Abstract
Double-stranded RNA (dsRNA) has been shown to play a key role as an inducer of different interference phenomena occurring in both the plant and animal kingdoms. Here, we show that dsRNA derived from viral sequences can interfere with virus infection in a sequence-specific manner by directly delivering dsRNA to leaf cells either by mechanical inoculation or via an Agrobacterium-mediated transient-expression assay. We have successfully interfered with the infection of plants by three viruses belonging to the tobamovirus, potyvirus, and alfamovirus groups, demonstrating the reliability of the approach. We suggest that the effect mediated by dsRNA in plant virus infection resembles the analogous phenomenon of RNA interference observed in animals. The interference observed is sequence specific, is dose dependent, and is triggered by dsRNA but not single-stranded RNA. Our results support the view that a dsRNA intermediate in virus replication acts as efficient initiator of posttranscriptional gene silencing (PTGS) in natural virus infections, triggering the initiation step of PTGS that targets viral RNA for degradation.
Gene sequences derived from different plant viruses have been introduced into a wide variety of plant species to produce transgenic plants protected against virus infection. In a number of cases, it is known that the mechanism of resistance is a posttranscriptional, RNA-mediated process that targets both the viral RNA and the transgene mRNA for degradation in a sequence-specific manner (11, 19). RNA-mediated virus resistance is a manifestation of posttranscriptional gene silencing (PTGS), a more general phenomenon which was first described as a coordinated and reciprocal inactivation of host gene and transgenes encoding the same sense RNA (reviewed in references 33 and 41). More recently, three components in the dynamics of PTGS have been proposed: initiation, propagation of a systemic silencing signal, and maintenance (27, 31). For the last step, a nuclear component sharing sequence homology with the target mRNA is absolutely required (8, 28). PTGS also takes place in nontransgenic plants as a natural defense mechanism against virus infection. According to this idea, following the onset of virus replication, viral RNA or a derivative would be perceived as a pathogenic agent by the host, triggering a process that could be responsible for the progressive slowdown in virus accumulation observed at late stages in the infection process of some viruses (7, 29, 30). In this scenario, viruses counteract the host response by encoding suppressors of PTGS in their genomes, which seems to be a widespread strategy used by RNA and DNA viruses of plants (42).
RNA interference (RNAi) was originally observed in Caenorhabditis elegans, where injection of double-stranded RNA (dsRNA) leads to PTGS of homologous sequences (13). dsRNA blocks specific gene expression even when expressed by bacteria fed to the worms (38) or transcribed from transgenes carrying an internal inverted repeat (36). Recently, RNAi has been reported in a wide variety of animals, and in these cases the organism exhibits gene-specific phenocopies of loss-of-function mutations (2, 14). A similar phenomenon in Neurospora crassa is termed quelling (5). Considering that the interference process occurs posttranscriptionally and involves mRNA degradation (22, 26), RNAi, quelling, and PTGS in plants have been proposed to be related phenomena that could play an important biological role in protecting the organism's genome against foreign nucleic acids. Moreover, the analysis of mutants defective in those processes has revealed the involvement of similar gene products in different organisms (6, 9, 12, 23, 34).
A growing body of evidence suggests that plants, animals, and yeasts share related mechanisms of specific degradation of RNAs in which double-stranded forms of RNA are involved (5, 33). It has been shown that PTGS in plants can be triggered at high efficiency by the presence of an inverted repeat in the transcribed region of a transgene (4, 15, 18). Moreover, tobacco plants transformed with constructs that produce RNAs capable of duplex formation induce virus immunity or gene silencing with almost 100% efficiency when targeted against virus or endogenous genes (35, 43). Globally, strong evidence supports a key role for dsRNA as an inducer of PTGS in both the plant and animal kingdoms. However, there is not yet a direct probe of the formation of dsRNA in vivo in transgenic plants expressing palindromic sequences. Here, we expanded previous findings on RNAi in animals by using dsRNA to specifically interfere with viral sequences in plants. We show that exogenously applied dsRNA can act as a trigger of RNA-mediated virus resistance and elicit a local response in nontransgenic plants. To assess the potential of dsRNA-mediated interference in plant virus infections, we used pepper mild mottle virus (PMMoV), tobacco etch virus (TEV), and alfalfa mosaic virus (AMV). These three viruses belong to distinct taxonomic groups of positive-strand RNA viruses (21). dsRNA-mediated interference in plants recapitulates many of the features of RNAi in animals: the interference observed is sequence specific and dose dependent, is triggered by dsRNA but not single-stranded RNA, and seems to require a minimum length of dsRNA.
MATERIALS AND METHODS
RNA synthesis and inoculation.
For the production of dsRNA, sense and antisense RNAs were synthesized in vitro from the corresponding DNA plasmids by using T3 and T7 RNA polymerase. Sense and antisense RNA strands (2.5 μM) in 25 mM sodium phosphate (pH 7) were heated at 95°C for 3 min and then cooled and annealed at 37°C for 30 min. Formation of dsRNA was confirmed by testing a shift in gel mobility of the annealed material compared to each single-stranded RNA and by resistance to RNase A under high-salt conditions. For PMMoV dsRNAs, fragments corresponding to positions 3411 to 4388, 5086 to 5682, and 3454 to 3769 in the PMMoV sequence (1) were subcloned into pT3T7 (Boehringer Mannheim Biochemicals), yielding after transcription and annealing 977-bp (54-kDa-protein segment), 596-bp (30-kDa-protein segment), and 315-bp (one-third of a 54-kDa-protein segment) dsRNAs, respectively. For TEV dsRNA, a fragment corresponding to positions 845 to 2328 in the TEV sequence (10) was subcloned into pBluescript SK(−) (Stratagene), yielding a 1,483-bp dsRNA (TEV-HC dsRNA). For AMV dsRNA, a fragment corresponding to positions 369 to 1493 in the AMV RNA 3 sequence (24) was subcloned into pBluescript SK(−), yielding a 1,124-bp dsRNA (AMV-3 dsRNA). Two nonviral dsRNAs were synthesized corresponding to the SstI fragment (300 bp) from the Nicotiana plumbaginifolia Cab-E gene and the AsuII-BamHI fragment (413 bp) from the binary plant transformation vector pGSJ780A cloned in pT3T7 (37). All plasmids were linearized with appropriate restriction enzymes and used as templates for in vitro transcription reactions to generate sense and antisense RNAs. An exact cDNA copy of the PMMoV 54-kDa-protein gene (nt 3499 to 4908) was produced by PCR using primers corresponding to the relevant positions of the 54-kDa-protein gene as previously described (37). This cDNA product was tested for its interfering activity on PMMoV infection.
