Abstract
Studies across various pathogens highlight the importance of pathogen genetic differences in disease manifestation. In the human fungal pathogen Cryptococcus neoformans, sequence type (ST) associates with patient outcome. We performed a meta-analysis of four genomic studies and identified overlapping gene regions associated with virulence, suggesting the importance of these gene regions in cryptococcal disease in diverse clinical isolates. We explored the relationship between virulence and strain genetic differences using the cryptococcosis mouse model and a closely related library of ST93 clinical isolates. We identified four in vivo virulence phenotypes: hypervirulence, typical virulence with CNS disease, typical virulence with non-CNS disease, and latent disease. Hypervirulent isolates were clade specific and associated with an interferon gamma (IFNγ) dominated immune response. Using a genome wide association study (GWAS), we identified nine genes with polymorphisms associated with IFNγ production, including the inositol sensor ITR4. The itr4Δ mutant recapitulated the hypervirulence phenotype and ITR4 affects expression of two IFNγ associated genes. Finally, we showed that IFNγ production is associated with SNPs that downregulate ITR4 and with SNP accumulation in other IFNγ associated genes. These data highlight the complex role of pathogen genetics in virulence and identify genes associated with hypervirulence and IFNγ in Cryptococcus neoformans.
Subject terms: Fungal pathogenesis, Fungal genetics, Fungal infection
Identification of virulence-associated genes in pathogens is important to understand mechanisms of disease. Here, Jackson et al. use a mouse model and clinical isolates of Cryptococcus neoformans to identify novel gene networks that impact virulence.
Introduction
Genetic differences between pathogens of the same species can have important consequences for disease manifestation. Powerful genetic and bioinformatic tools provide opportunities to identify these causative differences. Cryptococcus neoformans is a pathogenic yeast with a well-defined genome that is globally distributed, largely clonal, and primarily causes disease in patients with immunocompromising conditions, including advanced HIV disease1,2. In individuals with advanced HIV disease, the most common disease manifestation is cryptococcal meningitis (CM), a dangerous condition that kills an estimated 112,000 individuals annually3. Cryptococcal meningitis is the most common cause of meningitis in adults with advanced HIV disease and is responsible for 19% of all AIDS-related mortality3,4. Even with improvements in standard of care, CM mortality rates have plateaued in recent years3.
CM patient survival is related to both host-specific factors and the genetics of the infecting strain. Genomic sequencing technology revealed the population structure of C. neoformans5, including the major clades VNI, VNII, and VNB and subclades, referred to by their sequence type (ST)6–11. Variability in patient mortality was thought to be independent of genetic variability until Wiesner et al. showed that genetic differences across sub-clades were associated with patient mortality and host immune response12. Subsequent studies support this association between C. neoformans clades and patient outcome5,8,9,13–19.
To define the association between patient outcome and strain-specific genotype, several groups performed whole genome sequencing to identify genes and single nucleotide polymorphisms (SNPs) associated with differences in patient outcome19–21. Yet how these polymorphisms impact disease outcome remains largely unknown.
Our group sequenced clinical isolates from the ST93 genetic background, one of the most common ST found worldwide and the predominant ST in Uganda and Brazil9,20. We then performed a genome wide association study (GWAS) to identify SNPs and short insertion/deletions (INDELs) associated with human mortality, clinical parameters of human disease, immune response, and in vitro phenotypes20. From the GWAS, we identified 145 variants in 40 genes that were significantly associated with variation in one or more of these phenotypes. We also found that ST93 is comprised of two distinct subpopulations, ST93A and ST93B.
These previous results are intriguing, but variability in patient data makes further characterization of the C. neoformans candidate polymorphisms using human data challenging. In this study, we analyzed the results of previous genomic studies and found that, when diverse isolates and study designs are compared, overlaps in genes and gene regions associated with virulence can be identified across genetically diverse populations. To further refine our genomic studies and link gene polymorphisms to disease phenotypes, we decreased host variability by infecting inbred mice with the ST93 clinical isolates at a consistent initial fungal burden. A GWAS identified 32 genes associated with virulence and immune response, seven of which had been identified in the previous genomic studies. Further analysis of genes associated with IFNγ production revealed a compounding impact of polymorphisms in nine genes that resulted in increased IFNγ levels and mouse mortality.
Results
Analysis of five genomic studies revealed universal virulence-associated genes
A goal of C. neoformans research is to identify candidate genes involved in human disease that can be targeted for new treatments. Ideal candidates would be genes universally associated with disease across diverse clades and genetic backgrounds, especially if variants in these genes drive shifts in the immune response. In the last decade, four genomic studies were performed using C. neoformans clinical isolates: (1) a GWAS that compared polymorphisms in 38 clinical isolates from the ST93 clade to human disease phenotypes20, (2) a genomic study by Day et al. that identified genes with early truncation events distinguishing ST4 from ST519, (3) a GWAS by Sephton-Clark et al. that identified genes associated with fungal burden in patients in Malawi21, and (4) a bulk segregant analysis by Agustinho et al. that identified a region implicated in virulence in mice when an ST5 clinical isolate was crossed with the laboratory reference strain KN99α22.
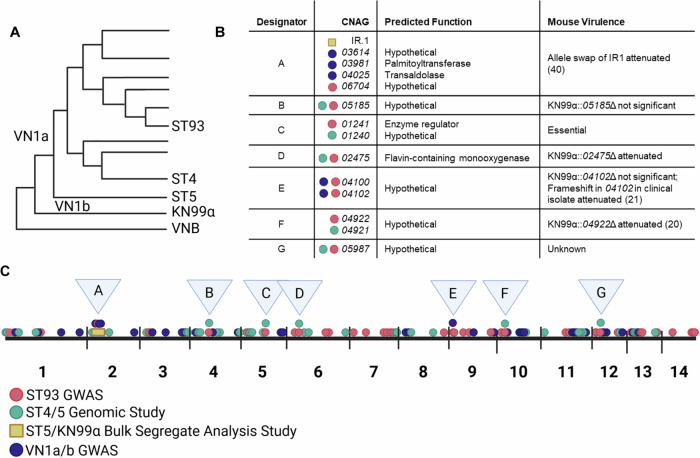
We hypothesized that similar genes and gene regions in these distantly related isolates would be associated with patient outcomes. To test this hypothesis, we compared the virulence-associated genes and gene regions from these four studies and identified several overlapping genomic regions (Fig. 1A). A 184,000 bp region on chromosome 2 implicated in mouse virulence by bulk segregant analysis22 contains three genes identified in the Malawi GWAS (CNAG_03614, CNAG_03981, CNAG_04025)21 and one identified in the ST93 GWAS (CNAG_06704)20 (Fig. 1B, C, Supplementary Data 1). Three genes were found in both the ST93 GWAS and the ST4/5 genomic study (CNAG_05185, CNAG_02475, CNAG_05987)19 while an additional two were only a single gene away (CNAG_01240 vs CNAG_01242 and CNAG_04922 vs CNAG_04921) (Fig. 1B, C). Two genes in the same region were identified by the ST93 and Malawi GWAS (CNAG_04100 and CNAG_04102) (Fig. 1B, C). Five of these overlapping genes or gene regions (IR.1, CNAG_05185, CNAG_02475, CNAG_04102, CNAG_04922) were previously deleted in KN99α and tested in mice, either by our group or others (Figs. 1B, and S1)20–22. All of these gene deletions except CNAG_05185 and CNAG_04102 revealed a role in virulence. A clinical isolate with a frameshift in CNAG_04102 had reduced virulence, suggesting this gene may also influence virulence21. Of the remaining two genes, CNAG_01241 is predicted to be essential23 and CNAG_05987 is hypothetical, specific to pathogenic Cryptococcus spp., and has an unknown function24 (Fig. 1B). Combined, these data show multiple genomic studies encompassing diverse clades identify overlapping genomic regions and suggest these regions may have universal impacts on disease.
Fig. 1. A comparison of four genetic studies reveals overlapping genomic regions associated with virulence.
A literature search was performed to identify C. neoformans genomic studies and the results from each study were compared. A Studies used isolates from ST9320, ST4 and ST519,22, and VN1a and VN1b21. B, C Overlapping genomic regions, designated A-G, contained genes identified in multiple studies. Genes or gene regions identified in at least two studies are highlighted. Figure made with BioRendor.
Mice infected with clinical isolates exhibit four disease manifestations
The above approach discovered overlapping genomic regions even in the context of diverse clinical isolate genetic backgrounds, host genetic variability, and dissimilar clinical study designs. We next asked if the opposite approach—i.e., reducing variability for both the host and the pathogen—would also reveal SNPs, genes, and gene regions associated with virulence. For these studies, we selected a collection of 38 closely related Ugandan ST93 clinical isolates we had used previously in a GWAS study that had known SNPs, INDELs, and population structure20.
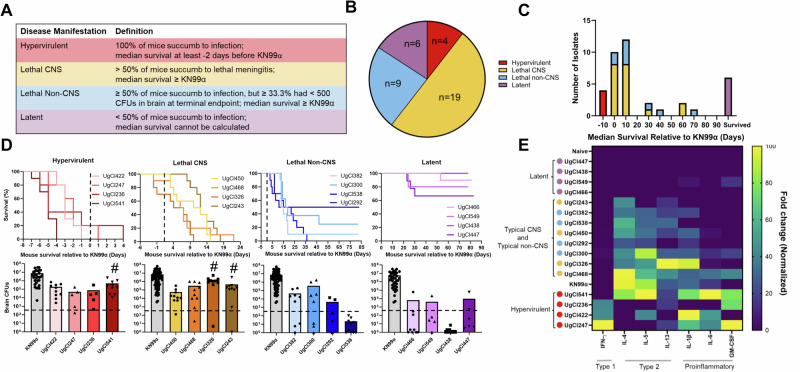
To further reduce variability in host genetic background and environment, and control the initial fungal burden, A/J mice were infected intranasally with the 38 C. neoformans ST93 clinical isolates. Using this mouse model, we identified four disease manifestations (Fig. 2A, B): (1) Lethal CNS disease (19/38) that led to mouse mortality due to meningitis symptoms and high fungal burden in the brain; (2) Lethal non-CNS disease (9/38), defined as mouse mortality with no meningitis symptoms and low or variable fungal burden in the brain; (3) Latency (6/38), with median survival unable to be determined due to most mice surviving to 100 days, as previously defined25; and (4) Hypervirulence (4/38), characterized as median survival that was at least 2 days shorter than the KN99α control. Overall, we observed a spectrum of disease within the closely related ST93 clinical isolates, with median survivals ranging from 5 days before to 77 days after the KN99α median survival (Figs. 2C, D, and S2).
