Abstract
The nuclear envelope is a specialized subdomain of the endoplasmic reticulum and comprises the inner and outer nuclear membranes. Despite the crucial role of the inner nuclear membrane in genome regulation, its lipid metabolism remains poorly understood. Phosphatidic acid (PA) is essential for membrane growth as well as lipid storage. Using a genome-wide lipid biosensor screen in S. cerevisiae, we identify regulators of inner nuclear membrane PA homeostasis, including yeast Seipin, a known mediator of nuclear lipid droplet biogenesis. Here, we show that Seipin preserves nuclear envelope integrity by preventing its deformation and ectopic membrane formation. Mutations of specific regions of Seipin, some linked to human lipodystrophy, disrupt PA distribution at the inner nuclear membrane and nuclear lipid droplet formation. Investigating the Seipin co-factor Ldb16 reveals that a triacylglycerol binding site is crucial for lipid droplet formation, whereas PA regulation can be functionally separated. Our study highlights the potential of lipid biosensor screens for examining inner nuclear membrane lipid metabolism.
Subject terms: Nuclear envelope, Nucleus, Nuclear pore complex, Endoplasmic reticulum
The inner nuclear membrane is crucial for genome regulation, but its lipid metabolism is poorly understood. Here, the authors use a genome-wide lipid biosensor screen in yeast to identify factors, including Seipin, that maintain nuclear envelope integrity.
Introduction
The nuclear envelope (NE) acts as a protective barrier separating the nucleoplasm from the cytoplasm. It consists of two membranes: the outer nuclear membrane (ONM) and the inner nuclear membrane (INM), which are spaced by only ~10–50 nm. The ONM is continuous with the endoplasmic reticulum (ER), while the INM connects to the ONM exclusively at sites where nuclear pore complexes (NPCs) are embedded1–3. While it is recognized that different organelles possess unique lipid compositions that dictate their specific functions4, our understanding of the lipid composition of the INM is limited, primarily due to difficulties in isolating this particular membrane.
The ONM and peripheral ER produce glycerophospholipids (PL) for membrane growth and triacylglycerol (TAG) for energy storage5. The INM, which directly contacts the genome, is also metabolically active, enabling the storage of fatty acids in nuclear lipid droplets (nLDs)6,7. The lipid storage metabolism of the NE can be reprogrammed in response to imbalances of lipid unsaturation, redirecting unsaturated fatty acids away from the INM8,9. Presumably due to an asymmetric lipid metabolism across the NE, the lipid composition of the INM differs from that of the ONM. For example, the INM is characterized by elevated levels of diacylglycerol (DAG), metabolized from phosphatidic acid (PA)7,10. The fundamental unanswered questions are how cells detect and regulate the lipid properties of the INM and what benefits this regulation provides.
PA plays a versatile role in lipid metabolism as it can generate precursors for both, membrane PLs and storage lipids. For storage, PA undergoes processing by a group of phosphatidate phosphatases (e.g. Pah1) to yield DAG, which is further metabolized into TAG5,11. Additionally, PA can be converted to cytidine diphosphate diacylglycerol (CDP-DAG) by Cds1, and CDP-DAG serves as a precursor for the synthesis of phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), or cardiolipin, which are all important components of membranes5,11 (Fig. 1a). At the INM, PA is metabolized by both, Pah1 and Cds1 (Fig. 1a)7. Elevated PA levels at the INM lead either to NE proliferation or to nLD formation, depending on the metabolic branch that is active7. Given the key role of PA in lipid metabolism, it is crucial to understand how cells regulate PA homeostasis at the INM in response to different metabolic states.
Fig. 1. High-throughput screening uncovers PA regulators at the INM.
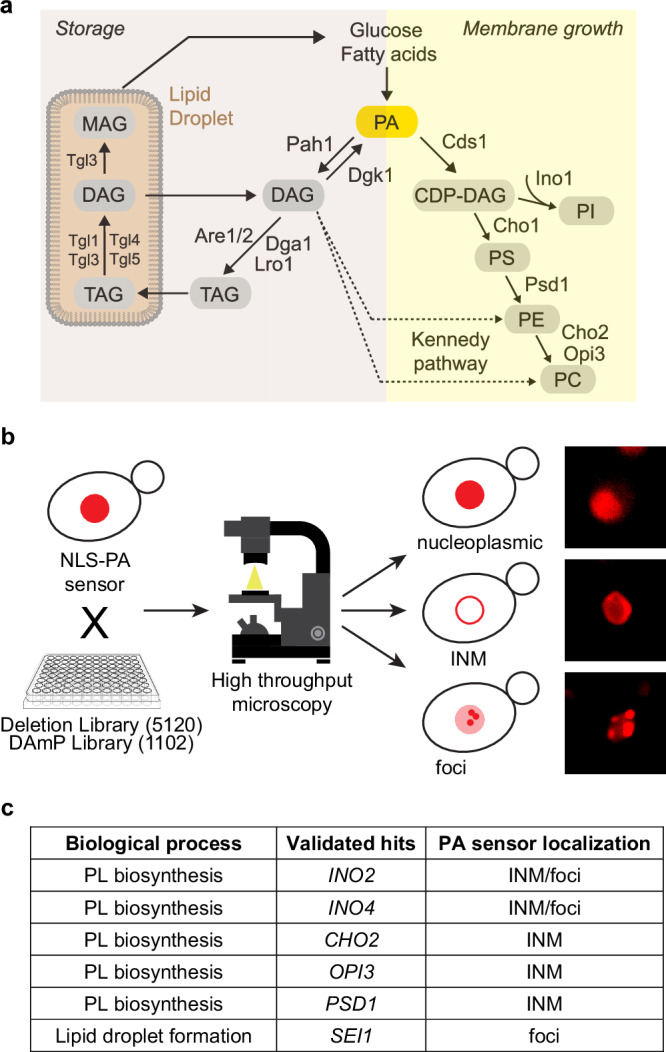
a Simplified scheme of yeast lipid biosynthesis depicting the two major branches leading to synthesis of phospholipids (Membrane growth) or triacylglycerol (TAG) (Storage). Phosphatidic acid (PA) is a central precursor. The Kennedy pathway (dashed line) channels diacylglycerol (DAG) into phospholipid production. CDP-DAG, cytidine diphosphate diacylglycerol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol; MAG, monoacylglycerol. b Scheme of the high-throughput screening approach used to identify new regulators of PA at the INM with examples of phenotypes of PA localization observed using high-throughput screening. The 96-well plate icon was obtained from Clipground under a Creative Commons license. c Table showing validated hits, their biological function and PA sensor localization. Note that foci in ino2∆ and ino4∆ cells likely reflect nuclear lipid droplets7. PL, phospholipid; INM, inner nuclear membrane.
In this study, we employed a genome-wide visual screen in S. cerevisiae (from here on simply called yeast) to identify regulators of PA homeostasis at the INM. The screen identified Seipin (Sei1) as essential for INM PA homeostasis and NE integrity. Sei1 is a key regulator of nLD formation and we identified 12 nLD-associated factors, thereby expanding the inventory of the nLD proteome. We dissected the role of specific regions within Sei1, including one implicated in lipodystrophy in humans, and identified mutations that caused aberrant distribution of PA at the INM and nLD biogenesis defects. Notably, Sei1 selectively influences specific lipid species at the INM, affecting PA and DAG without impacting PS. Additionally, we show that Sei1 and its co-factor Ldb16 differentially affect PA homeostasis and nLD formation at the INM. A model of the Sei1-Ldb16 complex provides a framework for understanding their functional separation, highlighting the TAG binding site of Ldb16 as a means of controlling LD formation without affecting INM PA.
Results
A genome-wide screen for PA regulators at the INM
To pinpoint factors affecting INM PA homeostasis, we employed a genome-wide microscopy screen based on a previously characterized INM-specific PA biosensor7. The NLS-PA-mCherry sensor contains the Q2 domain of the yeast transcription factor Opi1 that specifically recognizes PA12. The NLS of yeast nucleoporin Nup60 is attached for nuclear import and the mCherry fluorophore for visual detection. PA sensing depends on specific residues within a critical amphipathic helix (AH), which harbors two PA-selective three-finger grips13.
First, we generated a yeast strain with the INM PA sensor genomically integrated, which showed a nucleoplasmic sensor distribution, indicative of low INM PA levels. This parental strain was crossed with a library of 5120 strains carrying deletions of non-essential genes14 (Fig. 1b). In addition, essential genes were targeted by crossing the parental strain with 1102 strains of the DAmP (Decreased Abundance by mRNA Perturbation) library15. In this library, a gene’s 3′ untranslated region (UTR) is disrupted with an antibiotic resistance cassette, thereby destabilizing the corresponding transcript and reducing the mRNA amount typically two- to tenfold16.
Following automated image acquisition, all mutant strains were inspected for a relocalization of the PA sensor to the INM, indicative of high INM PA levels, or a focal accumulation of the sensor, indicative of inhomogeneities or intranuclear PA structures such as PA-rich nLDs. 19 strains of the deletion library and 7 strains of the DAmP library displayed an altered sensor localization (Supplementary Fig. 1a, b). The positive hits could be further categorized into strains with PA-positive foci (65.4%), PA-positive INM staining (19.2%), and varied PA inhomogeneities (15.4%), all indicative of an altered nuclear PA status. The positive hits included several regulators of INM PA metabolism previously identified by a candidate approach (INO2, INO4, OPI3, and CHO2)7,8, thus validating the screen. 8 of the identified genes are involved in lipid metabolism, either as transcription factors that regulate expression of PL biosynthetic genes (e.g. the Ino4-Ino2 complex), as enzymes that consume products from PA metabolism for PL synthesis (e.g. CHO2) (Fig. 1a), or as regulators of PL metabolism (e.g. SCS2). As is often the case in high-throughput screens, some slow-growing strains were lost during the procedure (11% in the deletion library, 30% in the DAmP library). Hence, some regulators of INM PA homeostasis may remain undetected.
To validate the results, we first re-examined positively tested strains under exponential growth conditions in liquid media. We then examined these strains in a different genetic background (BY4741) than the screening strain. After these two validation steps, we arrived at a consolidated list of 6 factors (Fig. 1c and Supplementary Fig. 1a–c), which included the previously known regulators INO2, INO4, OPI3, and CHO2.