In the experiments with PMMoV, the standard inoculum was 10 μg of purified virus per ml (1). The plasmid pTEV-7D (a generous gift from J. C. Carrington, Washington State University), containing a full-length clone of TEV, was linearized and transcribed in vitro with SP6 RNA polymerase as described previously (10). Transcription with T7 RNA polymerase of full-length clones of AMV RNA 1 (pUT17A), RNA 2 (pUT27A), and RNA 3 (pAL3) (kindly provided by J. F. Bol, Leiden University) was performed as described previously (25). Inoculation mixtures were made by adding 5 μl of each dsRNA to an equal volume of purified virus (PMMoV) or to 10 μl of viral transcripts (TEV and AMV). With AMV, 10 μg of purified AMV coat protein (CP) was added to the inoculation mixture. Inoculation of plants was done on two fully expanded leaves of at least two plants per assay by gently rubbing the leaf surface with the inoculum using Carborundum as an abrasive (21). For comparisons of sense RNA, antisense RNA, dsRNA, and cDNA effects on virus infection, equal molar concentrations of each molecule were used. The inoculated plants were kept in growth chambers with a 16-h light–8-h dark cycle at 25°C, and the development of symptoms of viral infection in systemic hosts was monitored for the duration of their life cycles. For local lesion hosts, inoculated leaves were photographed 5 days after inoculation.
Analysis of viral RNA in plants.
Total RNA was extracted from inoculated leaves between 6 and 10 days postinoculation (dpi) and from upper leaves 6 to 21 dpi as previously described (20). RNA samples (1 to 5 μg) were electrophoresed on 1 to 1.2% agarose formaldehyde gels and transferred to Hybond-N membranes. Ethidium bromide staining of the agarose gels prior to blotting was done to confirm integrity of the RNA and loading of similar amounts of RNA. Northern blot hybridization was carried out with digoxigenin (DIG)-labeled riboprobes (Boehringer Mannheim Biochemicals) as described previously (25). Virus-specific riboprobes were used to detect the respective viruses. PMMoV RNA was detected with a probe complementary to PMMoV nucleotides (nt) 3411 to 4388, which was transcribed from pT3T7/54 kDa (37). TEV RNA was detected with a probe complementary to TEV nt 845 to 2328, which was transcribed from pBluescript SK(−)/HC (a gift from C. Llave). AMV RNAs 3 and 4 were detected with a probe complementary to nt 369 to 1493 of AMV RNA 3, which was transcribed from pBluescript SK(−)/AMV-3 (see above).
Agrobacterium tumefaciens-mediated transient expression.
The region of PMMoV RNA encoding the 54-kDa protein and flanking regions (nt 3411 to 5016) were inserted in either sense or antisense orientation between the 35S promoter of cauliflower mosaic virus and the transcriptional terminator of TL-DNA gene 7 in binary vector pGSJ780A as previously described (37). These constructs were introduced into A. tumefaciens strain LBA 4404 by direct transformation. Recombinant A. tumefaciens was grown overnight at 28°C in tubes containing 10 ml of Luria-Bertani medium supplemented with 50 μg of rifampin and 40 μg of streptomycin per ml. Cells were precipitated and resuspended to a final concentration corresponding to an optical density at 600 nm of 0.5 in a solution containing 10 mM MgCl2, 10 mM MES (morpholinepropanesulfonic acid, pH 5.6), and 150 μM acetosyringone. Cultures were incubated at 28°C for 2 to 3 h before infiltration. Two leaves per plant were infiltrated in their entirety with a 1-ml syringe without a needle, and the whole plant was covered with a transparent plastic bag for 2 days. For the coinfiltration of Agrobacterium cultures carrying the PMMoV 54-kDa-protein construct in sense and antisense orientations, equal volumes of both cultures were mixed before infiltration.
RESULTS
dsRNA causes specific inhibition of PMMoV infection.
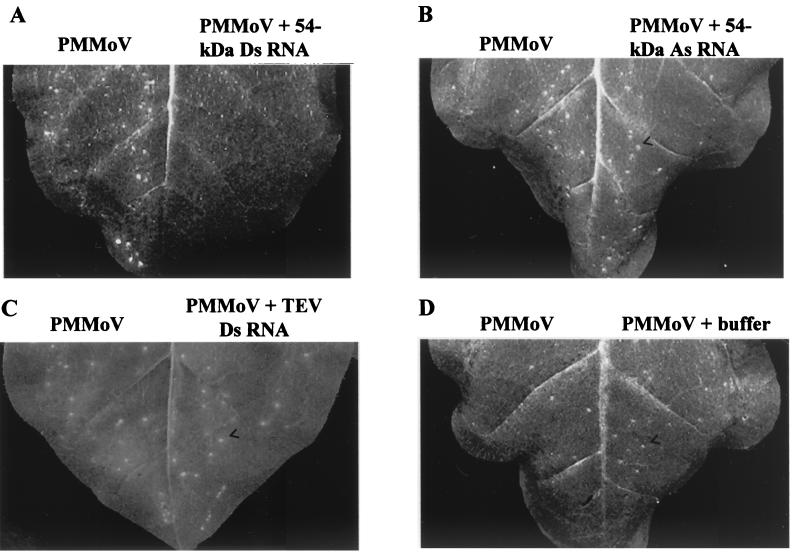
As mentioned above, dsRNA can confer extreme virus resistance when sense and antisense virus-derived transgenes are simultaneously expressed in a plant. Therefore, we investigated whether direct delivery by mechanical inoculation of dsRNA together with the virus could interfere with infection in a sequence-specific manner. Sense RNA and antisense RNA corresponding to part of the readthrough domain of the replicase gene of PMMoV were transcribed in vitro and annealed to each other to produce the dsRNA (54-kDa-protein dsRNA). The dsRNA and the single-stranded RNAs with either sense or antisense orientation were each tested for their ability to inhibit local lesion development elicited by PMMoV in Nicotiana tabacum cv. Xanthi nc, a hypersensitive host. Half-leaves were inoculated with virus alone, and opposite halves were inoculated with virus plus either sense RNA, antisense RNA, dsRNA (each at 0.62 μM, final concentration), or in vitro transcription buffer. Figure 1 shows a representative outcome taken as an example of several experiments carried out with RNAs produced in different transcription reactions and summarized in Table 1. Infectivity was completely blocked by coinoculation with a 977-bp dsRNA that includes part of the replicase gene of PMMoV, whereas the opposite half of the leaf inoculated only with an equivalent amount of PMMoV displayed more than 50 local lesions (Fig. 1A and Table 1). Neither antisense RNA nor sense RNA derived from the same region of PMMoV affected local lesion formation by the virus (Fig. 1B and Table 1). Furthermore, coinoculation with TEV dsRNA (see below), a dsRNA of viral origin but unrelated to PMMoV, did not have any effect on PMMoV infectivity (Fig. 1C and Table 1). We carried out experiments similar to those described above but using Capsicum chinense instead of N. tabacum. PMMoV induces a hypersensitive reaction on this pepper indicator plant that was completely prevented by the presence of 54-kDa-protein dsRNA in the inoculum (Table 1). Thus, PMMoV infection was specifically inhibited by its cognate dsRNA in two different local lesion hosts belonging to different genera.