Fig. 2. Mice infected with clinical isolates showed four distinct disease manifestations that associated with immune response.
5–10 mice were infected with each clinical isolate and monitored for 100 days. A Disease manifestation was categorized based on survival relative to KN99α controls and CFUs in the brain. B Lethal CNS was the most prevalent disease manifestation, followed by lethal non-CNS, then latent, with the hypervirulent manifestation accounting for 10% of isolates. C Histogram displaying the median survival of the 38 clinical isolates relative to KN99α. Color codes as in (B). D Top panel: four representative isolates are shown for each disease manifestation. The survival curves were normalized to an infection-matched KN99α control with the black dotted line indicating KN99α median survival (n = 5–10 mice). Bottom panel: Mouse CFUs in brains at terminal endpoint, or at 100 days post infection for latent isolates. Significance was determined using Kruskal–Wallis nonparametric test with Dunn’s multiple comparison correction (n = 5–10 mice; exact p-values from left to right; Hypervirulent: 0.0045, 0.0050, 0.0020, Typical CNS: <0.0001, 0.0024, Typical Non-CNS: 0.0006, 0.0067, 0.0039, <0.0001, Latent: <0.0001, 0.0207, <0.0001, 0.0182). # indicates isolates that are not significantly different from the KN99α control. E Normalized and scaled cytokine fold-change data. The fold-change from an uninfected control mouse was determined for each cytokine. The heatmap was populated with cytokines that had a significant increase compared to the naïve control. Significance was determined using a Kruskal–Wallis nonparametric test without Dunn’s multiple comparison correction. Sample clustering was performed using an average clustering linkage method and Euclidean distance measurement63. The clustered heatmap was normalized by column and the highest value for each cytokine was assigned a value of 100 and the lowest amount 0. Yellow indicates the greatest fold-change from naïve control for that cytokine and blue indicates the lowest fold-change. Four representative samples from each disease manifestation are shown. Latent isolates (purple) clustered with the uninfected control and showed a mostly undetectable immune response. Lethal CNS (yellow) and lethal non-CNS (blue) isolates showed a type-2 immune response, with increases in IL-4, IL-5, and IL-13, and clustered with KN99α. Hypervirulent isolates (red) showed an increase in the type-1 cytokine IFNγ and/or the proinflammatory cytokines, IL-1β, IL-6, and GM-CSF. Source data are provided as a Source Data file.
In vitro phenotypes do not associate with ST93 genetic background or disease manifestation
Several in vitro characteristics of C. neoformans are proposed to influence virulence. We previously showed these in vitro characteristics were not associated with human disease20. We examined the previously tested characteristics as well as additional cell growth (heat, pH, salt), cell wall (caffeine, calcofluor white, Congo red), and membrane (Congo red, SDS) stressors for their association with the four mouse disease manifestations. We also assayed cell morphology characteristics of each clinical isolate including ability to form titan cells in vitro, capsule size, urease activity, and melanin production. We used average linkage clustering paired with a Euclidean distance matrix to determine the correlation of phenotypes with disease manifestation and found no apparent association between any in vitro phenotype and disease manifestation (Fig. S3).
Immune response is associated with disease manifestation
Based on clinical studies showing an association between immune response and clinical outcome in humans26–31, we hypothesized that the mouse immune response could influence disease manifestation. We examined cytokine levels in lung homogenates of the clinical isolate infections to measure the immune response. Lung cytokine levels generally correlated with disease manifestation using the average linkage clustering and Euclidean distance matrix method. Mice infected with isolates that caused latent disease showed a non-detectable cytokine response that clustered with uninfected control mice (Figs. 2E, and S4a, b, Table S1). Mice infected with lethal isolates showed a type-2 immune response with an increase in IL-4, IL-5, and IL-13 and clustered with the KN99α control, regardless of CNS or non-CNS disease manifestation (Figs. 2E, and S4a, b, Table S1). Finally, mice infected with three of the four hypervirulent isolates had a significant increase in the type-1 cytokine IFNγ and/or a significant increase in proinflammatory cytokines including IL-6, IL-1β, and GM-CSF (Fig. 2E, and S4a, b, Table S1). The other hypervirulent isolate (UgCl541) had an enhanced type-2 response that was previously shown to be associated with enhanced virulence32.
The association between IFNγ and hypervirulence was surprising, as IFNγ is typically associated with a protective immune response. To further investigate this link, IFNγ was fully depleted during infection with the hypervirulent UgCl422 clinical isolate. No difference in survival was observed but the fungal cells disseminated more quickly with IFNγ depletion (Fig. S5). While inconclusive, these data could suggest that IFNγ is necessary to control C. neoformans during infection, as shown in previously33,34, but that too much IFNγ may be detrimental.
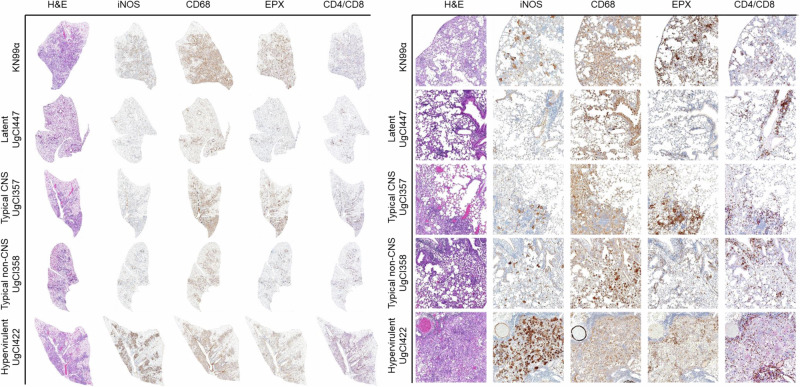
Histology of the lungs further supported the cytokine data. Each mouse lung was stained using hematoxylin and eosin (H&E) and immunohistochemistry stains, including iNOS and CD64 (macrophages), eosinophil protein X (EPX) (eosinophils), and CD4 and CD8 (T cells), to identify immune cells. KN99α infected mice had substantial immune cell infiltration, specifically eosinophils, neutrophils, lymphocytes, and phagocytes, and destruction of alveolar and interstitial tissue. Latently infected mice had lung histology that resembled normal tissue with localized granulomatous lesions. Typical CNS and non-CNS infections were similar to KN99α, though with decreased eosinophils and inflammation, as well as smaller areas of infected tissue in the non-CNS isolates. Mice infected with hypervirulent isolates had a complete loss of alveolar space, evidence of hemorrhage, and large populations of macrophages, eosinophils, neutrophils, phagocytes, and lymphocytes (Fig. 3).
Fig. 3. Histopathological analysis of clinical isolate pulmonary disease manifestations.
Three mice were infected with each of the clinical isolates and sacrificed at 17 days post infection. Lungs were inflated, excised, and fixed in 10% neutral-buffered formalin, and stored in 70% ethanol. Lungs were sectioned and stained with hematoxylin and eosin (H&E) or immunohistochemically with antibodies targeting iNOS, CD68, eosinophil protein X (EPX), or CD4/CD8 dual stain. Slides were scanned and a representative image was taken at 20x magnification (right) or of an entire lung lobe (left).
Disease manifestation is associated with ST93 population structure
Our previous study identified two major clades within ST93—ST93A and ST93B—when using all SNPs identified within a genetically diverse Ugandan strain set comprising 50 clinical isolates20. To analyze the relationships between the 38 ST93 isolates used in this study, we performed a principal component analysis (PCA) to cluster isolates using all SNPs (Fig. S6). Approximately 17% of the observed variance was explained in PC1 and PC2 (Fig. S7a). We observed two non-significant clusters (Fig. S8a), that trended towards associating with disease manifestation (Fig. S6).
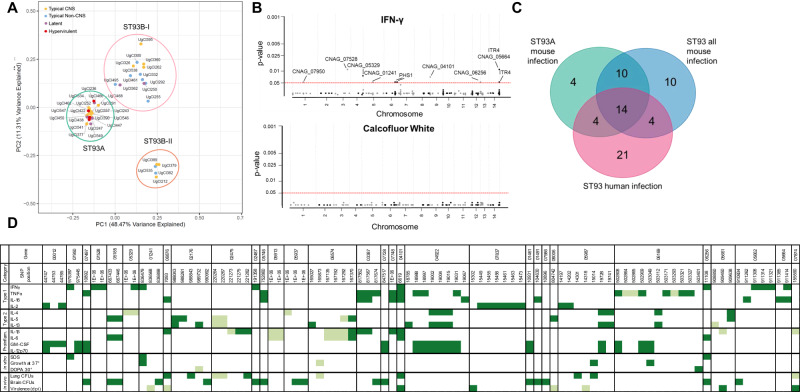
We hypothesized that silent, or non-effect, variants may mask the relationship between isolate-specific virulence and relatedness. To test this hypothesis, we focused on 652 variants predicted to impact gene regulation or protein function (hereafter referred to as effect variants). These variants included nonsynonymous SNPs and INDELs in the coding region of genes or within 1k upstream or downstream of genes20. A PCA using only these effect variants showed ~60% of the observed variance was explained in PC1 and PC2 (Fig. S7b), with the isolates clustered into three significant groups (Figs. 4A, and S8b). The largest cluster contained isolates in the previously defined ST93A subpopulation. The other two clusters contained isolates in the ST93B subpopulation that we further classified as ST93B-I and ST93B-II. Hypervirulent isolates occurred significantly more often in ST93A (4/4) (P = 0.0183). Lethal CNS isolates were more frequent in ST93A (12/19) (P = 0.0214), whereas lethal non-CNS isolates occurred more frequently in ST93B-I (7/9) (P = 0.0131). Latent isolates had a non-significant trend towards ST93A (4/6) (P = 0.1353) (Fig. 4A). Interestingly, both hypervirulent and latent isolates were most common in ST93A, despite having opposing virulence profiles, with some isolates having nearly identical genotypes, e.g., UgCl447 (latent) and UgCl247 (hypervirulent).
