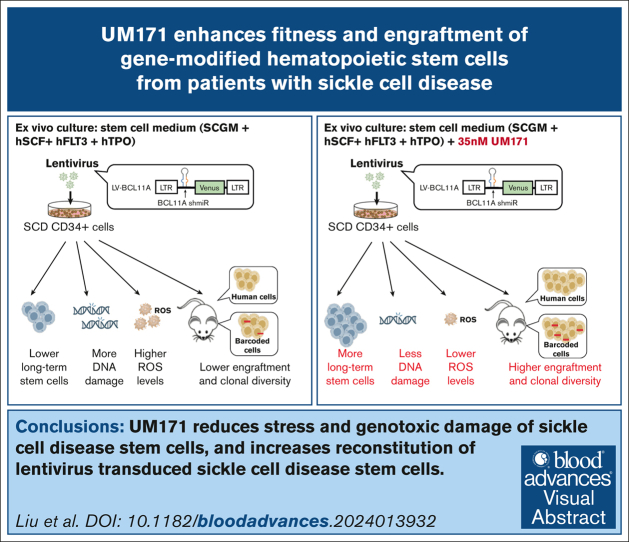
Key Points
-
•
UM171 reduces stress and genotoxic damage of SCD stem cells during ex vivo culture.
-
•
UM171 increases reconstitution of lentivirus-transduced SCD stem cells.
Visual Abstract
Abstract
Hematopoietic stem cell (HSC) transplantation with lentiviral vector (LVV)-transduced autologous cells has proven an effective therapeutic strategy for sickle cell disease (SCD). However, ex vivo culture or proliferative stress associated with in vivo reconstitution may amplify any underlying genetic risk of leukemia. We aimed to minimize culture-induced stress and reduce genomic damage during ex vivo culture and enhance stem cell fitness and reconstitution of SCD CD34+ cells transduced with BCL11A shmiR-encoding LVV. UM171, a pyrimidoindole derivative, can expand normal HSCs during in vitro culture and has been shown to be safe and effective using umbilical cord blood. We examined the effect of UM171 during ex vivo LVV transduction of SCD HSCs. Culture of SCD CD34+ HSCs with UM171 during transduction reduced DNA damage and reactive oxygen species, decreased apoptosis, and was associated with increased numbers of immunophenotypically defined long-term HSCs. UM171 increased the engraftment of LVV-transduced human HSCs in immunodeficient mice and barcode tracing revealed increased clonal diversity of engrafting cells. In competitive transplantation assays, analysis of bone marrow showed that cells transduced in the presence of UM171 consistently outcompeted those transduced under control conditions. In summary, exposure of SCD peripheral blood CD34+ cells to UM171 during LVV transduction enhances stem cell fitness. These findings suggest manufacturing of genetically modified HSCs in the presence of UM171 may improve efficacy, safety, and sustainability of gene therapy using ex vivo approaches. BCL11A shmiR-encoding LVV is in clinical trials to treat SCD (NCT03282656), UM171 is in clinical trials to culture umbilical cord blood (NCT02668315).
Introduction
Treatment of sickle cell disease (SCD) remains a challenge in the landscape of genetic disorders, affecting millions worldwide. SCD is characterized by the production of abnormal hemoglobin that leads to acute and chronic pain, hemolytic anemia, and end-organ damage with significantly reduced life expectancy.1,2 Autologous gene therapy has emerged as a promising disease-modifying therapy with significant strides in recent years, including 2 recently approved commercial products using distinct approaches to treatment (Casgevy and Lyfgenia).3,4
Ex vivo gene therapy using autologous hematopoietic stem cells (HSCs) involves harvesting a patient’s HSCs, genetically modifying these cells to mitigate the erythrocyte cellular phenotype of sickle hemoglobin polymerization, and subsequently transplanting them back into the patient.5 Several different lentiviral vectors (LVVs)-based approaches have proven effective in clinical trials.3,6,7 However, the occurrence of myeloid leukemia and myelodysplastic syndrome seemingly unrelated to vector insertional mutagenesis has prompted further scrutiny and adjustments in the manufacturing process, emphasizing the critical importance of rigorous safety measures in advancing genetic therapies for complex diseases such as SCD.8, 9, 10 Notably, the occurrence of oncogenesis has highlighted the still undefined possibility of an underlying predisposition to cancer in patients with SCD that may be amplified with stress of ex vivo manipulation and in vivo reconstitution.11,12 In the present work, we used a lentiviral shmiR vector targeting BCL11A,6,13 a major repressor of gamma-globin expression, to induce fetal hemoglobin. We modified cell transduction processes targeting HSCs from patients with SCD in an attempt to reduce stress during the manufacturing process to increase stem cell fitness.
The recent discovery of small molecules stimulating the expansion of human HSCs in vitro, such as StemRegenin1 (SR1),14 fenretinide (4HPR),15 or enhancing engraftment, such as Prostaglandin E2,16 raises the possibility of their application in the setting of ex vivo cell manufacturing for genetic modifications. The pyrimidoindole derivative UM171 has been described as one of the most potent small molecules that stimulates HSC expansion in vitro.17 UM171 expands Endothelial protein C receptor (EPCR)+ cord blood (CB) HSCs with sustained short- and long-term repopulation potential18 and mobilized peripheral blood HSCs after lentiviral transduction19,20 and Cas9 gene editing.21 UM171 has been reported to target multiple cellular processes involved in HSC expansion, including self-renewal, differentiation, cell metabolism, cell cycle regulation, and epigenetic regulation.17,22 UM171 has been used in clinical trials to expand human CB-derived HSCs ex vivo for allogeneic transplantation.23
We hypothesized that UM171 could reduce stress on HSCs during ex vivo culture required for genetic manipulations and mitigate the risk of accumulated genomic damage during these cultures, and better HSC fitness and numbers should reduce repopulation stress during hematopoietic reconstitution. Our findings demonstrate that transient exposure of SCD CD34+ cells to UM171 improves transduction efficiency, stem cell fitness, and reconstitution of SCD CD34+ cells transduced with LVV. Our study suggests a practical approach to enhancing the safety of ex vivo manipulations necessary for any genetic therapy in SCD, including LVV gene therapy and gene editing, and may have applications in other diseases.