FIG. 1.
Specific interference with PMMoV infection by dsRNA in a hypersensitive host. Response of N. tabacum cv. Xanthi nc to PMMoV (5 μg/ml) alone (left halves of leaves) or to a combination of PMMoV plus either 54-kDa-protein dsRNA (A), 54-kDa-protein antisense (As) RNA (B), TEV-HC dsRNA (C), or in vitro transcription buffer (D) (right halves of the leaves). Leaves were photographed at 5 dpi. Similar numbers of local lesions (arrowheads) were observed in both halves of the leaves in panels B, C, and D. No visible local response was observed in the half-leaf inoculated with PMMoV plus 54-kDa-protein dsRNA.
TABLE 1.
Interference with PMMoV infection by dsRNA in a hypersensitive hosta
| Expt | No. of lesions on leaves inoculation with:
|
||||
|---|---|---|---|---|---|
| PMMoV/PMMoV + 54-kDa dsRNA | PMMoV/PMMoV + 54-kDa S RNA | PMMoV/PMMoV + 54-kDa As RNA | PMMoV/PMMoV + TEV dsRNA | PMMoV/PMMoV + buffer | |
| 1 | 54/0 | 38/55 | 42/48 | 60/20 | 15/31 |
| 2 | 56/0 | 43/37 | 39/47 | 42/58 | 56/46 |
| 3 | 29/0 | 31/40 | |||
| 4 | 32/0 | ||||
| 5 | 51/0 | ||||
Values are the numbers of local lesions elicited by PMMoV (5 μg/ml) alone (left halves of leaves) or by a combination of PMMoV plus either 54-kDa-protein dsRNA, 54-kDa-protein sense (S) RNA, 54-kDa-protein antisense (As) RNA, TEV-HC dsRNA, or in vitro transcription buffer inoculated in the right halves of the leaves. Lesions were counted at 5 dpi. Experiments 1 to 4 were carried out on N. tabacum cv. Xanthi nc; experiment 5 was done on C. chinense.
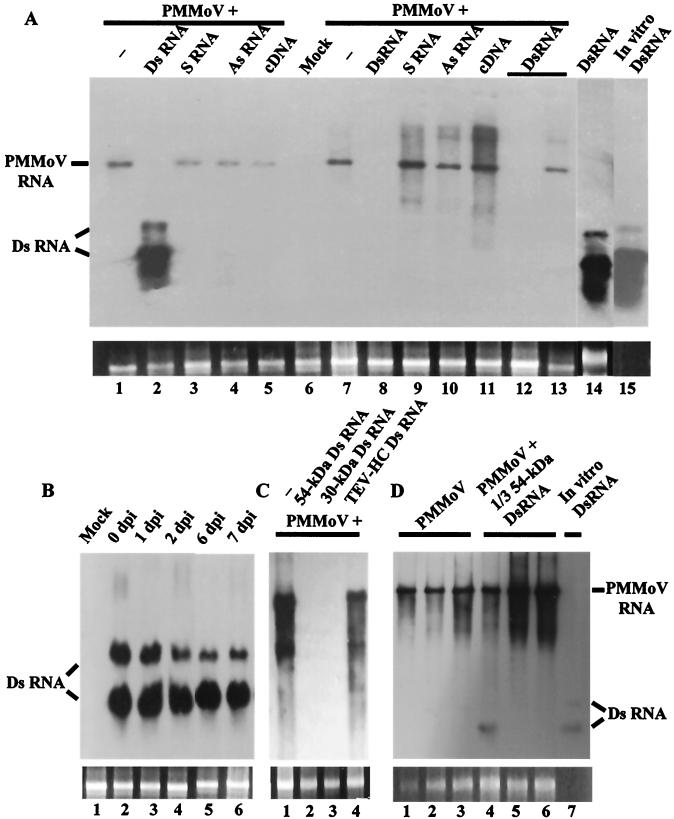
To evaluate the capability of dsRNA to interfere with PMMoV infection in a systemic host, Nicotiana benthamiana plants were inoculated with mixtures of PMMoV and one of the transcription products derived from PMMoV used above. In addition, a cDNA PCR product corresponding to the region of the virus transcribed in the in vitro reactions was included. By 7 dpi, most plants were susceptible to virus infection. Interestingly, only plants coinoculated with PMMoV plus 54-kDa-protein dsRNA were protected against infection, since they did not display disease symptoms. Consistent with the above results, Northern blot analysis of total RNA extracted from inoculated or systemic leaves at 7 dpi showed that plants coinoculated with either sense RNA, antisense RNA, or DNA molecules homologous to the virus accumulated PMMoV positive-strand RNA at levels that were comparable to those of the control plant (Fig. 2A). However, when dsRNA was present in the inoculum, PMMoV multiplication was apparently blocked in the inoculated leaves (Fig. 2A, lane 2) and the virus did not accumulate to detectable levels in upper leaves (lane 8), even after extended exposure of the autoradiogram. Furthermore, biologically active virus was not detected in homogenates of the upper leaves of these plants when used to back-inoculate the local lesion host, N. tabacum cv. Xanthi nc. In addition, there was no evidence of accumulation of PMMoV negative-strand RNA in these samples (data not shown). In total, more than 10 independent assays with 22 plants have been done, corroborating the interference with PMMoV infection by 54-kDa-protein dsRNA. However, in a low percentage (about 18%) of individuals, the virus overcame the protection conferred by dsRNA and plants displayed disease symptoms with a delay of 1 to 3 weeks compared to the control plants. PMMoV RNA accumulated in the uppermost leaves of these plants at moderate levels (Fig. 2A, lane 13). The remaining plants were free of symptoms until their life cycles were completed, and viral RNA did not accumulate at detectable levels up to 3 weeks postinoculation (Fig. 2A, lane 12), nor was virus infectivity recovered by back-inoculation up to 40 dpi.