Fig. 4. Population structure and SNPs associate with disease manifestation.
A The first two principal components based on the 652 variants predicted to influence gene regulation or protein structure. Three clusters were supported using K-means clustering. Cluster names are based on Gerstein et al. 20. Disease manifestation is indicated with a colored dot. Hypervirulent isolates (red) were significantly associated with ST93A (P = 0.0183), lethal CNS isolates (yellow) with ST93A (P = 0.0214), non-CNS isolates (blue) with ST93B-I (P = 0.0131), and latent isolates (purple) had a non-significant trend towards ST93A (P = 0.1353). Significance was determined using a two-sided Chi-squared test comparing expected versus observed values. B GWAS results for a trait (IFNγ) with multiple significant associations (Top Panel) and a trait with no statistically significant associatons (Calcofluor white, Bottom Panel). The ordinal categorical variables were analyzed with a proportional odds logistic mixed model and the quantitative variables were analyzed using a mixed linear model. Significant variants were identified by likelihood ratio test P values < 0.05 after a Bonferroni correction. Each dot represents a SNP, arranged by chromosome; significant SNPs are lableled with the gene name. C Venn diagram comparing the number of genes with significantly identified SNPs in the human GWAS20, the ST93 all mouse GWAS, and the ST93A mouse GWAS. 14 genes overlapped across all three GWAS studies. D Significant SNPs from the ST93A GWAS are arranged by chromosome order and colored for significant variants. A lighter color indicates differences from the H99 genome associated with a decrease in the trait whereas a darker color indicates variants associated with an increase in that phenotype.
GWAS results for ST93A had the least variability
We hypothesized that the differences in disease manifestations were due to a small number of variants between the ST93A clinical isolates. To test this, we performed two GWAS: the first GWAS examined all 652 effect variants in all ST93 isolates while the second GWAS examined only the 276 variants that were polymorphic only in the ST93A clade. Both GWAS compared variants using a proportional odds logistic mixed model35 for ordinal categorical variables and univariate linear mixed model36 for quantitative variables. Thirty-one traits were analyzed (Supplementary Data 2).
It is standard in human GWAS to define significant variants as having a P < 5 × 10−8 due to extensive variability in clinical data37. However, we removed much of this variability by normalizing the host genetic background, using equivalent inoculums, and using closely related ST93 isolates. Manhattan plots revealed a Bonferroni-corrected significance threshold of P < 0.05 was appropriate, with most trait-variant associations well below this threshold, therefore allowing easy identification of significant variants. For example, while no significant variants were associated with in vitro growth on calcofluor white, several variants were above the P < 0.05 threshold and showed association with in vivo IFNγ production (Fig. 4B).
For the GWAS of all ST93 isolates, 161 variants in 75 genes had P < 0.05 and were associated with at least one trait (Table S2, Supplementary Data 3, Table S3); in the ST93A-specific GWAS, 108 variants in 45 genes were associated with at least one trait. Our previous human disease GWAS using this same set of clinical isolates identified 207 variants associated with human immune response or survival (Table S3, Supplementary Data 4)20. To reduce the number of false positives, we removed genes that had a single variant associated with a single trait, as described previously20. For the GWAS with all ST93 isolates, 127 variants in 38 genes remained after the filter (Table S3); in the ST93A GWAS, 93 variants in 32 genes remained (Table S3). As previously described, 145 variants in 40 genes remained from the human GWAS after removing variants associated with only a single trait20 (Table S3).
When the GWAS using all ST93 isolates was compared to the human GWAS, 42 variants and 16 genes overlapped (Table S3, Supplementary Data 4); for the ST93A population, 34 variants and 17 genes overlapped (Table S3, Supplementary Data 4). We compared the genes found in all three GWAS and found that 14 genes overlapped (Fig. 4C). Finally, the ST93A GWAS had only four unique genes, suggesting that limiting the genetic diversity of the host and the pathogen can also emphasize universal variants.
The ST93A GWAS identified 32 genes associated with disease
Our overall goal in this study was to reduce the variability inherent to GWAS in human hosts. Because ST93A isolates had less genetic variability but still showed a diversity of disease manifestations, we chose to focus on the ST93A GWAS for our downstream genetic analysis. In the ST93A GWAS, 93 variants in 32 genes associated with one or more disease traits (Fig. 4D). These traits were grouped into five categories: type-1 cytokines, type-2 cytokines, proinflammatory cytokines, in vitro phenotype, and in vivo phenotype (fungal burden and survival). Only nine SNPs were associated with in vitro phenotypes and seven of those were also associated with an immune phenotype, further supporting the limited association between virulence and in vitro phenotypes. Of the 32 genes, 15 (47%) have no characterized function; six (19%) are involved in sugar metabolism or transport, four (13%) in nucleic acid binding, two in metal transport, and two in fatty acid synthesis (6%) (Table S4). Only three of the genes—APP1, PHS1, and ITR4 - are named and functionally characterized38–41, two of which (APP1 and ITR4) were also identified in our human GWAS20. Finally, we compared the 32 genes we identified with the gene regions from our meta-analysis (Fig. 1B) and found that our mouse GWAS identified six of the seven genes found in the meta-analysis (CNAG_05185, CNAG_01242, CNAG_02475, CNAG_04101, CNAG_04922, and CNAG_05987) along with an additional gene found in both our mouse GWAS and the Vietnam genomic study (CNAG_07728) (Fig. 1B). A 07728Δ deletion in KN99α had an extreme in vivo growth defect. These data show the mouse model and the genomic meta-analysis identified an overlapping set of genes and suggesting that reducing variability is also effective at identifying genes associated with human disease.
Genes associated with IFNγ produced differences in virulence, immune response, and capsule size when deleted
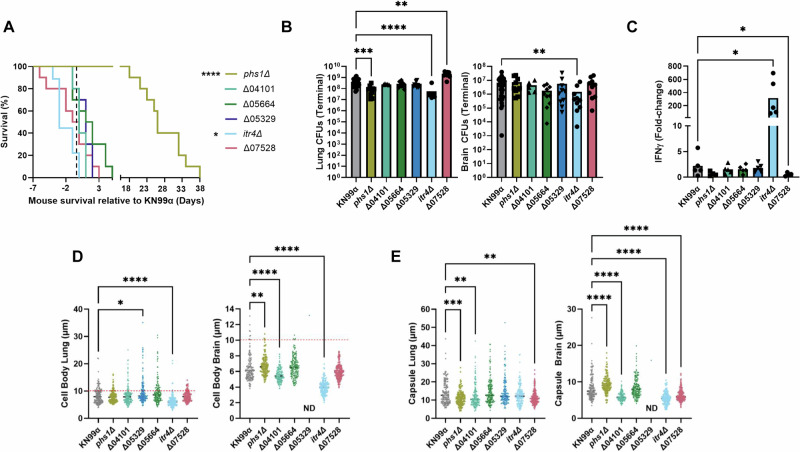
As previously noted, we were surprised by the association between hypervirulent isolates and increased IFNγ accumulation (Fig. 2E) because IFNγ is thought to result in beneficial immune responses and patient outcomes33,34. In the ST93A GWAS, nine genes had variants associated with IFNγ production in mice (Table 1). To assess the role of the IFNγ-associated genes in disease manifestation, we analyzed the virulence of deletion mutants in the KN99α control strain. Six genes were able to be analyzed via gene deletion, however, CNAG_01241 is an essential gene with a putative role in enzyme regulation23; CNAG_07950 is next to the centromere and silenced23; and CNAG_06256 is subtelomeric and has 14 paralogs (Table 1). A/J mice were infected with the six deletion mutants to determine murine survival, fungal burden in lungs and brain, and cytokine production. The itr4Δ mutant was hypervirulent (P = 0.0472) (Fig. 5A), with decreased fungal burden in both the lungs (P < 0.0001) and brain (P = 0.0074) compared to KN99α infection (Fig. 5B). There was additionally a 200-fold increase in IFNγ compared to the uninfected control and a significant increase compared to KN99α (P = 0.0387) (Fig. 5C). The phs1Δ mutant had attenuated virulence (P < 0.0001) (Fig. 5A), with significant decreases in lung fungal burden at terminal endpoint (P = 0.0006) (Fig. 5B) and IL-5 (P = 0.0009). The CNAG_07528Δ mutant had a non-significant trend toward increased virulence (P = 0.1509) (Fig. 5A), a significant increase in lung fungal burden (P = 0.0012) (Fig. 5B), and a decrease in IFNγ (P = 0.0417), as well as a type-2 immune response with increased IL-4 (P = 0.0449) (Figs. 5C, and S9). All other mutant strains had no significant impact on survival, fungal burden, or cytokine response patterns (Fig. 5A–C).
Table 1.