Materials and methods
Construction of lentivirus vector
The generation of LV-BCL11A (LV-LCR-mir223 BCL11A) vectors has been described previously,13,24 and the structure is shown in supplemental Figure 1A. The green fluorescent protein (GFP) transfer vector (pCCL.cPPT.PGK.MGMT-P140K.T2A.eGFP.pre) encodes a self-inactivating HIV-1–derived LVV genome consisting of a human phosphoglycerate kinase (PGK) promoter regulating GFP.25 A barcoded third-generation LVV library was generated by introducing an oligonucleotide containing 16 randomized nucleotides into the pCCL.PGK.MGMT-P140K.T2A.eGFP.pre plasmid upstream of the PGK promoter. The plasmid was digested with SphI and BsiWI, and the phosphorylated oligonucleotide 5′-CTACACGACGCTCTTCCGATCTCACCGGAGACGNNCACNNAGANNCTTNNCGANNCTANNGGANNCTTNNCGTCTCTTCGAAGATCGGAAGAGCACACGTCT-3′ was introduced by HiFi DNA cloning kit (New England Biolabs, Ipswich, MA) via a 21 nt compatible overhang, followed by transformation into ElectroMAX Stbl4 Competent Cells (Invitrogen, Waltham, MA),26,27 as shown in supplemental Figure 1B. Barcode diversity and distribution were confirmed by amplicon sequencing of plasmid maxi preparations.
Virus production and titration
Lentiviral supernatants were produced by transfection of third-generation lentiviral packaging plasmids (10 μg lentiviral transfer vector [LV-LCR-mir223 BCL11A or pCCL.cPPT.PGK.MGMT-P140K.T2A.eGFP.pre], 5 μg lentiviral gag-pol [pMDLg/pRRE],28 2.5 μg RSV-REV [pRSV-Rev],28 and 2.5 μg of VSVG [hCMV-VSVG]29) into HEK293T cells grown in 10 cm plates. Plasmids were mixed with 1 mL Dulbecco’s modified Eagle medium (Cytiva, Marlborough, MA) and 60 μL of 1 mg/mL linear polyethylenimine (Polysciences, Warrington, PA), incubated for 15 to 20 minutes at room temperature, and added to the culture dish. The medium was changed 7 hours later and viral supernatants were collected 48 hours after transfection, filtered through a 0.45 μm polyethersulfone filter (Corning, Corning, NY), and then concentrated by ultracentrifugation at 24 000 rpm for 2 hours in a Beckman XL-90 centrifuge with SW-28 rotor with swinging buckets. Infectious titers were determined on HEK293T cells by applying serial dilutions of vector supernatant followed by flow cytometric analyses 4 days later.
Transduction of SCD CD34+ cells
CD34+ HSCs from patients with SCD were isolated from unmobilized PB following a protocol approved by the Boston Children’s Hospital Institutional Review Board and an informed patient consent. The CD34+ HSCs were enriched using the Miltenyi CD34 Microbead kit (Miltenyi Biotec, Auburn, CA). CD34+ cells were prestimulated for 36 to 40 hours at 1 × 106 cells per mL in Stem Cell Growth Media (CellGenix, Portsmouth, NH) supplemented with human stem cell factor (hSCF, 100 ng/mL), human FMS-like tyrosine kinase 3 ligand (hFLT3L, 100 ng/mL), and human thrombopoietin (hTPO, 100 ng/mL), all from PeproTech (Rocky Hill, NJ) in the presence or absence of 35 nM UM171 provided from ExCellThera (Montreal, Canada), 500 nM StemRegenin1 (Stem Cell Technologies, Vancouver, Canada), or 4 μM 4HPR (MedChemExpress, Monmouth Junction, NJ). At the end of the prestimulation period, cells were then enumerated and transduced with the LVVs in presence of Poloxamer Synperonic F10830 at the indicated multiplicity of infection for 24 hours before downstream processing for in vivo or ex vivo studies.
Flow cytometry assay
After 3- or 7-day ex vivo culture, immunophenotyping of cells was performed by flow cytometry using a LSRII cytometer (BD Biosciences, Woburn, MA) and the following antibodies: CD34-PerCP/Cy5.5, CD90-APC, CD201-FITC, CD45RA-APC/Fire750, CD133-BV421, and CD38-PE/Cy5. Briefly, cells were stained with antibodies at 4°C for 30 minutes at dilutions recommended by the manufacturer. Then cells were washed with 1 mL phosphate-buffered saline (GIBCO, Grand Island, NY) and analyzed by flow cytometry.
DNA damage assay
After 3- or 7-day ex vivo culture, cells were washed in phosphate-buffered saline, pelleted, and then fixed and permeabilized using Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences), followed by staining with H2A.X-PE.
Vector copy number
After 7-day ex vivo culture, genomic DNA was extracted using the QIAGEN (Hilden, Germany) DNeasy protocol. Vector copy number (VCN) was assessed by quantitative reverse transcription polymerase chain reaction (PCR), performed with the use of TaqMan Fast Advanced Master Mix (Applied Biosystems, Waltham, MA) as per manufacturer’s instructions, and used the following primers and probes. The HIV Ψ RNA packaging signal (Psi), which is contained in lentivirus vector, was used as basis for quantitative PCR of the integrated vector in the genome (forward primer 5′-CAGGACTCGGCTTGCTGAAG-3′, reverse primer 5′-TCCCCCGCTTAATACTGACG-3′, probe FAM-5′-CGCACGGCAAGAGGCGAGG-3′), and the human Glycosyltransferase Like Domain Containing 1 gene (hGTDC1) as an internal reference standard (forward primer 5′-GAAGTTCAGGTTAATTAGCTGCTG-3′, reverse primer 5′-TGGCACCTTAACATTTGGTTCTG-3′, probe VIC-5′-ACGAACTTCTTGGAGTTGTTTGCT-3′) for comparison. A series of diluted plasmid containing both HIV Psi and hGTDC1 genes was used as standard curve to calculate the copies of GTCD1 and HIV in test samples. VCN was calculated as (copies HIV Psi/copiesGTDC1) × 2.
Cell cycle and reactive oxygen species expression
After 3- or 7-day ex vivo culture, cell cycle status was assessed by using the APC BrDU Flow Kit (BD Biosciences) followed by staining with Bromodeoxyuridine-Allophycocyanin and 7-aminoactinomycin D according to the manufacturer’s instructions. Reactive oxygen species (ROS) expression was analyzed by flow cytometry with CellROX Orange Flow Cytometry Assay Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions.
