees and has less traffic than other sampling places during sampling hours. At the Mukarram Hussain Bhaban, the sampling was conducted at the Atmospheric & Environmental Chemistry Research Laboratory, which was chosen to have a proper air ventilation system and to avoid the potential confounding effect of traffic congestion. Supplementary Table S4 provides an extensive overview of the features of the office structure and sampling site.
Sample collection
During the pre-monsoon season (February to June of 2023), air samples in the hospitals were collected using UV sterilized Quartz filter paper (Gelman, Membrane Filters, Type TISSU Quartz 2500QAT-UP, 47 mm diameter) with a 4.0-minute hold period between each measurement. Particulate matter was collected using a low-volume air sampler, in which the airflow rate (16.7 L per minute) was recorded by an orifice plate inserted between the filter and the vacuum pump. This design employs a filter cassette set up in a single-filter tray and a Partisol FRM® Model 2000 single-channel air sampler. The concentrations of particulate matter (PM1.0, PM2.5, and PM10) were determined using the AEROCET-531 (USA) air quality monitoring instrument. For three days in a row, each hospital’s working hours (8:00 am to 4:00 pm) were selected for the purpose of sampling. Samples of bioaerosol were taken at a height of approximately 1.5 m in order to replicate the human breathing zone’s aspiration. IGERESS air quality monitoring device was used to gather temperature and relative humidity data (Model: WP6930S, VSON Technology Co., Ltd, Guangdong, China).
Conditioning of filter paper
We used an ultraviolet irradiation process for 8 h to sterilize the blank quartz filter paper, either killing off any remaining microbiological particles or rendering them inactive. The autoclaved water was used to moisten the irradiated filter paper before it was immediately put in the low-volume air sampler’s filter holder. After the completion of the sampling, a pre-sterilized anti-cutter was used to cut the filter papers into small pieces, which were then added to the 100 mL nutrient broth. The material was completely dissolved in the broth after being agitated for 30 to 40 min on a hot plate (37 °C) magnetic stirrer. Next, using a sterile bent glass rod, 25 µL of the material was spread out over the nutritional agar medium plates. The plates were then incubated at 37 °C for 24 h. Then, the total colony forming units (CFU) were counted. Following sampling, the loaded filter paper was kept at 4 °C until further examination.
Calculation of the bioaerosol concentration
The concentration of bacteria in the bioaerosol was calculated by dividing the CFU by the measured air volume (CFU/m3)23.
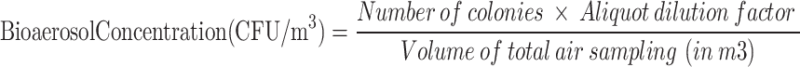 |
1 |
Identification of the bacterial bioaerosol species
Obtaining pure culture
Different bacterial colonies were preliminarily determined only by observing their colony characteristics. Using sterile loops, each colony was removed, and a streak plate experiment was conducted on nutritional agar medium. Following that, the plates were incubated at 37 °C for 24 to 48 h. After getting a pure culture, gram staining, antibiotic sensitivity test, and identification of bacteria were performed.
Gram staining method
At first, a smear was made of a bacterial culture on the glass side, heat-fixed and then applied a primary stain (crystal violet) for 60 s. It was washed gently to remove the dye and added the iodine solution for 60 s. After removing iodine through washing, ethanol was added for 15 s and gently washed the slide again. Then, a counter stain was added, safranin, and the slide was kept for 60 s. After washing, the slides were air-dried and observed under a light microscope with 100x magnification, and the morphology, arrangement, and distinguishing features of the bacterial cells were observed58.
Antibiotic resistivity of the isolates
The Kirby-Bauer disk diffusion method24 was used to assess the antibiotic susceptibility of the chosen bacterial isolates. For this, selected bacteria were cultured in liquid nutrient broth media, and from this culture media, 100 µl was taken and spread on Muller-Hinton agar (Difco, USA) plate. After that, antibiotic discs were positioned and incubated at 37ºC for the entire night. The sensitivity was then evaluated by measuring the inhibition zone in millimeters and comparing it with the reference chart24. Antibiotic discs (Oxoid, England) used for this experiment were tigecycline (15 µg), ciprofloxacin (5 µg), gentamicin (10 µg), imipenem (10 µg), azithromycin (15 µg), cloxacillin (1 µg), colistin (10 µg), chloramphenicol (30 µg), vancomycin (30 µg), cefepime (30 µg), cephalexin (30 µg), and meropenem (10 µg). The guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) were followed in determining the antibiotics’ susceptibility and resistance25.