Genes containing variants associated with IFNγ levels in clinical isolates
| Category | Chromosome | Gene | SNP location | Gene function |
|---|---|---|---|---|
| Fatty acid synthesis | 6 | PHS1 (CNAG_02487) | Upstream of coding region | 3-hydroxy acyl-CoA dehydratase involved in synthesis of branched chain fatty acids |
| 9 | CNAG_04101 | Coding region; non-synonymous | Hypothetical; orthologous to phytanoyl-CoA hydroxylase | |
| Amino acid metabolism | 14 | CNAG_05664 | Upstream of coding region | Branched-chain-amino-acid transaminase |
| Inositol sensing and metabolism | 14 | ITR4 (CNAG_05662) | Downstream of coding region | Membrane bound inositol signaler |
| 4 | CNAG_05329 | Upstream of coding region | myo-inositol 2-dehydrogenase | |
| Enzyme regulator | 5 | CNAG_01241 | Upstream of coding region | Enzyme regulator; putative role in protein signal transduction |
| DNA structure | 1 | CNAG_07950 | Upstream of coding region | Hypothetical protein; centromeric |
| 3 | CNAG_07528 | Coding region; non-synonymous | DNA binding protein; potential transposon | |
| 13 | CNAG_06256 | Upstream of coding region | Hypothetical protein; subtelomeric |
Fig. 5. Genes with SNPs associated with ITR4 have virulence, IFNγ, and capsule phenotypes.
A Survival curves of mice infected with each of the KN99α deletion mutants. Significance was determined using a two-sided Gehan-Breslow-Wilcoxon test (n = 5–10 mice; itr4Δ P = 0.0472; phs1Δ P < 0.0001). B Fungal CFUs in samples of lungs and brain at terminal endpoints. Significance was determined using a Kruskal–Wallis nonparametric test with Dunn’s multiple comparison correction (n = 5–10 mice; Lungs: itr4Δ P < 0.0001; phs1Δ P = 0.0006; CNAG_07528Δ P = 0.0012; Brain: itr4Δ P = 0.0074). C Fold-change of IFNγ levels in lungs relative to an uninfected control. Significance was determined using a Kruskal–Wallis nonparametric test (n = 5 mice; itr4Δ P = 0.0387; CNAG_07528Δ P = 0.0417). D Five mice were sacrificed at terminal endpoint and lungs and brains were removed. Cryptococcal cells were collected from each organ, stained with India ink, and imaged. Cell body was measured for at least 150 cells collected from brain or lung of three mice. Cells above dashed line are titan cells. Only one very large (30 µm) cell was collected from the brains of four mice infected with Δ05329. Significance was determined using a Kruskal–Wallis nonparametric test. (n = 150; Lung: CNAG_05329Δ 0.0418; itr4Δ P < 0.0001; Brain: phs1Δ P = 0.0036; CNAG_04101Δ < 0.0001; itr4Δ P < 0.0001). E Capsule size in exclusion of cell body was measured for at least 150 cells collected from brain or lung of three mice. Significance was determined using a Kruskal–Wallis nonparametric test (n = 150; Lung: phs1Δ P = 0.0005; CNAG_04101Δ P = 0.0025; CNAG_07528Δ P = 0.0056; Brain: phs1Δ P < 0.0001; CNAG_04101Δ < 0.0001; CNAG_07528Δ P < 0.0001; itr4Δ P < 0.0001). Source data are provided as a Source Data file. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
The capsule of Cryptococcus is widely accepted as being the most important virulence factor and impacts both disease and immune response42. The predicted functions of the genes associated with IFNγ include both inositol and amino acid metabolism, both of which have been linked to capsule size43,44, and so we measured the in vivo capsule and cell wall of each deletion mutant. Five of the six deletion mutants had significant changes in capsule size in the lungs and brain at terminal endpoints (Fig. 5D, E).
The itr4Δ deletion influences transcription of other IFNγ associated genes
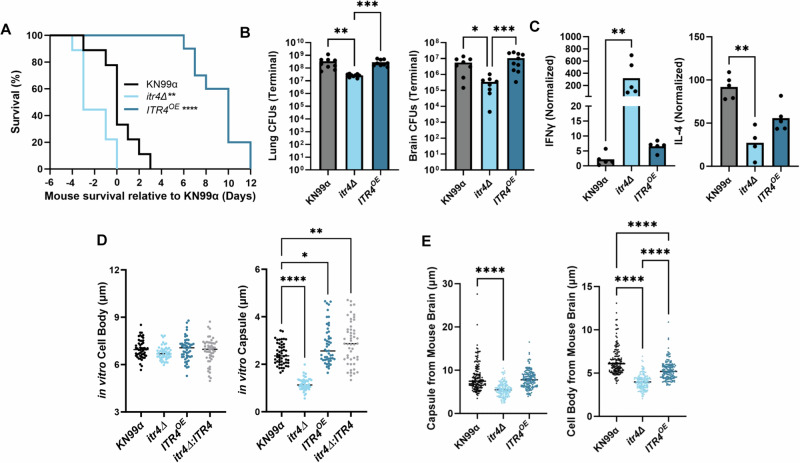
As itr4Δ was the only strain that recapitulated the IFNγ associated hypervirulent phenotype, we hypothesized that ITR4 influenced expression of the other IFNγ associated genes. Overexpression of ITR4 in KN99α (ITR4OE) attenuated virulence in mice compared to KN99α (P < 0.0001) (Fig. 6A) with no significant difference in lung or brain fungal burden (Fig. 6B). Additionally, the ITR4OE strain had equivalent IFNγ production to KN99α (Fig. 6C) and the capsule and cell body size was larger both in vitro and in vivo when compared to the itr4Δ deletion strain (Fig. 6D, E).
Fig. 6. ITR4OE is attenuated with lower IFNγ than itr4Δ.
A Mice were infected with 5 × 104 cells of itr4Δ, ITR4OE, and KN99α. Significance was determined using a two-sided Gehan-Breslow-Wilcoxon test (n = 10 mice; itr4Δ P = 0.0472; ITR4OE P < 0.0001). B Lungs and brain were collected at terminal endpoints. Significance was determined using a Kruskal–Wallis nonparametric test with Dunn’s multiple comparison correction (n = 10 mice; Lung: from left to right: P = 0.0012, P = 0.001; Brain, from left to right: P = 0.0233; P = 0.006). C Mice were sacrificed on day 17–20 post infection and lungs removed. The lung supernatant was collected, and cytokine levels determined. Fold-change was calculated relative to an uninfected control. Significance was determined using a Kruskal–Wallis nonparametric test (n = 5 mice; IFNγ: itr4Δ P = 0.0016; IL-4: itr4Δ P = 0.0065). D KN99α, itr4Δ, ITR4OE, and itr4Δ:ITR4 were grown for three days in media supplemented with DME, stained with India ink, and imaged (n = 50; Capsule, from left to right: P < 0.0001; 0.0189; 0.0017). E Mice were sacrificed at terminal endpoint and lungs and brains were removed. Cryptococcal cells were collected from each organ, stained with India ink, and imaged. Capsule and cell body size from at least 50 cells per mouse from 5 mice were measured from the brain. Significance was determined using ordinary one-way ANOVA (n = 150; Capsule, from left to right: P < 0.0001; Cell body, from left to right: P < 0.0001; P < 0.0001; P < 0.0001). Source data are provided as a Source Data file. (**P < 0.01; ****P < 0.0001).
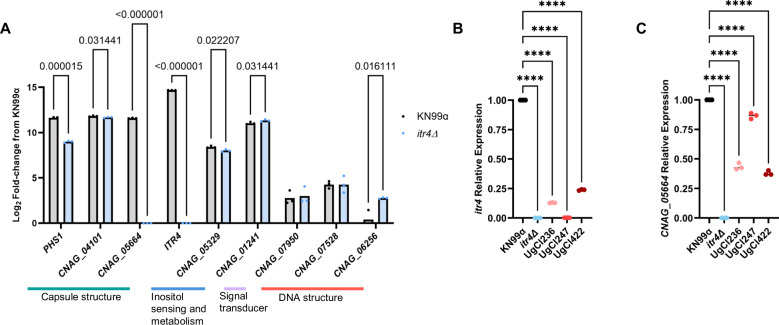
To characterize the relationship between ITR4 and the other genes associated with IFNγ, we performed RNA-seq to compare gene expression between KN99α and the itr4Δ deletion strain. Three of the IFNγ-associated genes (PHS1, CNAG_05664, CNAG_06526) had a greater than twofold change in expression in the itr4Δ strain compared to KN99α (Fig. 7A, Table S5). Interestingly, CNAG_05664 was not expressed in the absence of itr4Δ. While informative, these results only describe the result of full deletion of ITR4 in the KN99α control strain. We next wanted to understand ITR4 expression in clinical isolates. Using qPCR, we found that ITR4 was significantly downregulated in the three hypervirulent isolates that had SNPs in ITR4 (Fig. 7B). Finally, we asked if the decreased expression of ITR4 in the hypervirulent isolates also led to downregulation of CNAG_05664. Consistent with the deletion mutant RNAseq data, CNAG_05664 was downregulated in the hypervirulent isolates with low ITR4 expression (Fig. 7C).
Fig. 7. ITR4 is downregulated in clinical isolates with SNPs in ITR4.
A Cells were grown in media supplemented with dextrose and inositol and RNA-seq was performed to compare transcription differences between KN99α and itr4Δ. Significance was determined using multiple unpaired T-tests with two stage linear step-up (n = 3 biological replicates; from left to right: p = 0.000015; p = 0.031441; p < 0.000001; p < 0.000001; p = 0.022207; p = 0.031441; p = 0.016111). B qPCR showing the relative expression of ITR4 in KN99α, itr4Δ and three hypervirulent clinical isolates that have SNPs in ITR4. Significance was determined using a one-way ANOVA (n = 3 biological replicates; p values, from left to right: P < 0.0001, P < 0.0001, P < 0.0001, P < 0.0001). C qPCR showing the relative expression of CNAG_05664 in KN99α, itr4Δ and three hypervirulent clinical isolates that have SNPs in ITR4 (n = 3 biological replicates; from left to right: P < 0.0001, P < 0.0001, P < 0.0001, P < 0.0001). Significance was determined using a one-way ANOVA. Source data are provided as a Source Data file. (****P < 0.0001).
