Abstract
Copy number variations in the SMN1 gene on chromosome 5 are the primary cause of Spinal Muscular Atrophy (SMA) disease, characterized by muscle weakness and degeneration due to impaired alpha motor neurons in the spinal cord. To obtain a comprehensive molecular understanding of the SMA, including carriers, silent carriers, and patients in the Iranian population, we analyzed data from 5224 individuals referred to Kariminejad - Najmabadi Pathology & Genetics Center, Tehran, Iran, between 2006 and 2023 using MLPA and quantitative RT-PCR methods. The carrier frequency of SMA was estimated to be 5.55%. Furthermore, 3.06% of SMA parents (n = 24) had two copies of the SMN1 gene. Among 725 patients, those with an earlier onset of SMA were more likely to have two copies of the SMN2 gene (46.45%) and no copies of the NAIP gene (49.36%). Among the 654 fetal samples screened for SMA, 22.33% were found to be affected, while 3.46% of their parents tested normal. These findings are valuable for genetic counseling, carrier screening, and prenatal diagnosis of SMA in Iran. Furthermore, they underscore the importance of CNV analysis of SMN1, SMN2, and NAIP genes for accurate diagnosis and prognosis of SMA.
Keywords: Spinal muscular atrophy, Carrier frequency, Silent carriers, SMN, Copy numbers, Iran
Subject terms: Clinical genetics, Medical genetics, Population genetics
Introduction
Spinal Muscular Atrophy (SMA), resulting from the degeneration of motor neurons within the spinal cord, is characterized by muscle weakness stemming from progressive muscle degeneration and atrophy. The estimated incidence of this condition is approximately 1 in 10,000 individuals, with a prevalence ranging from 1 to 2 per 100,000 population1,2. SMA is an autosomal recessive disorder caused by mutations in the survival of motor neurons 1 (SMN1) gene, located in an inverted, duplicated region on chromosome 5 (locus 5q13). This gene encodes a 294 amino acid protein, which, along with other proteins, constructs the SMN complex. The SMN complex is essential in assembling spliceosomal small nuclear ribonucleoproteins (snRNPs)3,4. The majority of SMA patients (94%), have a homozygous deletion of the SMN1 gene, while the remaining cases exhibit inherited or de novo point mutations5. Most deletions in the SMN1 gene involve exons 7 and 8; however, in some cases, recombination between exon 7 of SMN2 and exon 8 of SMN1 can lead to the formation of a hybrid SMN gene, where exon 7 is deleted while exon 8 remains intact6. Although the condition primarily results from mutations in the SMN1 gene, other genes within the same genomic region, notably SMN2 and the neuronal apoptosis inhibitory protein (NAIP), play critical roles in influencing disease severity. SMN2 is a very similar gene to SMN1, with only a few nucleotide differences including two exonic variations (c.840 C > T in exon 7 and c.*239A > G in exon8). The change in exon 7 affects the splicing process, leading to truncation and instability of the SMN2 protein3,7. The distinct nucleotides in these genes are targets for developing molecular genetic methods to differentiate between genes, quantify their copy numbers, and detect SMN1 mutations. The severity of SMA is inversely related to the copy numbers of the SMN2 gene, as it can produce a limited amount of SMN protein. Additionally, severe types of SMA cases often display deletions in the exon 5 of the NAIP gene, possibly due to unequal crossover, while milder cases usually lack NAIP deletions. It’s important to note that SMN2 and NAIP mutations don’t cause SMA but can affect disease presentation8–11.
Regarding the recommendations from The American College of Medical Genetics (ACMG) advocating for population-based carrier screening for SMA12, several countries have contributed data regarding carrier frequency. Ethnicity emerges as a significant factor influencing allelic variations of SMN1, with Iran and Arabic countries demonstrating elevated carrier frequencies, while individuals of African descent exhibit a notably higher prevalence of duplicated alleles. This disparity implies a greater proportion of ‘2 + 0’ carriers within these populations, potentially resulting in a lower detection rate compared to other ethnic groups. Furthermore, previous research conducted in Iran has indicated a noteworthy elevation in carrier risk and ‘2-copy’ risk among Iranians2.
Given the absence of comprehensive molecular picture of SMA in Iran in the context of carrier frequency, silent carrier status, and patient diagnosis, we aimed to present the result from the molecular analysis of a large sample of individuals referred to Kariminejad - Najmabadi Pathology & Genetics Center for carrier detection, diagnosis and prenatal diagnosis which can facilitate informed decision-making and genetic counseling for families within multi-ethnic populations with a high prevalence of consanguineous marriage, such as Iran.