Identification of bacterial isolates using 16 S rRNA sequencing
For each isolate, a portion of the 16 S rRNA gene was amplified via the Polymerase Chain Reaction (PCR) technique. Total genomic DNA was isolated using the kit (Invitrogen™ PCR Master Mix Starter, UK). Primers 27 F-AGA GTT TGA TCM TGG CTC AG and 1492 R-TAC GGY TAC CTT GTT ACG ACTT were utilized to amplify the specific region of the 16 S rRNA sequence59. The PCR was run for 30 cycles and the condition of PCR was annealing at 57 °C for 45 s, primer extension at 72 °C for 2 min, and denaturation at 94 °C for 1 min. The last extension stage was carried out at 72 °C for 10 min. Gel electrophoresis (2% agarose gel) was performed to confirm PCR product amplification. The PCR product was then purified using a kit (FavorPrep™ GEL/PCR Purification Kit, Taiwan), and concentration was determined using nanodrop (Thermo Fisher Scientific, USA). The purified PCR products were then subjected to Sanger sequencing (3500 Genetic Analyzer, Thermo Fisher Scientific, USA). By using the online blast software interface, all sequences were compared to the 16 S rRNA database of bacteria and archaea. The top 10 sequences obtained from the blast findings were taken into consideration for creating phylogenetic trees for each isolate using the Maximum Likelihood procedure in MEGA version 5.25 software41. The most closely related sequences for each isolate were found by examining the resultant trees, and the alignment outcome was noted. The names of the species were allocated based on the best match.
Statistical analysis
All statistical analyses were performed using the MS Excel-2019 software. The variations in particulate matter concentrations were examined using one-way ANOVA (analysis of variance). Statistically significant alterations were determined using a paired t-test with a 95% confidence level (p-value = 0.05). The R2 value was used to measure the proportion of the variance in the bioaerosol concentration with the particulate matter concentration and with the meteorological parameter. The supplementary section contains all the applicable ANOVA (Analysis of variance) test equations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors greatly acknowledge the support from the Department of Chemistry and Department Genetic Engineering and Biotechnology, University of Dhaka for the analytical support for chemical and biological parameters.
Author contributions
B.A.K.: Experimental work, initial draft and editing. S.R. : Initial draft, review and editing. N.T. : Experimental and editing. K.B.: Experimental and editing. K.C.D.: Experimental, review and editing. M.S.I. : Conceptual, review and editing. N.A. : Conceptual, review and editing. A.S.: Conceptual and review and editing.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Després, V. et al. Primary biological aerosol particles in the atmosphere: a review. Tellus B Chem. Phys. Meteorol.64, 15598. 10.3402/tellusb.v64i0.15598 (2012). [Google Scholar]
- 2.Fröhlich-Nowoisky, J. et al. Bioaerosols in the Earth system: climate, health, and ecosystem interactions. Atmos. Res.182, 346–376. 10.1016/j.atmosres.2016.07.018 (2016). [Google Scholar]
- 3.Brandl, H. Bioaerosols in indoor environment - A review with special reference to residential and occupational locations. Open. Environ. Biol. Monit. J.4 (1), 83–96. 10.2174/1875040001104010083 (2011). [Google Scholar]
- 4.Humbal, C., Joshi, S. K., Trivedi, U. K. & Gautam, S. Evaluating the colonization and distribution of fungal and bacterial bio-aerosol in Rajkot, western India using multi-proxy approach. Air Qual. Atmos. Health. 12 (6), 693–704. 10.1007/s11869-019-00689-6 (2019). [Google Scholar]
- 5.Chen, Q. & Hildemann, L. M. The effects of human activities on exposure to Particulate Matter and Bioaerosols in Residential homes. Environ. Sci. Technol.43 (13), 4641–4646. 10.1021/es802296j (2009). [DOI] [PubMed] [Google Scholar]