SNPs in genes associated with IFNγ have a compounding impact on IFNγ production
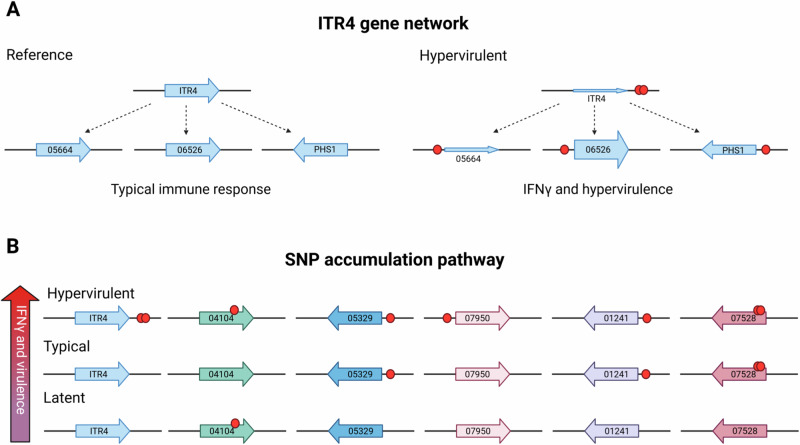
The itr4Δ deletion strain recapitulated the hypervirulent phenotype, however only increased IFNγ production 200-fold compared to the 800- to 2000-fold increase seen in the hypervirulent isolates (Fig. 6C). Our RNA-seq data suggest CNAG_05664, PHS1, and CNAG_06256 are associated with ITR4 and may have been identified in the GWAS due to this association (Fig. 8A). Of the other hypervirulent genes, only CNAG_07528, a putative transposon (Fig. S10), had an IFNγ phenotype (Fig. 5C).
Fig. 8. Model showing hypothesized SNP relationship to IFNγ and hypervirulence.
A Genes predicted to be in a network with ITR4. The size of the gene arrows represents the hypothesized transcription amount of each gene based on RNA-seq data. B Genes hypothesized to be in the SNP accumulation pathway, with the increase in number of SNPs correlating with an increase in IFNγ and virulence. Red circle represents the SNP location. Figure created with BioRendor.
It is also possible that the high IFNγ levels seen in the hypervirulent isolates were due to the compounding effect of multiple variants across these nine genes. To test this additional hypothesis, we determined the average number of variants in the IFNγ-associated genes in latent, typical, and hypervirulent isolates (Table 2). We found that hypervirulent isolates had significantly more SNPs, and genes with SNPs, associated with IFNγ than latent (P = 0.0095, P = 0.0031, respectively) or typical (P = 0.0261, P = 0.0062, respectively) isolates (Table 2). These data suggest accumulation of SNPs in these genes may have a compounding effect on IFNγ production (Fig. 8B).
Table 2.
SNPs and genes associated with IFNγ in hypervirulent, typical, and latent isolates
| Hypervirulent | Typical | Latent | All ST93A isolates | |
|---|---|---|---|---|
| Average variants associated with IFNγ (#) | 5.75 | 2.90 | 1.75 | 3.28 |
| Average genes with variant associated with IFNγ (#) | 4.50 | 2.40 | 1.75 | 2.72 |
| Average IFNγ fold change | 890.46 | 72.74 | 0.58 | 258.42 |
| Average normalized mean survival (days) | −3.75 | 14.15 | 100 | 29.25 |
Discussion
In the age of high throughput genetic and molecular tools, it is substantially easier to identify genes absolutely required for pathogen virulence. It remains a challenge, however, to define genes that modulate in vivo virulence or the interacting networks of genes that work in concert to impact virulence. Pathogen GWAS is one method used to link genetic polymorphisms in an infecting pathogen with host disease outcome45–48. Unlike genetic studies that identify genes required to cause disease, these pathogen GWAS studies identify genetic variation that modifies the outcome of the infection. Yet patient population variability can obscure the gene(s) associated with virulence, and many pathogen virulence factors are polygenic. For example, while the fungal pathogen C. neoformans requires a capsule to cause disease, there are at least 35 genes involved in capsule formation and modification42,49. In addition, there are antigenic proteins embedded in the capsule that also influence the host response42. Polymorphisms in these genes are likely to influence the amount and structure of the capsule which then affects host recognition and ultimately infection outcomes.
Variability and noise within a genomic study can be reduced in two ways: by removing variability from the host side, or by increasing the number of isolates being analyzed. In this study, we used both methods. We leveraged multiple previous genomic studies to identify overlapping genes. We used two GWAS comparing human disease outcome to C. neoformans genetics20,21, one comparative genomic study of isolates that cause differential disease in immunocompetent and immunocompromised patients19, and one bulk segregant analysis that identified genomic regions implicated in mouse virulence22. When the 158 genes collected from all four studies were compared, we found genomic hotspot regions that contained multiple gene hits across diverse strains, and seven genes or gene regions that were found in at least two sequence types (Fig. 1B). The overlap across these diverse genomic studies suggests that polymorphisms in the identified genomic regions are universal and likely influencing disease outcome irrespective of isolate genetic background. Further studies of the biological significance of polymorphisms in these universal hotspots with larger and more diverse populations of clinical isolates are needed to determine whether these regions are simply recombination hotspots more prone to mutation or whether polymorphisms in this region are biologically significant and under selection.
We additionally leveraged the mouse inhalation model of cryptococcal meningitis to reduce host genetic, environmental, and fungal burden variability. We also used a set of closely related ST93 clinical isolates to further reduce pathogen genetic variation. Seven of the 32 genes found in this highly controlled and low variability GWAS were in the hotspot regions identified in the GWAS meta-analysis, further suggesting that decreasing variability increases the chances of identifying genes important in virulence. Importantly, by reducing variability we were able to easily differentiate SNPs with impact and traits associated with virulence. The ST93A GWAS, with the lowest variability, also had the highest proportion of conserved SNPs found across multiple studies. We believe these conserved SNPS are far more likely to have a universal role in virulence. With this variability reduction, we were able to identify a previously unknown network of genes in C. neoformans that apparently impact virulence and host IFNγ production.
The 38 ST93 strains used in this study are closely related, yet we were able to identify four distinct disease manifestations in mice, including an unusual hypervirulent phenotype associated with high IFNγ production. The association between IFNγ production and hypervirulence was surprising, as IFNγ is typically associated with improved patient outcome33,34. When we depleted IFNγ, we saw no change in virulence but observed an increase in dissemination. While inconclusive, these data could suggest that while IFNγ is necessary to control C. neoformans during infection, as shown in previous studies33,34, too much IFNγ may be detrimental. In this scenario, IFNγ has a Goldilocks effect with too little resulting in an immune response that is unable to control the infection whereas too much generates a destructive inflammatory response. Another possibility is that IFNγ may work in concert with other immune responses during hypervirulence. The link between IFNγ and virulence in clinical isolates is very intriguing and should be more extensively studied in future investigations.
The other hypervirulent clinical isolate, UgCl541, also had increased dissemination and produced a notable type 2 immune response. Type 2 immune responses are known to be ineffective at controlling C. neoformans infections and can enhance disease32,50. Overall, these data suggest that hypervirulence could be the result of either an uncontrolled immune response or an infective immune response leading to unchecked dissemination.
When we more closely examined the genes associated with IFNγ production in the mouse model, we found that deletion mutants in the KN99α genetic background had variable phenotypes. A total gene deletion from the KN99α reference strain — which is distantly related to the ST93 lineage5—is not a perfect comparison to a SNP in a clinical isolate, as that SNP may be impacting the gene in a variety of ways, yet it is a good first step to understanding the role of the gene in a known lab strain. Two deletion mutants altered IFNγ production (itr4Δ, Δ07528), two mutants had altered virulence (phs1Δ, itr4Δ), and five of the six genes analyzed had a capsule phenotype (phs1Δ, Δ04101, Δ05329, itr4Δ, Δ07528). This link between IFNγ production and capsule production has not been previously identified, yet cell surface changes such as capsule structure and secretion are known to impact host immune recognition42,51 and can cause a detrimental neutrophil-mediated response52. The putative role of our IFNγ-associated genes in capsule size suggests that the IFNγ phenotype could be the result of polymorphisms causing capsule remodeling that then leads to altered host recognition and ultimately an uncontrolled type 1 response. It is also possible that the IFNγ change is due to cell surface variations, though these are challenging to detect due to capsule structure that prevents proper in vitro labeling.
One of the IFNγ-associated genes, ITR4, is an inositol sensor (Wang and Xue, unpublished data). Deletion of ITR4 enhanced IFNγ production, overexpression reduced IFNγ production, and corresponding changes in capsule size were also observed. These data led us to conclude that inositol sensing through ITR4 may influence the capsule surface and subsequent immune response. Our RNA-seq data shows that ITR4 deletion influences the expression of some, but not all, of the other IFNγ-associated genes. Thus, ITR4 only interacts with a subset of these genes (Fig. 8A). In concert, our SNP prevalence analysis shows that hypervirulence and IFNγ production is associated with accumulation of polymorphisms across all the IFNγ-associated genes (Fig. 8B). These data lead us to propose that hypervirulence due to increased IFNγ production is caused by polymorphisms in a network of inositol-associated genes and others that function in parallel pathways. These data highlight the utility of combining pathogen GWAS with classical gene deletion studies to identify and characterize networks of genes related to virulence.