SCD CD34+ HSC transplant and flow cytometry analysis
All animal experiments were approved by the Boston Children’s Hospital Institutional Animal Care and Use Committee. NOD.Cg-KitW-41J Tyr+ Prkdcscid Il2rgtm1Wjl/ThomJ (NBSGW) mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Nonirradiated NBSGW female mice (4-6 weeks of age) were infused by retro-orbital injection with 0.2 × 106 LVV expressing GFP (LV-GFP) virus–transduced SCD CD34+ HSCs. For competitive repopulation experiments, equal numbers of cells (total of 0.2 × 106 cells per animal) from different transduction groups (LV-BCL11A virus–transduced SCD CD34+ cells with or without UM171) were mixed before transplantation into NBSGW mice. Cell mixtures were analyzed via flow cytometry to confirm equal contributions of both competitor cell fractions and readjusted if required. PB samples were collected at weeks 4, 8, 12, and 16 to measure engraftment by flow cytometry analysis of human CD45 and to determine gene-marked cells. At week 16, mice were euthanized, and bone marrow (BM) was isolated for human xenograft analysis. A portion of the BM cells were used for erythroid differentiation in vitro and for genomic DNA isolation. For flow cytometric analyses of BM, the following antibodies were used: hCD45-APC, mCD45-PE/Cy7, fixable viability dye eFluor 780 (Thermo Fisher Scientific), hCD235a-AF700, hCD33-PE, hCD19-BV605, hCD34-AF700, and hCD3-PerCP/Cy5.5 (BioLegend) at dilutions recommended by the manufacturer.
In vitro erythroid differentiation of CD34+ cells
The erythroid differentiation protocol used is based on a 3-phase protocol adapted from Giarratana et al.31 Briefly, the CD34+ cells were cultured in erythroid differentiation medium (EDM) consisting of Iscove-modified Dulbecco’s medium (CellGro, Manassas, VA) supplemented with 1% L-glutamine and 2% penicillin-streptomycin (Thermo Fisher Scientific), 330 μg/mL human holo-transferrin, 10 μg/mL recombinant human insulin and 2 IU/mL heparin (Sigma-Aldrich, Burlington, MA), 5% human solvent detergent pooled plasma AB (Rhode Island Blood Center, Providence, RI), and 3 IU/mL erythropoietin (Amgen, Thousand Oaks, CA). During phase 1 of expansion (days 0-7), CD34+ cells were cultured in EDM in the presence of 10–6 mol/L hydrocortisone (Sigma-Aldrich), 100 ng/mL hSCF (PeproTech), and 5 ng/mL hIL-3 (R&D Systems, Minneapolis, MN), as EDM-1. In phase 2 (days 7-11), the cells were resuspended in EDM supplemented with 100 ng/mL hSCF, as EDM-2. For phase 3 (days 11-18), the cells were cultured in EDM without additional supplements, as EDM-3.
Barcode recovery and data analysis
Genomic DNA from transplanted BM cells was extracted with the DNeasy Kit (Qiagen) and 200 to 500 ng DNA or the whole cell lysis from each cell population was used for barcode retrieval. PCR amplification (35 cycles) of the barcode region was performed using primers (forward 5′-ATCGATCACGAGACTAGCCTCG-3′, reverse 5′-CCAACCCCGTGGAATTCGATATC-3′) with Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA). After gel purification of the PCR products, samples were prepared for barcode sequencing. Postsequencing data were processed using custom Python code to identify barcoded clones. Data analysis and plot generation were performed using Prism (GraphPad Software, San Diego, CA).
Colony-forming assay
Hematopoietic colony-forming potential was assessed using a methylcellulose assay by mixing 1000 cells of interest with 1 mL H4435 Methocult (StemCell Technologies, Vancouver, Canada). Cell suspensions was then transferred to 35-mm dishes (StemCell Technologies) and cultured for 12 to 14 days at 37°C. Triplicates was performed for each sample. Hematopoietic colonies were counted and classified according to cellular morphology.
Statistical analysis
All the data are reported as mean ± standard deviation unless otherwise stated. Statistical analyses were performed using GraphPad Prism version 10.2 (GraphPad Software). The statistical significance between averages was established using the unpaired t test. When the statistical significance among 3 or more averages were evaluated, analysis of variance test was applied. All the statistical tests were 2-tailed; statistical significance differences are indicated with asterisks (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001), and N.S. denotes P > .05.
Results
UM171 enhances expansion of phenotypically defined SCD HSCs during LVV transduction
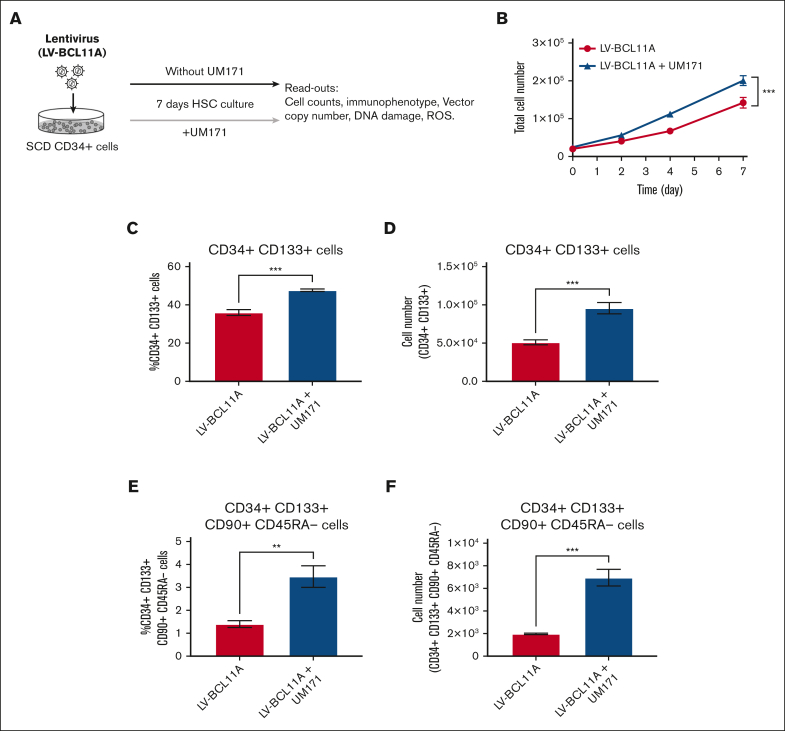
The small molecule UM171 has been validated as a novel and potent agonist for the renewal of human HSCs (supplemental Figure 2). In initial experiments, we sought to determine whether UM171 can preserve or expand HSCs from patients with SCD during ex vivo manipulation for gene modification. CD34+ cells obtained from the PB of patients with SCD were transduced with an LVV containing the BCL11A shmiR (LV-BCL11A) in the presence or absence of UM171 and cultured for 7 days (Figure 1A). Transduced cells were assayed for cell number, immunophenotype, VCN, DNA damage and ROS generation. In the presence of UM171, the expansion of transduced SCD CD34+ cells was significantly increased (Figure 1B). In additional analyses, we focused on various HSCs subsets defined by specific cell surface markers including CD34, EPCR, CD133, CD90, and CD45RA as shown in Figure 1C-F and supplemental Figures 3 and 4. Compared with the cells transduced with LV-BCL11A in the absence of UM171, cells transduced with LV-BCL11A + UM171 showed similar frequencies of live events and an increase in the frequencies of CD34+ CD133+ HSCs (47.7% vs 36.0%; P < .001), CD34+ CD133+ CD90+ CD45RA– long-term hematopoietic stem cells (LT-HSCs) (3.5% vs 1.4%; P < .01) (Figure 1C,E; supplemental Figure 5), and CD34+ EPCR+ cells (12.2% vs 0.38%; P < .0001) (supplemental Figure 3A). Differences in the absolute numbers of CD34+ CD133+ HSCs, CD34+ CD133+ CD90+ CD45RA– LT-HSCs, and CD34+ EPCR+ were even more pronounced (1.9-, 3.6-, and 45.4-fold expansions, respectively; Figure 1D,F and supplemental Figure 3B; P < .001). We also analyzed these populations 3 days after UM171 culture replicating our standard ex vivo transduction duration. The result showed a significantly higher CD34+ CD133+ CD90+ CD45RA– population. UM171 treated cells showed a trend toward higher CD34+ EPCR+ that was not significant (supplemental Figure 6). These findings indicate that UM171 promotes the ex vivo expansion of immunophenotypically defined LV-BCL11A-transduced CD34 HSCs and LT-HSCs from patients with SCD.