FIG. 2.
dsRNA-mediated interference with PMMoV infection in a systemic host. (A) Specificity of interference with PMMoV infection by dsRNA. Northern blot analysis of total RNA extracted from inoculated (lanes 1 to 6) or uppermost systemic (lanes 7 to 13) leaves of N. benthamiana. Plants were mock inoculated or were inoculated with PMMoV (5 μg/ml) alone (−), with 54-kDa-protein dsRNA alone, or with PMMoV plus either 54-kDa-protein dsRNA, sense (S) 54-kDa-protein RNA, antisense (As) 54-kDa-protein RNA, or 54-kDa-protein cDNA, as indicated. Leaf tissues were harvested at 7 dpi, except for the samples in lanes 12 and 13, which were harvested at 21 dpi. The samples in lanes 2, 8, and 12 were taken from the same plant, which did not display symptoms of infection until its life cycle was completed. The sample in lane 13 was taken from another individual showing disease symptoms at 21 dpi. The 54-kDa-protein dsRNA used in the inoculum was run in lane 15 for comparison. (B) Time course analysis of dsRNA stability on plant leaves. The 54-kDa-protein dsRNA (10 μl, 0.62 μM) was inoculated on fully expanded leaves of N. benthamiana (three- to four-leaf stage). After the inoculated leaves had been washed with Triton X-100 (0.05%) for 30 min, RNA was extracted at the indicated times. Mock, RNA extracted from a mock-inoculated plant. (C) Interference with PMMoV infection by homologous dsRNAs. RNA was extracted from upper leaf tissue of plants inoculated with PMMoV (5 μg/ml) alone (−) or with PMMoV plus the indicated RNAs. (D) Interference seems to require a minimum length of dsRNA. RNA was extracted from inoculated (lanes 1 and 4) or upper (lanes 2, 3, 5, and 6) leaf tissue of plants infected with PMMoV (5 μg/ml) alone or with PMMoV plus 1/3 54-kDa-protein dsRNA at 7 (lanes 1, 2, 4 and 5) or 12 dpi (lanes 3 and 6). The 1/3 54-kDa-protein dsRNA used in the inoculum was run in lane 7 for comparison. Similar amounts (1 μg) of RNA samples were fractionated by 1% agarose gel electrophoresis in all panels, and a DIG-labeled 54-kDa-protein RNA was used as a probe. The positions of PMMoV RNA and RNA species derived from partially denatured input dsRNA are indicated. Ethidium bromide staining of 25S rRNA was used as a loading control for the RNA blots (bottom panels).
Consistently, hybridization bands corresponding to what seems to be partially denatured 54-kDa-protein dsRNA were observed in RNA preparations obtained from the inoculated leaves of plants challenged with virus plus 54-kDa-protein dsRNA (Fig. 2A, lane 2) or with 54-kDa-protein dsRNA alone (lane 14), as judged by comparison with the behavior on gels of the dsRNA preparation used as the inoculum (Fig. 2A, lane 15). Furthermore, the same hybridization pattern was observed when RNA samples were treated with RNase A, which will degrade any ssRNA, and when a 54-kDa-protein RNA probe specific for the negative strand was used (data not shown). Several experiments have been done in order to determine the origin and location of these dsRNA molecules. They include an extended wash step of the inoculated leaves just before RNA extraction and an analysis of the stability of these molecules after delivery into leaves. The data support the idea that most of the input dsRNA was relatively stable and persisted in the leaf, probably inside the leaf cells, at detectable levels at least 7 days after inoculation (Fig. 2B).
The interference with viral infection exhibited by a dsRNA corresponding to the readthrough region of the gene encoding the replicase of PMMoV could reflect any kind of inhibitory effect of this sequence in particular on virus replication. We investigated whether dsRNA generated from a different region of the PMMoV genome could specifically block PMMoV infection when delivered simultaneously with the virus into the plant. To address this question, a dsRNA covering a 596-nt segment of the 30-kDa-protein gene of PMMoV was produced and its effect on virus infection was compared with that of 54-kDa-protein dsRNA derived from PMMoV or a nonhomologous, viral dsRNA. Like 54-kDa-protein dsRNA, the presence of 30-kDa-protein dsRNA in the inoculum prevented expression of viral symptoms on N. benthamiana plants at times when control plants displayed disease symptoms. Correspondingly, accumulation of viral RNA was not detectable in RNA extracted from upper leaf tissue of these plants (Fig. 2C, lanes 2 and 3). However, coinoculation of PMMoV together with a 1,483-bp dsRNA corresponding to most of the helper component gene of TEV (TEV-HC dsRNA) had no effect on symptom expression, and PMMoV RNA accumulated in upper leaves as in control plants (Fig. 2C, lanes 1 and 4). This failure of nonhomologous dsRNA to interfere with PMMoV infection has also been observed with different nonviral dsRNA segments of various lengths, precluding any effect concerning molar stoichiometry on the lack of interference observed with nonhomologous dsRNA (see Materials and Methods; also data not shown). Thus, we found interference with PMMoV infection only when dsRNA molecules shared sequence identity with the virus. At present, it is not known how effective the protection conferred by dsRNA is when the challenging virus and the protective molecules share a lower degree of sequence homology.
While the 54-kDa- and 30-kDa-protein dsRNAs (977 and 596 nt, respectively) were effective in protecting plants against PMMoV infection, a smaller dsRNA covering approximately one-third (315 nt) the length of 54-kDa-protein dsRNA (1/3 54-kDa-protein dsRNA) had a marginal effect. The appearance of viral symptoms on these plants was delayed by 1 to 2 days compared to plants inoculated only with PMMoV, and viral RNA accumulated at levels similar to that of the control (Fig. 2D). So this result may indicate that the ability of dsRNA to specifically interfere with PMMoV infection could be length dependent.
To evaluate the protective effect against PMMoV infection at different times after delivery of dsRNA to plant leaves, 54-kDa-protein dsRNA and PMMoV were sequentially inoculated into the same leaves. It was not necessary to mix PMMoV and 54-kDa-protein dsRNA in a tube just before mechanical coinoculation to observe inhibition of virus infection. Although the immediate, sequential inoculation of virus and dsRNA was also able to protect plants against infection, a interval of 24 h (and up to 96 h) between consecutive inoculations, did not result in interference with PMMoV infection (data not shown).
dsRNA inhibits infection by different plant viruses.
To determine whether the use of dsRNA molecules could be a general strategy to prevent infection by plant viruses other than PMMoV, we assessed the effects of specific dsRNA on the infectivity of two viruses unrelated to the Tobamovirus genus: TEV, which belongs to the family Potyviridae, and AMV, within the family Bromoviridae (21).