GWAS studies are highly variable and often noise can mask the gene networks that influence virulence and immune response. By reducing variability using a closely related population of isolates, mouse-models, and performing a meta-analysis of recent GWAS, we identified previously concealed genes and gene networks that can alter virulence. Here, we build the framework for developing future genetic and genomic studies that are definitively linked to the biology and virulence outcomes of pathogenic microbes by pairing an animal model with a closely related strain set. Ultimately this study demonstrates that for future pathogen GWAS limiting variation can be beneficial for identifying and understanding the biology underlying virulence outcomes in pathogenic microbes.
Methods
Ethics statement
Animal experiments were done in accordance with the Animal Welfare Act, United States federal law, and NIH guidelines. Mice were handled in accordance with guidelines defined by the University of Minnesota Animal Care and Use Committee (IACUC) under protocols 1908A37344, 2207A40205, and 2104A39016.
Strain selection
UgCl isolates used in this study were collected in Uganda as part of the Cryptococcal Optimal ART Timing (COAT) trial53. Single colony purified strains were obtained and sequenced with short reads in ref. 20. Sequence data is housed in the BioProject ID PRJNA549026. SNPs and INDELS were as identified previously20. In short, variant calling for each strain was adapted from the best practices described for the Genome Analysis Toolkit (GATK v3.3.0)54–56. KN99α57 was used as a control for mouse and in vitro phenotype experiments and the H99 reference genome was used for genomic analyses58. Strains were maintained as glycerol stocks at −80 °C and grown overnight on yeast peptone dextrose (YPD) plates with 0.04 g/l chloramphenicol.
Generation of the itr4∆ mutant, complemented strain, and ITR4 overexpression strain
To generate the itr4Δ mutant, the 1 kb 5′ upstream and 3′ downstream fragments of the ITR4 gene were amplified from KN99a genomic DNA with the primer pair CX2281 and CX2282, and the pair CX2283 and CX2284, respectively (Table S6). The dominant selectable marker (NEO) was amplified with the M13 primers (CX5 and CX6) from plasmid pJAF1. Each target gene replacement cassette was generated by overlap PCR with primers CX2281 and CX2284. Purified overlap PCR products were precipitated onto 10 μl of gold micro-carrier beads (0.6 μm; Bio-Rad) and into strain KN99a with biolistics. Stable transformants were selected on YPD medium containing G418 (200 mg/l). To screen for mutants, diagnostic PCR was performed by analyzing the 5′ junction of the disrupted mutant alleles with primers CX1009 and JH8994 and the 3′ junction of the disrupted mutant alleles with primers CX249 and CX2285 (Table S6). Positive transformants were identified by PCR screening with primers CX1010 and CX1011.
To generate complemented strains of the itr4Δ mutant, a genomic DNA fragment that contained a 1.5 kb upstream promoter region, the ITR4 ORF, and its 500-bp downstream region was amplified by PCR with primers CX1419/CX1420. The ITR4 PCR fragment was inserted into the BamHI site of vector pJAF1 using in-fusion cloning. The construct carrying ITR4 fragment was biolistically transformed in itr4Δ cells to generate the complemented strain. Phenotypic assays were performed to identify transformants in which the itr4Δ phenotype was complemented. The selecting colonies were further confirmed by PCR.
To generate ITR4 overexpression strain, the ITR4 full-length ORF genomic DNA was amplified with primers CX1800/CX1801 (Table S6) and cloned into the BamHI site of vector pCXU200 containing the Cryptococcus actin promoter to generate a plasmid which contains the ITR4:mCherry DNA cassette. Linearized plasmid was introduced into itr4Δ mutant strain to generate overexpression strain that express ITR4:mCherry protein.
Mouse model virulence studies
Five to ten 6-week-old A/J mice from Jackson laboratories were infected intranasally with 5 × 104 cells of each C. neoformans UgCl strain over the course of 12 experiments. Female mice were used for all experiments due to the length of the studies (survival was analyzed through 100 days post infection). Hormone fluctuations in older male mice are known to influence Cryptococcus pathogenesis59. Mice were kept on a 14:10 light cycle with lights turning on at 6 AM and off at 8PM, temperature maintained at 68–74 °F, and humidity at 30–70%.
Each of the 12 mouse experiments included a KN99α control that was used to normalize survival data across the experiments. Mice were weighed daily from day 14 to day 100. Mice were sacrificed with CO2 inhalation when they reached a terminal endpoint: loss of 20% body weight, loss of 2 g over 2 days, or visible symptoms of meningitis such as a domed head, paralysis, and disorientation; any surviving mice were sacrificed on day 100. Lungs and brains were removed from each mouse at sacrifice, homogenized in 4 ml or 2 ml phosphate buffered saline (Gibco, Waltham, MA), respectively, and plated on YPD plates with 0.04 g/l chloramphenicol. Plates were counted for colony forming units (CFU) after 2 days incubation at 30 °C to determine lung and brain fungal load. Terminal fungal burden was compared to KN99α with a Kruskal–Wallis nonparametric test with Dunn’s multiple comparison correction. When the median survival was undefined (i.e., latent isolates), only day 100 CFUs were analyzed. The median survival of each clinical isolate was calculated with Kaplan–Meier survival curves. Survival curves were normalized to the median survival of the KN99α control for each of the 12 experiments. KN99α median survival ranged between 17 and 23 days, with a mean survival of 21 days.
“Hypervirulent” isolates were defined as having a median survival of at least 2 days less than the median survival of the KN99α control. “CNS typical virulence” isolates were defined as having a median survival equal to or greater than KN99α and high CFUs in the brain. “Non-CNS typical virulence” isolates were defined as having a median survival equal to or greater than KN99α with at least 1/3rd of the mice succumbing to infection with fewer than 500 CFUs in the brain. Median survival was unable to be calculated for “Latent” isolates due to minimal mortality.
in vitro phenotype assays
For in vitro phenotype assays, each isolate was grown on solid YPD supplemented with various chemical stressors, as previously described12,60. In brief, each isolate was grown overnight in YPD broth, counted on a hemocytometer, and diluted to 106 cells/ml. Serial dilutions were performed to generate cultures with 106 through 101 cells/ml. 20 µl of each cell dilution were spot plated on YPD or YPD supplemented with: 1.5 M NaCl (Avantor, Radnor, PA), 0.5% Congo red (Chem-Impex International Inc., Wood Dale, IL), 0.06% Sodium Dodecyl Sulfate (SDS) (Sigma-Aldrich, Saint Louis, MO), 1 mg/ml caffeine (Ward’s Science, Rochester, NY), and 1 mg/ml Calcofluor white (Sigma-Aldrich, St. Louis, MO). Alkaline pH plates were made by adding 150 mM HEPES buffer (Fisher Bioreagents, Fair Lawn, NJ) to 1 l of YPD medium and adjusting the pH to 8.15 with NaOH prior to autoclaving. All inoculated plates were incubated at 30 °C for 2 days, except for the high temperature growth plates that were incubated at 37 °C.
Urease and melanin production, in vitro capsule size, and ability to form titan cells were also tested for each clinical isolate. Urease production was analyzed using Christensen urea agar61. Dilutions were plated as described above, incubated for 3 days, then relative urea production at each dilution was determined using numerical scores (−1 = no urease was produced at the 104 dilution, 0 = urease was produced at 104 dilution, 1 = urease was produced at the 103 dilution). Melanin production was measured using Niger seed and L-DOPA agar, as previously described12. Melanin plates were grown at 30 °C and 25 °C. Each isolate was compared to KN99α and given numerical scores based on color (0 = no color, 1 = tan, 2 = light brown, 3 = dark brown, 4 = same as KN99α, 5 = darker than KN99α for Niger Seed and 0 = no color, 1 = slightly gray, 2 = light gray, 3 = dark gray, 4 = same as KN99α, 5 = darker than KN99α for L-DOPA). In vitro, capsule induction and titan cell formation were performed as previously described62. In brief, cells were grown overnight in liquid YNB with amino acids (Sigma, Y1250) diluted to 106 cells/ml and grown for 3 days in liquid 0.5 ml sterile PBS supplemented with 10% fetal bovine serum (FBS) and 5% CO2. Cells were fixed in formaldehyde, stained with India ink, and imaged using a ZEISS Axioskop microscope using differential interference contrast (DIC). Capsule and cell body size were measured for at least 200 cells per strain. Titan cells were defined as having a cell body larger than 10 µm. The total cell was measured, then the cell body. The cell body was subtracted from the total cell to generate capsule size data. For each isolate, capsule size, cell body width, and total cell width were compared to KN99α and given a numeric score, according to size increase (−2 = 2.5 µm, −1 = −1.5 µm, 0 = +/- less than 1.5 µm, 1 = +1.5 µm, 2 = +3 µm, 3 = +4.5 µm).