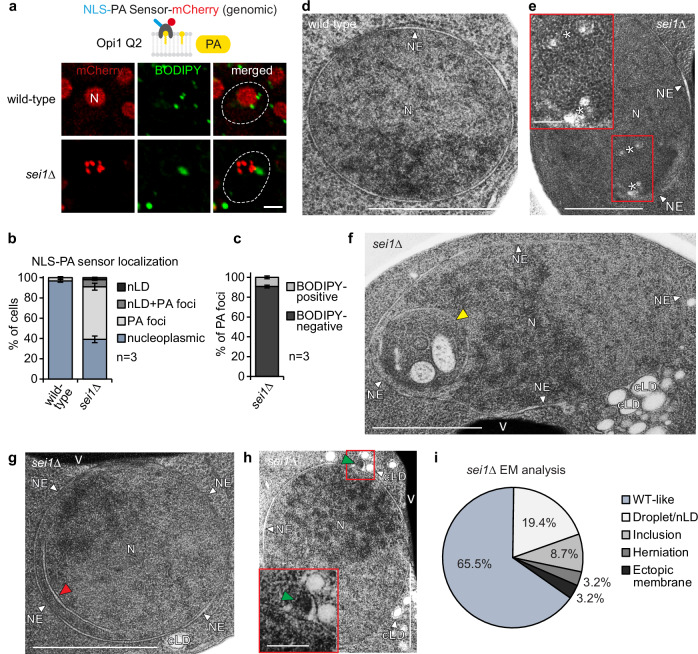
Notably, while the deletion of five of these factors, all involved in PA metabolism, led to an INM localization of the PA sensor, the deletion of the LD biogenesis factor SEI1 showed multiple PA-positive foci within the nucleus (Figs. 1c and 2a, b). Of note, we previously detected Seipin at the INM7 and Seipin has been proposed to bind PA17. These findings prompted us to examine Seipin’s role in INM PA homeostasis.
Fig. 2. Seipin regulates nuclear membrane architecture.
a Live imaging of wild-type or sei1∆ cells expressing a genomically integrated NLS-PA-mCherry sensor. BODIPY stains LDs. N, nucleus. Cell contours were marked with a dashed white line based on brightfield imaging. Scale bar, 2 μm. b Quantification of NLS-PA-mCherry sensor localization in (a). Mean value and standard deviation indicated. n, number of biological replicates. 480 cells for sei1∆ and 526 cells for wild-type analysed. Source data are provided as a Source Data file. c Analysis of NLS-PA-mCherry sensor foci co-localization with BODIPY in sei1∆ in (a). Mean value and standard deviation indicated. n, number of biological replicates. 591 foci in total analysed. Source data are provided as a Source Data file. d TEM analysis of a representative example of wild-type cell grown in SDC medium. N, nucleus; NE, nuclear envelope. Scale bar, 1 μm. e–h TEM analysis of representative examples of sei1∆ cells transformed with an empty vector and grown in SDC medium. Cells exhibit small lipid droplet-like structures marked with white asterisks in (e), intranuclear inclusions marked with a yellow arrowhead in (f), ectopic intranuclear membrane sheets marked with a red arrowhead in (g) and omega-shaped NE herniations marked with a green arrowhead in (h). See more TEM images in Supplementary Fig. 2. Insets show a magnified view of the marked areas. N, nucleus; NE, nuclear envelope; cLD, cytoplasmic lipid droplet; V, vacuole. Scale bar, 1 μm; 200 nm for insets. i Analysis of nuclear deformation frequency in nuclei of sei1∆ cells by TEM. sei1∆ cells were transformed with an empty vector and grown in SDC medium. Source data are provided as a Source Data file.
Seipin preserves nuclear membrane architecture
The PA-positive foci in sei1∆ cells frequently did not co-localize with the LD marker BODIPY (Fig. 2a), unlike nLDs induced by oleic acid overload or by genetically induced increase of lipid storage metabolism7,8. About 50% of sei1∆ cells showed foci that were positive for PA but did not stain with BODIPY; ~10% displayed a mixed phenotype with both PA-positive/BODIPY-negative foci and PA-positive/BODIPY-positive foci. In contrast, only a very small fraction of cells (~ 2%) contained PA-positive/BODIPY-positive structures which resemble properly matured LDs (Fig. 2b and Supplementary Fig. 3a). When analyzing individual foci, we found that ~90% of PA-positive foci in sei1∆ cells lacked a BODIPY signal (Fig. 2c). This suggests that, in the absence of Seipin, the PA-containing structures had not undergone TAG-enrichment as would be expected in LD formation.
This discrepancy raised questions about the ultrastructure of these foci. Thus, we employed transmission electron microscopy (TEM) to study the cell nucleus in the sei1∆ mutant, screening TEM images of 310 yeast cell nuclei. In comparison to wild-type cells (Fig. 2d), we noted the presence of small lipid droplet-like structures within the nucleus as the most common ultrastructural abnormality (Fig. 2e, i and Supplementary Fig. 2a–d), as has been observed previously18,19. These likely correspond to the PA-positive foci observed by fluorescence microscopy. It is plausible that these small lipid droplets have not acquired enough TAG to stain with BODIPY, even though they are easily detected by the PA sensor. Consistent with earlier TEM data, we observed cytoplasmic LDs of heterogeneous size in sei1∆ cells, including clusters of small LDs and supersized LDs (Supplementary Fig. 2e, j, l)18,20–22. In contrast, no supersized LDs were observed in the nucleus of sei1∆ cells in any of the TEM images examined.
We observed further nuclear membrane deformations, where portions of the nucleus engulfed cytoplasmic material, including membranes of unknown origin (Fig. 2f and Supplementary Fig. 2h, l). We also found ectopic intranuclear membranes, localized in proximity to the INM. While these membranes look like a thin tube in EM cross-section, they likely form a three-dimensional membrane sheet (Fig. 2g and Supplementary Fig. 2c, e–g). Lastly, we observed omega-shaped NE herniations23 in a fraction of sei1∆ cells (Fig. 2h), suggesting that Seipin also plays a role, either directly or indirectly, in maintaining NE integrity. These nuclear Sei1 deletion phenotypes are more diverse than previously appreciated and could reflect abnormalities in both nucleoplasmic lipid storage and INM membrane proliferation as well as NE stability.
Seipin differentially alters lipid dynamics at the INM
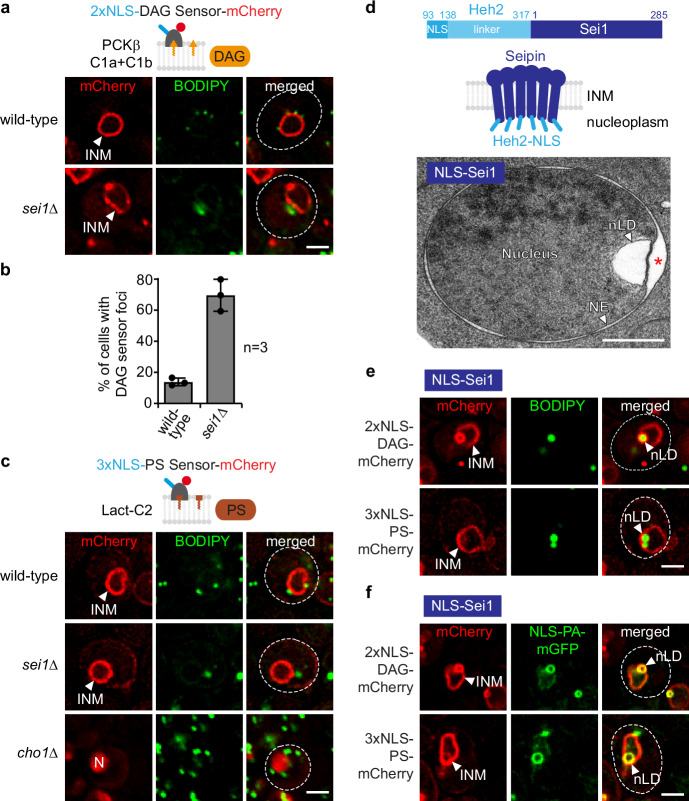
Because the deletion of Seipin had pronounced effects on membrane homeostasis, we wanted to know if PA is channelled towards membrane growth or lipid storage in sei1∆ cells (Fig. 1a). We therefore examined the relative distribution of PA, DAG, and PS at both the INM and nLDs in wild-type and sei1∆ cells. To identify DAG, we employed the DAG-specific recognition domains of protein kinase C (PKCβ C1a + C1b). These domains are specific for DAG both in vitro and in vivo7,24,25. Here, we used a modified sensor containing two tandem NLSs for nuclear import. The DAG sensor labels the INM smoothly in wild-type cells (Fig. 3a). In contrast, in sei1∆ cells the DAG sensor exhibited non-homogenous staining and detected nuclear foci in ∼70% of cells compared to ∼14% in the wild type (Fig. 3a, b). Using Sec62 as an NE marker, we determined that ~10% of DAG foci are present in the nucleoplasm of sei1∆ cells, with the remainder localizing to the INM. In contrast, ~30% of PA foci in sei1∆ cells are nucleoplasmic (Supplementary Fig. 3b, c). Collectively, this suggests that besides its role in maintaining nuclear PA homeostasis, Seipin is also required for a homogeneous distribution of DAG, the downstream metabolite of PA, at the INM. Since the absence of Seipin appears to differently impact the nuclear localization of PA and DAG, Seipin may have distinct effects on the nuclear sorting of these lipids.
Fig. 3. Seipin differentially alters lipid dynamics at the INM.
a Live imaging of wild-type or sei1∆ cells expressing plasmid-based 2xNLS-DAG-mCherry sensor and stained with BODIPY. INM, inner nuclear membrane. Scale bar, 2 μm. b Quantification of cells with 2xNLS-DAG-mCherry sensor foci in (a). Mean value and standard deviation indicated. n, number of biological replicates. 454 cells for sei1∆ and 628 cells for wild-type analysed. Source data are provided as a Source Data file. c Live imaging of wild-type, sei1∆ or cho1∆ cells expressing plasmid-based 3xNLS-PS-mCherry sensor and stained with BODIPY. INM, inner nuclear membrane; N, nucleus. cho1∆ cells were supplemented with ethanolamine and the sensor was expressed from the GPD promoter in cho1∆ cells. Scale bar, 2 μm. d Cartoon of the engineered NLS-Sei1 construct which contains the nuclear localization sequence (NLS) and the linker of the INM transmembrane protein Heh2 (aa93-317) appended to Sei1. Putative membrane topology of Sei1 is based on cryo-EM models. TEM analysis of a representative example of NLS-Sei1-expressing cells. Plasmid-based mGFP-NLS-SEI1 was expressed from the SEI1 promoter in a sei1∆ strain. N, nucleus; NE, nuclear envelope; nLD, nuclear lipid droplet. Asterisk marks a widened perinuclear space beneath an nLD. Scale bar, 0.5 μm. e Live imaging of sei1∆ cells expressing plasmid-based NLS-SEI1 and lipid sensors tagged with mCherry. Cells are stained with BODIPY. nLD, nuclear lipid droplet; INM, inner nuclear membrane. Scale bar, 2 μm. f Live imaging of sei1∆ cells expressing plasmid-based NLS-SEI1, NLS-PA-mGFP and lipid sensors tagged with mCherry. nLD, nuclear lipid droplet; INM, inner nuclear membrane. Scale bar, 2 μm.
PS was reported to localize to the cytoplasmic leaflet of the plasma membrane. Its detection there used a biosensor that contains the C2 domain of lactadherin (Lact-C2), a domain known for its ability to specifically bind PS26–29. Of note, a recent study has also detected PS at the INM using freeze-fracture EM30. To detect PS at the INM in live cells, we developed a nuclear mCherry-tagged Lact-C2-based PS biosensor by appending three NLSs in tandem to promote efficient import. Whereas the non-NLS version of the PS sensor mostly labeled the plasma membrane, as described before29, the NLS-variant labeled the INM in wild-type cells (Fig. 3c and Supplementary Fig. 3d).