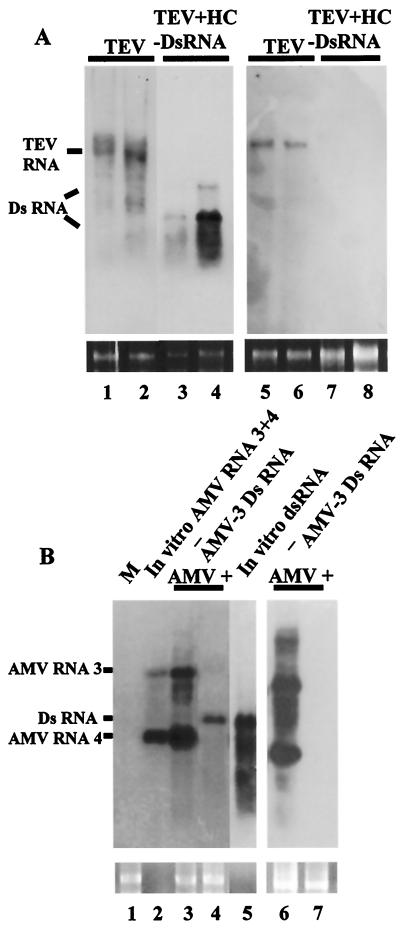
Potyviruses are a positive-strand RNA virus group with a monopartite genome organization similar to that of the picornavirus superfamily. N. tabacum plants were inoculated either with SP6 transcripts of a cDNA clone of TEV alone or with a mixture containing both TEV RNA transcripts and TEV-HC dsRNA. By 2 weeks postinoculation, neither localized lesions nor systemic symptoms appeared on the inoculated or upper leaves of 10 plants inoculated with the mixture, whereas plants inoculated only with TEV displayed typical disease symptoms of vein-clearing and etching at 6 dpi. Figure 3A shows a Northern blot analysis of total RNA extracted from two representative plants per treatment at 6 dpi. TEV RNA accumulated in both the inoculated leaf tissue (Fig. 3A, lanes 1 and 2) and the upper leaf tissue (lanes 5 and 6) of the control plants, whereas viral RNA levels were below the limit of Northern blot detection in plants coinoculated with the virus and the protective, homologous dsRNA (lanes 3, 4, 7, and 8), even after a longer exposure of the autoradiogram. As before, hybridization bands of variable intensity corresponding to TEV-HC dsRNA were observed in the inoculated leaves of plants challenged with virus plus dsRNA.
FIG. 3.
Interference of dsRNA with the infection of different plant viruses. (A) dsRNA-mediated interference with TEV. Northern blot analysis of total RNA extracted from inoculated (lanes 1 to 4) or systemic (lanes 5 to 8) leaves of N. tabacum plants challenged with TEV alone or with TEV plus TEV-HC dsRNA at 6 dpi. Similar amounts (5 μg) of each RNA sample were fractionated by 1% agarose gel electrophoresis, and the filter was hybridized with a DIG-labeled RNA probe specific for TEV. The positions of TEV RNA and RNA species derived from partially denatured input TEV-HC dsRNA are indicated. Differences in stability between different RNA samples could probably account for variations in the intensity of the dsRNA bands. Ethidium bromide staining of 25S rRNA was used as a loading control for the RNA gel blot (bottom panel). (B) dsRNA-mediated interference with AMV. Northern blot analysis of total RNA extracted at 6 dpi from inoculated (lanes 1, 3, and 4) or systemic (lanes 6 and 7) leaves of N. benthamiana plants challenged with AMV RNAs 1, 2, and 3 alone (−) or with this mixture plus AMV-3 dsRNA. M, RNA extracted from a mock-inoculated plant. In vitro-transcribed AMV RNAs 3 and 4 (lane 2) and AMV-3 dsRNA (lane 5) used in the inoculum were run for comparison. Similar amounts (1 μg) of RNA samples were fractionated by 1.2% agarose gel electrophoresis, and the filter was hybridized with a DIG-labeled RNA probe specific for AMV RNA3. The positions of AMV RNAs 3 and 4 and input AMV-3 dsRNA are indicated. Ethidium bromide staining of 25S rRNA was used as a loading control for the RNA blot (bottom panel).
Alfamoviruses are a positive-strand RNA virus group with a multipartite genome organization similar to that of members of the Sindbis-like virus superfamily. A distinctive property of the alfamoviruses is that a mixture of the three genomic RNAs of AMV is not infectious to plants unless AMV CP is added in the inoculum (3). Ten N. benthamiana plants were inoculated either with a mixture of T7 transcripts of AMV RNAs 1, 2, and 3 and AMV CP or with this mixture plus a dsRNA covering a 1,124-nt segment of AMV RNA 3 (AMV-3 dsRNA). Figure 3B shows a Northern blot analysis of total RNA extracted from inoculated or systemic leaves of these plants using a probe specific for both the genomic RNA 3 and the subgenomic RNA 4 of AMV. In vitro-transcribed RNAs 3 and 4 and AMV-3 dsRNA used in the inoculum were loaded as controls (Fig. 3B, lanes 2 and 5, respectively). By 6 dpi, none of the plants inoculated with the mixture of AMV RNAs plus AMV-3 dsRNA showed disease symptoms, whereas all of the plants inoculated only with AMV RNAs were susceptible to virus infection, showing severe stunting and mosaic on systemically infected leaves. In agreement with the observed symptoms, accumulation of AMV RNAs was not detectable in plants inoculated with the mixture of AMV RNAs plus dsRNA (Fig. 3B, lanes 4 and 7), whereas plants inoculated only with AMV RNAs accumulated RNAs 3 and 4 in both the inoculated (Fig. 3B, lane 3) and upper (lane 6) leaf tissue.
Inhibition by dsRNA is dose dependent and acts inside plant cells.