Results
Investigated individuals
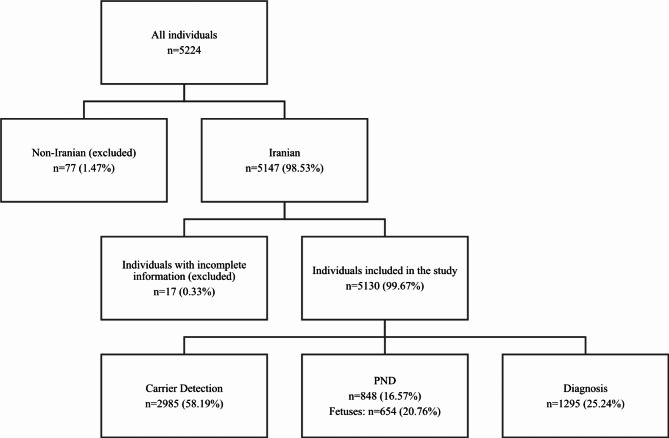
A total of 5224 individuals were referred to our laboratory and tested for SMA between 2006 and 2023. Out of all the individuals investigated in our study, 42.92% (n = 2242) had a positive history of SMA in their core and/or extended family. Of note, 39.28% (n = 2052) were born to consanguineous parents. Figure 1 illustrates the sequential pathway followed for the filtration and categorization of individuals.
Fig. 1.
Flowchart of studied individuals.
Carrier frequency of SMA
Out of 2985 individuals referred for carrier detection, analysis of SMN1 copy numbers among 1225 non-relative Iranian individuals (Mean age (SD) = 30 (± 7) years) with no prior history of SMA, showed that 5.55% (n = 68, CI = 0.95%, 4.36–7.02) had one copy of SMN1 being a carrier for the disease. Among the remaining individuals, the majority (86.05%, n = 1054) demonstrated two copies of SMN1, while the rest of the individuals exhibited an occurrence of more than two copies. We also assessed the copy numbers of SMN2 among the same population. Data on both SMN1 and SMN2 copy numbers is presented in Table 1.
Table 1.
Distribution of SMN1/SMN2 Copy numbers among 1225 Iranian individuals.
| SMN1 | SMN2 | ||
|---|---|---|---|
| Copy number | Frequency | Copy number | Frequency |
| Zero | 0 (0.00%) | Zero | 57 (4.65%) |
| One | 68 (5.55%) | One | 355 (28.98%) |
| Two | 1054 (86.04%) | Two | 641 (52.33%) |
| Three | 87 (7.10%) | Three | 80 (6.53%) |
| Four | 16 (1.31%) | Four | 10 (0.82%) |
| NA | 0 (0%) | NA* | 82 (6.69%) |
| Total | 1225 | Total | 1225 |
* Individuals who underwent testing using Real-time PCR did not have data on SMN2 copy numbers because only exon 7 of SMN1 was checked.
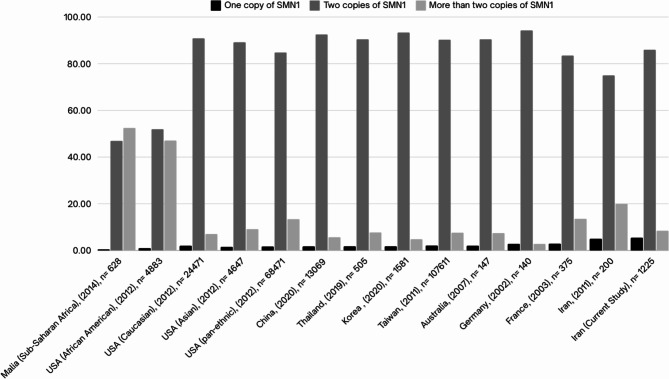
SMA carrier frequency across various countries is summarized in Table 2. Studies reporting the distribution of SMN1 copy numbers among healthy individuals are presented in Fig. 2.
Table 2.
Carrier frequency of SMA across various countries.
| Country | Carrier frequency | Sample size | Technique | Reference |
|---|---|---|---|---|
| Iran (current study) | 68 (5.55%) | 1225 | Real-time PCR/MLPA p021 | |
| Iran (previous study) | 10 (5.00%) | 200 | Real-time PCR | 13 |
| Saudi Arabia (2007) | 9 (4.81%) | 187 | Multiplex-PCR | 14 |
| Morocco | 6 (4.00%) | 150 | Real-time PCR | 15 |
| Qatar | 381 (2.84%) | 13,426 | The SMN copy number caller tool using WGS data | 16 |
| Saudi Arabia (2022) | 108 (2.57%) | 4198 | Multiplex PCR with Dral restriction fragment analysis | 17 |
| North India | 16 (2.64%) | 606 | MLPA p060 | 18 |
| Taiwan | 2262 (2.1%) | 107,611 | DHPLC/ Multiplex-PCR | 19 |
| Korea | 29 (1.83%) | 1581 | MLPA p460 A1 | 20 |
| China | 231 (1.77%) | 13,069 | Quantitative Real-time PCR | 21 |
| Thailand | 9 (1.78%) | 505 | Quantitative Real-time PCR | 22 |
| USA (Pan-ethnic) | 1162 (1.69%) | 68,471 | Quantitative Real-time PCR | 23 |
| Germany | 4 (2.86%) | 140 | Real-time PCR | 24 |
| France (2003) | 11 (2.93%) | 375 | Competitive PCR and primer extension | 25 |
| France (2012) | 13 (2.09%) | 621 | Quantitative multiplex PCR | 26 |
| Sweden | 9 (1.79%) | 502 | Quantitative multiplex PCR | 26 |
| Australia | 3 (2.04%) | 147 | Quantitative Real-time PCR | 27 |
| Sub-Saharan Africa | 3 (0.48%) | 628 | qPCR | 28 |
Fig. 2.