To confirm the specificity of the sensor in detecting changes in PS levels and distribution at the INM, we evaluated the sensor’s response to the deletion of Cho1, a phosphatidylserine synthase. Deletion of Cho1 is expected to lower cellular PS levels (Fig. 1a). Consequently, we observed decreased binding of the 3xNLS-PS sensor to the INM, resulting in a nucleoplasmic signal (Fig. 3c and Supplementary Fig. 3d, e). Likewise, a previously characterized mutation in the Lact-C2 domain (3 A mutant), known to reduce Lact-C2 affinity towards PS29, resulted in localization of the mutant sensor to the nucleoplasm (Supplementary Fig. 3f, g), confirming that PS recognition is specific. Unlike the DAG and PA sensors, the PS sensor did not exhibit a notable change between wild-type and sei1∆ cells (Fig. 3c and Supplementary Fig. 3h), indicating that the PA- and DAG-positive foci contain minimal amounts of PS. This suggests specificity in Seipin’s impact on INM lipids. Evidently, our comparative assessment is only qualitative as we lack information on the affinities of these biosensors to their target lipids in vivo.
In summary, our findings suggest that Sei1 regulates both, PA and DAG homeostasis at the INM but has little, if any, effect on PS distribution, at least as assessed by the PS sensor under the conditions tested.
Lipid features of Seipin-induced nLDs
These findings raised the question as to how Seipin normally affects the distribution of DAG and PS in nLDs. Because nLDs are rare in wild-type cells not overloaded with fatty acids, we targeted Seipin to the INM using a method developed earlier8, which involves appending the NLS and the linker region of the INM protein Heh2 (aa93-317)31 to Sei1 (abbreviated as NLS-Sei1). This approach targets Sei1 to the INM and leads to the formation of nLDs (Fig. 3d). These nLDs exhibit a notable enrichment in PA, forming a distinct outer shell around a BODIPY-positive core composed of neutral lipids (Fig. 3f). Based on these characteristics, they resemble the nLDs observed in wild-type cells after oleic acid supplementation or those induced by genetic modifications, such as INO4 deletion or CDS1 inactivation7. To address how DAG and PS partition between the INM and NLS-Sei1-induced nLDs, we first combined the mCherry-labeled NLS-DAG and NLS-PS sensors and the LD marker BODIPY (Fig. 3e). Subsequently, we combined the mGFP-tagged NLS-PA sensor with the mCherry NLS-DAG and NLS-PS sensors (Fig. 3f). These comparative assessments suggest that DAG is present on the surface of nLDs while still maintaining localization at the INM. Conversely, PA can be fully incorporated into the nLD lipid monolayer with minimal PA remaining at the INM. In contrast, PS exhibits enrichment at the INM but barely diffuses into the lipid monolayer of the nLD. Consequently, by employing a triple-INM sensor strategy, we elucidated the distinct distribution of lipids between the INM and nLDs. Our findings revealed significant differences between the abnormal droplet-like structures in sei1∆ cells and the lipid dynamics of nLDs formed by Seipin at the INM.
Localization of cytoplasmic LD biogenesis factors to nLDs
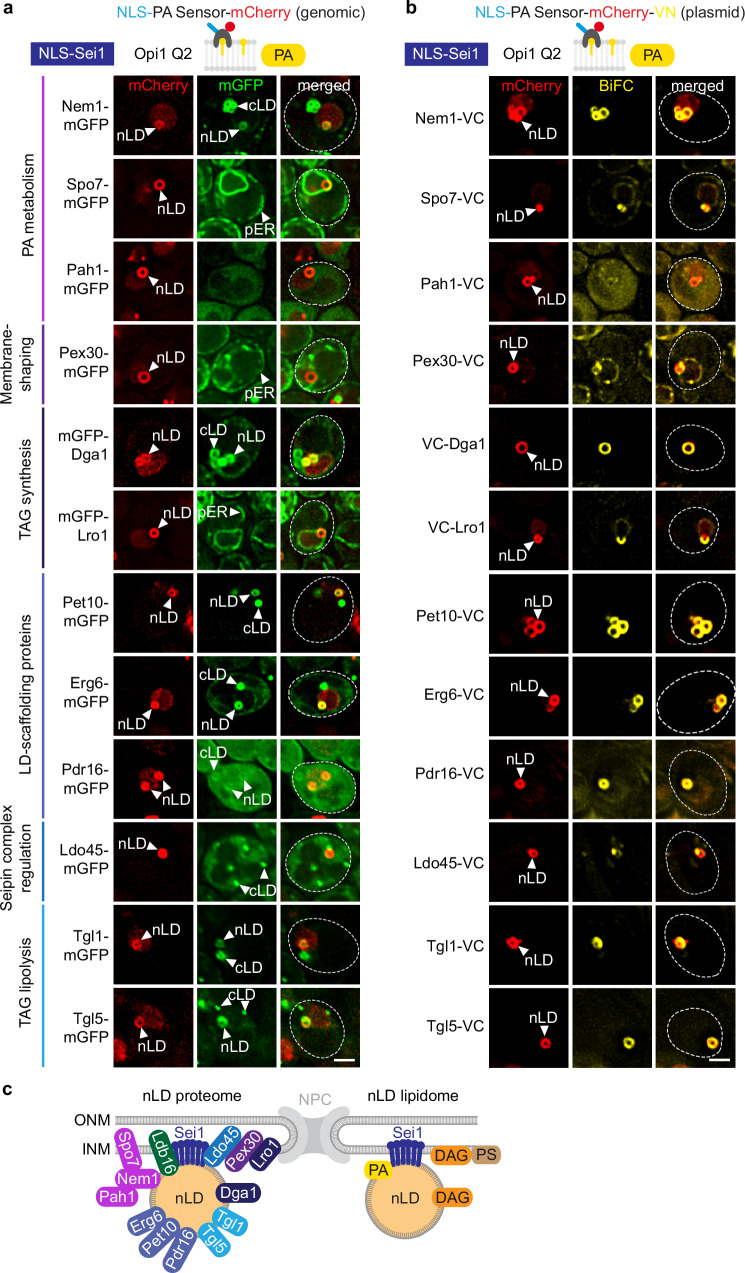
To understand how lipids are distributed to nLDs, a fundamental understanding of nLD formation is required. However, in contrast to cytoplasmic LD formation32–34, LD biogenesis within the nucleus is still poorly understood. In the cytoplasm, proteins and lipids required for LD formation undergo a sequential recruitment to specific sites of the ER35–39. Factors involved in nuclear LD formation, whether they are soluble or membrane-bound, need to pass through the NPC, which restricts the access of large non-NLS-containing cargo40. Whether LD biogenesis factors can reach the INM and have a role in nLD formation remains largely unknown. To determine the spectrum of nLD-associated factors, we induced nLD biogenesis via NLS-Sei1 (Fig. 3d), visualized nLDs with the NLS-PA sensor, and examined the co-localization of various mGFP-tagged proteins normally involved in cLD biogenesis (Fig. 4a and Supplementary Fig. 4a). Although NLS-Sei1 promotes LD formation predominantly in the nuclear compartment, we occasionally observed cLDs as well, allowing us to evaluate mGFP-tagged proteins on both nLDs and cLDs in the same cell.
Fig. 4. Lipid droplet factors are localized to Sei1-formed nLDs.
a Live imaging of sei1∆ cells expressing plasmid-based NLS-SEI1, the indicated mGFP-tagged constructs and genomically integrated NLS-PA-mCherry sensor. All mGFP constructs were expressed from the GPD promoter, except PET10 (PET10 promoter) and PEX30 (TPI1 promoter). nLD, nuclear lipid droplet; cLD, cytoplasmic lipid droplet; pER, peripheral endoplasmic reticulum. Scale bar, 2 μm. b Live imaging of sei1∆ cells expressing plasmid-based NLS-SEI1, NLS-PA-mCherry-VN and the indicated VC-tagged constructs. All VC-tagged constructs were expressed from the GPD promoter, except PET10 (ADH1 promoter) and PEX30 (TPI1 promoter). nLD, nuclear lipid droplet. Scale bar, 2 μm. c Summary of Sei1-formed nLD proteome and lipidome.
We first asked whether PA metabolic enzymes co-localize with nLDs. PA is converted to DAG by Pah1 (mammalian Lipin) (Fig. 1a), which requires the Nem1-Spo7 complex for activation41. Indeed, we detected a prominent colocalization of Nem1 with both nLDs and cLDs, as evidenced by a circular fluorescence signal, suggesting that Nem1 is able to integrate into the lipid monolayer of an LD (Fig. 4a). In contrast, Spo7, the regulator of Nem1, localized to the NE as well as the peripheral ER and did not show a strong enrichment with nLDs or cLDs. This suggests that the interaction between Nem1 and Spo7 in cells can be dynamic.
Pah1 did not exhibit a strong colocalization with nLDs, likely because Pah1 only transiently interacts with membranes when dephosphorylated by Nem1-Spo7 as shown earlier for cLDs42. Accordingly, earlier immunogold TEM showed that a major fraction of Pah1 localized to the nucleoplasm and cytoplasm in wild-type cells7.
At cLD biogenesis sites, the membrane-shaping protein Pex3043 has been reported to act downstream of Sei1 and Nem1 to recruit the TAG-synthases Lro1 and Dga135. Although we detected Pex30 enriched in cytoplasmic puncta37,39, likely associated with cLDs, no noticeable Pex30 signal was observed around nLDs (Fig. 4a). In contrast, we observed a strong Dga1 signal surrounding nLDs. Interestingly, Lro1 was present at the NE, yet it did not show enrichment around nLDs. Hence, Dga1 and Lro1 TAG synthases may differ in their ability to associate with nLDs.
When a nascent cLD grows at a Sei1-Nem1 site, the perilipin Pet10 and Erg6, a ∆(24)-sterol C-methyltransferase, are recruited to its surface35. We observed Pet10 and Erg6 localizing to the periphery of nLDs and confirmed their association with cLDs (Fig. 4a). Additionally, the phosphatidylinositol transfer protein Pdr16, which localizes to a subpopulation of LDs44,45, showed a faint signal around nLDs and weakly labelled cLDs as well.
Ldo45 (a homolog to human promethin/LDAF146) is another protein that decorates only a subset of LDs and regulates Seipin44,47. Ldo45 did not encircle nLDs but was often found in proximity to nLDs, possibly at their biogenesis sites (Fig. 4a). It also appeared as fluorescent puncta in the cytoplasm, indicating its association with cLDs48,49.
Stored TAG in LDs can be mobilized by TAG lipases, all of which are found on cLDs11. Tgl1 and Tgl5, the two candidate lipases tested, were found to associate with both nLDs and cLDs. This suggests that nLDs undergo lipase-dependent degradation and shrinkage similar to their cytoplasmic counterparts (Fig. 4a).