Figure 1.
UM171 enhances the in vitro expansion of CD34+ cells from patients with SCD. (A) Outline of in vitro manipulations and analysis. CD34+ cells from patients with SCD were prestimulated and transduced with LV-BCL11A in the presence or absence of UM171 and cultured 7 days for readouts. See “Material and methods” for details. (B) Total cell numbers of CD34+ cells transduced with LV-BCL11A in presence or absence of UM171. (C) The percentage of CD34+ CD133+ cells and (D) CD34+ CD133+ absolute cell numbers after transduction in the presence or absence of UM171. (E) The percentage of CD34+ CD133+ CD90+ CD45RA– cells and (F) absolute numbers of CD34+ CD133+ CD90+ CD45RA– after transduction in presence or absence of UM171. Data represent mean ± standard deviation (SD), n = 3, 3 independent experiments from cells of different patients with SCD. ∗∗P < .01; ∗∗∗P < .001.
UM171 enhances the efficiency of gene transfer
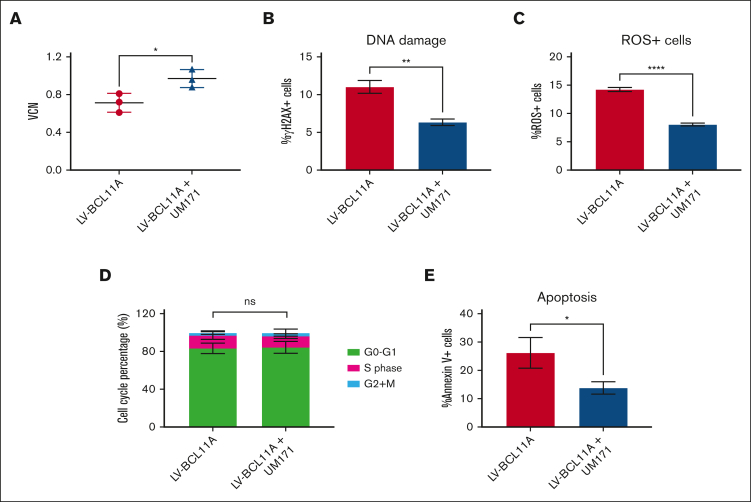
To investigate the impact of UM171 on LVV-mediated gene transfer specifically in SCD cells, VCN was determined by quantitative PCR after transduction and an additional 7-day culture period in cytokine-supplemented medium with or without UM171. As shown in Figure 2A, the presence of UM171 was associated with a modest but significant 1.4-fold increase in the VCN of transduced SCD CD34+ -derived cells compared with controls (0.97 vs 0.71; P < .05). Given that the LV-BCL11A contains the β-globin locus control region restricting expression of the BCL11A shmiR and Venus to erythroid cells, to facilitate additional analysis, we tested an alternative LV-GFP in all hematopoietic lineages. As shown in supplemental Figure 1B, the LV-GFP (pCCL.cPPT.PGK.MGMT-P140K.T2A.eGFP.pre) encodes a self-inactivating HIV-1-derived LVV genome consisting of a human PGK promoter driving expression of GFP. UM171 also significantly increased the transduction efficiency of LV-GFP in SCD CD34+ cells, as indicated by both the proportion of GFP+ cells and the VCN (supplemental Figure 7).
Figure 2.
UM171 enhances the efficiency of gene transfer into CD34+ from patients with SCD and reduces manufacturing stress in vitro. (A) Lentiviral VCN in CD34+ cells transduced with LV-BCL11A in the presence or absence of UM171 was measured by quantitative PCR. (B) DNA damage was assessed by the level of γH2AX staining. (C) Assessment of the level of ROS+ cells, (D) cell cycle, and (E) the percentage of Annexin V+ cells after transduction in the presence or absence of UM171. Readouts after 7 days in culture. Data represent mean ± SD, n = 3, 3 independent experiments from cells of different patients with SCD. ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.
To evaluate the effect of UM171 on the frequency and transduction rate of multilineage hematopoietic progenitor cells, we performed hematopoietic progenitor colony assays. We found that there is a significant increase in clonogenic cells, including burst-forming erythroid progenitor cells, granulocyte-macrophage colony-forming unit (CFU) progenitor cells, and multipotent granulocyte, erythroid, macrophage, and megakaryocyte CFU progenitor cells, as well as higher numbers of transduced colonies, in the LV-GFP + UM171 group compared with the LV-GFP group (supplemental Figure 8). These findings suggest that the expanded SCD CD34+ cells cultured with UM171 retain the capacity for multilineage differentiation. In summary, these initial experiments demonstrated that prestimulation, transduction, and culture of SCD HSCs in the presence of UM171 lead to substantial increases in the efficiency of LVV gene transfer while preserving multilineage differentiation capacity.