Copy number(s) of SMN1 in normal individuals from different countries with no family history of SMA.
SMA parents with two or more SMN1 copies
From a total of 785 parents with at least one SMA-affected child, who were considered to be obligate carriers of SMA, 96.69% (n = 759) had one copy, 3.06% (n = 24) had two copies and 0.25% (n = 2) had three copies of SMN1. Table 3 presents the proportion of SMA parents with two copy numbers of SMN1 from various countries.
Table 3.
Frequency of SMA parents with two copies of SMN1 among various countries.
| Country of study | Number of SMA parents with two SMN1 copies (%) | Sample size | Reference |
|---|---|---|---|
| Spain | 21 (4.30%) | 488 | 29 |
| Australia (2007) | 7 (5.98%) | 117 | 27 |
| China | 2 (4.54%) | 40 | 30 |
| Japan | 3 (4.61%) | 65 | 31 |
| Australia (2023) | 9 (7.62%) | 118 | 32 |
| France | 9 (4.45%) | 202 | 25 |
| Saudi Arabia | 8 (5.33%) | 150 | 14 |
| North America | 4 (4.00%) | 100 | 33 |
| Current study | 24 (3.06%) | 785 |
SMA diagnosis
A total of 1295 individuals presenting with symptoms or signs indicative of SMA were referred for diagnosis. Among them, 725 individuals were confirmed to have SMA through the identification of homozygous deletion in exon 7 of the SMN1 gene, resulting in a diagnostic rate of 56%. Of note, 3.63% of cases (n = 47) exhibited heterozygous deletion of SMN1, while the remaining cases (40.39%) showed normal results regarding SMA.
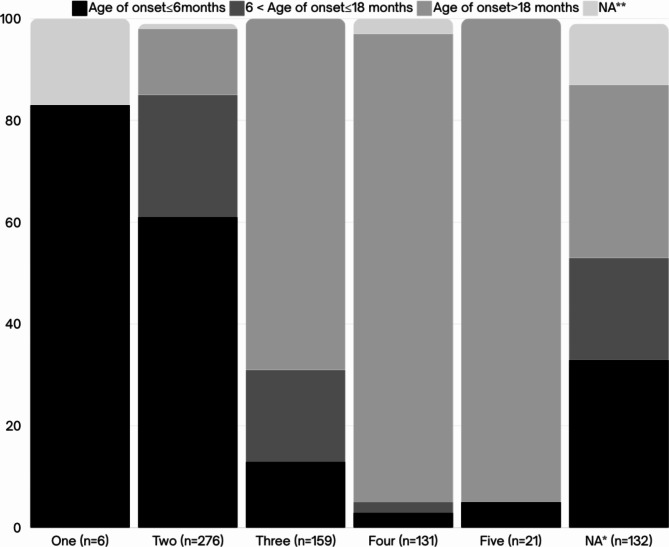
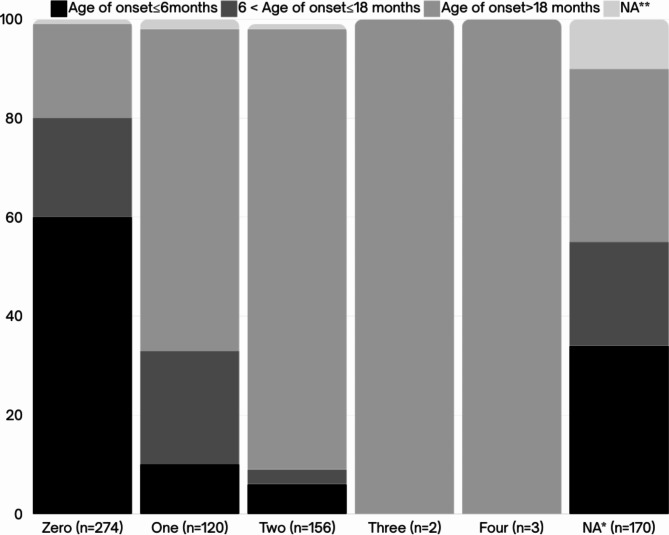
Of all 725 patients, 81.79% (n = 593) were tested with MLPA and 18.21% (n = 132) were tested using Real-time PCR. The majority of cases (56%) were born to consanguineous marriages. Figure 3 depicts the SMN2 gene copy numbers for affected individuals with different ages of onset, indicating that the majority of patients had two copies of SMN2. Figure 4, on the other hand, displays the NAIP gene copy numbers for patients with varying onset ages, revealing that most patients had zero copies of the NAIP gene.
Fig. 3.