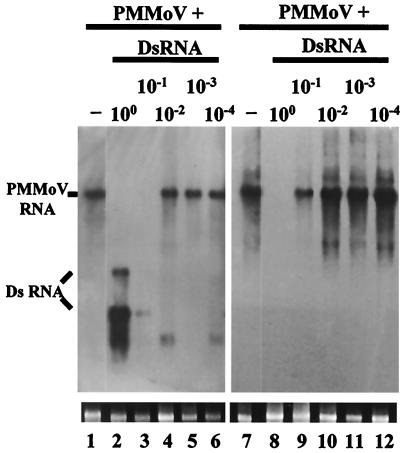
To obtain a semiquantitative assessment of the relationship between dsRNA dose and inhibition of virus infection, a series of log dilutions (100 to 10−4) were made from the 54-kDa-protein dsRNA preparation (0.8 μg/μl) typically used in the experiments with PMMoV. The dilutions were mixed in equal volumes with a fixed concentration of PMMoV (10 μg/ml), and the mixtures were tested for their ability to inhibit PMMoV infection on N. benthamiana plants. At the highest dose tested (undiluted dsRNA), the amount of dsRNA corresponds to a 5,240-fold molar excess of dsRNA over viral RNA. In accord with previous results, PMMoV accumulation was completely inhibited by a concentration of 54-kDa-protein dsRNA corresponding to undiluted dsRNA preparation, whereas lowering the dsRNA concentration 10-fold had a slight effect on viral infection (Fig. 4). Symptom expression in these plants was delayed by only 3 days compared to that in plants inoculated with PMMoV alone. PMMoV RNA accumulated at very low levels in the inoculated leaves of these plants at 6 dpi (Fig. 4, lane 3) (visible after longer exposure of the blot) but reached a moderate level in upper leaf tissue at 15 dpi (Fig. 4, lane 9). The dsRNA diluted 100-fold or more showed no effect on virus infection, and PMMoV RNA accumulated in both the inoculated and upper leaves of these plants at levels comparable to that of the control plant. Thus, the ability of 54-kDa-protein dsRNA to provoke inhibition of PMMoV multiplication was dose dependent.
FIG. 4.
Dose-dependent interference with PMMoV infection by dsRNA. Northern blot analysis of total RNA extracted from inoculated (lanes 1 to 6) or uppermost systemic (lanes 7 to 12) leaves of N. benthamiana at 6 and 15 dpi, respectively. Plants were inoculated with PMMoV (5 μg/ml) alone (−) or with PMMoV plus a series of log dilutions (100 to 10−4) of the 54-kDa-protein dsRNA, as indicated. Similar amounts (1 μg) of RNA samples were fractionated by 1% agarose gel electrophoresis, and the filter was hybridized with a DIG-labeled 54-kDa-protein RNA probe. The positions of PMMoV RNA and RNA species derived from partially denatured, input 54-kDa-protein dsRNA are indicated. The minor, low-molecular-weight bands in lanes 4 and 6 are probably degradation products of genomic RNA. Ethidium bromide staining of 25S rRNA was used as a loading control for the RNA blot (bottom panel).
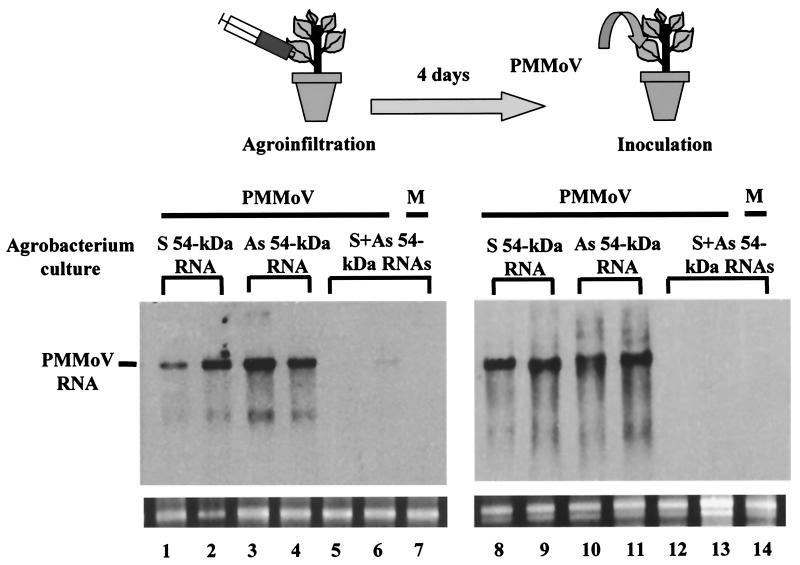
One possible explanation for the observation that coinoculation of dsRNA with either viral RNA or virus particles specifically prevents infection by different plant viruses is that some kind of inhibitory interaction occurs in the mixture before the virus penetrates into the cell. We used an Agrobacterium-mediated transient-expression assay (agroinfiltration) (40) to test if dsRNA could inhibit virus infection when expressed directly inside plant cells. We cloned the entire 54-kDa-protein coding region and flanking sequences of PMMoV (nt 3411 to 5016) in either the sense or antisense orientation under the control of the cauliflower mosaic virus 35S constitutive promoter in a binary plasmid vector. A. tumefaciens cultures carrying the sense and antisense 54-kDa-protein RNA-expressing vectors were mixed in a 1:1 ratio and coinfiltrated into N. benthamiana leaves in their entirety. For comparative purpose, plants were agroinfiltrated with single cultures carrying plasmids encoding the sense or the antisense 54-kDa-protein RNA. At 4 days postinfiltration, plants were challenged with PMMoV that was directly inoculated on the entire infiltrated leaves. In three independent experiments, all the plants transiently expressing sense or antisense 54-kDa-protein RNA displayed disease symptoms in upper leaves at 10 days after inoculation, whereas plants that were agroinfiltrated with vectors expressing the sense-antisense mixture showed no symptoms or symptoms that were delayed 1 to 3 weeks compared to the controls. Figure 5 shows a Northern blot analysis of the accumulation of PMMoV RNA in total RNA preparations extracted from two individuals per treatment at 15 dpi. PMMoV accumulated at very low level, if any, in the inoculated leaves of plants infiltrated with the mixture of single-stranded 54-kDa-protein RNAs, which could have annealed to each other to form a dsRNA structure inside the plant cell. There was no signal of PMMoV RNA in upper leaf tissue of these plants. In contrast, neither sense nor antisense 54-kDa-protein RNA expressed by Agrobacterium prevented PMMoV accumulation in either the inoculated or systemic leaves of the control plants. Thus, the specific interference with virus infection exhibited by homologous dsRNA seems to operate inside the plant cell and to not be due to some other inhibitory effect occurring in vitro. Furthermore, the interference effect on PMMoV infection took place even when a longer interval (up to 7 days) between agroinfiltration with 54-kDa-protein dsRNA and virus inoculation was tested. However, cointroduction of dsRNA and virus into the same leaves seemed critical, because dsRNA-agroinfiltrated plants challenged with PMMoV in upper, noninfiltrated leaves became susceptible to virus infection without any apparent delay in symptom expression (data not shown).
FIG. 5.