- 6.Wan, G. H., Chung, F. F. & Tang, C. S. Long-term surveillance of air quality in medical center operating rooms. Am. J. Infect. Control. 39 (4), 302–308. 10.1016/j.ajic.2010.07.006 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Wang, A., Daneman, N., Tan, C., Brownstein, J. S. & MacFadden, D. R. Evaluating the relationship between Hospital Antibiotic Use and Antibiotic Resistance in Common Nosocomial pathogens. Infect. Control Hosp. Epidemiol.38 (12), 1457–1463. 10.1017/ice.2017.222 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Hosseini, S., Samadi Kafil, H., Mousavi, S. & Gholampour, A. Seasonal and spatial variations of bioaerosols and antibiotic resistance bacteria in different wards of the hospital. Journal of Air Pollution and Health, DOI: (2022). 10.18502/japh.v7i4.11387
- 9.Bessonneau, V. et al. VOC Contamination in Hospital, from Stationary Sampling of a large panel of compounds, in View of Healthcare Workers and patients exposure Assessment. PLoS ONE. 8 (2). 10.1371/journal.pone.0055535 (2013). e55535. [DOI] [PMC free article] [PubMed]
- 10.El-Sharkawy, M. & Noweir, M. E. H. Indoor air quality levels in a University Hospital in the Eastern Province of Saudi Arabia. J. Family Community Med.21 (1), 39. 10.4103/2230-8229.128778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Alessandro, D. & Fara, G. M. Hospital environments and epidemiology of Healthcare-Associated infections. SpringerBriefs Public. Health. 9783319491592, 41–52. 10.1007/978-3-319-49160-8_4 (2017). [Google Scholar]
- 12.Liu, H. et al. Effect of air pollution on the total bacteria and pathogenic bacteria in different sizes of particulate matter. Environ. Pollut.233, 483–493. 10.1016/j.envpol.2017.10.070 (2018a). [DOI] [PubMed] [Google Scholar]
- 13.Smets, W., Moretti, S., Denys, S. & Lebeer, S. Airborne bacteria in the atmosphere: Presence, purpose, and potential. Atmos. Environ.139, 214–221. 10.1016/j.atmosenv.2016.05.038 (2016). [Google Scholar]
- 14.Fernandes, J. J. D., Aguiar, P. A. D. F., Mendes-Rodrigues, C. & Martins, C. H. G. Assessing bacterial bioaerosol and environmental variables of critical hospitalization units of a tertiary hospital. Aerobiologia, DOI: (2023). 10.1007/s10453-023-09792-9
- 15.Fan, X. Y. et al. More obvious air pollution impacts on variations in bacteria than fungi and their co-occurrences with ammonia-oxidizing microorganisms in PM2.5. Environ. Pollut.251, 668–680. 10.1016/j.envpol.2019.05.004 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Yan, D. et al. Diversity and Composition of Airborne Fungal Community Associated with particulate matters in Beijing during Haze and Non-haze days. Front. Microbiol.710.3389/fmicb.2016.00487 (2016). [DOI] [PMC free article] [PubMed]
- 17.Valdez-Castillo, M. & Arriaga, S. Response of bioaerosol cells to photocatalytic inactivation with ZnO and TiO2 impregnated onto Perlite and Poraver carriers. Front. Environ. Sci. Eng.15 (3). 10.1007/s11783-020-1335-9 (2021). [DOI] [PMC free article] [PubMed]
- 18.Yan, X. et al. Distribution characteristics and noncarcinogenic risk assessment of culturable airborne bacteria and fungi during winter in Xinxiang, China. Environ. Sci. Pollut. Res.26 (36), 36698–36709. 10.1007/s11356-019-06720-8 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Gandolfi, I., Bertolini, V., Ambrosini, R., Bestetti, G. & Franzetti, A. Unravelling the bacterial diversity in the atmosphere. Appl. Microbiol. Biotechnol.97 (11), 4727–4736. 10.1007/s00253-013-4901-2 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Chretien, J. P. et al. Global Climate Anomalies and Potential Infectious Disease Risks: 2014–2015. PLoS Currents, DOI: (2015). 10.1371/outbreaks.95fbc4a8 [DOI] [PMC free article] [PubMed]
- 21.Pedersen, M. K. et al. Occupational Tuberculosis in Denmark through 21 years analysed by Nationwide Genotyping. PLOS ONE. 11 (4). 10.1371/journal.pone.0153668 (2016). e0153668. [DOI] [PMC free article] [PubMed]
- 22.Berger, T., Kassirer, M. & Aran, A. A. Injectional anthrax - new presentation of an old disease. Eurosurveillance19 (32). 10.2807/1560-7917.ES2014.19.32.20877 (2014). [DOI] [PubMed]
- 23.Morgado-Gamero, W. B., Parody, A., Medina, J., Rodriguez-Villamizar, L. A. & Agudelo-Castañeda, D. Multi-antibiotic resistant bacteria in landfill bioaerosols: environmental conditions and biological risk assessment. Environ. Pollut.29010.1016/j.envpol.2021.118037 (2021). [DOI] [PubMed]