Distribution of SMA patients with various numbers of exon 7 of the SMN2 gene. * Patients who underwent testing using Real-time PCR did not have data on SMN2 copy numbers because only exon 7 of SMN1 was checked. ** The age of onset for some of the patients has not been recorded.
Fig. 4.
Distribution of SMA patients with various numbers of exon 5 of the NAIP gene. * Data on NAIP copy numbers for 170 SMA patients has not been documented due to either undergoing Real-time PCR testing or the absence of registered NAIP data. ** The age of onset for some of the patients has not been recorded.
Of note, 24 patients were found to possess a deletion in exon 7 of SMN1 while retaining exon 8, indicating the presence of a hybrid SMN gene. The majority of patients with the hybrid SMN1 gene had an age of onset above 18 months (66.66%, n = 16), with a smaller proportion showing symptoms under 6 months of age (20.83%, n = 5), and the remaining cases experiencing onset between 6 and 18 months (12.5%, n = 3). Information regarding the copy number of SMN2 and NAIP for these patients can be found in Table 4.
Table 4.
SMN2 and NAIP copy numbers of patients with hybrid SMN gene (n = 24).
| Zero copies | One copy | Two copies | Three copies | Four copies | Five copies | Total | |
|---|---|---|---|---|---|---|---|
| SMN2 copy numbers |
0 (0.00%) |
1(4.17%) |
2 (8.33%) |
13 (54.17%) | 7 (29.17%) |
1 (4.17%) |
24 |
| NAIP copy Numbers | 8 (33.33%) | 8 (33.33%) | 8 (33.33%) |
0 (0.00%) |
0 (0.00%) |
0 (0.00%) |
24 |
Prenatal diagnosis
Data from a total of 654 fetuses were examined in our analysis. Among them, 22.33% (n = 146) were found to be affected, 50.15% (n = 328) were identified as carriers, and 27.52% (n = 180) tested normal.
The test results for the parents of the 146 affected fetuses from 130 families have been presented in Table 5. Notably, 3.46% of parents (n = 9) were tested normal for SMA.
Table 5.
Test results for parents of affected fetuses.
| Test result of parents with affected fetuses | Carrier parents | Normal parents | NA* | Total |
|---|---|---|---|---|
| Frequency | 233 (89.62%) | 9 (3.46%) | 18 (6.92%) | 260 |
* Data regarding parents’ test results have not been recorded in some cases.
Discussion
In this study, we investigated individuals from across Iran, including those referred to carrier detection, parents of SMA patients, SMA patients themselves, and fetuses undergoing prenatal diagnosis. Given the diversity of our population34, we included carrier frequency data and information from the parents of SMA patients from various countries to provide a broader perspective on SMA across different populations. Our selection aimed to represent diverse ethnic groups, as SMA carrier frequency vary significantly among populations. We chose countries with similar cultural practices, such as high rates of consanguinity, to facilitate relevant comparisons with Iran. Additionally, we selected countries based on the availability of well-documented carrier frequency data.
After comparing our data to that of other countries to assess our status among diverse populations worldwide, we noted a higher carrier frequency rate compared to the United States, European, and East Asian populations, which confirms our earlier findings13. The carrier frequency in our population aligns more closely to Middle East countries which ranges from 2.57 to 4.81% (Table 2). This study highlights the significance of establishing a well-structured referral system for genetic counseling, not only in Iran but also in countries sharing a similar cultural background, particularly those with a high prevalence of consanguineous marriages. The analysis also revealed that 4.65% of normal individuals lack the SMN2 gene (Table 1), which falls within the range of previous studies conducted elsewhere (5–15%)3,35.
To assess the status of silent carriers, we evaluated SMA parents to identify those with two copies of the SMN1 gene. We found that the frequency of SMA parents possessing two SMN1 copies is 3.06%, which may indicate the silent carrier frequency in Iran. This is also supported by our finding obtained from the investigation of the parents of the affected fetuses (3.46%) (Table 5). This result is more consistent with the reports from European, American and Asian countries (Table 3) but was lower than the previous finding by Sharifi, et al. which reported a silent carrier frequency of 11.4%36. A high frequency of silent carriers has been uniquely reported in African populations, which is attributed to the high copy numbers of the SMN1 gene in this population2,23,28. However, a considerable dissimilarity has been noted between our population and African populations in the current study (Fig. 2) and a previous report by Mehrjoo et al.34, supporting the claim that our findings provide a more accurate representation of the SMA silent carriers’ status in Iran. Typically, healthy couples where one parent is a carrier are not referred to prenatal diagnosis, which poses a potential risk that should be taken into consideration by genetic counselors. Unfortunately, access to additional family members was not available in the families under investigation to discern the precise allelic phases and accurately identify silent carrier cases.
From the perspective of patient diagnosis, the second causative variant was not detected approximately in 4% of the investigated affected, indicating the need for conducting additional analysis to detect point mutations which would be beneficial in determining the precise genetic cause of the disease.