Because enzyme-substrate interactions are often transient, we additionally employed a variation of the bimolecular fluorescence complementation (BiFC)50 technique to visualize low-affinity interactions of cLD biogenesis factors with nLDs. To this end, we tagged the NLS-PA-mCherry sensor with the N-terminal half of the Venus fluorophore (VN) and LD biogenesis factors with the C-terminal half of the fluorophore (VC). We then tested for the occurrence of a BiFC Venus signal and co-localization with NLS-PA-mCherry on nLDs (Fig. 4b). This assay revealed robust fluorescence complementation on nLDs for Nem1, Dga1, Pet10, Erg6, Pdr16, and the Tgl1 and Tgl5 lipases, consistent with the results of the co-localization assay (Fig. 4a). mGFP-Pah1 did not exhibit significant enrichment on the nLDs; however, a faint BiFC signal suggests that Pah1 might be in transient contact with these structures. Of note, Spo7, Pex30 and Lro1 which were not enriched on nLDs in the co-localization assay (Fig. 4a), exhibited BiFC signal around nLDs, which often was not completely homogenous. This could indicate that these proteins are in contact with nLDs and their biogenesis sites but cannot shuttle onto the nLD surface. An only partial co-localization with nLDs was also observed with mGFP-tagged lipases (Fig. 4a).
Collectively, these findings indicate that many factors involved in the formation of cLDs can reach the INM and associate with nLDs (Fig. 4c). For NLS-Sei1-induced nLDs, only two factors (Dga1 and Pet10) were previously identified as associated with nLDs8, while this study has identified ten additional factors (Nem1, Spo7, Pah1, Pex30, Lro1, Erg6, Pdr16, Ldo45, Tgl1, Tgl5). The presence of PA metabolic enzymes and TAG synthase can mediate nLD growth, while lipases catalyze nLD degradation and TAG mobilization51, indicating both nLD formation and turnover in the nucleus. Consequently, the processes governing nLD and cLD formation in yeast share the same core molecular machinery, despite occurring in distinct cellular compartments. However, differences in enzyme abundance and other yet-to-be-identified factors may create compositional differences between nLDs and cLDs.
Specific Sei1 residues are important for INM PA homeostasis
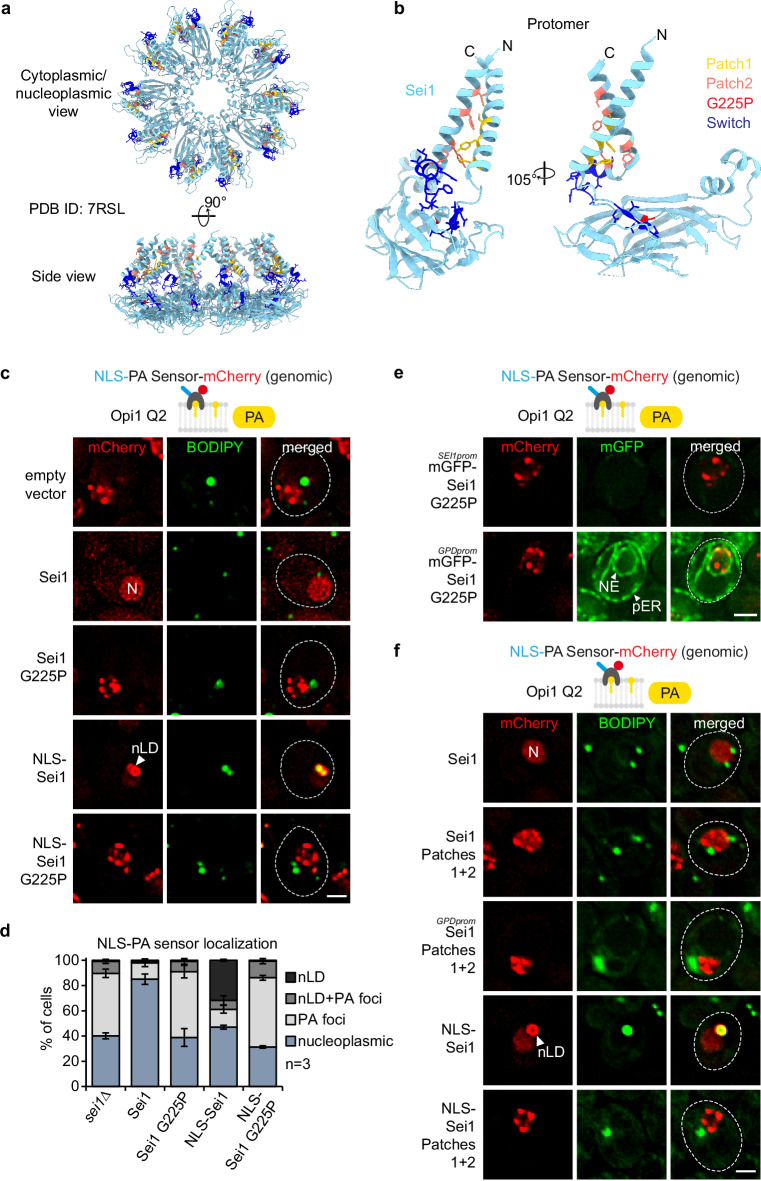
Yeast Seipin forms a decameric, cage-like structure, where the domains located in the ER lumen create a ring at the base, while the transmembrane (TM) segments form the sides of the cage (Fig. 5a)17,52–54. Interactions between TM segments of adjacent subunits occur in two distinct conformations (termed A and B)52, which stem from conformational differences in “switch regions” positioned between the lumenal domains and the TM segments. Current models posit that, initially, a closed cage conformation facilitates the phase separation of TAG, followed by a transition to an open conformation that facilitates LD expansion and budding52.
Fig. 5. PA defects at the INM in Sei1 lipodystrophy and TM contact mutants.
a Cartoon representation of S. cerevisiae Sei1 homodecamer (PDB ID: 7RSL) with select amino acid residues shown in atom (stick) representation. The amino acid residues, which were either substituted or deleted in corresponding Sei1 mutants, are colour-coded (yellow: Patch1, salmon: Patch2, red: G225P, blue: Switch). Note that the mutated amino acid residues do not map to the inter-subunit interfaces, and are therefore unlikely to disrupt the formation of the Sei1 ring assembly. b Close-up view of the Sei1 protomer (PDB ID: 7RSL, B conformation) shown in hybrid cartoon-atom representation as in Fig. 5a. c Live imaging of sei1∆ cells expressing genomically integrated NLS-PA-mCherry sensor and the indicated plasmid-based mGFP-SEI1 constructs. BODIPY stains LDs. nLDs have a BODIPY-positive core surrounded by a PA-rich shell. Note that even though cells contain mGFP-Sei1, the green BODIPY fluorescence signal is significantly brighter, hence Sei1 fluorescence remains undetectable when the settings for BODIPY imaging are applied. N, nucleus; nLD, nuclear lipid droplet. Scale bar, 2 μm. d Quantification of NLS-PA-mCherry sensor localization in (c). Mean value and standard deviation indicated. n, number of biological replicates. More than 435 cells analysed for each condition. Source data are provided as a Source Data file. e Live imaging of sei1∆ cells expressing genomically integrated NLS-PA-mCherry sensor and plasmid-based mGFP-sei1 G225P constructs from the endogenous SEI1 or a strong GPD promoter. Scale bar, 2 μm. f Live imaging of sei1∆ cells expressing genomically integrated NLS-PA-mCherry sensor and indicated plasmid-based mGFP-SEI1 constructs from the endogenous SEI1 or a strong GPD promoter. BODIPY stains LDs. N, nucleus; nLD, nuclear lipid droplet. Scale bar, 2 μm.
To investigate the role of specific Sei1 regions on INM PA homeostasis and nLD formation, we conducted a mutational analysis (Fig. 5a, b). Mutations in the human Seipin gene, such as the A212P missense mutation in the lumenal domain, are associated with Berardinelli-Seip congenital lipodystrophy55–57. This condition is characterized by a lack of body fat, severe insulin resistance, and various other abnormalities. Whereas human Seipin can effectively complement the SEI1 deletion in yeast, the human pathogenic A212P mutant fails to do so22. An analogous G225P mutant of the yeast protein, predicted to correspond to the human A212P mutant22, also failed to complement the SEI1 deletion22 and is unstable in cells58. A superposition of yeast Sei1 and human Seipin cryo-EM structures reveals differences in the yeast G225 and human A212 location and the overall region (e.g., the yeast protein has two β-strands while the human protein only one) (Supplementary Fig. 5a). Nevertheless, the yeast “disease” mutation likely has a similarly detrimental effect on protein stability and assembly as the human disease mutation.
To test different mutants, we employed a complementation assay. In the absence of Sei1, numerous PA-positive, BODIPY-negative foci formed in the nucleus (Fig. 5c, d; also see Fig. 2a), which could be largely restored by introducing a plasmid-based SEI1 (expressed from the endogenous SEI1 promoter) (Fig. 5c). Introducing Sei1 G225P did not rescue the phenotype, showing aberrant PA-foci similar to sei1∆ cells, indicating that this region of Seipin is functionally important from yeast to human (Fig. 5c, d). Since the mutant has decreased protein expression levels58 (Supplementary Fig. 5b), we tested whether its overexpression from a strong GPD promoter would rescue its function. However, even the overexpressed G225P mutant exhibited numerous PA-positive foci lacking BODIPY staining (Fig. 5e and Supplementary Fig. 5c, d). We further probed whether this mutant, when targeted to the INM, could generate nLDs. Once more, it resembled sei1∆ cells and, unlike wild-type NLS-Sei1, was unable to form properly matured nLDs which are characterized by PA- and BODIPY-positive staining (Fig. 5c, d and Supplementary Fig. 5e, f).
A recent study of the yeast Sei1 structure identified additional functionally important residues52 (Fig. 5a, b). Within a Sei1 subunit, the N- and C- terminal TM segments interact with each other (Fig. 5b). Hence, we tested whether mutations in the N-terminal TM segment (Patch 1 = S33A, Y37A, Y41A) and the C-terminal TM segment (Patch 2 = M240G, Y248I, F255R, I259K), which are expected to disrupt intra-subunit interactions between the TM helices, affect PA homeostasis at the INM. The combined mutant (Patches1 + 2) exhibited an abnormal PA distribution in the nucleus and could not form nLDs when targeted to the INM with an NLS (Fig. 5f and Supplementary Fig. 5g, h). This defect could not be compensated by overexpressing the mutant from the strong GPD promoter (Fig. 5f and Supplementary Fig. 5h, i). Thus, Seipin’s transmembrane architecture is critical for INM PA homeostasis and nLD formation at the INM.
Conformational changes of Sei1 may facilitate the transition from a membrane-contained TAG lens to an LD bud59,60. Specifically, Sei1 monomers can adopt two different conformations. These arise from changes in switch regions, positioned between the lumenal domains and the TM segments52. When the Sei1 switch region was mutated, we observed defects in PA metabolism and nLD formation, underscoring its structural and functional importance (Supplementary Fig. 6)
Overall, our findings highlight the critical role of Seipin’s TM segments, conformational changes of the complex, and a region with links to Berardinelli-Seip lipodystrophy in regulating PA homeostasis, consistent with the outcome of our initial PA biosensor screen (Fig. 1c).