Agrobacterium-mediated transient expression of 54-kDa-protein dsRNA interferes with PMMoV infection. Leaves of N. benthamiana plants were initially infiltrated as indicated with A. tumefaciens cultures carrying either the sense (S) or the antisense (As) 54-kDa-protein RNA-expressing vector, or a mixture of both cultures. After 4 days, the agroinfiltrated leaves of these plants were challenge inoculated with PMMoV or were mock inoculated (M). After another 15 days, accumulation of PMMoV RNA was assessed on inoculated (lanes 1 to 7) and upper (lanes 8 to 14) leaves of two plants per treatment by Northern blot analysis. Similar amounts (1 μg) of RNA samples were fractionated by 1% agarose gel electrophoresis, and the filter was hybridized with a DIG-labeled 54-kDa-protein RNA probe. A faint degradation product of genomic RNA is observed in RNA extracts containing PMMoV RNA. Ethidium bromide staining of 25S rRNA was used as a loading control for the RNA blot (bottom panel).
DISCUSSION
We have shown that dsRNA derived from viral sequences can interfere with virus infection in a sequence-specific manner by directly delivering dsRNA to leaf cells by mechanical inoculation. This approach differs from strategies based on transgenic expression of RNAs with the potential to form duplexes which confer protection against potato virus Y through a PTGS mechanism (35, 43). Our results support the view suggested by others that the dsRNA intermediate in virus replication acts as an efficient initiator of PTGS in natural virus infections (29, 30). In our study, we targeted three viruses (PMMoV, TEV, and AMV) that represent extremes in the evolution of positive-strand RNA viruses in plants with the corresponding dsRNAs. We did not detect any reduction of virus accumulation in plants inoculated in the presence of either sense single-stranded RNA, antisense single-stranded RNA, DNA, or nonhomologous dsRNA. This specificity in both sequence and structure of the interfering molecule argues against hypothetical artifacts produced by the inoculation procedure. Also, the interference phenotype was not restricted to a particular plant species but was expressed in different host and nonhost plants, demonstrating the reliability of the phenomenon. Furthermore, we have been able to demonstrate interference with virus infection by two different strategies: (i) direct, mechanical inoculation of dsRNA together with the virus and (ii) delivery of plasmid constructs via Agrobacterium whose transcription products are predicted to form dsRNA in vivo in a process separate from virus entry. Both approaches protected plants against virus infection, making it very unlikely that in vitro interactions between dsRNA and virus inocula were the cause of the protective effect. Therefore, we suggest that the effects mediated by dsRNA in plant virus infection reflect the phenomenon of RNAi, a particular case of PTGS, observed in animals (2, 14). In support of this view, the helper component proteinase of potyviruses, a well-known suppressor of PTGS in plants (42), prevents the interfering activity on PMMoV infection by dsRNA when both this proteinase and 54-kDa-protein dsRNA are delivered into plant cells via Agrobacterium (unpublished data). In addition, preliminary data on interference with PMMoV infection observed here using RNA duplexes of different lengths is in agreement with results obtained with RNAi in C. elegans (13) and Drosophila (16).
The results obtained in a dilution series of 54-kDa-protein dsRNA suggest that a 500-fold excess of dsRNA over PMMoV RNA is the threshold required for limited interference with virus multiplication. Threshold concentrations of dsRNA required for RNAi have also been reported for plants (32) and animals (39). In our case, this may reflect a minimum amount of dsRNA required for the onset of RNAi associated with a highly replicating pathogen. Alternatively, because input dsRNA itself cannot move between cells at detectable levels in the inoculated leaves (unpublished data), this dose effect probably reflects a minimum amount of dsRNA required to ensure that most viral RNA penetrates into the cell in combination with dsRNA. Taking into account the interference of dsRNA with PMMoV infection on a hypersensitive host, we favor the hypothesis that dsRNA interferes with virus infection at the cell level in the inoculated leaves. Indeed, in cases where the virus is able to overcome the protection conferred by dsRNA in a systemic host, there is a delay in symptom expression from 1 to 3 weeks, suggesting a reduced number of competent infection foci in the inoculated leaves. In plants, analysis of the dynamics of virus-induced gene silencing, a particular case of PTGS, revealed that there are separate initiation and maintenance stages (31). In our scenario, dsRNA delivered by inoculation or expressed directly inside plant cells would act as the dsRNA intermediate involved in viral replication, triggering the initiation step of PTGS, which targets viral RNA for degradation. As the maintenance step of PTGS requires a nuclear component (transgene or DNA homologous to the target) that is absent in our plants, the sequence-specific signal involved in PTGS does not propagate to upper parts of the plant, and PTGS would not progress beyond the initiation stage (8, 17). It has been shown that biolistic introduction of 35S nitrate reductase cDNA constructs induces local but not systemic PTGS in nontransgenic plants (28). Thus, in our case, once a few infectious particles penetrate into the cells alone, without dsRNA, the virus spreads and moves to the upper leaves, the infection progresses, and the plant become susceptible to the virus. In support of this lack of a PTGS systemic signal, when dsRNA-agroinfiltrated plants were challenged with PMMoV in upper, noninfiltrated leaves, they became susceptible to virus infection.
Although coinoculation and rapid sequential inoculation of dsRNA and virus produced interference with PMMoV infection, a long interval (24 h) between sequential inoculations resulted in a complete susceptibility to virus infection. Therefore, it seems likely that some kind of interaction must occur between the protective dsRNA, or derivatives, and the viral RNA before dsRNA becomes degraded or inaccessible to the RNAi machinery. One question arises regarding the fate of dsRNA delivered on the leaf. It has been reported that in Drosophila, dsRNA is processed efficiently to 21- to 23-nt fragments that may be the active agents in initiating gene silencing (44). In our work, evidence indicating partial persistence of undegraded, input dsRNA on the leaf for at least 7 days after inoculation has been obtained. However, initial attempts to determine whether input dsRNA is cleaved to small RNAs did not yield positive results. The absence of a nuclear component and/or a replicating virus in our system could preclude the accumulation of detectable levels of these RNA species (8).
The direct delivery of dsRNA by mechanical inoculation or via Agrobacterium adds to the tools available for induction of RNAi in plants. Our approach provides an alternative to genetic transformation of plant species with dsRNA-expressing constructs able to produce interference with endogenous plant genes (4, 18). In principle, it should be possible to silence specific host sequences by direct delivery of dsRNA with homology to the target gene, as has been done recently in cereals by a biolistic procedure (32). In this case, the maintenance stage of PTGS would operate because of the presence of a nuclear component (endogenous gene), and the dsRNA-mediated genetic interference would be expressed in the whole plant. In addition, this strategy would also allow the study of the initiation stage of PTGS in cases involving transgenic resistance against virus infection. Our results suggest that homologous dsRNA could serve as protective molecules against virus infections, provided that inexpensive and effective means of production and delivery of adequate interference products onto plant surfaces are developed. Further research will help to identify the limits and uses of such strategies.