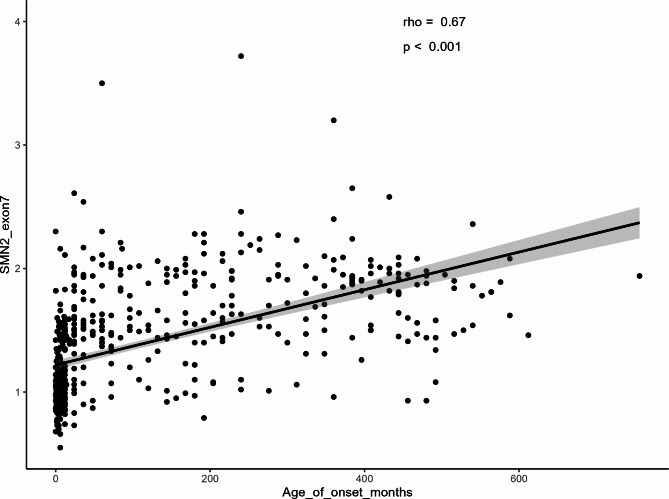
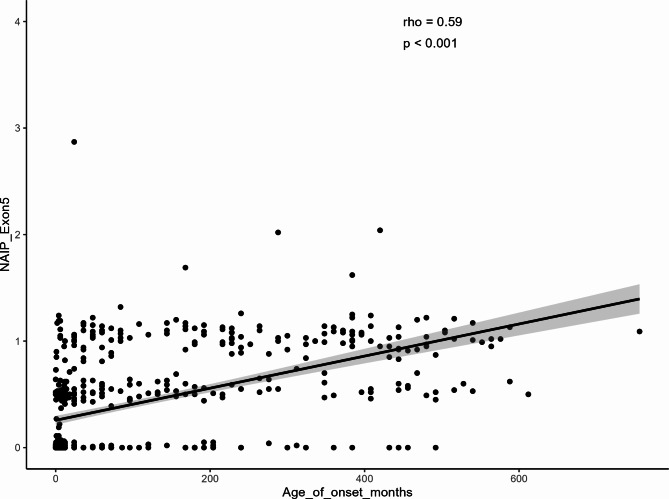
Furthermore, SMN2 and NAIP copy numbers observed in patients, demonstrated a positive correlation with the onset of the disease (Figs. 5 and 6), remaining largely consistent with previous studies2,37,38.
Fig. 5.
Correlation between SMN2 copy numbers (ratio of exon7) and age of onset of the disease.
Fig. 6.
Correlation between NAIP copy numbers (ratio of exon 5) and age of onset of the disease.
The findings regarding the hybrid SMN gene in our patients indicate that, according to prior reviews and previous research conducted in Iran39 the prevalence of individuals with the hybrid SMN1 gene falls within the low-frequency category40 In contrast to earlier research39, our study did not show a notable correlation between the hybrid SMN gene and disease severity, as measured by the age of onset (Correlation coefficient: 0.071, P-Value: 0.088). This may be attributed to the fact that both studies had a limited sample size, with only 24 samples included in each study.
This report represents the largest sample size reported in Iran to date, providing valuable insights into the frequency of carriers and silent carriers, as well as the results observed in patients with SMA. These findings underscore the significance of promoting genetic counseling and carrier screening for SMA before pregnancy in Iran, along with acknowledging the potential risk of being a silent carrier. Subsequently, it’s essential to prioritize prenatal testing for couples at risk to prevent the birth of children affected by SMA. Additionally, it’s crucial to identify the copy number variations of SMN2 and NAIP genes in patients, as they play a vital role in predicting prognosis and characterizing the disease phenotype.
More extensive genotype-phenotype correlations are limited due to the insufficient clinical characteristics’ data. Additionally, limited access to the investigated families prevented us from confirming the status of silent carriers.
Materials and methods
Subject
We analyzed data of 5224 individuals referred to Kariminejad - Najmabadi Pathology & Genetics Center for SMA carrier detection, and patient diagnosis between 2006 and 2023 by physicians. Data from non-Iranian individuals (n = 77, 1.47%) and individuals with inadequate information (n = 17, 0.33%) were excluded from the study dataset. The flowchart depicting the studied population has been shown in Fig. 1.