Ldb16 and Sei1 have distinguishable functions at the INM
Seipin is widely conserved, however, in S. cerevisiae and other yeasts, Seipin function is encoded by two proteins - Sei1 and Ldb1661,62. Earlier studies proposed that the function of Sei1 fully depends on Ldb16, because the LD abnormalities observed in cells lacking SEI1, LDB16, or both, were indistinguishable62,63. Whereas human Seipin was proposed to directly interact with TAG, promoting its concentration and phase separation into a lens-like structure64–66, this function in yeast appears to be outsourced to Ldb1653. However, the stoichiometry and structure of the Ldb16-Sei1 complex is unknown and the division of labor between Ldb16 and Sei1 is unclear. Whether and how Ldb16 is relevant for INM PA homeostasis is unknown.
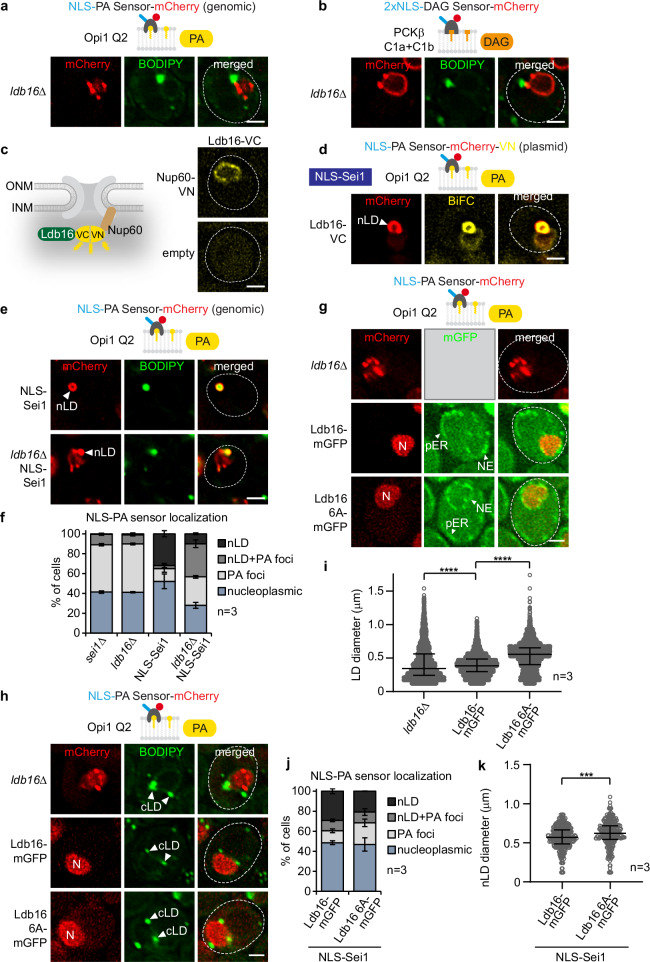
In our high-throughput PA screen, the ldb16∆ well contained no viable cells, preventing us from assessing the impact of Ldb16 on INM PA. We therefore generated ldb16∆ strains with either the NLS-PA or NLS-DAG sensor. Both sensors recognized PA- and DAG-containing nuclear foci, respectively, as seen in sei1∆ cells (Fig. 6a, b). A detailed analysis revealed that 50% of ldb16∆ cells had PA-positive, but BODIPY negative foci; ~9% displayed a mixed phenotype with both PA-positive/BODIPY-negative foci and PA-positive/BODIPY-positive foci, while very few cells (~ 1%) contained typical nLD structures with PA-positive/BODIPY-positive staining (Fig. 6f). This indicates that Sei1 and Ldb16 cooperate to regulate PA and DAG at the INM, consistent with them forming a complex (Fig. 7a).
Fig. 6. Ldb16’s hydroxyl residues govern proper cellular TAG accumulation but not PA metabolism at the INM.
a Live imaging of ldb16∆ cells expressing plasmid-based NLS-PA-mCherry sensor. BODIPY stains LDs. Scale bar, 2 μm. b Live imaging of ldb16∆ cells expressing plasmid-based 2xNLS-DAG-mCherry sensor and stained with BODIPY. Scale bar, 2 μm. c Experimental design for BiFC (bimolecular fluorescence complementation). VN, VC, complementary Venus fragments. Live imaging of wild-type cells expressing Ldb16 fused with VC and Nup60 fused with VN. Ldb16 is expressed from the GPD promoter. Nup60 is a basket nucleoporin, exclusively localized on the nuclear face of the nuclear pore complex. Empty vector is used as a control. Scale bar, 2 µm. d Live imaging of sei1∆ cells expressing plasmid-based NLS-PA-mCherry-VN, LDB16-VC and NLS-SEI1 constructs. Ldb16 is expressed from the GPD promoter. nLD, nuclear lipid droplet. Scale bar, 2 μm. e Live imaging of sei1∆ or sei1∆ ldb16∆ cells expressing plasmid-based mGFP-NLS-SEI1 and genomically integrated NLS-PA-mCherry sensor. Cells are stained with BODIPY. nLD, nuclear lipid droplet. Scale bar, 2 μm. f Quantification of NLS-PA-mCherry sensor localization in (e). Mean value and standard deviation indicated. n, number of biological replicates. More than 380 cells analysed for each condition. Source data are provided as a Source Data file. g Live imaging of ldb16∆ cells expressing plasmid-based NLS-PA-mCherry sensor and indicated plasmid-based LDB16-mGFP constructs from the ADH1 promoter. N, nucleus. Scale bar, 2 μm. h Live imaging of ldb16∆ cells expressing plasmid-based NLS-PA-mCherry sensor and indicated plasmid-based LDB16-mGFP constructs expressed from the ADH1 promoter. BODIPY stains LDs. Note that even though cells contain Ldb16-mGFP, the green BODIPY fluorescence signal is significantly brighter, hence Ldb16 fluorescence remains undetectable when the settings for BODIPY imaging are applied. cLD, cytoplasmic lipid droplet. Scale bar, 2 μm. i Automated quantification of cellular LD diameter in (h). n, number of biological replicates. Median and interquartile range indicated. P value (****P < 0.0001) determined by two-sided Kolmogorov-Smirnov test. Source data are provided as a Source Data file. j Quantification of NLS-PA-mCherry sensor localization in sei1∆ ldb16∆ cells expressing genomically integrated NLS-PA-mCherry, plasmid-based mGFP-NLS-SEI1 and genomically integrated LDB16-mGFP constructs. LDB16 was expressed from the ADH1 promoter. Mean value and standard deviation indicated. n, number of biological replicates. 611 cells for LDB16-mGFP and 663 cells for ldb16 6A-mGFP were analysed. Source data are provided as a Source Data file. k Automated quantification of nLD diameter in sei1∆ ldb16∆ cells expressing genomically integrated NLS-PA-mCherry, plasmid-based mGFP-NLS-SEI1 and genomically integrated LDB16-mGFP constructs. LDB16 was expressed from the ADH1 promoter. n, number of biological replicates. Median and interquartile range indicated. P value (***P < 0.001) was determined by two-sided Kolmogorov-Smirnov test. 302 nLDs for LDB16-mGFP and 252 nLDs for ldb16 6A-mGFP were analysed. Source data are provided as a Source Data file.
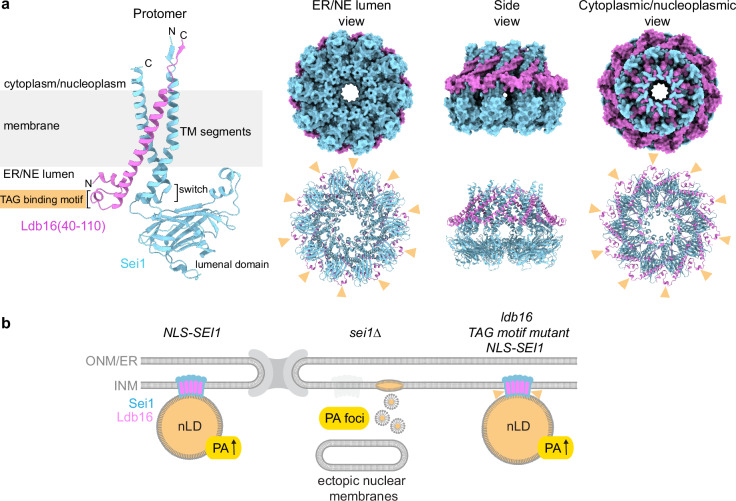
Fig. 7. Sei1 positions the TAG-binding domain of Ldb16 outside of the decameric ring.
a Cartoon representation of AlphaFold 3 model of S. cerevisiae Sei1·Ldb16(40-110) protomer (left), and both surface (right top) and cartoon (right bottom) representations of AlphaFold 3 model of Sei1·Ldb16(40-110) 10:10 ring assembly. The models are coloured by chain. Yellow arrowheads indicate putative serine/threonine-rich TAG binding motifs of Ldb16. For Sei1·Ldb16 prediction and confidence scores, see Supplementary Fig. 7. b A model illustrating the role of the Sei1-Ldb16 complex in regulating nLDs and INM lipid composition. When Sei1 is localized to the INM, nLDs with PA-enriched surfaces are formed. In the absence of Sei1, abnormal PA-rich but TAG-deficient droplets arise, accompanied by diverse defects in nuclear membrane structure. Mutations in the TAG-binding domain of Ldb16 (yellow arrowheads) reduce nLD numbers but slightly enlarge nLDs, suggesting impaired nLD biogenesis.
Since Sei1 localizes to the INM7, we asked whether Ldb16 is also present in this location. We performed BiFC with Ldb16 fused to the C-terminal half of the Venus fluorophore (VC) and an INM protein, Nup60, fused to the N-terminal half of the fluorophore (VN) (Fig. 6c). Nup60 is a basket nucleoporin, exclusively localized on the nucleoplasmic side of the NPC. Ldb16 showed fluorescence complementation with Nup60 specifically at the nuclear rim consistent with the presence of Ldb16 at the INM (Fig. 6c). We also confirmed the presence of Ldb16 at the site of nLD formation. As before, nLDs were induced by directing NLS-Sei1 to the INM. We then employed BiFC to probe whether INM-localized Ldb16 and PA-rich nLDs physically interact. Ldb16-VC showed fluorescent complementation with the NLS-PA sensor-VN, indicating a co-localization of Ldb16 and nLDs (Fig. 6d). Thus, Ldb16 is present in proximity to nLDs, likely reflecting the formation of a Sei1-Ldb16 complex at the INM.
Next, we aimed to test whether the function of Sei1 and Ldb16, specifically at the INM, can be functionally separated. We induced nLDs by NLS-Sei1 and monitored the nuclear PA distribution in NLS-SEI1 and NLS-SEI1 ldb16∆ cells (Fig. 6e). NLS-SEI1 cells exhibited ∼30% of nLDs with a BODIPY-positive core and a PA-rich shell. Contrary to expectation, the NLS-SEI1 ldb16∆ strain still produced ~10% of cells with PA- and BODIPY-positive nLDs and ~30% of cells displayed a mixed phenotype with both PA-positive/BODIPY-positive foci and PA-positive/BODIPY-negative foci, thus differing from ldb16∆ cells, which produce almost no nLDs (Fig. 6f). This suggests, that NLS-Sei1 is partially capable of forming nLDs on its own, however, it does require Ldb16 to mature them into larger TAG-rich structures. Hypothetically, Sei1 may be sufficient to promote TAG lens formation, but require Ldb16 to channel TAG into a growing nLD.