UM171 reduces stress associated with ex vivo manipulation of SCD CD34+
Given the potential toxicity associated with ex vivo cultures and cell expansion, we sought to assess the capacity of UM171 to mitigate genome damage and minimize stress during cell manufacturing. As shown in Figure 2B and supplemental Figure 9A, transduction with LV-BCL11A in the presence of UM171 was associated with a reduction in DNA damage as measured by γH2AX+ staining compared with the control cells (6.3% vs 11.0%; P < .01). In addition, the transduction in the presence of UM171 resulted in a significant decrease in ROS expressed cells (8.0% vs 14.2%, with UM171 vs without UM171; P < .0001) (Figure 2C; supplemental Figure 9B). We tested SCD CD34+ cells from different sources, including clinically used BM CD34+ cells and mobilized peripheral blood CD34+ cells from patients with SCD. In CD34+ cells from all of these sources, UM171 reduced DNA damage and ROS generation and increased total number of cells and CFU clones (supplemental Figure 10). In contrast, the addition of UM171 had no significant effect on the proportion of cells in the G0 to G1, G2 + M, and S phases of the cell cycle (Figure 2D) but the presence of UM171 led to a significant reduction in apoptosis in LVV-transduced SCD CD34+ cells (13.8% vs 26.2%; P < .05) (Figure 2E). These results collectively suggest that UM171 mitigates stress on HSCs from patients with SCD during ex vivo manipulations, increasing the efficiency of manufacturing and potentially enhancing the safety of the manufacturing process. We also compared UM171 with the compounds SR114 and 4HPR,15 which have been reported to support ex vivo HSC expansion. The result showed that UM171 performed superior to these compounds in terms of cell expansion and reduction of DNA damage, ROS levels, and apoptosis (supplemental Figure 11).
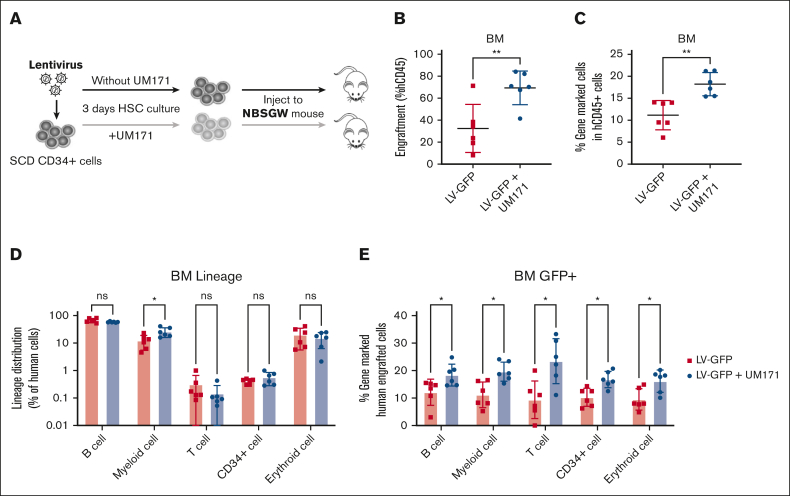
UM171 increases engraftment of transduced SCD CD34+ cells in xenografts
To validate the findings in immunophenotypically defined HSCs, we sought to assess the in vivo engraftment and differentiation capabilities of cultured HSCs. To evaluate this, we conducted xenotransplantation of transduced cells in NBSGW mice, as illustrated in Figure 3A. SCD CD34+ cells were transduced with or without UM171, and 200K cells were injected into NBSGW mice. Mice were bled every 4 weeks after transplantation and BM was collected at week 16 to analyze the engraftment of human cells. Engraftment was calculated as a percentage of human CD45+ cells in the total human and murine CD45+ cell populations. Cells transduced in the presence of UM171 showed 70% engraftment in the BM compared with 35% engraftment in the absence of UM171 (Figure 3B) representing a nearly twofold increase in human cell engraftment. UM171 also enhanced the engraftment of gene-marked cells in the BM (18% vs 11%; P < .01; Figure 3C), as well as in PB (supplemental Figure 12). To investigate whether UM171 preserves the multilineage differentiation potential of HSCs in vivo, we analyzed the lineage distribution of BM cells obtained at week 16 of engraftment. As shown in Figure 3D, although a moderately higher percentage of myeloid cells was observed (25.9% vs 12.3%; P < .05) in the UM171 group, the overall lineage distribution was similar in both groups (supplemental Figure 13). In addition, a higher proportion of gene-marked human cells was found across all lineages after UM171 treatment (Figure 3E; P < .05). These data demonstrate that exposure of SCD CD34+ cells to UM171 not only enhances LVV-mediated transduction efficiency with repopulating potential but also maintains their normal differentiation into lineage cells.
Figure 3.
UM171 increases engraftment of CD34+ cells in xenografts. (A) Outline of assay. CD34+ cells from patients with SCD were prestimulated and transduced with LV-GFP in the presence or absence of UM171 and transduced cells were injected into NBSGW mice. (B) Engraftment of CD34-derived CD45+ cells and (C) gene-marked CD45+ cells in BM of NBSGW mice at week 16 after transplant. (D) Lineage distribution and (E) the percentage of gene-marked human cells from BM. Data represent mean ± SD, n = 6, each data point represents an individual mouse. ∗P < .05; ∗∗P < .01.
UM171 enhances repopulation of SCD CD34+ cells expressing BCL11A shmiR in NBSGW mice
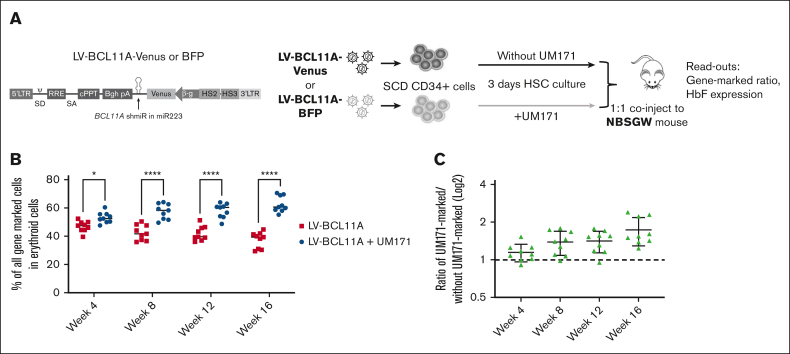
To better assess the impact of UM171 on functionally defined HSCs, we used a competitive repopulation assay in which equal numbers of SCD CD34+ cells transduced separately with color coded LV-BCL11A virus (Venus or blue fluorescent protein [BFP]) in the presence or absence of UM171 were mixed and cotransplanted into recipient NBSGW mice (Figure 4A). After transplant, mice were bled every 4 weeks and BMs were collected at week 16 for analysis. Analyses of the PB demonstrated that cells transduced in the presence of UM171 consistently outcompeted those transduced under control conditions for up to 16 weeks of follow-up. The proportion of engrafted SCD cells transduced in the presence of UM171 group increased over time (Figure 4B-C) suggesting a larger impact on a more primitive hematopoietic HSC population. Specifically at the time of euthanasia at 16 weeks, the erythroid cell population derived from SCD CD34+ cells transduced in the presence of UM171 was significantly higher than that derived from those transduced under control conditions, 63.6% vs 37.4% (P < .001; Figure 4B). Overall, there was a 1.8-fold increase for the UM171-treated group of mice compared with the control group (Figure 4C). We next assessed the output of gene-marked erythroid cells derived from transplanted BM CD34+ cells. These data confirmed that the LV-BCL11A + UM171 treated group had significantly higher levels of gene-marked erythroid cells than the LV-BCL11A group (supplemental Figure 14A). In addition, given that the BCL11A shmiR targets a repressor of γ-globin expression, we evaluated γ-globin induction in sorted Venus+ and BFP+ erythroid cells derived from transplanted BM CD34+ cells. γ-globin expression was comparable as expected, demonstrating no untoward effects on the expression and function of the integrated vector (supplemental Figure 14B). In addition, the human cells frequency in PB was normal (5%-10%) and reached 80% in BM (supplemental Figure 15A), and BM cells demonstrated the expected multilineage differentiation potential (supplemental Figure 15B-C). Taken together, these results suggest that UM171 enhances the reconstitution of SCD CD34+ cells expressing BCL11A shmiR in vivo with no discernible impact on the functionality of the BCL11A shmiR or γ-globin expression in erythroid cells.