Methods of testing
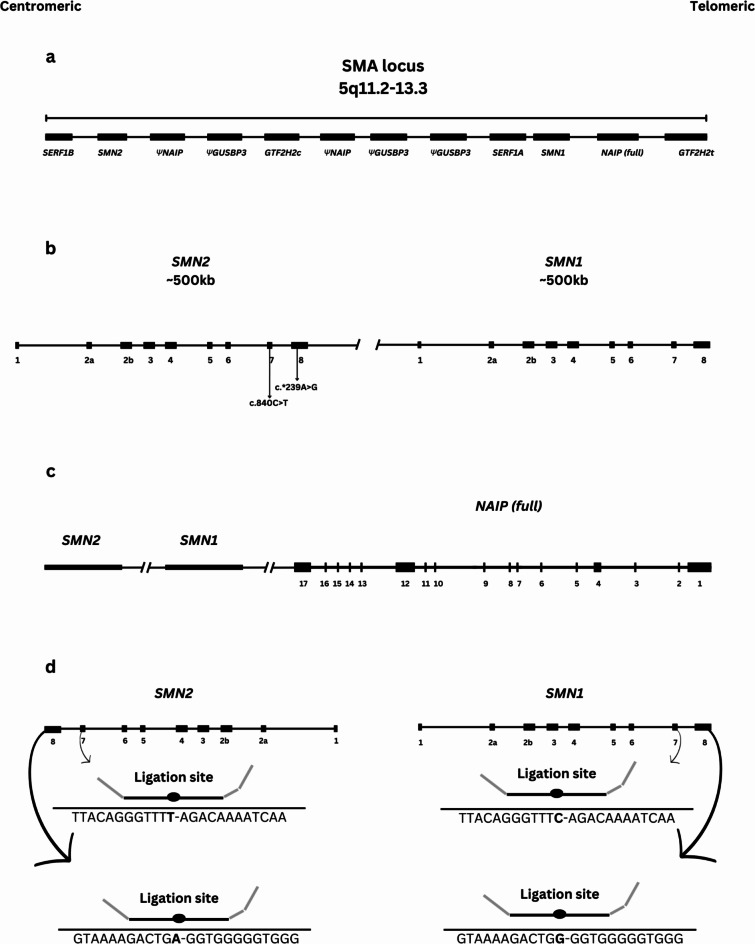
The majority of the samples (n = 4095, 78.38%) were tested using the Multiplex Ligation-dependent Probe Amplification (MLPA) technique, following the manufacturer’s instructions as outlined in the MRC Holland website (https://www.mrcholland.com/product/P021/634). The MLPA analysis utilizes a set of 32 specific probes targeting different regions of the SMA locus. Specifically, two probes target exon 7 and exon 8 of the SMN1 and SMN2 genes. Detection accuracy for copy number variations and gene conversions is remarkably reliable, with both analytical sensitivity and specificity exceeding 99%. The ligation site of these probes is located at different nucleotides between the two genes. This difference in the ligation site allows the MLPA assay to distinguish SMN1 from SMN2, as the probes will only ligate and amplify their respective target sequences (Fig. 7). The PCR products obtained from the MLPA analysis were subsequently analyzed using Coffalyser.NET software, which is also developed by MRC-Holland. The final ratios were determined by comparing each sample to reference samples to compute the copy numbers. Internal validation with 16 DNA samples from healthy individuals was conducted to ensure a standard deviation of ≤ 0.10 for all reference probes.
Fig. 7.
Schematic diagram of the SMA locus. a: expansion of the SMA locus on the long arm of chromosome 5, including SMN1, SMN2, and NAIP genes, as well as nearby genes and pseudogenes in this region42. b: SMN1 and SMN2 genes and two nucleotide differences in exons 7 and 8 of SMN1 and SMN2*. c: the NAIP gene and its position near the SMN1 and SMN2. d: Mechanism for detecting SMN1 and SMN2 copy numbers using the MLPA technique. The ligation site of each probe is located in the different variants of the two genes on exons 7 and 8. * The exon numbering in this schematic diagram follows the traditional format, rather than the sequential 1 to 9 numbering used in online databases for the exons of the SMN1 and SMN2 genes.
Before to the widespread adoption of MLPA for copy number variation analysis, the earlier samples (n = 1129, 22.61%) were tested using Real-time PCR with the delta-delta Ct method to determine the copy numbers of exon 7 of the SMN1 gene with the specificity of 100% and a sensitivity of 96.2%41. with SYBR green I dye. The real-time PCR assay utilized primers specifically designed to amplify the SMN1 gene. To differentiate SMN1 from the highly similar SMN2 gene, the 3’ ends of the primers are designed to target SMN1-specific sequences - in exon 7 (Forward primer: 5’-CCTTTTATTTTCCTTACAGGGTTTC-3’, reverse primer: 5’-GATTGTTTTACATTAACCTTTCAACTTTT-3’). The specificity of the SMN1 primers was confirmed with the Albumin gene in samples from both patients and normal individuals (Forward primer: 5’-AGCTATCCGTGGTCCTGAAC-3’, reverse primer: 5’-TTCTCAGAAAGTGTGCATATATCTG-3‘). To validate our test results, 20 SMA patients, 20 obligate carrier couples, and 20 healthy individuals from the normal population were tested. The methods employed in this study followed the relevant guidelines and regulations.
Statistical analyses
Data filtration and statistical analyses were performed in RStudio software, version 2023.09.1 + 494. Statistical significance was determined at a threshold of p < 0.05. To estimate the carrier frequency among the Iranian population, a confidence interval (CI) of 95% was employed. Non-parametric Spearman’s correlation coefficient (rho) was employed to examine the relationship between copy numbers of SMN2 and NAIP genes and the age of onset of the disease as well as assess the correlation between the presence of a hybrid SMN gene and the age of onset of the disease.