To identify a separation-of-function mutant of Ldb16 that could decouple Ldb16’s role in concentrating TAG into nascent LDs from its role in INM PA homeostasis, we examined an Ldb16 mutant with a known defect in TAG binding. Ldb16 features a short helical motif with several hydroxyl residues (T52/S53/S55/T61/S62/T63), important for TAG binding. Mutating these six residues to alanine (Ldb16 6 A) resulted in an abnormal variation in LD sizes53. Notably, this Ldb16 6 A mutant showed a nucleoplasmic PA sensor localization similar to wild-type Ldb16, yet it exhibited the aberrant cellular LD phenotype as previously reported (Fig. 6g–i). To explore the effect of the hydroxyl residues of Ldb16 specifically on nLD formation, we induced nLDs via NLS-Sei1 in the Ldb16 6 A mutant. Interestingly, this led to a reduced number of nLDs, which were significantly larger compared to those generated in Ldb16 wild-type cells (Fig. 6j, k). This finding reveals an intriguing separation of function for Ldb16’s hydroxyl residues: they are crucial for proper TAG accumulation in nLDs and cLDs but do not affect PA homeostasis at the INM. This contrasts with the tested Seipin mutants, which all disrupted PA homeostasis at the INM, supporting the idea that functions of the Sei1-Ldb16 complex are distributed between the two subunits.
In silico structure of the Ldb16-Sei1 complex
We took advantage of the AlphaFold 3 deep learning algorithm67 to investigate the Sei1-Ldb16 complex in silico. Based on this analysis, Sei1 is predicted to form a 1:1 complex with Ldb16 with the interaction primarily mediated by the hydrophobic packing of transmembrane helices (Supplementary Fig. 7a-c). This results in an architecture composed of three tightly bundled TM helices, two provided by Sei1 and one from Ldb16. Notably, AlphaFold 3 predicts Sei1 in the previously described A conformation, which may reflect the fact that the A conformation of Sei1 was the only experimental structure available at the time of AlphaFold 3 training53,67.
To predict the homodecamer, we needed to conform with the upper limit of total amino acids per prediction currently imposed by the AlphaFold 3 server. We therefore truncated Ldb16 so that only the highest confidence TM helix and adjacent regions (i.e. aa40-110) were included. The Sei1 homodecamer together with 10 copies of Ldb16(40–110) revealed a belt of Sei1 and Ldb16 TM helices surrounding a hollow cavity as a putative site of LD biogenesis (Fig. 7a, see Supplementary Fig. 7d, e for confidence scores and surface representations of the protomer). The ER/NE lumenal face of the assembly features the putative serine- and threonine-rich TAG binding helical motifs of Ldb16 (aa52-63) (Fig. 7a) that were proposed to facilitate the incorporation of TAG molecules into growing LDs53. According to this prediction, the orientation of these residues would suggest that Ldb16 plays a role in attracting TAG to the outside of the Sei1 ring. Although a mechanistic understanding of how TAG molecules penetrate tightly packed TM helices of Sei1 and Ldb16 is lacking, it is conceivable that A-B conformational changes of Sei1 may create gaps for TAG entry between TM segments. This model contrasts with the proposed TAG-enrichment mechanisms for human Seipin64,66. Here, a hydrophobic helix, which is positioned at the center of the human Seipin ring, concentrates TAG molecules and therefore facilitates lens formation and LD budding. In contrast to human Seipin, the yeast Sei1 lumenal ring does not contain this hydrophobic helix. Our model would predict that a TAG enrichment step in yeast may occur on the outside of the Seipin cage rather than within its cavity.
In summary, AlphaFold3 predictions, earlier structural data17,52–54 and our analysis of Ldb16’s impact on nLD formation suggest that Ldb16 enhances the ability of Sei1 to channel TAG into growing LDs. This is achieved by locally concentrating TAG around the Sei1-Ldb16 complex. The proposed model remains to be tested in future studies, also with regards to cytoplasmic LD formation.
Discussion
The close proximity of the INM and ONM makes it difficult to isolate them into pure fractions suitable for lipidomics68. To identify INM regulators of the key precursor lipid PA, we conducted a genome-wide screen to detect changes in yeast INM lipid dynamics. Our screen successfully identified changes in INM PA levels, with Seipin emerging as a key regulator. We found that Seipin is essential for maintaining nuclear envelope integrity and controlling nLD biogenesis, and identified specific residues, including those analogous to human lipodystrophy mutations, as critical for INM PA homeostasis. We also examined Seipin’s co-factor Ldb16, demonstrating that TAG enrichment and INM PA regulation are distinct functions. Our mapping of nLD-associated factors reveals that nLDs and cLDs share core machinery for biogenesis and turnover. However, variations in some factors may indicate differences in the lipid and protein composition between nLDs and cLDs.
Seipin and PA homeostasis
Earlier reports suggested that PA plays a role in the formation of cytoplasmic LDs21, however, the precise mechanism is still unclear. PA, being a cone-shaped lipid, promotes negative membrane curvature due to its small anionic phosphomonoester head group lying relatively close to the lipid bilayer’s hydrophobic interior. PA can therefore trigger various membrane fusion and fission events, possibly because of its capacity to create non-bilayer phases4. At the INM, PA might play a role in nLD biogenesis by inducing nascent nLD membrane curvature and bud neck remodelling. The extent to which PA at the INM acts as a precursor for localized TAG synthesis also requires further clarification. Enzymes involved in converting PA into TAG, such as Pah1, its regulators Nem1 and Spo7, and the TAG-synthase Dga1 are in proximity to nLDs, as shown in this study. While Seipin has been reported to interact with Lipin (the mammalian ortholog of Pah1) in adipocytes69, this connection has yet to be validated in yeast.
The identification of Seipin and Ldb16 as regulators of INM PA homeostasis (Fig. 7b) prompted us to ask whether and how INM PA homeostasis and nLD formation are connected. Deletion of SEI1 and LDB16, or mutations of functionally important Sei1 residues, consistently caused aberrant nLDs and disrupted INM PA distribution, making it difficult to distinguish between the two phenotypes. However, a key finding of this study is that a mutation in the TAG binding motif of Ldb16 specifically affects LD formation while leaving INM PA levels largely unchanged. This intriguing separation of function in the mutant suggests that PA regulation, or potentially its binding, is governed by a different part of the Sei1-Ldb16 complex. Indeed, it has been suggested that the lumenal domain of human Seipin directly binds PA17. Currently, the exact PA binding site in human or yeast Seipin remains unidentified, which will be crucial for examining its influence on INM PA levels, including the mode of PA transfer onto nLDs in the future. The binding site in yeast could, in principle, be present in either Sei1 or Ldb16, which are predicted to form a complex (Fig. 7a).
Yeast Seipin exhibits a comparable lumenal domain structure as human Seipin17,52,53, although the yeast oligomer does not feature analogous TAG-binding hydrophobic helices positioned at the center of the ring. In an unexpected twist, AlphaFold3 positions the TAG binding motif of yeast Ldb16 on the exterior of the Sei1-Ldb16 complex, suggesting differences in how TAG is concentrated and how it enters the Seipin cage when compared to human Seipin. Evidently, an experimental structure of the Sei1-Ldb16 complex is necessary to clarify the topological relationship between the TAG and potential PA binding sites and their role in nLD biogenesis. Our experimental system, inducing nLD formation of INM-targeted Sei1, is invaluable in this context, enabling the analysis of LD formation distinct from the intricate ER environment.
Access of Seipin and nLD factors to the INM
Studying Seipin function specifically at the INM poses experimental challenges due to the proximity of the INM and ONM. This is further exacerbated because Seipin is estimated to be expressed at low levels (~ 850 molecules in yeast70), which translates to only about 85 decameric complexes in the entire yeast ER/NE network. This low abundance makes immunogold EM, which has low sensitivity, unreliable to confirm Seipin localization to the INM. In contrast, techniques such as BiFC7 and split-GFP71 detect Seipin at the INM. A previous study, using a human osteosarcoma cell line, arrived at the conclusion that nLD formation does not depend on Seipin72. This conclusion was based on the inability to detect Seipin at the INM through immunogold EM and on the assumption that uncontrolled phase separation of TAG from membranes in the absence of Seipin equates to properly matured LDs72. However, without Seipin, a conserved factor from yeast to human, the phase transition of neutral lipids becomes irregular, resulting in formation of many small and few supersized LDs20,22. Therefore, the presence of LD-like structures alone does not indicate that LD formation is independent of Seipin. Although TAG phase separation can occur without Seipin, proper LD biogenesis requires it. Seipin is essential for the coordinated transfer of lipids and proteins onto LDs51. An imbalance of phospholipid surfactants, an increase in fusogenic lipids such as PA and ripening defects might cause LDs to merge into larger supersized LDs21,73,74 or prevent the accumulation of sufficient TAG into an nLD, as shown in this study.
Leveraging the sensitivity of BiFC enabled the detection of Sei1 at the INM7. We have now added additional proteins involved in Seipin regulation, PA metabolism, membrane remodeling, TAG synthesis, LD scaffolding, and TAG lipolysis to the list of nLD-proximal factors (Fig. 4c). Our study suggests that the core machinery of Seipin-dependent LD formation is shared between nLDs and cLDs. However, the stoichiometry and abundance of these components may vary, as NPCs might hinder or restrict access to the INM for some factors. This could influence the biogenesis kinetics, number, and morphology of LDs formed in each compartment. Consequently, variations in the stoichiometry and abundance of lipid metabolism enzymes and other LD-associated factors in the nucleus could result in a distinct nLD monolayer composition and protein inventory, potentially leading to functional specializations of nLDs that are tailored to the nuclear environment.
A question raised by our data is how a subset of cytoplasmic LD biogenesis factors from the ER gain access to the INM. In general, membrane proteins with extraluminal domains of up to 90 kDa in molecular mass can enter the nucleus40, probably through either passive diffusion via peripheral channels in the NPC75 or through NLS-dependent transport31,76. However, size selection alone cannot explain INM localization, as demonstrated in a previous study where many small soluble and ER membrane proteins failed to reach the nucleus71. Therefore, the proteome of the INM is probably established by a combination of protein diffusion, transport, retention, and degradation mechanisms. Understanding how Seipin and other LD biogenesis factors enter the nucleus, as well as the conditions under which the Sei1-Ldb16 complex assembles at the INM will be an important subject of future investigations.
Sei1-Ldb16 complex safeguards NE integrity
A notable finding from our study is the presence of a highly irregular NE architecture in sei1∆ cells. These defects extend beyond the reported proliferation of ER membranes or supersized LDs in the vicinity of the NE19. They encompass NE herniations, anomalous intranuclear membrane sheets, and distorted nuclei containing engulfed cytoplasmic material. These findings suggest that Seipin has a role in preserving the structural integrity of the cell nucleus. Although we cannot determine the specific contribution of either the INM or ONM pool of Sei1 to these phenotypes, it appears likely that Sei1 deficiency affects the NE through its involvement in PA and TAG metabolism, possibly in combination. In bilayer membranes, TAG is soluble up to a concentration of about 3 mol%77. Beyond this concentration, oil lenses form spontaneously64,78, which might disrupt the NE membrane and the function of NE resident proteins. The irregular LD budding processes themselves could potentially form weak points in the NE. The accumulation of PA as a non-bilayer lipid4 may additionally destabilize the NE.