- 24.James, J. B. M. D. Antimicrobial susceptibility testing by the Kirby-Bauer Disc Diffusion Method. Ann. Clin. Lab. Sci.3 (2), 135–140 (1973). [PubMed] [Google Scholar]
- 25.James, H. J. F. H. & Janet New consensus guidelines from the clinical and laboratory standards institute for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria. Clini Infec Disea. 44, 280–286 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Zaman, S. U., Yesmin, M., Pavel, M. R. S., Jeba, F. & Salam, A. Indoor air quality indicators and toxicity potential at the hospitals’ environment in Dhaka. Bangladesh Environ. Sci. Pollution Res.28 (28), 37727–37740. 10.1007/S11356-021-13162-8/FIGURES/5 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Wang, K. et al. Seasonal concentration distribution of PM1.0 and PM2.5 and a risk assessment of bound trace metals in Harbin, China: Effect of the species distribution of heavy metals and heat supply. Sci. Rep.10 (1), 8160. 10.1038/s41598-020-65187-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grange, S. K. et al. Switzerland’s PM10 and PM2.5 environmental increments show the importance of non-exhaust emissions. Atmospheric Environment: X. 12, 100145. 10.1016/j.aeaoa.2021.100145 (2021). [Google Scholar]
- 29.Roy, S. et al. Impact of fine particulate matter and toxic gases on the health of school children in Dhaka, Bangladesh. Environ. Res. Commun.5 (2), 025004. 10.1088/2515-7620/ACB90D (2023). [Google Scholar]
- 30.World Health Organization. WHO Global air Quality Guidelines: Particulate Matter (PM2.5and PM10), Ozone, Nitrogen Dioxide, Sulfur Dioxide and Carbon Monoxide (World Health Organization, 2021). https://apps.who.int/iris/handle/10665/345329 [PubMed]
- 31.Cabo Verde, S. et al. Microbiological assessment of indoor air quality at different hospital sites. Res. Microbiol.166 (7), 557–563. 10.1016/j.resmic.2015.03.004 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Mousavi, M. S. et al. Investigating the effect of several factors on concentrations of bioaerosols in a well-ventilated hospital environment. Environ. Monit. Assess.191 (7), 407. 10.1007/s10661-019-7559-0 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Bolookat, F. et al. Assessment of bioaerosol particle characteristics at different hospital wards and operating theaters: a case study in Tehran. MethodsX5, 1588–1596. 10.1016/j.mex.2018.11.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, H. et al. Effect of air pollution on the total bacteria and pathogenic bacteria in different sizes of particulate matter. Environ. Pollut.233, 483–493. 10.1016/j.envpol.2017.10.070 (2018b). [DOI] [PubMed] [Google Scholar]
- 35.Hoeksma, Aarnink, A. J. A., Venglovsky, J., Gregová, G. & Čornejová, T. Eff. Temp. Relative Humidity Survival Airborne Bacteria (2015).
- 36.RS, A. et al. Multi-drugs resistant bacteria associated particulate matter in the ambient atmosphere of Dhaka, Bangladesh. J. Biodivers. Conserv. Bioresource Manage.7 (2), 1–12. 10.3329/jbcbm.v7i2.60145 (2022). [Google Scholar]
- 37.Chandra, P., Venkata, M. S. & Jayarama, R. S. Assessment of microbial concentration at ambient air of semi-urban region. Appl. Ecol. Environ. Res.3 (2), 139–149. 10.15666/aeer/0302_139149 (2005). [Google Scholar]
- 38.Gulshan, J. E. et al. Seasonal variations of microbes in particulate matter obtained from Dhaka City in Bangladesh. Environ. Pollutants Bioavailab.33 (1), 122–134. 10.1080/26395940.2021.1940302 (2021). [Google Scholar]
- 39.Knudsen, S. M., Gunnarsen, L. & Madsen, A. M. Airborne fungal species associated with mouldy and non-mouldy buildings – effects of air change rates, humidity, and air velocity. Build. Environ.122, 161–170. 10.1016/j.buildenv.2017.06.017 (2017). [Google Scholar]
- 40.Li, Y., Lu, R., Li, W., Xie, Z. & Song, Y. Concentrations and size distributions of viable bioaerosols under various weather conditions in a typical semi-arid city of Northwest China. J. Aerosol. Sci.106, 83–92. 10.1016/j.jaerosci.2017.01.007 (2017). [Google Scholar]