Acknowledgements
We extend our heartfelt gratitude to all SMA families for their invaluable cooperation, as well as physicians, healthcare centers, and genetic laboratories throughout Iran who have referred families to our laboratory.
Author contributions
Hossein Najmabadi: Idea, study design and supervisionAli Khanbazi: Data gathering, analysis, and manuscript writingMaryam Beheshtian: Study design, supervision, and manuscript writingMaryam Azad, Masoumeh Akbari Kelishomi: Conducting genetic testingFariba Afroozan, Fatemeh Fatehi, Khadijeh Noudehi, Shima Zamanian Najafabadi, Mohammadamin Omrani, Haleh Habibi, Maryam Taghdiri, Isa Abdi Rad, Shahriar Nafissi, Aria Jankhah, Hilda Yazdan, Parvaneh Daneshmand, Seyed Hosseinali Saberi, Kimia Kahrizi, Ariana Kariminejad: Genetic counseling and clinical characterization of individuals.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethical statements
This study has been confirmed by the ethical committee of the University of Social Welfare and Rehabilitation Sciences in Iran (Ethics code: IR.USWR.REC.1401.247). Prior to conducting any testing, it was imperative to obtain informed consent from all individuals and/or their legal guardian(s).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ogino, S., Leonard, D. G., Rennert, H., Ewens, W. J. & Wilson, R. B. Genetic risk assessment in carrier testing for spinal muscular atrophy. Am. J. Med. Genet.110, 301–307. 10.1002/ajmg.10425 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Verhaart, I. E. C. et al. Prevalence, incidence and carrier frequency of 5q–linked spinal muscular atrophy – A literature review. Orphanet J. Rare Dis.12, 124. 10.1186/s13023-017-0671-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre, S. et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 80, 155–165. 10.1016/0092-8674(95)90460-3 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Paushkin, S., Gubitz, A. K., Massenet, S. & Dreyfuss, G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell. Biol.14, 305–312. 10.1016/s0955-0674(02)00332-0 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Wirth, B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum. Mutat.15, 228–237. 10.1002/(sici)1098-1004(200003)15:3%3C228::Aid-humu3%3E3.0.Co;2-9 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Hahnen, E., Schönling, J., Rudnik-Schöneborn, S., Zerres, K. & Wirth, B. Hybrid survival motor neuron genes in patients with autosomal recessive spinal muscular atrophy: New insights into molecular mechanisms responsible for the disease. Am. J. Hum. Genet.59, 1057–1065 (1996). [PMC free article] [PubMed] [Google Scholar]
- 7.Blasco-Pérez, L. et al. Beyond copy number: A new, rapid, and versatile method for sequencing the entire SMN2 gene in SMA patients. Hum. Mutat.42, 787–795. 10.1002/humu.24200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre, S. et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet.16, 265–269. 10.1038/ng0797-265 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Burlet, P. et al. Large scale deletions of the 5q13 region are specific to Werdnig-Hoffmann disease. J. Med. Genet.33, 281–283. 10.1136/jmg.33.4.281 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy, N. et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 80, 167–178. 10.1016/0092-8674(95)90461-1 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Velasco, E., Valero, C., Valero, A., Moreno, F. & Hernández-Chico, C. Molecular analysis of the SMN and NAIP genes in Spanish spinal muscular atrophy (SMA) families and correlation between number of copies of cBCD541 and SMA phenotype. Hum. Mol. Genet.5, 257–263. 10.1093/hmg/5.2.257 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Prior, T. W. Carrier screening for spinal muscular atrophy. Genet. Med.10, 840–842. 10.1097/GIM.0b013e318188d069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasanzad, M. et al. Carrier frequency of SMA by quantitative analysis of the SMN1 deletion in the Iranian population. Eur. J. Neurol.17, 160–162. 10.1111/j.1468-1331.2009.02693.x (2010). [DOI] [PubMed] [Google Scholar]
- 14.Al Jumah, M., Majumdar, R., Rehana, Z., Al Rajeh, S. & Eyaid, W. A pilot study of spinal muscular atrophy carrier screening in Saudi Arabia. J. Pediatr. Neurol.5, 221–224 (2007). [Google Scholar]
- 15.Lyahyai, J., Sbiti, A., Barkat, A., Ratbi, I. & Sefiani, A. Spinal muscular atrophy carrier frequency and estimated prevalence of the disease in Moroccan newborns. Genet. Test. Mol. Biomarkers. 16, 215–218. 10.1089/gtmb.2011.0149 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Ibrahim, F. et al. Studying carrier frequency of spinal muscular atrophy in the state of Qatar and comparison to other ethnic groups: Pilot study. Mol. Genet. Genomic Med.11, e2184. 10.1002/mgg3.2184 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al Jumah, M. et al. Spinal muscular atrophy carrier frequency in Saudi Arabia. Mol. Genet. Genomic Med. 10, 2049. 10.1002/mgg3.2049 (2022). [DOI] [PMC free article] [PubMed]