For the itr4Δ mutant studies, C. neoformans KN99α, itr4Δ mutant, ITR4 overexpression strain (itr4Δ:ITR4OE) and ITR4 complemented strain (itr4Δ:ITR4) were grown overnight at 30 °C in YPD liquid medium. Cells were collected, washed twice with ddH2O, and suspended in ddH2O with a final concentration of 107 cells/ml. 30 µl of suspension of each strain was incubated on Dulbecco modified Eagles (DME) medium at 37 °C for three days for assessing capsule production. Capsule sizes and cell body sizes of more than 100 cells were measured and calculated.
in vivo immune response
To measure cytokine production during infection, five A/J mice were Infected intranasally with 5 × 104 cells of each C. neoformans UgCl strain over the course of 11 experiments. Mice were weighed daily from day 14 to either day 17 or day 21. Mice were sacrificed when they reached a terminal endpoint (see above) or at 17–21 days. It was confirmed that there are no significant differences between cytokine production in the lungs of mice euthanized at days 17, 19, 20, and 21 days post infection. Lungs and brains were isolated and added to 4 ml or 2 ml PBS (Gibco) on ice, respectively. Each organ was homogenized and the homogenate split in half. Half the homogenate was plated on YPD-chlorophenol to determine CFUs. The remaining half of the homogenate was combined with protease inhibitor cocktail (Roche, Basal, Switzerland) on ice and centrifuged for 10 min at 9188 × g. The supernatant was transferred to a fresh tube and flash frozen with liquid nitrogen. The supernatant was stored at −80 °C until cytokine analysis. Briefly, 25 µl of supernatant was added to the Mouse Th1/Th2 Cytokine 11-Plex Mouse ProcartaPlex™ Panel (Thermo Fischer Scientific, Waltham, MA) and analyzed per the manufacturer’s instructions on a Luminex MAGPIX (Luminex Corportation, Austin, TX). Each of the mouse experiments included a KN99α control. Cytokines for each clinical isolate were compared to a naïve mouse control using a non-parametric Kruskal–Wallis analysis, with Dunn’s multiple comparison correction. ROUT outlier analysis was used to identify and remove outliers. Fold change from naïve was determined for each strain. Heat map clusters were generated using Heatmapper63 with the average linkage clustering method and Euclidean distance measurement. Four representative isolates were chosen from each disease manifestation classification that aligned with the overall trend of disease manifestation and immune response.
IFNγ depletion
30 A/J mice were intranasally infected with 5 × 104 cells of the C. neoformans clinical isolate UgCl422 and five with KN99α, as previously described. 15 of the mice infected with UgCl422 were injected bi-weekly with 500 μg of anti–IFN-γ (Bio X Cell, Lebanon, New Hampshire, clone R4-6A2) or its isotype control (Bio X Cell, clone HRPN) twice a week for a total of eight injections, including one at day 1 after infection. Five mice from each group were sacrificed on day 17 for cytokines; the remaining mice were sacrificed at terminal endpoint as previously described. Cytokines levels were determined as previously described and significance determined using a Mann-Whitney non-parametric T-test.
Histopathology
Three mice were infected with 5 × 104 cells of KN99α, UgCl447, UgCl357, UgCl538, and UgCl422 and were sacrificed at 17 days post infection, as described previously. Whole lungs were collected and fixed in 10% neutral-buffered formalin (Fisher Scientific, Waltham, MA). Fixed organs were paraffin-embedded, sectioned, slide mounted, and stained at the Pathology Services Core at University of North Carolina-Chapel Hill. Slide mounted sections were stained with hematoxylin and eosin (H&E) for standard histological visualization or with immunohistochemical antibodies for CD4 (1:1000), CD8 (1:100), iNOS (1:175), CD68 (1:100), and EPX (1:1000). Digital image analysis was performed using QuPath (https://qupath.github.io) and Aperio ImageScope (Leica Biosystems, Nußloch, Germany).
Principal component analysis
The variants used in the PCA were those previously identified as effect variants20. A custom Python script was used to transform the genotype data into a PLINK binary format. Variant sites with a minor allele count of less than three were excluded from the dataset. The first 10 principal components were then calculated from the filtered genotype data with PLINK 1.964. A scree plot was used to determine the number of principal components that should be included as covariates to the association analyses: an “elbow” in the proportion of variance explained by each principal component was chosen as the threshold of inclusion. Isolates were clustered using K-means clustering and Euclidean distance to determine the distance between points. Gap statistic65 and silhouette score66 were used to compute the optimal number of clusters. PC1 and PC2 plot was generated with the color of the points corresponding to their K = 3 assignments.
Genome wide association study
A total of 31 phenotypes were analyzed for genotype-to-phenotype associations with a GWAS framework. Phenotypes were divided into two categories, ordinal categorical and quantitative (Supplementary Data 1). The ordinal categorical variables were analyzed with a proportional odds logistic mixed model (POLMM)35 implemented in the ‘GRAB’ package for R. The quantitative variables were analyzed using a mixed linear model implemented in the GEMMA software package36. A total of 562 variants survived the minor allele count filter and were used in the association analyses. The first two principal components of the genotype data were used as covariates to control for relatedness or population structure in the sample. Significant variants were identified by likelihood ratio test P values < 0.05 after a Bonferroni correction, where the number of variants (562) was used as the number of independent hypothesis tests.
Deletion mutant studies
15 6-week-old A/J mice were infected intranasally with 5 × 104 cells of each deletion strain. CNAG_05664, CNAG_04101, CNAG_05329, and PHS1Δ were acquired from the Hiten Madhani strain deletion library67. CNAG_07528 was generated in the Heitman lab and ITR4Δ was generated as described above. Ten mice were used for survival infections, as described above. The remaining 5 mice were used to determine cytokine response, as described above.
in vivo cell-size and capsule imaging
Lungs and brains were removed from each mouse at sacrifice and homogenized in 4 ml or 2 ml phosphate buffered saline (Gibco), respectively. Organs were transferred to a tube containing 1 mg/ml Collagenase Type 1 (Gibco) and incubated for 1 h at 37 °C. Cells were washed 3x in 0.05% SDS and filtered using a 70 µm filter. Cells were fixed in 3.7% formaldehyde and stained with India ink. Capsule and cell measurements were performed as described above, using 50 cells from the lungs of 3 mice each and at least 50 Cryptococcus cells from the brain, when successful.
RNA-seq and qPCR
C. neoformans cells of KN99α and itr4∆ mutant strains were cultured overnight at 30 °C in YPD liquid medium and then inoculated in triplicate in YNB medium containing 1% glucose and 1% inositol and grown for 4 h. The cells were collected and total RNA extracted using TRIzol (Invitrogen) and purified with NucleoSpin RNA Clean-up (MACHEREY-NAGEL, New Hampshire), following the manufacturer’s instructions. The purified RNAs were used as templates for PCR amplification with primers of the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) to determine potential genomic DNA contamination. Purified RNAs were quantified using a Nanodrop (Fisher Scientific).
For the RNAseq, cDNA library preparation, Illumina sequencing, and data analysis were performed by Novogene (Sacramento, CA). Sequencing was performed using Illumina NovoSeq 6000 to generate 150 bp paired-end reads. Approximately 50 million raw reads were generated. Adapter and low-quality reads, or reads with reads with uncertain nucleotides constitute more than 10% of either read (N > 10%), and reads with low-quality nucleotides (base quality less than 5) constitute more than 50% of the read68, were removed. Clean reads were mapped to the annotated genome of C. neoformans H99 (NCBI:txid235443) using HISAT2 software (version 2.0.5)69. Differential expression analyses were conducted using the DESeq2 package (version 1.20.0)70. Significance was determined using multiple unpaired T-tests with two stage linear step-up. The RNAseq dataset is available in the NCBI database under BioProject ID PRJNA1142147.
For the qPCR, first-strand cDNAs were synthesized using a SMARTScribe Reverse Transcriptase (Takara Bio, San Jose, CA, USA) following the manufacturer’s instructions. Gene expression was analyzed using TB Green Advantage qPCR Premix (Takara Bio). Primer efficiency was determined by serially diluting the cDNA and monitoring DNA amplification by real-time PCR. Gene expression levels were normalized using the endogenous control gene GAPDH, and the relative levels were determined using the comparative threshold cycle (CT) method. Real-time PCRs were performed using AriaMx Real-Time PCR system (Agilent, Santa Clara, CA, USA) (Table S6). The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis.
Statistics and reproducibility
Sample sizes are noted in the figure legends; mouse survival studies were performed with either 5 or 10 mice per isolate and all analyses were done with at least 3 biological and technical replicates. Data were processed using GraphPad Prism 9 software. ROUT outlier analysis was performed to remove outliers from the cytokines and fungal burden in the organs. While all the cytokines were run in technical triplicates, if one replicate from the triplicates was substantially different from the other two replicates across all the cytokines, that replicate was excluded. IL-2 results were excluded because IL-2 was not significantly increased in any of the clinical isolates we studied. Mouse data was excluded if there was a subjective reason for euthanasia (ie-mouse in distress), but no fungal burden in lungs or brain. Mouse data was also excluded if the mouse died for reasons unrelated to fungal infection (ie- tumor). Mechanisms of exclusion were determined in advance, with the exception of IL-2. We opted to exclude IL-2 once we saw that it was universally not significant. The experiments were not randomized or blinded as we used clinical isolates that had never been used before and had no expectation for outcome.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
Funding was provided by National Institutes of Health grants F31AI148047 to KMJ, R21NS108715, R01NS118538, and R01AI134636 to KN, and R01AI123315 and R01AI155647 to CX. We are appreciative and grateful to the patients and their families who participated in Cryptococcus-related clinical trials and research studies to enhance our understanding of cryptococcosis and improve treatment of future patients. We are immensely thankful for the sustained effort put forth by all healthcare professionals providing care to these patients with cryptococcal disease. We thank the Heitman lab for provided us with the CNAG_07528 deletion strain.
Author contributions
K.M.J. performed most experiments, data analysis, and data interpretation. T.J.Y.K. and K.D.K. performed bioinformatic analysis. J.B., Y.W., M.D., P.K., G.H., J.M.Y., S.R.F., L.M., A.G., and R.B.B. contributed to experiments and data analysis. M.D., P.T., T.J.Y.K., and C.X. provided critical input and supported data interpretation. D.B.M. provided critical clinical isolates. C.X. and Y.N. provided critical strains. K.N. supervised the study, performed data interpretation, and acquired funding. K.M.J and K.N. wrote the paper with input from all co-authors. All authors read and approved the paper.
Peer review
Peer review information
Nature Communications thanks Rebecca Drummond, Jason Sahl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The RNAseq data generated in this study have been deposited in the NCBI BioProject database under accession code PRJNA1142147. The original sequence data is housed in the BioProject ID PRJNA549026. Source data are provided with this paper.