The appearance of ectopic intranuclear membranes suggests a potential increase in membrane production. Multiple studies across different model organisms have detected heightened cellular PA levels following Seipin depletion21,69,79,80. However, it remains uncertain whether this results from an inhibition of proper LD formation, causing a buildup of precursors (Fig. 1a). Regardless of the origin, increased cellular PA levels could prompt a diversion of PA toward PL synthesis, thereby boosting membrane production. The precise location where these membranes might form (bearing in mind that S. cerevisiae has a closed mitosis) poses an intriguing question, possibly indicating a misregulation of localized lipid synthesis at the INM. Regarding the NE herniations in sei1∆ cells, it is noteworthy that the ESCRT factor Chm7, which plays a role in NE membrane surveillance, specifically recognizes PA and is thought to repair these PA-rich NE defects81. If such nuclear irregularities occur in patients with Berardinelli-Seip lipodystrophy, this would offer new insights into the disease pathology.
In summary, using a genome-wide screen we have made progress in characterizing key factors of INM PA homeostasis. This approach could be expanded to other lipid species in the future, contingent on the availability of NLS-lipid biosensors, such as our NLS-PS sensor.
Methods
Strains and media
All yeast strains used in this study are listed in Supplementary Table 1. Genes in yeast were tagged/deleted by a standard one-step PCR-based technique82. For integrating plasmids into the genome, plasmid digestion using the appropriate restriction enzymes was performed, followed by cell transformation with the digested product. Microbiological techniques followed standard procedures. Cells were grown in standard yeast extract peptone dextrose (YPD) prior to transformation or in synthetic dextrose complete (SDC)+all amino acids for experiments, or when transformed with plasmids in selective SDC drop-out media at 30 °C. Where indicated, ethanolamine (Sigma-Aldrich) was added to the growth medium to 2 mM.
Lipid sensor and Sei1 construct design
All plasmids used in this study are listed in Supplementary Table 2. For the PA sensor, the Q2 domain of yeast Opi1(103-191) was used. The DAG sensor contains the C1a + C1b domains of rat PKCβ(31-158). The PS sensor is based on the Lact-C2 domain of bovine Lactadherin MFGE8(270-427). Sensor NLS-sequences correspond to yeast nucleoporin Nup60(1-24), whereas NLS-Sei1 contains the NLS and the linker of the INM transmembrane protein Heh2 (aa93-317) attached to Sei131. The Sei1 mutants Patches1 + 2, ∆Switch and shuffled-Switch were constructed according to52. Plasmid-based Ldo45-mGFP and Ldo45-VC contains the sequence of Ldo45 according to the description of splicing in ref. 44: transcript corresponds to most of the YMR147W sequence excluding the last 90 nucleotides, and 210 nucleotides of the annotated YMR148W promoter, and the full YMR148W sequence.
Library preparation and high-throughput screening
To construct the query strain for the screen (NLS-Q2-mCherry Erg6-mNeonGreen), Erg6 was tagged and NLS-Q2-mCherry was genomically integrated into a strain with markers for automated mating, sporulation and haploid selection (YMS721)15, and was then crossed with the yeast deletion and hypomorphic allele collections14,15 using automated approaches83,84. In short: A RoToR bench-top colony array instrument (Singer Instruments) was used to handle libraries. Cells were mated on rich medium plates and diploids were selected in SDCMSG-Ura containing Geneticin (200 µg/ml) (Formedium) and Nourseothricin (200 µg/ml) (WERNER BioAgents “ClonNAT”). Sporulation was induced by transferring cells to nitrogen starvation media plates for 7 days. Haploid cells were selected by transferring cells to SDCMSG-Ura plates containing Geneticin (200 µg/ml) and Neurseothricin (200 µg/ml), alongside the toxic amino-acid derivatives Canavanine and Thialysine (Sigma-Aldrich) to select against remaining diploids, and lacking leucine to select an alpha mating type. A set of randomly selected strains were verified by PCR.
The generated libraries were screened using an automated microscopy setup. Cells were transferred from agar plates into 384-well plates for growth in liquid media using the RoToR arrayer. Liquid cultures were grown in a LiCONiC incubator overnight at 30 °C in SDC-Ura. A JANUS liquid handler (PerkinElmer) connected to the incubator was used to dilute the strains to an OD600 of ~0.2 into plates containing SDC-Ura medium, and plates were incubated at 30 °C for 5 h. Strains were then transferred by the liquid handler into glass-bottom 384-well microscope plates (Matrical Bioscience) coated with Concanavalin A (Sigma-Aldrich). After 20 min, wells were washed twice with SDC-Ura-Riboflavin media to remove non-adherent cells and to obtain a cell monolayer. The plates were then transferred to an Olympus automated inverted fluorescent microscope system using a robotic swap arm (Hamilton). Cells were imaged in SDC-Ura-Riboflavin at room temperature using a × 60 air lens (NA 0.9) and with an ORCA-ER charge-coupled device camera (Hamamatsu), using the ScanR software. Images were acquired in two channels: GFP (excitation filter 490/20 nm, emission filter 535/50 nm) and mCherry/RFP (excitation filter 572/35 nm, emission filter 632/60 nm). After acquisition, images were manually reviewed using ImageJ. Each strain was analyzed based on its PA sensor localization. A strain was considered a ‘hit’ if the PA sensor localization was not uniformly nucleoplasmic (e.g., foci, INM localization) in at least 25% of cells analyzed. For all identified hits, over 100 cells were examined, unless stated otherwise in Supplementary Fig. 1a. To validate the results, we first re-examined positively tested strains under exponential growth conditions in liquid media and then examined these strains in a different genetic background (BY4741) than the screening strain. Some expected hits were absent from our screen, likely due to technical issues during automated library preparation or strain propagation, such as the missing pah1∆ and ldb16∆ strains. Although a cds1 allele was included in the DAmP library, the PA sensor was nucleoplasmic. In contrast, a different cds1-ts allele that we previously studied7 showed robust PA sensor accumulation at the INM and nLD formation. This discrepancy is likely because the cds1-ts allele has a stronger inactivation phenotype than the DAmP variant.
Manual live-cell imaging of yeast
Exponentially growing cells were immobilized on microscope slides with agarose pads and imaged on a DeltaVision Elite microscope (GE Healthcare). Images were acquired with a 60x oil immersion objective and recorded with a CoolSNAP HQ2 CCD camera (Photometrics). Deconvolution was carried out using softWoRx software (GE Healthcare). Images were processed with ImageJ. Cell contours were marked with a dashed white line based on brightfield imaging. To stain lipid droplets, BODIPY 493/503 (final concentration 5.7 μM, Thermo Fisher Scientific) was added and cells were imaged after 20 min.
AlphaFold 3 modelling of Sei1·Ldb16 complex
AlphaFold 3-based structural modelling of S. cerevisiae Sei1·Ldb16 complex was performed via the public AlphaFold Server (https://golgi.sandbox.google.com/). Initial modelling was carried out using full-length sequences of Sei1 and Ldb16 proteins. Ldb16 was later truncated to remove low-confidence regions, retaining the Ldb16(40–110) sequence. The assessment of prediction confidence was guided by the local confidence score (pLDDT), global confidence scores (pTM, ipTM), PAE maps and the degree of superposition between 5 models produced by AlphaFold 3 model. Additionally, Sei1 chains were superimposed with previously determined experimental structures in order to assess the validity of the prediction (root mean square deviation (RMSD) between >190 pruned atom pairs was ≤ 0.80 Å).
Transmission electron microscopy (TEM)
Wild-type, sei1∆ and NLS-Sei1 cells were grown in SDC medium. Pelleted cells were mixed 1:1 with 10% BSA, used as a filler, and transferred into the 100 μm cavity of a 3 mm aluminum specimen carrier. This carrier was sandwiched with a flat 3 mm aluminum carrier and immediately high pressure frozen in an HPF Compact 01 (both carriers and high-pressure freezer from Engineering Office M. Wohlwend GmbH). The frozen samples were subsequently transferred into a Leica EM AFS-2 freeze substitution unit (Leica Microsystems). Over a period of 4 days, samples were substituted in a medium of acetone containing 2% osmium tetroxide (Agar Scientific), 0.2% uranyl acetate and 5% water. Freeze substitution was performed according to the following protocol: 40 hr at −90 °C, warm up at a rate of 2 °C per hour to −54 °C, 8 h at −54 °C, warm up at a rate of 5 °C per hour to −24 °C, 15 hr at −24 °C, warm up at a rate of 5 °C per hour to 0 °C, 2 h at 0 °C. At 0 °C samples were taken out and washed 3 times in anhydrous acetone (on ice) and infiltrated with Agar 100 Epoxy resin (Agar Scientific) in a graded series of acetone and resin over a period of 3 days. Polymerization took place at 60 °C. Ultra-thin sections with a nominal thickness of 70 nm were cut using a Leica UCT ultramicrotome (Leica Microsystems). Regions on the sections were randomly selected and inspected with a FEI Morgagni 268D (FEI) operated at 80 kV. Digital images were acquired using an 11 megapixel Morada CCD camera (Olympus-SIS).
Statistics and reproducibility
The number of biological replicates is indicated in the figures, and sample size in the figure legends. All microscopy experiments were repeated at least 3 times, except screen validation and Supplementary Figs. 5g and 6c, d which were repeated 2 times. The screen and EM-based experiments were done once. All attempts to replicate the data were successful. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
To quantify the NLS-PA sensor localization, the following criteria were used: “nLDs” are defined as spherical, BODIPY-positive structures surrounded by the fluorescent NLS-PA sensor. “nLD+PA foci” are defined as spherical, BODIPY-positive structures surrounded by the fluorescent NLS-PA sensor with additional NLS-PA foci in the nucleus which are not stained by BODIPY. “PA foci” are defined as NLS-PA sensor-labelled foci in the nucleoplasm that do not co-localize with BODIPY. When none of the above-mentioned criteria were met, the sensor was classified as “nucleoplasmic”.
To quantify cells with the 2xNLS-DAG sensor foci, the following criteria were used: if the 2xNLS-DAG sensor forms a roundish spot which exhibits intensity that is two times higher than the intensity of the INM, then the cell was counted as having a focus of the 2xNLS-DAG sensor.
To quantify foci localization of the NLS-PA and 2xNLS-DAG sensors in sei1∆ cells using Sec62-mNeonGreen as a marker for the NE, the following criteria were used: foci are classified as “INM” if they overlap with the Sec62 signal and foci are classified as “nucleoplasmic” if they do not overlap with the Sec62 signal.
To quantify 3xNLS-PS sensor localization, the following criteria were used: “INM” is defined as a fluorescent labeling of the INM with a peak fluorescence intensity at least 1.5 times higher than the nucleoplasm; otherwise “nucleoplasmic”.