ACKNOWLEDGMENTS
We thank J. F. Bol for the generous gift of full-length clones of AMV. The kind gift of pTEV-7D by J. C. Carrington is acknowledged. We also thank J. J. López-Moya for his comments on the manuscript, C. Llave for providing pBluescript SK(−)/HC, and D. Hermán for technical assistance.
This work was supported by grants BIO97-0615-C02-01, BIO98-0849, BIO2000-1605-C02-02, and BIO2000-0914 from Comisión Interministerial de Ciencia y Tecnología (CICYT) and by grant 07M/0123/2000 from the Dirección General de Investigación de la Comunidad de Madrid. F.T. is a recipient of a contract from Consejo Superior de Investigaciones Científicas.
REFERENCES
- 1.Alonso E, García-Luque I, De la Cruz A, Wicke B, Avila-Rincón M J, Serra M T, Castresana C, Díaz-Ruíz J R. Nucleotide sequence of the genomic RNA of pepper mild mottle virus, a resistance-breaking tobamovirus in pepper. J Gen Virol. 1991;72:2875–2884. doi: 10.1099/0022-1317-72-12-2875. [DOI] [PubMed] [Google Scholar]
- 2.Bass B L. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 3.Bol J F. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J Gen Virol. 1999;80:1089–1102. doi: 10.1099/0022-1317-80-5-1089. [DOI] [PubMed] [Google Scholar]
- 4.Chuang C-F, Meyerowitz E M. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogoni C, Macino G. Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr Opin Microbiol. 1999;2:657–662. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- 6.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 7.Covey S N, Al-Kaff N S, Langara A, Turner D S. Plants combat infection by gene silencing. Nature. 1997;385:781–782. [Google Scholar]
- 8.Dalmay T, Hamilton A, Mueller E, Baulcombe D C. Potato Virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell. 2000;12:369–379. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe D C. An RNA-dependent RNA polymerase in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 10.Dolja V V, McBride H J, Carrington J C. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase into the viral polyprotein. Proc Natl Acad Sci USA. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.English J J, Mueller E, Baulcombe D C. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagard M, Boutet S, Morel J-B, Bellini C, Vaucheret H. AGO-1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton A J, Brown S, Yuanhai H, Ishizuka M, Lowe A, Solis A-G A, Grierson D. A transgene with repeat DNA causes high frequency, post-transcriptional suppression of ACC-oxidase gene expression in tomato. Plant J. 1998;15:737–746. doi: 10.1046/j.1365-313X.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- 16.Hammond S, Bernstein E, Beach D, Hannon G. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 17.Jones A L, Hamilton A J, Voinnet O, Thomas C L, Maule A J, Baulcombe D C. RNA-DNA interactions and DNA methylation on post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2302. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin J Z, Framond A J, Tuttle A, Bauer M W, Heifetz P B. Methods of double-stranded RNA-mediated gene inactivation in Arabidopsis and their use to define an essential gene in methionine biosynthesis. Plant Mol Biol. 2000;44:759–775. doi: 10.1023/a:1026584607941. [DOI] [PubMed] [Google Scholar]
- 19.Lindbo J A, Silva-Rosales L, Proebsting W M, Dougherty W G. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissue. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 21.Matthews R E F. Plant virology. 3rd ed. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 22.Montgomery M D, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J B, Jouette D, Lacombe A M, Nikic S, Picault N, Remoue K, Sanial M, Vo T A, Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 24.Neeleman L, van der Kuyl A, Bol J F. Role of alfalfa mosaic virus coat protein in symptom formation. Virology. 1991;181:687–693. doi: 10.1016/0042-6822(91)90902-n. [DOI] [PubMed] [Google Scholar]
- 25.Neeleman L, Bol J F. cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation. Virology. 1999;254:324–333. doi: 10.1006/viro.1998.9568. [DOI] [PubMed] [Google Scholar]
- 26.Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palauqui J-C, Vaucheret H. Transgenes are dispensable for the RNA degradation step of cosuppression. Proc Natl Acad Sci USA. 1998;95:9675–9680. doi: 10.1073/pnas.95.16.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palauqui J-C, Balzergue S. Activation of systemic acquired silencing by localised introduction of DNA. Curr Biol. 1999;9:59–66. doi: 10.1016/s0960-9822(99)80016-5. [DOI] [PubMed] [Google Scholar]
- 29.Ratcliff F, Harrison B D, Baulcombe D C. A similarity between viral defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 30.Ratcliff F, MacFarlane S, Baulcombe D C. Gene silencing without DNA: RNA-mediated cross protection between viruses. Plant Cell. 1999;11:1207–1215. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz M T, Voinnet O, Baulcombe D C. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweizer P, Pokorny J, Schulze-Lefert P, Dudler R. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 2000;24:895–903. doi: 10.1046/j.1365-313x.2000.00941.x. [DOI] [PubMed] [Google Scholar]
- 33.Sijen T, Kooter J M. Post-transcriptional gene-silencing: RNAs on the attack or on the defense? BioEssays. 2000;22:520–531. doi: 10.1002/(SICI)1521-1878(200006)22:6<520::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 34.Smardon A, Spoerke J M, Stacey S C, Klein M E, Mackin N, Maine E M. EGO-1 is related to RNA-directed RNA polymerase and function in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 35.Smith N, Singh S, Wang M-B, Stoutjesdijk P, Green A, Waterhouse P. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- 36.Tavernarakis N, Wang S L, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 37.Tenllado F, García-Luque I, Serra M T, Díaz-Ruíz J. Nicotiana benthamiana plants transformed with the 54 kDa region of the pepper mild mottle tobamovirus replicase gene exhibit two types of resistance responses against viral infection. Virology. 1995;211:170–183. doi: 10.1006/viro.1995.1389. [DOI] [PubMed] [Google Scholar]
- 38.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 39.Tuschl T, Zamore P, Bartel D, Sharp P. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaucheret H. Promoter-dependent trans-inactivation in transgenic tobacco plants: kinetic aspects of gene silencing and gene reactivation. C R Acad Sci Ser III. 1994;317:310–323. [Google Scholar]
- 41.Vaucheret H, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Mourrain P, Palauqui J-C, Vemhettes S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 42.Voinnet O, Pinto Y M, Baulcombe D C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse P, Gramham M, Wang M-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamore P D, Tuschl T, Sharp P A, Bartel D P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]