Figure 4.
UM171 enhances reconstitution of CD34+ cells expressing a BCL11A shmiR in NBSGW mice. (A) Outline of competitive assay, CD34+ cells from patients with SCD (n = 3) were prestimulated and transduced in the presence or absence of UM171 with a Venus- or a BFP-expressing vector. Equal numbers of Venus-marked and BFP-marked cells were mixed 1:1 and coinjected into NBSGW mice. (B) Competitive repopulation between cells transduced in presence or absence of UM171. Both vectors expressed the BCL11A shmiR. (C) Ratio of engrafted gene-marked cells after transduction in the presence or absence of UM171. Data represent mean ± SD, n = 9, each data point represents an individual mouse. ∗P < .05; ∗∗∗∗P < .0001.
Transduction in the presence of UM171 increases clonality of engrafted BM cells
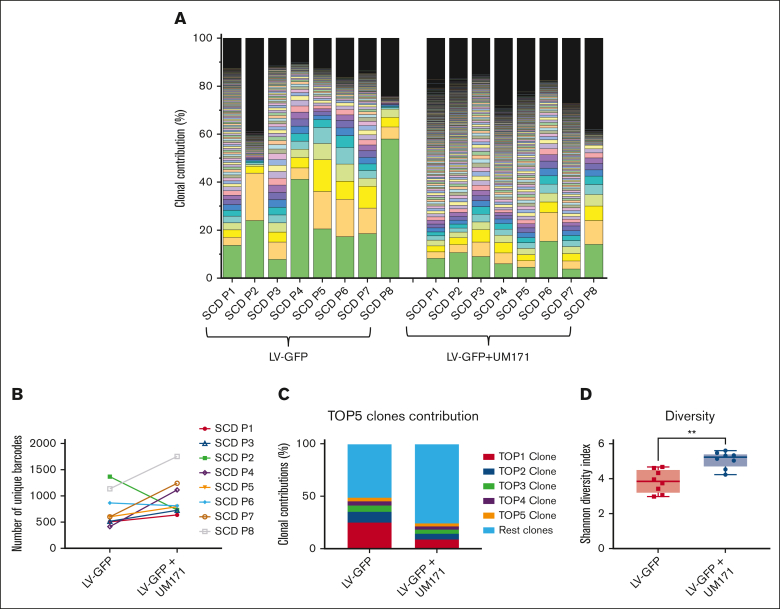
We next examined whether UM171 influenced the clonal composition of engrafting cells in vivo. CD34+ cells from patients with SCD were transduced with an LVV containing random barcodes as unique molecular identifiers in the presence or absence of UM171 and transplanted into NBSGW mice (supplemental Figure 16). Amplicon sequencing was conducted on BM cells collected at week 16 of engraftment to identify and quantify the barcodes. Engrafted SCD CD34+ cells transduced both in the presence or absence of UM171 displayed a high degree of clonal diversity, as shown in supplemental Figure 17, with a median of 600 to 800 different barcodes per animal. Figure 5A-B demonstrates the unique barcode and clonal contributions from each mouse transplanted with CD34+ cells of different patients with SCD. Six of 8 engrafted samples exhibited more unique barcodes after transduction in the presence of UM171 (supplemental Figure 17).
Figure 5.
Effect of UM171 on clonality of barcoded engrafted BM cells. (A) Distribution of barcode frequencies detected in BM cells from transplanted NBSGW mice. Each color represents a distinct barcode. (B) Assessment of the number of unique clones from NBSGW mouse BM cells transplanted with CD34+ cells from different patients with SCD (noted P1-P8). Each data point represents an individual mouse transplanted with different patient’s cells. (C) The percentage of contributions of top 5 clones to engrafted BM harvested from transplanted mouse. (D) Shannon diversity index of barcodes from transplanted BM cells. Data represent mean ± SD, n = 8. ∗∗P < .01.
As another measure of the clonal balance of transduced cells, we compared the contributions of the top 5 clones from the CD34+ cells engrafted from the LV-GFP + UM171 group with control groups. As shown in Figure 5C, the top 5 highest contributing clones with the LV-GFP + UM171 groups showed aggregated contribution of 24% of total clones, whereas the top 5 clones of the CD34+ cells without UM171 exposure contributed 49%, indicating that UM171 exposure favors engraftment of more HSC clones. The clonal contributions of all barcoded cells, as shown in Figure 5A, demonstrated that the UM171 treatment groups consistently exhibited fewer large clones than the cells transduced in the absence of UM171. As an aggregate measure of clonal diversity and clonal balance, the Shannon diversity index further confirmed that the UM171-treated group performed significantly better than the control group (Figure 5D). Taken together, exposure to UM171 for a short duration increases clonal diversity and clonal balance of transduced engrafting populations.
Discussion
A primary finding of the studies presented here is that UM171 improves the fitness of SCD HSCs transduced with LV-BCL11A. This enhanced fitness includes reduction of DNA damage, ROS formation, and apoptosis—key cellular assays measuring stress associated with ex vivo cell manufacturing. UM171 also enhances the proliferative capacity of SCD CD34+ cells as we demonstrate increased numbers of immunophenotypically defined HSCs. Overall, these effects culminate in improved competitive repopulation of transduced SCD HSCs after a brief UM171 exposure.