Code availability
The bioinformatic pipeline scripts are available through GitHub at https://github.com/TomJKono/C_neoformans_Association and is deposited in Zenodo with the following 10.5281/zenodo.1403901771.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54729-6.
References
- 1.Jackson K. M., Ding M., Nielsen K. Importance of clinical isolates in Cryptococcus neoformans research. J. Fungi9, 364 (2023). [DOI] [PMC free article] [PubMed]
- 2.Maziarz, E. K. & Perfect, J. R. Cryptococcosis. Infect. Dis. Clin. N. Am.30, 179–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajasingham, R. et al. The global burden of HIV-associated cryptococcal infection in adults in 2020: a modelling analysis. Lancet Infect. Dis.22, 1748–1755 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajasingham, R. et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis.17, 873–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashton, P. M. et al. Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat. Commun.10, 2035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litvintseva, A. P., Thakur, R., Vilgalys, R. & Mitchell, T. G. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics172, 2223–2238 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khayhan, K. et al. Geographically structured populations of Cryptococcus neoformans Variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS ONE8, e72222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira-Paim, K. et al. MLST-Based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV Patients in southeastern Brazil. PLoS Negl. Trop. Dis.11, e0005223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrade-Silva, L. E. et al. Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PLoS ONE13, e0193237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer, W. et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol.47, 561–570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altamirano S., Jackson K. M., Nielsen K. The interplay of phenotype and genotype in Cryptococcus neoformans disease. Biosci. Rep.40, BSR20190337 (2020). [DOI] [PMC free article] [PubMed]
- 12.Wiesner D. L., et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio3, e00196-12 (2012). [DOI] [PMC free article] [PubMed]
- 13.Beale, M. A. et al. Genotypic diversity is associated with clinical outcome and phenotype in Cryptococcal meningitis across southern Africa. PLoS Negl. Trop. Dis.9, e0003847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bive, B. Z. et al. Clinical epidemiology and high genetic diversity amongst Cryptococcus spp. isolates infecting people living with HIV in Kinshasa, Democratic Republic of Congo. PLoS ONE17, e0267842 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponzio, V. et al. Genotypic diversity and clinical outcome of cryptococcosis in renal transplant recipients in Brazil. Emerg. Microbes Infect.8, 119–129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J. et al. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis.14, 755–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chau, T. T. et al. A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam - high prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect. Dis.10, 199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day, J. N. et al. Most cases of cryptococcal meningitis in HIV-uninfected patients in Vietnam are due to a distinct amplified fragment length polymorphism-defined cluster of Cryptococcus neoformans var. grubii VN1. J. Clin. Microbiol.49, 658–664 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day, J. N. et al. Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLoS Negl. Trop. Dis.11, e0005628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerstein A. C., et al. Identification of pathogen genomic differences that impact human immune response and disease during Cryptococcus neoformans infection. mBio10, e01440-19 (2019). [DOI] [PMC free article] [PubMed]
- 21.Sephton-Clark, P. et al. Genomic variation across a clinical Cryptococcus population linked to disease outcome. mBio13, e0262622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agustinho D. P. et al. Unbiased discovery of natural sequence variants that influence fungal virulence. Cell Host Microbe (2023) 10.1016/j.chom.2023.10.002. [DOI] [PMC free article] [PubMed]
- 23.Billmyre R. B. et al. 2024. Saturation transposon mutagenesis enables genome-wide identification of genes required for growth and fluconazole resistance in the human fungal pathogen Cryptococcus neoformans. Preprint at bioRxivhttps://pubmed.ncbi.nlm.nih.gov/39131341/ (2024).
- 24.Basenko E. Y. et al. FungiDB: An integrated bioinformatic resource for fungi and Oomycetes. J. Fungi4, 39 (2018). [DOI] [PMC free article] [PubMed]
- 25.Ding, M. et al. Use of clinical isolates to establish criteria for a mouse model of latent. Front. Cell Infect. Microbiol.11, 804059 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lofgren, S. et al. Differences in immunologic factors among patients presenting with altered mental status during cryptococcal meningitis. J. Infect. Dis.215, 693–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meya D. B. et al.. Monocyte phenotype and IFN-γ-inducible cytokine responses are associated with cryptococcal immune reconstitution inflammatory syndrome. J. Fungi3, 28 (2017). [DOI] [PMC free article] [PubMed]
- 28.Meya, D. B. et al. Cellular immune activation in cerebrospinal fluid from ugandans with cryptococcal meningitis and immune reconstitution inflammatory syndrome. J. Infect. Dis.211, 1597–1606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musubire, A. K. et al. Blood neutrophil counts in HIV-infected patients with cryptococcal meningitis: association with mortality. PLoS ONE13, e0209337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulware, D. R. et al. Human immune response varies by the degree of relative cryptococcal antigen shedding. Open Forum Infect. Dis.3, ofv194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scriven, J. et al. Early ART after cryptococcal meningitis is associated with cerebrospinal fluid pleocytosis and macrophage activation in a multisite randomized trial. J. Infect. Dis.212, 769–778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiesner, D. L. et al. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog.11, e1004701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarvis, J. N. et al. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS26, 1105–1113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wormley, F. L. Jr., Perfect, J. R., Steele, C. & Cox, G. M. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect. Immun.75, 1453–1462 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi, W. et al. Efficient mixed model approach for large-scale genome-wide association studies of ordinal categorical phenotypes. Am. J. Hum. Genet.108, 825–839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet.44, 821–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panagiotou, O. A. & Ioannidis, J. P. Genome-Wide Significance Project What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J. Epidemiol.41, 273–286 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Alanio, A., Desnos-Ollivier, M. & Dromer, F. Dynamics of Cryptococcus neoformans-macrophage interactions reveal that fungal background influences outcome during cryptococcal meningoencephalitis in humans. mBio2, e00158-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luberto, C. et al. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Invest.112, 1080–1094 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue C. et al. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio1, e00084-10 (2010). [DOI] [PMC free article] [PubMed]
- 41.Jin, J. H. et al. Genome-wide functional analysis of phosphatases in the pathogenic fungus Cryptococcus neoformans. Nat. Commun.11, 4212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadevall, A. et al. The capsule of Cryptococcus neoformans. Virulence10, 822–831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y. et al. Inositol metabolism regulates capsule structure and virulence in the human pathogen Cryptococcus neoformans. mBio12, e0279021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Do, E. et al. Leu1 plays a role in iron metabolism and is required for virulence in Cryptococcus neoformans. Fungal Genet Biol.75, 11–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, F. et al. Reciprocal adaptation of rice and Xanthomonas oryzae pv. oryzae: cross-species 2D GWAS reveals the underlying genetics. Plant Cell33, 2538–2561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genestet, C. et al. Mycobacterium tuberculosis genetic features associated with pulmonary tuberculosis severity. Int J. Infect. Dis.125, 74–83 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Martin R. M. et al. Identification of pathogenicity-associated loci in Klebsiella pneumoniae from hospitalized patients. mSystems3, e00015-18 (2018). [DOI] [PMC free article] [PubMed]
- 48.Sassi, M. et al. Forecasting Staphylococcus aureus infections using genome-wide association studies, machine learning, and transcriptomic approaches. mSystems7, e0037822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang, E. H., Kim, J. S., Yu, S. R. & Bahn, Y. S. Unraveling capsule biosynthesis and signaling networks in Cryptococcus neoformans. Microbiol. Spectr.10, e0286622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiesner, D. L. et al. Regulatory T cell induction and retention in the lungs drives suppression of detrimental type 2 Th cells during pulmonary cryptococcal infection. J. Immunol.196, 365–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Meara, T. R. & Alspaugh, J. A. The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol Rev.25, 387–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Meara T. R., Holmer S. M., Selvig K., Dietrich F., Alspaugh J. A. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio4, e00522-12 (2013). [DOI] [PMC free article] [PubMed]
- 53.Boulware, D. R. et al. Timing of antiretroviral therapy after diagnosis of cryptococcal meningitis. N. Engl. J. Med370, 2487–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet.43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res.20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform.43, 11 10 1–11 10 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen, K. et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun.71, 4831–4841 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janbon, G. et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet10, e1004261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lortholary, O., Improvisi, L., Fitting, C., Cavaillon, J. M. & Dromer, F. Influence of gender and age on course of infection and cytokine responses in mice with disseminated Cryptococcus neoformans infection. Clin. Microbiol Infect.8, 31–37 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Mukaremera, L. et al. The mouse inhalation model of Cryptococcus neoformans infection recapitulates strain virulence in humans and shows closely related strains can possess differential virulence. Infect. Immun.87, e00046–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christensen, W. B. Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J. Bacteriol.52, 461–466 (1946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dambuza, I. M. et al. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog.14, e1006978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babicki, S. et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res.44, W147–W153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tibshirani, R., Walther, G. & Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc.63, 411–423 (2001). [Google Scholar]
- 66.Rousseeuw, P. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math.20, 53–65 (1986). [Google Scholar]
- 67.Chun, C. D. & Madhani, H. D. Applying genetics and molecular biology to the study of the human pathogen Cryptococcus neoformans. Methods Enzymol.470, 797–831 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol.20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol.11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kono, T. 2024. Single nucleotide polymorphisms are associated with strain-specific virulence differences among isolates of Cryptococcus neoformans. TomJKono/C_neoformans_Association: 1.0-pub (1.0-pub). Zenodo. 10.5281/zenodo.14039018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The RNAseq data generated in this study have been deposited in the NCBI BioProject database under accession code PRJNA1142147. The original sequence data is housed in the BioProject ID PRJNA549026. Source data are provided with this paper.
The bioinformatic pipeline scripts are available through GitHub at https://github.com/TomJKono/C_neoformans_Association and is deposited in Zenodo with the following 10.5281/zenodo.1403901771.