- 41.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215 (3), 403–410. 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Venkatesh, M. P., Placencia, F. & Weisman, L. E. Coagulase-negative staphylococcal infections in the neonate and child: an update. Semin. Pediatr. Infect. Dis.17 (3), 120–127. 10.1053/j.spid.2006.06.005 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Memon, B. A., Bhutto, G. H. & Rizvi, W. H. Measurement of air contamination in different wards of public sector hospital, Sukkur. Pak. J. Pharm. Sci.29 (6), 2015–2021 (2016). [PubMed] [Google Scholar]
- 44.Suleyman, G. & Alangaden, G. J. Nosocomial fungal infections: epidemiology, infection control, and Prevention. Infect. Disease Clin.30 (4), 1023–1052. 10.1016/J.IDC.2016.07.008 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Allocati, N., Masulli, M., Alexeyev, M. & Di Ilio, C. Escherichia coli in Europe: an overview. Int. J. Environ. Res. Public Health. 10 (12), 6235–6254. 10.3390/ijerph10126235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomon, F. B., Wadilo, F. W., Arota, A. A. & Abraham, Y. L. Antibiotic resistant airborne bacteria and their multidrug resistance pattern at University teaching referral Hospital in South Ethiopia. Ann. Clin. Microbiol. Antimicrob.16 (1), 29. 10.1186/s12941-017-0204-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Worku, T., Derseh, D. & Kumalo, A. Bacterial Profile and Antimicrobial susceptibility pattern of the isolates from Stethoscope, Thermometer, and Inanimate surfaces of Mizan-Tepi University Teaching Hospital, Southwest Ethiopia. Int. J. Microbiol. 1–7. 10.1155/2018/9824251 (2018). [DOI] [PMC free article] [PubMed]
- 48.Sedighi, M., Salehi-Abargouei, A., Oryan, G. & Faghri, J. Epidemiology of VIM-1-imipenem resistant Pseudomonas aeruginosa in Iran: a systematic review and meta-analysis. J. Res. Med. Sciences: Official J. Isfahan Univ. Med. Sci.19 (9), 899–903 (2014). [PMC free article] [PubMed] [Google Scholar]
- 49.He, Y. et al. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. Npj Clean. Water. 3 (1). 10.1038/s41545-020-0051-0 (2020).
- 50.Wu, D. et al. Inhalable antibiotic resistomes emitted from hospitals: metagenomic insights into bacterial hosts, clinical relevance, and environmental risks. Microbiome10 (1). 10.1186/s40168-021-01197-5 (2022). [DOI] [PMC free article] [PubMed]
- 51.Al-Wrafy, F., Brzozowska, E., Górska, S. & Gamian, A. Pathogenic factors of Pseudomonas aeruginosa – the role of biofilm in pathogenicity and as a target for phage therapy. Postępy Higieny i Medycyny Doświadczalnej. 71 (1), 78–91. 10.5604/01.3001.0010.3792 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L. & Fowler, V. G. Staphylococcus aureus infections: Epidemiology, Pathophysiology, Clinical manifestations, and management. Clin. Microbiol. Rev.28 (3), 603–661. 10.1128/CMR.00134-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emam, A. M., Haridy, M. & Hossam Eldin Ahmed, N. Pathogenicity of newly emerged bacterial pathogens, Pseudomonas stutzeri and P. oleovorans, in the Red Sea Seabream Diplodus noct. Egypt. J. Aquat. Res.48 (2), 169–174. 10.1016/j.ejar.2022.02.001 (2022). [Google Scholar]
- 54.Qin, S. et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal. Transduct. Target. Therapy. 7 (1). 10.1038/s41392-022-01056-1 (2022). 199, DOI. [DOI] [PMC free article] [PubMed]
- 55.WHO. The 2019 WHO AWaRe Classification of Antibiotics for Evaluation and Monitoring of Use. (2019a).
- 56.Kim, K. Y. & Kim, C. N. Airborne microbiological characteristics in public buildings of Korea. Build. Environ.42 (5), 2188–2196. 10.1016/j.buildenv.2006.04.013 (2007). [Google Scholar]
- 57.Browne, A. J. et al. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planet. Health. 5 (12), e893–e904. 10.1016/S2542-5196(21)00280-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartholomew, J. W. & Mittwer, T. The Gram stain. Bacteriological Reviews. 16 (1), 1–29. 10.1128/br.16.1.1-29.1952 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sung, J. et al. Utility of conventional culture and MALDI-TOF MS for identification of microbial communities in bronchoalveolar lavage fluid in comparison with the GS Junior next-generation sequencing system. Annals Lab. Med.38 (2), 110–118. 10.3343/alm.2018.38.2.110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.