Automated quantification of cellular LDs was performed in Fiji using the plugin “Trainable Weka Segmentation”. The segmentation classifier was trained with two classes - to recognize LDs (class 1) and to recognize the background (class 2). The “Watershed” plugin was used to separate adjacent LDs. Next, particle analysis was performed and the diameter of each LD was quantified. Data normality was determined by the Shapiro-Wilk test using the GraphPad Prism software. Statistical significance was evaluated by two-tailed Kolmogorov-Smirnov test using the GraphPad Prism software.
Automated quantification of nLDs was performed as described above, except nLDs were manually selected and their diameter quantified. If the “Watershed” function did not clearly separate two adjacent nLDs, the total area was divided in half to estimate the average area for each nLD.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank M. Brandstetter and S. Jacob for assistance with TEM (Vienna Biocenter Core Facilities, VBCF). We thank Lihi Gal and Amir Fadel for help with the creation of the tailor-made libraries and for the microscopy screen, respectively. A.K. was funded by ERC-COG (772032; NPC BUILD), SFB F 7914-B, and by the NOMIS Foundation. Work in the Schuldiner lab is supported by the CZI grant (2023-331952). The robotic system of the Schuldiner lab was purchased through the support of the Blythe Brenden-Mann Foundation. MS is an Incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
Author contributions
A.K. and A.R. conceived the study. A.R. performed experiments. E.S. performed AlphaFold 3 modeling. M.S. oversaw the screening procedures. A.K. and A.R. wrote the paper with input from the coauthors. All authors read and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data reported in this article are available in the main text and its supplemental material. Plasmids and yeast strains generated in this study are available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54811-z.
References
- 1.Bahmanyar, S. & Schlieker, C. Lipid and protein dynamics that shape nuclear envelope identity. Mol. Biol. Cell31, 1315–1323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mannino, P. J. & Lusk, C. P. Quality control mechanisms that protect nuclear envelope identity and function. J. Cell Biol.221, e202205123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ungricht, R. & Kutay, U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol.10.1038/nrm.2016.153 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Harayama, T. & Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol.19, 281–296 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Carman, G. M. & Han, G. S. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem.80, 859–883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa, A. D. et al. Compartmentalized Synthesis of Triacylglycerol at the Inner Nuclear Membrane Regulates Nuclear Organization. Dev. cell50, 755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romanauska, A. & Kohler, A. The Inner Nuclear Membrane Is a Metabolically Active Territory that Generates Nuclear Lipid Droplets. Cell174, 700–715 e718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romanauska, A. & Kohler, A. Reprogrammed lipid metabolism protects inner nuclear membrane against unsaturated fat. Dev. cell56, 2562–2578 e2563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanauska, A. & Kohler, A. Lipid saturation controls nuclear envelope function. Nat. Cell Biol.25, 1290–1302 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, S., Carrasquillo Rodri Guez, J. W., Merta, H. & Bahmanyar, S. A membrane-sensing mechanism links lipid metabolism to protein degradation at the nuclear envelope. J. Cell Biol.222, e202304026 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry, S. A., Kohlwein, S. D. & Carman, G. M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics190, 317–349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loewen, C. J. et al. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science304, 1644–1647 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Hofbauer, H. F. et al. The molecular recognition of phosphatidic acid by an amphipathic helix in Opi1. J. Cell Biol.217, 3109–3126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature418, 387–391 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Breslow, D. K. et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods5, 711–718 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuldiner, M. et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell123, 507–519 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Yan, R. et al. Human SEIPIN Binds Anionic Phospholipids. Dev. Cell47, 248–256 e244 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Cartwright, B. R. et al. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. cell26, 726–739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolinski, H. et al. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochimica et. biophysica acta1851, 1450–1464 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Fei, W. H. et al. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol.180, 473–482 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fei, W. et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet.7, e1002201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szymanski, K. M. et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA104, 20890–20895 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thaller, D. J. & Patrick Lusk, C. Fantastic nuclear envelope herniations and where to find them. Biochemical Soc. Trans.46, 877–889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucic, I., Truebestein, L. & Leonard, T. A. Novel Features of DAG-Activated PKC Isozymes Reveal a Conserved 3-D Architecture. J. Mol. Biol.428, 121–141 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Oancea, E. & Meyer, T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell95, 307–318 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Del Vecchio, K. & Stahelin, R. V. Investigation of the phosphatidylserine binding properties of the lipid biosensor, Lactadherin C2 (LactC2), in different membrane environments. J. Bioenerg. Biomembr.50, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leventis, P. A. & Grinstein, S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev. Biophys.39, 407–427 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Maeda, K. et al. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature501, 257–261 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Yeung, T. et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Tsuji, T. et al. Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc. Natl. Acad. Sci. USA116, 13368–13373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meinema, A. C. et al. Long Unfolded Linkers Facilitate Membrane Protein Import Through the Nuclear Pore Complex. Science333, 90–93 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Thiam, A. R. & Ikonen, E. Lipid Droplet Nucleation. Trends Cell Biol.31, 108–118 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Schneiter, R. & Choudhary, V. Seipin collaborates with the ER membrane to control the sites of lipid droplet formation. Curr. Opin. Cell Biol.75, 102070 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathiowetz, A. J. & Olzmann, J. A. Lipid droplets and cellular lipid flux. Nat. Cell Biol.26, 331–345 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choudhary, V., El Atab, O., Mizzon, G., Prinz, W. A. & Schneiter, R. Seipin and Nem1 establish discrete ER subdomains to initiate yeast lipid droplet biogenesis. J. Cell Biol.219, e201910177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung, J. et al. LDAF1 and Seipin Form a Lipid Droplet Assembly Complex. Dev. Cell51, 551–563 e557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi, A. S. et al. Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun.9, 2940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassan, A. et al. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol.203, 985–1001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, S. et al. Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun.9, 2939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popken, P., Ghavami, A., Onck, P. R., Poolman, B. & Veenhoff, L. M. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. cell26, 1386–1394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khondker, S., Han, G. S. & Carman, G. M. Phosphorylation-mediated regulation of the Nem1-Spo7/Pah1 phosphatase cascade in yeast lipid synthesis. Adv. Biol. Regul.84, 100889 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karanasios, E. et al. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. cell24, 2124–2133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi, A. S. et al. A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol.215, 515–529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisenberg-Bord, M. et al. Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol.217, 269–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren, J. et al. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Mol. Biol. cell25, 712–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro, I. G. et al. Promethin Is a Conserved Seipin Partner Protein. Cells8, 268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teixeira, V. et al. Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol.217, 127–138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez-Guerra, I. et al. LDO proteins and Vac8 form a vacuole-lipid droplet contact site to enable starvation-induced lipophagy in yeast. Dev. Cell59, 759–775 e755 (2024). [DOI] [PubMed] [Google Scholar]
- 49.Diep, D. T. V. et al. A metabolically controlled contact site between vacuoles and lipid droplets in yeast. Dev.Cell59, 740–758 e710 (2024). [DOI] [PubMed] [Google Scholar]
- 50.Kerppola, T. K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev. Biophys.37, 465–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olzmann, J. A. & Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arlt, H. et al. Seipin forms a flexible cage at lipid droplet formation sites. Nat. Struct. Mol. Biol.29, 194–202 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klug, Y. A. et al. Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex. Nat. Commun.12, 5892 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sui, X. et al. Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. cell Biol.217, 4080–4091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berardinelli, W. An undiagnosed endocrinometabolic syndrome: report of 2 cases. J. Clin. Endocrinol. Metab.14, 193–204 (1954). [DOI] [PubMed] [Google Scholar]
- 56.Magre, J. et al. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet.28, 365–370 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Seip, M. Lipodystrophy and gigantism with associated endocrine manifestations. A new diencephalic syndrome? Acta Paediatr. (Stock.)48, 555–574 (1959). [PubMed] [Google Scholar]
- 58.Binns, D., Lee, S., Hilton, C. L., Jiang, Q. X. & Goodman, J. M. Seipin is a discrete homooligomer. Biochemistry49, 10747–10755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walther, T. C., Kim, S., Arlt, H., Voth, G. A. & Farese, R. V. Jr Structure and function of lipid droplet assembly complexes. Curr. Opin. Struct. Biol.80, 102606 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klug, Y. A., Ferreira, J. V. & Carvalho, P. A unifying mechanism for seipin-mediated lipid droplet formation. FEBS Lett.598, 1116–1126 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grippa, A. et al. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol.211, 829–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, C. W., Miao, Y. H. & Chang, Y. S. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci.127, 1214–1228 (2014). [DOI] [PubMed]
- 63.Han, S., Binns, D. D., Chang, Y. F. & Goodman, J. M. Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(DeltaNterm) only in combination with Ldb16p. BMC Cell Biol.16, 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prasanna, X. et al. Seipin traps triacylglycerols to facilitate their nanoscale clustering in the endoplasmic reticulum membrane. PLoS Biol.19, e3000998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Renne, M. F., Corey, R. A., Ferreira, J. V., Stansfeld, P. J. & Carvalho, P. Seipin concentrates distinct neutral lipids via interactions with their acyl chain carboxyl esters. J. Cell Biol.221, e202112068 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zoni, V. et al. Seipin accumulates and traps diacylglycerols and triglycerides in its ring-like structure. Proc. Natl. Acad. Sci. USA118, e2017205118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature10.1038/s41586-024-07487-w (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monneron, A., Blobel, G. & Palade, G. E. Fractionation of the nucleus by divalent cations. Isolation of nuclear membranes. J. Cell Biol.55, 104–125 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sim, M. F. et al. The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol. Metab.2, 38–46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho, B., Baryshnikova, A. & Brown, G. W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst.6, 192–205 e193 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Smoyer, C. J. et al. Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol.215, 575–590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soltysik, K. et al. Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol.220, e202005026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salo, V. T. et al. Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev. cell50, 478–493 e479 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Thiam, A. R., Farese, R. V. Jr. & Walther, T. C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. cell Biol.14, 775–786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akey, C. W. et al. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell185, 361–378 e325 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.King, M. C., Lusk, C. P. & Blobel, G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature442, 1003–1007 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Hamilton, J. A. & Small, D. M. Solubilization and localization of triolein in phosphatidylcholine bilayers: a 13C NMR study. Proc. Natl. Acad. Sci. USA78, 6878–6882 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim, S. et al. Seipin transmembrane segments critically function in triglyceride nucleation and lipid droplet budding from the membrane. eLife11, e75808 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang, M. et al. Lack of testicular seipin causes teratozoospermia syndrome in men. Proc. Natl. Acad. Sci. USA111, 7054–7059 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian, Y. et al. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet.7, e1001364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thaller, D. J. et al. Direct binding of ESCRT protein Chm7 to phosphatidic acid-rich membranes at nuclear envelope herniations. J. Cell Biol.220, e202004222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast21, 947–962 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Cohen, Y. & Schuldiner, M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol.781, 127–159 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Tong, A. H. & Boone, C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol.313, 171–192 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this article are available in the main text and its supplemental material. Plasmids and yeast strains generated in this study are available from the corresponding author upon request. Source data are provided with this paper.