Stem cell autologous transplantation, crucial for successful gene therapy, is challenged by the complexities of prolonged ex vivo culture, leading to stem cell loss, genomic instability, and senescence.32 Several small molecules, including UM171, have been reported to preserve or expand HSCs during culture. Various other small molecule compounds such as SR1,14 4HPR,15 resveratrol,33 and eupalinilide E34 have been previously investigated. To date, most research examining the effects of additives on expanding human pluripotent stem cells has focused on using UM171 and SR1, and some studies have even combined UM171 and SR1 or other reagents to expand stem cells.15,34, 35, 36, 37, 38 Indeed, clinical use of UM171 has demonstrated its feasibility, safety, and ability to expand CB-derived HSCs through successful engraftment, and studies have shown UM171 to be one of the most effective agents.23,39,40 Our focus on UM171 is also underpinned by its putative mechanism of action, involving the activation of the E3 ligase complex CRL3KBTBD4, which influences epigenetic modulation and maintains the lympho-myeloid potential of expanded cell populations.22,41, 42, 43
Our findings demonstrate UM171’s efficacy in enhancing the expansion of LV-BCL11A-transduced SCD HSCs, including various CD34+ cell subtypes, aligning with previous research demonstrating the epigenetic impact and effect on HSC populations.18,20,44 UM171 has been reported to target multiple cellular processes involved in HSC expansion, including self-renewal, differentiation, cell metabolism, cell cycle regulation, and epigenetic regulation, which is consistent with our result about UM171 work on SCD stem cells.17,22
UM171 is currently in clinical testing focusing on the ex vivo expansion of CB-derived HSCs. Although UM171 has proven effective in expanding human CB HSCs, its impact on HSCs derived from PB of patients with SCD has previously not been explored. These results are likely relevant from a clinical translation standpoint given ongoing concerns about transformation events in SCD gene therapy trials and the use of PB in current US Food and Drug Administration–approved therapies and ongoing clinical trials. Furthermore, we investigated UM171’s influence on clonal diversity in vivo, which has not previously been explored. UM171 increased the diversity of transplanted stem cells, which may be a result of the combined effect of UM171 on improving transduction efficiency as previously reported and expanding stem cells.20 This result underscores UM171’s potential in mitigating clonal selection concerns postgene therapy and fostering a diverse and robust stem cell pool.
In conclusion, UM171 may prove a significant factor in enhancing SCD gene therapy. Our study validates that UM171 improves fitness, transduction, and numbers of SCD CD34+ cells during ex vivo manipulation. This improved fitness includes reduced DNA damage, apoptosis, and ROS expression, coupled with increased engraftment and clonal diversity in xenograft models. These findings position UM171 as a clinically viable approach to optimize ex vivo manufacturing for SCD autologous transplantation, aligning with current safety and efficacy standards in gene therapy. Ultimately, UM171’s role in this therapeutic landscape needs clinical testing with long-term follow-up to validate the relevance of these preclinical findings in the large-scale setting.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors thank Meaghan McGuinness and Chad Harris for technical assistance, and Teresa Ortiz and Maria Donis for administrative assistance.
The funding sources included the Bill and Melinda Gates Foundation (INV-021791) and ExCellThera (79315).
Authorship
Contribution: B.L. performed the experimental work; B.L., C.B., and D.A.W. designed the experiments; Y.Z. and D.P. analyzed the sequence data; J.P.M. provided the patient samples; D.K. provided technical assistance and advice; G.S. is a founder and chief scientific officer of ExCellThera, a small biotechnology company that owns an exclusive license to UM171; and B.L. and D.A.W. wrote and edited the manuscript.
Footnotes
Sequence data have been deposited to http://www.ncbi.nlm.nih.gov/bioproject/1142540 (BioProject ID: PRJNA1142540). Additional data or information are available on request from the corresponding author, David A. Williams (david.williams2@childrens.harvard.edu).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Kato GJ, Piel FB, Reid CD, et al. Sickle cell disease. Nat Rev Dis Primers. 2018;4 doi: 10.1038/nrdp.2018.10. [DOI] [PubMed] [Google Scholar]
- 2.Demirci S, Uchida N, Tisdale JF. Gene therapy for sickle cell disease: an update. Cytotherapy. 2018;20(7):899–910. doi: 10.1016/j.jcyt.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanter J, Walters MC, Krishnamurti L, et al. Biologic and clinical efficacy of LentiGlobin for sickle cell disease. N Engl J Med. 2022;386(7):617–628. doi: 10.1056/NEJMoa2117175. [DOI] [PubMed] [Google Scholar]
- 4.Frangoul H, Locatelli F, Sharma A, et al. Exagamglogene autotemcel for severe sickle cell disease. N Engl J Med. 2024;390(18):1649–1662. doi: 10.1056/NEJMoa2309676. [DOI] [PubMed] [Google Scholar]
- 5.Morgan RA, Gray D, Lomova A, Kohn DB. Hematopoietic stem cell gene therapy: progress and lessons learned. Cell Stem Cell. 2017;21(5):574–590. doi: 10.1016/j.stem.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esrick EB, Lehmann LE, Biffi A, et al. Post-transcriptional genetic silencing of BCL11A to treat sickle cell disease. N Engl J Med. 2021;384(3):205–215. doi: 10.1056/NEJMoa2029392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson AA, Walters MC, Kwiatkowski J, et al. Gene therapy in patients with transfusion-dependent beta-thalassemia. N Engl J Med. 2018;378(16):1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh MM, Bonner M, Pierciey FJ, Jr., et al. Myelodysplastic syndrome unrelated to lentiviral vector in a patient treated with gene therapy for sickle cell disease. Blood Adv. 2020;4(9):2058–2063. doi: 10.1182/bloodadvances.2019001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spencer Chapman M, Cull AH, Ciuculescu MF, et al. Clonal selection of hematopoietic stem cells after gene therapy for sickle cell disease. Nat Med. 2023;29(12):3175–3183. doi: 10.1038/s41591-023-02636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyal S, Tisdale J, Schmidt M, et al. Acute myeloid leukemia case after gene therapy for sickle cell disease. N Engl J Med. 2022;386(2):138–147. doi: 10.1056/NEJMoa2109167. [DOI] [PubMed] [Google Scholar]