- 18.Nilay, M., Moirangthem, A., Saxena, D., Mandal, K. & Phadke, S. R. Carrier frequency of SMN1-related spinal muscular atrophy in north Indian population: the need for population based screening program. Am. J. Med. Genet. A. 185, 274–277. 10.1002/ajmg.a.61918 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Su, Y. N. et al. Carrier screening for spinal muscular atrophy (SMA) in 107,611 pregnant women during the period 2005–2009: A prospective population-based cohort study. PLoS One. 6, e17067. 10.1371/journal.pone.0017067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park, J. E. et al. Carrier frequency of spinal muscular atrophy in a large-scale Korean population. Ann. Lab. Med.40, 326–330. 10.3343/alm.2020.40.4.326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, J. et al. Carrier screening and prenatal diagnosis for spinal muscular atrophy in 13,069 Chinese pregnant women. J. Mol. Diagn.22, 817–822. 10.1016/j.jmoldx.2020.03.001 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Dejsuphong, D. et al. Carrier frequency of spinal muscular atrophy in Thailand. Neurol. Sci.40, 1729–1732. 10.1007/s10072-019-03885-5 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Sugarman, E. A. et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: Clinical laboratory analysis of > 72,400 specimens. Eur. J. Hum. Genet.20, 27–32. 10.1038/ejhg.2011.134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldkötter, M., Schwarzer, V., Wirth, R., Wienker, T. F. & Wirth, B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet.70, 358–368. 10.1086/338627 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusin, V., Clermont, O., Gérard, B., Chantereau, D. & Elion, J. Prevalence of SMN1 deletion and duplication in carrier and normal populations: Implication for genetic counselling. J. Med. Genet.40, e39. 10.1136/jmg.40.4.e39 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corcia, P. et al. Homozygous SMN2 deletion is a protective factor in the Swedish ALS population. Eur. J. Hum. Genet.20, 588–591. 10.1038/ejhg.2011.255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, M. et al. Population screening and cascade testing for carriers of SMA. Eur. J. Hum. Genet.15, 759–766. 10.1038/sj.ejhg.5201821 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Sangaré, M. et al. Genetics of low spinal muscular atrophy carrier frequency in sub-saharan Africa. Ann. Neurol.75, 525–532. 10.1002/ana.24114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alías, L. et al. Improving detection and genetic counseling in carriers of spinal muscular atrophy with two copies of the SMN1 gene. Clin. Genet.85, 470–475. 10.1111/cge.12222 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Sheng-Yuan, Z. et al. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. Eur. J. Hum. Genet.18, 978–984. 10.1038/ejhg.2010.54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ar Rochmah, M. et al. Spinal muscular atrophy carriers with two SMN1 copies. Brain Dev.39, 851–860. 10.1016/j.braindev.2017.06.002 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Davidson, J. E. et al. The Carrier frequency of two SMN1 genes in parents of symptomatic children with SMA and the significance of SMN1 exon 8 in carriers. Genes (Basel). 14. 10.3390/genes14071403 (2023). [DOI] [PMC free article] [PubMed]
- 33.Mailman, M. D. et al. Hybrids monosomal for human chromosome 5 reveal the presence of a spinal muscular atrophy (SMA) carrier with two SMN1 copies on one chromosome. Hum. Genet.108, 109–115. 10.1007/s004390000446 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Mehrjoo, Z. et al. Distinct genetic variation and heterogeneity of the Iranian population. PLoS Genet.15, e1008385. 10.1371/journal.pgen.1008385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mailman, M. D. et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med.4, 20–26. 10.1097/00125817-200201000-00004 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Sharifi, Z. et al. Comprehensive Comprehensive mutation analysis and report of 12 novel mutations in a cohort of patients with spinal muscular atrophy in Iran. J. Mol. Neurosci.71, 2281–2298. 10.1007/s12031-020-01789-0 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Calucho, M. et al. Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul. Disord. 28, 208–215. 10.1016/j.nmd.2018.01.003 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Savad, S. et al. A comprehensive overview of SMN and NAIP copy numbers in Iranian SMA patients. Sci. Rep.13, 3202. 10.1038/s41598-023-30449-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niba, E. T. E. et al. Clinical phenotypes of spinal muscular atrophy patients with hybrid SMN gene. Brain Dev.43, 294–302. 10.1016/j.braindev.2020.09.005 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Omrani, O., Bonyadi, M. & Barzgar, M. Molecular analysis of the SMN and NAIP genes in Iranian spinal muscular atrophy patients. Pediatr. Int.51, 193–196. 10.1111/j.1442-200X.2008.02665.x (2009). [DOI] [PubMed] [Google Scholar]
- 41.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 (2001). [DOI] [PubMed]
- 42.Ruhno, C. et al. Complete sequencing of the SMN2 gene in SMA patients detects SMN gene deletion junctions and variants in SMN2 that modify the SMA phenotype. Hum. Genet.138, 241–256. 10.1007/s00439-019-01983-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.