- 11.Brunson A, Keegan THM, Bang H, Mahajan A, Paulukonis S, Wun T. Increased risk of leukemia among sickle cell disease patients in California. Blood. 2017;130(13):1597–1599. doi: 10.1182/blood-2017-05-783233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seminog OO, Ogunlaja OI, Yeates D, Goldacre MJ. Risk of individual malignant neoplasms in patients with sickle cell disease: English national record linkage study. J R Soc Med. 2016;109(8):303–309. doi: 10.1177/0141076816651037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brendel C, Guda S, Renella R, et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J Clin Invest. 2016;126(10):3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie SZ, Garcia-Prat L, Voisin V, et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell. 2019;25(5):639–653.e7. doi: 10.1016/j.stem.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goessling W, Allen RS, Guan X, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8(4):445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fares I, Chagraoui J, Gareau Y, et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345(6203):1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fares I, Chagraoui J, Lehnertz B, et al. EPCR expression marks UM171-expanded CD34+ cord blood stem cells. Blood. 2017;129(25):3344–3351. doi: 10.1182/blood-2016-11-750729. [DOI] [PubMed] [Google Scholar]
- 19.Zonari E, Desantis G, Petrillo C, et al. Efficient ex vivo engineering and expansion of highly purified human hematopoietic stem and progenitor cell populations for gene therapy. Stem Cell Rep. 2017;8(4):977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngom M, Imren S, Maetzig T, et al. UM171 enhances lentiviral gene transfer and recovery of primitive human hematopoietic cells. Mol Ther Methods Clin Dev. 2018;10:156–164. doi: 10.1016/j.omtm.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlesworth CT, Camarena J, Cromer MK, et al. Priming human repopulating hematopoietic stem and progenitor cells for Cas9/sgRNA gene targeting. Mol Ther Nucleic Acids. 2018;12:89–104. doi: 10.1016/j.omtn.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chagraoui J, Girard S, Mallinger L, Mayotte N, Tellechea MF, Sauvageau G. KBTBD4-mediated reduction of MYC is critical for hematopoietic stem cell expansion upon UM171 treatment. Blood. 2024;143(10):882–894. doi: 10.1182/blood.2023021342. [DOI] [PubMed] [Google Scholar]
- 23.Cohen S, Bambace N, Ahmad I, et al. Improved outcomes of UM171-expanded cord blood transplantation compared with other graft sources: real-world evidence. Blood Adv. 2023;7(19):5717–5726. doi: 10.1182/bloodadvances.2023010599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Brendel C, Vinjamur DS, et al. Development of a double shmiR lentivirus effectively targeting both BCL11A and ZNF410 for enhanced induction of fetal hemoglobin to treat beta-hemoglobinopathies. Mol Ther. 2022;30(8):2693–2708. doi: 10.1016/j.ymthe.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raoul C, Abbas-Terki T, Bensadoun JC, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med. 2005;11(4):423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 26.Gerrits A, Dykstra B, Kalmykowa OJ, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115(13):2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 27.Bystrykh LV. Generalized DNA barcode design based on Hamming codes. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dull T, Zufferey R, Kelly M, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart SA, Dykxhoorn DM, Palliser D, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masiuk KE, Zhang R, Osborne K, Hollis RP, Campo-Fernandez B, Kohn DB. PGE2 and poloxamer synperonic F108 enhance transduction of human HSPCs with a beta-globin lentiviral vector. Mol Ther Methods Clin Dev. 2019;13:390–398. doi: 10.1016/j.omtm.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giarratana MC, Rouard H, Dumont A, et al. Proof of principle for transfusion of in vitro-generated red blood cells. Blood. 2011;118(19):5071–5079. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trounson A, McDonald C. Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell. 2015;17(1):11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Heinz M, Graeber D, Zak D, Zwirnmann E, Gelbrecht J, Pusch MT. Comparison of organic matter composition in agricultural versus forest affected headwaters with special emphasis on organic nitrogen. Environ Sci Technol. 2015;49(4):2081–2090. doi: 10.1021/es505146h. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Huang X, Guo B, et al. Effects of Eupalinilide E and UM171, alone and in combination on cytokine stimulated ex-vivo expansion of human cord blood hematopoietic stem cells. Blood Cells Mol Dis. 2020;84 doi: 10.1016/j.bcmd.2020.102457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pineault N, Abu-Khader A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp Hematol. 2015;43(7):498–513. doi: 10.1016/j.exphem.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Ren Y, Cui Y, Feng J, et al. Synergistic effect and molecular mechanism of PVA and UM171 in ex vivo expansion of primitive hematopoietic stem cells. J Cell Biochem. 2024;125(1):79–88. doi: 10.1002/jcb.30505. [DOI] [PubMed] [Google Scholar]
- 37.Sakurai M, Ishitsuka K, Ito R, et al. Chemically defined cytokine-free expansion of human haematopoietic stem cells. Nature. 2023;615(7950):127–133. doi: 10.1038/s41586-023-05739-9. [DOI] [PubMed] [Google Scholar]
- 38.Wen R, Dong C, Xu C, et al. UM171 promotes expansion of autologous peripheral blood hematopoietic stem cells from poorly mobilizing lymphoma patients. Int Immunopharmacol. 2020;81 doi: 10.1016/j.intimp.2020.106266. [DOI] [PubMed] [Google Scholar]
- 39.Cohen S, Roy J, Lachance S, et al. Hematopoietic stem cell transplantation using single UM171-expanded cord blood: a single-arm, phase 1-2 safety and feasibility study. Lancet Haematol. 2020;7(2):e134–e145. doi: 10.1016/S2352-3026(19)30202-9. [DOI] [PubMed] [Google Scholar]
- 40.Elnaggar M, Al-Mohannadi A, Hasan W, et al. CD14+/CD31+ monocytes expanded by UM171 correct hemophilia A in zebrafish upon lentiviral gene transfer of factor VIII. Blood Adv. 2023;7(5):697–711. doi: 10.1182/bloodadvances.2022009014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chagraoui J, Girard S, Spinella JF, et al. UM171 preserves epigenetic marks that are reduced in ex vivo culture of human HSCs via potentiation of the CLR3-KBTBD4 complex. Cell Stem Cell. 2021;28(1):48–62.e6. doi: 10.1016/j.stem.2020.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Subramaniam A, Zemaitis K, Talkhoncheh MS, et al. Lysine-specific demethylase 1A restricts ex vivo propagation of human HSCs and is a target of UM171. Blood. 2020;136(19):2151–2161. doi: 10.1182/blood.2020005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zemaitis K, Ghosh S, Hansson J, Subramaniam A. The stem cell-supporting small molecule UM171 triggers Cul3-KBTBD4-mediated degradation of ELM2 domain-harboring proteins. J Biol Chem. 2023;299(5) doi: 10.1016/j.jbc.2023.104662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Xia C, Wang T, et al. Pyrimidoindole derivative UM171 enhances derivation of hematopoietic progenitor cells from human pluripotent stem cells. Stem Cell Res. 2017;21:32–39. doi: 10.1016/j.scr.2017.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.